Abstract
Salmonella enterica serotype Typhimurium is able to expand in the lumen of the inflamed intestine through mechanisms that have not been fully resolved. Here we utilized streptomycin-pretreated mice and dextran sodium sulfate (DSS)-treated mice to investigate how pathways for S. Typhimurium iron acquisition contribute to pathogen expansion in the inflamed intestine. Competitive infection with an iron uptake-proficient S. Typhimurium strain and mutant strains lacking tonB feoB, feoB, tonB or iroN in streptomycin pretreated mice demonstrated that ferric iron uptake requiring IroN and TonB conferred a fitness advantage during growth in the inflamed intestine. However, the fitness advantage conferred by ferrous iron uptake mechanisms was independent of inflammation and was only apparent in models where the normal microbiota composition had been disrupted by antibiotic treatment.
Keywords: Salmonella, Fe2+, Fe3+, inflammation, animal models
Introduction
Salmonella Pathogenicity Island 1 (SPI-1) encodes a type III secretion system (T3SS-1) that is required for invasion of epithelial cells and induction of the host inflammatory response (Tsolis et al., 1999; Barthel et al., 2003; Altier, 2005; Bruno et al., 2009; Raffatellu et al., 2009). Salmonella-elicited intestinal inflammation generates an environment that favors growth of the pathogen while commensal microorganisms are suppressed (Stecher et al., 2007). The expansion of Salmonella enterica serovar Typhimurium (S. Typhimurium) in the inflamed gut is essential for transmission of the pathogen by the fecal-oral route (Lawley et al., 2008).
S. Typhimurium requires iron as an essential nutrient for growth. Approximately 7% of the S. Typhimurium genome is regulated by levels of iron under in vitro conditions (Bjarnason et al., 2003). In vertebrates, the availability of the ferric (Fe3+) and ferrous (Fe2+) forms of iron are very limited extracellularly. One of the mechanisms by which the host limits iron in the inflamed intestine is by secreting lipocalin-2 (Raffatellu et al., 2009), which has a bacteriostatic effect (Flo et al., 2004) by binding to enterochelin, a high affinity iron chelator (siderophore) produced by bacteria of the Enterobacteriaceae family (Goetz et al., 2002). Similarly to other Enterobacteriaceae, S. Typhimurium secretes enterochelin, but it also produces a glycosylated derivative of enterochelin, termed salmochelin, which is not bound by lipocalin-2 and therefore confers lipocalin-2 resistance (Hantke et al., 2003; Crouch et al., 2008; Raffatellu et al., 2009).
Iron acquisition through salmochelin is mediated by the iroBCDE and iroN genes, which are responsible for the biosynthesis (iroB), export (iroC), and absorption (iroN) of salmochelin (Hantke et al., 2003; Bäumler et al., 1996). The process of internalization of this siderophore is an energy dependent process, which requires the TonB/ExbB/ExbD protein-complex located at the inner membrane (Hannavy et al., 1990). TonB energizes transport of ferric iron (Fe3+)-siderophore complexes into the periplasm of Gram-negative bacteria by interacting with specific outer membrane transporters (reviewed by Braun & Hantke, 2011), such as IroN. An additional source of iron that may be available in the anaerobic intestine is soluble ferrous iron (Fe2+), which enters the periplasm through porins and is transported accross the cytoplasmic membrane by a membrane transporter encoded by the feoABC operon. FeoB is the inner membrane Fe2+ transporter (Tsolis et al., 1996; Boyer et al., 2002).
Considering that iron acquisition is a key step in growth of S. Typhimurium, the aim of this study was to evaluate the role of acquiring Fe2+ and Fe3+ for expansion of S. Typhimurium in the lumen of the inflamed and non-inflamed intestine.
Material and Methods
Ethics statement
This study was approved by the Committee for Ethical Animals Use (CEUA) of the Universidade Federal de Minas Gerais (Brazil) under protocol numbers 197/2008, 254/2012 and 386/2013.
Bacterial strains and culture conditions
Bacterial strains are listed in Table 1. IR715 is a nalidixic acid-resistant S. Typhimurium strain derived from wild type ATCC14028. The invA feoB (SW710), invA tonB (SW711), and invA tonB feoB mutants (SW712) were constructed by introducing the invA::pGP704 mutation from SW399 into AJB15 (feoB mutant in IR715 strain), AJB36 (tonB mutant in IR715 strain), and AJB62 feoB tonB mutant in IR715 strain), respectively, by generalized phage transduction (P22 HT int-105) (Schmieger, 1972). Confirmation of mutagenesis in invA feoB (SW710), invA tonB (SW711), and invA tonB feoB (SW712) was confirmed by PCR, with primers listed in Table S1. Mutation of tonB was confirmed by visualization of a fragment of 2.2 kb corresponding to a fragment of 0.9 kb of the band in the wild type strain plus the 1.3 kb of kanamycin resistance cassette. Mutation of feoB was confirmed by visualization of a fragment of 5 kb, which corresponds to the 3 kb fragment of wild type strain plus the 2 kb fragment from tetracycline resistance cassette. Mutation in invA was confirmed by visualization of a fragment of 0.5 kb in invA feoB (SW710), invA tonB (SW711), and invA tonB feoB (SW712) mutants, which is consistent with the insertion of invA::pGP704 (Fig. S1).
Table 1.
Salmonella enterica serotype Typhimurium strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| AJB15 | IR715 feoB::TetR | Tsolis et al., 1996 |
| AJB36 | IR715 tonB::KanR | Tsolis et al., 1996 |
| AJB52 | IR715 iroN::pGP704-AmpR | Bäumler et al.,1998 |
| AJB62 | IR715feoB::TetRtonB::KanR | Tsolis et al., 1996 |
| AJB715 | IR715 phoN::KanR | Kingsley et al., 2003 |
| IR715 | ATCC 14028 NalR | Stojiljkovic et al., 1995 |
| SPN452 | IR715 invA::tetRAspiB::KSAC-KanR | Raffatellu et al., 2009 |
| SPN454 | IR715 invA::tetRAspiB::KSAC-KanRiroN::pGP704-AmpR | Raffatellu et al., 2009 |
| SW399 | IR715 invA::pGP704 | Winter et al., 2009 |
| SW710 | IR715 invA::pGP704feoB::TetR | This study |
| SW711 | IR715 invA::pGP704 tonB::KanR | This study |
| SW712 | IR715 invA::pGP704feoB::TetRΔtonB::KanR | This study |
| TH199 | IR715 phoN::KanRinvA::tetra | Winter et al., 2014 |
TetR: Tetracycline resistance (tetRA); KanR: Kanamycin resistance; AmpR: Ampicillin resistance; NalR: Nalidixic acid resistance.
Bacteria were cultured aerobically for 18 hours at 37ºC under agitation in Luria-Bertani (LB) broth with appropriate antibiotics, followed by dilution 1:50 in LB broth (without antibiotic) and incubation for 3 hours at 37°C under agitation. Bacterial concentration was estimated by measuring the optical density of cultures at 600 nm (OD600). Antibiotics were added to LB broth cultures and LB agar plates at the following concentrations: 50 mgL−1 nalidixic acid, 12.5 mgL−1 tetracycline, 100 mgL−1 kanamycin, and 100 mgL−1 ampicillin. Competitive indices were obtained by dividing the output ratio (CFU of the phoN strain over CFU of the iron-uptake-deficient mutant) by the input ratio (CFU of phoN strain over CFU of the iron-uptake-deficient mutant).
For in vitro growth, iron-uptake proficient S. Typhimurium strain (phoN mutant) and mutant strains of S. Typhimurium were cultured overnight at 37ºC under agitation in Nutrient Broth (NB) with 0.2 mM 2,2′-dipyridyl (Sigma Aldrich). Microbiological growth was estimated by measuring OD600. Cultures were inoculated with an initial 0.01 OD600 in NB, NB supplemented with 40 μM FeSO4, 40 μM FeCl3 or 0.2 mM dipyridyl, and OD600 was measured after 24 hours.
Streptomycin-pretreated mouse model
Six to eight-week-old female C57BL/6 mice were pretreated with streptomycin as previous described (Barthel et al., 2003) and intragastrically inoculated with 0.1 mL of a 1:1 mixture containing 1×107 CFU of S. Typhimurium IR715 tagged with phoN mutation (AJB715) and either one of the following mutant strains: tonB feoB (AJB62), or feoB (AJB15), or tonB (AJB36), or iroN (AJB52). In parallel, mice were infected under the same conditions with a phoN invA S. Typhimurium mutant strain (TH199) and either one of the following mutant strains: invA tonB feoB (SW712), or invA feoB (SW710), or invA tonB (SW711), or invA iroN (SPN454). Samples were collected at 48 hours post infection (hpi) for bacteriologic culture on LB + X-Phos to distinguish between wild type, which yields white colonies on X-Phos-containing medium and iron uptake mutants, which (since they express the acid phosphatase encoded by phoN) grow as blue colonies on X-Phos medium. Further, tissue samples were collected for histopathology and quantitative real time RT-PCR (qRT-PCR). Fecal pellets were homogenized in 2 mL of sterile PBS and plated on LB agar containing nalidixic acid and on LB agar containing nalidixic acid and other appropriate antibiotics to the respective mutant strain. Cecum was collected for histopathological and qRT-PCR analysis. Total RNA was extracted from cecum using the Tri-reagent (Molecular Research Center) and processed as described (Raffatellu et al., 2008). qRT-PCR was performed using SYBR Green (Applied Biosystems), 7900HT Fast Real-Time PCR System and data were analyzed using the comparative delta-Ct method (Livak & Schmittgen, 2001) normalized by transcriptional level of murine gene Gapdh. Primers used for qRT-PCR are listed in Table S2.
Dextran sulfate sodium (DSS) mouse model of enteritis
Six to eight-week-old female C57BL/6 mice received oral administration of 3% DSS diluted in sterile water ad libitum. This concentration of DSS was selected because preliminary experiments (not shown) showed that it elicits significant submucosal inflammation, submucosal edema and mucosal inflammation. After 120 h of DSS administration, mice were submitted to 4 h of food and water withdrawal, and then inoculated with 0.1 mL of a 1:1 mixture containing 1×107 CFU of S. Typhimurium phoN mutant (AJB715), and one of the following mutant strains: tonB feoB (AJB62), or feoB (AJB15), or tonB (AJB36), or iroN (AJB52). DSS treatment was maintained until euthanasia. Cecum and colon were collected at 48 hpi for bacteriologic culture and histopathology.
Bovine ligated ileal loop model
Four healthy 4-week-old male Salmonella-free Holstein calves were used. Ligated ileal loops were surgically made as previously described (Santos et al., 2002). Loops were inoculated with intraluminal injection of 3 mL of sterile LB broth or with 3 mL of a 1:1 mixture containing 1 x 109 CFU of the reference strain phoN (AJB715), and one of the following mutant strains: tonB feoB (AJB62), or feoB (AJB15), or tonB (AJB36), or iroN (AJB52). In parallel, loops were inoculated under the same conditions with a phoN invA mutant (TH199), and one of the following mutant strains: invA tonB feoB (SW712), or invA feoB (SW710), or invA tonB (SW711), or invA iroN (SPN454). Samples were collected at 8 hpi for bacteriology and histopathology. Intestinal fluid and homogenates from fragments of the intestinal mucosa obtained with 6 mm biopsy punches in 2 mL of sterile phosphate-buffered saline (PBS) were plated on LB agar plates containing nalidixic acid and appropriate antibiotics.
Statistical analysis
Bacteriology and mRNA levels measured by qRT-PCR were logarithmically transformed and analyzed by ANOVA followed by Tukey’s test. Histopathology scores were analyzed by Mann Whitney test.
Results and discussion
To investigate to role of Fe2+ and Fe3+ uptake systems during S. Typhimurium pathogenesis, we generated S. Typhimurium strains defective for Fe3+ uptake (tonB mutant), defective for Fe3+-salmochelin uptake (iroN mutant), a mutant strain lacking the Fe2+ uptake system (feoB mutant), and a mutant strain incapable of acquiring both Fe2+ and Fe3+ (tonB feoB mutant) (Fig. S1). The fitness of these strains was compared to a S. Typhimurium strain that was marked by an antibiotic resistance marker inserted in the phoN gene (phoN mutant), to facilitate recovery from animals. Inactivation of the phoN gene does not reduce the ability of S. Typhimurium to colonize organs or the cecum of mice (Weening et al., 2005). We first compared the growth of the iron-uptake proficient strain (phoN mutant, AJB715) to the growth of strains carrying mutations in genes involved in iron-acquisition (the iroN, tonB, feoB and tonB feoB, mutant strains) in iron-replete medium (nutrient broth [NB] supplemented with FeSO4 or FeCl3), low-iron medium (NB) or iron-deplete medium (NB + dipiridyl) (Fig. 1). As expected, all strains grew equally well in iron-replete medium. Only the mutant strain incapable of acquiring both Fe2+ and Fe3+ (tonB feoB mutant) exhibited impairment growth in low-iron medium. In iron-deplete medium, both the tonB mutant and the tonB feoB mutant exhibited reduced growth. These data suggested that iron uptake systems were not required under iron-replete conditions. Either intact Fe2+ or intact Fe3+ uptake mechanisms were sufficient for supporting growth in low-iron medium. However, under conditions of severe iron limitation, both Fe2+ and Fe3+ uptake mechanisms were required for growth.
Fig. 1.
Growth of an iron-uptake proficient S. Typhimurium strain (tagged with phoN mutation) and iron uptake-deficient mutants in iron-replete, low-iron and iron-deplete media. An iron-uptake proficient S. Typhimurium strain (tagged with phoN mutation) and tonB feoB, feoB, tonB and iroN mutants were inoculated at an initial optical density at 600 nm (OD600) of 0.01 in NB, NB supplemented with 40 μM FeSO4 (+FeSO4), FeCl3 40 μM (+FeCl3) or 0.2 mM dipyridyl (−Fe dipyridyl) and OD600 measurement was performed after 24 hours. Bars represent means ± SD of three independent experiments. Asterisks indicate significant differences of treatments compared to NB media in the same strain, Tukey test, * P < 0.001. Number signs indicate significant differences between mutant strains compared to the iron-uptake proficient S. Typhimurium strain in the same treatment, Tukey test, # P< 0.001.
Next, we wanted to determine whether intestinal inflammation alters the requirement for Fe2+ and Fe3+ uptake mechanisms during growth of S. Typhimurium in the intestinal lumen. To this end, we utilized the streptomycin-pretreated mouse model. Streptomycin-pretreated mice develop acute neutrophilic typhlitis after infection with S. Typhimurium, and this inflammatory response is dependent on the invasion-associated T3SS-1 (Barthel et al., 2003). Therefore, to evaluate the role of Fe2+ and Fe3+ uptake mechanisms in intestinal colonization of S. Typhimurium, we compared the fitness of iroN, tonB, feoB, or tonB feoB mutant strains with that of an iron-uptake proficient S. Typhimurium strain (phoN mutant) in streptomycin-pretreated mice, using a competitive colonization assay. This experimental design was chosen to ensure that the iron uptake mutants, some of which may have colonization defects, are exposed to the same host inflammatory response that is induced by the iron uptake-proficient strains, since different levels of colonization are known to elicit different levels of inflammation and of lipocalin-2, a major iron-limiting response of the host in the inflamed intestine. To investigate the effect of intestinal inflammation on the outcome of these competitions, we used mutants that lack a functional T3SS-1 due to a mutation in invA (Raffatellu et al., 2009). To this end we compared the fitness of invA iroN, invA tonB, invA feoB, or invA tonB feoB mutant strains with that of an iron-uptake proficient S. Typhimurium strain (invA phoN mutant) in streptomycin-pretreated mice. As expected (Barthel et al., 2003), histopathological analysis (Fig. 2A and Fig. S2A) and quantification of Cxcl2 and Cxcl1 transcript levels (encoding the neutrophil chemoattractants Mip-2 and KC, respectively) by real time PCR (Fig. 2B and 2C) revealed that the intestinal inflammatory response was muted in mice infected with strain mixtures that lacked a function T3SS-1 (due to a mutation in invA), compared to mice infected with T3SS-1-proficient strains. Importantly, lcn2 (encoding lipocalin-2) transcription was blunted in mice infected with strain mixtures that lacked a functional T3SS-1 (Fig. 2D). Consistent with this observation, strains that lacked lipocalin-resistance (iroN mutant and tonB mutant) were recovered in lower numbers than an iron-uptake proficient, phoN-tagged S. Typhimurium when T3SS-1 was intact (Fig. 2E). However, reduced Lcn2 expression caused by a mutation in invA (Fig. 2D) abrogated the fitness advantage of an iron-uptake proficient S. Typhimurium strain (invA phoN mutant) over strains lacking lipocalin-resistance (i.e. the invA iroN mutant and the invA tonB mutant). These results confirmed a previous study showing that lipocalin resistance confers a fitness advantage upon S. Typhimurium during growth of in the inflamed gut (Raffatellu et al., 2009). Our results are also consistent with a previous study that demonstrated a modest effect of a tonB mutation on intestinal colonization in mice infected intragastrically (IG) in the absence of streptomycin (Tsolis et al., 1996). Fe2+ uptake conferred a fitness advantage in the mouse colitis model, but the magnitude of this fitness advantage was similar in strains with functional T3SS-1 (phoN vs. feoB) and defective T3SS-1 (invA phoN vs. invA feoB). These data suggested that Fe2+ uptake is equally required under both inflammatory and non-inflammatory conditions in the intestine. These data were in agreement with a previous study showing a contribution of FeoB to fitness of S. Typhimurium in the non-inflamed intestine (Deriu et al., 2013). In the setting of inflammation, TonB-dependent iron uptake was also required for the probiotic activity of E. coli Nissle 1917 (Deriu et al., 2013), suggesting that Fe3+ is limiting to both E. coli and Salmonella in the inflamed intestine. Our data suggest that Fe2+ and Fe3+ uptake mechanisms cooperated during intestinal inflammation, since the phoN mutant was recovered 361-fold higher numbers than the tonB feoB mutant from feces. This fitness advantage was markedly reduced in the absence of a functional T3SS-1 (invA phoN vs. invA tonB feoB).
Fig. 2.
Intestinal colonization of C57BL/6 mice pre-treated with streptomycin and coinfected with an iron-uptake proficient S. Typhimurium strain (tagged with a phoN mutation) and iron acquisition-deficient mutant strains. Mice were inoculated with 1:1 mixture of 1 x 107 CFU of the iron-uptake proficient S. Typhimurium phoN strain and one of the mutant strains tonB feoB, feoB, tonB, or iroN. Alternatively, mice were inoculated with iron-uptake proficient T3SS-1-deficient S. Typhimurium strain (invA phoN mutant) and one of the mutant strains: invA tonB feoB, invA feoB, invA tonB or invA iroN. Histological score of the cecum at 48 hours after infection (A). Bars represent the average of histopathological scores for submucosal inflammation (SI), mucosal inflammation (MI), and submucosal edema (SE). Scores for individual mice are shown in Fig. S2A. Transcription levels of Cxcl2 (B), Cxcl1 (C) and Lcn2 (D) measured by qRT-PCR in cecum collected at 48 hours after infection. Bars represent fold change over mock inoculated tissues normalized by transcriptional level of Gapdh. Competitive index of iron-uptake proficient S. Typhimurium phoN strain and iron uptake-deficient mutants or invA mutant and invA iron defective mutants in pellet fecal at 48 hours after infection (E). Bars represent geometric means ± SEM. Significance of differences observed was analysed using a Mann-Whitney test (A) or ANOVA followed by Tukey’s test (B–E) * P < 0.05; ** P < 0.01.
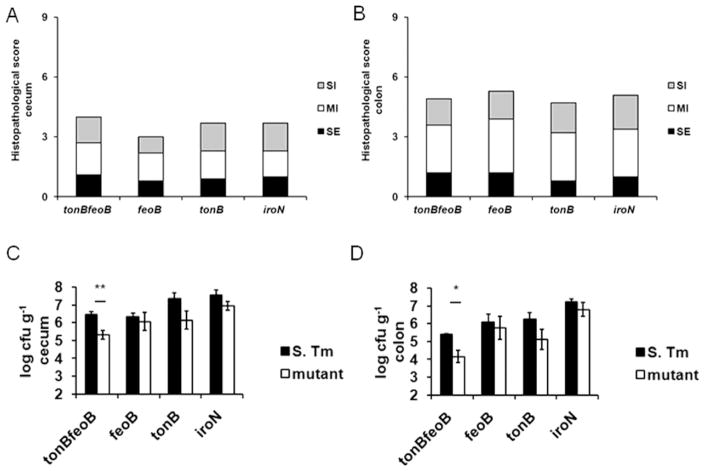
Streptomycin induces marked changes in the composition of the intestinal microbiota (dysbiosis), with a decrease in the diversity of microbial communities in the cecum and ileum of mice infected with S. Typhimurium (Garner et al., 2009). Furthermore, streptomycin treated mice develop mild cecal inflammation even in the absence of S. Typhimurium infection (Spees et al., 2013). In order to investigate the general influence of inflammation on iron uptake mechanisms in the intestine in the absence of confounding effect of antibiotic-mediated microbiota disruption, competitive infections of S. Typhimurium and the mutant strains included in this study were performed in mice with chemically induced (by 3% DSS) acute intestinal inflammation. DSS treatment induces acute and chronic colitis in mice. The acute phase is characterized by a neutrophilic ulcerative colitis, body weight loss, and diarrhea (Okayasu et al., 1990). As expected, mice treated with 3% DSS in the absence of S. Typhimurium infection developed inflammatory changes in the colon (not shown). DSS treated mice inoculated with mixtures of wild-type S. Typhimurium (phoN mutant) and feoB, tonB, iroN or tonB feoB mutants developed inflammatory changes in the cecum and colon after 48h that were similar to mice treated with 3% DSS only. This similarity was expected, since in untreated mice development of intestinal inflammation in response to S. Typhimurium infection takes several days (Rivera-Chávez et al., 2016). Histopathologic analysis of cecum (Fig. 3A and Fig. S2B) and colon (Fig. 3B and Fig. S2C) revealed moderate neutrophilic and lympho-histio-plasmacytic infiltration in the mucosa and submucosa, and moderate submucosal edema.
Fig. 3.
Intestinal colonization of C57BL/6 mice treated with 3% DSS and intragastrically coinfected with an iron-uptake proficient S. Typhimurium strain (phoN mutant) and iron acquisition-deficient mutant strains. Mice received DSS 3% diluted in water for 120 hours and were inoculated with 1:1 mixture of 1 x 107 CFU of the iron-uptake proficient S. Typhimurium strain (phoN mutant) and mutants tonB feoB, feoB, tonB, or iroN. Histological score of the cecum (A) and colon (B) at 48 hpi. Bars represent the average of histopathological scores for submucosal inflammation (SI), mucosal inflammation (MI), and submucosal edema (SE). Scores for individual mice are shown in Figs S2B and S2C. Colony-forming units (CFU) recovered in cecum (C) and colon (D) at 48 hpi. Bars represent mean and standard error (n=5). Significance of differences observed was analysed using a Mann-Whitney test (A–B) or ANOVA followed by Tukey’s test (C–D). * P < 0.05; ** P < 0.01.
Competitive indices between bacterial strains in intestinal tissues obtained at 48 hours after infection from 3% DSS-treated mice demonstrated that the iron-uptake proficient S. Typhimurium strain (phoN mutant) was recovered in 15- and 12-fold higher CFU numbers than the tonB mutant in the cecum and colon, respectively (Fig. 3C and 3D). Similar results were obtained with the tonB feoB mutant (Fig. 3C and 3D). IroN-mediated, also conferred a fitness advantage, as the iron-uptake proficient S. Typhimurium strain (phoN mutant) was recovered in 5- and 6-fold higher CFU numbers than a iroN mutant in the cecum (Fig. 3C) and colon (Fig. 3D), respectively. These results indicated that intestinal inflammation in DSS colitis model has similar influence in defective growth of mutant with loss of ferric iron acquisition system. However, the iron-uptake proficient S. Typhimurium strain (phoN mutant) was recovered in similar numbers as the feoB mutant strain from the cecum (Fig. 3C), and in only 3-fold higher numbers than feoB mutant from the colon tissues (Fig. 3D). The absence of an overt fitness advantage conferred by FeoB in the intestinal lumen was also observed in previous studies, in which mice were infected with S. Typhimurium in the absence of antibiotic treatment (Nagy et al., 2014; Tsolis et al., 1996).
The competitive advantage of Salmonella with intact iron acquisition pathways was also evaluated in cattle, a host species that naturally responds to S. Typhimurium with acute intestinal inflammation (Costa et al., 2012), paralleling the clinical and pathological manifestations of S. Typhimurium infection in human patients. The T3SS-1 effector proteins encoded in SPI-1 were demonstrated to mediate influx of neutrophils and fluid accumulation in the bovine ligated ileal loop model (Santos et al, 2002; Zhang et al., 2002; Costa et al., 2012). In the present study, fluid contents and bovine ileal tissues were collected at 8 hpi from loops inoculated with a 1:1 mixture of the reference strain and one of the mutant strains, similarly to mouse experiments described above. S. Typhimurium reference strain and the mutant strains were recovered in similar CFU numbers from intestinal fluid and the ileal mucosa (Fig. S3). There were also similar CFU numbers of S. Typhimurium invA and the invA tonB feoB, invA feoB, invA tonB or invA iroN mutants in the intestinal fluid and ileal mucosa (Fig. S3). Importantly, coinfection of the reference strain and tonB feoB, feoB, tonB or iroN mutants elicited a prominent inflammatory response at 8 hpi in the bovine ileal mucosa (not shown). These results are in agreement with previous report in which S. Typhimurium induced marked inflammatory changes at 5 hpi (Santos et al., 2002). As expected, coinfection with S. Typhimurium mutant defective in invasion invA and the invA tonB feoB, invA feoB, invA tonB or invA iroN mutants caused markedly less pathology when compared to the S. Typhimurium reference strain (not shown). This was the first attempt to use bovine ileal ligated loop model to elucidate iron uptake of S. Typhimurium, but this experimental model has an intrinsic limitation, which is that the experiment can be conducted only for a few hours since the calf remains under anesthesia during the course of the experiment (Alves et al., 2003). The number of doublings that can be accomplished during 8 h of infection in bovine ileal ligated loops is limited, which may explain why this assay did not reveal any competitive advantage to strains that carry intact iron uptake pathways in comparison to mutants.
In summary, ferric iron uptake mediated by salmochelin and by a TonB energy carrier of S. Typhimurium provided a fitness advantage in the inflamed intestine of streptomycin pretreated mice and during DSS-induced colitis. In contrast, ferrous iron uptake mediated by FeoB only provided benefit during intestinal colonization in streptomycin pretreated mice, but this advantage was not dependent on an inflamed intestinal environment, because it was neither altered by inactivation of T3SS-1 (Fig. 2E) nor observed in mice with DSS-induced colitis (Fig. 3C and 3D). Furthermore, in the absence of antibiotic treatment, inactivation of feoB does not result in an overt loss of fitness for colonizing the mouse intestine (Nagy et al., 2014; Tsolis et al., 1996). Collectively, these data suggest that a disruption of the microbiota might increase the availability of (or decrease competition for) Fe2+ in the gut environment, thereby increasing the importance of Fe2+ for growth in this environment.
Supplementary Material
Confirmation of S. Typhimurium mutants by PCR. (A) Schematic representation of the target genes and mutagenesis approach. The tonB and feoB coding sequence was interrupted by inserting a kanamycin (KanR) or tetracycline (TetR) resistance gene cassette. The invA gene was disrupted by insertion of a derivative of pGP704. Approximate location of primer binding sites is indicated by small arrows. (B) The indicated S. Typhimurium strains served as templates for the amplification of the various gene targets. PCR products were separated by agarose gel electrophoresis. Approximate size markers are indicated on the left side of the panel.
(A) Histologic scores of individual C57BL/6 mice pre-treated with streptomycin and coinfected with an iron-uptake proficient S. Typhimurium strain (tagged with a phoN mutation) and iron acquisition-deficient mutant strains. Mice were inoculated with 1:1 mixture of 1 x 107 CFU of the iron-uptake proficient S. Typhimurium phoN strain and one of the mutant strains tonB feoB, feoB, tonB, or iroN. Alternatively, mice were inoculated with iron-uptake proficient T3SS-1-deficient S. Typhimurium strain (invA phoN mutant) and one of the mutant strains: invA tonB feoB, invA feoB, invA tonB or invA iroN. Bars represent histopathological scores for submucosal inflammation (SI), mucosal inflammation (MI), and submucosal edema (SE) from individual mice. Summary data are shown in Fig. 2A. (B and C) Histological scoring of cecum (B) and colon (C) from individual C57BL/6 mice treated with 3% DSS and intragastrically coinfected with an iron-uptake proficient S. Typhimurium strain (phoN mutant) and iron acquisition-deficient mutant strains. Scoring was performed as in (A). Summary data are shown in Fig. 3A and 3B.
Intestinal colonization of bovine ligated ileal loops coinfected with S. Typhimurium and defective iron acquisition mutant strains. Each loop was inoculated with a 1 x 109 CFU containing a 1:1 mixture of the S. Typhimurium reference strain (AJB715) with either the tonB feoB; feoB; tonB; or iroN mutant (one loop per pair of strains). Competitive indices in fluid (A) and ileal tissue (B) were determined at 8 hpi. Columns represent geometric mean and standard error obtained from 4 calves used (4 independent experiments) for the ligated ileal loop experiment.
Table S1. Primers used in PCR to confirm mutagenesis in invA tonB feoB, invA feoB, and invA tonB of Salmonella Typhimurium.
Table S2. Primers used for qRT-PCR.
Acknowledgments
Work in RLS lab is supported by CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil), and FAPEMIG (Fundação de Amparo à Pesquisa do Estado de Minas Gerais, Brazil). This study was supported by a PNPD fellowship for JPSM from CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior), and a fellowship from the John Simon Guggenheim Memorial Foundation to RLS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altier C. Genetic and environmental control of Salmonella invasion. J Microbiol. 2005;43:85–92. [PubMed] [Google Scholar]
- Alves GES, Hartsfield SM, Carroll GL, Santos DAML, Zhang S, Tsolis RM, Bäumler AJ, Adams LG, Santos RL. Emprego do propofol, isofluorano e morfina para a anestesia geral de longa duração em bezerros. Arq Bras Med Vet Zootec. 2003;55:411–420. [Google Scholar]
- Barthel M, Hapfelmeier S, Quintanilla-Martínez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Rüssmann H, Hardt WD. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun. 2003;71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäumler AJ, Norris TL, Lasco T, Voight W, Reissbrodt R, Rabsch W, Heffron F. IroN, a novel outer membrane siderophore receptor characteristic of Salmonella enterica. J Bacteriol. 1998;180:1446–1453. doi: 10.1128/jb.180.6.1446-1453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäumler AJ, Tsolis RM, van der Velden AW, Stojiljkovic I, Anic S, Heffron F. Identification of a new iron regulated locus of Salmonella typhi. Gene. 1996;183:207–213. doi: 10.1016/s0378-1119(96)00560-4. [DOI] [PubMed] [Google Scholar]
- Bjarnason J, Southward CM, Surette MG. Genomic profiling of iron-responsive gene in Salmonella enterica serovar Typhimurium high-throughput screening of a random promoter library. J Bacteriol. 2003;185:4973–4982. doi: 10.1128/JB.185.16.4973-4982.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer E, Bergevin I, Malo D, Gros P, Cellier MF. Acquisition of Mn (II) in addition to Fe (II) is required for full virulence of Salmonella enterica serovar Typhimurium. Infect Immun. 2002;70:6032–6042. doi: 10.1128/IAI.70.11.6032-6042.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun V, Hantke K. Recent insights into iron import by bacteria. Curr Opin Chem Biol. 2011;15:328–334. doi: 10.1016/j.cbpa.2011.01.005. [DOI] [PubMed] [Google Scholar]
- Bruno VM, Hannemann S, Lara-Tejero M, Flavell RA, Kleinstein SH, Galán JE. Salmonella Typhimurium type III secretion effectors stimulate innate immune responses in cultured epithelial cells. Plos Pathog. 2009;5:e1000538. doi: 10.1371/journal.ppat.1000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa LF, Paixão TA, Tsolis RM, Bäumler AJ, Santos RL. Salmonellosis in cattle: advantages of being an experimental model. Res Vet Sci. 2012;93:1–6. doi: 10.1016/j.rvsc.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Crouch ML, Castor M, Karlinsey JE, Kalhorn T, Fang FC. Biosynthesis and IroC-dependent export of the siderophore salmochelin are essential for virulence of Salmonella enterica serovarTyphimurium. Mol Microbiol. 2008;67:971–983. doi: 10.1111/j.1365-2958.2007.06089.x. [DOI] [PubMed] [Google Scholar]
- Deriu E, Liu JZ, Pezeshki M, Edwards RA, Ochoa RJ, Contreras H, Libby SJ, Fang FC, Raffatellu M. Probiotic bacteria reduce Salmonella Typhimurium intestinal colonization by competing for iron. Cell Host Microbe. 2013;14:26–37. doi: 10.1016/j.chom.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, Akira S, Aderem A. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432:917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- Frawley ER, Fang FC. The ins and outs of bacterial iron metabolism. Mol Microbiol. 2014;93:609–616. doi: 10.1111/mmi.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner CD, Antonopoulos DA, Wagner B, Duhamel GE, Keresztes I, Ross DA, Young VB, Altier C. Perturbation of the small intestine microbial ecology by streptomycin alters pathology in a Salmonella entericaserovarTyphimurium murine model of infection. Infect Immun. 2009;77:2691–2702. doi: 10.1128/IAI.01570-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz DH, Holmes MA, Borregaard N, Bluhm ME, Raymond KN, Strong RK. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophores mediated iron acquisition. Mol Cell. 2002;10:1033–1043. doi: 10.1016/s1097-2765(02)00708-6. [DOI] [PubMed] [Google Scholar]
- Hannavy K, Barr GC, Dorman CJ, Adamson J, Mazengera LR, Gallagher MP, Evans JS, Levine BA, Trayer IP, Higgins CF. TonB Protein of Salmonella Typhimurium. A model for signal transduction between membranes. J MolBiol. 1990;216:897–910. doi: 10.1016/S0022-2836(99)80009-6. [DOI] [PubMed] [Google Scholar]
- Hantke K, Nicholson G, Rabsch W, Winkelmann G. Salmochelins, siderophore of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc Natl Acad Sci USA. 2003;100:3677–3682. doi: 10.1073/pnas.0737682100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley RA, Humphries AD, Weening EH, De Zoete MR, Winter S, Papaconstantinopoulou A, Dougan G, Bäumler AJ. Molecular and phenotypic analysis of the CS54 island of Salmonella enterica serotype Typhimurium: identification of intestinal colonization and persistence determinants. Infect Immun. 2003;71:629–640. doi: 10.1128/IAI.71.2.629-640.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley TD, Bouley DM, Hoy YE, Gerke C, Relman DA, Monack DM. Host transmission of Salmonella enterica serovar Typhimurium is controlled by virulence factors and indigenous intestinal microbiota. Infect Immun. 2008;76:403–416. doi: 10.1128/IAI.01189-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak J, Schmittgen D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Nagy TA, Moreland SM, Detweiler CS. Salmonella acquires ferrous iron from haemophagocytic macrophages. Mol Microbiol. 2014;93:1314–1326. doi: 10.1111/mmi.12739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okayasu I, Hatakeyama S, Yamada M, Ohkusa T, Inagaki Y, Nakaya R. A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology. 1990;98:694–702. doi: 10.1016/0016-5085(90)90290-h. [DOI] [PubMed] [Google Scholar]
- Raffatellu M, George MD, Akiyama Y, Hornsby MJ, Nuccio SP, Paixao TA, Butler BP, Chu H, Santos RL, Berger T, Mak TW, Tsolis RM, Bevins CL, Solnick JV, Dandekar S, Bäumler AJ. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe. 2009;5:476–486. doi: 10.1016/j.chom.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffatellu M, Santos RL, Verhoeven D, George MD, Wilson RP, Winter SE, Godinez I, Sankaran S, Paixao TA, Gordon MA, Kolls JK, Dandekar S, Bäumler AJ. Simian immunodeficiency virus–induced mucosal IL–17 deficiency promotes Salmonella dissemination from the gut. Nat Med. 2008;14:421–428. doi: 10.1038/nm1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paixão TA, Roux CM, den Hartigh AB, Sankaran-Walters S, Dandekar S, Santos RL, Tsolis RM. Establishment of systemic Brucella melitensis infection through the digestive tract requires urease, the type IV secretion system, and lipopolysaccharide O antigen. Infect Immun. 2009;77:4197–4208. doi: 10.1128/IAI.00417-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Chavez F, Zhang LF, Faber F, Lopez CA, Byndloss MX, Olsan EE, Xu G, Velazquez EM, Lebrilla CB, Winter SE, et al. Depletion of Butyrate-Producing Clostridia from the Gut Microbiota Drives an Aerobic Luminal Expansion of Salmonella. Cell Host Microbe. 2016;19:443–454. doi: 10.1016/j.chom.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos RL, Zhang S, Tsolis RM, Bäumler AJ, Adams LG. Morphologic and molecular characterization of Salmonella typhimurium infection in neonatal calves. Vet Pathol. 2002;39:200–215. doi: 10.1354/vp.39-2-200. [DOI] [PubMed] [Google Scholar]
- Schmieger H. Phage P22-mutants with increased or decreased transduction abilities. Mol Gen Genet. 1972;119:75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- Spees AM, Wangdi T, Lopez CA, Kingsbury DD, Xavier MN, Winter SE, Tsolis RM, Bäumler AJ. Streptomycin-induced inflammation enhances Escherichia coli gut colonization through nitrate respiration. Mbio. 2013;4:e00430–13. doi: 10.1128/mBio.00430-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, Dougan G, von Mering C, Hardt WD. Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. Plos Biol. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojiljkovic I, Bäumler AJ, Heffron F. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchAcchBeutEeutJeutGeutH gene cluster. J Bacteriol. 1995;177:1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsolis RM, Adams LG, Ficht TA, Bäumler AJ. Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect Immun. 1999;67:4879–4885. doi: 10.1128/iai.67.9.4879-4885.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsolis RM, Bäumler AJ, Heffron F, Stojiljkovic I. Contribution of TonB- and Feo-mediated iron uptake to growth of Salmonella typhimurium in the mouse. Infect Immun. 1996;64:4549–4556. doi: 10.1128/iai.64.11.4549-4556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weening EH, Barker JD, Laarakker MC, Humphries AD, Tsolis RM, Bäumler AJ. The Salmonella enterica serotype Typhimurium lpf, bcf, stb, stc, std, and sth fimbrial operons are required for intestinal persistence in mice. Infect Immun. 2005;73:3358–3366. doi: 10.1128/IAI.73.6.3358-3366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Thiennimitr P, Nuccio SP, Haneda T, Winter MG, Wilson RP, Russell JM, Henry T, Tran QT, Lawhon SD, Gomez G, Bevins CL, Rüssmann H, Monack DM, Adams LG, Bäumler AJ. Contribution of flagellin pattern recognition to intestinal inflammation during Salmonella enterica serotype Typhimurium infection. Infect Immun. 2009;77:1904–1916. doi: 10.1128/IAI.01341-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Winter MG, Poon V, Keestra AM, Sterzenbach T, Faber F, Costa LF, Cassou F, Costa EA, Alves GE, Paixão TA, Santos RL, Bäumler AJ. Salmonella enterica serovar Typhi conceals the invasion associated type three secretion system from the innate immune system by gene regulation. Plos Pathog. 2014;10:e1004207. doi: 10.1371/journal.ppat.1004207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Santos RL, Tsolis RM, Stender S, Hardt WD, Bäumler AJ, Adams LG. The Salmonella enterica serotype Typhimurium effector proteins SipA, SopA, SopB, SopD, and SopE2 act in concert to induce diarrhea in calves. Infect Immun. 2002;70:3843–3855. doi: 10.1128/IAI.70.7.3843-3855.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Confirmation of S. Typhimurium mutants by PCR. (A) Schematic representation of the target genes and mutagenesis approach. The tonB and feoB coding sequence was interrupted by inserting a kanamycin (KanR) or tetracycline (TetR) resistance gene cassette. The invA gene was disrupted by insertion of a derivative of pGP704. Approximate location of primer binding sites is indicated by small arrows. (B) The indicated S. Typhimurium strains served as templates for the amplification of the various gene targets. PCR products were separated by agarose gel electrophoresis. Approximate size markers are indicated on the left side of the panel.
(A) Histologic scores of individual C57BL/6 mice pre-treated with streptomycin and coinfected with an iron-uptake proficient S. Typhimurium strain (tagged with a phoN mutation) and iron acquisition-deficient mutant strains. Mice were inoculated with 1:1 mixture of 1 x 107 CFU of the iron-uptake proficient S. Typhimurium phoN strain and one of the mutant strains tonB feoB, feoB, tonB, or iroN. Alternatively, mice were inoculated with iron-uptake proficient T3SS-1-deficient S. Typhimurium strain (invA phoN mutant) and one of the mutant strains: invA tonB feoB, invA feoB, invA tonB or invA iroN. Bars represent histopathological scores for submucosal inflammation (SI), mucosal inflammation (MI), and submucosal edema (SE) from individual mice. Summary data are shown in Fig. 2A. (B and C) Histological scoring of cecum (B) and colon (C) from individual C57BL/6 mice treated with 3% DSS and intragastrically coinfected with an iron-uptake proficient S. Typhimurium strain (phoN mutant) and iron acquisition-deficient mutant strains. Scoring was performed as in (A). Summary data are shown in Fig. 3A and 3B.
Intestinal colonization of bovine ligated ileal loops coinfected with S. Typhimurium and defective iron acquisition mutant strains. Each loop was inoculated with a 1 x 109 CFU containing a 1:1 mixture of the S. Typhimurium reference strain (AJB715) with either the tonB feoB; feoB; tonB; or iroN mutant (one loop per pair of strains). Competitive indices in fluid (A) and ileal tissue (B) were determined at 8 hpi. Columns represent geometric mean and standard error obtained from 4 calves used (4 independent experiments) for the ligated ileal loop experiment.
Table S1. Primers used in PCR to confirm mutagenesis in invA tonB feoB, invA feoB, and invA tonB of Salmonella Typhimurium.
Table S2. Primers used for qRT-PCR.