Abstract
As a transcription factor, localization to the nucleus and the recruitment of co-factors to regulate gene transcription is essential. Nuclear localization and nucleosome remodeling and histone deacetylase (NuRD) complex binding are required for the zinc finger transcription factor CASZ1 to function as neuroblastoma (NB) tumor suppressor. However, the critical amino acids (AAs) that are required for CASZ1 interaction with NuRD complex and the regulation of CASZ1 subcellular localization have not been characterized. Through alanine scanning, immunofluorescence cell staining and co-immunoprecipitation we define a critical region at the CASZ1 N-terminus (AA23-40) that mediates the CASZ1b nuclear localization and NuRD interaction. Furthermore, we identify a nuclear export signal (NES) at the N-terminus (AA176-192) that contributes to CASZ1 nuclear-cytoplasmic shuttling in a chromosomal maintenance 1 (CRM1)-dependent manner. An analysis of CASZ1 protein expression in a primary neuroblastoma tissue microarray shows that high nuclear CASZ1 staining is detected in tumors from NB patients with good prognoses. In contrast, cytoplasmic-restricted CASZ1 staining or low nuclear CASZ1 staining is found in tumors from patients with poor prognoses. These findings provide insight into mechanisms by which CASZ1 regulates transcription, and suggests that regulation of CASZ1 subcellular localization may impact its function in normal development and pathologic conditions such as neuroblastoma tumorigenesis.
Keywords: CASZ1, NLS, NES, NuRD, Transcription factor, Tumor suppressor
INTRODUCTION
CASZ1 is a transcription factor that plays a central role in neural and cardiac developmental programs and has been increasingly been implicated in pathologic conditions including cancer (1–9). In humans, CASZ1 localizes to chromosome band 1p36, and loss of heterozygosity of this region is implicated in many types of cancers (6). A clinical case report showed that CASZ1 gene expression was disrupted as a consequence of human papillomavirus DNA integration into the CASZ1 gene locus in a case of cervical cancer (10). In colorectal cancer, CASZ1 and MASP2 fusion transcript was detected, and this fusion product encodes a N-terminal truncated MASP2 that under the control of CASZ1 promoter (11). Our studies have shown that the CASZ1 gene is silenced in poor prognosis neuroblastoma (NB) patient tumor samples through loss of heterozygosity and/or EZH2-mediated epigenetic silencing (12). The human CASZ1 gene encodes two major isoforms: isoform CASZ1a with 1759 AA with 11 TFIIIA class C2H2 zinc fingers and the more evolutionarily conserved isoform CASZ1b with 1166 AA that are identical to CASZ1a but lacks the last 6 zinc finger, and we found that both of them suppress NB tumor growth (7, 13–15).
With next generation sequencing approaches and the emerging sequence variations in the human genome, structure and function analyses of CASZ1 are critical for the ultimate understanding of benign or pathologic alterations in the CASZ1 tumor suppressor gene. In recent studies we identified multiple critical regions that are important for CASZ1 transcriptional activity, and found that CASZ1 interacts with the NuRD complex and the deletion of amino acids (AAs) 31–185 disrupts this interaction (15, 16). However, the critical AAs that mediate NuRD binding and are required for transcriptional activity have not been analyzed in detail. Moreover, nuclear-cytoplasmic shuttling of classic tumor suppressors such as BRCA1, p53 and Rb during tumor progression results in changes in their tumor suppressor capability. This phenomenon highlights the importance of identifying critical regions that mediate CASZ1 subcellular localization and investigating the subcellular localization of CASZ1 in NB tumors from patients.
In this report, we map the nuclear localization signal (NLS1) of the evolutionarily conserved CASZ1b isoform to AAs 23–40, and identify the critical AAs in NLS1 that are essential for CASZ1b transcriptional activity and NuRD complex binding. Moreover, we identify a novel CRM1 (chromosome region maintenance 1, also called XPO1, exportin 1) dependent nuclear export signal (NES) at the N-terminus of CASZ1b. Importantly, we find that localization of CASZ1 in the cytoplasm is associated with poor prognoses in NB patients. The identification of AAs mediating CASZ1 localization provides insights into the molecular mechanisms regulating CASZ1 function in normal development and human disease.
RESULTS
N-terminus of CASZ1b mediates its transcription activity and subcellular localization
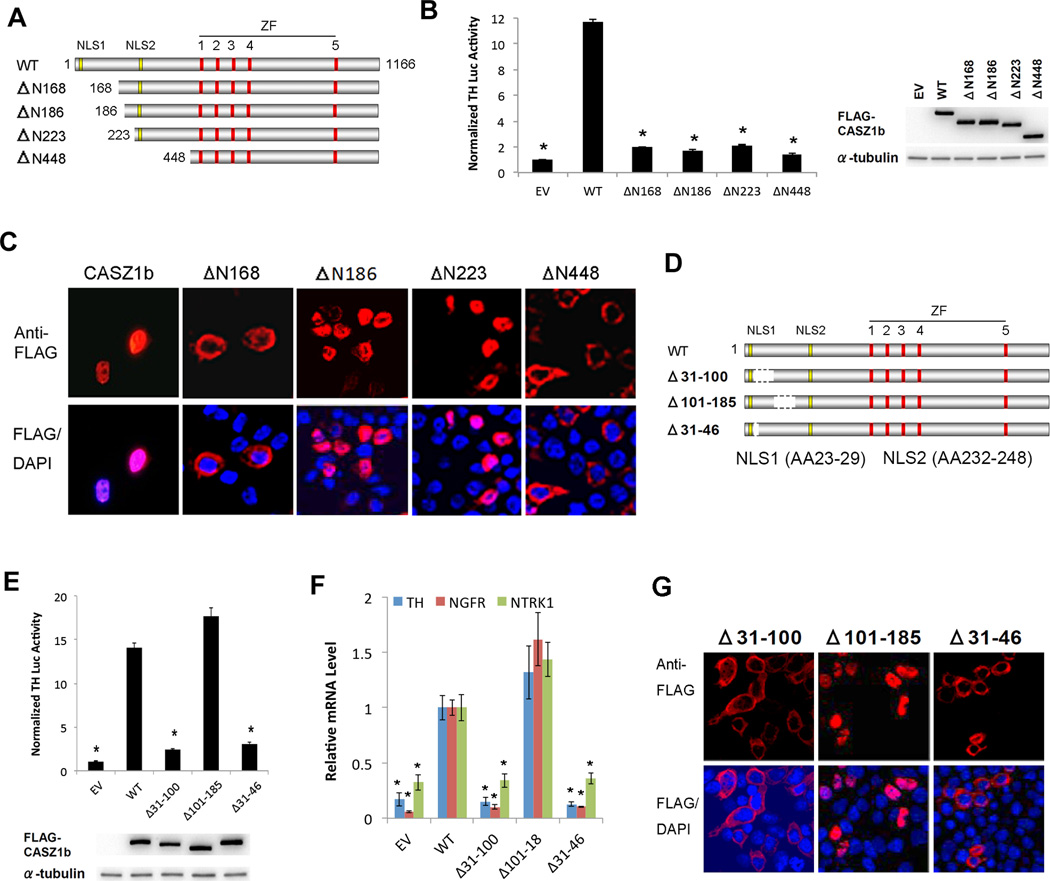
CASZ1b is the more evolutionarily conserved isoform of the CASZ1 gene(14). In a previous report we identified the N-terminus of CASZ1b (AA31–185) to be critical for the regulation of gene transcription and NuRD complex binding (15, 16). However, we did not have single amino acid resolution to show which AAs were critical for CASZ1b transcriptional activity and NuRD complex binding. To refine our understanding of the transcription regulation domain as well as to assess whether other domains in CASZ1 have a role in the regulation of its subcellular localization, we made additional variant N-terminal deletion constructs in an N-terminal FLAG-tagged CASZ1b (Fig. 1A). To investigate the transcription activity of these CASZ1b mutants, 293T cells that have low endogenous CASZ1 expression were transfected. We utilized a Tyrosine Hydroxylase luciferase (TH-luc) reporter assay, as we know TH is transcriptionally regulated by CASZ1 (15). We found that the deletion of amino acids (AA) 1–168, AA1–185, AA1–223 or AA1–448 leads to a significant decrease in the transcriptional activity of wild type (WT) CASZ1b based on the TH-luc reporter assay (Fig. 1B).
Figure 1. N-terminus is important for CASZ1b transcriptional activity and subcellular localization.
(A) Schematic of CASZ1b wild type protein (NLS1-nuclear localization signal 1; NLS2 - nuclear localization signal 2; ZF-zinc finger) and N-terminal deletion proteins. (B) Left panel: Assessment of CASZ1b N-terminal deletions transcriptional activity using TH-Luc reporter (data are shown as means ± S.E.M., *: p<0.001, compared to wild type CASZ1b); right panel: western blot showing steady-state levels of wild type (WT), Δ168, Δ186, Δ223 and Δ448 of CASZ1b protein levels 24 hr after transfection into 293T cells. (C) Assessment of cellular localization of CASZ1b and N-terminus deletion proteins. FLAG-tagged CASZ1b WT and N-terminus deletion plasmids were transfected into 293T cells and stained with anti-FLAG antibody and DAPI 24 hr after. (D) Schematic of CASZ1b wild type and mutant proteins with deletion between NLS1 and NLS2. (E) Top panel: activation of the TH-Luciferase construct 24 hr after transfection of 293T cells by CASZ1b WT, Δ31–100, Δ101–185 and Δ31–46 (data are shown as means ± S.E.M., *: p<0.001, compared to wild type CASZ1b); bottom panel: western blot showing steady-state levels of WT, Δ31–100, Δ101–185 and Δ31–46 of CASZ1b proteins 24 hr after transfection into 293T cells. (F) Realtime PCR to determine CASZ1b mutants transcriptional activity at regulating endogenous TH, NGFR and NTRK1 mRNA transcription, the variants with nucleus localization marked with (+) under the graph (data are shown as means ± S.E.M., *: p<0.005, compared to wild type CASZ1b). (G) Assessment of cellular localization of CASZ1b variants. Cells transfected with FLAG-tagged CASZ1b N-terminus deletion constructs were stained with an anti-FLAG antibody and DAPI.
To assess whether the loss of transcription activity is due to a change in its subcellular localization, FLAG-CASZ1b mutants transfected into 293T cells were stained with anti-FLAG antibody (Fig. 1C). In 293T cells transfected with wild type CASZ1b, the cells have nuclear staining (Fig. 1C). In 293T cells transfected with CASZ1bΔN168 (deletion of AA 1–168), CASZ1 predominantly localizes to the cytoplasm (Fig. 1C), which is not surprising since the NLS1 (AA23–29) is lost. However deletion of an additional 18AA in CASZ1bΔN186 resulted in a nuclear localization presumably utilizing NLS2. This suggests that a nuclear export signal (NES) in AA168–186 may be responsible for the cytosolic localization of CASZ1bΔN168, which counteracts the NLS2 signal. The CASZ1bΔN448 which loses both NLS1 (AA23–29) and NLS2 (AA232–248) localizes only in the cytoplasm (Fig. 1C). The finding that CASZ1bΔN186 transcriptional activity is significantly decreased (Fig. 1B), even though it predominantly localizes to the nucleus (Fig. 1 C), indicates that this region contains transcription regulation domain.
To further define the critical regions required for transcriptional activity of CASZ1b without disrupting nuclear localization, we made a series CASZ1b deletions between NLS1 and NLS2 within the AA31–185 region (Fig. 1D) and transfected these constructs into 293T cells. Deletion of AA101–185 resulted in retention of nuclear localization and transcriptional activity indicating that this region is not required for CASZ1b transcriptional activity (Fig. 1E). The deletion of AA31–46 or AA31–100 leads to a significant loss of transcriptional activity compared with wild type CASZ1b but this may be due to their predominant cytoplasmic localization (Fig. 1E and G). Consistently, realtime PCR results showed that deletion of AA31–46 or AA31–100 but not AA101–185 leads to a significant decrease of endogenous TH, NGFR and TRKA mRNA levels compared with wild type CASZ1b (Fig. 1F). This suggests that although the previously defined NLS1 (AA23–29) can play a role as nuclear localization signal (15), additional AAs after AA23–29 are also critical for proper nuclear retention of CASZ1b.
Alanine scanning to define transcriptional domain and NuRD binding sites
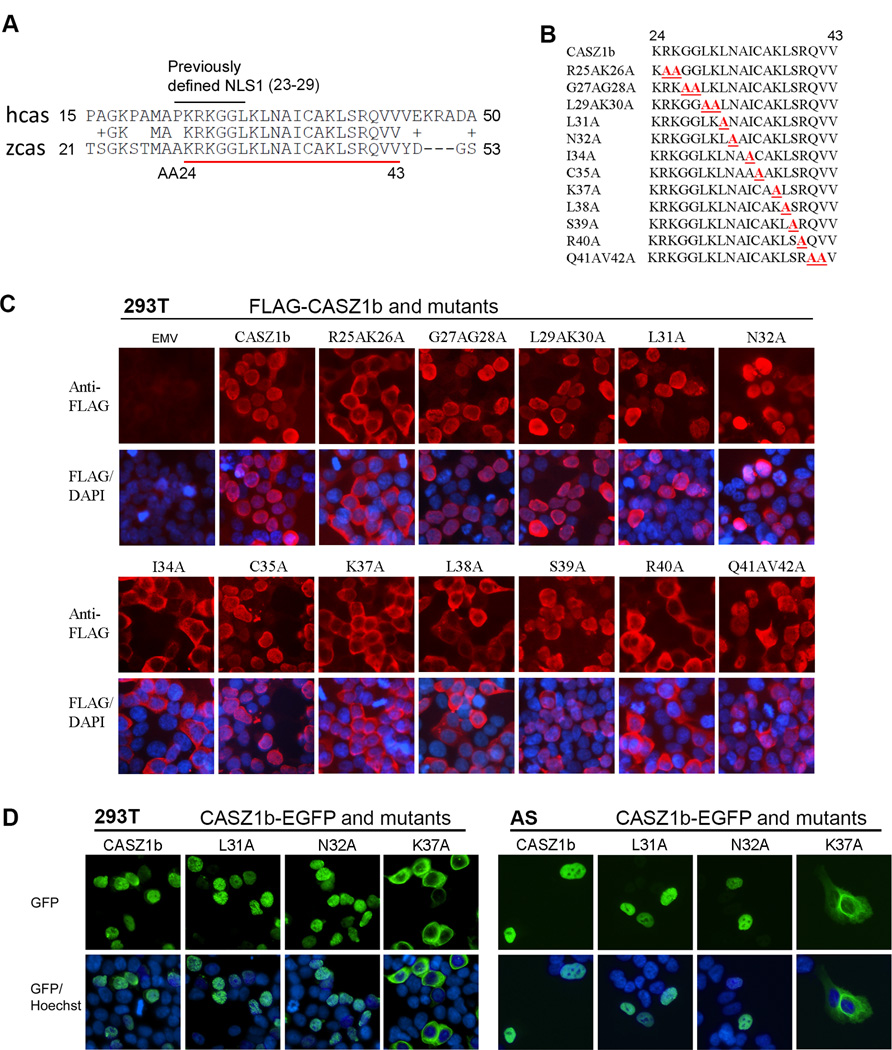
Deletion of AA31–46 led to CASZ1b cytoplasmic localization, which hindered assessment of the critical AAs required for NuRD complex binding and transcriptional activity of CASZ1b. In CASZ1, AA23–43 are evolutionarily conserved (Fig. 2A), so we used an alanine scanning mutation approach in this region to define AAs critical for NuRD binding and transcriptional activity (Fig. 2B). These FLAG-CASZ1b mutants were transiently transfected into 293T cells and stained using anti-FLAG antibody. CASZ1b R25AK26A, I34A, K37A, L38A, R40A mutants localize to the cytoplasm while G27AG28A, L29AK30A, L31A, N32A, C35A, S39, Q41AV42A mainly localize to the nucleus (Fig. 2C). To confirm the subcellular localization of CASZ1b mutants detected through immunofluorescence staining, we also created CASZ1b mutant-EGFP fusion constructs and transfected them into 293T cells and neuroblastoma SK-N-AS (AS) cells. AS cells have loss of heterozygosity on chromosome 1p and the endogenous CASZ1 level is low. Consistent with immunofluorescence staining, the CASZ1L31A–EGFP, and CASZ1N32A–EGFP localize to the nucleus, while the CASZ1-K37A–EGFP localize to the cytoplasm in both of the 293T and AS cell lines (Fig. 2D). These results indicate that the region encompassing AA23–40 is important for CASZ1b nuclear localization.
Figure 2. Alanine scanning to characterize previously defined NLS1 and the conserved region close to NLS1.
(A) The alignment of human CASZ1 (hcas) and zebrafish CASZ1 (zcas) at the N-terminus shows that AA23–42 are evolutionarily conserved. The previously defined NLS1 is marked on top and conserved AAs are underlined in red. (B) The alanine scanning mutations generated in the N-terminus conserved region of CASZ1. (C) Assessment of cellular localization of CASZ1b variants. FLAG-tagged CASZ1b WT and N-terminus deletion constructs were transfected into 293T cells and stained with anti-FLAG antibody and DAPI 24 hr after. (D) Assessment of cellular localization of CASZ1b variants. CASZ1b–EGFP and mutant constructs were transiently transfected into 293T cells (top panel) or SK-N-AS cells (bottom panel) overnight, then the cells were stained with Hoechst.
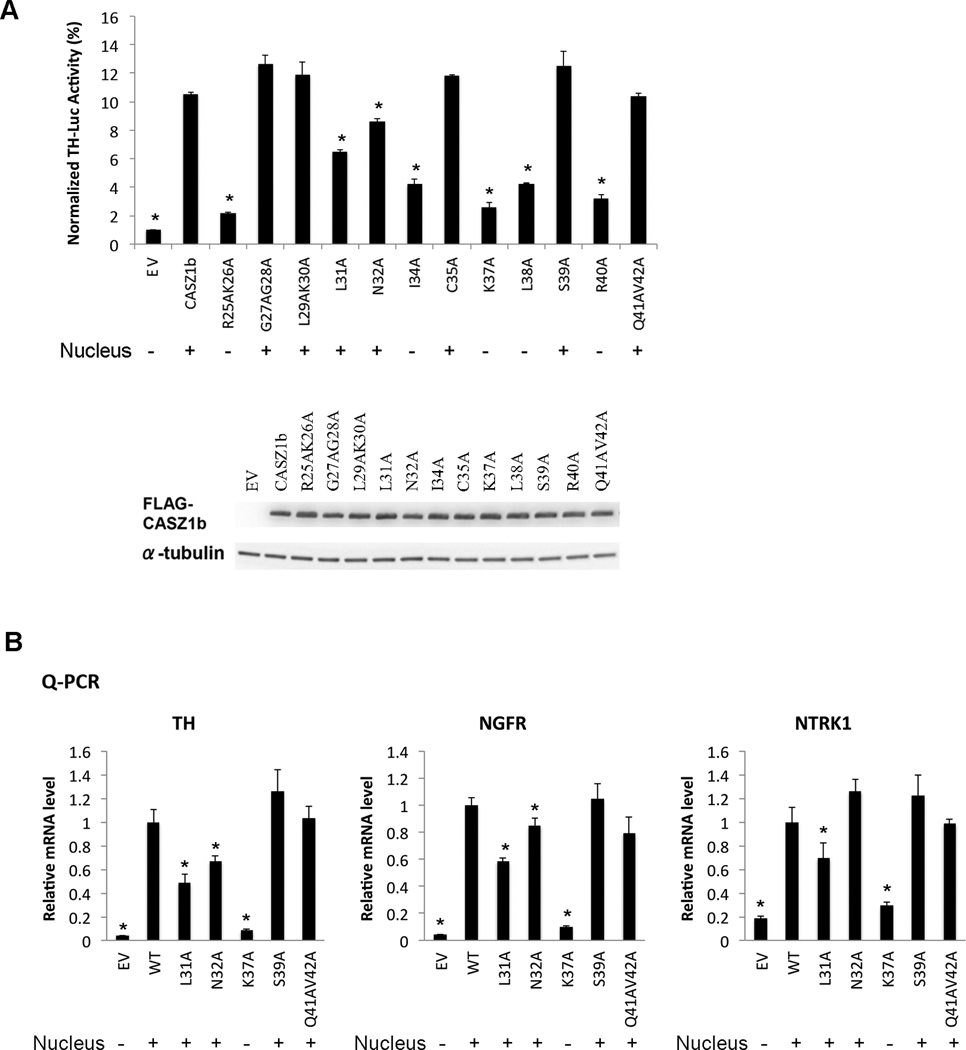
To determine the transcriptional activity of CASZ1b mutants, TH-Luc reporter assay was performed. Results showed that the transcriptional activity of CASZ1b–R25AK26A, -I34A, -K37A, -L38A and -R40A mutants was significantly decreased compared to wild type CASZ1b (Fig. 3A). These results correlated well with their cytoplasmic localization. CASZ1b–G27AG28A, -L29AK30A, -C35A, -S39 and -Q41AV42A mutants localized to the nucleus but retained transcriptional activity (Fig. 3A). However even though CASZ1b–L31A and -N32A mutants localized to the nucleus their transcriptional activity was impaired. L31A had a ~40% decreased transcriptional activity compared to CASZ1b (Fig. 3A). Consistently, realtime PCR results showed that the nuclear localized mutants L31A and cytoplasmic localized K37A also significantly decreased CASZ1b activity in regulating endogenous TH, NGFR and NTRK1 transcription (Fig. 3B). N32A mutant had only a ~25% decreased transcriptional activity at regulating TH-Luc reporter as well as endogenous TH, but had subtle effect on endogenous NGFR and no effect on endogenous NTRK1. This indicates that at the region AA23–40, L31 is the most critical AA for CASZ1b transcriptional activity that does not impact regulation of subcellular localization.
Figure 3. Identification of the critical AAs around the NLS1 region that mediate CASZ1b transcriptional activity.
A. Upper panel: assessment of CASZ1b NLS1 variants transcriptional activity using TH-Luc reporter. The variants with a nuclear localization are marked with (+) under the graph (data are shown as means ± S.E.M., *: p<0.01, compared to wild type CASZ1b); lower panel, western blot showing steady-state levels of CASZ1b and mutants 24 hr after transfection into 293T cells. B. Realtime PCR to determine CASZ1b NLS1 variants transcriptional activity at regulating endogenous TH, NGFR and NTRK1 mRNA transcription, the variants with nucleus localization marked with (+) under the graph (data are shown as means ± S.E.M., *: p<0.05, compared to wild type CASZ1b).
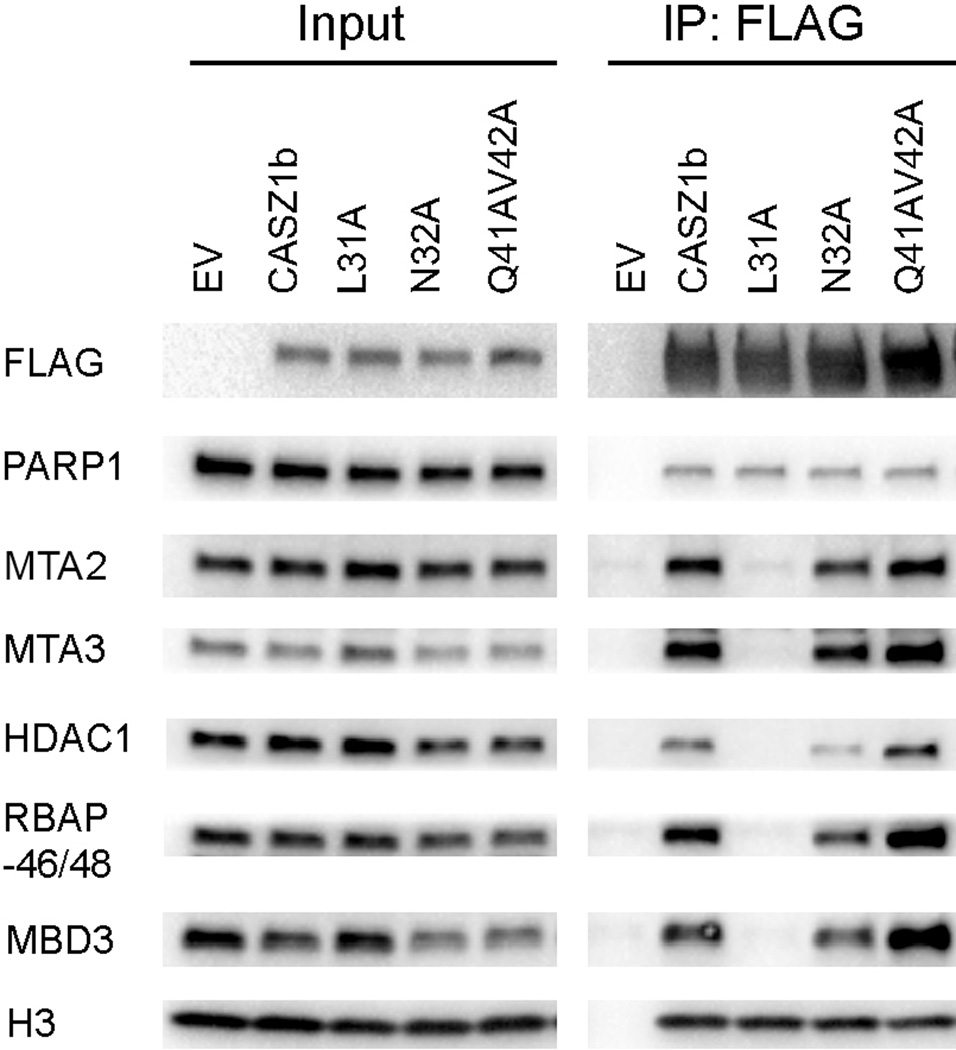
Previously we have shown that CASZ1 transcriptional activity is dependent on NuRD binding (16). To determine whether the decrease of the transcriptional activity of CASZ1b–L31A is associated with NuRD complex binding ability, EV, CASZ1b, L31A, N32A and Q41V42A constructs were transfected into 293T cells, and co-immunoprecipitation was performed. We found that while CASZ1b pulled down NuRD subunits, only the L31A mutant completely loses the ability to pull down NuRD subunits such as MTA2, MTA3, HDAC1, RBAP46/48 and MBD3 (Fig. 4). The disruption of the interaction between CASZ1b mutant L31A and NuRD subunits was specific since L31A retained its interaction with PARP1 and histone H3 (Fig. 4), this is expected since CASZ1b interacts with PARP1 and histone H3 through a PAR binding motif in the Zn-finger region of CASZ1b protein (16).
Figure 4. NLS1L31 is critical for NuRD complex binding.
Co-IP of FLAG-CASZ1b and mutant constructs transfected 293T cells using anti-FLAG antibody and the blot were probed with anti-FLAG, PARP1, MTA2, HDAC1, RBAP46/48, MBD3 and histone H3 antibody.
CASZ1b regulates genes that are important for cell growth and developmental processes in neuroblastoma
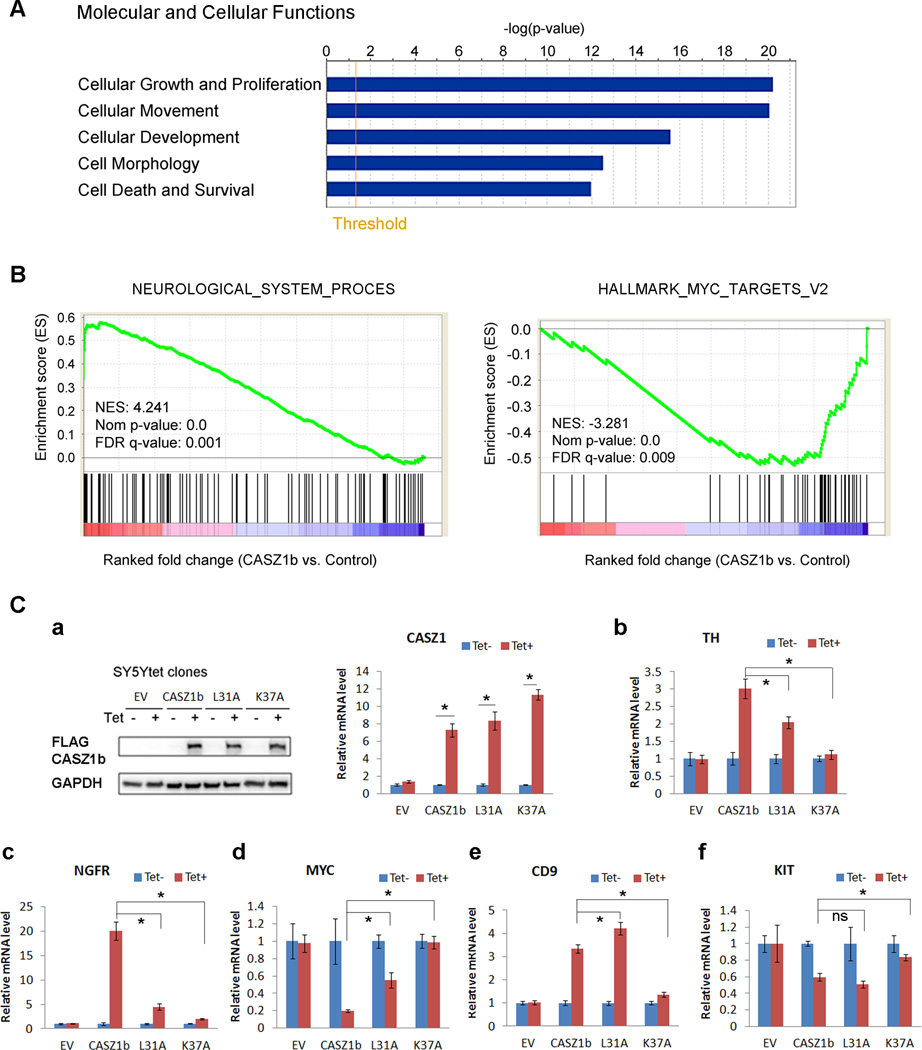
As the transcriptome regulated by CASZ1b isoform has not been investigated in neuroblastoma to date, we evaluated the global transcriptome regulated by CASZ1b to gain insight into potential targets and mechanisms of action. We used Agilent whole human genome oligo microarray analyses (G4112F, Agilent, Santa Clara, CA) to investigate the genome wide transcriptional consequences of expression of CASZ1b in NB cells (SY5YtetCASZ1b) in which CASZ1b gene expression is regulated by tetracycline (Tet). After Tet-induction for 24 hr (Tet+), 483 genes were commonly regulated by CASZ1b in SY5YtetCASZ1b three replicates (Supplementary Table 1, increased/decreased >2-fold, p<0.05). Among those regulated genes, clusterin (CLU) and NGFR also have NB tumor suppressor activity (17, 18). Molecular and cellular function assay of these 483 differently expressed genes using Ingenuity Pathways Analysis (IPA) tool, indicated that the top 5 classes of the altered genes involved in cell growth and proliferation, cellular movement, cellular development, cell morphology as well as cell death and survival (Fig. 5A, Supplementary Table 2). Physiological system development and function assay using IPA tool showed that genes involved in nervous system and cardio-vasculature development were significantly enriched in the genes regulated by CASZ1b (data not shown). To further gain insights from the microarray data, we performed gene set enrich analysis (GSEA) (19, 20) using the whole microarray gene list (Tet+ vs. Tet-). GSEA is able to determine whether an a priori defined set of genes shows a statistically significant difference between CASZ1b induced and control SY5Y cells (Tet+ vs. Tet-). The results showed that the”neurological_system_process” genes (genes involved in the process pertaining to the functions of the nervous system of an organism) were positively enriched in CASZ1b overexpressed cells (Fig. 5B, left panel; Supplementary Table 3). Moreover, the”hallmark_MYC_targets_v2” genes (a subgroup of genes regulated by MYC) were negatively enriched in CASZ1b overexpressed cells (Fig. 5B, right panel; Supplementary Table 4). Similarly, the “hallmark_MYC_targets_v1” genes (another subgroup of genes regulated by MYC) were also negatively enriched in CASZ1b overexpressed cells (data not shown). These results are consistent with CASZ1 being a neural fate-determination gene and tumor suppressor.
Figure 5. Transcriptome regulated by CASZ1b in NB cells.
(A) IPA assay of CASZ1b targets in SY5Y cells determined by microarray showed gene enrichment in the categories of molecular and cellular functions with a key subcategory being cell growth and proliferation (threshold represents p=0.05). (B) GSEA assay indicated the positive enrichment of genes involved neurological system process (left panel), and negative enrichment of genes regulated by MYC (right panel). NES, normalized enrichment score; Nom, nominal; FDR, false discovery rate. (C) Transcriptional activity assay of cytoplasmic localized K37A mutant and loss of NuRD binding mutant L31A in SY5Y cells by realtime PCR. a, Left panel: western blot analysis of CASZ1b and mutant proteins from SY5YtetCASZ1b and CASZ1b mutant cells after 24 h Tet treatment; right panel: realtime PCR analysis of CASZ1b and mutant mRNA levels from SY5YtetCASZ1b and CASZ1b mutant cells after 24 h Tet treatment. b-f, realtime PCR result verified the regulation of TH, NGFR, CD9, MYC and KIT by CASZ1b in SY5Y cells after 24 h Tet treatment. Compared to wild type CASZ1b, K37 almost completely lost transcriptional activity at regulating all these genes in SY5Y cells (data are shown as means ± S.E.M., *: p<0.05). Compared to wild type CASZ1b, L37 significantly decreased transcriptional activity at regulating TH, NGFR and MYC, but had a slight increase at regulating CD9 and no effect on KIT (data are shown as means ± S.E.M., *: p<0.05).
To investigate whether cytoplasmic sequester or loss of NuRD complex binding affects CASZ1b transcriptional activity in NB cells, we generated SY5YtetCASZ1b K37A stable cell line and SY5YtetCASZ1b L31A stable cell line and investigated the transcriptional activity of CASZ1b, L31A and K37A at regulating genes identified from microarray analysis. Consistent with the microarray data, we found that the induction of CASZ1b by Tet significantly up-regulated TH, NGFR, CD9, and significantly down-regulated MYC and MYC expression (Fig. 5C). The gene expression changes were not due to Tet treatment since their expression did not change when SY5YtetEV cells were treated with Tet (Fig. 5C). We found that the cytoplasmic localized K37A mutant almost completely lost transcriptional activity at regulating all the tested endogenous genes that have been regulated by wild type CASZ1b in SY5Y cells, which include TH, NGFR, CD9, MYC and KIT (Fig. 5C). The L31 mutant that fails to bind NuRD complex partially defective at regulating TH, NGFR and MYC transcription compared to wild type CASZ1b, but did not decrease transcriptional activity at regulating CD9 and KIT, instead there was a slight increase at regulating CD9 compared to wild type CASZ1b (Fig. 5C). This result indicates that cytoplasm retention of CASZ1b in neuroblastoma cells disrupt its downstream transcriptional pathways. Additionally the failure of CASZ1b to recruit NuRD complex may partially affect its ability to regulate a subset of CASZ1b target genes.
Identification of nuclear export signal (NES) in CASZ1b
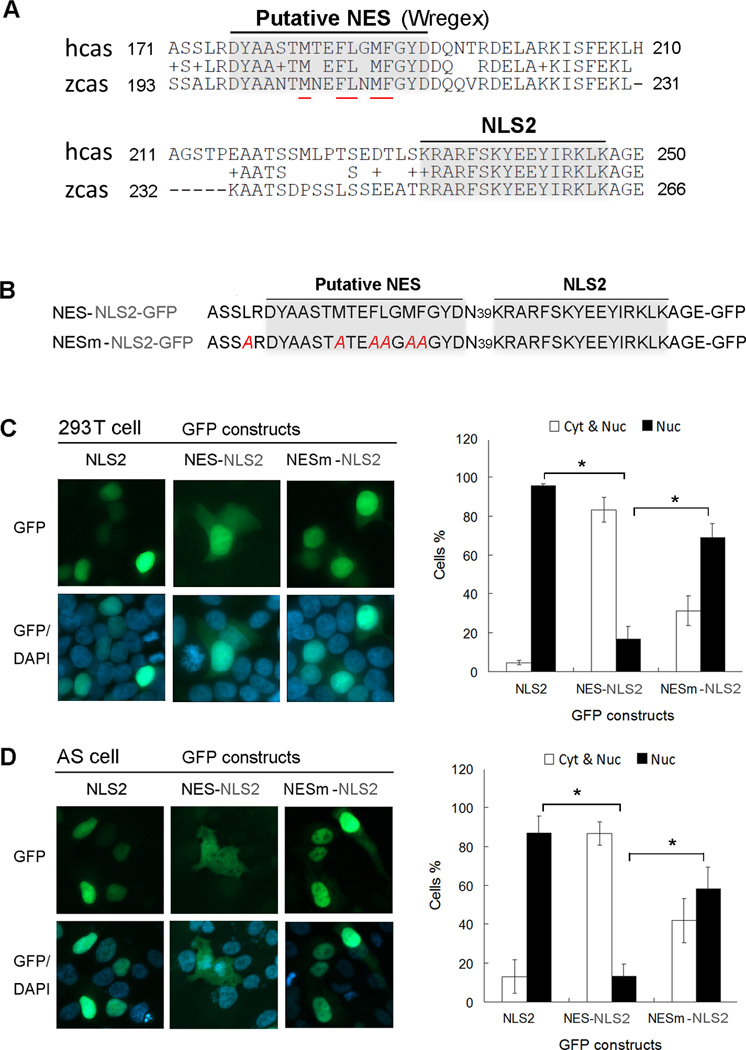
The CASZ1bΔN168 whose deletion encompasses NLS1 (Fig. 1A and C) has a cytosolic localization despite the retention of NLS2 (AA232–247). However the loss of an additional 18AA in the CASZ1bΔN186 mutant enables CASZ1 nuclear localization (Fig. 1A, C). These findings suggest the existence of a putative NES around AA168-AA186 region that functions to export CASZ1 when the NLS1 is deleted, but when the putative NES is lost or disrupted (CASZ1bΔN186), it allows NLS2 to function to retain the truncated CASZ1b in the nucleus. To test this hypothesis, we performed in silico bioinformatics analysis (http://wregex.ehubio.es/) of CASZ1b using Wregex (weighted regular expression) 1.1 program that has been developed for motif scanning (21). It revealed a CRM1 dependent NES motif in CASZ1b (AA176–192) (Fig. 6A). Hydrophobic AAs are critical for NES activity (22, 23). Of the 17 AAs in this region, 5 AAs are hydrophobic and they are all evolutionarily conserved between CASZ1 homologs in human and zebrafish (Fig. 6A, hydrophobic AAs are underlined in red).
Figure 6. CASZ1b contains nuclear export signal (NES) at the N-terminus.
(A) The alignment of human CASZ1 (hcas) and zebrafish CASZ1 (zcas) at the N-terminal region that contains a putative NES with 5 conserved hydrophobic AAs (underlined in red) between AA175 to AA191. (B) The NES-NLS2-GFP and NES mutant-NLS2-GFP constructs. CASZ1b AA170–249 region that contains both putative NES and NLS2 was fused to GFP. NESm-NLS2-GFP is a NES mutant constructed by replacing the 5 conservative hydrophobic AAs within putative NES and one hydrophobic AA close to NES with alanine (highlighted in red). (C) 293T cells and (D) AS cells were transiently transfected with NLS2-GFP, NES-NLS2-GFP and NESm-NLS2-GFP constructs, chromatin was stained with DAPI. A minimum of 300 cells were screened for each group from at least 3 different fields and scored for nuclear and cytoplasmic (Nuc & Cyt) or nuclear only (Nuc) staining (data are shown as means ± S.D., *: p<0.001). None of the cells have cytoplasmic only GFP signal, thus Cyt was not shown in the graph.
We hypothesized that in CASZ1b the putative NES can compete with NLS2. When the NES is intact (as in CASZ1bΔ168), it may over-ride the NLS2 nuclear localization signal, thus CASZ1bΔ168 stays in the cytoplasm. When the NES is disrupted (as in CASZ1bΔ186), only NLS2 plays a role, so CASZ1bΔ186 has a nuclear location. To test this hypothesis, we generated NLS2 (AA232–247) or NES-NLS2 (AA171–250) GFP fusion proteins (Fig. 6A and B). NLS2-GFP contains the NLS2 of CASZ1b, and NES-NLS2-GFP contains both the putative NES and the NLS2 of CASZ1b. To investigate the subcellular localization of these constructs, we transfected the NLS2-GFP or NES-NLS2-GFP plasmids into 293T cells (Fig. 6C). In over 90% of the transfected cells, NLS2-GFP localized exclusively to the nucleus. In contrast less than 20% of cells transfected with the NES-NLS2-GFP localized solely to the nucleus, which is significantly lower than NLS2-GFP (Fig. 6C, right panel, black columns, p<0.001). This is consistent with the putative NES having a nuclear export function. We further mutated the putative NES in the NES-NLS2-GFP construct by replacing the conserved hydrophobic AAs with alanine (Fig. 6B) and named this construct as NESm-NLS2-GFP. In contrast to NES-NLS2-GFP, NESm-NLS2-GFP mainly localizes to the nucleus in over 60% of the transfected 293T cells, which is significantly higher than NES-NLS2-GFP (Fig. 6C, right panel, black columns, p<0.001). Similar results were found when NES-NLS2-GFP and its mutant were transfected into the neuroblastoma AS cell line (Fig. 6D). We also generated NES-EGFP and NESm-EGFP constructs. We found that EGFP alone has a more nuclear localization perhaps due to diffusion (24), however, the GFP signal in NES-EGFP transfected cells is more evenly distributed between the cytoplasm and nucleus. When the NES is mutated, there are more cells with a stronger nuclear GFP signal (supplementary Fig. 1). Our results are consistent with this NES region having a nuclear export role in this reporter system.
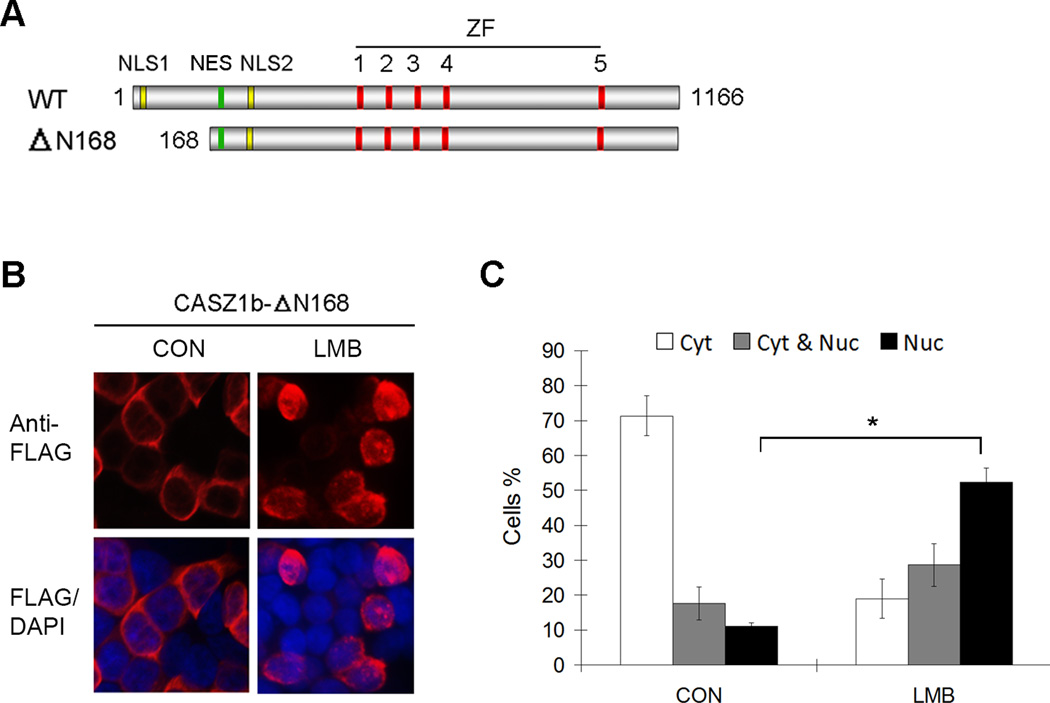
Most of the identified NESs regulate protein nuclear export in a CRM1-dependent manner. Leptomycin B (LMB) treatment can inactivate CRM-1 through covalent modification at a cysteine residue of CRM1 (23, 25, 26). To determine whether the NES in CASZ1b is CRM1-dependent, we utilized the CASZ1bΔN168 construct since it contains the NES and NLS2 (Fig. 7A), and it localizes to the cytoplasm in 293T cells. FLAG-CASZ1bΔN168 transfected 293T cells were treated with 20 ng/ml LMB for 3.5 hr, fixed and stained with anti-FLAG antibody (Fig. 7B). In untreated cells, approximately 10% of the cells contained solely nuclear CASZ1bΔN168. In contrast, in LMB treated cells there was an almost 5-fold increase in cells with nuclear CASZ1bΔN168 compared to controls (Fig. 7C, black columns, p<0.005).
Figure 7. CASZ1b nuclear export is CRM1 dependent.
(A) Schematic of CASZ1b wild type protein (NLS1-nuclear localization signal 1; NLS2 - nuclear localization signal 2; NES - nuclear export signal; ZF-zinc finger) and CASZ1b Δ168. (B) CASZ1b Δ168 was transfected into 293T cells for 24 hr, and treated the cells with or without the CRM1 inhibitor LMB (20 ng/ml) for 3.5 hr. The cells were stained with an anti-FLAG antibody and DAPI. (C) To generate the graph, a minimum of 300 cells were screened for each group from at least 3 different fields and scored for nuclear and cytoplasmic (Nuc & Cyt) or nuclear only (Nuc) staining (data are shown as means ± S.D., *: p<0.005).
The subcellular localization of CASZ1 differs in a tissue dependent manner
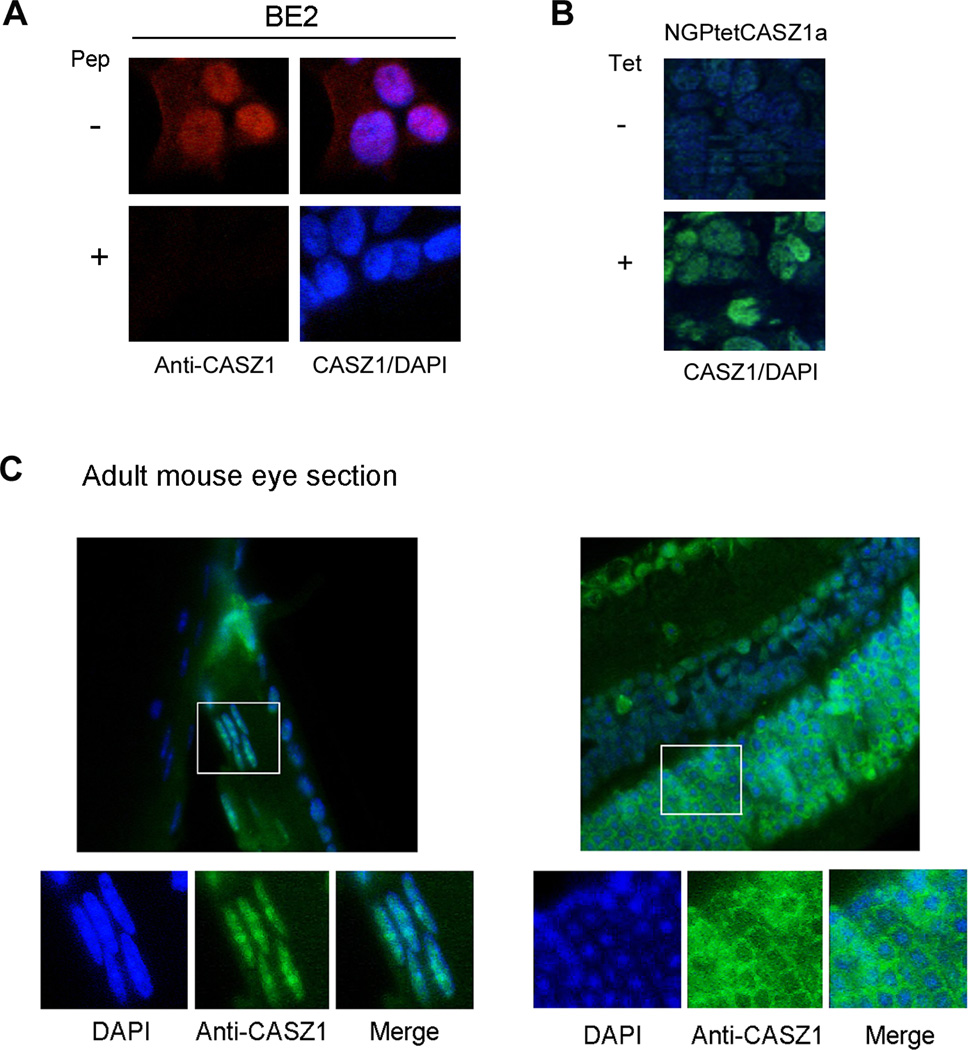
The existence of NLS and NES in CASZ1 indicates nuclear-cytoplasmic shuttling of the CASZ1 protein may occur during normal development and this shuttling may regulate CASZ1 function. To investigate the subcellular localization of CASZ1 in different cell or tissue types, an antibody specific for human and murine CASZ1 was used, and this antibody recognize the common epitope on both CASZ1a and CASZ1b isoforms. The anti-CASZ1 antibody was first used to stain SK-N-BE2 NB cells, which have high endogenous CASZ1 expression, and we found that CASZ1 was predominantly expressed in the nucleus (Fig. 8A, top panel). Prior incubation of the anti-CASZ1 antibody with the immunizing peptide that was used to generate CASZ1 antibody completely blocked CASZ1 immunoreactivity in SK-N-BE2 cells (Fig. 8A, bottom panel). To test whether anti-CASZ1 antibody can be used on formalin fixed tumor tissue, NGPtetCASZ1a xenograft tumor samples were incubated with anti-CASZ1 antibody. NGPtetCASZ1a is a cell model in which CASZ1a expression can be induced by tetracycline. Staining results showed that specific CASZ1 immunoreactivity is detected in NGPtetCASZ1a tumor xenograft from a tetracycline treated mouse (Fig. 8B, top) but not in the NGPtetCASZ1a tumor xenograft from a control treated mouse (Fig. 8B, bottom). These studies indicate that the anti-CASZ1 antibody is specific and can be used in formalin fixed tissue.
Figure 8. The subcellular localization of CASZ1 is tissue type dependent.
(A) Endogenous CASZ1 subcellular localization in BE2 cells was determined by staining the cells with rabbit anti-CASZ1 antibody. The anti-CASZ1 antibody was pre-incubated with (bottom panel) or without (top panel) CASZ1 antigen peptide (Pep) that used to generate CASZ1 antibody. The chromatin was stained with DAPI. (B) Mouse xenograft tumor of human NB cell line that has been stably transfected with tetracycline inducible CASZ1 (NGPtetCASZ1) was dissected and stained with rabbit anti-CASZ1 antibody and DAPI. (C) Endogenous CASZ1 in the eye of adult mouse was stained with rabbit anti-CASZ1 antibody and DAPI.
We next investigated subcellular localization of CASZ1 in normal murine ocular tissue since CASZ1 is highly expressed in this tissue at both the mRNA and protein level (14, 27). By staining adult murine ocular tissue sections for CASZ1, we found that CASZ1 localizes to the nucleus in lens epithelia but localizes primarily to the cytoplasm in photoreceptor cells (Fig. 8C). This is consistent with the finding that CASZ1 localizes to the cytoplasm of photoreceptor cells during retinogenesis (8). The findings of distinct nuclear and cytoplasmic localization of CASZ1 in different ocular tissues suggest that the subcellular localization of CASZ1 may play a role in mediating its functional activity during normal development.
Expression and subcellular localization of CASZ1 protein in NB patients’ tumor samples
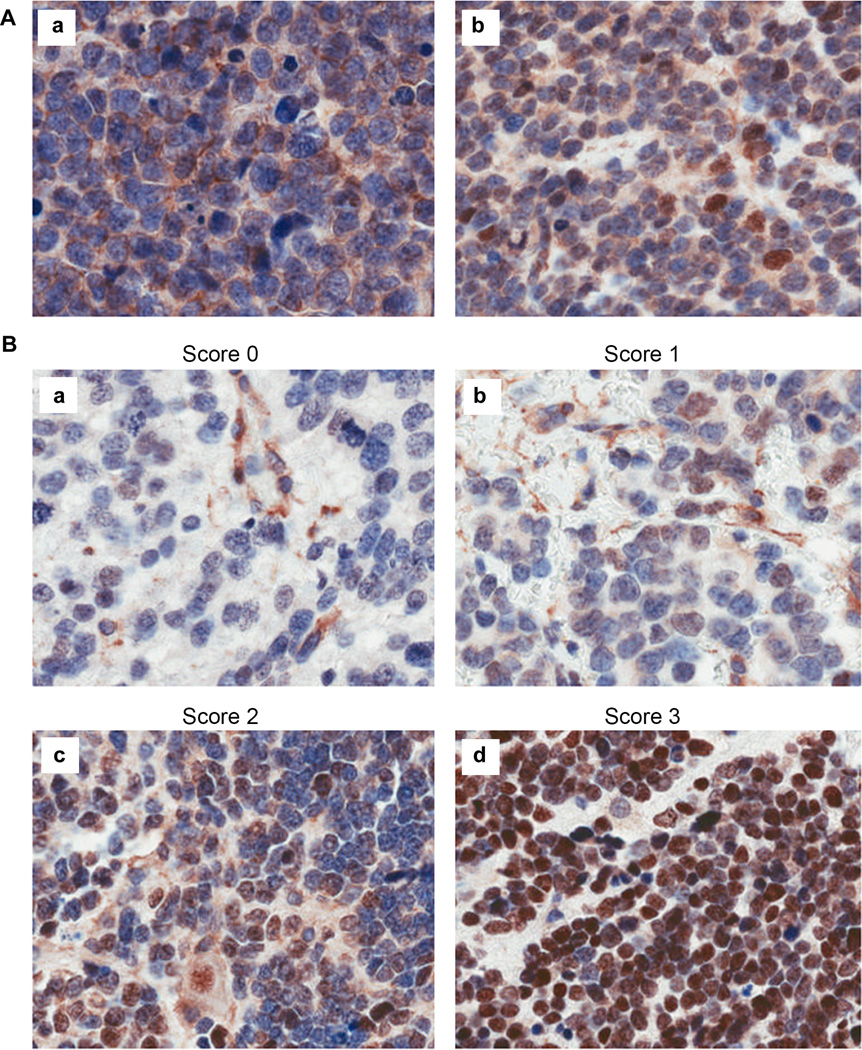
Although we have previously reported that high levels of CASZ1 mRNA are found in primary NB tumors from patients who have good prognoses (7, 14), the expression of CASZ1 protein in primary NB tumor tissue has never been assessed. To determine the association of CASZ1 protein levels and NB patients’ risk factors, we stained a tissue microarray (TMA) containing primary NB tumor samples with an anti-CASZ1 antibody. The TMA contained duplicate cores for each sample, and only those samples with consistent results in the duplicates were used for data analysis. Among the 112 evaluable patient tumor samples, 94 samples (84%) were found to have detectable CASZ1 expression. CASZ1 protein localized to the nucleus in 80% of the 94 samples while in 10% of the tumor samples, CASZ1 localized to both the nucleus and cytoplasm. In the remaining 10%, CASZ1 localized only to the cytoplasm of the tumor cells. Representative images of cytoplasmic only and nuclear plus cytoplasmic staining patterns of CASZ1 are shown in Fig. 9A.
Figure 9. CASZ1 is differentially expressed in NB patient tumor samples and may localize to the nucleus, the cytoplasm or both.
(A) a. CASZ1 localized exclusively in the cytoplasm showed by staining the tumor samples with anti-CASZ1 antibody; b. CASZ1 localized to both the nucleus and the cytoplasm. Nucleus was counterstained with hematoxylin solution showed in blue, and CASZ1 was probed with rabbit anti-CASZ1 antibody and showed in brown. (B) Examples of nuclear staining: a. Score 0 - a rare positive nuclei; b. Score 1 (1–10% positive) equivocal/uninterpretable; c. Score 2 (10–50%) weak positive, or d. Score 3 (> 50% positive cells)-strong positive.
We next evaluated the association of nuclear CASZ1 protein expression with the prognostic factors of NB patients. Representative images are shown in Fig. 9B for the four scoring categories of CASZ1 immunoreactivity detected in the TMA: (1) Score 0: negative, no reactivity to <1% positive nuclei; (2) score 1: 1–10% positive nuclei; (3) score 2: 10–50% positive nuclei; (4) score 3: strong positive, greater than 50% positive nuclei, respectively. Using 10% positive nuclear staining as a cut off, Chi-square analysis showed that the samples scoring 0 and 1 with low nuclear CASZ1 expression are associated with MYCN amplification, unfavorable Shimada histology and high risk (Table 1, all p<0.05; chromosome 1p status not available for this archival case series). Amplification of the MYCN gene (greater than 5 copies) or unfavorable Shimada histology (a histopathological classification system) are molecular and histologic parameters that are found in high-risk neuroblastoma patients (28–30). Among the 27 tumor samples scoring 0, nine samples (33%) had CASZ1 expression with the protein localizing only to the cytoplasm. Interestingly for the samples with CASZ1 restricted to the cytoplasm, 90% (7/8samples) of the ones with available information on Shimada pathology showed unfavorable Shimada histology. This contrasts with the tumors that had strong nuclear CASZ1 staining, scoring 2 and 3, in which 57% (38/67) showed significant favorable Shimada histology (Table 1, p=0.044). While the sample size is small, these data are consistent with a model in which the retention of CASZ1 in the cytosol may contribute to loss of CASZ1 function in the tumors of some NB patients who have poor prognoses.
Table 1.
Association of high nuclear CASZ1 protein level (score 2,3) with Prognostic Factors (n=112)
| Factor# | n | Number (%) of patients with high CASZ1 expression |
p-value* |
|---|---|---|---|
| Age | |||
| < 18 mo | 54 | 38 (70) | 0.0567 |
| ≥ 18 mo | 57 | 29 (51) | |
| Unknown | 1 | ||
| INSS Stage | |||
| 1,2,3,4s | 83 | 56 (67) | 0.0632 |
| 4 | 23 | 10 (43) | |
| Unknown | 6 | ||
| MYCN | |||
| Not amplified | 95 | 62 (65) | 0.0294 |
| Amplified | 9 | 2 (22) | |
| Unknown | 8 | ||
| Shimada histology | |||
| Favorable | 41 | 38 (93) | 0.0440 |
| Unfavorable | 54 | 29 (54) | |
| Unknown | 7 | ||
| Risk Group | |||
| Low/Intermediate | 69 | 51 (74) | 0.0167 |
| High | 30 | 14 (47) | |
| Unknown | 13 |
From the International Neuroblastoma Risk Group (INRG) classification system (29)
From Chi-square Test
DISCUSSION
The transcriptional activity of transcription factors is modulated by their subcellular localization and the co-factors they recruit. In this study, we defined AA23–40 in CASZ1b as critical for nuclear localization as well as NuRD complex interaction and regulation of gene transcription. We also identified and functionally characterized a CRM1- dependent NES domain at the N-terminus of CASZ1b that may be relevant to its subcellular localization in different cell types. Moreover, we found that high nuclear expression of CASZ1 in neuroblastoma tumors is associated with good prognostic features while tumors with cytoplasmic localization of CASZ1 are more frequently found in NB patients with poor prognoses.
Nuclear-cytoplasmic shuttling of transcription factors is an important way to control their activity in response to stress or extracellular environmental signals and this is regulated through NES and NLS. Through four approaches including (1) bioinformatic in silico assay of CASZ1b (Fig. 6A), (2) NES-NLS2-GFP subcellular localization study (Fig. 6C), (3) CASZ1bΔ168/ CASZ1bΔ186 subcellular localization study (Fig. 1C), and (4) CASZ1bΔ168 subcellular study with or without CRM1 inhibitor treatment (Fig. 7), we demonstrate that there is a NES signal within AA168–192 in the CASZ1 protein. The existence of both NLS and NES may provide a rapid mechanism to regulate CASZ1 transcriptional activity during development. Normal tissue staining revealed that CASZ1 could localize either to the nucleus or the cytoplasm (Fig. 8), indicating that CASZ1 subcellular localization is regulatable in normal tissues. In fact many critical cell fate determination transcription factors are tightly regulated by multiple NLS and NES motifs including SOX protein and T-box protein (31–33). For example, the SOX proteins contain both NLS and NES that are regulated by a nuclear export mechanism during murine embryonic development (33). Transcription factors with tumor suppressor activity such as BRAC1, p53 and RB also have both NLS and NES and can be shuttled between the nucleus and cytoplasm under stress or during tumor progression resulting in changes in their transcriptional activity and tumor suppressor capability (34–39). The p53 protein has two NESs and three NLSs (38, 40, 41): the N-terminal NES is regulated by phosphorylation events in response to DNA damage (40), and the NES in the tetramerization domain is masked upon p53 oligomerization (38). An area of future investigations is an assessment of which signal transduction pathways regulate CASZ1 nuclear-cytoplasmic localization.
Previously we have found that low levels of CASZ1mRNA are found in NB tumors that have a poor prognosis and high levels are found in those that have a better prognosis (7, 14). The TMA study shows tumors enriched in nuclear CASZ1 are from patients that have a low- and medium-risk or favorable Shimada histology. Yet in 10% of CASZ1 positive tumor samples, the CASZ1 protein was completely localized to the cytoplasm and these tumors have unfavorable Shimada histology and these patients have a poor prognosis. We have shown that when CASZ1b localizes to the cytoplasm it dramatically decreases its ability to suppress neuroblastoma cell growth (15). In this study, the transcriptome analysis of CASZ1b in NB indicates that CASZ1b regulates genes involved in cell growth and movement (Fig. 5A), and GSEA showed that CASZ1b represses MYC target genes (Fig. 5B), which supports CASZ1b function as a tumor suppressor. Importantly, the cytoplasmic localized mutant K37A almost completely lost the ability to regulate the transcription of all the tested CASZ1b target genes in NB cells , indicating that the decreased nuclear accumulation of CASZ1 disrupts its downstream transcriptional program. Thus under certain conditions, the shuttling of CASZ1 from the nucleus to the cytoplasm decreases its transcriptional activity and consequently impairs its tumor suppressing function. This functional loss of CASZ1 transcriptional activity may contribute to tumor progression. CRM1-dependent cytosolic localization has been shown to functionally reduce the activity of cancer-related proteins such as p53, Rb, APC, c-Abl and FOXO-3A. For this reason a number of nuclear export inhibitors have been developed and some show antitumor activity in different mouse tumor xenograft models, and are in early phase clinical testing (42–47). Whether this is a mechanism relevant for CASZ1 remains an area of future investigation.
The CASZ1 NES is not required for its transcription activity since CASZ1bΔN101–185, which has disrupted the NES has the same transcription activity as wild type CASZ1b (Fig. 1D–F). However our data reveal that the re-defined NLS1 is required for its nuclear localization as well as its transcriptional activity. Consistently, co-IP results demonstrate that CASZ1b recruits NuRD complex through this NLS1 region to regulate gene transcription (Fig. 4). The L31A mutant disrupts CASZ1 interaction with NuRD complex decreasing its transcriptional activity, supporting our model in which NuRD mediates CASZ1b transcriptional activity (16). We observed that CASZ1b L31A fails to bind the NuRD complex but only partially decreased transcriptional activity. NuRD has been reported to remodel chromatin to regulate gene transcription, or interact with p-TEFb to facilitate transcription elongation (48, 49). As a co-factor, NuRD complex can either enhance or decrease basal gene transcription regulation of a transcription factor, thus although CASZ1b L31A fails to bind NuRD, it may still retain the ability to regulate basal gene transcription via interacting with other co-factors and transcriptional machinery. In fact, we observed that CASZ1b L31A retains PARP1 binding (Fig. 4), a DNA repair protein that being reported to regulate chromatin structure and gene transcription (50).
In summary, we have identified and characterized critical AAs that regulate CASZ1 nuclear-cytoplasmic localization and co-factors interactions that modulate its transcriptional activity. Nuclear-cytoplasmic shuttling would be an important way to regulate the CASZ1 gene regulatory network during normal development and its disruption may contribute to pathological conditions.
MATERIALS AND METHODS
Cell culture
Human embryonic kidney cells (HEK293T) were obtained from ATCC, and were maintained in Dulbecco’s modified Eagle’s media. Neuroblastoma SK-N-AS and SK-N-BE2 cell lines were maintained in RPMI1640 media. All the cell culture medium was supplemented with 10% fetal calf serum as well as 100µg/mL streptomycin, 100U/mL penicillin, and L-glutamine. Cells were grown at 37°C with 5% CO2. Neuroblastoma NGPtetCASZ1 cells are NGP cells that were stably transfected with CASZ1a and its expression is tetracycline inducible (7). The cell lines used have been authenticated and are mycoplasma free.
Construction of missense, deletion, and fusion CASZ1b constructs and NES-NLS-GFP constructs
To make the NES-NLS2-GFP construct, the NES-NLS2 fragment of CASZ1b (AA171–250) was generated by PCR amplification prior to cloning, the primer used was synthesized with an attached 5’ EcoRI site and a 3’ BamHI site and was cloned into the corresponding sites of pmaxFP-Green-N vector (Lonza). To make the NES-EGFP construct, the NES fragment of CASZ1b (AA171–210) was generated by PCR amplification prior to cloning, the primer used was synthesized with an attached 5’ EcoRI site and a 3’ BamHI site and was cloned into the corresponding sites of pEGFP-N3. To make the CASZ1b–EGFP constructs, full length of CASZ1b was generated by PCR, and the primer used was synthesized with an attached 5’ EcoRI site and a 3’ BamHI site and was cloned into the corresponding sites of pEGFP-N3. The QuikChange XL Site-Directed Mutagenesis Kit (Agilent Technologies) was utilized to introduce various missense mutations into the wild type FLAG-CASZ1b plasmid (13), NES-EGFP or the NES-NLS2-GFP construct as per the manufacturer’s protocol. To generate CASZ1b–EGFP mutant constructs, the FLAG-CASZ1b mutants were sub-cloned into CASZ1b–EGFP through SbfI and PshAI enzyme digestion.
Luciferase assay
Plasmids were transiently transfected into HEK293T cells using the Lipofectamine 2000 cationic lipid reagent (Thermo Scientific) according to the manufacturer’s protocol. In luciferase experiments, the TH promoter – pGL4.1-luc construct (TH-Luc), which contains the proximal 2kb of the TH promoter, was generously provided by Dr. Gregory Wray (51). A CMV-driven β-galactosidase construct was co-transfected with TH-Luc and different CASZ1b mutants in order to provide an internal control for transfection efficiency. Luciferase activity was quantified after 24 hr using the Luciferase Reporter Assay System (Promega), and beta-galactosidase activity was measured concomitantly with the Luminescent Beta-galactosidase Detection Kit II (Clontech) to normalize the luciferase signals.
Realtime PCR
Total mRNA was collected 24 hours after transfection using the RNeasy Mini Kit (Qiagen) as per the manufacturer’s protocol. Endogenous transcription of known CASZ1 target gene Tyrosine Hydroxylase (TH), NGFR, NTRK1 (7), and some other newly identified CASZ1b target genes from this study were used to assess transcriptional activity of CASZ1b variants. Quantitative measurements of total β-actin and target gene levels were obtained using the BIO-RAD CFX Touch Real-Time PCR detection system and performed in triplicate. Ct values were standardized to β-actin levels, and the fold change in mRNA was calculated compared to the empty vector transfected control samples.
Stable clones
The neuroblastoma stable clone was made as reported previously (15). In brief, wild type CASZ1b, the CASZ1b L31A mutant and the CASZ1b mutant K37A cloned in pT-Rex-DEST30 were transfected into SH-SY5Ytet (SY5Ytet) cells that had been previously stably cloned with pcDNA6/TR . The transfected cells were cultured in selective medium containing both blasticidin S and G418. After selection, individual colonies were selected and treated with tetracycline (Tet) in order to induce CASZ1b transcription. Stable clones expressing CASZ1b or CASZ1 mutations are labeled SY5YtetCASZ1b, SY5YtetCASZ1bL31A (L31A) and SY5YtetCASZ1bK37A (K37A). The stable clone with empty vector are labeled SY5YtetEV. Pilot experiment results indicated that the protein levels of wild type CASZ1b and mutant CASZ1b stay the same when SY5YtetCASZ1b cells have been treated with 3.3 ng/ml Tet and L31A or K37A cells have been treated with 1 µg/ml Tet. Thus when compare the transcriptional activity of wild type CASZ1b and mutant CASZ1b in SY5Y cells, 3.3 ng/ml Tet was used for SY5YtetCASZ1b cells and 1 µg/ml Tet was used for SY5YtetEV, L31A and K37A cells. Otherwise the stable clones will be treated with 1 µg/ml Tet to induce CASZ1b expression. The cells used have been authenticated and are mycoplasma free.
The oligo microarray
Agilent whole human genome oligo microarray kit (G4112F, Agilent) was used to determine the CASZ1-induced transcriptional profiles in SY5YtetCASZ1b cells. Gene expression with or without Tet induction (24 h) was analyzed in biological triplicates. Array hybridization, chemiluminescence detection and image acquisition and analysis were performed by GenUs Biosystems (Northbrook, IL, USA). Intensity values are normalized to the 75th percentile of each array. The list of differentially expressed genes with 2-fold change, p<0.05 was also generated by GenUs Biosystems (Northbrook, IL, USA) using GeneSpring GX v7.3.1 software. P-values are based on paired T-test of three ratios of each pair of the replicates. The 2-fold differentially expressed gene list was uploaded to IPA (Ingenuity http://www.ingenuity.com/), and these were used to generate unbiased molecular and cellular functional analyses. Gene set enrich analysis (GSEA) (19, 20) was performed using the whole gene list generated by uploading the raw microarray data to Partek Genomics Suite 6.6. Intensity values were normalized to the 75th percentile of each array and the genes with intensity values smaller than 1 were excluded. A one-way ANOVA was used to detect differentially expressed genes. This whole gene list was pre-ranked based on fold change, then uploaded to GSEA software.
Protein isolation, co-immunoprecipitation and western blot analysis
For assessment of protein levels, cells were lysed 24 hours after transfection using RIPA buffer, and 10µg of total protein was separated and electroblotted as described previously (14). Anti-FLAG M2 monoclonal antibody (Sigma-Aldrich, catalog #F1804), anti-GAPDH antibody (Santa Cruz Biotechnology, catalog #sc-25778), anti-α-tubulin antibody (Cell Signaling, catalog #2144) were used as primary antibody. Protein bands were detected using a goat anti-mouse or rabbit IgG-HRP conjugated secondary antibody (Santa Cruz Biotechnology, catalog #sc-2030 and sc-2031) and visualized using enhanced chemiluminescence (Amersham Biosciences). Co-immunoprecipitation was performed as previously described (16). In brief, HEK293T cells in 10 cm dishes were transiently transfected with empty vector (EV) or FLAG-CASZ1b or mutant constructs for 24 hr and whole cell extracts were incubated with ANTI-FLAG M2 Magnetic Beads (Sigma-Aldrich, catalog #M8823) and agitated at 4°C for 4 hr. The co-IP products were eluted by incubating with 2x SDS loading buffer and boiling for 3 min. Rabbit anti-HDAC1, RBAP46/48, MBD3, and PARP1 antibody (Cell Signaling, catalog #2062, #4633, #14540 and #9532), anti- MTA3 antibody (Bethyl, A300-160A) and anti-histone H3 (active motif, catalog #39163) were used as primary antibody.
Analysis of CASZ1 subcellular localization
Different cell lines were transiently transfected with FLAG-CASZ1a, FLAG-CASZ1b, CASZ1b–EGFP, NLS2-GFP, NES-NLS2-GFP as well as deletion or mutation constructs as described above in 8-well Lab-Tek Chamber Slides or regular 12-well plates. To inhibit nuclear export, 20 hr after transfection, the transfected cells were treated with 20 ng/ml of leptomycin B (LMB) for 3.5 hr before fixation. Cells were fixed, permeabilized, blocked, and stained as described previously (14). For indirect immunofluorescent cell staining, an anti-FLAG M2 monoclonal antibody (Sigma, catalog: #F1804) and an Alexa Fluor 594-conjugated goat anti-mouse antibody (ThermoFisher Scientific, catalog: #A-11005) were used to detect FLAG-CASZ1b. Adult mouse eye sections and mouse xenograft tumors of human NB cell line that has been stably transfected with tetracycline inducible CASZ1 (NGPtetCASZ1) were dissected and stained as following the previously described protocol (4). A rabbit anti-CASZ1 antibody (1:1000) (made in collaboration with Rockland antibodies & assays) was used to detect CASZ1 protein. Localization was analyzed and imaged using a Zeiss LSM 710 confocal microscope (Zeiss), a Zeiss microscope ApoTome 2 or a Nikon Eclipse TE300 microscope. To quantify the subcellular localization of NLS2-GFP, NES-NLS2-GFP and NESm-NLS2-GFP, as well as CASZ1bΔ168 treated with or without LMB, 100 cells were counted from each filed, and three fields were counted for each experiment.
Tissue microarray (TMA) was performed as previously reported (52). Cases for the neuroblastoma tissue microarray were selected from a review of peripheral neuroblastic tumors accessioned to the Pathology Department of the Children’s Hospital of Philadelphia from 1987 to 2004. Selection of specimens and construction of the TMA followed approval by the Children’s Hospital of Philadelphia Institutional Review Board. All tumors were reviewed by a pediatric pathologist (BP) for adequacy, and classified pathologically using INPC criteria. The resultant TMA comprised two paraffin blocks and contained duplicate tumor cores from 187 samples obtained from 167 patients, 20 of who had multiple specimens. Cores (0.6 mm) of representative tumor tissue from each case were used to construct the tissue microarray blocks using a manual arrayer (Beecher Instruments, Inc., Sun Prairie, WI). Cores from normal tissues were placed in each of the tissue microarray blocks as controls. Mouse xenograft tumors of human NB cell line that has been stably transfected with tetracycline inducible CASZ1 (NGPtetCASZ1) was used as control to optimize the staining protocol. The TMA was stained with anti-rabbit CASZ1 (Rockland antibodies) at a dilution of 1:1000. Only those samples with consistent results in the duplicates were used for data analysis. Chi-square test was used for TMA data analysis.
Statistical analyses
Statistical analyses of the TMA data were performed using chi-square test with p < 0.05 considered significant. Statistical analyses of other data were performed using t-test with p < 0.05 considered significant. The statistical tests were two-sided. IPA of microarray data was analyzed by Fisher’s exact test (http://www.ingenuity.com/). Kolmogorov-Smirnov statistic was used by GSEA for the microarray data analysis. Values in the graphs are expressed as means ± S.E.M. or means ± S.D. from technical triplicates, and the data presented are the representative results from one of the two or three reproducible biological replicates.
Supplementary Material
Acknowledgments
We would like to thank: Dr. Gregory Wray from Institute for Genome Sciences & Policy, Duke University for generously providing the tyrosine hydroxylase promoter-pGL4.1-luc (TH-Luc) construct; Nisha Pawer and Neeraj Prasad of the POB, NCI for making the CASZ1b and mutant – EGFP constructs. We appreciate the insightful discussions with Drs. Dinah Singer and David Levens of the CCR, NCI on this study. This work was supported by the Intramural Research Program of the NIH, National Cancer Institute and Center for Cancer Research.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
SUPPLEMENTARY DATA
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
REFERENCES
- 1.Cui X, Doe CQ. ming is expressed in neuroblast sublineages and regulates gene expression in the Drosophila central nervous system. Development. 1992;116(4):943–952. doi: 10.1242/dev.116.4.943. [DOI] [PubMed] [Google Scholar]
- 2.Mellerick DM, Kassis JA, Zhang SD, Odenwald WF. castor encodes a novel zinc finger protein required for the development of a subset of CNS neurons in Drosophila. Neuron. 1992;9(5):789–803. doi: 10.1016/0896-6273(92)90234-5. [DOI] [PubMed] [Google Scholar]
- 3.Charpentier MS, Christine KS, Amin NM, Dorr KM, Kushner EJ, Bautch VL, et al. CASZ1 promotes vascular assembly and morphogenesis through the direct regulation of an EGFL7/RhoA-mediated pathway. Developmental cell. 2013;25(2):132–143. doi: 10.1016/j.devcel.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Z, Li W, Ma X, Ding N, Spallotta F, Southon E, et al. Essential role of the zinc finger transcription factor Casz1 for mammalian cardiac morphogenesis and development. The Journal of biological chemistry. 2014;289(43):29801–29816. doi: 10.1074/jbc.M114.570416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jordan VK, Zaveri HP, Scott DA. 1p36 deletion syndrome: an update. Appl Clin Genet. 2015;8:189–200. doi: 10.2147/TACG.S65698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henrich KO, Schwab M, Westermann F. 1p36 tumor suppression--a matter of dosage? Cancer research. 2012;72(23):6079–6088. doi: 10.1158/0008-5472.CAN-12-2230. [DOI] [PubMed] [Google Scholar]
- 7.Liu Z, Yang X, Li Z, McMahon C, Sizer C, Barenboim-Stapleton L, et al. CASZ1, a candidate tumor-suppressor gene, suppresses neuroblastoma tumor growth through reprogramming gene expression. Cell death and differentiation. 2011;18(7):1174–1183. doi: 10.1038/cdd.2010.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mattar P, Ericson J, Blackshaw S, Cayouette M. A conserved regulatory logic controls temporal identity in mouse neural progenitors. Neuron. 2015;85(3):497–504. doi: 10.1016/j.neuron.2014.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takeuchi F, Isono M, Katsuya T, Yamamoto K, Yokota M, Sugiyama T, et al. Blood pressure and hypertension are associated with 7 loci in the Japanese population. Circulation. 2010;121(21):2302–2309. doi: 10.1161/CIRCULATIONAHA.109.904664. [DOI] [PubMed] [Google Scholar]
- 10.Schmitz M, Driesch C, Beer-Grondke K, Jansen L, Runnebaum IB, Durst M. Loss of gene function as a consequence of human papillomavirus DNA integration. International journal of cancer Journal international du cancer. 2012;131(5):E593–E602. doi: 10.1002/ijc.27433. [DOI] [PubMed] [Google Scholar]
- 11.Hoff AM, Johannessen B, Alagaratnam S, Zhao S, Nome T, Lovf M, et al. Novel RNA variants in colorectal cancers. Oncotarget. 2015 doi: 10.18632/oncotarget.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang C, Liu Z, Woo CW, Li Z, Wang L, Wei JS, et al. EZH2 Mediates epigenetic silencing of neuroblastoma suppressor genes CASZ1, CLU, RUNX3, and NGFR. Cancer research. 2012;72(1):315–324. doi: 10.1158/0008-5472.CAN-11-0961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Z, Yang X, Tan F, Cullion K, Thiele CJ. Molecular cloning and characterization of human Castor, a novel human gene upregulated during cell differentiation. Biochem Biophys Res Commun. 2006;344(3):834–844. doi: 10.1016/j.bbrc.2006.03.207. [DOI] [PubMed] [Google Scholar]
- 14.Liu Z, Naranjo A, Thiele CJ. CASZ1b, the short isoform of CASZ1 gene, coexpresses with CASZ1a during neurogenesis and suppresses neuroblastoma cell growth. PloS one. 2011;6(4):e18557. doi: 10.1371/journal.pone.0018557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Virden RA, Thiele CJ, Liu Z. Characterization of critical domains within the tumor suppressor CASZ1 required for transcriptional regulation and growth suppression. Molecular and cellular biology. 2012;32(8):1518–1528. doi: 10.1128/MCB.06039-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Z, Lam N, Thiele CJ. Zinc finger transcription factor CASZ1 interacts with histones, DNA repair proteins and recruits NuRD complex to regulate gene transcription. Oncotarget. 2015 doi: 10.18632/oncotarget.4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chayka O, Corvetta D, Dews M, Caccamo AE, Piotrowska I, Santilli G, et al. Clusterin, a haploinsufficient tumor suppressor gene in neuroblastomas. J Natl Cancer Inst. 2009;101(9):663–677. doi: 10.1093/jnci/djp063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schulte JH, Pentek F, Hartmann W, Schramm A, Friedrichs N, Ora I, et al. The low-affinity neurotrophin receptor, p75, is upregulated in ganglioneuroblastoma/ganglioneuroma and reduces tumorigenicity of neuroblastoma cells in vivo. International journal of cancer Journal international du cancer. 2009;124(10):2488–2494. doi: 10.1002/ijc.24204. [DOI] [PubMed] [Google Scholar]
- 19.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature genetics. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 21.Prieto G, Fullaondo A, Rodriguez JA. Prediction of nuclear export signals using weighted regular expressions (Wregex) Bioinformatics. 2014;30(9):1220–1227. doi: 10.1093/bioinformatics/btu016. [DOI] [PubMed] [Google Scholar]
- 22.la Cour T, Gupta R, Rapacki K, Skriver K, Poulsen FM, Brunak S. NESbase version 1.0: a database of nuclear export signals. Nucleic Acids Res. 2003;31(1):393–396. doi: 10.1093/nar/gkg101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.la Cour T, Kiemer L, Molgaard A, Gupta R, Skriver K, Brunak S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel. 2004;17(6):527–536. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- 24.Seibel NM, Eljouni J, Nalaskowski MM, Hampe W. Nuclear localization of enhanced green fluorescent protein homomultimers. Anal Biochem. 2007;368(1):95–99. doi: 10.1016/j.ab.2007.05.025. [DOI] [PubMed] [Google Scholar]
- 25.Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. cell. 1997;90(6):1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 26.Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B, et al. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci U S A. 1999;96(16):9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vacalla CM, Theil T. Cst, a novel mouse gene related to Drosophila Castor, exhibits dynamic expression patterns during neurogenesis and heart development. Mech Dev. 2002;118(1–2):265–268. doi: 10.1016/s0925-4773(02)00282-4. [DOI] [PubMed] [Google Scholar]
- 28.Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nature reviews cancer. 2003;3(3):203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 29.Cohn SL, Pearson AD, London WB, Monclair T, Ambros PF, Brodeur GM, et al. The International Neuroblastoma Risk Group (INRG) classification system: an INRG Task Force report. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27(2):289–297. doi: 10.1200/JCO.2008.16.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maris JM. Recent advances in neuroblastoma. The New England journal of medicine. 2010;362(23):2202–2211. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rehberg S, Lischka P, Glaser G, Stamminger T, Wegner M, Rosorius O. Sox10 is an active nucleocytoplasmic shuttle protein, and shuttling is crucial for Sox10-mediated transactivation. Molecular and cellular biology. 2002;22(16):5826–5834. doi: 10.1128/MCB.22.16.5826-5834.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kulisz A, Simon HG. An evolutionarily conserved nuclear export signal facilitates cytoplasmic localization of the Tbx5 transcription factor. Molecular and cellular biology. 2008;28(5):1553–1564. doi: 10.1128/MCB.00935-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malki S, Boizet-Bonhoure B, Poulat F. Shuttling of SOX proteins. The international journal of biochemistry & cell biology. 2010;42(3):411–416. doi: 10.1016/j.biocel.2009.09.020. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez JA, Henderson BR. Identification of a functional nuclear export sequence in BRCA1. The Journal of biological chemistry. 2000;275(49):38589–38596. doi: 10.1074/jbc.M003851200. [DOI] [PubMed] [Google Scholar]
- 35.Jiang J, Yang ES, Jiang G, Nowsheen S, Wang H, Wang T, et al. p53-dependent BRCA1 nuclear export controls cellular susceptibility to DNA damage. Cancer research. 2011;71(16):5546–5557. doi: 10.1158/0008-5472.CAN-10-3423. [DOI] [PubMed] [Google Scholar]
- 36.Foo RS, Nam YJ, Ostreicher MJ, Metzl MD, Whelan RS, Peng CF, et al. Regulation of p53 tetramerization and nuclear export by ARC. Proc Natl Acad Sci U S A. 2007;104(52):20826–20831. doi: 10.1073/pnas.0710017104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nie L, Sasaki M, Maki CG. Regulation of p53 nuclear export through sequential changes in conformation and ubiquitination. The Journal of biological chemistry. 2007;282(19):14616–14625. doi: 10.1074/jbc.M610515200. [DOI] [PubMed] [Google Scholar]
- 38.Stommel JM, Marchenko ND, Jimenez GS, Moll UM, Hope TJ, Wahl GM. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18(6):1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiao W, Datta J, Lin HM, Dundr M, Rane SG. Nucleocytoplasmic shuttling of the retinoblastoma tumor suppressor protein via Cdk phosphorylation-dependent nuclear export. The Journal of biological chemistry. 2006;281(49):38098–38108. doi: 10.1074/jbc.M605271200. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, Xiong Y. A p53 amino-terminal nuclear export signal inhibited by DNA damage-induced phosphorylation. Science. 2001;292(5523):1910–1915. doi: 10.1126/science.1058637. [DOI] [PubMed] [Google Scholar]
- 41.Liang SH, Clarke MF. A bipartite nuclear localization signal is required for p53 nuclear import regulated by a carboxyl-terminal domain. The Journal of biological chemistry. 1999;274(46):32699–32703. doi: 10.1074/jbc.274.46.32699. [DOI] [PubMed] [Google Scholar]
- 42.Mutka SC, Yang WQ, Dong SD, Ward SL, Craig DA, Timmermans PB, et al. Identification of nuclear export inhibitors with potent anticancer activity in vivo. Cancer research. 2009;69(2):510–517. doi: 10.1158/0008-5472.CAN-08-0858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ranganathan P, Yu X, Na C, Santhanam R, Shacham S, Kauffman M, et al. Preclinical activity of a novel CRM1 inhibitor in acute myeloid leukemia. Blood. 2012;120(9):1765–1773. doi: 10.1182/blood-2012-04-423160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakakibara K, Saito N, Sato T, Suzuki A, Hasegawa Y, Friedman JM, et al. CBS9106 is a novel reversible oral CRM1 inhibitor with CRM1 degrading activity. Blood. 2011;118(14):3922–3931. doi: 10.1182/blood-2011-01-333138. [DOI] [PubMed] [Google Scholar]
- 45.Nguyen KT, Holloway MP, Altura RA. The CRM1 nuclear export protein in normal development and disease. International journal of biochemistry and molecular biology. 2012;3(2):137–151. [PMC free article] [PubMed] [Google Scholar]
- 46.Gerecitano J. SINE (selective inhibitor of nuclear export)--translational science in a new class of anti-cancer agents. J Hematol Oncol. 2014;7:67. doi: 10.1186/s13045-014-0067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ishizawa J, Kojima K, Hail N, Jr, Tabe Y, Andreeff M. Expression, function, and targeting of the nuclear exporter chromosome region maintenance 1 (CRM1) protein. Pharmacol Ther. 2015;153:25–35. doi: 10.1016/j.pharmthera.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26(37):5433–5438. doi: 10.1038/sj.onc.1210611. [DOI] [PubMed] [Google Scholar]
- 49.Bottardi S, Mavoungou L, Pak H, Daou S, Bourgoin V, Lakehal YA, et al. The IKAROS interaction with a complex including chromatin remodeling and transcription elongation activities is required for hematopoiesis. PLoS Genet. 2014;10(12):e1004827. doi: 10.1371/journal.pgen.1004827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krishnakumar R, Kraus WL. PARP-1 regulates chromatin structure and transcription through a KDM5B–dependent pathway. Mol cell. 2010;39(5):736–749. doi: 10.1016/j.molcel.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warner LR, Babbitt CC, Primus AE, Severson TF, Haygood R, Wray GA. Functional consequences of genetic variation in primates on tyrosine hydroxylase (TH) expression in vitro. Brain Res. 2009;1288:1–8. doi: 10.1016/j.brainres.2009.06.086. [DOI] [PubMed] [Google Scholar]
- 52.Winter C, Pawel B, Seiser E, Zhao H, Raabe E, Wang Q, et al. Neural cell adhesion molecule (NCAM) isoform expression is associated with neuroblastoma differentiation status. Pediatr Blood cancer. 2008;51(1):10–16. doi: 10.1002/pbc.21475. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.