Abstract
Flap endonuclease-1 (FEN1) is a multifunctional, structure-specific nuclease that has a critical role in maintaining human genome stability. FEN1 mutations have been detected in human cancer specimens and have been suggested to cause genomic instability and cancer predisposition. However, the exact relationship between FEN1 deficiency and cancer susceptibility remains unclear. In the current work, we report a novel colorectal cancer-associated FEN1 mutation, L209P. This mutant protein lacks the FEN, exonuclease (EXO) and gap endonuclease (GEN) activities of FEN1 but retains DNA-binding affinity. The L209P FEN1 variant interferes with the function of the wild-type FEN1 enzyme in a dominant-negative manner and impairs long-patch base excision repair in vitro and in vivo. Expression of L209P FEN1 sensitizes cells to DNA damage, resulting in endogenous genomic instability and cellular transformation, as well as tumor growth in a mouse xenograft model. These data indicate that human cancer-associated genetic alterations in the FEN1 gene can contribute substantially to cancer development.
Introduction
Genomic DNA is constantly exposed to endogenous and exogenous insults, which cause DNA damage. The damage to DNA, if not repaired, could lead to genetic mutations and subsequent genome instability and cancer initiation.1, 2 Removal of DNA damage and maintenance of genomic integrity depend on robust cellular DNA repair systems. Base excision repair (BER) is one of the major repair pathways in eukaryotic cells for processing DNA base damage caused by endogenous and exogenous agents.3, 4 It is estimated that BER is responsible for repairing about 104 damaged/modified bases per cell per day.5, 6, 7 BER can be divided into two types: short-patch BER (SP-BER) and long-patch BER (LP-BER).8, 9 LP-BER is the major pathway used to repair oxidized bases in the nuclei and mitochondria.10, 11 Furthermore, previous reports have shown that defects in LP-BER can lead to DNA double-strand breaks (DSBs) and genomic instability.12, 13
BER is initiated with the excision of the damaged base by a specific DNA glycosylase, followed by incision of the DNA backbone by apurinic/apyrimidinic (AP) endonuclease 1 (APE1) to produce a nicked abasic intermediate.14 This intermediate structure can be processed through either the SP-BER or the LP-BER pathway.8, 9 In SP-BER, DNA polymerase β (Pol β) adds only one nucleotide to the 3′-end of the nicked AP site, and then the dRP lyase activity of Pol β catalyzes the β-elimination of the 5′-sugar phosphate residue, resulting in a ligatable nick that can then be sealed by X-ray repair cross-complementing protein 1 and Ligase IIIa (XRCC1/Ligase IIIa).15, 16 However, if the 5′-deoxyribose phosphate moiety is reduced or oxidized, the 5′-deoxyribose phosphate lyase of Pol β cannot remove the modified sugar residue and LP-BER is initiated.17, 18 In LP-BER, a few nucleotides are added by Pol β to produce a 2–10 nucleotide (nt)-long short flap, which is subsequently removed by FEN1 in complex with proliferating cell nuclear antigen (PCNA).19, 20, 21 DNA ligase I then seals the nick.8
FEN1 has a central role in LP-BER and therefore is indispensable for maintaining genome stability and integrity.17, 22 FEN1 is a structure-specific nuclease with 5′ flap endonuclease (FEN), 5′–3′ exonuclease (EXO) and gap endonuclease (GEN) activities.23 Deletion of FEN1 in Saccharomyces cerevisiae (rad27) results in a high level of sensitivity to DNA damage reagents such as ultraviolet irradiation and methyl methane sulfonate.24, 25 The complete removal of FEN1 activity via homozygous knockout causes early embryonic lethality in mice.26 Furthermore, knock-in of a FEN1 mutation with deficiency in GEN and EXO activity results in a high incidence of lung adenoma in a mouse model.27 The data suggest a linkage between functional defects in FEN1 and increased cancer risk in humans.28, 29, 30 However, it is still unclear whether and how these variations in the FEN1 gene impact cancer initiation and progression.
Even though somatic mutations have been found in many other DNA repair genes in a wide range of cancers,31 FEN1 mutation is very rare,32 suggesting that FEN1 is important for normal DNA metabolism. Recently, four somatic, non-synonymous, heterozygous single-nucleotide substitutions in FEN1 were identified in human colon cancer samples.33 These are E20D, L209P, R245G and S329N. It will be important to determine whether these mutations affect FEN1's function and contribute to cancer initiation and development. Among the four FEN1 mutations, L209P is particularly interesting because the residue at position 209 is conserved from archaea, such as the hyperthermophile Pyrococcus furiosus, to humans.34
In the current study, we found that the FEN1 L209P mutation significantly reduces the FEN, EXO and GEN activities of FEN1 but does not impair its DNA substrate-binding affinity. We also found impaired LP-BER efficiency when the L209P FEN1 variant was tested in a reconstitution assay with purified proteins or with cellular extracts prepared from cells harboring L209P FEN1. Compared with wild-type (WT) FEN1-containing cells, cells expressing the L209P FEN1 variant were more susceptible to DNA-damaging agents and accumulated more DNA damage lesions in the genome. Consequently, L209P FEN1-containing cells displayed various chromosomal aberrations, along with a higher cellular transformation rate and increased tumor growth in a mouse xenograft model. This evidence demonstrates, for the first time, that FEN1 mutations found in human cancer are capable of causing cellular transformation and may therefore be important determinants of human cancer.
Results
Occurrence of the L209P variant in colorectal cancer
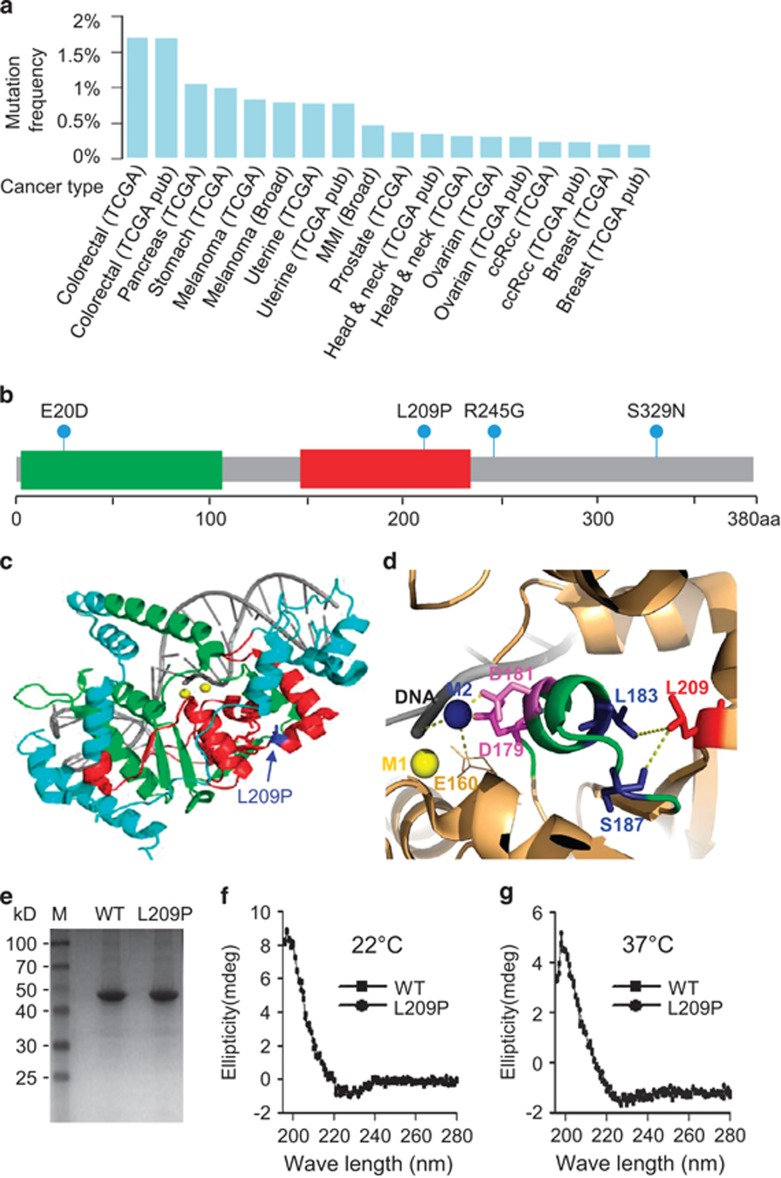
Because FEN1 is important for maintaining genomic DNA stability and integrity,35, 36 we speculated that FEN1 loss-of-function mutations might lead to cancer onset and development. We searched The Cancer Genome Atlas database (https://tcga-data.nci.nih.gov/tcga/) and found that heterozygous FEN1 somatic mutations are present in the tumor tissues of most cancer types,33 with colorectal cancer showing the highest FEN1 mutation frequency (Figure 1a). Four FEN1 mutations have been detected in colorectal cancers. They are E20D, L209P, R245G and S329N (Figure 1b). The conservation of the FEN1 mutations was described by Hosfield et al.34, 37 From Pyrococcus furiosus to human, amino acids E20, L209 and R245, which are indicated in Figure 1b, are conserved, whereas S329 is not conserved. We focused on the L209P mutation because it is in a core nuclease domain (Figure 1c) and is evolutionarily conserved.34
Figure 1.
The FEN1 L209P mutation in cancer. (a) FEN1 gene mutation frequency from the The Cancer Genome Atlas (TCGA) data set. (b) The sites of four colorectal cancer-associated mutations in the FEN1 gene from the TCGA data set: E20D, L209P, R245G, and S329N. Green indicates XPG_N. Red indicates XPG_I. (c) 3D structure of FEN1 showing the site of the L209P mutation. (d) ‘Allosteric effect' of L209P by analyzing the crystal structure of the FEN1–DNA complex (PDB code 3Q8K). L209 (red stick) closely interacts with L183 and S187 (blue sticks), which are on the helix containing amino acids 179–187 (green cartoon). The FEN1 active center comprises the metal atoms M1 (yellow sphere) and M2 (blue sphere). The M2 atom is surrounded by the DNA backbone (gray cartoon), E160 (brown line) and D179/D181 residues (magenta sticks). (e) SDS–PAGE of WT and L209P FEN1 (42 KDa). 6xHis-tagged FEN1 was expressed in E. coli and purified using a Ni-NTA column. Circular dichroism spectra of 1 μm WT or L209P FEN1 in 10 mm K2PO4 at 22 °C (f) or 37 °C (g). Measurements were collected in 1-nm steps from 190 to 280 nm.
To predict the possible effects of the L209P mutation on FEN1 activity and DNA substrate binding, we performed bioinformatics analysis based on human FEN1 crystal structures with DNA (PDB code 3Q8K).38 The behavior of the L209P mutation may be explained as an ‘allosteric effect' on the FEN1 active center. As shown in Figure 1d, the FEN1 active center is predicted to comprise two magnesium metal atoms, M1 and M2. The M2 atom is positioned by a strong electrostatic interaction with negative charges on the DNA backbone and the E160/D179/D181 residues. Meanwhile, the L209 residue is predicted to closely contact S187 and form a hydrophobic interaction with L183. The L209P mutation is also likely to disrupt side-chain interactions between the L209 and S187/L183 residues. Therefore, the helix around the 179–187 amino acid region is likely to lose part of its anchor and become more flexible and unstable. Consequently, the stability of the D179/D181 residues at the other end of the 179–187 amino-acid helix may be weakened. Eventually, the interactions between D179/D181 and the M2 metal atom may be disrupted, leading to FEN1 activity loss. On the other hand, L209P mutation does not significantly change the FEN1/DNA interaction surface, consistent with the observation that it does not affect DNA binding.
To partly test the prediction, FEN1 L209P and WT were expressed in Escherichia coli and purified to homogeneity (Figure 1e). We first performed circular dichroism analysis at 22 °C. The results showed that the L209P FEN1 protein had almost the same conformation as the WT FEN1 protein at 22 °C (Figure 1f), suggesting that the proline-to-leucine transition had no obvious effects on protein conformation. To investigate whether the L209P mutation affects the thermostability of FEN1, circular dichroism analysis was also carried out at 37 °C (Figure 1g). The results showed that L209P FEN1 has a similar overall structure to WT FEN1, indicating that the thermostability of FEN1 is not affected by the L209P mutation.
FEN1 L209P mutant lacks all three nuclease activities
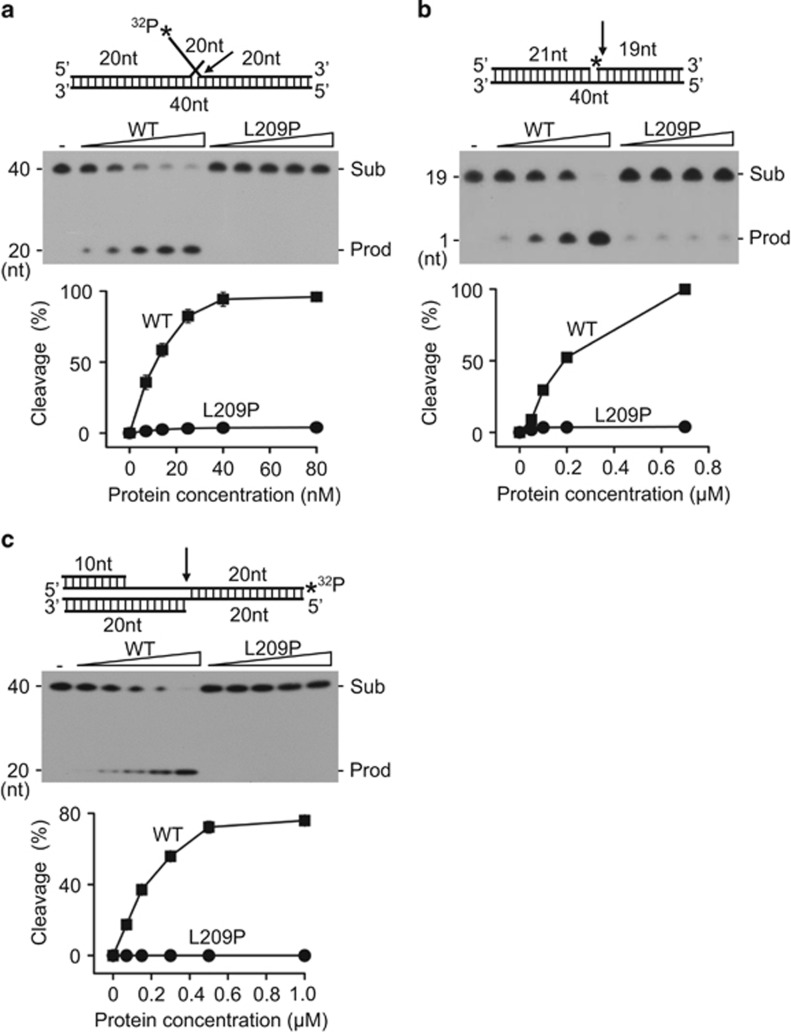
To further verify the prediction and evaluate the effects of the L209P mutation on FEN1 biochemical activity, we assayed the FEN, EXO and GEN activities of L209P FEN1, in vitro, using synthetic DNA substrates (Table 1). Indeed, as predicted in Figure 1d, L209P mutation almost completely eliminated all three activities of FEN1 (Figures 2a–c) when assayed at 37 °C. Because L209P and WT FEN1 have the same overall structure at both 22 and 37 °C, it is not likely that the defect in L209P FEN1 activity is due to the sensitivity of the L209P protein to temperature. Activity assays at 22 °C also showed that L209P FEN1 is inactive (Supplementary Figure S1). This type of mutation has never been characterized previously and further strengthened our enthusiasm for studying the L209P FEN1 variant.39
Table 1. Oligonucleotides and applications.
| Name | Oligonucleotide sequence | Application |
|---|---|---|
| FEN1 L209P F | 5′-CCAGGAATTCCACCCGAGCCGGATTCTGC-3′ | Mutagenesis |
| FEN1 L209P R | 5′-GCAGAATCCGGCTCGGGTGGAATTCCTGG-3′ | |
| FEN1-21-T | 5′-ATGCGATACAGTCCGATAGCT-3′ | FEN1 FEN activity assay |
| FEN1-40 | 5′-32P-CCATGAGCAACTACGATATGCGTACTAAGCCTAATCCGAC-3′ | |
| FEN1-A-F-G-40 | 5′-GTCGGATTAGGCTTAGTACGGCTATCGGACTGTATCGCAT-3′ | |
| FEN1-21-EXO | 5′-ATGCGATACAGTCCGATAGCC-3′ | FEN1 EXO activity assay |
| FEN1-19-EXO | 5′-32P-GTACTAAGCCTAATCCGAC-3′ | |
| FEN1-A-F-G-40 | 5′-GTCGGATTAGGCTTAGTACGGCTATCGGACTGTATCGCAT-3′ | |
| FEN1-10-GAP | 5′-TTGCTCATGG-3′ | |
| FEN1-20-GAP | 5′-ATGCGATACAGTCCGATAGC-3′ | FEN1 GAP activity assay |
| FEN1-40 | 5′-32P-CCATGAGCAACTACGATATGCGTACTAAGCCTAATCCGAC-3′ | |
| FEN1-A-F-G-40 | 5′-GTCGGATTAGGCTTAGTACGGCTATCGGACTGTATCGCAT-3′ | |
| FEN1-F | 5′-CTTACACGTTGACTACCTTFTTTGAGGCAAGAGTGGATCC-3′ | LP-BER assay |
| 3′-GAATGTGCAACTGATGGCAGAAACTCCGTTCTCACCTAGG-5′ |
Abbreviations: EXO, exonuclease; FEN1, flap endonuclease-1; GAP, gap endonuclease; LP-BER, long-patch base excision repair.
Figure 2.
Activities of WT and mutant FEN1 on double-flap, nick and gap substrates. Labeled substrates (1 pmol) were incubated with various amounts of purified WT or L209P FEN1 at 37 °C for 30 min. (a) FEN activity on a 5′-labeled double-flap substrate. The FEN1 concentration is 0–80 nM. (b) EXO activity on a 5′-labeled nick substrate. The FEN1 concentration is 0–0.8 μm. (c) GEN activity on a 3′-labeled gap substrate. The FEN1 concentration is 0–1.0 μm. The top panel shows the schematic structure of the corresponding DNA substrates. The middle panel shows the PAGE gel separating the DNA substrates (Sub) and the cleavage products (Prod). The graph on the bottom represents the quantification of the PAGE image. Arrows indicate the cleavage site of DNA by FEN1 and asterisks indicate the position of DNA labeled by 32P.
In previous studies, we found that FEN1 mutations could affect either the interaction of FEN1 with its partner proteins or its biochemical activity.13, 27, 40, 41, 42, 43, 44, 45 We have reported that the S187D and F343A/F344A FEN1 mutations disrupt the FEN1–PCNA interaction and the E160D mutation impairs the EXO and GEN activities of FEN1. More recently, we found that the FEN1 E359K mutation lacks both Werner syndrome ATP-dependent helicase (WRN)-binding affinity and GEN activity. To test whether the L209P mutation affects FEN1 protein-binding affinity, we compared the binding abilities of WT and L209P FEN1 with the most common partner proteins, including PCNA, APE1 and Pol β.46, 47, 48, 49, 50 We found that the L209P mutation does not affect the FEN1 interaction with these proteins (Supplementary Figure S2).
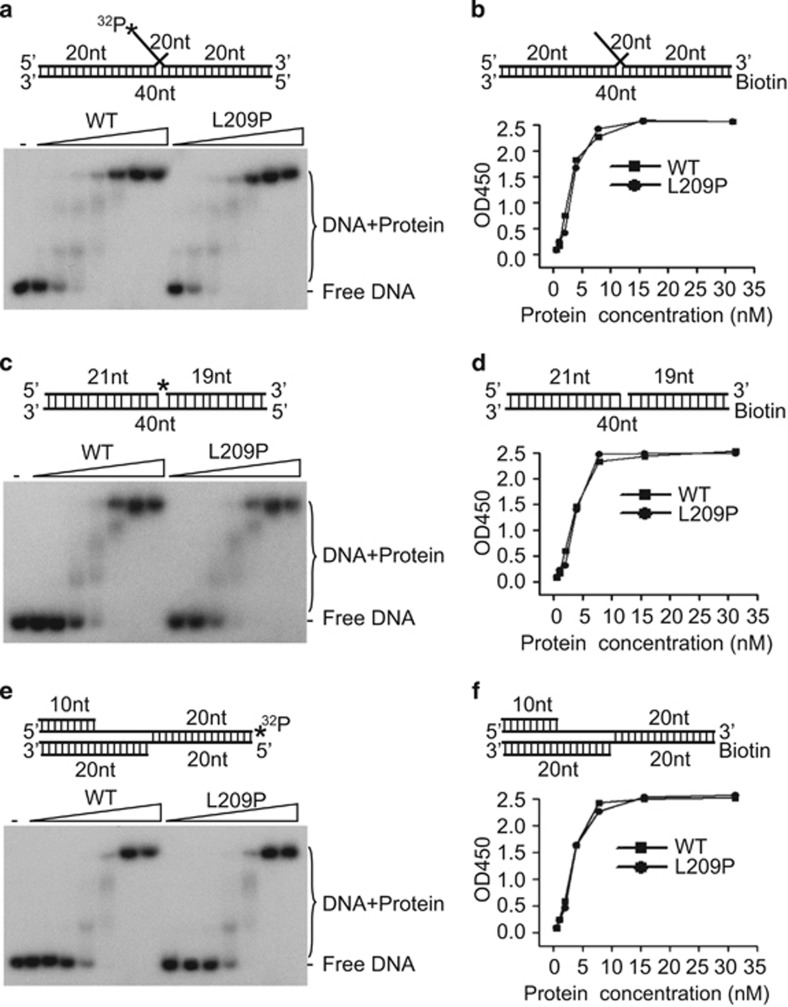
L209P FEN1 retains intact DNA substrate-binding capacity
Because L209P FEN1 has the same overall structure as WT FEN1 (Figure 1f), it is unlikely that a global structural defect is the reason for the decreased activity of L209P FEN1. We therefore tested whether the FEN1 L209P variant affects DNA substrate binding. We used gel shift (Figures 3a, c and e) and enzyme-linked immunosorbent (ELISA)-based assays (Figures 3b, d and f) to determine the FEN1–DNA-binding affinity. Both assays showed no difference between the DNA-binding affinities of the L209P variant and WT FEN1. This result is consistent with the prediction in Figure 1d and our previous observation that only positively charged amino acids, such as arginine in positions 70, 245 and 327 and lysine in positions 244, 252 and 326, are directly involved in FEN1–DNA interactions.51, 52
Figure 3.
The substrate-binding affinity of FEN1 is not affected by the L209P mutation. (a, c, e) Gel shift assay showing the DNA-binding affinity of L209P and WT FEN1. The DNA substrates (panels a, c and e for the FEN, EXO and GEN substrates, respectively) were labeled by 32P as shown with an asterisk (*) in the figure. (b, d, f) ELISA-based isotherm adsorption assay for DNA-binding affinity of L209P and WT FEN1. The DNA substrate was biotin-labeled as shown in the figure (panels b, d and f for the FEN, EXO and GEN substrates, respectively).
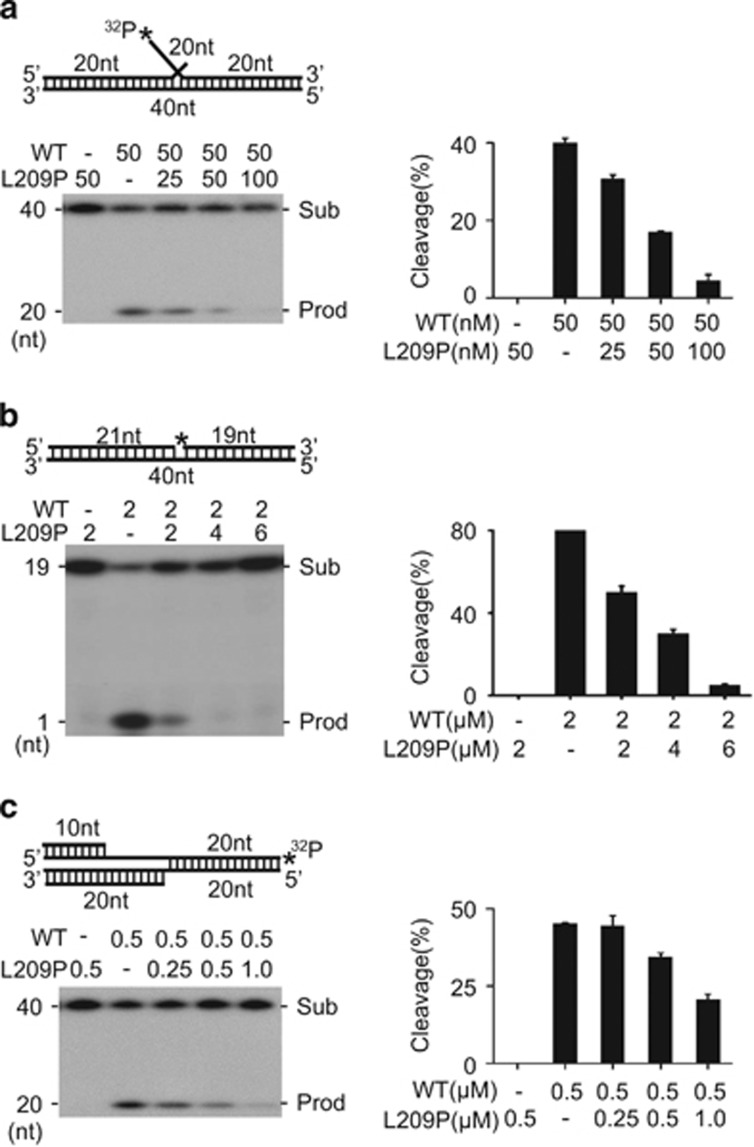
L209P FEN1 interferes with WT FEN1 nuclease activity
Because L209P FEN1 shows intact DNA substrate-binding affinity, we speculated that the presence of L209P FEN1 might interfere with the activity of WT FEN1. To evaluate this supposition, we mixed various amounts of each protein with DNA substrate and assayed their FEN (Figure 4a), EXO (Figure 4b) and GEN (Figure 4c) activities. When incubated with L209P FEN1, the cleavage activity of WT FEN1 was repressed. The inhibitory effect of L209P FEN1 on WT FEN1 was dose dependent. These results indicate that L209P interferes with WT FEN1 activity in a dominant-negative manner.
Figure 4.
L209P FEN1 interferes with the activity of WT FEN1. (a) FEN activity assay with purified WT and L209P FEN1. (b) EXO activity assay with purified WT and L209P FEN1. (c) GEN activity assay with purified WT and L209P FEN1. In all the panels, the DNA substrate is the same as that used in Figure 2. 32P labeling of DNA is indicated by *. The left panel shows the PAGE gel separating the DNA substrate and the cleavage products. The graph in the right panel represents quantification analysis of the image in the left panel.
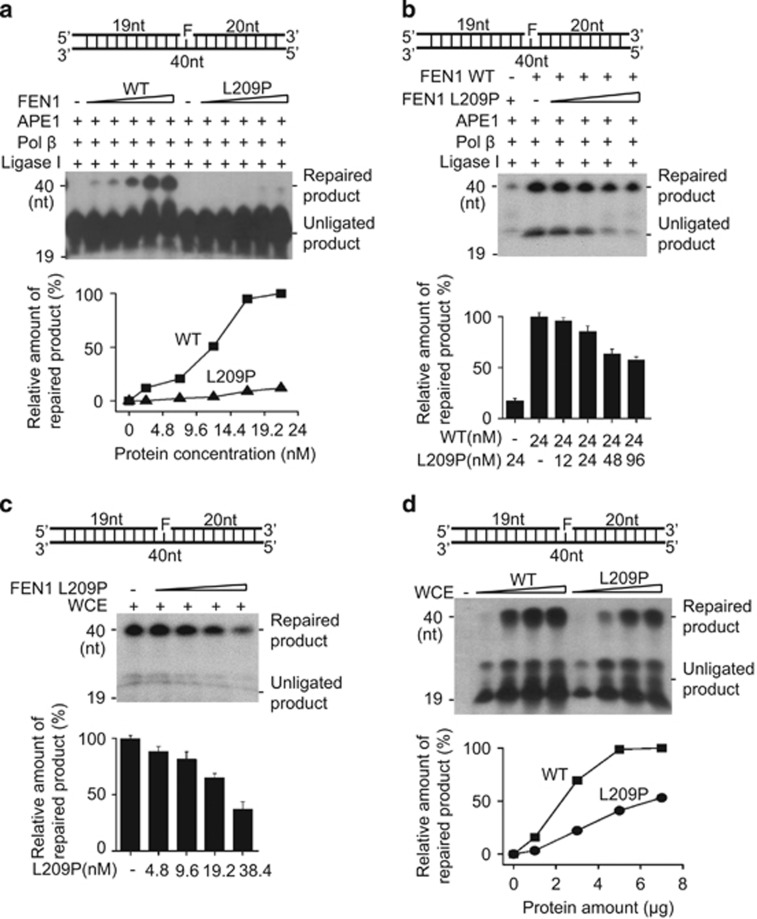
The L209P FEN1 mutation interferes with BER efficiency
The removal of flap structures is a key step for completing LP-BER. The impaired activity of L209P FEN1 suggests that this mutation likely affects LP-BER function. To test this hypothesis, the LP-BER assay was carried out using purified proteins.50 Tetrahydrofuran (THF)-containing substrates (FEN1-F) were used as LP-BER substrates (Table 1). The THF lesions were efficiently repaired in the presence of WT FEN1 but not the L209P FEN1 variant (Figure 5a). As shown in Figure 4, L209P interferes with FEN1 activities; we therefore speculated that L209P FEN1 could also interfere with the overall BER efficiency of WT FEN1. To test this, purified L209P FEN1 was mixed with WT FEN1 protein to perform the LP-BER assay. Our result showed that BER efficiency is reduced in the presence of the L209P FEN1 protein (Figure 5b). Furthermore, we performed LP-BER using SW480 cell extracts with or without additional L209P FEN1 protein to determine whether L209P FEN1 could impair BER in a cell extract context. The amount of L209P FEN1 added to the assay, relative to the WT protein within the whole-cell extract, was determined by quantitative western blotting (Supplementary Figure S3). As shown in Figure 5c, although SW480 cell extracts could repair THF lesions efficiently, the addition of L209P FEN1 to the cell extracts decreased BER efficiency. To mimic the heterozygous L209P somatic mutation of FEN1 detected in human colon cancer cells, we overexpressed L209P FEN1 or WT FEN1 in SW480 cells that also contain endogenous WT FEN1 (Supplementary Figure S4A). Our results showed that the BER efficiency of cell extracts expressing L209P FEN1 is much lower than that of WT FEN1-expressing cell extracts (Figure 5d). Altogether, the data above indicate that L209P FEN1, either purified or expressed in SW480 cells, interferes with the BER activity of WT FEN1.
Figure 5.
L209P reduces LP-BER efficiency. (a) Comparison of LP-BER efficiency by purified WT or L209P FEN1. L209P FEN1 interferes with WT FEN1 LP-BER in an in vitro assay with purified protein (b) or whole-cell extracts (WCE, c). (d) Comparison of the LP-BER efficiency of cell extracts with WT or L209P FEN1 expression. The substrates in all panels are not labeled. Instead, α-32P-dCTP is included in the reaction buffer. The top part of each panel shows the schematic structures of the corresponding DNA substrates. The middle part shows the PAGE-separated products and the bottom shows the relative amount of repaired product.
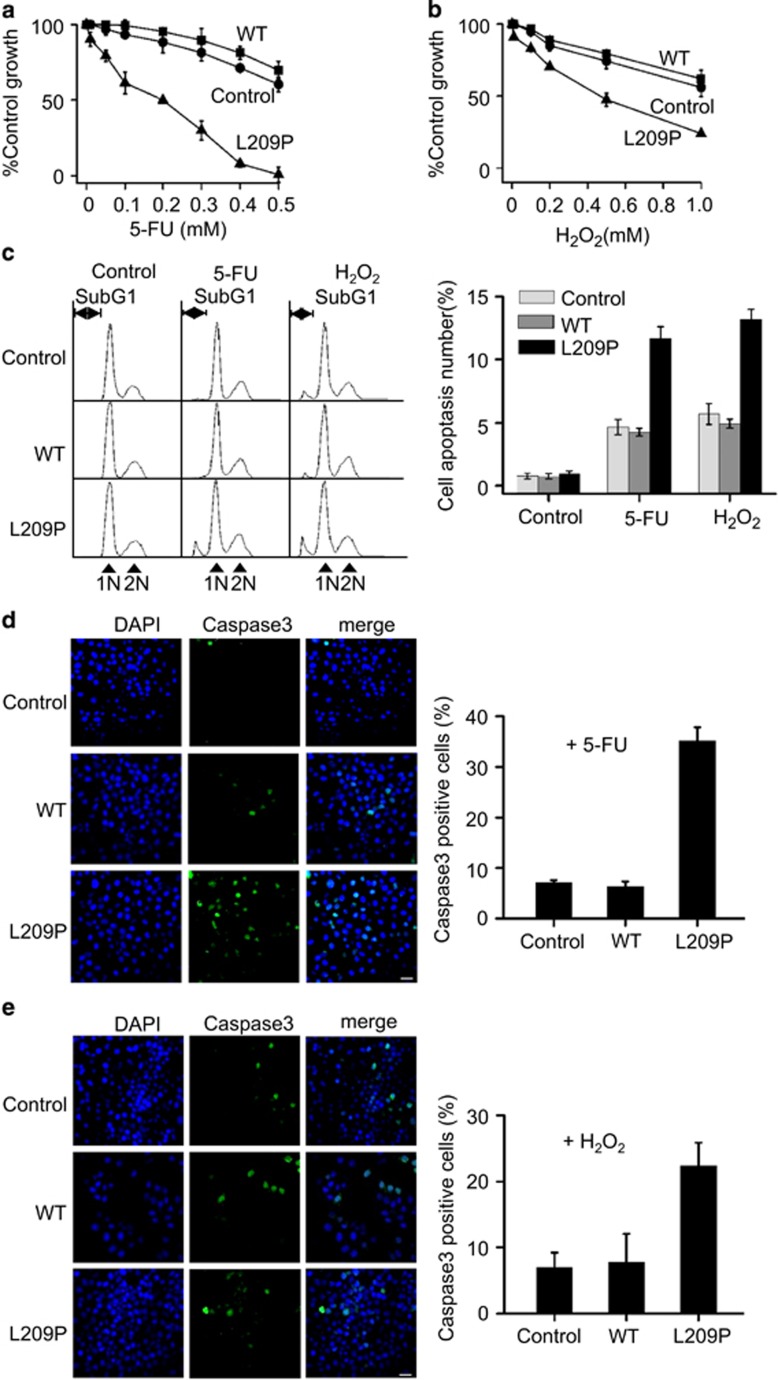
Expression of L209P FEN1 sensitizes cells to DNA damage
To investigate the possible biological function of the L209P FEN1 variant in cells, we transfected SW480 colorectal cancer cells with lentivirus-vectors containing L209P or WT FEN1 gene. Cells with similar expression levels of L209P FEN1 and WT FEN1 were chosen for our experiments (Supplementary Figure S4A). Because L209P interferes with LP-BER, we speculated that expression of L209P could sensitize cells to DNA damage. Cells harboring exogenous L209P or WT FEN1 were treated with 5-FU (Fluorouracil) or H2O2, which have been reported to cause DNA base oxidation, a lesion mainly repaired by LP-BER. We found that cells transfected with L209P FEN1 are more sensitive to 5-FU or H2O2 than cells transfected with WT FEN1 or transfected parental control SW480 cells, indicating that LP-BER repair is defective in L209P FEN1-containing cells (Figures 6a and b). Then fluorescence-activated cell sorting analysis was carried out to compare the percentage of apoptotic cells induced by 5-FU or H2O2. As shown in Figure 6c, after treatment with 5-FU or H2O2, the L209P FEN1-expressing cells showed a much higher percentage of apoptotic cells than the cells expressing WT FEN1 or parental SW480 cells. Cell staining with an antibody against Caspase-3, a marker of cell apoptosis, also corroborated the fluorescence-activated cell sorting result (Figures 6d and e). These data support previous reports that DNA damage, if not repaired, could lead to cellular apoptosis.53, 54, 55 However, the non-apoptotic treated cells may be more prone to transformation owing to accumulation of DNA lesions and genome instability.
Figure 6.
Overexpression of L209P FEN1 sensitizes cells to 5-FU and H2O2 and induces cellular apoptosis. Cells harboring L209P FEN1 are sensitive to 5-FU (a) and H2O2 (b). (c) Fluorescence-activated cell sorting analysis of cells expressing L209P and WT FEN1 after 5-FU or H2O2 treatment. The apoptotic cells are indicated as ‘SubG1'. The graph on the right panel represents the percentage of apoptotic cells. Immunostaining for Caspase-3 after 5-FU (d) or H2O2 (e) treatment. Green indicates Caspase-3, blue indicates DNA counterstaining with DAPI. Scale bars, 30 μm. The graph on the right panel shows the percentage of cells that are Caspase-3 positive. In all panels, values are the mean±s.d. of three independent experiments. P<0.05 (Student's t-test).
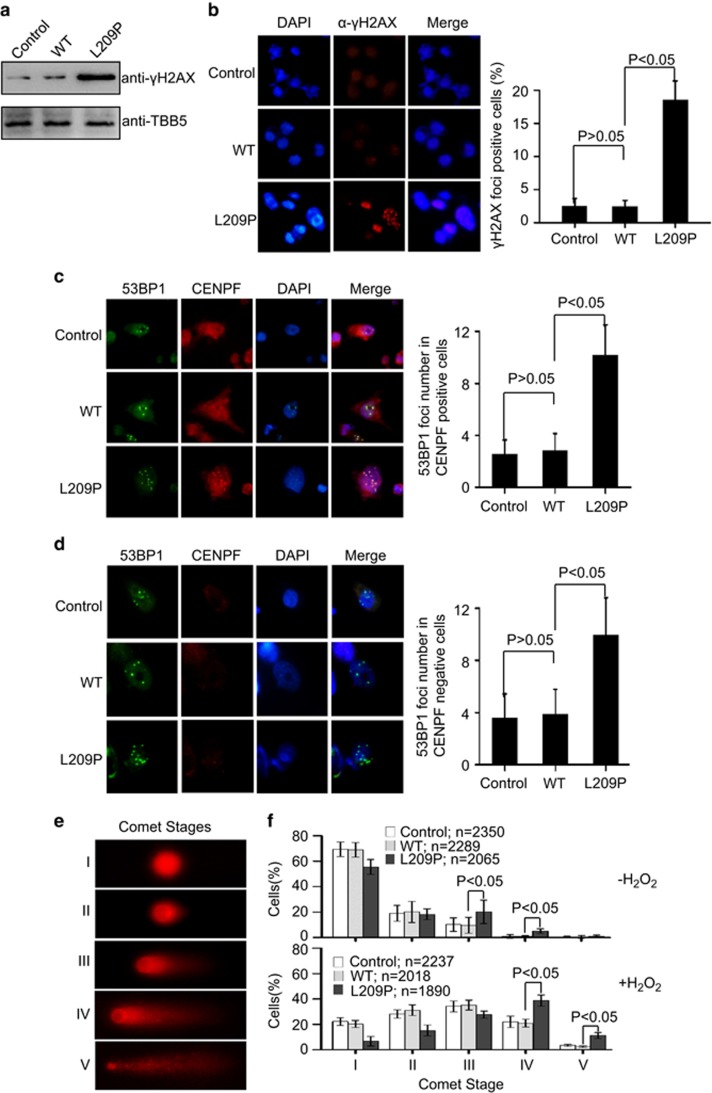
L209P causes endogenous DNA damage and chromosomal aberrations
In addition to damage from exogenous agents, DNA damage can occur naturally in response to metabolic or hydrolytic processes. Naturally occurring oxidative DNA damage arises at least 10 000 times per cell per day in humans.5, 6 Therefore, we expected that the L209P cells would show a higher level of endogenous DNA damage than WT cells. To verify this hypothesis, we examined the levels of the phosphorylated form of the H2AX histone protein (γH2AX), an established early marker of the cellular response to DNA breaks. In the absence of an exogenous source of DNA damage, the basal levels of phosphorylated γH2AX in L209P cells were higher than those in WT cells (Figure 7a). Furthermore, DSBs were detected as nuclear foci in cells stained with the γH2AX antibody. The results showed a greater number of γH2AX-positive nuclei in the L209P FEN1-containing cells than that in WT cells (Figure 7b). The data indicate that L209P FEN1 expression leads to the accumulation of DNA DSBs.
Figure 7.
The FEN1 L209P mutation induces spontaneous DNA damage. (a) Western blotting assay of γH2AX in L209P FEN1-containing cells. (b) Representative picture of γH2AX immunofluorescence in L209P FEN1-containing SW480 cells. Red indicates γH2AX and blue indicates DAPI. The right panel shows the percentage of nuclei that contain γH2AX foci. (c and d) Representative picture of 53BP1 and CENPF immunofluorescence in L209P FEN1-containing SW480 cells. Green indicates 53BP1, red indicates CENPF and blue indicates DAPI. The graph in the right panel shows the 53BP1 foci number in the CENPF positive (c) or negative (d) cells. A total of 60 CENPF-positive (G2) or -negative (G1) cells were scored for each cell line. (e) Typical examples of cell nuclei representing the comet stages I–V. (f) Distribution and statistical evaluation of the Comet stages of the L209P FEN1, WT FEN1 or parental SW480 cells. Cell distributions are shown before (up panel) and 24 h after H2O2 treatment (bottom panel). In all panels, values are the mean±s.d. of three independent experiments.
γH2AX is not a specific marker of DNA DSBs but can also be an indication of other types of damage that occur during DNA replication. Because of the critical role of FEN1 in DNA replication, it is important to clarify whether the chromosomal instability is due to DNA single-strand breaks (SSBs) converting to DNA DSBs during S phase or whether the chromosomal instability is caused by other replication defects that result in the persistence of under-replicated DNA throughout the cell cycle. To distinguish these two mechanisms, cells were co-stained with antibodies against 53BP1 (tumor-suppressor p53-binding protein 1) and CENPF (centromere protein F). 53BP1 is a key regulator of DSB repair.56, 57 53BP1 rapidly forms large foci near DNA lesions where DNA damage signaling is induced.58, 59, 60
The CENPF staining allows G1 and S-phase/G2 cells to be distinguished (G1 cells lack any CENPF staining, while CENPF staining increases in intensity from late S phase to G2 phase).61, 62 The formation of 53BP1 foci is suggestive of inefficient/defective DNA replication.63 The results showed that, in the absence of an exogenous source of DNA damage, a significant increase in 53BP1 foci occurred in G2 (CENPF-positive cells) (Figure 7c), indicating that spontaneous damage arises from unrepaired SSBs colliding with replication forks. However, an increase in 53BP1 foci was also observed in G1 (CENPF-negative cells) (Figure 7d), suggesting that L209P FEN1 also causes DNA replication defects.63 This data set is consistent with the dual role of FEN1 in both DNA replication Okazaki fragment maturation and LP-BER.
To test whether LP-BER is retarded in L209P-expressing cells in vivo, we performed the alkaline comet assay before and after H2O2 treatment. Using the same set of control and SW480-expressing L209P or WT FEN1 cell lines, we assessed steady-state levels of DNA fragmentation after exposure of cells to H2O2. To detect all possible intermediates of BER, that is, abasic sites and DNA SSBs and DSBs, as quantitatively as possible, we performed the comet assays under alkaline conditions.64, 65, 66, 67 We then evaluated the overall distribution of comet events into the defined stages of DNA fragmentation (Figure 7e). These stages range from cells with no DNA fragmentation (stage I, <5% fragmentation) to cells with heavy DNA fragmentation (stage V, >95% fragmentation).67, 68 Experiments without H2O2 treatment revealed that SW480 cells expressing L209P FEN1 had relatively higher levels of spontaneous DNA strand breaks than WT cells (P<0.05, Student's t-test, stages III and IV, Figure 7f). After H2O2 exposure, all cell lines showed severe amounts of DNA damage (Figure 7f). However, cells expressing L209P FEN1 showed a stronger DNA damage than those expressing WT FEN1 (P<0.05, Student's t-test, stages IV and V, Figure 7f). The major damage status of SW480 cells expressing WT FEN1 occurred in stage III but the major damage status of SW480 cells expressing L209P FEN1 occurred in stage IV after H2O2 exposure (Figure 7f). The examination of the comet stage distributions confirmed that the L209P FEN1 mutation causes defects in LP-BER, as well as Okazaki fragment maturation.
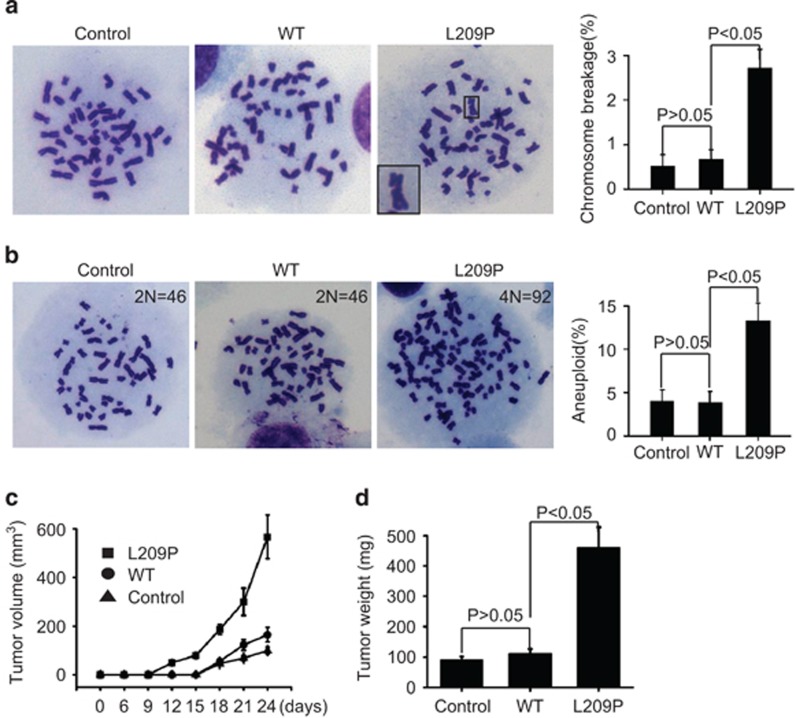
Defects in BER lead to significant accumulation of BER intermediates (Figures 7a–d), which will consequently cause chromosomal aberrations.69, 70 To test whether the L209P FEN1-expressing cells accumulate more chromosomal breaks than the WT FEN1-expressing and parental SW480 control cells, we analyzed metaphase nuclei for chromosomal aberrations. Cells expressing L209P FEN1 exhibited significantly increased the levels of chromosomal fragments and breaks, when compared with WT FEN1-expressing and control parental cells (Figure 8a). This result indicates that the expression of L209P FEN1 indeed results in genomic instability in cells. However, under normal culture conditions, the growth rate of L209P FEN1-expressing cells is similar to that of the WT FEN1-expressing or parental control cells (Supplementary Figures S4B–D). Based on the increase in chromosomal aberrations, we hypothesized that the L209P FEN1-expressing cells are likely to become transformed. Therefore, we next determined whether L209P FEN1 cells are susceptible to cellular transformation and tumorigenesis.
Figure 8.
The FEN1 L209P mutation induces chromosomal instability and cellular transformation. (a) Spontaneous chromosomal breaks in SW480 cells expressing L209P or WT FEN1. Representative Giemsa-stained WT and L209P FEN1-containing SW480 metaphase cells are shown. The right panel represents the percentage of cells containing chromosomal breaks for WT and L209P FEN1-containing SW480 cells. A total of 50 metaphase cells was scored for each cell line. (b) Representative chromosome counting image of WT and L209P FEN1-containing SW480 metaphase cells, showing the chromosomal instability and aneuploidy. A total of 60 metaphase cells was scored for each cell line. (c) Tumorigenesis assay in vivo. The cells (10 million cells per mouse) were injected subcutaneously into 6-week-old female mice. The tumor volume was measured every 3 days. (d) The tumor weights on day 24 are shown.
L209P induces cellular transformation
It has been reported that aneuploidy is a hallmark of cancer cells.71 Therefore, we first determined the aneuploidy rate in WT and mutant L209P FEN1-expressing cells. We found that the aneuploidy rate of L209P FEN1-expressing cells is nearly fourfold greater than the rate in WT FEN1-expressing cells (12.1 vs 3.2%) (Figure 8b). These data indicate that L209P FEN1-expressing cells spontaneously accumulate chromosomal fragments, which leads to the chromosomal instability and aneuploidy. These cellular abnormalities are likely to contribute to cellular transformation and lead to clonal expansion. To determine whether the L209P mutation promoted tumorigenesis, we performed focus formation and soft agar anchorage-independent growth assays. In both assays, the number of colonies formed by L209P FEN1-expressing cells was significantly higher than by the WT FEN1-expressing or parental control cells, suggesting that L209P FEN1 cells contain more transformed cells than WT FEN1 cells (Supplementary Figure S5).
To further address whether L209P FEN1-expressing cells are more tumorigenic than WT cells in vivo, we injected L209P FEN1-expressing, WT FEN1-expressing and parental control SW480 cells into the flank regions of adult immunodeficient mice. Figure 8c illustrates the tumorigenic capacity of each cell type. No tumors were detected after 15 days in animals inoculated with WT FEN1-expressing and parental SW480 cells, whereas all of the five animals inoculated with L209P FEN1-expressing cells had tumors. The tumor growth rate of the L209P FEN1-expressing tumors was substantially higher than the growth rates in WT FEN1-containing and parental control groups (Figure 8c). The animals were killed on day 24, and the tumors were weighed. Figure 8d shows that the average tumor weight from the cells expressing L209P FEN1 is significantly higher than the tumor weights of the WT FEN1-expressing and parental control cells. These data suggest that cells harboring the L209P FEN1 mutation are more oncogenic than WT cells.
Discussion
Because of the fundamental functions of FEN1 in the maintenance of genome stability, complete elimination of FEN1 activity in mice results in early embryonic lethality.26, 72 However, we propose that partial deficiency of FEN1 causes defects in DNA replication and repair, leading to the accumulation of DNA intermediates, which then induce genomic instability and tumorigenesis. Consistent with this hypothesis, we have detected FEN1 heterozygous mutations in various human cancers, including lung, breast and colon cancers.27 However, most of the identified mutations impair either the EXO or GEN activity of FEN1 but do not impair the FEN activity. For example, the E160D FEN1 mutation abrogates the EXO activity, leading to frequent spontaneous mutations and accumulation of incompletely digested DNA fragments in apoptotic cells. The E160D FEN1 mutant mice are predisposed to autoimmunity, chronic inflammation and cancers.13, 42 Recently, we reported a germline FEN1 mutation, E359K, from a breast cancer family. This variant abolishes the GEN activity and the interaction of FEN1 with WRN, an interaction that is critical for resolving stalled DNA replication forks. Cells harboring this E359K FEN1 mutation accumulate more spontaneous chromosomal anomalies than WT cells and show higher frequencies of transformation.40 However, no mutation disrupting the FEN activity of FEN1 has been ever reported and its role in tumorigenesis was unknown until now.39 L209P is the first mutation that completely abolishes the FEN activity of FEN1, along with the EXO and GEN activities. Therefore, the L209P FEN1 mutation represents a novel mechanism by which FEN1 dysregulation promotes cancer development.
The importance of the FEN, GEN and EXO activities of FEN1 in normal cells leads to the question: how do cells harboring the FEN1 L209P mutation survive? The cellular lethality of FEN1 deficiency is most likely due to a failure of FEN1's function in DNA replication.39 In the current case, the identified mutation exists in the heterozygous state in human somatic cells and in our experimental system. We used a lentiviral expression system to express L209P FEN1 in the endogenous WT expression background. During S phase, the endogenous WT FEN1 is highly expressed,73 allowing for successful DNA replication to occur in L209P FEN1 cells, though the cell morphology and cell cycle progression are slightly affected (Supplementary Figures S4B and D). However, in the G1 and G2 phases, when DNA repair occurs, the WT FEN1 protein expression level is reduced, whereas L209P FEN1 is expressed at constant levels. The mutant protein would then bind to the DNA replication fork or damage foci but fail to remove the RNA primer in Okazaki fragment or DNA flap structure in LP-BER, thus blocking the function of the WT protein.
FEN1 is a key enzyme for maintaining genome integrity and stability. FEN1 mutation is rare in both the normal population and in cancer patients. Sato et al.32 sequenced seven small-cell lung cancers and nine non-small-cell lung cancers. No FEN1 mutation was found, suggesting that FEN1 is an essential gene. We have previously identified a set of mutations in cancer that eliminate non-essential exonuclease and gap-dependent endonuclease activities and demonstrated that the loss of those activities causes inflammation and cancer.74 In the current manuscript, we found a FEN1 L209P mutation in colorectal cancer from the TGCA database. Currently, there is not enough data to calculate the prevalence of the L209P mutation in colorectal and other types of cancers. However, based on the observation that FEN1 L209P mutation leads to the accumulation of DNA lesions owing to its functional defects in DNA replication and repair, we predict that people carrying the L209P mutation should be more sensitive to treatment with DNA damage-inducing drugs.
LP-BER is a major pathway used to repair oxidized DNA bases in mammals. Successful repair would require three sequential steps: (i) Pol β-dependent polymerization to form a DNA flap, (ii) removal of the flap by FEN1, and (iii) ligation of the upstream and downstream DNA strand by Ligase I. FEN1 is a key enzyme in LP-BER. L209P FEN1 interrupts the WT FEN1 activity and perturbs the overall LP-BER efficiency, generating unligated DNA intermediates. As shown in Figure 5, a fully repaired product is 40 nt long, whereas non-repaired intermediates are 20–40 nt long. Furthermore, THF lesions were efficiently repaired by WT FEN1 but not the L209P variant (Figure 5a). At the same time, with the decrease in the levels of repaired product, more unligated intermediates accumulated in the L209P FEN1-containing assay than in the WT FEN1-containing assay. This accumulation is mainly due to the inability of L209P FEN1 to cleave the flap structures of the intermediate, leading to failure to generate nicked DNA duplex substrates for Ligase I, and resulting in unfilled DNA gaps. DSBs could be generated when the replication fork reaches these unfilled gaps. Besides, in LP-BER, the DSBs could also be formed owing to the failure in Okazaki fragment maturation in cells carrying FEN1 L209P mutation. These DNA DSBs, if left unrepaired or repaired aberrantly, can lead to genome instability. Indeed, L209P FEN1-containing cells have more γH2AX and 53BP1 foci and, consequently, more chromosome breaks. Chromosome breaks can then lead to loss of heterozygosity, activation of oncogenes and loss of tumor-suppressor genes.75, 76 In addition, L209P FEN1 cells are prone to develop aneuploidy. We have previously revealed that aneuploidy is associated with epigenetic alterations that promote the ability of cancer cells to evade cellular senescence and apoptosis pathways.77
Indeed, the levels of endogenous DNA DSBs in L209P-expressing cells, as combination results of failure in both DNA replication and repair, are significantly higher than those in WT cells, as shown in Figures 7a and b. However, L209P expression does not alter the cell cycle profile and baseline level of apoptosis without DNA damaging agent treatment. This may be because the DSBs in the cells are rapidly repaired by the homology-directed repair (HDR) pathway. The remaining and persistent DSBs could be in the range of the cellular tolerance but not sufficient to trigger DNA fragmentation and cell apoptosis. Therefore, the mutant cells appear to grow normally. However, the persistently elevated DSBs could lead to the accumulation of DNA mutations and eventually cellular transformation. A recent report has shown that inhibition of FEN1 leads to the accumulation of DSBs in cells. When the HDR pathway is also blocked, the cells are much more sensitive to a FEN1 inhibitor, suggesting that HDR supports cell growth in FEN1-deficient cells.78
Materials and methods
Antibodies
The anti-FEN1 (sc-56675), PCNA (sc-7907), APE1 (sc-55498), c-Myc (sc-40), Tubulin (sc-23950), 53BP1 (sc-22760), CENP-F (sc-22791), goat anti-rabbit IgG-HRP (sc-2004) and goat anti-mouse IgG-HRP (sc-2005) antibodies were purchased from Santa Cruz Biotechnology (Dallas, TX, USA). The Pol β-specific antibody (ab26343) and anti-γH2AX (ab2893) were purchased from Abcam (Cambridge, UK).
Cell lines and cell culture
SW480 colorectal cancer cell lines were obtained from ATCC (Manassas, VA, USA) and maintained in Dulbecco's modified Eagle's medium (Invitrogen, Shanghai, China) supplemented with 10% fetal bovine serum (Invitrogen) and penicillin–streptomycin (Invitrogen) at 37 °C in a humidified 5% CO2 incubator. Cells are tested for mycoplasma contamination.
Protein expression and purification
The WT human FEN1 cDNA was described previously.79 The 6xHis-tagged WT FEN1 and the L209P mutant FEN1 cDNAs were constructed as previously described.79 The QuikChange Site-directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA) was used to generate the L209P mutant FEN1. The primers used for mutagenesis are shown in Table 1. The pET28b vectors containing the WT and mutant genes were transformed into E. coli BL21 cells for expression. Protein expression was performed as previously described.43 The proteins were purified from inclusion bodies under denaturing conditions, and refolding was induced using a method described previously.80, 81 To purify the 6xHis-tagged proteins, the harvested cells (150 ml of culture) were lysed in 3 ml of lysis buffer (50 mm NaH2PO4, 300 mM NaCl (pH 8.0)) containing 6 m guanidinium chloride. The cell lysate was sonicated until clear, followed by centrifugation at 18 000 g for 30 min. The supernatant was then loaded onto PrepEase columns (USB Corporation, Cleveland, OH, USA). The 6xHis-tagged proteins were eluted and refolded. The protein purity was determined by sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis (PAGE), and the concentration was quantified using the Bradford Assay Kit (Bio-Rad Laboratories, Hercules, CA, USA).
Lentiviral and stable cell line preparation
Lentivirus particles expressing the L209P FEN1 or WT FEN1 gene were generated by transfecting 293T cells with the L209P FEN1 or WT FEN1 plasmids, together with packaging plasmids. The virus-containing medium was collected every 24 h for 3 days. The cells were incubated with the lentivirus-containing medium plus 4 μg/ml polybrene for 24 h and then selected for 72 h in 1.5 μg/ml puromycin. All the lentivirus particles were prepared by Guangzhou Fitgene Biotechnology CO., LTD (Guangzhou, China).
Immunofluorescence
Cells were cultured in six-well plates containing acid-treated cover slides and incubated overnight. The cover slides were then washed with phosphate-buffered saline (PBS), fixed with 4% formaldehyde in PBS for 30 min and then washed again with PBS. Triton X-100 (0.05%) was added for 5 min to permeabilize the cells. The slides were blocked with 2% bovine serum albumin and then incubated with the primary antibody. The slides were washed with PBS and then incubated with secondary antibody conjugated with fluorescein isothiocyanate. The slides were then washed again with PBS and stained with DAPI (4′,6-diamidino-2-phenylindole). The mounted slides were viewed with a Zeiss Axioscope (Oberkochen, Baden-Wuerttemberg, Germany), and the images were captured with a charge-coupled-device camera (Oberkochen).
Alkaline comet assay
Alkaline comet assay analysis was performed on SW480 cells expressing L209P or WT FEN1. The cells were exposed to 0.3 mm H2O2 for 1 h, then washed with PBS, replated in normal medium and incubated for 24 h. For the comet assay, we applied the procedure according to previous reports.64, 65, 66 Comets were visualized by PI (propidium iodide) staining and examined at × 400 magnification with a fluorescence microscope (Axiovert 200 M, Zeiss, Jena, Germany). DNA spots from each sample were classified into five categories, corresponding to the amount of DNA in the tail.67, 68
Drug-sensitivity assay
As described previously,50 cells were seeded in 12-well plate, treated with multiple dilutions of drugs to be tested and incubated for 48 h under normal growth conditions. The number of viable cells was determined by the CellTiter 96 AQueous One Solution Cell Proliferation Assay (MTS, Promega, Madison, WI, USA). At least three replicate experiments for each clone were averaged. The data are expressed as the percentage of growth relative to untreated controls.
FEN1 nuclease activity assay
The cleavage of DNA substrates by FEN1 was determined under the same conditions as those previously published.43 Briefly, 32P-labeled DNA substrates were incubated with purified FEN1 in a buffer solution containing 50 mm Tris-HCl (pH 8.0), 50 mm NaCl and 5 mm MgCl2. The reactions were carried out at 37 °C for 30 min and were terminated with stop solution (95% formamide, 20 mm EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol). The product and substrate were then separated by 15% SDS–PAGE and visualized by autoradiography.
Reconstituted BER assay
The BER assay was performed as described previously.50 Complete repair reactions were carried out in 20 μl of reaction buffer (40 mm HEPES–KOH (pH 7.8), 70 mm KCI, 7 mm MgCl2, 1 mm dithiothreitol, 0.5 mm EDTA, 2 mm ATP, 200 U creatine-phosphokinase, 0.5 mm NAD and 5 mm phosphocreatine, 50 μM each of dATP, dTTP and dGTP and 8 μCi (α-32P)-dCTP]. For BER reconstitution with purified proteins, the substrate was incubated with a mixture of FEN1 (0–24 nm) and other purified BER proteins. For cell extract reconstitution, the LP-BER DNA substrate was incubated with the whole-cell extracts (0–7 μg). Reactions (30 min, 37 °C) were then stopped by adding an equal volume of the gel-loading buffer and visualized by autoradiography.
DNA-binding assay
Gel shift and ELISA-based assays were used to compare the DNA-binding affinity.50 For the gel shift assays, various concentrations of FEN1 protein (0.5–2000 nm) were incubated (15 min, room temperature) with 0.5 nm radio-labeled DNA substrates in a buffer containing 50 mm Tris-HCl (pH 8.0), 100 mm NaCl, 10% glycerol and 0.1% NP-40. The samples were run on a 5% native polyacrylamide gel and visualized by autoradiography. For ELISA-based affinity assays, a biotin-labeled DNA substrate (1 pmol) was immobilized on a streptavidin-coated 96-well ELISA plate and washed three times with binding buffer. Bound FEN1 was detected using a mouse anti-FEN1 antibody and goat anti-mouse IgG linked to horseradish peroxidase (HRP). Color was developed by adding tetramethylbenzidine and stopped by the addition of 1 n HCl. The optical density at 450 nm was read on a microplate reader.
Metaphase spread preparation and chromosome counting
As described previously,13 the cells were collected and treated with colcemid to arrest the cells at metaphase. The cells were then incubated (20 min, room temperature) with hypotonic solution (75 mm KCl), placed in a 37 °C water bath (5 min) and fixed with Carnoy's solution. The fixation process was repeated three times and a dropper was used to place the cells onto a clean slide. The cell spread was incubated (55 °C overnight), stained with Giemsa solution and scanned under a microscope for mitotic cells. The images were recorded and analyzed with ImagePro 7.0 (MediaCybernetics, Bethesda, MD, USA), and the chromosomes in each metaphase cell were counted.
Cellular transformation assay
The focus formation assay was conducted according to a previous report.40 The presence of foci was visually evaluated after staining the cells with 0.05% crystal violet. The anchorage-independent growth assay in soft agar was described previously.82 Briefly, the cells (3000 cells per 35 mm well) were suspended in complete medium containing 0.35% agarose. The cells were grown on tissue culture dishes containing a 2-ml layer of solidified 0.7% agar in a complete medium. After 10 days, the number of colonies was quantified from micrographs taken at random locations on the plate (original magnification, × 20).
In vivo tumorigenesis assays
Xenograft growth of tumors in null mice is as described previously.83 Female null mice were purchased from Model Animal Research Center of Nanjing University. Mice were procured and the study was conducted according to the guidelines as well as protocol approved by the Institutional Animal Ethics Committee, Nanjing Normal University, China. Mice were randomly divided into three groups (n=5). SW480 cells transfected with vector control, L209P or WT FEN1 (1 × 106 cells per mouse) were suspended in 100 μl Dulbecco's modified Eagle's medium and held on ice. The entire 100-μl sample was injected into subcutaneous tissue of 6–8-week-old female null mice. Five mice per group were used. The mice were checked daily for tumor appearance by palpation, and the tumor volume was measured every 3 days and recorded in mm3 (length × width2). The tumors were removed on day 25 and weighed.84, 85
Statistical analysis
Phenotypic and molecular differences, owing to genetic differences or environmental treatments, in cultured cells or mice were assessed by Student's t-test, using the n-Query program (Statistical Solutions, Saugus, MA, USA).
Acknowledgments
The work is supported by Ministry of Science and Technology of China (2013CB911600), National Natural Science Foundation of China (31271449), Jiangsu Provincial Natural Science Foundation (BK20130044, BK20130061, BK2011783), The Research Fund for the Doctoral Program of Higher Education of China (RFDP) (20133207110005), The Program for New Century Excellent Talents in University of Ministry of Education of China (NCET-13- 0868) and the Priority Academic Program Development Award for Jiangsu Higher Education Institutions. The work was also partially supported by an NIH grant RO1 CA085344 to BHS and NCI-designated cancer center support grant P30 CA033572. We also thank Dr Nancy Linford for her professional editing of the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 2005; 434: 864–870. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature 2001; 411: 366–374. [DOI] [PubMed] [Google Scholar]
- Zharkov DO. Base excision DNA repair. Cell Mol Life Sci 2008; 65: 1544–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature 2007; 447: 941–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T. Instability and decay of the primary structure of DNA. Nature 1993; 362: 709–715. [DOI] [PubMed] [Google Scholar]
- Lindahl T, Wood RD. Quality control by DNA repair. Science 1999; 286: 1897–1905. [DOI] [PubMed] [Google Scholar]
- Nakamura J, Walker VE, Upton PB, Chiang SY, Kow YW, Swenberg JA. Highly sensitive apurinic/apyrimidinic site assay can detect spontaneous and chemically induced depurination under physiological conditions. Cancer Res 1998; 58: 222–225. [PubMed] [Google Scholar]
- Pascucci B, Stucki M, Jonsson ZO, Dogliotti E, Hubscher U. Long patch base excision repair with purified human proteins. DNA ligase I as patch size mediator for DNA polymerases delta and epsilon. J Biol Chem 1999; 274: 33696–33702. [DOI] [PubMed] [Google Scholar]
- Frosina G, Fortini P, Rossi O, Carrozzino F, Raspaglio G, Cox LS et al. Two pathways for base excision repair in mammalian cells. J Biol Chem 1996; 271: 9573–9578. [DOI] [PubMed] [Google Scholar]
- Szczesny B, Tann AW, Longley MJ, Copeland WC, Mitra S. Long patch base excision repair in mammalian mitochondrial genomes. J Biol Chem 2008; 283: 26349–26356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler U, Frit P, Salles B, Calsou P. Long-patch DNA repair synthesis during base excision repair in mammalian cells. EMBO Rep 2003; 4: 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascucci B, Russo MT, Crescenzi M, Bignami M, Dogliotti E. The accumulation of MMS-induced single strand breaks in G1 phase is recombinogenic in DNA polymerase beta defective mammalian cells. Nucleic Acids Res 2005; 33: 280–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Zheng L, Dai H, Zhou M, Hua Y, Shen B. Chemical-induced cancer incidence and underlying mechanisms in Fen1 mutant mice. Oncogene 2011; 30: 1072–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demple B, Sung JS. Molecular and biological roles of Ape1 protein in mammalian base excision repair. DNA Repair 2005; 4: 1442–1449. [DOI] [PubMed] [Google Scholar]
- Srivastava DK, Berg BJ, Prasad R, Molina JT, Beard WA, Tomkinson AE et al. Mammalian abasic site base excision repair. Identification of the reaction sequence and rate-determining steps. J Biol Chem 1998; 273: 21203–21209. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y, Kim K. Excision of deoxyribose phosphate residues by DNA polymerase beta during DNA repair. Science 1995; 269: 699–702. [DOI] [PubMed] [Google Scholar]
- Klungland A, Lindahl T. Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1). EMBO J 1997; 16: 3341–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Qian L, Sung JS, de Souza-Pinto NC, Zheng L, Bogenhagen DF et al. Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol Cell Biol 2008; 28: 4975–4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Beard WA, Shock DD, Prasad R, Hou EW, Wilson SH. DNA polymerase beta and flap endonuclease 1 enzymatic specificities sustain DNA synthesis for long patch base excision repair. J Biol Chem 2005; 280: 3665–3674. [DOI] [PubMed] [Google Scholar]
- Gary R, Kim K, Cornelius HL, Park MS, Matsumoto Y. Proliferating cell nuclear antigen facilitates excision in long-patch base excision repair. J Biol Chem 1999; 274: 4354–4363. [DOI] [PubMed] [Google Scholar]
- Gembka A, Toueille M, Smirnova E, Poltz R, Ferrari E, Villani G et al. The checkpoint clamp, Rad9-Rad1-Hus1 complex, preferentially stimulates the activity of apurinic/apyrimidinic endonuclease 1 and DNA polymerase beta in long patch base excision repair. Nucleic Acids Res 2007; 35: 2596–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Biade S, Matsumoto Y. Involvement of flap endonuclease 1 in base excision DNA repair. J Biol Chem 1998; 273: 8842–8848. [DOI] [PubMed] [Google Scholar]
- Zheng L, Zhou M, Chai Q, Parrish J, Xue D, Patrick SM et al. Novel function of the flap endonuclease 1 complex in processing stalled DNA replication forks. EMBO Rep 2005; 6: 83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reagan MS, Pittenger C, Siede W, Friedberg EC. Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J Bacteriol 1995; 177: 364–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff DX, Filosi N, Gaida GM, Kolodner RD. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell 1997; 88: 253–263. [DOI] [PubMed] [Google Scholar]
- Kucherlapati M, Yang K, Kuraguchi M, Zhao J, Lia M, Heyer J et al. Haploinsufficiency of Flap endonuclease (Fen1) leads to rapid tumor progression. Proc Natl Acad Sci USA 2002; 99: 9924–9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Dai H, Zhou M, Li M, Singh P, Qiu J et al. Fen1 mutations result in autoimmunity, chronic inflammation and cancers. Nat Med 2007; 13: 812–819. [DOI] [PubMed] [Google Scholar]
- Figueroa JD, Malats N, Real FX, Silverman D, Kogevinas M, Chanock S et al. Genetic variation in the base excision repair pathway and bladder cancer risk. Hum Genet 2007; 121: 233–242. [DOI] [PubMed] [Google Scholar]
- Yang M, Guo H, Wu C, He Y, Yu D, Zhou L et al. Functional FEN1 polymorphisms are associated with DNA damage levels and lung cancer risk. Hum Mutat 2009; 30: 1320–1328. [DOI] [PubMed] [Google Scholar]
- Chang JS, Wrensch MR, Hansen HM, Sison JD, Aldrich MC, Quesenberry CP Jr. et al. Base excision repair genes and risk of lung cancer among San Francisco Bay Area Latinos and African-Americans. Carcinogenesis 2009; 30: 78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev 2002; 11: 1513–1530. [PubMed] [Google Scholar]
- Sato M, Girard L, Sekine I, Sunaga N, Ramirez RD, Kamibayashi C et al. Increased expression and no mutation of the Flap endonuclease (FEN1) gene in human lung cancer. Oncogene 2003; 22: 7243–7246. [DOI] [PubMed] [Google Scholar]
- Zhang B, Jia WH, Matsuda K, Kweon SS, Matsuo K, Xiang YB et al. Large-scale genetic study in East Asians identifies six new loci associated with colorectal cancer risk. Nat Genet 2014; 46: 533–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosfield DJ, Mol CD, Shen B, Tainer JA. Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity. Cell 1998; 95: 135–146. [DOI] [PubMed] [Google Scholar]
- Balakrishnan L, Bambara RA. Flap endonuclease 1. Annu Rev Biochem 2013; 82: 119–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Kao HI, Bambara RA. Flap endonuclease 1: a central component of DNA metabolism. Annu Rev Biochem 2004; 73: 589–615. [DOI] [PubMed] [Google Scholar]
- Hosfield DJ, Frank G, Weng Y, Tainer JA, Shen B. Newly discovered archaebacterial flap endonucleases show a structure-specific mechanism for DNA substrate binding and catalysis resembling human flap endonuclease-1. J Biol Chem 1998; 273: 27154–27161. [DOI] [PubMed] [Google Scholar]
- Tsutakawa SE, Classen S, Chapados BR, Arvai AS, Finger LD, Guenther G et al. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell 2011; 145: 198–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Jia J, Finger LD, Guo Z, Zer C, Shen B. Functional regulation of FEN1 nuclease and its link to cancer. Nucleic Acids Res 2011; 39: 781–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung L, Onyango D, Guo Z, Jia P, Dai H, Liu S et al. The FEN1 E359K germline mutation disrupts the FEN1-WRN interaction and FEN1 GEN activity, causing aneuploidy-associated cancers. Oncogene 2015; 34: 902–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank G, Qiu J, Somsouk M, Weng Y, Somsouk L, Nolan JP et al. Partial functional deficiency of E160D flap endonuclease-1 mutant in vitro and in vivo is due to defective cleavage of DNA substrates. J Biol Chem 1998; 273: 33064–33072. [DOI] [PubMed] [Google Scholar]
- Wu Z, Lin Y, Xu H, Dai H, Zhou M, Tsao S et al. High risk of benzo[alpha]pyrene-induced lung cancer in E160D FEN1 mutant mice. Mutat Res 2012; 731: 85–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Qian L, Liu R, Dai H, Zhou M, Zheng L et al. Nucleolar localization and dynamic roles of flap endonuclease 1 in ribosomal DNA replication and damage repair. Mol Cell Biol 2008; 28: 4310–4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneke G, Koundrioukoff S, Hubscher U. Phosphorylation of human Fen1 by cyclin-dependent kinase modulates its role in replication fork regulation. Oncogene 2003; 22: 4301–4313. [DOI] [PubMed] [Google Scholar]
- Zheng L, Dai H, Qiu J, Huang Q, Shen B. Disruption of the FEN-1/PCNA interaction results in DNA replication defects, pulmonary hypoplasia, pancytopenia, and newborn lethality in mice. Mol Cell Biol 2007; 27: 3176–3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank G, Qiu JH, Zheng L, Shen BH. Stimulation of eukaryotic flap endonuclease-1 activities by proliferating cell nuclear antigen (PCNA) is independent of its in vitro interaction via a consensus PCNA binding region. J Biol Chem 2001; 276: 36295–36302. [DOI] [PubMed] [Google Scholar]
- Zheng L, Dai HF, Hegde ML, Zhou M, Guo ZG, Wu XW et al. Fen1 mutations that specifically disrupt its interaction with PCNA cause aneuploidy-associated cancer. Cell Res 2011; 21: 1052–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dianova II, Bohr VA, Dianov GL. Interaction of human AP endonuclease 1 with flap endonuclease 1 and proliferating cell nuclear antigen involved in long-patch base excision repair. Biochemistry 2001; 40: 12639–12644. [DOI] [PubMed] [Google Scholar]
- Prasad R, Dianov GL, Bohr VA, Wilson SH. FEN1 stimulation of DNA polymerase beta mediates an excision step in mammalian long patch base excision repair. J Biol Chem 2000; 275: 4460–4466. [DOI] [PubMed] [Google Scholar]
- Guo Z, Zheng L, Dai H, Zhou M, Xu H, Shen B. Human DNA polymerase beta polymorphism, Arg137Gln, impairs its polymerase activity and interaction with PCNA and the cellular base excision repair capacity. Nucleic Acids Res 2009; 37: 3431–3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapados BR, Hosfield DJ, Han S, Qiu J, Yelent B, Shen B et al. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell 2004; 116: 39–50. [DOI] [PubMed] [Google Scholar]
- Qiu J, Liu R, Chapados BR, Sherman M, Tainer JA, Shen B. Interaction interface of human flap endonuclease-1 with its DNA substrates. J Biol Chem 2004; 279: 24394–24402. [DOI] [PubMed] [Google Scholar]
- Roos WP, Kaina B. DNA damage-induced cell death: from specific DNA lesions to the DNA damage response and apoptosis. Cancer Lett 2013; 332: 237–248. [DOI] [PubMed] [Google Scholar]
- Roos WP, Kaina B. DNA damage-induced cell death by apoptosis. Trends Mol Med 2006; 12: 440–450. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Coffman CR. DNA damage-induced programmed cell death: potential roles in germ cell development. Ann NY Acad Sci 2005; 1049: 9–16. [DOI] [PubMed] [Google Scholar]
- Noon AT, Goodarzi AA. 53BP1-mediated DNA double strand break repair: Insert bad pun here. DNA Repair 2011; 10: 1071–1076. [DOI] [PubMed] [Google Scholar]
- Adams MM, Carpenter PB. Tying the loose ends together in DNA double strand break repair with 53BP1. Cell Div 2006; 1: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 Binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol 2000; 151: 1381–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappold I, Iwabuchi K, Date T, Chen JJ. Tumor suppressor p53 binding protein 1 (53BP1) is involved in DNA damage-signaling pathways. J Cell Biol 2001; 153: 613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson L, Henderson C, Adachi Y. Phosphorylation and rapid relocalization of 53BP1 to nuclear foci upon DNA damage. Mol Cell Biol 2001; 21: 1719–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H, RJW, Mack G, Rattner JB, Yen TJ. CENP-F is a protein of the nuclear matrix that assembles onto kinetochores at late G2 and is rapidly degraded after mitosis. J Cell Biol 1995; 130: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du JA, Zhang YR, Liu Y, Li Y, Zhu XL. Involvement of Cenp-F in interphase chromatin organization possibly through association with DNA-dependent protein kinase. Acta Biochim Biophys Sin 2010; 42: 839–846. [DOI] [PubMed] [Google Scholar]
- Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Pedersen RS et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol 2011; 13: 243–253. [DOI] [PubMed] [Google Scholar]
- Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res 1988; 175: 184–191. [DOI] [PubMed] [Google Scholar]
- Singh NP, Tice RR, Stephens RE, Schneider EL. A microgel electrophoresis technique for the direct quantitation of DNA damage and repair in individual fibroblasts cultured on microscope slides. Mutat Res 1991; 252: 289–296. [DOI] [PubMed] [Google Scholar]
- Bajpayee M, Dhawan A, Parmar D, Pandey AK, Mathur N, Seth PK. Gender-related differences in basal DNA damage in lymphocytes of a healthy Indian population using the alkaline Comet assay. Mutat Res 2002; 520: 83–91. [DOI] [PubMed] [Google Scholar]
- El-Andaloussi N, Valovka T, Toueille M, Steinacher R, Focke F, Gehrig P et al. Arginine methylation regulates DNA polymerase beta. Mol Cell 2006; 22: 51–62. [DOI] [PubMed] [Google Scholar]
- Ivancsits S, Diem E, Pilger A, Rudiger HW, Jahn O. Induction of DNA strand breaks by intermittent exposure to extremely-low-frequency electromagnetic fields in human diploid fibroblasts. Mutat Res 2002; 519: 1–13. [DOI] [PubMed] [Google Scholar]
- van Gent DC, Hoeijmakers JH, Kanaar R. Chromosomal stability and the DNA double-stranded break connection. Nat Rev Genet 2001; 2: 196–206. [DOI] [PubMed] [Google Scholar]
- Soza S, Leva V, Vago R, Ferrari G, Mazzini G, Biamonti G et al. DNA ligase I deficiency leads to replication-dependent DNA damage and impacts cell morphology without blocking cell cycle progression. Mol Cell Biol 2009; 29: 2032–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Storchova Z, Pellman D. Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev 2007; 17: 157–162. [DOI] [PubMed] [Google Scholar]
- Larsen E, Gran C, Saether BE, Seeberg E, Klungland A. Proliferation failure and gamma radiation sensitivity of Fen1 null mutant mice at the blastocyst stage. Mol Cell Biol 2003; 23: 5346–5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Kanjanapangka J, Liu N, Liu S, Liu C, Wu Z et al. Sequential posttranslational modifications program FEN1 degradation during cell-cycle progression. Mol Cell 2012; 47: 444–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Dai HF, Zhou M, Li M, Singh P, Qiu JZ et al. Fen1 mutations result in autoimmunity, chronic inflammation and cancers. Nat Med 2007; 13: 812–819. [DOI] [PubMed] [Google Scholar]
- Nussenzweig A, Nussenzweig MC. Origin of chromosomal translocations in lymphoid cancer. Cell 2010; 141: 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Putnam CD, Kane MF, Zhang W, Edelmann L, Russell R et al. Mutation in Rpa1 results in defective DNA double-strand break repair, chromosomal instability and cancer in mice. Nat Genet 2005; 37: 750–755. [DOI] [PubMed] [Google Scholar]
- Zheng L, Dai H, Zhou M, Li X, Liu C, Guo Z et al. Polyploid cells rewire DNA damage response networks to overcome replication stress-induced barriers for tumour progression. Nat Commun 2012; 3: 815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrmann RS, Karjalainen JM, Krajewska M, Westra HJ, Maloney D, Simeonov A et al. Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat Genet 2015; 47: 115–125. [DOI] [PubMed] [Google Scholar]
- Guo Z, Chavez V, Singh P, Finger LD, Hang H, Hegde ML et al. Comprehensive mapping of the C-terminus of flap endonuclease-1 reveals distinct interaction sites for five proteins that represent different DNA replication and repair pathways. J Mol Biol 2008; 377: 679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling C, Zhang J, Lin D, Tao A. Approaches for the generation of active papain-like cysteine proteases from inclusion bodies of Escherichia coli. World J Microbiol Biotechnol 2015; 31: 681–690. [DOI] [PubMed] [Google Scholar]
- Gao J, Wang H. Prokaryotic expression, refolding and purification of high-purity mouse midkine in Escherichia coli. Appl Biochem Biotechnol 2015; 176: 454–466. [DOI] [PubMed] [Google Scholar]
- Bose R, Kavuri SM, Searleman AC, Shen W, Shen D, Koboldt DC et al. Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov 2013; 3: 224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka N, Wang L, Kishimoto K, Tsuji T, Hu GF. A therapeutic target for prostate cancer based on angiogenin-stimulated angiogenesis and cancer cell proliferation. Proc Natl Acad Sci USA 2006; 103: 14519–14524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet 2007; 39: 673–677. [DOI] [PubMed] [Google Scholar]
- Narita K, Staub J, Chien J, Meyer K, Bauer M, Friedl A et al. HSulf-1 inhibits angiogenesis and tumorigenesis in vivo. Cancer Res 2006; 66: 6025–6032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.