Abstract
Invasive growth and apoptosis resistance of breast cancer cells are associated with metastasis and disease relapse. Here we identified that the lysine-specific demethylase KDM3A played a dual role in breast cancer cell invasion and apoptosis by demethylating histone and the non-histone protein p53, respectively. While inducing pro-invasive genes by erasing repressive histone H3 lysine 9 methylation, KDM3A promotes chemoresistance by demethylating p53. KDM3A suppressed pro-apoptotic functions of p53 by erasing p53-K372me1 as this methylation site is crucial for the stability of chromatin-bound p53. Unexpectedly, depletion of KDM3A was capable of reactivating mutated p53 to induce the expression of pro-apoptotic genes in breast cancer with mutant p53. Moreover, KDM3A knockdown also potently inhibited tumorigenic potentials of breast cancer stem-like cells and rendered them sensitive to apoptosis induced by chemotherapeutic drugs. Taken together, our results suggest that KDM3A might be a potential therapeutic target for human breast cancer treatment and prevention.
Keywords: Histone demethylase, Invasion, Chemoresistance, p53, Breast Cancer
Breast cancer is the most common malignant disease affecting the women worldwide and is the second leading cause of cancer related death in women.1 Invasive growth, metastasis and development of cancer therapy resistance are the critical events associated with high recurrence rate and poor prognosis in breast cancer.2 Invasive growth plays a critical role in breast cancer progression and is refers to the ability of cancer cells to invade the adjacent tissues, degrade the surrounding matrix, survive in foreign compartments, and move to the locations distant from the site of origin.3,4 In addition to metastasis, it also plays a critical role in the development and progression of invasive breast cancer from epithelial dysplasia and carcinoma in situ. To initiate invasion and metastasis, various signaling pathways are activated to render breast cancer cells with the motile and invasive phenotype.5 As a result, breast cancer cells undergo extensive modifications in the expression and distribution of cell junction and cell adherence components as well as epigenetic changes.6,7 Once the tumor cells invade the adjacent tissues, they can intravasate into blood or lymphatic vessels and subsequently disseminated to the distant organs by extravasation.8 In the target tissue, the metastatic tumor cells undergo proliferation; promote angiogenesis from secondary cancer8.
Because invasive cells lose their cadherin-based intercellular junction and matrix attachment, cell resistance to apoptosis appears to be essential during breast cancer cell invasive growth. Invasion and apoptosis resistance are biologically distinct but interrelated process as they share some common signaling networks in cancer development and progression.9 For example, the tumor suppressor protein, p53 negatively regulates these processes in various cancers and therefore loss of expression and/or functions of p53 due to genetic mutations are strongly associated with highly invasive and resistant phenotypes of breast cancers.10,11 Unlike other tumor suppressor genes, mutant p53 not only loses its tumor suppressor function, but also gains oncogenic potentials.12 Breast cancer cells expressing mutant p53 have impaired apoptotic responses and increased invasiveness and migration and metastasis.11 Moreover, breast cancer cells with mutant p53 have been found to have stem-cell-like transcriptional signature.13 Mutant p53 might directly regulate metabolic signaling pathways to control self-renewal and drug resistance of breast cancer stem cells (CSCs) that are highly tumorigenic and invasive.14,15 Therefore, targeting mutant p53 might enhance cancer therapy-induced apoptosis and help to prevent tumor relapse by eliminating CSCs.
Despite the substantial progress that has been made in understanding the molecular regulation of breast cancer invasive growth, apoptosis resistance and stemness, how these processes are epigenetically controlled is poorly understood. Emerging evidence suggests that epigenetic histone modification by lysine methyltransferases (KMTs) and demethylases (KDMs) play a key role in controlling gene expressions.16 A large group of Jumonji-C domain (JmjC) containing KDMs have been discovered to regulate diverse biological and pathological processes including embryonic development, stem cell self-renewal and differentiation, genome integrity and oncogenesis.17,18 Acquiring CSC properties and chemo resistance are the critical events strongly integrated with cancer metastasis that begins with cell invasion. Therefore, to explore epigenetic mechanisms regulating these processes, we conducted functional in vitro small interfering RNA (siRNA) screening in a highly invasive breast cancer cell line MDA-MB-231 to identify potential histone demethylases that are required for invasion. We found KDM3A as a key epigenetic factor activating the expression of pro-invasive genes in breast cancer cells by removing the repressive dimethylation of histone H3 lysine 9 (H3K9me2) on their promoters. The depletion of KDM3A inhibited breast cancer invasive growth in vitro and in vivo. Unexpectedly, KDM3A also was a lysine demethylase for the non-histone protein p53 and could inhibit p53-proapoptotic functions. Inhibiting KDM3A reactivated mutant p53 to induce the expression of the pro-apoptotic genes and inhibited oncogenic potentials and chemoresistance of breast CSCs.
RESULTS
KDM3A promotes breast cancer cell invasion
In an attempt to identify potent histone demethylases that were associated with breast cancer cell invasion, we transfected a highly invasive p53-mutated breast cancer cell line MDA-MB-231 with siRNA targeting 16 different histone demethylases and confirmed the knockdown efficiency by Real-time reverse transcription polymerase chain reaction (RT-PCR) (Figure 1a). Next we examined the invasive ability of the cells depleted for individual histone demethylases along with scramble siRNA transfected cells by Matrigel invasion assays. Our initial screening showed that depletion of KDM3A, KDM6B and FBXL11 significantly inhibited the invasion, while knockdown of JHDM1D, KDM3B, KDM4C, PHF2, PHF8 and UTX modestly enhanced the invasiveness of MDA-MB-231 cells (Figure 1b). Even though depletion of KDM6B and FBXL11 also significantly inhibited invasion, the magnitude of inhibition was very dramatic with KDM3A knockdown. Therefore, we focused our further studies only on KDM3A. In order to validate our results from KDM3A siRNA and to further explore the underlying mechanism of KDM3A-mediated invasion, we generated two lentivirus-based short hairpin RNAs (shRNAs; KDM3Ash1 and KDM3Ash2) to target different sequences on KDM3A mRNA. As demonstrated by Western blot (Figure 1c), both KDM3Ash1 and KDM3Ash2 were able to deplete KDM3A protein in MDA-MB-231 cells (MDA/KDM3Ash1 and MDA/KDM3Ash2 cells). Indeed, stable knockdown of KDM3A by both shRNAs consistently inhibited the invasion of MDA-MB-231 cells, recapitulating the effect of KDM3A siRNA on cell invasion (Figures 1d and e). To exclude the possibility of proliferation differences that could be reflected in the invasion results, we simultaneously seeded equal number of cells in a separate well and quantified the cell number at the end of invasion assay. There was no significant difference in cell proliferation rates between MDA/Scrsh and MDA/KDM3Ash1 or MDA/KDM3Ash2 during the period of invasion assays (data not shown). Further, to rule out the off-target effects, we stably depleted KDM3A in MDA-MB-231 cells using KDM3Ash3 targeting 3’-prime untranslated region of KDM3A mRNA and restored KDM3A expression using retroviral KDM3A plasmid. KDM3Ash3 depleted more than 90% of KDM3A proteins in MDA-MB-231 cells (Supplementary Figure 1a). Consistently, MDA/KDM3Ash3 cells showed less invasiveness as compared to MDA/Scrsh cells (Supplementary Figure 1b and c). Conversely, restoration of KDM3A expression in MDA/KDM3Ash3 cells rescued cell invasion (Supplementary Figure 1d, e and f), indicating that KDM3A shRNAs-mediated inhibition of cell invasion is specifically due to KDM3A knockdown.
Figure 1. KDM3A knockdown inhibits breast cancer cell invasion.
(a) Quantitative RT-PCR analysis of the relative expression of histone demethylases in MDA-MB-231 cells transfected with targeted siRNA relative to their expression in cells treated with scrambled (Scr) siRNA.
(b) Analysis of invasion in MDA-MB-231 cells transfected with the indicated histone demethylase siRNA relative to Scrsi control. Invasive cells from 5 random fields were quantified. Data are means ± SD from a representative experiment of 3 independent experiments.
(c) Western blot analysis of the abundance of KDM3A in MDA-MB-231 cells stably expressing Scrsh (MDA/Scrsh) or KDM3Ash (MDA/KDM3Ash1 and MDA/KDM3Ash2).
(d and e) Knockdown of KDM3A inhibited MDA-MB-231 cell invasion. Invaded cells were counted in 5 random fields per well. Data are means ± SD of triplicate samples from a representative experiment of 3 independent experiments.
(f) Knockdown of KDM3A in Hs578T cells by shRNA. Hs/Scrsh, Hs578T cells expressing scramble shRNA; Hs/KDM3Ash1, Hs578T cells expressing KDM3A shRNA1; Hs/KDM3Ash2, Hs578T cells expressing KDM3A shRNA2.
(g and h) Knockdown of KDM3A inhibited Hs578T cell invasion. Invaded cells were counted in 5 random fields per well. Data are means ± SD of triplicate samples from a representative experiment of 3 independent experiments.
(i) Immunostaining of KDM3A, JUN and MMP-9 proteins in human invasive breast cancers and normal breast tissues.
Next, we asked whether KDM3A was required for the invasiveness of other breast cancer cells. We depleted KDM3A in another highly invasive and p53-mutated breast cancer cell line Hs578T. As expected, both KDM3Ash1 and KDM3Ash2 reduced the expression of KDM3A proteins in Hs578T cells (Figure 1f). Similar to MDA-MB-231 cells, KDM3A knockdown also significantly inhibited Hs578T cell invasion (Figures 1g and h). To determine whether KDM3A is associated with the development and progression of human invasive breast cancer, we examined the expression of KDM3A proteins in human breast cancer tissue array containing invasive breast cancer, lymph node metastasized breast cancer and normal breast tissues. KDM3A was highly expressed in invasive breast cancers and lymph node metastasized breast cancers as compared to normal breast tissues (Figure 1i and Table 1). However, there was no significant difference in KDM3A expression between primary tumors and metastatic breast cancer tissues although KDM3A was strongly expressed in some metastatic breast cancer tissues, suggesting that KDM3A might be involved in early invasive growth of breast cancer cells.
Table 1.
KDM3A is highly expressed in human invasive breast cancer tissues
| KDM3 |
MMP-9 |
JUN |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | + | ++ | +++ | 0 | + | ++ | +++ | 0 | + | ++ | +++ | |
| Normal | 30% (3/10) |
70% (7/10) |
0% (0/10) |
0% (0/10) |
40% (4/10) |
30% (3/10) |
20% (2/10) |
10% (1/10) |
40% (4/10) |
60% (6/10) |
0% (0/10) |
0% (0/10) |
| Cancer** | 6% (3/50) |
30% (15/50) |
42% (21/50) |
22% (11/50) |
8% (4/50) |
30% (15/50) |
34% (16/50) |
30% (15/50) |
6% (3/50) |
28% (14/50) |
42% (21/50) |
24% (12/50) |
| LN** | 2.5% (1/40) |
32.5% (13/40) |
30% (12/40) |
35% (14/40) |
5% (2/40) |
35% (14/40) |
32.5% (13/40) |
27.5% (11/40) |
10% (4/40) |
22.5% (9/40) |
32.5% (13/40) |
35% (14/40) |
Normal human breast tissues (Normal; n = 10), human invasive primary breast cancer without lymph node metastasis (cancer; n = 50), and lymph node metastatic breast cancer (LN; n = 40) were stained for KDM3A, MMP-9 and JUN. The staining intensity was scored as follows: 0, negative staining; +, weak staining; ++, moderate staining; +++ strong staining.
P < 0.01 breast cancer versus normal, Wilcoxon rank sum test;
P < 0.01 LN versus normal. P > 0.05 breast cancer versus LN.
KDM3A promotes chemoresistance in breast cancer cells by inhibiting apoptosis
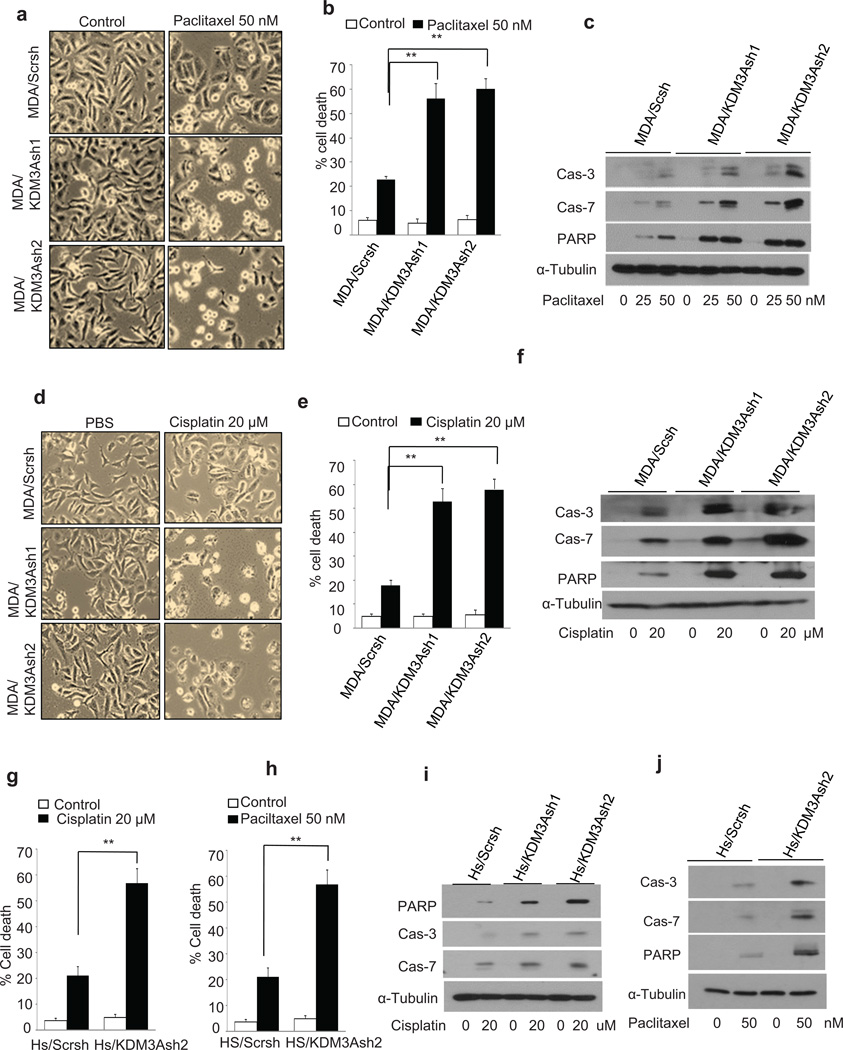
Invasive growth and chemoresistance are interlinked processes in breast cancer development.2 MDA-MB-231 cells express mutant p53 and are very resistant to apoptosis induced by chemotherapeutic drugs. Because KDM3A promotes the breast cancer cells invasion, we hypothesized that KDM3A might promote chemoresistance. To test this hypothesis, we treated MDA/Scrsh, MDA/KDM3Ash1 and MDA/KDM3Ash2 cells with two widely used chemotherapeutic drugs, paclitaxel and cisplatin. Interestingly, KDM3A depletion significantly sensitized MDA-MB-231 cells to paclitaxel-induced cell death (Figures 2a and b). To determine that increased cell death in KDM3A knockdown cells was due to the induction of apoptosis, we examined caspase activation in MDA/Scrsh, MDA/KDM3Ash1 and MDA/KDM3Ash2 cells upon paclitaxel treatment. Western blot analysis revealed that KDM3A knockdown potently enhanced the activation of caspase-3 and −7 in MDA-MB-231 cells induced by paclitaxel (Figure 2c). Consistently, KDM3A knockdown also enhanced the cleavage of PARP, a substrate of caspase-3 and −7 (Figure 2c). Similarly, we found that KDM3A knockdown also enhanced apoptosis and the activation of caspase-3 and −7 induced by cisplatin in MDA-MB-231 cells (Figures 2d, e and f). Also, we examined whether KDM3A expression promoted apoptosis resistance in Hs578T cells. Indeed, KDM3A knockdown in Hs578T cells significantly enhanced cell death following cisplatin and paclitaxel exposure (Figures 2g and h). In line, KDM3A knockdown also enhanced the activation of caspase-3 and −7 and the cleavage of PARP induced by cisplatin and paclitaxel in Hs578T cells (Figures 2i and j).
Figure 2. KDM3A knockdown sensitizes breast cancer cells to apoptosis induced by paclitaxel and cisplatin.
(a and b) Image (a) and quantification (b) of paclitaxel induced cell death in MDA/Scrsh, MDA/KDM3Ash1 and MDA/KDM3Ash2 cells. The cells were treated with paclitaxel (50 nM) for 24 hrs. Data are means ± SD of triplicate samples from a representative experiment of 3 independent experiments.
(c) Immunoblot analysis of caspase activation induced by paclitaxel in MDA/Scrsh, MDA/KDM3Ash1 and MDA/KDM3Ash2 cells. The cells were treated with paclitaxel (25 and 50 nM) for 24 hrs.
(d and e) Image (d) and quantification (e) of cisplatin induced cell death in MDA/Scrsh, MDA/KDM3Ash1 and MDA/KDM3Ash2 cells. The cells were treated with cisplatin (20 µM) for 24 hrs. Data are means ± SD of triplicate samples from a representative experiment of 3-independent experiments.
(f) Immunoblot analysis of caspase activation induced by cisplatin in MDA/Scrsh, MDA/KDM3Ash1 and MDA/KDM3Ash2 cells. The cells were treated with cisplatin (20 µM) for 24 hrs.
(g) KDM3A knockdown enhanced cisplatin-induced cell death in Hs578T cells. The cells were treated with cisplatin (20 µM) for 24 hrs. Data are means ± SD of triplicate samples from a representative experiment of 3-independent experiments.
(h) KDM3A knockdown enhanced paclitaxel-induced cell death in Hs578T cells. The cells were treated with paclitaxel (50 nM) for 24 hrs. Data are means ± SD of triplicate samples from a representative experiment of 3 independent experiments.
(i) Immunoblot analysis of caspase activation induced by cisplatin in Hs/Scrsh and Hs/KDM3Ash2 cells. The cells were treated with cisplatin (20 µM) for 24 hrs.
(j) Immunoblot analysis of caspase activation induced by paclitaxel in Hs/Scrsh and Hs/KDM3Ash2 cells. The cells were treated with paclitaxel (20 µM) for 24 hrs.
**p < 0.01, unpaired two-tailed Student’s t test.
KDM3A epigenetically activates pro-invasive genes to induce invasion
It is known that KDM3A can promote gene transcription by removing repressive H3K9me2 and H3K9me1 marks from the target gene promoter which are often associated with gene silencing.18 To gain the insight into how KDM3A epigenetically promoted breast cancer invasive growth and chemoresistance, we performed gene expression profiling to screen for potential genes that were affected by KDM3A knockdown in MDA-MB-231 cells. Microarray analysis revealed 332 genes that had decreased expression (Supplementary Table 1) and 186 genes that had increased expression (Supplementary Table 2) by at least twofold after KDM3A knockdown. Gene enrichment analysis revealed that knockdown of KDM3A significantly reduced the expression of genes associated with wound healing, cell migration and cell motility, including matrix metallopeptidase 9 (MMP-9) and S100A family proteins. Next we validated the microarray results by Real-time RT-PCR. Consistently, both expressions of MMP-9 and S100A4 were downregulated in MDA/KDM3Ash1 and MDA/KDM3Ash2 cells as compared to MDA/Scrsh cells (Figure 3a). To gain mechanistic insight into how KDM3A regulates the expression of MMP-9 and S100A4, we examined whether KDM3A occupied on the promoters of MMP-9 and S100A4 by ChIP assays in MDA-MB-231 cells. ChIP assays revealed that KDM3A was significantly enriched on the MMP-9 promoter in MDA/Scrsh cells compared to MDA/KDM3Ash1 and MDA/KDM3Ash2 cells (Figure 3b). Since KDM3A removes the repressive H3K9me2 marks to prime the gene transcription, we next examined the level of H3K9me2 on the same regions where KDM3A binding was detected on the MMP-9 promoter. Indeed, the abundance of H3K9me2 was increased in MDA/KDM3Ash1 and MDA/KDM3Ash2 cells compared to MDA/Scrsh cells (Figure 3c). As a negative control, we found that anti-IgG control did not enrich signals. Also, KDM3A was not detected on 6 kb upstream of the MMP-9 promoter. Our ChIP assays also found that KDM3A was present on the S100A4 promoter to erase H3K9me2 marks in MDA-MB-231 cells (Figures 3d and e). Moreover, MMP-9 expression was also inhibited in the Hs578T cells expressing KDM3Ashs as determined by Real-time RT PCR (Figure 3f). Interestingly, KDM3A knockdown did not affect the S100A4 expression in Hs578T cells (Figure 3f). Similarly, our ChIP assays showed that KDM3A was also present on the MMP-9 promoter in Hs578T cells and KDM3A knockdown increased the abundance of H3K9me2 in the MMP-9 promoter (Figure 3g and h).
Figure 3. KDM3A activates the expression of pro-invasive genes in breast cancer cells by demethylating H3K9me2.
(a) Real-time RT-PCR of MMP-9 and S100A4 expressions in MDA-MB-231 cells. Data are means ± SD of triplicate samples from a representative experiment.
(b and c) ChIP assays of KDM3A (b) and H3K9me2 (c) occupancies on the MMP-9 promoter in MDA/Scrsh, MDA/KDM3Ash1 and MDA/KDM3Ash2 cells. Data are means ± SD of triplicate assays from a representative experiment.
(d and e) ChIP assays of KDM3A (e) and H3K9me2 (f) occupancies on the S100A4 promoter in Hs/Scrsh and Hs/KDM3Ash2 cells. Data are means ± SD of triplicate assays from a representative experiment.
(f) Real-time RT-PCR of MMP-9 and S100A4 expressions in Hs578T cells. Data are means ± SD of triplicate samples from a representative experiment.
(g and h) ChIP assay of KDM3A (g) and H3K9me2 (h) binding on the MMP-9 promoter in HS/Scrsh and HS/KDM3Ash2 cells. Data are means ± SD of triplicate assays from a representative of three independent experiments.
(i) Western blotting of JUN and FOSL-1 expressions in MDA-MB-231 and Hs578T cells.
(j) Real-time RT-PCR of JUN and FOSL-1 expressions in MDA/Scrsh and MDA/KDM3Ashs cells.
(k) ChIP assay of KDM3A and H3K9me2 binding on the JUN promoter in MDA/Scrsh and MDA/KDM3Ash2 cells. Data are means ± SD of triplicate assays from a representative experiment.
*p < 0.05, **p < 0.01, unpaired two-tailed Student’s t test.
It is well known that MMP-9 and S100A family genes are transcriptionally regulated by the transcription factor activator protein 1 (AP-1), which plays a critical role in tumor invasion and metastasis.4 Moreover, we noticed that several KDM3A-dependent genes identified by microarray, including CSF2, PLAU and EGR1, are also well-known AP-1 target genes. The AP-1 family members including JUN and FOSL1 have been found to be associated with breast cancer invasion and metastasis in vitro and in vivo.3 Moreover, the region on the MMP-9 promoter that KDM3A bound to contains the AP-1 site. Although our microarray did not reveal that KDM3A knockdown affected JUN and FOSL1 expression, we first performed Western blot to determine whether KDM3A knockdown modulated their expression. Interestingly, KDM3A depletion completely inhibited JUN expression in both MDA-MB-231 and Hs578T cells, while FOSL1 expression was partially inhibited (Figure 3i). Real-time RT-PCR also confirmed that KDM3A knockdown significantly inhibited JUN expression (Figure 3j). To further confirm our results, we found that the restoration of KDM3A restored the expression of JUN and MMP-9 in KDM3A knockdown cells (Supplementary Figure 2). To explore whether KDM3A epigenetically controlled JUN transcription, we also performed ChIP assays. Indeed, ChIP assays revealed that KDM3A was present on the JUN promoter, and KDM3A knockdown leads to the enrichment of its substrate, H3K9me2 on the JUN promoter (Figure 3k). Taken together, our results suggest that KDM3A epigenetically promotes the expression of invasive genes by erasing H3K9me2 marks in breast cancer cells.
To further confirm that KDM3A regulated the expression of MMP-9 and JUN expressions in human breast cancer development, we examined whether KDM3A expression was correlated with the level of MMP-9 and JUN using human breast cancer tissue arrays. The expression of both MMP-9 and JUN was significantly higher in the primary and metastatic breast cancer tissues compared to normal breast tissues (Figure 1i and Table 1). Pearson correlation analysis suggested that KDM3A was positively correlated with the level of MMP-9 and JUN in breast cancer tissues (Table 2 and 3).
Table 2.
The abundance of KDM3A correlates with the abundance of MMP-9 in human breast cancer
| KDM3A ** |
||||||
|---|---|---|---|---|---|---|
| 0 | + | ++ | +++ | Total | ||
| MMP-9 | 0 | 7 | 3 | 0 | 0 | 10 |
| + | 0 | 25 | 4 | 3 | 32 | |
| ++ | 0 | 4 | 23 | 4 | 31 | |
| +++ | 0 | 3 | 6 | 18 | 27 | |
| Total | 7 | 35 | 33 | 25 | 100 | |
Normal human breast tissues (n=10), invasive primary breast tumor (n=50), and lymph node metastatic tissues (n=40) were stained for KDM3A and MMP-9. The staining intensity was scored as detailed in Table 1. Pearson correlation, R2 = 0.759
P < 0.01.
Table 3.
The abundance of KDM3A correlates with the abundance of JUN in human breast cancer.
| KDM3A ** |
||||||
|---|---|---|---|---|---|---|
| 0 | + | ++ | +++ | Total | ||
| JUN | 0 | 7 | 4 | 0 | 0 | 11 |
| + | 0 | 21 | 5 | 3 | 29 | |
| ++ | 0 | 5 | 26 | 2 | 33 | |
| +++ | 0 | 3 | 3 | 20 | 26 | |
| Total | 7 | 35 | 33 | 25 | 100 | |
Normal human breast tissues (n=10), invasive primary breast tumor (n=50), and lymph node metastatic tissues (n=40) were stained for KDM3A and JUN. The staining intensity was scored as detailed in Table 1. Pearson correlation, R2 = 0.78
P < 0.01.
Inhibiting KDM3A reactivates mutant p53 by lysine demethylation
Our microarray profiling failed to provide sufficient information on how KDM3A epigenetically promotes chemoresistance in breast cancer cells. We decided to directly profile the expression of the pro-apoptotic genes in MDA-MB-231 cells. Real-time RT-PCR revealed that KDM3A knockdown significantly induced the expression of PUMA and NOXA while other pro-apoptotic genes, including BIK, BIM, BAK, and BAX, were weakly upregulated (Figure 4a). Consistently, Western blot analysis showed that the expression of PUMA and NOXA proteins was increased by KDM3A knockdown (Figure 4b). Since Hs578T cells also express mutant p53 and KDM3A knockdown rendered them sensitive to apoptosis induced by cisplatin and paclitaxel, we examined whether KDM3A knockdown induced the expression of pro-apoptotic genes in Hs578T cells. Interestingly, KDM3A knockdown induced the expression of PUMA and BAX, but not NOXA, in Hs578T cells probably due to difference in cellular contexts (Figure 4c). To examine whether KDM3A knockdown affected the expression of p53 target genes in breast cancer cells expressing wild type p53, we knocked down KDM3A in MCF-7 cells (Figure 4d). Real-time PCR revealed that KDM3A knockdown increased the expression of PUMA, NOXA and BAX in MCF-7 cells (Figure 4e).
Figure 4. Inhibiting KDM3A reactivates mutant p53 to induce pro-apoptotic genes by demethylating p53-K372me1.
(a) Real-time RT-PCR of p53 target genes expression in MDA-MB-231 cells. Data are means ± SD of triplicate samples from a representative experiment.
(b) Immunoblotting of PUMA and NOXA in MDA/Scrsh, MDA/KDM3Ash1 and MDA/KDM3Ash2 cells.
(c) Quantitative RT-PCR of p53 target genes expression in Hs578T expressing Scrsh or KDM3Ash. Data are means ± SD of triplicate samples from a representative experiment.
(d) Knockdown of KDM3A in MCF-7 cells by shRNA
(e) Quantitative RT-PCR of p53 target genes expression in MCF-7 cells expressing Scrsh or KDM3Ash. Data are means ± SD of triplicate samples from a representative experiment.
(f) Quantitative RT-PCR of MDM2 expression in MDA-MB-231, Hs578T and MCF-7 cells. Data are means ± SD of triplicate samples from a representative experiment.
(g) Western blot of p53 proteins in MDA-MB-231 cells.
(h and i) Knockdown of KDM3A increased p53-K372me1 and p53 interacted with KDM3A as determined by co-immunoprecipitation (Co-IP) and Western blot in Hs578T cells and MDA-MB-231 cells.
(j) Knockdown of KDM3A increased p53-K372me1 and p53 interacted with KDM3A as determined by co-immunoprecipitation (Co-IP) and Western blot in MCF-7 cells.
(k) p53-K372me1 was demethylated by recombinant KDM3A proteins in vitro
*p < 0.05, **p < 0.01, unpaired two-tailed Student’s t test.
These observations raise the question of how KDM3A negatively regulates gene expression, while its function is to epigenetically activate gene expression by histone demethylation.18 Interestingly, we noticed that KDM3A knockdown upregulated MDM2 from microarray analysis which is a well-known p53 target gene (Supplementary Table 2). Real-time RT-PCR confirmed that KDM3A knockdown upregulated MDM2 expression in MDA-MB-231 and Hs578T cells as well as MCF-7 cells (Figure 4f). Since MDA-MB-231 and Hs578 cells express oncogenic mutant p53, it was totally unexpected that KDM3A knockdown upregulated MDM2. We explored whether KDM3A might target non-histone proteins to confer the apoptosis resistance in breast cancer cells. Since MDM2, PUMA, NOXA and BAX are p53 target genes,19 we hypothesized that KDM3A might inhibit p53 transcriptional activities to inhibit their expression by demethylating p53. Western blot analysis showed that KDM3A knockdown did not affect p53 protein levels in MDA-MB-231 cells (Figure 4g). Previously, it has been shown that lysine methylation of p53 occurs at the C-terminal regulatory domain, and four lysine residues including K370, K372, K373 and K382 have been identified. In general, while methylation of p53 at K370, K373 and K382 represses p53 transcriptional activities, methylation at K372 enhances p53 activities.20–23 Since KDM3A knockdown activated the expression of p53-dependent genes, we reasoned that increasing p53 methylation enhanced its transcriptional activities. Mono-methylation of p53 at K372 (p53-K372me1) mediated by Set9 histone-lysine N-methyltransferase has been reported to activate and stabilize the p53 protein.23 This unique modification is required for the nuclear localization and stabilization of the chromatin-associated p53 for the activation of target genes.23 Therefore, we hypothesized that KDM3A might inhibit p53 transcriptional activity by demethylating K372, thus impeding p53-mediated transcription and pro-apoptotic functions. To test this possibility, we took advantage of anti-p53-K372me1 antibodies and determined whether KDM3A knockdown increased the level of p53-K732me1. We utilized immunoprecipitation (IP) to enrich the p53-K372me1 from the whole cell lysates and subsequently detected them using the anti-p53 antibodies. IP-Western blot revealed that KDM3A knockdown dramatically increased the level of p53-K372me1 in Hs578T cells (Figure 4h, top panel). Similarly, KDM3A knockdown also increased the level of p53-K372me1 in MDA-MB-231 and MCF-7 cells (Figures 4i and j top panel). If KDM3A catalyzed the removal of the methyl group from p53-K372me1, these two proteins could physically interact with each other. Co-IP assays revealed that KDM3A could pull down p53 in Hs578T/Scrsh cells, but not in Hs578/KDM3Ash2 cells (Figure 4h). Furthermore, Co-IP also found that KDM3A interacted with p53 in MDA-MB-231 cells and MCF-7 cells (Figure 4i and j). To further confirm that KDM3A demethylated p53-K372me1 directly, we performed in vitro demethylation assays using human recombinant KDM3A proteins. Equal amounts of immunoprecipitated p53 proteins from MDA/KDM3Ash2 cell lysates were incubated with or without recombinant KDM3A proteins. After arresting the reaction, the incubation mixtures were subjected to Western blot analysis to probe p53-K372me1. Consistent with the in vivo results, recombinant KDM3A proteins drastically reduced the level of p53-K372me1 (Figure 4k). Collectively, these results suggest that KDM3A demethylates p53-K372me1 to inhibit its transcriptional activity.
Inhibiting KDM3A promotes p53 binding to the PUMA and NOXA promoter
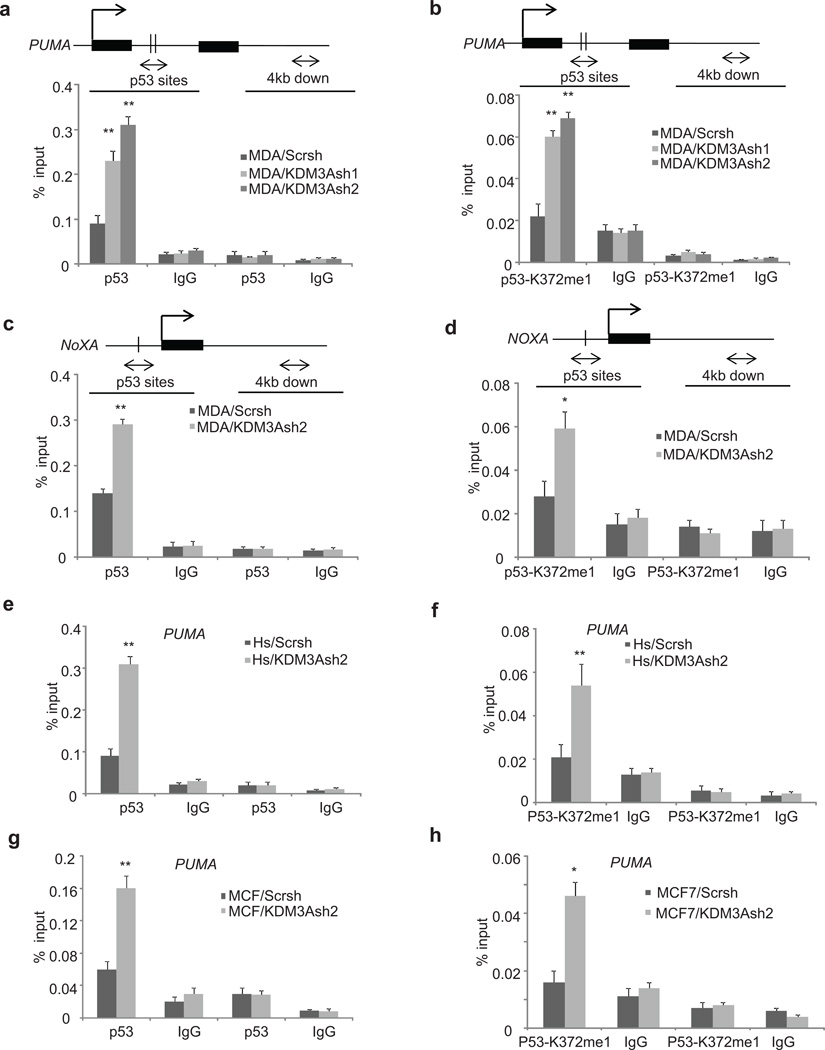
We noticed that the effect of p53 methylation on its transcriptional activities was mainly examined in cells with wild type p53.23 Since both MDA-MB-231 and Hs587T cells solely express mutant p53, KDM3A knockdown unexpectedly revealed that increasing p53-K372me1 could reactivate mutant p53 to induce the expression of pro-apoptotic genes. Interestingly, KDM3A knockdown did not dramatically affect p21 expression in MDA-MB-231 and Hs587T cells which is associated with cell cycle arrest. Since KDM3A knockdown robustly increased the expression of PUMA in three different breast cancer cell lines and p53 binding sites on the PUMA promoter are well characterized, we first examined how KDM3A modulated p53 binding to the PUMA promoter by demethylating p53. We hypothesized that KDM3A knockdown would enhance the p53 occupancy on the PUMA promoter, thereby stimulating PUMA transcription. ChIP assays revealed that p53 occupancies on the PUMA promoter were significantly increased in MDA/KDM3Ash1 and MDA/KDM3Ash2 cells as compared to MDA/Scrsh cells (Figure 5a). To further check whether the increased level of p53 on the PUMA promoter was due to the stabilization of chromatin associated p53-K372me1, we also performed ChIP assays using anti-p53-K372me1 antibodies. Indeed, KDM3A knockdown led to increased binding of p53-K372me1 on the PUMA promoter (Figures 5b). Similarly, we found that KDM3A knockdown enhanced p53-K372me1 binding to the NOXA promoter (Figures 5c and d). To further confirm these observations we also determined the extent of p53 and p53-K372me1 occupancy on the PUMA promoter in Hs578T and MCF-7 cells. Consistently, KDM3A depletion promoted the enrichment of p53 as well as p53K372me1 on the PUMA promoter in Hs578T and MCF-7 cells (Figures 5e, f, g and h). As a control, we used normal IgG enriched chromatin and the negative primer designed to amplify 4 kb downstream of p53 binding sites on the PUMA and NOXA promoters. Finally, to confirm that the effects of KDM3A depletion on p53 target genes are mediated through p53, we knocked down p53 in MDA/KDM3Ash2 cells and examined the expression of p53 target genes. The knockdown of p53 in MDA/KDM3Ash2 cells abolished the induction of apoptotic gene expression caused by KDM3A depletion (Supplementary Figure 3a). Consistently, the knockdown of p53 also rescued the paclitaxel-induced apoptosis in MDA/KDM3Ash2 cells (Supplementary Figure 3b).
Figure 5. KDM3A knockdown increases the occupancies of p53 and p53-K372me1 on the promoter of PUMA and NOXA.
(a and b) ChIP assays of p53 and p53K372me1 occupancy on the PUMA promoter in MDA-MB-231 cells using anti-p53 and anti-p53-K372me1.
(c and d) ChIP assays of p53 and p53-K372me1 occupancy on the NOXA promoter in MDA-MB-231 cells using anti-p53 and anti-p53-K372me1.
(e and f) ChIP assays of p53 and p53-K372me1 occupancy on the PUMA promoter in Hs578T cells using anti-p53 and anti-p53-K372me1.
(g and h) ChIP assays of p53 and p53K372me1 occupancy on the PUMA promoter in MCF-7 cells using anti-p53 and anti-p53-K372me1.
Data are means ± SD of triplicate assays from a representative experiment. *p < 0.05, **p < 0.01, unpaired two-tailed Student’s t test.
KDM3A is required for oncogenic potentials of breast CSCs
Because KDM3A promotes breast cancer invasion and survival and is highly expressed in human invasive breast cancer tissues, we examined whether KDM3A knockdown affected tumor growth in vivo. Both MDA/Scrsh and MDA/KDM3Ash2 cells were subcutaneously inoculated into the flank of athymic nude mice. KDM3A knockdown significantly inhibited tumor growth of MDA-MB-231 cells in vivo as determined by the periodic measurement of tumor volume (Figure 6a). We further explored whether KDM3A was necessary for in vivo metastatic colonization of breast cancer cells by utilizing the mouse model of tail vein injection. KDM3A knockdown also significantly inhibited lung metastasis of MDA-MB-231 cells in nude mice (Figure 6b).
Figure 6. KDM3A is required for breast cancer stem cell (CSCs) self-renewal and tumor growth.
(a) Photograph and tumor growth curve of subcutaneously injected MDA/Scrsh or MDA/KDM3Ash2 cells in nude mice. Data are means ± SD of tumor volume measured from five mice. **p < 0.01, unpaired two-tailed Student’s t test.
(b) Representative photograph and number of lung colonization formed by tail vein injection of MDA/Scrsh or MDA/KDM3Ash2 cells in nude mice. *p < 0.05, unpaired two-tailed Student’s t test.
(c) Flow cytometry analysis of the abundance of CSCs in MDA/Scrsh and MDA/KDM3Ash2 cells.
(d) Real-time RT-PCR of indicated gene expression in CSCs. CSCs/Scrsh, CSCs from MDA-MB-231 cells expressing Scrsh; CSCs/KDM3Ash2, CSCs from MDA-MB-231 cells expressing KDM3Ash2. Data are mean ± SD of triplicate samples from a representative experiment.
(e) KDM3A knockdown enhanced Paclitaxel-induced cell death in CSCs. Data are means ± SD of triplicate samples from a representative experiment.
(f and g) Image (f) and quantification (g) of CSCs/Scrsh or CSCs/KDM3Ash2 cell invasion. Invasive cells were counted in 5 random fields. Data are means ± SD of triplicate samples from a representative experiment.
(h and i) Representative image (h) and quantification (i) of tumorsphere formation by CSCs/Scrsh and CSCs/KDM3Ash2 cells. Data are means ± SD of triplicate samples from a representative experiment.
(j) Tumor growth curve of subcutaneously injected CSCs/Scrsh or CSCs/KDM3Ash2 cells in nude mice. Data are mean ± SD, n = 5.
*p < 0.05, **p < 0.01, unpaired two-tailed Student’s t test.
Invasive growth, metastasis and development of chemoresistance are intimately wired to CSCs as these processes rely on the CSCs populations.24,25 The CSCs are proposed to drive tumor initiation and progression in vivo as reported in human cancer xenograft mouse models.26 Moreover, while wild-type p53 negatively regulates CSCs,27 mutant p53 promotes self-renewal of CSCs.12 Since KDM3A knockdown inhibited breast cancer tumor growth and metastasis, we further asked whether KDM3A could regulate breast CSC activities. Previously, it has been shown that MDA-MB-231 cells contained functional CSCs or cancer stem-like cells.28 First we assessed the relative abundance of CSCs between MDA/Scrsh and MDA/KDM3Ash2 cells based on aldehyde dehydrogenase (ALDH) activity, as assessed by fluorescent ALDEFLUOR assays. Strikingly, the percentage of CSCs was significantly lower in MDA/KDM3Ash2 cells than in MDA/Scrsh cells (Figure 6c), indicating the requirement of KDM3A for the maintenance of CSCs. Since CSCs constitute only a small fraction of the entire cancer cell population, based on these results it is difficult to conclude that KDM3A played a decisive role in determining the CSCs-driven tumor progression in breast cancer. Therefore, we addressed this issue by a more straightforward manner by utilizing the enriched fraction of CSCs. We sorted CSCs from MDA-MB-231 cells by Flow cytometry based on ALDH activity. The enriched cell preparations were allowed to recover in standard culture conditions for at least 12 hr before being used for further experiments. Subsequently, we knocked down KDM3A in CSCs as confirmed by Real-time RT-PCR (Figure 6d). Next we examined whether the KDM3A depletion in CSCs recapitulated the effect observed in parental MDA-MB-231 cells. Indeed, similar to parental MDA-MB-231 cells, KDM3A knockdown in CSCs also inhibited the expression of MMP-9, S100A4 and JUN, while PUMA expression was upregulated (Figure 6d). Consistently, KDM3A knockdown in CSCs also enhanced the paclitaxel-induced apoptosis and inhibited cell invasion (Figure 6e, f and g). Similar to the embryonic stem cells, CSCs also rely on the self-renewal mechanism to replenish the stem cell populations in cancer.29 In general, self-renewal capacity of CSCs can be assessed by their ability to propagate and form a sphere in the suspension culture. Since we noted significant reduction in the percentage of CSCs in KDM3A depleted MDA-MB-231 cells, it was possible that KDM3A might be required for the self-renewal of CSCs to maintain the CSCs pool in breast cancer. To test this possibility, we examined whether KDM3A knockdown impaired self-renewal of CSCs by assessing their capacity to form and propagate tumorspheres in vitro. KDM3A knockdown significantly inhibited CSCs-mediated tumorsphere formation (Figures 6h and i). Consistently, KDM3A knockdown in CSCs also inhibited tumor growth in nude mice (Figure 6j), suggesting that KDM3A mediates tumorigenic potentials of CSCs.
DISCUSSION
The findings presented here revealed the oncogenic functions of KDM3A in promoting and integrating the crucial events of breast cancer cell invasion, drug resistance and stemness. Intriguingly, we demonstrated that KDM3A employs two distinct mechanisms to promote the breast cancer cells to acquire invasive and chemo-resistance phenotype along with the stem cell properties. On one hand, KDM3A activates the expression of pro-invasive genes by histone demethylation and on the other hand it indirectly represses the expression of pro-apoptotic genes by demethylating the non-histone protein p53. Unexpectedly, we found that inhibition of KDM3A was able to reactivate mutant p53 to induce the expression of pro-apoptotic genes, thereby restoring apoptotic sensitivity to chemotherapeutic drugs.
KDM3A controls numerous biological and pathological processes, including sexual differentiation, germ cell development, and obesity resistance and cancer progression by epigenetically controlling gene expression.30–33 All these studies consistently showed that KDM3A activates gene transcription through the lysine-specific demethylation of chromatin. Indeed, our results also demonstrated that KDM3A directly binds to the promoter of the pro-invasive genes such as MMP-9, S100A4 and JUN and activate their transcription by erasing H3K9me2. Although the role of MMP-9, S100A4 and JUN in breast cancer invasion and metastasis is well established,34–36 our study for the first time, provides evidence on how these genes are epigenetically controlled in breast cancer. While S100A4 expression in MDA-MB-231 cells is epigenetically regulated by KDM3A, no such regulation was found in Hs578T cells. Since the origin and genetic background of these two cell lines are different, it is possible that the epigenetic mechanisms regulating S100A4 expression might be governed by distinct histone demethylases. Previously, KDM3A was found to regulate ER-dependent cell proliferation in ER positive breast cancer cells.37 Our study uncovered ER-independent functions for KDM3A in promoting the invasive growth and therapeutic resistance of triple negative breast cancer cells (MDA-MB-231 and Hs578T). Taken together, KDM3A could serve as a common therapeutic target for hormone dependent as well as independent breast cancers.
Apart from the epigenetic control of breast cancer cell invasion by KDM3A, unexpectedly, we found that KDM3A demethylates a non-histone protein p53 and inhibits its transcriptional activity, inducing chemoresistance in breast cancer cells. The tumor suppressor p53 acts as “guardian of the genome” by preventing the cellular transformations resulting from the oncogenic and genotoxic stress.38,39 Besides the important role in cancer, p53 also controls numerous biological and pathological processes including fertility, stem cells pluripotency and dedifferentiation, neurodegenerative disorders, diabetes and myocardial infarction. Therefore, our study elucidating the KDM3A mediated regulation of p53 function may also have implications in other biological and pathological process that are governed by p53. C-terminal lysine residues of p53 are methylated in vivo at K370, K372, K373, and K382 by histone lysine methyl transferases, Smyd2, SET9, G9a/Glp, and Set8, respectively.20–23 While K372 methylation by SET9 in human cells enhanced p53 transcriptional activities, the role of p53 methylation in the mouse by Set7/9 was controversial.40–42 However, it should be pointed out that the results from mouse embryonic fibroblasts in the mouse study might be inappropriate to apply to human epithelial cells. Importantly, our results from a new and different aspect further confirm that K372 methylation enhances p53 transcriptional activities.
Nearly 50% of all human cancers have p53 mutations,43 and accumulating evidences suggest that mutant p53 protein not only loses tumor-suppressive functions, but also acquires oncogenic functions.12 While MCF-7 cells express a wild-type p53, mutant p53 are expressed in MDA-MB-231 cells (p53-R280K) and Hs578T cells (p53-V157F). It is well known that mutant p53 mediates chemoresistance in breast cancer cells and other tumor cells. While it is not unexpected that KDM3A knockdown activates p53-mediated induction of pro-apoptotic genes in MCF-7 cells, surprisingly, KDM3A knockdown also promotes the expression of p53-dependent pro-apoptotic genes in MDA-MB-231 and Hs578T cells. Our results suggest that the tumor suppressor function of mutant p53 can be reactivated by K372 methylation. Previously, acetylation of the mutant p53 has been found to restore the DNA binding ability and the growth suppression function.44 Since p53 methylation at K372 promotes its subsequent acetylation,40,45 it is possible that KDM3A knockdown might reactivate mutant p53 by increasing p53 acetylation. Growing evidence suggests that breast CSCs are chemoresistant and highly invasive which might be associated with disease relapse.11 Since p53 is frequently mutated in human breast cancer, mutant p53 might promote the development of CSCs and help to maintain tumorigenic potentials of CSCs. Interestingly, mutant p53 was found to promote the reprogramming of somatic cells. Breast cancer cells with mutant p53 expressed stem-cell-like transcriptional signature.13 Our results showed that reactivation of p53 tumor suppressor activities by KDM3A knockdown in breast CSCs promoted apoptosis and inhibited tumorsphere formation, suggesting that mutant p53 might promote tumorigenic potentials and self-renewal of CSCs. Targeting mutant p53 might not only shrink the bulk of the tumor, but also eliminate CSCs, thereby preventing tumor relapse.
In general, aberrant activation of intracellular signaling alters the transcriptional machineries and reprogram the gene expression pattern in cancer cells. The development of small molecule inhibitors targeting the transcriptional machinery is very challenging. However, the discovery of epigenetic modifying enzymes that contain catalytic domains opens up new avenues to develop small molecule inhibitors to epigenetically reverse the transcriptional state of cancer cells. Since KDM3A plays a crucial role in altering the gene expressing pattern through epigenetic and non-epigenetic mechanisms in breast cancer, targeting KDM3A might offer new strategies for treating breast cancer.
MATERIALS AND METHODS
Cell culture and viral transduction
MDA-MB-231, MCF-7 and Hs578T cells were purchased from ATCC and were cultured in DMEM containing 10% heat-inactivated fetal bovine serum (FBS) and antibiotics (streptomycin and penicillin) at 37 °C in a 5% CO2, 95% air atmosphere. Cells were tested without mycoplasma contamination. To stably deplete KDM3A, lentiviruses expressing human KDM3A shRNAs were packaged and generated in 293T cells as described previously.7,46 Breast cancer cells were seeded in the 6-well plates overnight and then infected with lentiviral particles. 24 h after infection, the cells were selected with puromycin (1 µg/ml) for at least 1 week. The sequences of shRNAs targeting KDM3A were: sh1 5’-TAAATGCTTCACAATCAAAGC-3’; sh2 5’-ATAAGCATTATACATCTTGGG-3’; and sh3 5’-GCATAGGACTGGCATTATA-3’. For the rescue experiments, the retroviral vector expressing KDM3A, generously provided by Dr. Yi Zhang (Department of Genetics, Harvard Medical School, Boston, USA), was packaged in 293T cells as described previously.7 Cell death was quantified using Vi-cell XR cell viability analyzer (Beckman Coulter, Miami, USA).
siRNA transfection and invasion screening
siRNA pools targeting histone demethylases were purchased from Santa Cruz Biotechnology. MDA-MB-231 cells were transfected with siRNAs using Lipofectamine RNAiMax (Invitrogen). 48 hr post transfection, the knockdown efficiency was determined by Real-time RT-PCR. Primers used for the human histone demethylases were described previously.4 To screen tumor cell invasion, 1×105 siRNA-transfected cells were seeded in triplicate on BD BioCoat Matrigel Invasion Chambers. After 24 hours, matrigel was removed, and the invaded cells were stained with the HEMA 3 kit (Fisher). The invaded cells were counted from at least 5 random fields and averaged.
RNA extraction, human affymetrix microarray and real-time RT-PCR
Total RNA was extracted with miRNeasy kit according to the manufacturer’s instruction (Qiagen). Five µg of total RNA were transcribed from each sample into double-stranded cDNA using SuperScript II RT (Invitrogen) with an oligo(dT) primer and then used to synthesize single-stranded RNAs. The biotin-labeled RNAs were fragmented and hybridized with an Affymetrix Human Genome U133 Plus 2.0 Array at the Clinical Microarray Facility of University of California, Los Angeles (UCLA). The arrays were scanned with a GeneArray scanner (Affymetrix). The robust multichip average (RMA) method was used to normalize the raw data. For Real-time RT-PCR, total RNA was isolated from cells using TRIzol reagent, and complementary DNA (cDNA) was synthesized with oligo(dT) primers using SuperScript III (Invitrogen). Real-time RT-PCR analysis was carried out with iQ SYBR Green Supermix (Bio-Rad) on an iCycler iQ real-time PCR detection system (Bio-Rad). Primers used for quantitative RT-PCR were listed in the supplementary methods.
Immunoprecipitation, Western blot analysis and ChIP assays
Cells grown in 10 cm dishes were lysed with 500 µl of IP lysis buffer (1% NP40) for 30 min on ice. After centrifugation at 10,600 × g at 4°C, the supernatants were incubated with antibodies at 4°C overnight, followed by incubation with protein G-Sepharose (GE Healthcare) for 1 h. Immunoprecipitates were washed three times with lysis buffer at 4°C. Proteins bound to the beads were eluted with SDS loading buffer at 98°C for 2 min and then subjected to SDS-PAGE. For Western blotting, cells were lysed in RIPA (radioimmuno precipitation assay) buffer and 25 µg of total proteins were resolved on SDS-PAGE. The gel was transferred onto PVDF membrane for 50 min at 15 V using the Bio-Rad semidry transfer system. The blot transferred membranes were blocked for 2 hours in 5% milk and incubated with primary antibodies overnight. The immunecomplexes were detected with horseradish peroxidase (HRP)–conjugated anti-rabbit or anti-mouse immunoglobulin G (IgG) (Promega) and envisioned with SuperSignal reagents (Pierce) as previously described.7 Source of primary antibodies and the dilutions used for the Western blotting were indicated in supplementary methods.
ChIP assays were performed using a ChIP assay kit following the manufacturer’s Protocol (Upstate Biotechnology). Cells (2×106) were preincubated with a dimethyl 3,3′-dithiobispropionimidate-HCl (Pierce) solution (5 mmol) for 30 min on ice and then treated with formaldehyde. The ChIP-enriched DNA samples were quantified by Real-time PCR, and the data are expressed as a percentage of input. The primer pairs used for ChIP assays were described in the supplementary methods.
ALDH activity assays and isolation of BCSCs, in vitro tumorsphere formation assay
The ALDEFLUOR kit (Stem Cell Technologies) was used to identify and isolate MDA-MB-231 cells with high Aldehyde Dehydrogenase 1 (ALDH) activity. Briefly, trypsinized cells were rinsed with PBS and re-suspended in ADEFLUOR® assay buffer (2×105/ml) and incubated with ADEFLUOR substrate with or without ALDH inhibitor, diethylaminobenzaldehyde (DEAB) for 30 minutes at 37°C. Fluorescence-activated cell sorting (FACS) was adopted to analyze and sort the ALDH high cell (CSCs) population. After overnight culture, ALDH-positive MDA-MB-231 cells were infected with Scrsh or KDM3Ash2 lentiviruses and selected with puromycin (1 µg/ml) for 48 hrs. Afterwards, cells were trypsinized and seeded (2000 cells / well) into ultra-low attachment 6-well culture plates containing 3 ml of sphere forming media ((DMEM/F12 50:50 containing 1% supplement B, 10 ng/ml EGF and 10 ng/ml FGF) and cultured for two weeks. The tumor sphere measuring > 70 µm in diameter were counted using the inverted microscope.
In vivo tumor growth and lung metastatic colonization assays
All animal experiments were performed in accordance with a protocol approved by the UCLA Committee on Animal Care. For tumor xenograft studies, MDA/Scrsh or MDA/KDM3Ash2 cells (1×106) were injected subcutaneously into the flanks of 6 to 8 weeks-old nude female mice. To evaluate the impact of KDM3A knockdown on in vivo growth of CSCs, CSCs/Scrsh or CSCs/KDM3Ash2 cells (5,000 cells) were suspended in Matrigel (BD bioscience, San Diego, CA, USA) and injected subcutaneously into nude mice. Mice were randomly injected with tumor cells. Sample size was determined based on our past experiences, and no statistical method was used. The investigator was not blinded to the group allocation during the experiment. Once the palpable tumor was noticed, tumor growth was assessed periodically and tumor volume (mm3) was calculated using the formula 0.5 × length × width2. Animals were euthanized when the tumors reached approximately 1.0 cm in diameter. To analyze the metastatic colonization in lungs, MDA-MB-231 cells (1×106) were injected into the tail vein of mice (five mice per group). After eight weeks, the mice were euthanized, the lungs were resected, and nodules were counted.
Immunohistochemical staining of human breast cancer tissue arrays
Human tissue array (Cat No. BR1008) containing primary and lymph node metastastic breast cancer along with normal breast tissues was purchased from US Biomax, USA. The tissue array slides were deparaffinized with xylene twice and then rehydrated with distilled water through an ethanol series step by step. Tissue antigens were retrieved as described previously.47 The slides were stained with polyclonal antibodies against KDM3A (1:100; Bethyl), JUN (1:200; Cell Signaling Technology), and MMP-9 (1:100; Santa Cruz Biotechnology) or control IgG (Santa Cruz Biotechnology) at 4°C overnight. We then incubated the sections with HRP-labeled polymer for 60 min, detected the immunocomplexes with AEC+ chromogen (Dako EnVision System), and counterstained with hematoxylin QS. The intensity of immunostaining was scored as follows: 0, no staining; +, weak staining; ++, moderate staining; +++, strong staining. The Wilcoxon rank sum test was applied to test the significant differences in immunohistochemical staining intensity between different groups. The Pearson correlation coefficient of linear regression was used to determine the correlation between different proteins. All statistical analyses were performed with SPSS 17.0 software.
Supplementary Material
Acknowledgments
This work was supported by NIH grants R3713848 and DE15964, and the Shapiro Family Charitable Foundation. The authors thank Dr. Yi Zhang for the generous gift of KDM3A plasmid.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interests.
AUTHOR CONTRIBUTIONS
S. R. and C.-Y.W. conceived ideas. S. R., C.-Y. W. and G.G. designed experiments, S. R. and G.G. performed experiments. S. R. and C.-Y.W. wrote the manuscript.
REFERENCES
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Acharyya S, Oskarsson T, Vanharanta S, Malladi S, Kim J, Morris PG, et al. A CXCL1 paracrine network links chancer chemoresistance and metastasis. Cell. 2012;150:165–178. doi: 10.1016/j.cell.2012.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Comoglio PM, Trusolino L. Invasive growth: from development to metastasis. J Clin Invest. 2002;109:857–862. doi: 10.1172/JCI15392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding X, Pan H, Li J, Zhong Q, Chen X, Dry SM, et al. Epigenetic activation of AP1 promotes squamous cell carcinoma metastasis. Sci Signal. 2013;6:ra28.1–ra28.13. doi: 10.1126/scisignal.2003884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwon M. Epithelial-to-mesenchymal transition and cancer stem cells: emerging targets for novel cancer therapy. Cancer Gene Ther. 2014;21:179–180. doi: 10.1038/cgt.2014.17. [DOI] [PubMed] [Google Scholar]
- 6.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Ramadoss S, Chen X, Wang CY. Histone demethylase KDM6B promotes epithelial-mesenchymal transition. J Biol Chem. 2012;287:44508–44517. doi: 10.1074/jbc.M112.424903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scully OJ, Bay BH, Yip G, Yu Y. Breast cancer metastasis. Cancer Genomics Proteomics. 2012;9:311–320. [PubMed] [Google Scholar]
- 9.Alexander S, Friedl P. Cancer invasion and resistance: interconnected processes of disease progression and therapy failure. Trends Mol Med. 2012;18:13–26. doi: 10.1016/j.molmed.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 10.Muller PA, Vousden KH, Norman JC. p53 and its mutants in tumor cell migration and invasion. J Cell Biol. 2011;192:209–218. doi: 10.1083/jcb.201009059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shetzer Y, Solomon H, Koifman G, Molchadsky A, Horesh S, Rotter V. The paradigm of mutant p53-expressing cancer stem cells and drug resistance. Carcinogenesis. 2014;35:1196–1208. doi: 10.1093/carcin/bgu073. [DOI] [PubMed] [Google Scholar]
- 12.Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol. 2013;15:2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- 13.Mizuno H, Spike BT, Wahl GM, Levine AJ. Inactivation of p53 in breast cancers correlates with stem cell transcriptional signatures. Proc Natl Acad Sci U S A. 2010;107:22745–22750. doi: 10.1073/pnas.1017001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ginestier C, Charafe-Jauffret E, Birnbaum D. p53 and cancer stem cells: the mevalonate connexion. Cell Cycle. 2012;11:2583–2584. doi: 10.4161/cc.21092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freed-Pastor WA, Mizuno H, Zhao X, Langerød A, Moon SH, Rodriguez-Barrueco R, et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell. 2012;148:244–258. doi: 10.1016/j.cell.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 18.Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- 19.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53 mediated tumor suppression. Nat Rev Cancer. 2014;14:359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, et al. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444:629–632. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- 21.Huang J, Dorsey J, Chuikov S, Pérez-Burgos L, Zhang X, Jenuwein T, et al. G9a and Glp methylate lysine 373 in the tumor suppressor p53. J Biol Chem. 2010;285:9636–9641. doi: 10.1074/jbc.M109.062588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi X, Kachirskaia I, Yamaguchi H, West LE, Wen H, Wang EW, et al. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell. 2007;27:636–646. doi: 10.1016/j.molcel.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, et al. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 24.Malanchi I, Santamaria-Martínez A, Susanto E, Peng H, Lehr HA, Delaloye JF, et al. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature. 2011;481:85–89. doi: 10.1038/nature10694. [DOI] [PubMed] [Google Scholar]
- 25.Baccelli I, Schneeweiss A, Riethdorf S, Stenzinger A, Schillert A, Vogel V, et al. Identification of population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograftassay. Nat Biotechnol. 2013;31:539–544. doi: 10.1038/nbt.2576. [DOI] [PubMed] [Google Scholar]
- 26.Cheng L, Ramesh AV, Flesken-Nikitin A, Choi J, Nikitin AY. Mouse models for cancer stem cell research. Toxicol Pathol. 2010;38:62–71. doi: 10.1177/0192623309354109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang CJ, Chao CH, Xia W, Yang JY, Xiong Y, Li CW, et al. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat Cell Biol. 2011;13:317–323. doi: 10.1038/ncb2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C, et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147:759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 29.O’Brien CA, Kreso A, Jamieson CH. Cancer stem cells and self-renewal. Clin Cancer Res. 2010;16:3113–3120. doi: 10.1158/1078-0432.CCR-09-2824. [DOI] [PubMed] [Google Scholar]
- 30.Okada Y, Scott G, Ray MK, Mishina Y, Zhang Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature. 2007;450:119–123. doi: 10.1038/nature06236. [DOI] [PubMed] [Google Scholar]
- 31.Tateishi K, Okada Y, Kallin EM, Zhang Y. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature. 2009;458:757–761. doi: 10.1038/nature07777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krieg AJ, Rankin EB, Chan D, Razorenova O, Fernandez S, Giaccia AJ. Regulation of the histone demethylase JMJD1A by hypoxia-inducible factor 1 alpha enhances hypoxic gene expression and tumor growth. Mol Cell Biol. 2010;30:344–353. doi: 10.1128/MCB.00444-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuroki S, Matoba S, Akiyoshi M, Matsumura Y, Miyachi H, Mise N, et al. Epigenetic regulation of mouse sex determination by the histone demethylase Jmjd1a. Science. 2013;341:1106–1109. doi: 10.1126/science.1239864. [DOI] [PubMed] [Google Scholar]
- 34.Boye K, Maelandsmo GM. S100A4 and metastasis: a small actor playing many roles. Am J Pathol. 2010;176:528–535. doi: 10.2353/ajpath.2010.090526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiao X, Katiyar S, Willmarth NE, Liu M, Ma X, Flomenberg N, et al. c-Jun induces mammary epithelial cellular invasion and breast cancer stem cell expansion. J Biol Chem. 2010;285:8218–8226. doi: 10.1074/jbc.M110.100792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vandooren J, Van den Steen PE, Opdenakker G. Biochemistry and molecular biology of gelatinase B or matrix metalloproteinase-9 (MMP-9): the next decade. Crit Rev Biochem Mol Biol. 2013;48:222–272. doi: 10.3109/10409238.2013.770819. [DOI] [PubMed] [Google Scholar]
- 37.Wade MA, Jones D, Wilson L, Stockley J, Coffey K, Robson CN, et al. The histone demethylase enzyme KDM3A is a key estrogen receptor regulator in breast cancer. Nucleic Acids Res. 2014;43:196–207. doi: 10.1093/nar/gku1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 39.Lane D, Levine A. p53 Research: the past thirty years and the next thirty years. Cold Spring Harb Perspect Biol. 2010;2:a000893. doi: 10.1101/cshperspect.a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurash JK, Lei H, Shen Q, Marston WL, Granda BW, Fan H, et al. Methylation of p53 by Set7/9 mediates p53 acetylation and activity in vivo. Mol Cell. 2008;29:392–400. doi: 10.1016/j.molcel.2007.12.025. [DOI] [PubMed] [Google Scholar]
- 41.Campaner S, Spreafico F, Burgold T, Doni M, Rosato U, Amati B, et al. The methyltransferase Set7/9 (Setd7) is dispensable for the p53-mediated DNA damage response in vivo. Mol Cell. 2011;43:681–688. doi: 10.1016/j.molcel.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 42.Lehnertz B, Rogalski JC, Schulze FM, Yi L, Lin S, Kast J, et al. p53-dependent transcription and tumor suppression are not affected in Set7/9-deficient mice. Mol Cell. 2011;43:673–680. doi: 10.1016/j.molcel.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 43.Joerger AC, Fersht AR. Structural biology of the tumor suppressor p53 and cancer associated mutants. Adv Cancer Res. 2007;97:1–23. doi: 10.1016/S0065-230X(06)97001-8. [DOI] [PubMed] [Google Scholar]
- 44.Perez RE, Knights CD, Sahu G, Catania J, Kolukula VK, Stoler D, et al. Restoration of DNA-binding and growth suppressive activity of mutant forms of p53 via a PCAF-mediated acetylation pathway. J Cell Physiol. 2010;225:394–405. doi: 10.1002/jcp.22285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ivanov GS, Ivanova T, Kurash J, Ivanov A, Chuikov S, Gizatullin F, et al. Methylation-acetylation interplay activates p53 in response to DNA damage. Mol Cell Biol. 2007;27:6756–6769. doi: 10.1128/MCB.00460-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramadoss S, Li J, Ding X, Al Hezaimi K, Wang CY. Transducin β-like protein 1 recruits nuclear factor κB to the target gene promoter for transcriptional activation. Mol Cell Biol. 2011;31:924–934. doi: 10.1128/MCB.00576-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeng Q, Li S, Chepeha DB, Giordano TJ, Li J, Zhang H, et al. Crosstalk between tumor and endothelial cells promotes tumor angiogenesis by MAPK activation of Notch signaling. Cancer Cell. 2005;8:13–23. doi: 10.1016/j.ccr.2005.06.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.