ARABIDOPSIS: A MODEL PLANT FOR STUDYING PLANT BIOLOGY
After almost a century since its first appearance in the scientific literature, Arabidopsis has now been widely adopted as a model plant of choice for biological research (Somerville and Koornneef, 2002). Its many advantages include a small genome, short life cycle, small stature, prolific seed production, and ease of transformation. In addition, a wealth of genomics resources exists, such as a completely sequenced genome, a near saturation insertion mutant collection, a genome array that contains the entire transcriptome, and more than 50,000 molecular markers. Add in a vibrant collaborative community, and it is easy to see why Arabidopsis is often the system of choice for plant research. While the culminating achievement of Arabidopsis research—the release of its complete genome sequence—is still fresh in memory, a new 10-year project of comparable importance has already begun. The ambitious goal of this program is to know the function of all Arabidopsis genes by 2010 (Somerville and Dangl, 2000). This knowledge will facilitate the development of a virtual plant—a computer model that will use information about each gene product to simulate the growth and development of a plant under many environmental conditions. However, it has not escaped many plant biologists that another goal is to use knowledge gained from research on Arabidopsis to facilitate the understanding of biological phenomena in other plant species. One specific challenge is how to utilize information gained from Arabidopsis research to produce new commercial plant varieties. In this perspective, we reflect on the critical role Arabidopsis is playing in unraveling abiotic stress signal transduction (focusing primarily on salt, cold, and drought stress), and how these new insights on the mechanisms of tolerance to these stresses are suggesting novel approaches to engineer the next generation of biotech crops.
ARABIDOPSIS AND BEYOND: FROM CONCEPTS TO ENGINEERING STRESS TOLERANCE IN CROP PLANTS
Salt Stress in Arabidopsis
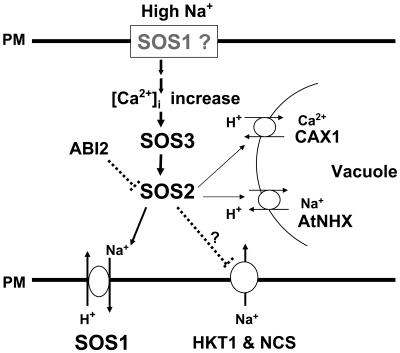
The potential of manipulating ion transporters to improve ion homeostasis is well recognized. For example, several classes of transporters are required in regulating sodium homeostasis under salt stress (Fig. 1). The influx of Na+ is controlled by AtHKT1, a low affinity Na+ transporter (Schachtman and Schroeder, 1994; Rus et al., 2001), and other nonselective cation channels, while the efflux is controlled by Salt Overly Sensitive1 (SOS1), a plasma membrane Na+/H+ antiporter (Shi et al., 2000). Vacuolar membrane ion transporters, such as the tonoplast Na+/H+ antiporter AtNHX1(Apse et al., 1999; Gaxiola et al., 1999), also play a vital role in regulating cytoplasm Na+ homeostasis by sequestering Na+ ions in the vacuole. The sensing of Na+ in plant cells is still unknown but is speculated to involve SOS1 (Fig. 1). It has been suggested that the long C-terminal tail predicted to reside in the cytoplasm might be a sensor of cytoplasm Na+ (Shi et al., 2000; Zhu, 2003). Similarly, evidence suggests that the C terminus of AtNHX1, present in the vacuolar lumen, may also have a regulatory role (Yamaguchi et al., 2003). It is speculated that Na+ sensors regulate the cytoplasmic Ca2+ level, which triggers the SOS signal transduction chain: SOS3, a myristoylated calcium binding protein, interacts with and activates the SOS2 kinase, which phosphorylates and activates SOS1, the plasma membrane Na+/H+ antiporter (Zhu, 2002). There is also evidence that the SOS2 kinase positively regulates the activities of AtNHX1 and CAX1 (a vacuolar Ca2+/ H+ exchanger) and may negatively regulate AtHKT1 (Cheng et al., 2004; Qiu et al., 2004). Therefore, coordination exists between the transporters in the tonoplast and plasma membranes in part through the SOS2 kinase (Cheng et al., 2004; Qiu et al., 2004).
Figure 1.
The SOS signaling pathway for the regulation of Na+ homeostasis and salt tolerance in Arabidopsis. High Na+ stress triggers a calcium signal that activates the SOS3-SOS2 protein kinase complex, which then stimulates the Na+/H+ exchange activity of SOS1 at the plasma membrane. SOS2 also activates Na+/H+ (AtNHX) and Ca2+/H+ (CAX1) exchangers on the vacuolar membrane. The protein phosphatase ABI2 has been shown to physically interact with SOS2 and is proposed to inactivate SOS2. The SOS pathway may down-regulate the activity of Na+ influx transporters (AtHKT1 and NCS). SOS1 in gray indicates that this transporter may also have a sensory role. Dotted lines indicate possible regulation.
Readers are referred to a recent publication for an extensive review of the regulation of ion homeostasis under salt stress (Zhu, 2003). It is noteworthy that Arabidopsis has played a vital role in many investigations of the basic processes that occur during salt stress. For example, SOS1/2/3 were identified by Arabidopsis mutants and position-based cloning (Liu and Zhu, 1998; Liu et al., 2000; Shi et al., 2000); AtNHX1 was discovered from Arabidopsis genome sequences (Apse et al., 1999; Gaxiola et al., 1999); and HKT1, initially identified in wheat (Triticum aestivum; Schachtman and Schroeder, 1994), has been shown through the study of Arabidopsis knockout mutants to be an important Na+ influx system in plant roots (Rus et al., 2001) and to play a critical role in controlling the long distance transport of Na+ (Maser et al., 2002; Berthomieu et al., 2003). These and other examples illustrated how mutant isolation, position-based cloning, and gene expression analysis in Arabidopsis can be combined with biochemical and physiological studies to unravel the inner workings of salt stress signal transduction (Zhu, 2002).
Translation of Arabidopsis Models to Engineer Salt Tolerance in Crop Plants
It is generally accepted that maintaining a low cytosolic Na+ concentration is essential to achieve salt tolerance and can be achieved by restricting inflow, increasing outflow, or increasing vacuole sequestration of Na+. Intuitively, increasing plasma membrane Na+ exporters and tonoplast Na+ importers and/or restricting the amount of Na+ influx by lowering the amount of plasma membrane Na+ importers should suffice. Indeed, a number of successes have ensued when these strategies were used (see Table I). For example, increased expression of the Arabidopsis tonoplast membrane Na+/ H+ antiporter, AtNHX1, under a strong constitutive promoter was reported to result in salt-tolerant Arabidopsis (Apse et al., 1999), Brassica napus (Zhang et al., 2001), and tomato (Lycopersicon esculentum; Zhang and Blumwald, 2001). AgNHX1, a AtNHX1 ortholog from the halophytic plant Atriplex gmelini (Hamada et al., 2001), when overexpressed in rice (Oryza sativa) plants, improved salt tolerance of the transgenic rice (Ohta et al., 2002). AtNHX1 orthologs from many plant species have been isolated, mostly based on their sequence homology to the Arabidopsis gene. Examples include NHX1 genes from rice (Fukuda et al., 1999), wheat (Chen et al., 2002), Japanese morning glory (Yamaguchi et al., 2001), A. gmelini (Hamada et al., 2001), ice plant (Chauhan et al., 2000), and sugar beet (Xia et al., 2002). Thus, the NHX1 system seems to be highly conserved between many different plant species, and manipulation of this system in crop species will likely result in improved salt tolerance. In addition to NHX1, other transporters have also been used successfully. Overexpression of the plasma membrane Na+ / H+ antiporter SOS1 in Arabidopsis (Shi et al., 2003), increased expression of vacuolar H+-ATPase AVP1 in Arabidopsis (Gaxiola et al., 2001), and antisense of the wheat high affinity K+ transporter HKT1 in wheat (Laurie et al., 2002) also have resulted in improved salt tolerance (Table I).
Table I.
Engineered abiotic stress tolerance: from ion transporters to transcription factors
| Gene Name | Gene Source | Transgenic Species | Intervention Method | Traits | Reference |
|---|---|---|---|---|---|
| Ion Transporters | |||||
| AtNHX1 (vacuolar Na+/H+ antiporter) | Arabidopsis | Arabidopsis | Overexpression | Salt tolerance | Apse et al. (1999) |
| AtNHX1 (vacuolar Na+/H+ antiporter) | Arabidopsis | B. napus | Overexpression | Salt tolerance | Zhang et al. (2001) |
| AtNHX1 (vacuolar Na+/H+ antiporter) | Arabidopsis | Tomato | Overexpression | Salt tolerance | Zhang and Blumwald (2001) |
| AtNHX1 (vacuolar Na+/H+ antiporter) | A. gmelini | Rice | Overexpression | Salt tolerance | Ohta et al. (2002) |
| SOS1 (plasma membrane Na+/H+ antiporter) | Arabidopsis | Arabidopsis | Overexpression | Salt tolerance | Shi et al. (2003) |
| AVP1 (vacuolar H+-ATPase) | Arabidopsis | Arabidopsis | Overexpression | Salt tolerance | Gaxiola et al. (2001) |
| HKT1 (high affinity K+ transporter) | Wheat | Wheat | Antisense | Salt tolerance | Laurie et al. (2002) |
| Transcription Factors | |||||
| CBF1, DREB1a (CBF3) | Arabidopsis | Arabidopsis | Overexpression | Freezing, salt, and drought tolerance | Jaglo-Ottosen et al. (1998); Liu et al. (1998); Haake et al. (2002) |
| CBF1, CBF2, CBF3 | Arabidopsis | B. napus | Overexpression | Freezing and drought tolerance | Jaglo et al. (2001); Mendel Biotechnology (unpublished data) |
| CBF1 | Arabidopsis | Tomato | Overexpression | Drought, chilling, and oxidative stress tolerance | Hsieh et al. (2002a, 2002b); Lee et al. (2003) |
| CBF1 | Arabidopsis | Strawberry | Overexpression | Freezing tolerance | Owens et al. (2002) |
| ZmCBF | Maize | Maize | Overexpression | Cold tolerance | Chaiappetta (2002) |
| DREB1a (CBF3) | Arabidopsis | Wheat | Overexpression | Drought tolerance | Pellegrineschi et al. (2002) |
| OsDREB1A | Rice | Arabidopsis | Overexpression | Drought, salt, and freezing tolerance | Dubouzet et al. (2003) |
| CBF3 | Arabidopsis | Rice | Overexpression | Stress tolerance | J. K. Kim (personal communication) |
| ICE1 | Arabidopsis | Arabidopsis | Overexpression | Freezing tolerance | Chinnusamy et al. (2003) |
| ABF3/4 | Arabidopsis | Arabidopsis | Overexpression | Drought tolerance | Kang et al. (2002) |
| Tsi1 | Tobacco | Tobacco | Overexpression | Salt tolerance | Park et al. (2001) |
| SCOF-1 | Soybean | Arabidopsis, Tobacco | Overexpression | Low temperature stress tolerance | Kim et al. (2001) |
| AtMYC2/AtMYB2 | Arabidopsis | Arabidopsis | Overexpression | Drought tolerance | Abe et al. (2003) |
Cold and Drought Stress in Arabidopsis
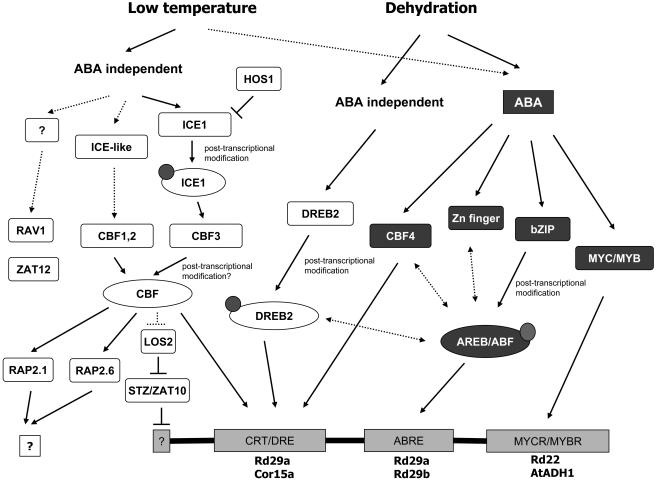
The signal transduction events that occur during cold and drought stress have recently been reviewed (Shinozaki et al., 2003). In contrast to ion homeostasis, a plant's adaptation to cold and drought is to a greater extent under transcriptional control—some processes are regulated by abscisic acid (ABA), while others are ABA independent (Shinozaki et al., 2003). In this perspective, we will look at the transcription factors (TFs) involved in the adaptation to cold and drought stress (Fig. 2).
Figure 2.
Transcriptional cascades of low temperature and dehydration signal transduction. ABA-dependent transcriptional factors are shaded, while ABA independent factors are not. Small circles indicate posttranscriptional modification, such as phosphorylation. Transcription factor binding sites are represented as rectangles at the bottom of the figure, with the representative promoters listed below. Dotted lines indicate possible regulation. Double arrow lines indicate possible cross talk.
The regulation of gene expression by ABA has been reviewed in great detail elsewhere (Finkelstein et al., 2002; Himmelbach et al., 2003; Kuhn and Schroeder, 2003). Mediators of ABA-triggered gene expression include TFs belonging to many different classes, including the bZIP, MYC/MYB, homeodomain Leu zipper (or HD-Zip), Zn finger, and ABI3/VP1 families. The bZIP TFs bind as dimers to ABA response elements (ABREs), with optimal ABA responsiveness usually requiring a second cis-element or coupling element (CE; Shen and Ho, 1995). The CE element is sometimes similar to a dehydration-responsive element (CRT/DRE), thus ABRE-binding bZIPs and CRT/DRE-binding AP2s possibly interact to control ABA-regulated gene expression (Narusaka et al., 2003). ABI5 is active mainly during seed maturation and early seedling development (Finkelstein and Lynch, 2000), whereas the AREB/ABF appears to function later in development (Uno et al., 2000; Kang et al., 2002). For some bZIPs, other TFs may be necessary for optimum transcriptional activation. For example, ABI5 is able to form a complex with ABI3 (Nakamura et al., 2001) and thereby recruit ABI3 to ABRE-containing promoters, even though ABI3 does not bind ABREs directly (Lopez-Molina et al., 2002). In another example, the binding efficiency of the bZIP TF SGBF-1 to ABREs is enhanced by another protein, a C2H2 Zn finger protein, SCOF-1, even though SCOF-1 does not bind directly to ABREs (Kim et al., 2001). The activity of some bZIPs may also be regulated by phosphorylation. For example, the phosphorylation of ABI5 stabilizes this TF (Lopez-Molina et al., 2001), perhaps by blocking the ABI5 BINDING PROTEIN-mediated degradation by the 26S proteasome (Lopez-Molina et al., 2003). The homeodomain TF AtHB6 (Himmelbach et al., 2002), which interacts with ABI1, can also heterodimerize with AtHB5 and possibly with other HD-Zip TFs (Johannesson et al., 2003), while AtMYC2 and AtMYB2 act cooperatively to activate the expression of ABA-inducible genes such as RD22 (Abe et al., 2003).
One class of AP2 TFs that plays a central role in both the ABA-dependent and ABA-independent pathways is the CRT binding factors (CBFs; also called DREB1s). Expression of all CBF genes in Arabidopsis is low under normal growth condition but increases within several minutes after cold (CBF1-3; Gilmour et al., 1998) or drought stress (CBF4; Haake et al., 2002). In addition, the DNA-binding activity of some CBFs can also be modulated by temperature—it was reported that the binding affinity of a barley (Hordeum vulgare) CBF to the CRT/DRE element at 0°C was more than 10 times higher than that at 25°C (Xue, 2003). The cold induction of the Arabidopsis CBF1-3 genes is ABA independent, while the dehydration induced expression of the CBF4 gene is controlled by ABA (Haake et al., 2002).
In order to identify regulators of the CBF genes, Arabidopsis mutants that either impact cold-inducible gene expression under stress or its ability to survive freezing have been isolated and studied. One study identified a bHLH TF, ICE1, which binds specifically to the MYC recognition sequences in the CBF3 promoter and activates CBF3 expression in the cold (Chinnusamy et al., 2003). It is likely that other ICE-like proteins exist because mutational analysis of the CBF2 promoter identified two segments, designated ICEr1 and ICEr2, that work in concert to impart cold-regulated CBF2 expression (Zarka et al., 2003). The sfr6 mutant of Arabidopsis was identified on the basis of its inability to gain freezing tolerance after cold acclimation (Warren et al., 1996; Knight et al., 1999). Transcriptome analysis indicates that the sfr6 mutant is deficient in CRT/DRE-regulated COR gene expression during cold, osmotic stress, or exogenous ABA (Boyce et al., 2003), indicating that SFR6 could interact with the CBFs or DREB2s. In contrast to the sfr6 mutation, the hos1 mutant has higher levels of CBF2 and CBF3 (and downstream COR gene) induction under stress (Lee et al., 2001). HOS1 encodes a RING finger protein that may function in the ubiquitin-mediated degradation of nuclear proteins. It is possible that the elevated CBF transcript levels in hos1 mutant could be a consequence of decreased turnover of ICE proteins (Lee et al., 2001).
Other TFs that either operate in parallel pathways or downstream of CBF have also been identified from Arabidopsis (Fig. 2). AP2 TF DREB2 plays a role in drought adaptation in an ABA-independent manner (Liu et al., 1998; Nakashima et al., 2000). Two transcription factors, RAV1 (AP2; Kagaya et al., 1999) and ZAT12 (Zn finger; Meissner and Michael, 1997) had patterns of expression that were similar to those of CBF1-3, and because neither RAV1 nor ZAT12 transcript levels were affected in CBF1-3 overexpressing plants, they probably operate in pathways that are parallel to those of the CBFs (Fowler and Thomashow, 2002). By contrast, two AP2 domain proteins, RAP2.6 and RAP2.1 (Okamuro et al., 1997), were induced after the initial wave of CBF1-3, RAV1, and ZAT12 inductions. The RAP2.1 promoter contains two copies of the CCGAC core sequence of the CRT/DRE elements, and RAP2.1 expression was high in transgenic Arabidopsis plants that constitutively express CBF1-3, suggesting that it might be a target of the CBF activators (Fowler and Thomashow, 2002). LOS2, identified from an Arabidopsis mutant screen, may repress expression of a zinc finger transcriptional repressor STZ/ZAT10, which in turn regulates COR/RD gene expression (Lee et al., 2002).
Engineering Cold and Drought Tolerance in Crop Plants Based on Arabidopsis Models
Because many aspects of the cold and drought adaptation process are under transcriptional control, it is not surprising that transcription factors represent one of the best targets for engineering plants to achieve enhanced cold and drought tolerance. Even so, not all TFs involved in the cold and drought signal transduction are suitable targets for biotechnological intervention. For example, even though the DREB2s may play a major role in drought-regulated gene expression, overexpression of the DREB2 cDNAs in transgenic plants only caused weak induction of the downstream genes and did not result in more stress tolerance (Liu et al., 1998). It is speculated that posttranslational alterations may be needed for these proteins to be active in transgenic plants (Shinozaki and Yamaguchi-Shinozaki, 2000).
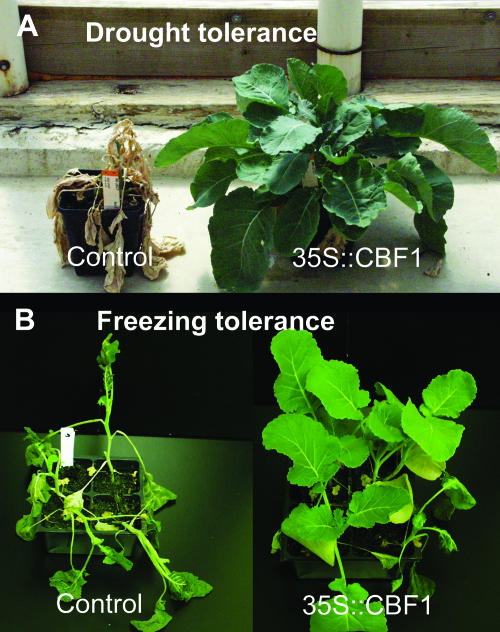
The CBF genes have been successfully used to engineer abiotic stress tolerance in a number of different species (Table I). The orthologous genes of CBF have been found in most crop plants examined so far, including canola (B. napus), soybean (Glycine max), broccoli (Brassica oleracea), tomato, alfalfa (Medicago sativa), tobacco (Nicotiana tabacum), cherry (Prunus avium), strawberry (Fragaria spp.), wheat, rye (Secale cereale), corn (Zea mays), rice, and barley (Jaglo et al., 2001; Choi et al., 2002; Gao et al., 2002; Owens et al., 2002; Dubouzet et al., 2003; Francia et al., 2004; Shen et al., 2003a, 2003b; Vagujfalvi et al., 2003; Xue, 2003). Many of the putative orthologs have been functionally tested, indicating conservation of the pathway in those plant species. Constitutive overexpression of the Arabidopsis CBF genes in canola results in increased freezing tolerance (Jaglo et al., 2001) and drought tolerance (J.Z. Zhang, unpublished data; Fig. 3). Similarly, constitutive overexpression of CBF orthologs from rice (OsDREB1) in transgenic Arabidopsis resulted in salt, cold, and drought tolerance (Dubouzet et al., 2003). Likewise, the overexpression of a ZmCBF gene in maize also resulted in increased cold tolerance (Chaiappetta, 2002). In these and other studies, it was discovered that ectopic overexpression of the CBF genes in plants produced, in addition to increased stress tolerance, dark-green, dwarfed plants with higher levels of soluble sugars and Pro (Liu et al., 1998; Gilmour et al., 2000). To overcome these problems, stress-inducible promoters that have low background expression under normal growth condition have been used in conjunction with the CBF genes to achieve increased stress tolerance without the retarded growth (Kasuga et al., 1999; Lee et al., 2003).
Figure 3.
Engineered drought and freezing tolerance in transgenic B. napus through constitutive expression of CBF1 (Jaglo et al., 2001). A, Three-week-old plants were frozen at −6°C for 2 d and then let to recover for 2 d at 28°C before pictures were taken. B, Seven-week-old greenhouse grown plants were withheld water for 1 week and then rewatered for 2 weeks before picture was taken.
CONCLUSIONS AND PERSPECTIVES
As examples from the above discussions have shown, Arabidopsis has been an excellent model plant for the studying of abiotic stress responses and biotechnology applications. In many cases, not only are structural proteins such as the ion transporters conserved between Arabidopsis and other plant species, but also regulatory proteins such as CBF/DREB1 and entire transcriptional regulons can be conserved as well. Only after we thoroughly understand how plants respond to stress—in many cases first in Arabidopsis and then applying the Arabidopsis model to crop plants—will we be able to begin engineering stress tolerance. A case in point: While the overexpression of Na+/H+ antiporters is sufficient to achieve measurable improvement in salt tolerance in plants, the engineering of robust cold and drought tolerance requires the coordinated expression of many genes through the altered expression of global regulators such as CBFs. Therefore, Arabidopsis will continue to play a critical role in the foreseeable future, not only in the understanding of biological mechanisms of abiotic stress tolerance, but also in providing a facile means for testing in biotechnology.
Acknowledgments
We thank Mike Thomashow for discussions and comments.
This work was supported by the U.S. Department of Agriculture National Research Initiative, the National Science Foundation (NSF), and the National Institutes of Health (grants to J.-K.Z.) and in part by the NSF Small Business Innovation Research (to J.Z.Z.).
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285: 1256–1258 [DOI] [PubMed] [Google Scholar]
- Berthomieu P, Conejero G, Nublat A, Brackenbury WJ, Lambert C, Savio C, Uozumi N, Oiki S, Yamada K, Cellier F, et al (2003) Functional analysis of AtHKT1 in Arabidopsis shows that Na(+) recirculation by the phloem is crucial for salt tolerance. EMBO J 22: 2004–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce JM, Knight H, Deyholos M, Openshaw MR, Galbraith DW, Warren G, Knight MR (2003) The sfr6 mutant of Arabidopsis is defective in transcriptional activation via CBF/DREB1 and DREB2 and shows sensitivity to osmotic stress. Plant J 34: 395–406 [DOI] [PubMed] [Google Scholar]
- Chaiappetta L (2002) Ottenimento di piante transegniche di mais tolleranti al freddo. PhD thesis. University of Bologna, Bologna, Italy
- Chauhan S, Forsthoefel N, Ran Y, Quigley F, Nelson DE, Bohnert HJ (2000) Na+/myo-inositol symporters and Na+/H+-antiport in Mesembryanthemum crystallinum. Plant J 24: 511–522 [DOI] [PubMed] [Google Scholar]
- Chen S, Guo B, He S, Tian A, Wang Z, Zhang J (2002) Cloning and characterization of the Na+/H+ antiport genes from Triticum aestivum. Acta Bot Sin 44: 1203–1208 [Google Scholar]
- Cheng NH, Pittman JK, Zhu JK, Hirschi KD (2004) The protein kinase SOS2 activates the Arabidopsis H+/Ca2+ antiporter CAX1 to integrate calcium transport and salt tolerance. J Biol Chem 279: 2922–2926 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK (2003) ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DW, Rodriguez EM, Close TJ (2002) Barley Cbf3 gene identification, expression pattern, and map location. Plant Physiol 129: 1781–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J 33: 751–763 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell 14 (suppl.): S15–S45 [DOI] [PMC free article] [PubMed]
- Finkelstein RR, Lynch TJ (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14: 1675–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francia E, Rizza F, Cattivelli L, Stanca AM, Galiba G, Toth B, Hayes PM, Skinner JS, Pecchioni N (2004) Two loci on chromosome 5H determine low-temperature tolerance in a ‘Nure’ (winter) x ‘Tremois’ (spring) barley map. Theor Appl Genet 108: 670–680 [DOI] [PubMed] [Google Scholar]
- Fukuda A, Nakamura A, Tanaka Y (1999) Molecular cloning and expression of the Na+/H+ exchanger gene in Oryza sativa. Biochim Biophys Acta 1446: 149–155 [DOI] [PubMed] [Google Scholar]
- Gao MJ, Allard G, Byass L, Flanagan AM, Singh J (2002) Regulation and characterization of four CBF transcription factors from Brassica napus. Plant Mol Biol 49: 459–471 [DOI] [PubMed] [Google Scholar]
- Gaxiola RA, Li J, Undurraga S, Dang LM, Allen GJ, Alper SL, Fink GR (2001) Drought- and salt-tolerant plants result from overexpression of the AVP1 H+-pump. Proc Natl Acad Sci USA 98: 11444–11449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaxiola RA, Rao R, Sherman A, Grisafi P, Alper SL, Fink GR (1999) The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proc Natl Acad Sci USA 96: 1480–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF (2000) Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol 124: 1854–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF (1998) Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J 16: 433–442 [DOI] [PubMed] [Google Scholar]
- Haake V, Cook D, Riechmann JL, Pineda O, Thomashow MF, Zhang JZ (2002) Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol 130: 639–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada A, Shono M, Xia T, Ohta M, Hayashi Y, Tanaka A, Hayakawa T (2001) Isolation and characterization of a Na+/H+ antiporter gene from the halophyte Atriplex gmelini. Plant Mol Biol 46: 35–42 [DOI] [PubMed] [Google Scholar]
- Himmelbach A, Hoffmann T, Leube M, Hohener B, Grill E (2002) Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J 21: 3029–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach A, Yang Y, Grill E (2003) Relay and control of abscisic acid signaling. Curr Opin Plant Biol 6: 470–479 [DOI] [PubMed] [Google Scholar]
- Hsieh TH, Lee JT, Charng YY, Chan MT (2002. b) Tomato plants ectopically expressing Arabidopsis CBF1 show enhanced resistance to water deficit stress. Plant Physiol 130: 618–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TH, Lee JT, Yang PT, Chiu LH, Charng YY, Wang YC, Chan MT (2002. a) Heterology expression of the Arabidopsis C-repeat/dehydration response element binding factor 1 gene confers elevated tolerance to chilling and oxidative stresses in transgenic tomato. Plant Physiol 129: 1086–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglo KR, Kleff S, Amundsen KL, Zhang X, Haake V, Zhang JZ, Deits T, Thomashow MF (2001) Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol 127: 910–917 [PMC free article] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF (1998) Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280: 104–106 [DOI] [PubMed] [Google Scholar]
- Johannesson H, Wang Y, Hanson J, Engstrom P (2003) The Arabidopsis thaliana homeobox gene ATHB5 is a potential regulator of abscisic acid responsiveness in developing seedlings. Plant Mol Biol 51: 719–729 [DOI] [PubMed] [Google Scholar]
- Kagaya Y, Ohmiya K, Hattori T (1999) RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res 27: 470–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JY, Choi HI, Im MY, Kim SY (2002) Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14: 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17: 287–291 [DOI] [PubMed] [Google Scholar]
- Kim JC, Lee SH, Cheong YH, Yoo CM, Lee SL, Chun HJ, Yun DJ, Hong JC, Lee SY, Lim CO, et al (2001) A novel cold-inducible zinc finger protein from soybean, SCOF-1, enhances cold tolerance in transgenic plants. Plant J 25: 247–259 [DOI] [PubMed] [Google Scholar]
- Knight H, Veale EL, Warren GJ, Knight MR (1999) The sfr6 mutation in Arabidopsis suppresses low-temperature induction of genes dependent on the CRT/DRE sequence motif. Plant Cell 11: 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JM, Schroeder JI (2003) Impacts of altered RNA metabolism on abscisic acid signaling. Curr Opin Plant Biol 6: 463–469 [DOI] [PubMed] [Google Scholar]
- Laurie S, Feeney KA, Maathuis FJ, Heard PJ, Brown SJ, Leigh RA (2002) A role for HKT1 in sodium uptake by wheat roots. Plant J 32: 139–149 [DOI] [PubMed] [Google Scholar]
- Lee H, Guo Y, Ohta M, Xiong L, Stevenson B, Zhu JK (2002) LOS2, a genetic locus required for cold-responsive gene transcription encodes a bi-functional enolase. EMBO J 21: 2692–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Xiong L, Gong Z, Ishitani M, Stevenson B, Zhu JK (2001) The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes Dev 15: 912–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT, Prasad V, Yang PT, Wu JF, Ho THD, Charng YY, Chan MT (2003) Expression of Arabidopsis CBF1 regulated by an ABA/stress promoter in transgenic tomato confers stress tolerance without affecting yield. Plant Cell Environ 26: 1181–1190 [Google Scholar]
- Liu J, Ishitani M, Halfter U, Kim CS, Zhu JK (2000) The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci USA 97: 3730–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhu JK (1998) A calcium sensor homolog required for plant salt tolerance. Science 280: 1943–1945 [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua NH (2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA 98: 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Kinoshita N, Chua NH (2003) AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev 17: 410–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, McLachlin DT, Chai BT, Chua NH (2002) ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J 32: 317–328 [DOI] [PubMed] [Google Scholar]
- Maser P, Eckelman B, Vaidyanathan R, Horie T, Fairbairn DJ, Kubo M, Yamagami M, Yamaguchi K, Nishimura M, Uozumi N, et al (2002) Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett 531: 157–161 [DOI] [PubMed] [Google Scholar]
- Meissner R, Michael AJ (1997) Isolation and characterisation of a diverse family of Arabidopsis two and three-fingered C2H2 zinc finger protein genes and cDNAs. Plant Mol Biol 33: 615–624 [DOI] [PubMed] [Google Scholar]
- Nakamura S, Lynch TJ, Finkelstein RR (2001) Physical interactions between ABA response loci of Arabidopsis. Plant J 26: 627–635 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Shinwari ZK, Sakuma Y, Seki M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K (2000) Organization and expression of two Arabidopsis DREB2 genes encoding DRE-binding proteins involved in dehydration- and high-salinity-responsive gene expression. Plant Mol Biol 42: 657–665 [DOI] [PubMed] [Google Scholar]
- Narusaka Y, Nakashima K, Shinwari ZK, Sakuma Y, Furihata T, Abe H, Narusaka M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J 34: 137–148 [DOI] [PubMed] [Google Scholar]
- Ohta M, Hayashi Y, Nakashima A, Hamada A, Tanaka A, Nakamura T, Hayakawa T (2002) Introduction of a Na+/H+ antiporter gene from Atriplex gmelini confers salt tolerance to rice. FEBS Lett 532: 279–282 [DOI] [PubMed] [Google Scholar]
- Okamuro JK, Caster B, Villarroel R, Van Montagu M, Jofuku KD (1997) The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc Natl Acad Sci USA 94: 7076–7081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens CL, Thomashow MF, Hancock JF, Iezzoni AF (2002) CBF1 orthologs in sour cherry and strawberry and the heterologous expression of CBF1 in strawberry. J Am Soc Hortic Sci 127: 489–494 [Google Scholar]
- Park J, Park C, Lee S, Ham B, Shin R, Paek K (2001) Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell 13: 1035–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrineschi A, Ribaut JM, Trethowan R, Yamaguchi-Shinozaki K, Hoisington D (2002) Progress in the genetic engineering of wheat for water-limited conditions. JIRCAS Working Report: 55–60
- Qiu QS, Guo Y, Quintero FJ, Pardo JM, Schumaker KS, Zhu JK (2004) Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the SOS pathway. J Biol Chem 279: 207–215 [DOI] [PubMed] [Google Scholar]
- Rus A, Yokoi S, Sharkhuu A, Reddy M, Lee BH, Matsumoto TK, Koiwa H, Zhu JK, Bressan RA, Hasegawa PM (2001) AtHKT1 is a salt tolerance determinant that controls Na(+) entry into plant roots. Proc Natl Acad Sci USA 98: 14150–14155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman DP, Schroeder JI (1994) Structure and transport mechanism of a high-affinity potassium uptake transporter from higher plants. Nature 370: 655–658 [DOI] [PubMed] [Google Scholar]
- Shen Q, Ho TH (1995) Functional dissection of an abscisic acid (ABA)-inducible gene reveals two independent ABA-responsive complexes each containing a G-box and a novel cis-acting element. Plant Cell 7: 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen YG, Zhang WK, He SJ, Zhang JS, Liu Q, Chen SY (2003. a) An EREBP/AP2-type protein in Triticum aestivum was a DRE-binding transcription factor induced by cold, dehydration and ABA stress. Theor Appl Genet 106: 923–930 [DOI] [PubMed] [Google Scholar]
- Shen YG, Zhang WK, Yan DQ, Du BX, Zhang JS, Liu Q, Chen SY (2003. b) Characterization of a DRE-binding transcription factor from a halophyte Atriplex hortensis. Theor Appl Genet 107: 155–161 [DOI] [PubMed] [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu JK (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA 97: 6896–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Lee BH, Wu SJ, Zhu JK (2003) Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat Biotechnol 21: 81–85 [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K (2000) Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol 3: 217–223 [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K, Seki M (2003) Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol 6: 410–417 [DOI] [PubMed] [Google Scholar]
- Somerville C, Dangl J (2000) Genomics: plant biology in 2010. Science 290: 2077–2078 [DOI] [PubMed] [Google Scholar]
- Somerville C, Koornneef M (2002) A fortunate choice: the history of Arabidopsis as a model plant. Nat Rev Genet 3: 883–889 [DOI] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97: 11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagujfalvi A, Galiba G, Cattivelli L, Dubcovsky J (2003) The cold-regulated transcriptional activator Cbf3 is linked to the frost-tolerance locus Fr-A2 on wheat chromosome 5A. Mol Genet Genomics 269: 60–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G, McKown R, Marin A, Teutonico R (1996) Isolation of mutations affecting the development of freezing tolerance in Arabidopsis thaliana (L.) Heynh. Plant Physiol 111: 1011–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia T, Apse MP, Aharon GS, Blumwald E (2002) Identification and characterization of a NaCl-inducible vacuolar Na+/H+ antiporter in Beta vulgaris. Physiol Plant 116: 206–212 [DOI] [PubMed] [Google Scholar]
- Xue GP (2003) The DNA-binding activity of an AP2 transcriptional activator HvCBF2 involved in regulation of low-temperature responsive genes in barley is modulated by temperature. Plant J 33: 373–383 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Apse MP, Shi H, Blumwald E (2003) Topological analysis of a plant vacuolar Na+/H+ antiporter reveals a luminal C terminus that regulates antiporter cation selectivity. Proc Natl Acad Sci USA 100: 12510–12515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Fukada-Tanaka S, Inagaki Y, Saito N, Yonekura-Sakakibara K, Tanaka Y, Kusumi T, Iida S (2001) Genes encoding the vacuolar Na+/H+ exchanger and flower coloration. Plant Cell Physiol 42: 451–461 [DOI] [PubMed] [Google Scholar]
- Zarka DG, Vogel JT, Cook D, Thomashow MF (2003) Cold induction of Arabidopsis CBF genes involves multiple ICE (Inducer of CBF Expression) promoter elements and a cold-regulatory circuit that is desensitized by low temperature. Plant Physiol 133: 910–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HX, Blumwald E (2001) Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat Biotechnol 19: 765–768 [DOI] [PubMed] [Google Scholar]
- Zhang HX, Hodson JN, Williams JP, Blumwald E (2001) Engineering salt-tolerant Brassica plants: characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proc Natl Acad Sci USA 98: 12832–12836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK (2003) Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol 6: 441–445 [DOI] [PubMed] [Google Scholar]