Fluctuations in the length of the day affect developmental processes and behaviors of many organisms. Mammals and birds reproduce in spring in response to lengthening days and insects pupate in autumn when daylength shortens. These phenomena, called photoperiodism, allow detection of seasonal changes and anticipation of environmental conditions such as low temperatures and desiccation. Photoperiodism was first described in detail by Garner and Allard in 1920 through the demonstration that many plants flower in response to changes in daylength (Garner and Allard, 1920). Subsequently, they showed that some plant species promote flowering when daylength falls below a critical daylength, whereas other plants accelerate flowering in response to daylengths longer than a critical daylength. These plants are called short-day (SD) and long-day (LD) plants, respectively. During the last decade, molecular-genetic approaches were applied to understanding the control of flowering time, mainly in the LD plant Arabidopsis, and notable progress has been made in identifying the molecular mechanisms by which Arabidopsis recognizes daylength and promotes flowering specifically under LDs. Also, recent genetic studies in rice enabled the mechanisms of the daylength response in this SD plant to be compared with those of Arabidopsis. Here we review the recent advances in understanding the regulatory mechanisms for daylength response of flowering in Arabidopsis and compare them with those of rice.
MODEL OF DAYLENGTH MEASUREMENT FOR CONTROL OF FLOWERING TIME
Erwin Bünning first proposed that the photoperiodic time-keeping mechanism is associated with the circadian clock (Bünning, 1936), an autonomous mechanism that generates biological rhythms with a period of approximately 24 h. This model proposes that the circadian clock generates a rhythm with an approximate 24-h period that controls flowering and is sensitive to light at a particular phase of the rhythm. Consequently, if a plant is grown under a specific daylength that causes it to be exposed to light at this particular phase, then flowering is induced if the plant shows a LD response, or repressed if the plant shows a SD response. This model, called the external coincidence model (Pittendrigh and Minis, 1964), has been supported by a number of physiological studies for the control of flowering time, indicating that the basis of daylength measurement is the interaction of an external light signal with a circadian rhythm (Thomas and Vince-Prue, 1997). In contrast, another model, called the internal coincidence model, proposes that the floral response occurs under conditions in which two differentially entrained rhythms are brought into the same phase under daylengths that promote flowering, but that under other daylengths these two rhythms are out of phase. Studies of photoperiodism in insects supported this model (Vaz Nunes and Saunders, 1999), but detailed analyses have not yet been carried out to test it in plants.
CIRCADIAN CLOCK FUNCTION IN ARABIDOPSIS
Genetic studies in Arabidopsis support the involvement of the circadian clock in the control of flowering by daylength. Most mutants that were initially isolated based on an altered circadian rhythm phenotype, such as alterations in period length and/or amplitude of clock-controlled gene expression, also exhibit changes in flowering time. In addition, some mutants originally isolated based on a defect in the control of flowering by daylength also exhibit changes in circadian rhythms. The circadian clock system is often divided into three general parts (Dunlap, 1999). The central oscillator is the core of the system, responsible for driving 24-h rhythms. The oscillator is entrained to day-night or temperature cycles through a mechanism involving input pathways that transmit light or temperature signals to the core oscillator. Output pathways are controlled by the core oscillator and represent a wide range of biochemical and developmental pathways. The control of flowering by daylength is assumed to be regulated by one or more of these output branches. In this way, the core oscillator can determine the activity of diurnal rhythms in output genes, and these genes can set the light sensitive phase for triggering the floral transition.
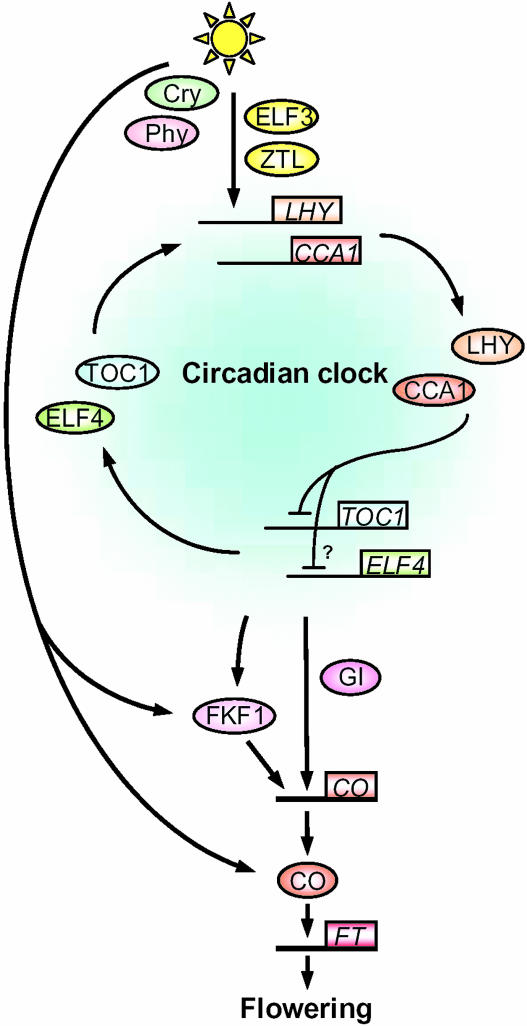
Molecular-genetic studies of circadian-clock function in mammals and cyanobacteria reveal that the core oscillator is composed of an autoregulatory transcriptional and translational negative-feedback loop. In Arabidopsis, CIRCADIAN CLOCK ASSOCIATED1 (CCA1), LATE ELONGATED HYPOCOTYL (LHY), TIMING OF CAB EXPRESSION1 (TOC1), and EARLY FLOWERING4 (ELF4) are the candidate genes that may form the feedback loop (Fig. 1; Schaffer et al., 1998; Wang and Tobin, 1998; Strayer et al., 2000; Alabadi et al., 2001; Doyle et al., 2002). Molecular studies of these genes reveal that TOC1, whose mRNA abundance peaks in the evening, functions as a positive regulator to raise LHY and CCA1 transcript abundance in the morning. This idea is based on the observation that loss of TOC1 function severely reduces the transcript levels of LHY and CCA1. The strong reduction of these transcripts is also observed in elf4 mutants. Furthermore, ELF4 transcript oscillates with a phase similar to that of TOC1, which indicates that ELF4 could act together with TOC1 to induce LHY/CCA1. TOC1 belongs to a novel family of pseudo response regulators, and has a CCT (CO, COL, and TOC1) domain that may be responsible for protein-protein interaction and nuclear localization, whereas ELF4 encodes a small nuclear protein with no similarity to other proteins.
Figure 1.
Model of the circadian system of Arabidopsis and its relationship to the flowering-time gene CO. Phytochromes and cryptochromes perceive light and are involved in resetting of the circadian clock. ELF3 and ZTL mediate between photoreceptors and the circadian clock. LHY/CCA1 and TOC1/ELF4 form a negative feedback loop within the circadian oscillator. LHY/CCA1 act as negative regulators of TOC1 and ELF4, which positively regulate the transcription of LHY/CCA1. The oscillator functions to determine the phase of CO transcription, a key gene that mediates between the circadian clock and flowering. The transcription of CO is regulated by FKF1 and GI, whose transcription is under the control of the circadian clock. FKF1 protein is directly regulated by light, and this allows FKF1 to increase CO transcript under LDs. CO protein is also directly activated by light, and this allows CO to generate a LD signal and activate a flowering-time gene FT for the promotion of flowering specifically under LDs.
Reciprocally, overexpression of either LHY or CCA1 strongly suppresses the expression of TOC1, and lhy cca1 double mutants exhibit increased TOC1 mRNA levels (Alabadi et al., 2001; Mizoguchi et al., 2002). LHY and CCA1 encode MYB-related transcription factors, and suppression of TOC1 by these proteins may be mediated directly through the cis-acting evening element, which was identified in the promoter regions of several clock-controlled genes whose transcripts peak in the evening (Harmer et al., 2000; Alabadi et al., 2001). Thus, LHY/CCA1 are proposed to act as negative regulators to generate the TOC1 rhythm, with a circadian phase opposite to that of LHY/CCA1. Therefore, as LHY/CCA1 rise in the morning, TOC1 expression falls. This eventually causes a reduction in expression of LHY and CCA1 leading in turn to the reactivation of TOC1 in the evening, and the second cycle then begins with the activation of LHY and CCA1.
Genes that are involved in light input to the clock have also been isolated from Arabidopsis (Fig. 1). Phytochromes and cryptochromes are involved in red- and blue-light input to the clock, respectively (Somers et al., 1998; Devlin and Kay, 2000). Although the molecular mechanism that transmits light signals to the clock is not yet clear, recent genetic studies have allowed several genes involved in this process to be identified. EARLY FLOWERING 3 (ELF3) functions to repress or gate the light input pathway (McWatters et al., 2000). ELF3 protein levels are regulated by the circadian clock and accumulate to high level during the evening. This makes the clock insensitive to light during the evening, ensuring that it is reset predominantly during the morning. ELF3 encodes a nuclear protein with no similarity to other proteins. This protein binds to PhyB in vitro, consistent with the idea that ELF3 suppresses the input pathway through binding to PhyB and restricting its activity (Liu et al., 2001). ZEITLUPE (ZTL) is also proposed to be involved in the input pathway to the clock, and this protein binds to PhyB and CRY1 in vitro (Somers et al., 2000; Jarillo et al., 2001). ZTL protein contains an F-box and repeated kelch motifs, suggesting that this protein functions in the degradation of a specific protein via the proteasome (Somers et al., 2000). Recent analysis reveals that the target protein is TOC1, and that ZTL degrades TOC1 especially during the night to generate a robust diurnal rhythm of this protein in light/dark cycles (Mas et al., 2003).
THE CONSTANS GENE AND DAYLENGTH MEASUREMENT IN ARABIDOPSIS
Transcriptional Regulation of CO by the Circadian Clock
The external coincidence model proposes that the circadian clock sets a light-sensitive phase within the day-night cycle, and that floral responses occur under a particular daylength that exposes plants to light during the light-sensitive phase. Recent molecular-genetic studies of the flowering-time gene CONSTANS (CO) suggest that the interaction between circadian rhythms and light signaling may occur at the level of CO transcription and CO protein stability (Figs. 1 and 2). CO was originally isolated using a mutant that exhibits late flowering specifically under LDs (Putterill et al., 1995). The gene encodes a nuclear protein that contains a CCT motif and two B-box type zinc-finger domains, which were originally identified in several animal proteins and are believed to mediate protein-protein interaction. The transcript levels of this gene show a circadian rhythm under continuous light. However, CO overexpression does not alter the circadian rhythm in CAB gene expression in continuous light, suggesting that it does not have a general effect on circadian rhythms (Ledger et al., 2001), but it does result in dramatic early flowering (Putterill et al., 1995). This indicates that CO acts as a clock-output gene and mediates between the circadian clock and flowering (Suarez-Lopez et al., 2001). The important role of CO in acting as a clock output to control flowering is also suggested by studying several mutations that alter both flowering time and circadian rhythms, and showing that these affect CO expression in ways that are correlated with their effects on flowering time (Suarez-Lopez et al., 2001). Moreover, CO directly induces the expression of FLOWERING LOCUS T (FT), which was originally isolated using a late-flowering mutant, and whose transcript is induced specifically under LDs (Samach et al., 2000). This strongly suggests that in Arabidopsis CO plays a key role in integrating circadian rhythms and the light signal to measure daylength.
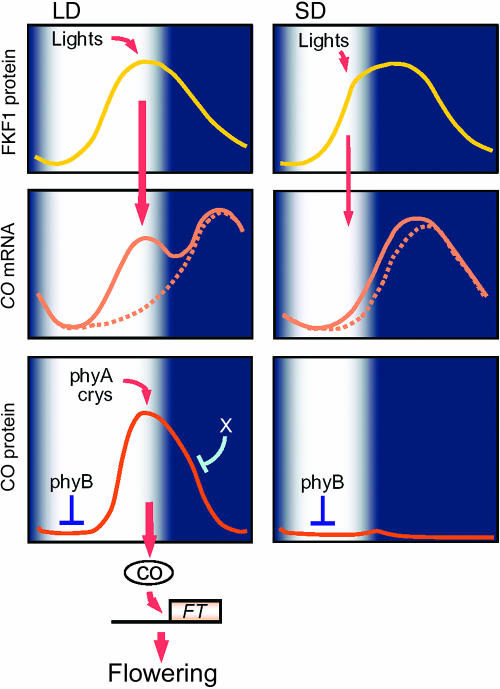
Figure 2.
A model of daylength measurement in Arabidopsis. Expression of FKF1 protein is regulated by the circadian clock and exhibits a diurnal rhythm under LD and SD. FKF1 protein functions as a photoreceptor, and accumulates at high levels during mid to end of the day under an LD. FKF1 is regulated by light and functions under LDs to increase CO mRNA abundance. CO mRNA levels in an fkf1 mutant are indicated by the dotted line, whereas the solid line illustrates CO mRNA levels in wild-type plants. CO protein is thereafter activated by light, because blue and far-red light stabilize CO through the action of cryptochromes and phyA and darkness destabilizes it. phyB antagonizes the activity of phyA and cryptochromes and promotes the degradation of CO especially in the morning, allowing CO protein to be expressed with a more refined waveform under LDs. The combination of phyB activity that promotes degradation of CO in the morning and FKF1 activity raising CO mRNA levels during the day under LDs results in robust FT induction and floral promotion specifically under LDs in Arabidopsis.
Under the normal day-night cycle, CO transcripts show a diurnal rhythm. Under SDs, high levels of CO mRNA only occur during the night, whereas under LDs high CO levels occur at the end of and during the night (Fig. 2; Suarez-Lopez et al., 2001). This observation suggested that CO mRNA level determines the light-sensitive phase, and flowering is promoted specifically under LDs because only under these conditions are plants exposed to light at times when CO is highly expressed. The importance of these CO patterns in daylength measurement is also supported by the analyses of toc1-1 mutants, which exhibit early flowering with decreased sensitivity to daylength and a shortened circadian period in CAB mRNA expression (Millar et al., 1995; Somers et al., 1998). The toc1-1 mutation does not change the photomorphogenic phenotype of Arabidopsis seedlings (Somers et al., 1998), although more severe toc1 alleles do (Mas et al., 2003), and this decreases the possibility that the toc1-1 mutation causes early flowering by affecting light signal transduction. In the toc1-1 mutant, the phase of the CO rhythm is advanced both under LD and SD, leading to high levels of CO mRNA at times at which plants are exposed to light under SDs. Furthermore, the daylength response of FT induction in toc1-1 is recovered under light-dark cycles with a total duration of 21 h, which is the circadian period of CAB gene expression in this mutant, suggesting that the early flowering of toc1-1 under SDs is suppressed when the circadian period and the diurnal cycle are synchronized (Yanovsky and Kay, 2002).
The regulatory mechanism generating the diurnal patterns of CO transcription under day-night cycles is still not completely clear. However, recent molecular-genetic studies of a flowering time gene FLAVIN-BINDING, KELCH REPEAT, F-BOX (FKF1) provided more information about this mechanism (Imaizumi et al., 2003). FKF1 generates high levels of CO mRNA observed in mid to late day under LDs. In the fkf1 mutant, the high levels of CO mRNA observed during the day under LD are strongly reduced, and the daytime peak is completely abolished, although a peak in the night remains (Fig. 2). FKF1 was identified based on a late-flowering mutant under LDs (Nelson et al., 2000). Circadian rhythms in the expression of clock-output genes such as CAB and CCR2 are not affected by overexpression of FKF1 or by fkf1 mutations, but both mRNA and protein levels of FKF1 oscillate, indicating that this gene is a clock-output that promotes flowering under LDs. FKF1 encodes a protein containing a LOV domain, a light sensing module that was originally found in the blue-light receptor phototropin, suggesting that FKF1 acts as a photoreceptor. In support of this, a FMN chromophore was detected from purified fusion protein containing the FKF1 LOV domain. Importantly, FKF1 protein levels exhibit a diurnal pattern with a peak in the late day under LD, allowing these proteins to accumulate at high levels during the day, whereas under SD this protein is expressed at peak levels during the early to mid nighttime, with low FKF1 protein levels during the day. Thus, this model proposes that high levels of FKF1 protein accumulation and the direct activation of FKF1 protein by light occur simultaneously under LDs, and this eventually generates the daytime peak of CO mRNA under these conditions. How FKF1 protein generates the peak in CO mRNA during the day is unknown. However, FKF1 protein contains an F-box and repeated kelch motifs as well as a LOV domain, suggesting that this protein might recruit for degradation by the proteasome a specific transcription factor that regulates CO, and that this may be influenced by light.
Posttranscriptional Regulation of CO by Light
Studies of CO reveal that the post-transcriptional activation of CO by light is a key event for daylength measurement. This activation was predicted to be mediated by the photoreceptors PhyA and CRY2, because loss-of-function in either gene caused late flowering under LDs and reduced FT levels (Johnson et al., 1994; Guo et al., 1998; Yanovsky and Kay, 2002). Particularly, loss of CRY2 function delays flowering without affecting the diurnal pattern of CO mRNA (Yanovsky and Kay, 2002). In contrast, phyB mutations result in early flowering (Goto et al., 1991), indicating that these photoreceptors have different roles in the control of flowering time despite their common roles in triggering photomorphogenesis in response to light.
Recently, studies of CO protein suggested how posttranscriptional regulation of CO generates a LD signal through light-mediated activation and also verified the direct regulation of CO by light (Fig. 2; Valverde et al., 2004). Analyses of CO protein were carried out using 35S::CO transgenic plants, in which CO mRNA levels are constantly high independently of the effect of the circadian clock and exposure to light. In these transgenic plants, CO protein accumulates under continuous white light, whereas CO levels are strongly reduced under continuous dark. Light dependent accumulation of CO was also observed using the fluorescence of the GFP:CO fusion protein, which exists at high levels in the nucleus during the day and disappears in the dark. The dark-dependent reduction of CO protein is derived from ubiquitin-dependent active degradation of CO protein by the proteasome, as CO protein accumulates to high abundance in the dark in vivo in the presence of proteasome inhibitors and is detected attached to ubiquitin in vitro. Furthermore, CO protein accumulates to high levels in continuous blue and far-red light, whereas CO protein disappears in continuous red light, consistent with flowering time under these light conditions. PhyA and cryptochromes are involved in far-red and blue light-dependent accumulation of CO, whereas PhyB is involved in red-light-dependent reduction of CO protein abundance. Thus, these results confirm that CO is a direct target of light signals and identify the cognate photoreceptors.
However, analyses of CO protein in light-dark cycles provided the unexpected observation that CO protein levels in 35S::CO plants are strongly dependent on daylength; CO protein under LD exhibits a diurnal pattern with a strong peak in abundance at the end of the day, whereas under SD CO protein is diurnally expressed with a much weaker peak in expression at the early nighttime. This daylength dependent CO protein accumulation must be regulated independently of the transcriptional control that drives diurnal patterns of CO mRNA in wild-type plants. Notably, the time of the strong CO protein peak under LDs in 35S::CO plants coincides with that of CO mRNA peak detected in the evening in wild-type plants. Thus, a combination of the transcriptional and posttranscriptional diurnal patterns in CO expression could enhance each other and drive a high amplitude of CO activity under LDs, allowing FT to be induced at high levels specifically under these conditions.
How is the diurnal pattern of CO protein generated? The regulation of CO protein abundance during the day is mediated by phyB, which promotes reduction of CO protein especially early in the morning. This was demonstrated by the observation that phyB mutations cause constantly high CO protein accumulation during the day. In contrast, phyA and cryptochromes stabilize CO protein at the end of the day, as loss-of-function mutations impairing these photoreceptors decreases the levels of CO protein abundance in the day under LD. The phyA and cryptochrome photoreceptors seem to stabilize CO independently of phyB, because loss of these photoreceptors decreases CO protein abundance under continuous blue or far-red light, where phyB would not be activated. Thus, these observations reveal that phyA and cryptochromes act to stabilize CO during the day in response to far-red and blue light, respectively, whereas phyB is activated especially in the morning and antagonizes the activity of phyA and cryptochromes to promote the degradation of CO. Towards the end of a LD the balance between these activities favors stabilization of CO, which eventually allows CO protein levels to be increased until the end of the day. During the night CO protein is degraded, probably via an independent mechanism similar to those proposed for other light-stabilized transcription factors.
Control of Flowering Time in an SD Plant, Rice
Photoperiodic control of flowering is widespread among the Angiosperms, and whether the molecular mechanism controlling the LD promotion of flowering in Arabidopsis is conserved in other plant species exhibiting different responses to daylength is of importance. CO and FT homologous genes have been identified in many species suggesting conservation of the components of the Arabidopsis photoperiod pathway (Yano et al., 2000; Liu et al., 2001; Kojima et al., 2002; Griffiths et al., 2003). In addition, recent molecular-genetic studies in the SD plant, rice (Oryza sativa), as well as the completion of its whole genome sequence, have allowed us to compare the molecular mechanisms controlling flowering time between a SD and a LD plant.
Conservation of the Molecular Mechanisms Controlling Daylength Response of Flowering in Rice and Arabidopsis
The genetic mechanisms controlling photoperiodic flowering in rice and Arabidopsis appear to be closely related (Fig. 3). For example, Heading-date1 (Hd1), Heading-date3a (Hd3a), and Heading-date6 (Hd6) have been recently isolated as quantitive trait loci responsible for the different flowering times of rice cultivars, and found to encode proteins similar to CO, FT, and the α-subunit of casein kinase 2, respectively (Yano et al., 2000; Takahashi et al., 2001; Kojima et al., 2002). PHOTOPERIOD SENSITIVITY5 (Se5) is also a flowering-time gene and encodes a protein similar to Arabidopsis HY1, a heme oxygenase which participates in biosynthesis of phytochrome chromophore. The rice se5 mutant exhibits severe early flowering in continuous light as well as under LDs and SDs and shows no flowering response to daylength, indicating that phytochrome is an essential photoreceptor for the regulation of daylength responses of flowering in rice (Izawa et al., 2000). The GI homolog of rice (OsGI) was also isolated, as a gene whose mRNA abundance is altered in se5 mutants, and the flowering behaviors of transgenic plants with increased expression of OsGI or reduced OsGI expression levels showed its participation in the control of the daylength response of flowering (Hayama et al., 2002, 2003). Control of flowering time by OsGI may be mediated by regulation of Hd1 activity, perhaps through a mechanism similar to that in Arabidopsis (Hayama et al., 2003).
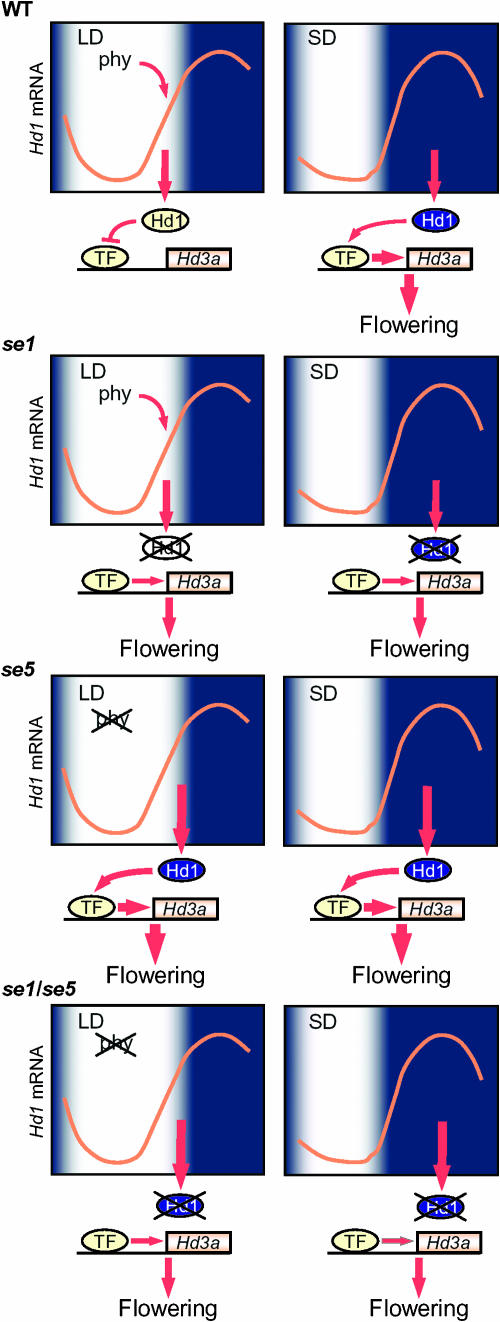
Figure 3.
A model of daylength measurement in rice. Hd1, a CO homolog in rice, has the independent functions of inhibiting and promoting flowering under LDs and SDs, respectively. Hd1 mRNA exhibits diurnal rhythms under both SD and LD with their phases similar to those of CO in Arabidopsis. Hd1 mRNA highly accumulates at the mid to the end of the day under LD, and the coincidence of Hd1 expression and exposure to light suppresses the transcription of Hd3a and inhibits flowering under these conditions. Hd1 is proposed to inhibit Hd3a by suppressing the function of a transcription factor that autonomously activates Hd3a. Phytochrome modifies Hd1 function so that it can act to inhibit Hd3a. Without phytochrome activity, Hd1 induces Hd3a. Therefore, under SDs, when Hd1 accumulates at high levels during the night and phytochrome is inactive, Hd1 induces Hd3a and promotes flowering. The se1 mutant is deficient in Hd1 and exhibits early flowering under LDs due to the lack of Hd1 during the day. This mutant also shows late flowering under SDs due to the absence of Hd1 during the night. The se5 mutant is defective in phytochrome activity and shows early flowering irrespective of the daylength, because Hd1 is constitutively in the dark form. The double mutant shows later flowering than the se5 mutant due to the lack of the dark form of Hd1.
Comparison of the genome sequences of rice and Arabidopsis also suggests wider conservation of the molecular mechanisms controlling flowering time in response to daylength. CCA1- and TOC1-like genes are found in the rice genome, and a CCA1-like gene was reported to exhibit circadian rhythms with a phase similar to that of CCA1 of Arabidopsis (Izawa et al., 2002, 2003). Furthermore, genes similar to Arabidopsis ZTL and ELF3, involved in light input to the clock, are also found in the rice genome (Izawa et al., 2003). These observations suggest that the components of the genetic network that controls flowering time of Arabidopsis in response to daylength are highly conserved in rice and that similar underlying mechanisms are likely to occur in both species.
Daylength Measurement in Rice
If rice utilizes similar molecular mechanisms to those of Arabidopsis to control flowering time in response to daylength, how is the reverse response to daylength generated in rice? Recent studies have suggested an answer to this question. Transgenic plants overexpressing Hd3a mRNA exhibit strong early flowering, indicating that Hd3a, similar to FT in Arabidopsis, acts as a floral promoter in rice (Kojima et al., 2002). However, Hd3a expression is induced specifically under SDs, and therefore shows the reverse regulation to that of FT in Arabidopsis (Kojima et al., 2002).
The daylength dependent regulation of Hd3a is mediated by Hd1. In Arabidopsis, CO induces FT expression under LDs and promotes flowering. In contrast, Hd1 was proposed to have two independent and opposite functions in the control of flowering time. This idea is based on the observation that loss of Hd1 function causes early flowering under LDs and late flowering under SDs (Yano et al., 2000). Transcription of Hd3a is altered in the se1 mutant (a loss-of-function mutant of Hd1) in ways consistent with its flowering phenotype; under LDs the transcript levels of Hd3a are increased in this mutant, whereas under SDs they are decreased (Izawa et al., 2002; Kojima et al., 2002). Notably, transcripts of Hd1 exhibit diurnal patterns under LDs and SDs in a phase similar to those of CO (Izawa et al., 2002; Kojima et al., 2002; Hayama et al., 2003). Thus, the mechanism by which Hd1 suppresses Hd3a and inhibits flowering under LDs may be explained in a similar way to the function of CO in Arabidopsis; under LDs, Hd1 is expressed at high levels at the mid to end of the day, and a coincidence between Hd1 expression and exposure to light may generate LD signals that inhibit Hd3a transcription and suppress flowering. Activation of Hd1 by light under LDs could be mediated by phytochrome, because loss of Se5 function does not largely alter the diurnal pattern of Hd1 or the circadian rhythms of several clock output genes such as CAB and CCA1-like genes, despite the severe early-flowering phenotype of se5 mutants under LDs (Izawa et al., 2002). Furthermore, the double mutant se5 se1 never flowers earlier than each single mutant under LDs, indicating that under LDs they inhibit flowering within the same genetic pathway. In contrast, the double mutant flowers later than se5 mutant, indicating that in the absence of Se5 function Hd1 promotes flowering under LDs (Izawa et al., 2002).
These observations provide a model of how Hd1 acts in wild-type plants to inhibit or promote flowering dependent on the daylength. Under LDs, Hd1 protein that is expressed at the end of the day is activated by phytochrome to inhibit flowering through inactivating Hd3a expression. In contrast, under SDs, Hd1 is not expressed during the day but is expressed during the night, when phytochrome is proposed to be inactivated, and this allows Hd1 to induce Hd3a expression and promote flowering under these conditions. The se1 mutant therefore exhibits early flowering under LDs because Hd1 is not present during the day to inhibit flowering, while this mutant exhibits late flowering under SDs because Hd1 is not expressed in the dark when it would promote flowering. The strong early-flowering phenotype of the se5 mutant irrespective of the daylength conditions may be explained because in this mutant, Hd1 is in a form that promotes flowering irrespective of the length of day or night, due to the lack of phytochrome activity (Izawa et al., 2002).
The critical molecular differences between rice and Arabidopsis that generate the differences in Hd3a/FT regulation are not yet clear. However, loss of Hd1 results in an increase in Hd3a mRNA levels under LDs, indicating that in rice an additional transcription factor is responsible for general up-regulation of Hd3a expression independent of Hd1 activity. Hd1 may suppress Hd3a through the inactivation of this transcription factor under LDs, but could induce Hd3a transcription in the dark through the enhancement of the activity of the transcription factor or through another mechanism. In contrast, a transcription factor that can activate FT autonomously may not be required in Arabidopsis. Furthermore, the opposite roles of Hd1 and CO in the control of Hd3a and FT transcription, respectively, may not be caused by differences in the proteins themselves, because examples have been described where the same transcriptional complex can induce or inhibit the transcription of genes directly, dependent on external signals (Eastburn and Han, 2004).
CONCLUSIONS AND PERSPECTIVES
Recent molecular-genetic studies of the daylength response of flowering in Arabidopsis have suggested mechanisms by which the LD signal is perceived during floral induction. For example, studies of FKF1 provided a mechanism for increasing transcript levels of CO at the critical time under LDs, so that CO activated by light can generate high levels of a LD signal. Moreover, studies of CO protein suggest a synegistic mechanism of amplifying a LD signal to a high level by the interaction of the diurnal regulation of CO mRNA by the circadian clock and the LD induction of CO protein by photoreceptors that occurs independently of the regulation of CO mRNA. These studies imply that in Arabidopsis, the LD signal for promotion of flowering is not generated by a simple interaction between circadian rhythms and light, suggested in the external coincidence model, but by complex interactions between several mechanisms. This machinery may enable plants to induce flowering effectively in response to a small change in daylength. Elaboration of the basic mechanisms identified so far will be necessary to fully understand this process. For example, analyses of CO protein have not yet identified the molecular mechanisms that are directly involved in light-dependent stability or degradation of CO protein. Isolation of mutants in which CO activation by light is altered or of proteins that physically interact with CO protein, may allow us to understand these mechanisms further. Studies of flowering time in rice have demonstrated that this plant utilizes similar genetic pathways to Arabidopsis for controlling flowering time and that the difference in the function of particular genes in a pathway contributes to the reverse response to daylength observed between LD and SD plants. Although the critical differences in the regulatory mechanisms between Arabidopsis and rice are not yet clear, several reciprocal experiments, in which for example the promoters of FT and Hd3a are exchanged between these plant species, may provide important information to understand this general question.
Finally, whether or not the molecular mechanism controlling the daylength response of flowering is conserved among plants that exhibit the same response to daylength has not been addressed. Several plant genera, such as Nicotiana and Lemna, include both SD and LD plants, suggesting that a daylength response can diverge rapidly during evolution. Therefore, the regulatory mechanism for flowering time could be different even in plants that exhibit the same response to daylength. Recently, CO homologs in Pharbitis were isolated and their roles in the control of flowering time tested (Liu et al., 2001). The molecular studies in Pharbitis, as well as other plant species, may help us to understand how the diversity in the photoperiodic pathways was generated during the evolution of the molecular mechanisms for daylength response of flowering in plants.
References
- Alabadi D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Bünning E (1936) Die endogene Tagesrhythmik als Grundlage der photoperiodischen Reaktion. Ber Dtsch Bot Ges 54: 590–607 [Google Scholar]
- Devlin PF, Kay SA (2000) Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell 12: 2499–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle MR, Davis SJ, Bastow RM, McWatters HG, Kozma-Bognar L, Nagy F, Millar AJ, Amasino RM (2002) The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419: 74–77 [DOI] [PubMed] [Google Scholar]
- Dunlap JC (1999) Molecular bases for circadian clocks. Cell 96: 271–290 [DOI] [PubMed] [Google Scholar]
- Eastburn D, Han M (2004) When Ras signaling reaches the mediator. Dev Cell 6: 158–159 [DOI] [PubMed] [Google Scholar]
- Garner WW, Allard HA (1920) Effect of the relative length of day and night and other factors of the environment on growth and reproduction in plants. J Agric Res 18: 553–606 [Google Scholar]
- Goto N, Kumagai T, Koornneef M (1991) Flowering responses to light-breaks in photomorphogenic mutants of Arabidopsis thaliana, a long-day plant. Physiol Plant 83: 209–215 [Google Scholar]
- Griffiths S, Dunford RP, Coupland G, Laurie DA (2003) The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol 131: 1855–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Yang H, Mockler TC, Lin C (1998) Regulation of flowering time by Arabidopsis photoreceptors. Science 279: 1360–1363 [DOI] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113 [DOI] [PubMed] [Google Scholar]
- Hayama R, Izawa T, Shimamoto K (2002) Isolation of rice genes possibly involved in the photoperiodic control of flowering by a fluorescent differential display method. Plant Cell Physiol 43: 494–504 [DOI] [PubMed] [Google Scholar]
- Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K (2003) Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422: 719–722 [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Tran HG, Swartz TE, Briggs WR, Kay SA (2003) FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426: 302–306 [DOI] [PubMed] [Google Scholar]
- Izawa T, Oikawa T, Sugiyama N, Tanisaka T, Yano M, Shimamoto K (2002) Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev 16: 2006–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T, Oikawa T, Tokutomi S, Okuno K, Shimamoto K (2000) Phytochromes confer the photoperiodic control of flowering in rice (a short-day plant). Plant J 22: 391–399 [DOI] [PubMed] [Google Scholar]
- Izawa T, Takahashi Y, Yano M (2003) Comparative biology comes into bloom: genomic and genetic comparison of flowering pathways in rice and Arabidopsis. Curr Opin Plant Biol 6: 113–120 [DOI] [PubMed] [Google Scholar]
- Jarillo JA, Capel J, Tang RH, Yang HQ, Alonso JM, Ecker JR, Cashmore AR (2001) An Arabidopsis circadian clock component interacts with both CRY1 and phyB. Nature 410: 487–490 [DOI] [PubMed] [Google Scholar]
- Johnson E, Bradley M, Harberd NP, Whitelam GC (1994) Photoresponces of light grown phyA mutants of Arabidopsis. Plant Physiol 105: 141–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M (2002) Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol 43: 1096–1105 [DOI] [PubMed] [Google Scholar]
- Ledger S, Strayer C, Ashton F, Kay SA, Putterill J (2001) Analysis of the function of two circadian-regulated CONSTANS-LIKE genes. Plant J 26: 15–22 [DOI] [PubMed] [Google Scholar]
- Liu J, Yu J, McIntosh L, Kende H, Zeevaart JA (2001) Isolation of a CONSTANS ortholog from Pharbitis nil and its role in flowering. Plant Physiol 125: 1821–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XL, Covington MF, Fankhauser C, Chory J, Wagner DR (2001) ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 13: 1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas P, Alabadi D, Yanovsky MJ, Oyama T, Kay SA (2003) Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 15: 223–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas P, Kim WY, Somers DE, Kay SA (2003) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426: 567–570 [DOI] [PubMed] [Google Scholar]
- McWatters HG, Bastow RM, Hall A, Millar AJ (2000) The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature 408: 716–720 [DOI] [PubMed] [Google Scholar]
- Millar AJ, Carre IA, Strayer CA, Chua NH, Kay SA (1995) Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267: 1161–1163 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song HR, Carre IA, Coupland G (2002) LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell 2: 629–641 [DOI] [PubMed] [Google Scholar]
- Nelson DC, Lasswell J, Rogg LE, Cohen MA, Bartel B (2000) FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101: 331–340 [DOI] [PubMed] [Google Scholar]
- Pittendrigh CS, Minis DH (1964) The entrainment of circadian oscillations by light and their role as photoperiodicclocks. Am Nat 108: 261–295 [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857 [DOI] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carre IA, Coupland G (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219–1229 [DOI] [PubMed] [Google Scholar]
- Somers DE, Devlin PF, Kay SA (1998) Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282: 1488–1490 [DOI] [PubMed] [Google Scholar]
- Somers DE, Schultz TF, Milnamow M, Kay SA (2000) ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101: 319–329 [DOI] [PubMed] [Google Scholar]
- Somers DE, Webb AA, Pearson M, Kay SA (1998) The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development 125: 485–494 [DOI] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Mas P, Panda S, Kreps JA, Kay SA (2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771 [DOI] [PubMed] [Google Scholar]
- Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120 [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Shomura A, Sasaki T, Yano M (2001) Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the α subunit of protein kinase CK2. Proc Natl Acad Sci USA 98: 7922–7927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B, Vince-Prue D (1997) Photoperiodism in Plants, Ed 2. Academic Press, London
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- Vaz Nunes M, Saunders D (1999) Photoperiodic time measurement in insects: a review of clock models. J Biol Rhythms 14: 84–104 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207–1217 [DOI] [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, et al (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12: 2473–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky MJ, Kay SA (2002) Molecular basis of seasonal time measurement in Arabidopsis. Nature 419: 308–312 [DOI] [PubMed] [Google Scholar]