The bodies of seed plants are comprised of two classes of organs with contrasting growth and symmetry attributes. Stems and roots are indeterminate organs that exhibit apical growth at apical meristems and radial growth at the vascular cambium, a pattern of growth that results in primarily radially symmetric organs. Lateral organs of the shoot, for example cotyledons, leaves, and floral organs, are determinate and exhibit localized planer growth resulting in breaking of radial symmetry and asymmetric development. Localized planer growth in the leaf generates the leaf blade, the principle site of photosynthesis in most plants.
The seed plant body may be described with reference to defined axes of symmetry (Fig. 1). The primary axis of the seed plant body is the apical-basal axis. The apical-basal axis is established during embryogenesis and runs from the center of the primary shoot apical meristem to the center of the primary root apical meristem (Fig. 1A, Axis 1). Distinct organ types are generated at specific positions along the apical-basal axis (root meristem, root, hypocotyl, cotyledon, stem-internode, and shoot apical meristem). Radially symmetric organs possess a second axis, the central-peripheral axis, which extends from the apical-basal axis at the center of the organ outward to the organ epidermis (Fig. 1, A and B, Axis 2). Asymmetric development along the central-peripheral axis is evident in the pattern of vascular tissue placement, with xylem developing in closer proximity to the apical-basal axis relative to phloem (Fig. 1B).
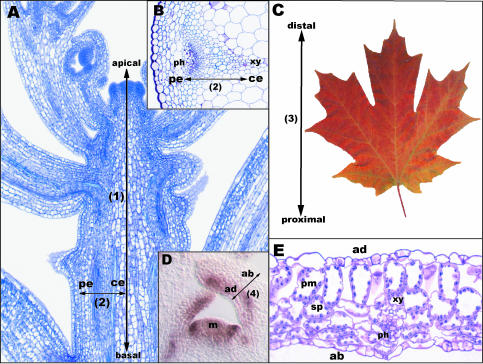
Figure 1.
Four axes of development in seed plants. The apical-basal axis (1) of the plant represents a polarity established in embryogenesis with the shoot apical meristem being at the apical end and the root meristem residing at basal tip shown here in a longitudinal section of an Arabidopsis shoot (A). The other 3 axes depicted are defined with reference to this apical-basal axis. The central (ce)-peripheral (pe) axis (2) in the stem (B) is analogous to the adaxial (ad)-abaxial (ab) axis (4) of lateral organs (D) with the central/adaxial end being adjacent to the center of the shoot and the peripheral/abaxial end being away from the center of the shoot meristem (m). Lateral organs such as leaves also exhibit a proximal-distal axis (3) that can also be defined with respect to the stem, with the distal end away from the stem and the proximal end attached to the stem, as shown in the Acer leaf in C. Positions of tissues within vascular bundles develop with respect to the central-peripheral axis with the phloem (ph) positioned peripherally, the xylem (xy) positioned centrally in the stem (B), and the xylem positioned adaxially and phloem abaxially in lateral organs (E). In addition to polar differentiation of vascular tissues, leaf asymmetry is evident with palisade mesophyll (pm) differentiating adaxially and spongy mesophyll (sp) positioned abaxially, as shown in E, a longitudinal section of an Arabidopsis leaf.
Lateral organs of the shoot arise from the flanks of shoot meristems and consequently possess an intrinsic positional relationship with the meristem from which they are derived. Two axes of lateral organs, the proximal-distal (Fig. 1C, Axis 3) and adaxial-abaxial (Fig. 1D, Axis 4) axes, may be readily defined with respect to the position of the lateral organ in relationship to the meristem from which the organ is derived. Both these axes are exploited as references for the regulation of asymmetrical development. The proximal-distal axis runs from the base of the organ (proximal, i.e. nearest to the meristem) to the tip (distal, i.e. furthest from the meristem; Fig. 1C). Asymmetric development along the proximal-distal axis is most evident in leaves, in that the amount of lateral growth frequently correlates with location along the proximal-distal axis in a manner that is highly predictable for a given species. The adaxial-abaxial axis runs from the surface of the lateral organ closest to the meristem (ad-adjacent) to the surface of the organ furthest from the meristem (ab-away; Fig. 1D). Asymmetric development in the adaxial-abaxial axis is most evident in many species in the leaf blade, where the adaxial epidermis and underlying mesophyll differentiate to develop characteristics specialized for efficient light capture, while the abaxial epidermis and mesophyll develop characteristics specialized for efficient gas exchange (Fig. 1E). Proper asymmetric development along the adaxial-abaxial axis is therefore often critical for development of a leaf architecture optimized for photosynthesis.
It should be noted that the central-peripheral axis of radially symmetric organs and the adaxial-abaxial axis of shoot lateral organs are equivalent. Both axes are defined with respect to the same reference, the apical-basal axis at the center of the shoot meristem. Moreover, the pattern of vascular tissue placement in lateral organs (xylem-adaxial, phloem-abaxial; Fig. 1E) is equivalent to that in radially symmetric organs (xylem-central, phloem-peripheral; Fig. 1B).
The ultimate basis of asymmetrical development in all multicellular organisms is the ability of cells to interpret their position with respect to an external reference and transduce this information into asymmetric patterns of cellular differentiation, a process termed polarity establishment. The positional relationship of lateral organs to the meristem from which they are derived indicates that the meristem could in principle serve as the reference for polarity establishment in lateral organs. That this is in fact the case is demonstrated by surgical experiments performed roughly 50 years ago (Warlaw, 1949; Sussex, 1955; Snow and Snow, 1959). When an incision is placed between the meristem and the site where a lateral organ primordia will next emerge (the P0 site), a lateral organ primordia emerges but develops as a radially symmetrical structure with apparently abaxial characteristics. Several tentative conclusions may be drawn from this simple experiment. First, some form of signal emanating from the meristem, transmission of which is interrupted by the presence of the incision, is required for interpretation of the adaxial-abaxial axis. Second, adaxial-abaxial polarity establishment is necessary for blade growth. Third, in the absence of adaxial-abaxial polarity establishment, abaxial identity may be the default state.
The surgical experiments provide a conceptual framework in which the results of subsequent genetic analyses of polarity establishment may be interpreted. Beginning in 1995 with characterization of the role of PHANTASTICA (PHAN) in promoting adaxial cell fate in Antirrhinum, the genetic basis of adaxial-abaxial polarity establishment in lateral organs has begun to be elucidated in Antirrhinum, maize, and Arabidopsis (Waites and Hudson, 1995; Tsiantis et al., 1999; Bowman et al., 2002). Results of these studies largely reinforce the conclusions derived from the early surgical experiments.
THE GENETIC BASIS OF POLARITY ESTABLISHMENT IN ARABIDOPSIS
Genetic studies in Arabidopsis have identified several families of genes that play a role in promoting proper adaxial-abaxial development (Table I; for review, see Bowman et al., 2002). The class III HD-Zip genes, in particular the PHABULOSA (PHB), PHAVOLUTA (PHV), and REVOLUTA (REV) genes (collectively referred to as PHAB genes) promote adaxial identity and meristem maintenance (McConnell and Barton, 1998; McConnell et al., 2001; Emery et al., 2003). The KANADI1-3 (KAN1-3) genes and members of the YABBY gene family promote abaxial identity (Eshed et al., 1999, 2001; Sawa et al., 1999; Siegfried et al., 1999; Kerstetter et al., 2001). Members of all three families are believed on the basis of homology to function as transcription factors. However, there remain significant gaps in our understanding of how interactions between these gene families direct polarity establishment and differentiation in the developing lateral organ. If these genes encode transcription factors as predicted, upon what promoters do they act? Do PHAB, KANADI, and YABBY proteins act in a linear polarity-promoting pathway or in parallel polarity-promoting pathways? Do PHAB, YABBY, and KANADI proteins function to interpret cellular position along the adaxial-abaxial axis (axis establishment), subsequently in regulating asymmetrical differentiation along a previously established axis (polar differentiation), or perhaps in both processes? As our understanding of polarity establishment deepens, it should even become apparent whether the distinction we have just drawn, of axis establishment and polar differentiation as discreet processes, accurately reflects the actual biological process of polarity establishment.
Table I.
Partial list of Arabidopsis genes referenced in the text with expression profiles
| Protein Family
|
Arabidopsis Gene
|
Expression in the Shoot
|
|
|---|---|---|---|
| Meristem | Lateral Organs | ||
| Class III HD-ZIP | PHABULOSA | Apical meristems | Adaxial |
| PHVOLUTA | Apical meristems | Adaxial | |
| REVOLUTA | Apical meristems | Adaxial | |
| ATHB8 | Procambium | Not detected | |
| ATHB15 | Procambium | Not detected | |
| YABBY | FILAMENTOUS FLOWER | Not detected | Abaxial |
| YABBY2 | Not detected | Abaxial | |
| YABBY3 | Not detected | Abaxial | |
| GARP/KANADI | KANADI1 | Not detected | Abaxial |
| KANADI2 | Not detected | Abaxial | |
| KANADI3 | Not detected | Abaxial | |
| MYB | ASYMMETRIC LEAVES1 | Not detected | Uniform |
| LOB-Domain | ASYMMETRIC LEAVES2 | Not detected | Abaxial |
| Class I KNOX | SHOOT MERISTEMLESS | Apical meristems | Not detected |
| BREVIPEDICELLUS | Apical meristems | Not detected | |
| KNAT2 | Apical meristems | Detected | |
| KNAT6 | Apical meristems | Detected | |
Conspicuously missing from the field of Arabidopsis polarity mutants is the equivalent of the phan mutant of Antirrhinum, in which a single loss-of-function mutation generates a dramatic adaxial to abaxial conversion in lateral organs (Waites and Hudson, 1995). Loss-of-function mutations of the Arabidopsis PHAN ortholog, ASYMMETRIC LEAVES1 (AS1), result in only a weakly abaxialized leaf phenotype (Byrne et al., 2000). What is the underlying basis for this apparent mechanistic divergence in polarity establishment between Arabidopsis and Antirrhinum?
In this update, we discuss several recent publications that provide insight into the questions stated above and consider how the new findings might best be incorporated into a framework of adaxial-abaxial polarity establishment in Arabidopsis.
CLASS III HD-ZIP GENES, VASCULATURE PATTERNING, AND MicroRNA REGULATION
The class III HD-ZIP genes in Arabidopsis are transcription factors that belong to a family composed of five members: PHB, PHV, and REV which comprise one clade and ATHB8 and ATHB15 which form a separate clade (Emery et al., 2003). All five proteins are characterized by an amino-terminal homeodomain/Leu zipper (HD-ZIP) followed by a region exhibiting sequence similarity to mammalian sterol/lipid-binding domain (START domain) (McConnell et al., 2001). Previous work has shown that PHB, PHV, and REV are involved in establishment of adaxial identity of lateral organs, as well as in the development of the apical meristem and the vascular bundles (McConnell and Barton, 1998; McConnell et al., 2001; Emery et al., 2003). All three genes exhibit similar mRNA expression patterns in apical and floral meristems, vasculature, and the adaxial domain of lateral organ primordia (McConnell et al., 2001; Otsuga et al., 2001; Emery et al., 2003). Gain-of-function mutations in PHB, PHV, and REV suggest that these genes might play slightly different roles in plant development, with PHB and PHV more important for lateral organ patterning and REV in vascular patterning. The other two family members, ATHB8 and ATHB15, are also likely to direct vascular development, although their precise roles are yet to be uncovered (Baima et al., 2001; Ohashi-Ito and Fukuda, 2003). Dominant phb and phv gain-of-function mutations result in an apparent abaxial to adaxial conversion and disruption of lateral growth manifested by filamentous and radially symmetric leaves (McConnell and Barton, 1998; McConnell et al., 2001). In contrast, dominant rev-10d mutations result in defects in vascular patterning in the stem, characterized by radialized amphivasal bundles consisting of xylem surrounding the phloem (Emery et al., 2003). Since these changes in vascular patterning are in the stem, they are independent of patterning events in the leaves.
Single loss-of-function mutations in PHB or PHV show no conspicuous effects on lateral organ polarity (McConnell et al., 2001; Emery et al., 2003), while rev loss-of-function plants fail to generate axillary meristems but exhibit normal leaf polarity and stem vascular patterning (Talbert et al., 1995). Since PHB, PHV, and REV exhibit a similar expression pattern it was postulated that these genes act redundantly to promote meristem development and adaxial fate in lateral organs. Indeed, the homozygous triple loss of function mutant, phb phv rev, exhibits a severe loss-of-polarity phenotype, with the apical part of the plant consisting of a single radialized cotyledon and lacking an apical meristem (Emery et al., 2003). Consistent with the hypothesis that these genes promote adaxial fates in lateral organs, the vascular pattern in the radialized cotyledon represents a mirror image of that observed in rev-10d gain-of-function mutant, with phloem surrounding the xylem, the predicted arrangement in abaxialized organs.
Molecular and genetic evidence suggests that PHB and KAN genes mutually repress each other's activity in leaf primordia giving rise to two adjacent distinct regions of expression (Eshed et al., 2001). The juxtaposition of adaxial-abaxial domains is proposed to promote lamina growth (Waites and Hudson, 1995). KANADIs and PHABs exhibit a complementary expression pattern not only in the leaf primordia but also in the vasculature. PHABs are expresses in the developing xylem (previously interpreted as the adaxial domain of the vascular bundle), while KANADIs are expressed in the phloem (the abaxial domain; Baima et al., 2001; Emery et al., 2003; Ohashi-Ito and Fukuda, 2003). Complementary loss- and gain-of-function leaf phenotypes of KANADI and PHAB genes indicate that these expression patterns represent functional domains. Also consistent with expression patterns, the stem vasculature phenotype of rev-10d gain-of-function and KANADI triple loss-of-function, kan1 kan2 kan3 are similar. In both mutants the stem vascular bundles are radialized with xylem surrounding phloem (Emery et al., 2003). Thus, the same genetic program controlling polarity in leaves generated from the apical meristem also patterns vascular tissues generated from the procambium in the stem. Since the spatial position of these tissues relative to the central axis of the shoot system is similar, a signal derived from the meristem could activate the PHBs both in the adaxial regions of the leaves and the central region of the stem, thus patterning tissues generated from both meristems (Emery et al., 2003).
All identified gain-of-function mutations in PHB, PHV, and REV occur in the START domain, indicating the importance of this region in regulating the expression of these genes. The START domain is predicted on the basis of sequence homology to encode a putative sterol/lipid binding domain, leading McConnell et al. (2001) to propose that PHB may be activated upon binding of a sterol-like molecule ligand, possibly a component of the meristem to leaf primordium signal. According to this model, low levels of PHB are expressed throughout lateral-organ anlagen, but as the leaf primordia develops, a ligand emanating from the meristem activates PHB in the adaxial side of primordia, which in turn promotes adaxial leaf fate. Positive feedback loops help to maintain PHB activity as well as the ligand stability and/or synthesis. Once the positive feedback activation is established, the developing leaf is self-sufficient and does not depend on the meristem for PHB activation and polarity maintenance. Following this model, the gain-of-function mutations in the START domain were initially proposed to render the proteins active in the absence of the ligand (McConnell et al., 2001). However, recent work has identified a different basis of the phb gain-of-function mutant phenotype: altered regulation by miRNAs.
MicroRNAs (miRNAs) are small RNAs 21 to 25 nucleotides in length that have the capacity to regulate gene expression at the posttranscriptional level (for review, see Bartel and Bartel, 2003; Carrington and Ambros, 2003). miRNAs were identified in plants more-or-less simultaneously by four different groups (Llave et al., 2002; Mette et al., 2002; Park et al., 2002; Reinhart et al., 2002). Recent work has shown that the PHABs are among the genes that are regulated by miRNA. Reinhart et al. (2002) isolated miRNAs from Arabidopsis by using techniques aimed at cloning RNAs 20 to 24 nucleotides long. They isolated 16 miRNAs, among them miRNA165 and miRNA166, derived from the precursor genes MIR165 and MIR166. Subsequently, Rhoades et al. (2002) searched for the Arabidopsis mRNAs that were complementary to those 16 previously identified miRNAs. Among the predicted miRNA targets were all five members of the class III HD-ZIP family, with miRNA165 potentially targeting PHB, PHV, REV, and ATHB8 mRNAs, and miRNA166 potentially targeting ATHB15 mRNA. Since both miRNA165 and miRNA166 exhibit nearly complete identity to a region within the START domains of all five PHABs, it is not yet clear whether the interactions listed above are specific.
The putative miRNA binding region overlaps with phb, phv, and rev gain-of-function mutations, suggesting that these genes could be regulated by miRNA and that alterations in the miRNA binding site are responsible for the gain-of-function phenotypes. A biochemical approach taken by Tang et al. (2003) provided experimental data supporting the theoretical predictions. PHV and PHB mRNAs incubated with wheat germ extract are cleaved at the sequence complementary to miR165, while the dominant gain-of-function phv mRNA remains intact. Efficient cleavage of the mutant mRNA can be achieved only when a synthetic miRNA with perfect complementarity to the mutant sequence is added to the extract, suggesting that the failure of the wild-type miRNA165 to cleave the mutant mRNA was a direct consequence of its reduced complementarity. Transgenic plants provide support for the role of miRNA165/166 in the regulation of PHABs in planta. Arabidopsis plants transformed with a REV cDNA carrying mutations altering the miRNA165/166 complementary site but not the translated REV protein, exhibit a phenotype similar to that of the rev-10d gain-of-function mutant (Emery et al., 2003). It was hypothesized that the disruption of the miRNA binding site leads to derepression of REV expression that in turn causes the adaxiallized phenotype.
Loss-of-function alleles of HASTY (hst) are affected in several processes in Arabidopsis and are weakly adaxialized (Bollman et al., 2003). In a kan1 pickle (pkl) background, mutations in hst enhances the loss of abaxial identity with the triple mutant characterized by development adaxial tissues, placenta and ovules, in abaxial positions in the carpel (Eshed et al., 2001). HST encodes a protein similar to the mammalian karyopherin exportin 5 (Xpo5), a nucleocytoplasmic transport factor (Bollman et al., 2003). Recently it has been demonstrated that Xpo5 mediates efficient export of miRNA precursors (pre-miRNA) from the nucleus to the cytoplasm where they are processed by RNAase III family members (DICER) to give the mature approximately 22-nucleotide miRNAs (Yi et al., 2003; Lund et al., 2004). Thus, one possible explanation for the adaxialized phenotype of hst mutants is that HST acts to promote abaxial identity through its postulated role in miRNA biogenesis. In this scenario, loss of HST activity would lead to a reduction in miRNA165/166 and derepression of the PHABs. HST belongs to the importin β family consisting of 17 predicted members in Arabidopsis. Redundancy in the importin β family might explain the weak polarity phenotype in the single hst mutant and the conspicuous adaxialized phenotype in the triple pkl kan1 hst mutant. In the absence of other genes promoting abaxial fates, such as KAN1, the polarity defect in HST is enhanced.
Despite new insights regarding miRNA regulation of the PHAB genes via the START domain, the previous sterol-ligand theory cannot be excluded since there is no data arguing against the function of the START domain as a ligand-binding site. On the contrary, the START domain might take part in both processes and serve as a receptor for a ligand emanating from the meristem, as well as an miRNA-binding site, which enables negative regulation of the PHABs.
INSIGHTS FROM FILAMENTOUS FLOWER EXPRESSION STUDIES
The role of the YABBY genes in promoting adaxial-abaxial polarity establishment is less clear than that of the PHAB and KANADI genes. This may be largely the result of apparent functional redundancy among YABBY family members, which to date has precluded analysis of a complete loss of YABBY function mutant, and to the fact that ectopic expression of YABBY genes generates only a partial adaxial to abaxial conversion of cellular identity (Sawa et al., 1999; Siegfried et al., 1999). The Arabidopsis YABBY family consists of six genes. Two of these, CRABS CLAW and INNER NO OUTER, show comparatively limited expression, being restricted to the carpel and ovule integuments, respectively (Bowman and Smyth, 1999; Villanueva et al., 1999). Three of the remaining YABBY genes, FILAMENTOUS FLOWER (FIL), YABBY3 (YAB3), and YAB2, are expressed more broadly in abaxial regions of the shoot lateral organs (Sawa et al., 1999; Siegfried et al., 1999). FIL and YAB3 are paralogous genes and possess substantial identity not only in their protein-encoding sequences but also over a sizeable stretch of their promoters.
In a recent study, Watanabe and Okada (2003) constructed FIL::GREEN FLUORESCENT PROTEIN (FIL::GFP) transcriptional fusions to provide a means of assaying FIL expression. Using a full-length FIL promoter, sufficient for complementation of a fil mutant phenotype, GFP fluorescence is restricted to the three most abaxial cell layers of leaf primordia with expression slightly expanded in the midrib, a pattern similar to that observed from in situ hybridization studies (Sawa et al., 1999; Siegfried et al., 1999). In principle, restriction of FIL expression to abaxial cell layers could be accomplished by several distinct mechanisms: induction in the abaxial region, repression in the adaxial region, or autonomous sorting of FIL into the abaxial region in response to a polarized signal. Watanabe and Okada (2003) proceeded to employ promoter deletion analysis to define several major cis elements regulating FIL expression, and the structure of the FIL promoter strongly favors a mechanism of adaxial repression over abaxial induction.
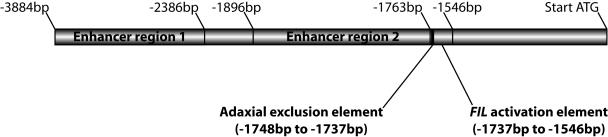
Successive deletions from the 5′-end of the full-length FIL promoter first result in stepwise reduction in GFP fluorescence intensity, consistent with the removal of promoter elements required for enhancement of basal transcription (Fig. 2). Further deletion identified a promoter element that when deleted results in GFP fluorescence throughout organ primordia rather than being restricted to the abaxial regions. A series of three nucleotide substitutions within this region were employed to define a 12-nucleotide sequence as necessary for restricting FIL expression to the three abaxial most cell layers of leaf primordia. (Fig. 2) Here we will refer to the 12-nucleotide sequence as the adaxial exclusion (AdEx) element. The AdEx element lies within a region of the FIL promoter with substantial identity to the corresponding region of the YAB3 promoter. Expansion of GFP fluorescence resulting from constructs lacking the AdEx element relative to constructs with an intact AdEx element is evident as early as GFP fluorescence can be detected, in the organ anlagen prior to primordia emergence (Fig. 2). Deletion of a second promoter element, located 3′ of the AdEx element and also within the region of high FIL-YAB3 promoter identity, abolishes GFP fluorescence. This promoter element remains to be defined as precisely as the AdEx element, although stretches of near-perfect identity between FIL and YAB3 promoters provide targets for nucleotide substitution experiments (Fig. 2).
Figure 2.
Schematic representation of the FIL promoter as elucidated by Watanabe and Okada (2003). The start-codon in the first FIL exon is located at the far right. Position 5′ of the Start-codon is indicated above the promoter. Regions containing the two enhancer elements and the FIL activation element are delineated by solid bars. The 12-bp Adaxial exclusion element is defined by the solid box. Relative sizes of all regions are depicted to scale.
What is the adaxial repressor of FIL? Under the regulation of the full-length promoter, FIL appears to be restricted to the abaxial regions of the anlagen, indicating that axis establishment occurs early in the establishment of the anlagen and precedes FIL induction. Consistent with this interpretation, deletion of the AdEx element results in the expansion of FIL expression within the anlagen, suggesting FIL, and possibly other YABBYs, are more likely to play a role in asymmetrical differentiation in response to a previously established adaxial-abaxial axis than to play a role in establishment of the axis itself. This favors a model where repression of FIL in adaxial regions is under the control of a regulator that itself is adaxially localized prior to the induction of FIL. FIL expression is greatly reduced as early as the organ anlagen stage in the gain-of-function phb-1d mutant (Siegfried et al., 1999), making PHB, whose expression domain corresponds to the region of primordia in which FIL is excluded, an attractive candidate for regulating adaxial repression of FIL. Alternatively, Watanabe and Okada (2003) suggest that the AdEx element is similar to target promoter elements of the Drosophila transcriptional repressor Kruppel and may be the target of an Arabidopsis Kruppel homolog.
YABBY MEDIATED REPRESSION OF KNOX HOMEOBOX GENES
While fil and yab3 mutants individually have no conspicuous aberrant leaf phenotype, fil yab3 double mutants exhibit partial loss of abaxial characteristics, most evident in the abaxial epidermis, and partial radialization of floral organs (Siegfried et al., 1999; Kumaran et al., 2002). Leaves of fil yab3 double mutants also exhibit occasional bifurcation and initiation of ectopic shoot meristems, either at the site of bifurcation or on the adaxial leaf surface, most commonly in the more proximal regions of the blade and close to the leaf margins. Initiation of meristems on the leaf lamina is not a normal feature of either adaxial or abaxial leaf domains, nor is it observed in the most severe loss-of-polarity mutants. These observations indicate that formation of ectopic meristems in the leaf is not strictly a polarity defect per se, but rather reflects a partial loss of organ determinacy. However, that ectopic meristems arise only at points of bifurcation or on the adaxial surface and not on the abaxial leaf surface suggests a link between organ polarity and organ determinacy. This is consistent with earlier observations that expansion of adaxial identity promotes ectopic meristem initiation, while expansion of abaxial identity is antagonistic to meristem initiation and maintenance (McConnell and Barton, 1998; Eshed et al., 2001; Kerstetter et al., 2001). Production of ectopic meristems on the adaxial leaf surface is also characteristic of plants that constitutively express class I Knox genes (Chuck et al., 1996; Byrne et al., 2000). In Arabidopsis, the class I Knox gene family is comprised of the SHOOTMERISTEMLESS (STM), BREVIPEDICELLUS (BP, formerly KNAT1), KNAT2, and KNAT6 genes. Class I Knox genes are homeobox containing transcription factors that promote meristem indeterminacy and are down-regulated in the organ anlagen (Vollbrecht et al., 1991; Long et al., 1996; Byrne et al., 2002).
The phenotypes of 35S::BP and fil yab3 plants indicate that class I Knox genes promote and YABBY genes repress meristem initiation in the leaf. The possibility that class I Knox genes are repressed by YABBY genes is the focus of a recent investigation by Kumaran et al. (2002) who employed reverse transcription (RT)-PCR to analyze transcript levels of three class I Knox genes, STM, BP, and KNAT2, in leaves of wild-type, fil, yab3, and fil yab3 plants (Kumaran et al., 2002). Transcripts of STM and BP are not detected in wild-type leaves but are present in fil, yab3, and fil yab3 leaves. Transcripts of KNAT2 are detected at low levels in wild-type leaves but at higher levels in fil, yab3, and fil yab3 leaves. The fil yab3 leaves selected for analysis did not possess ectopic meristems, a fact verified by the absence of detectable WUSCHEL transcripts, precluding the possibility that ectopic meristems themselves are the source of elevated Knox gene transcript levels. These results are consistent with a model whereby FIL and YAB3 repression of meristem initiation in the leaf is mediated by repression of class I Knox genes.
The Kumaran et al. (2002) RT-PCR analysis also yielded intriguing indications of at least partial nonredundancy of function between FIL and YAB3. STM is detected in fil leaves at higher levels than in yab3 leaves and at the highest levels in fil yab3 leaves. Conversely, BP is detected at higher levels in yab3 relative to fil leaves and at the highest levels in fil yab3 leaves. KNAT2 is detected at similar levels in both fil and yab3 leaves but at a higher level in fil yab3 leaves. If these results accurately reflect Knox gene levels in the leaf, they indicate that in the context of Knox repression, YABBY genes act with only partial redundancy. However, a caveat in advancing these interpretations is that Kumaran et al. (2002) did not employ quantitative analysis of Knox gene expression levels and therefore these interpretations must be regarded as preliminary, pending confirmation of relative Knox gene levels by quantitative analysis.
Do FIL and YAB3 repress Class I Knox gene expression throughout leaf primordia or only within their domain of abaxial expression? Introduction of a BP::GUS reporter construct into a fil mutant background resulted in GUS expression restricted to the abaxial margin of leaf primordia, consistent with YABBY mediated repression of class I Knox genes being limited to the YABBY expression domain and possibly occurring in a cell-autonomous manner. That KNOX proteins may act non-cell-autonomously by moving from the abaxial to the adaxial regions of the leaf (Wu et al., 2002; Kim et al., 2003), and that a combination of KNOX and PHAB activity may be required for the de novo formation of shoot meristems (Emery et al., 2003), could explain why ectopic meristems develop exclusively from the adaxial leaf surface although ectopic Knox gene expression in fil yab3 plants is restricted to the abaxial regions of leaves.
AS1, which we have previously noted as the Arabidopsis ortholog of PHANTASTICA, encodes a Myb family transcription factor and is expressed throughout organ primordia (Byrne et al., 2000). AS2 encodes a LOB-domain family transcription factor and is expressed in the adaxial region of leaf primordia (Iwakawa et al., 2002). as1 and as2 mutants have similar leaf morphology phenotypes, suggesting that AS1 and AS2 act in the same genetic pathway. This idea has been further bolstered by yeast two-hybrid experiments demonstrating that AS1 and AS2 dimerize in yeast cells, suggesting that they act in a single protein complex (Xu et al., 2003). Ectopic expression of class I Knox genes in the leaf is characteristic of both as1 and as2 mutants, and as in fil yab3 mutants, strong as1 mutants occasionally produce ectopic meristems on the adaxial leaf surface (Byrne et al., 2000), although with a notably lower frequency than in fil yab3 mutants. However the effects of fil yab3 compared to as1 and as2 on leaf morphology are quite dissimilar, with fil yab3 plants exhibiting partially adaxialized leaves with reduced lateral growth, and as1 and as2 plants exhibiting lobed, rumpled leaves with an expanded midvein.
What is the significance of class I Knox gene repression in leaf primordia being under the control of polarly localized factors? Is failure to repress class I Knox genes the basis of the as1 and as2 leaf morphology phenotype or is AS1/AS2 multifunctional along the lines of the YABBY genes? Returning to a question proposed earlier, what is the basis of the apparent mechanistic divergence between the PHAN gene of Antirrhinum and its ortholog AS1 in Arabidopsis? Some perspective on the last two questions has recently been provided by results indicating that AS1/AS2 promote adaxial identity in the leaf.
PROMOTION OF ADAXIAL IDENTITY BY ASYMMETRIC LEAVES2
Recent studies have employed constitutive expression of AS2 under the regulation of the 35S promoter to examine the function of AS2 in leaf development (Iwakawa et al., 2002; Lin et al., 2003; Xu et al., 2003). This approach partially circumvents the problem of functional redundancy complicating interpretation of loss-of-function phenotypes. 35S::AS2 transgenic plants exhibit a range of phenotypic severity with some lines exhibiting striking similarities to kan1 kan2 plants (Eshed et al., 2001), including partially radialized cotyledons and leaves and the formation of outgrowths on the abaxial leaf surface, indicating the possible repression of KANADI by AS1/AS2. More severe phenotypes exhibit arrest of shoot and root meristems. The arrangement of the vascular tissues in the leaf is altered in 35S::AS2 plants with xylem cells largely surrounding associated phloem. Alterations are also evident in the mesophyll where cells resembling palisade mesophyll, normally restricted to the layer of cells adjacent to the adaxial epidermis, are located around the entire circumference of the leaf. Collectively, these phenotypes are consistent with an expansion of adaxial leaf identity, implicating a role for AS2 in adaxial-abaxial leaf polarity.
Epidermal cells of the adaxial and abaxial leaf surfaces display characteristic features of size, shape, and distribution of stomata and leaf hairs. Although the size and shape of epidermal cells in the leaves and petals of 35S::AS2 plants are subtly altered, there is no clear abaxial to adaxial conversion. This pattern of tissue conversion is essentially the opposite of that observed in 35S::YAB3 plants, where adaxial to abaxial conversions are most apparent in the epidermis but are not conspicuous in the mesophyll (Siegfried et al., 1999). These results may be interpreted as indicating that the epidermis is more sensitive to YABBY activity than internal tissues and conversely that internal tissues are more sensitive to AS1/AS2 activity than epidermal tissues.
Consistent with the phenotype of ectopic expression of class I Knox genes in as1 and as2 mutants, Lin et al. (2003) observed reduced levels of BP, KNAT2, and KNAT6 in 35S::AS2 seedlings by RT-PCR. Changes in STM expression levels are not apparent in 35S::AS2 seedlings. This profile of AS2 regulation of class I Knox genes agrees with earlier studies demonstrating enhanced expression of BP, KNAT2, and KNAT6 but not STM in as2 leaves (Semiarti et al., 2001). These results highlight a potential divergence between YABBY and AS1/AS2 repression of class I Knox genes, in that FIL/YAB3 appear to be required for repression of STM in the leaf while AS1/AS2 does not. As we noted earlier, there is an apparent link between polarity and regulation of leaf determinacy in that ectopic meristems are formed on the adaxial but not the abaxial leaf surface. Restriction of meristem initiation to the adaxial leaf surface may reflect a requirement for PHB (Emery et al., 2003), itself restricted to the adaxial domain of the leaf, in meristem initiation. This interpretation is consistent with the initiation of axillary meristems in the abaxial base of the leaf in semidominant gain-of-function phb-1d mutants (McConnell and Barton, 1999).
Lin et al. (2003) examined the expression of KAN1, KAN2, FIL, YAB3, and PHB in the as2 mutant and in 35S::AS2 transgenic plants. No changes in level of expression are detectable for any of the polarity genes examined in as2 relative to wild type, indicating that AS1/AS2 alone is dispensable for normal regulation of these polarity genes, a finding consistent with the weak polarity phenotypes of as1 and as2 mutants. In contrast, expression of the abaxial genes, KAN1, KAN2, FIL, and YAB3, is reduced in 35S::AS2 while the expression level of the adaxial promoting PHB is increased, consistent with the interpretation of the 35S::AS2 phenotype as adaxialized. Ectopic expression of AS2 is therefore sufficient to induce partial abaxial to adaxial conversion at both the molecular and morphological levels.
Until recently, one possible explanation for the strongly abaxialized phenotype of phan mutants and the comparatively weakly abaxialized phenotype of as1 mutants is that promotion of adaxial identity is not a conserved function of AS1. This possibility is effectively eliminated by the results of the Lin et al. (2003) study that clearly implicate the AS1/AS2 complex as a promoter of adaxial identity. An alternative, and still viable, explanation for the weak polarity phenotype of as1 and as2 mutants is that AS1/AS2 activity is redundant for the promotion of adaxial identity with other factors. Candidates for this role include the PHAB genes, which along with AS1/AS2 are predicted to repress the KANADIs. Perhaps the Antirrhinum ortholog of PHB is less effective in repression of the Antirrhinum KANADI orthologs than is observed in Arabidopsis. Alternatively, factors acting redundantly with AS1/AS2 may remain to be identified or recognized as functioning in promoting adaxial identity. The latter possibility seems plausible given that the loss-of-function phenotype of such genes would be predicted to be only weakly abaxialized.
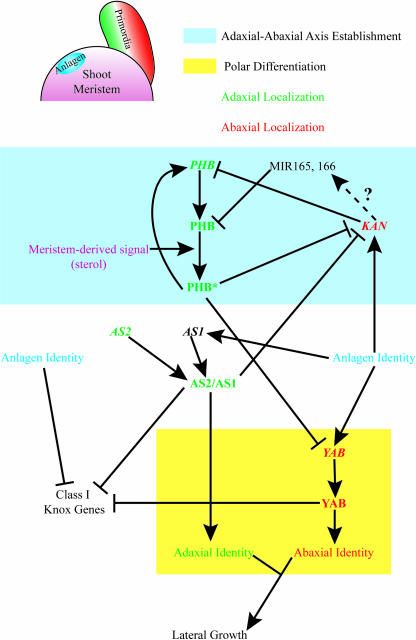
Where does AS1/AS2 act in the genetic network of polarity establishment? Lin et al. (2003) examined the relative levels of AS2 in wild-type plants relative to polarity mutants in an effort to place AS2 within the pathway of genetic regulation of adaxial-abaxial polarity establishment. No change in AS2 expression is evident in kan1 kan2 seedlings relative to wild type. However fil yab3 seedlings exhibit higher levels of AS2 relative to wild type. This result is consistent with several possible scenarios, including negative regulation of AS2 by YABBY genes or positive feedback on AS2 transcription by elevated levels of class I Knox genes resulting from fil yab3 loss-of-function. AS2 levels are also higher in gain-of-function phb-1d mutants relative to wild-type plants, an interesting result in light of the fact that levels of PHB are increased in the 35S::AS2 transgenic background. One interpretation of these results is that AS2 and PHB act in parallel adaxial promoting pathways that mutually reinforce one another. In Figure 3 we summarize our current interpretation of adaxial-abaxial polarity establishment in Arabidopsis.
Figure 3.
Conceptual framework of adaxial-abaxial polarity establishment in lateral organs of Arabidopsis. Organs arise from the flanks of shoot meristems. The organ anlage is marked by the expression of AS1 and concomitant repression of class I Knox genes. The apical meristem is the source of a putative signal(s). Reception of the signal activates the PHB protein (PHB*). Activated PHB* promotes PHB transcription and represses transcription of the YABBY and KANADI genes. PHB in turn is negatively regulated at the transcriptional and posttranscriptional levels by the KANADI genes and microRNAs 165,166, respectively. The dashed line linking KAN to MIR165,166 reflects the hypothesis that negative regulation of PHB expression by KANADIs could occur in part or in whole through KANADI activation of MIR165,166 transcription. PHB and KANADI are predicted to sort into adaxial and abaxial domains of expression, respectively, and this may establish the adaxial-abaxial axis of the lateral organ. Expression of YABBY genes is predicted based on the structure of the FIL promoter, to be localized to the abaxial domain of the organ anlagen by a combination of transcriptional activation throughout the anlage coupled with repression in the adaxial region. YABBY genes can promote differentiation of abaxial cell fates. AS2 is predicted to be expressed in the adaxial organ domain where AS2 forms a complex with AS1 (AS2/AS1) to promote repression of KANADI and differentiation of adaxial cell fates. Interactions between AS1/AS2 and other components of polarity establishment remain unclear, as does the extent to which AS1/AS2 promotes axis establishment, polar differentiation, or both processes. Both YABBY genes and AS1/AS2 repress expression of class I Knox genes. Juxtaposition of adaxial and abaxial organ domains is a requirement for the establishment of lateral growth.
STILL SOMETHING TO BE LEARNED FROM SURGICAL EXPERIMENTS
The earliest insights into the mechanism of adaxial-abaxial polarity establishment in vascular plants were obtained from experiments employing surgical modification of the apical meristem (Warlaw, 1949; Sussex, 1955; Snow and Snow, 1959). These experiments have provided an important conceptual framework for interpreting the results of genetic studies. The reverse is also true. “It is of considerable interest to interpret the surgical experiments in the framework of the recent molecular models, and vice versa” (Reinhardt et al., 2003). Reinhardt et al. (2003) recently employed microsurgical and laser ablation removal of meristem regions coupled with analysis of the expression of the LeWUSCHEL and LeT6 genes to elucidate the molecular basis of meristem regeneration and the function of the L1 meristem layer in regulating meristem growth and organ production. Precise ablation of meristem cells coupled with gene expression analysis could be similarly applied to answering important questions of adaxial-abaxial polarity establishment. For example, PHB and PHV are modeled as receptors of a putative meristem-derived sterol signal and are predicted to autoregulate their expression. How does ablation of cells between the meristem and the P0 site, known to disrupt adaxial-abaxial polarity establishment, affect PHB expression and PHB localization? Is ablation of the L1 layer between meristem and the P0 site sufficient to disrupt adaxial-abaxial polarity establishment or are deeper ablations involving multiple meristem layers required? Do the radially symmetrical, abaxialized organs that result from incision between the meristem and P0 exhibit expanded expression of KANADI and/or YABBY genes? The time may be ripe for the consolidation of surgical and molecular approaches to the study of lateral organ polarity.
Acknowledgments
We thank Sandra Floyd and Stuart Baum for contributing images to Figure 1.
This work was supported by the NSF (grant nos. IBN 9986054 and IBN 0077984 to J.L.B.), by the NRICGP (project no. CALR–2003–02600 to E.M.E.), and by BARD (postdoctoral fellowship no. FI–314–2001 to A.I.).
References
- Baima S, Possenti M, Matteucci A, Wisman E, Altamura MM, Ruberti I, Morrelli G (2001) The Arabidopsis ATHB-8 HD-ZIP protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol 126: 643–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel B, Bartel DP (2003) MicroRNAs: at the root of plant development? Plant Physiol 132: 709–717 [DOI] [PMC free article] [PubMed]
- Bollman KM, Aukerman MJ, Park M, Hunter C, Berardini TZ, Poethig RS (2003) Hasty, the Arabidopsis ortholog of exportin 5/MSN5, regulates phase change and morphogenesis. Development 130: 1493–1504 [DOI] [PubMed] [Google Scholar]
- Bowman J, Eshed Y, Baum SF (2002) Establishment of polarity in angiosperm lateral organs. Trends Genet 18: 134–141 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR (1999) CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126: 2387–2396 [DOI] [PubMed] [Google Scholar]
- Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA (2000) Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408: 967–971 [DOI] [PubMed] [Google Scholar]
- Byrne ME, Simorowski J, Martienssen RA (2002) ASYMMETRIC LEAVES1 reveals Knox gene redundancy in Arabidopsis. Development 129: 1957–1965 [DOI] [PubMed] [Google Scholar]
- Carrington JC, Ambros V (2003) Role of microRNAs in plant and animal development. Science 310: 336–338 [DOI] [PubMed] [Google Scholar]
- Chuck G, Lincoln C, Hake S (1996) KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8: 1277–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL (2003) Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol 13: 1768–1774 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Bowman JL (1999) Abaxial cell fate in the carpels is established by two distinct mechanisms. Cell 99: 199–209 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Perea JV, Bowman JL (2001) Establishment of polarity in lateral organs of plants. Curr Biol 11: 1251–1260 [DOI] [PubMed] [Google Scholar]
- Iwakawa H, Ueno Y, Semiarti E, Onouchi H, Kojima S, Tsukaya H, Hasebe M, Soma T, Ikezaki M, Machida C, et al (2002) The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol 43: 467–478 [DOI] [PubMed] [Google Scholar]
- Kerstetter RA, Bollman K, Taylor RA, Bomblies K, Poethig RS (2001) KANADI regulates organ polarity in Arabidopsis. Nature 411: 706–709 [DOI] [PubMed] [Google Scholar]
- Kim JY, Yuan Z, Jackson D (2003) Developmental regulation and significance of KNOX protein trafficking in Arabidopsis. Development 130: 4351–4362 [DOI] [PubMed] [Google Scholar]
- Kumaran MK, Bowman JL, Sundaresan V (2002) YABBY polarity genes mediate the repression of KNOX homeobox genes in Arabidopsis. Plant Cell 14: 2761–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Shuai B, Springer PS (2003) The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell 15: 2241–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C, Kasschau KD, Rector MA, Carrington JC (2002) Endogenous and silencing-associated small RNAs in plants. Plant Cell 14: 1605–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK (1996) A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U (2004) Nuclear export of miRNA precursors. Science 303: 95–98 [DOI] [PubMed] [Google Scholar]
- McConnell JR, Barton MK (1998) Leaf polarity and meristem formation in Arabidopsis. Development 125: 2935–2942 [DOI] [PubMed] [Google Scholar]
- McConnell JR, Emery JF, Eshed Y, Bao N, Bowman J, Barton MK (2001) Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411: 709–713 [DOI] [PubMed] [Google Scholar]
- Mette MF, van der Winden J, Matzke AJ (2002) Short RNA can identify new candidate transposable element families in Arabidopsis. Plant Physiol 130: 6–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi-Ito K, Fukuda H (2003) HD-ZIP III homeobox genes that include a novel member, ZeHB-13 (Zinnia)/ATHB-15 (Arabidopsis), are ivolved in procambium and xylem cell differentiation. Plant Cell Physiol 44: 1350–1358 [DOI] [PubMed] [Google Scholar]
- Otsuga D, DeGuzman B, Prigge MJ, Drews GN, Clark SE (2001) REVOLUTA regulates meristem initiation at lateral positions. Plant J 25: 223–236 [DOI] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X (2002) CARPEL FACTORY, a dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis Thaliana. Curr Biol 12: 1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Frenz M, Mandel T, Kuhlemeier C (2003) Microsurgical and laser ablation analysis of interactions between the zones and layers of the tomato shoot apical meristem. Development 130: 4073–4083 [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) MicroRNA in plants. Genes Dev 16: 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP (2002) Prediction of plant microRNA targets. Cell 110: 513–520 [DOI] [PubMed] [Google Scholar]
- Sawa S, Watanabe K, Goto K, Liu YG, Shibata D, Kanaya E, Morita EH, Okada K (1999) FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev 13: 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semiarti E, Ueno Y, Tsukaya H, Iwakawa H, Machida C, Machida Y (2001) The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128: 1771–1783 [DOI] [PubMed] [Google Scholar]
- Siegfried KR, Eshed Y, Baum SF, Otsuga D, Drews GN, Bowman JL (1999) Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126: 4117–4128 [DOI] [PubMed] [Google Scholar]
- Snow M, Snow R (1959) The dorsoventrality of leaf primordia. New Phytol 58: 188–207 [Google Scholar]
- Sussex IM (1955) Morphogenesis in Solanum tuberosum L.: experimental investigation of leaf dorsoventrality and orientation in the juvenile shoot. Phytomorphology 5: 286–300 [Google Scholar]
- Talbert PB, Adler HT, Parks DW, Comai L (1995) The REVOLUTA gene in necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development 121: 2723–2735 [DOI] [PubMed] [Google Scholar]
- Tang G, Reinhart BJ, Bartel DP, Zamore P (2003) A biochemical framework for RNA silencing in plants. Genes Dev 17: 49–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiantis M, Schneeberger R, Golz JF, Freeling M, Langdale JA (1999) The maize rough sheath2 gene and leaf development programs in monocot and dicot plants. Science 284: 154–156 [DOI] [PubMed] [Google Scholar]
- Villanueva JM, Broadhvest J, Hauser BA, Meister RJ, Schneitz K, Gasser CS (1999) INNER NO OUTER regulates abaxial-adaxial patterning in Arabidopsis ovules. Genes Dev 13: 3160–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht E, Veit B, Sinha N, Hake S (1991) The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature 350: 241–243 [DOI] [PubMed] [Google Scholar]
- Waites R, Hudson A (1995) phantastica: a gene required for dorsiventrality of leaves in Antirrhinum majus. Development 121: 2143–2154 [Google Scholar]
- Warlaw CW (1949) Experiments on organogenesis in ferns. Growth 9: 93–131 [Google Scholar]
- Watanabe K, Okada K (2003) Two discrete cis elements control the abaxial side-specific expression of the FILAMENTOUS FLOWER gene in Arabidopsis. Plant Cell 15: 2592–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Weigel D, Wigge PA (2002) Signaling in plants by intercellular RNA and protein movement. Genes Dev 16: 151–158 [DOI] [PubMed] [Google Scholar]
- Xu L, Xu Y, Dong A, Sun Y, Pi L, Xu Y, Huang H (2003) Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development 130: 4097–4107 [DOI] [PubMed] [Google Scholar]
- Yi R, Oin Y, Macara IG, Cullen BR (2003) Exportin-5 mediates the nuclear export of pre-miRNAs and short hairpin RNAs. Genes Dev 17: 3011–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]