The discovery that one's kitchen has been invaded by mice is often made indirectly. The holes chewed in the muesli bag and the teeth marks on the corn flakes box are a dead give away. Although you have not seen the mouse, you deploy your defensive weapons, and if successful, succeed in protecting your valuable goods from the invasion, hopefully before all of your food has been eaten. Recent research results indicate that plants also make use of such indirect surveillance systems to protect themselves from being consumed by pathogens. Rather than wait for a direct observation of the pest, plants appear to activate their defenses as soon as pathogen-induced damage is detected. As I describe in this Update, recent work in Arabidopsis has provided compelling evidence that such indirect surveillance may be the rule, rather than the exception, when it comes to plant alarm systems.
PATHOGEN RECOGNITION AND GENE-FOR-GENE INTERACTIONS
The idea that plants may detect pathogens by indirect mechanisms is still relatively new (van der Biezen and Jones, 1998b) and goes against dogma that has pervaded the plant pathology field for decades. This dogma posited that plants detected pathogens using receptors that directly bound pathogen-derived elicitor molecules (Gabriel and Rolfe, 1990). This receptor-ligand model grew out of classical genetic work performed by H.H. Flor over 50 years ago (Flor, 1956). Flor studied interactions between flax (Linum usitatissimum) and flax rust. He observed that the difference between flax varieties that were resistant versus those that were susceptible to specific rust strains could usually be attributed to a single dominant resistance (R) gene present in the resistant plant. Likewise, the difference between pathogen strains that were avirulent versus virulent on specific flax varieties could usually be attributed to a single dominant avirulence (avr) gene in the avirulent strain. Resistance was observed only when a specific R gene in flax was matched with a corresponding avr gene in the pathogen. Based on his genetic analyses, Flor likened such plant-pathogen interactions to antibody-antigen interactions in mammals, leading to speculation that Avr genes either encoded the antigens or encoded enzymes that produced the antigens. Such gene-for-gene interactions have been subsequently described in numerous crop and wild plant species, including Arabidopsis.
MOLECULAR ANALYSES OF PLANT R GENES
Over 40 R genes with recognition specificity for specific pathogen strains have been isolated from 10 plant species, including both monocots and dicots (for review, see Martin et al., 2003). The proteins encoded by these genes can be grouped into four general classes based on predicted structures: transmembrane receptor kinases, transmembrane receptors without a kinase domain, cytosolic kinases, and cytosolic proteins that contain a putative nucleotide binding site (NB). With the exception of the cytosolic kinase class, all of these proteins contain Leu rich repeats (LRRs). In other proteins, LRRs are known to participate in protein to protein interactions, including binding of peptide ligands by receptors (Kobe and Kajava, 2001). Careful sequence analysis of R gene families revealed that the putative ligand binding surface of LRR domains in many families are undergoing diversifying selection (Parniske et al., 1997; Meyers et al., 1998), meaning that mutations that give rise to amino acid substitutions outnumber silent mutations. This observation is consistent with idea that the LRR domains of R proteins might directly bind pathogen-derived proteins.
Of the four classes of plant R genes, the NB-LRR class is by far the largest. R genes in this class have been shown to confer resistance to viral, bacterial, oomycete, and fungal pathogens, and even to nematodes and aphids (Martin et al., 2003). Ten Arabidopsis R genes of known function have been cloned and all belong to the NB-LRR class. Analysis of the complete Arabidopsis genomic sequence revealed 149 genes containing both an NB and LRR domain (Meyers et al., 2003). NB-LRR genes can be further subdivided into two major subclasses based on their N-terminal domains. TIR-NB-LRR genes have similarity to Toll and Interleukin 1 receptors, while the non-TIR class typically contains a coiled-coil (CC) domain (Meyers et al., 2003). Phylogenetic trees based on only the NB domain separate the TIR and non-TIR classes from each other, indicating that these classes do not recombine (Meyers et al., 1999). Even with the addition of transmembrane receptor-type R genes, Arabidopsis is unlikely to contain more than 250 functional R genes. An interesting question to ponder is how Arabidopsis can detect the multitude of potentially infectious pathogens with less than 250 R genes. If the above indirect surveillance hypothesis is correct, plants do not need to detect a multitude of specific pathogen molecules but only the damage caused by them.
NB-LRR proteins lack obvious transmembrane domains or signal peptides and are presumed to be located in the plant cell cytoplasm. A cytoplasmic location for plant R proteins came as a surprise, initially, as most models had predicted them to be cell surface receptors. However, it was soon established that plant bacterial pathogens use a type III secretion system (TTSS) to translocate avirulence proteins into host cells (Alfano and Collmer, 1996). Several groups subsequently showed that expression of bacterial avirulence genes in plant cells, either transiently or in stable transgenic plants, induces cell death in an R gene-specific manner. This programmed cell death response is essentially the same as that induced by avirulent pathogens and is referred to as the hypersensitive response (HR). The HR is believed to be the endpoint of a complex suite of physiological changes that limit pathogen growth by producing toxic metabolites and depriving the pathogen of nutrients and water.
Most recently, similar experiments with Avr proteins from Flor's flax rust pathogen have revealed that these are also detected from inside plant cells (Dodds et al., 2004), indicating that at least some fungal pathogens also translocate Avr/virulence proteins across host cell membranes. This latter work is particularly noteworthy, because coexpression of the flax NB-LRR protein L6 and the fungal Avr protein AvrL567 in tobacco (Nicotiana tabacum) induced an HR-like response, while expression of either protein alone did not. This result implies that the flax L6 protein can activate the cell death program in tobacco in an Avr-dependent manner. This represents the first example of an NB-LRR protein retaining functional specificity across plant families.
THE BIOLOGICAL FUNCTIONS OF BACTERIAL AVIRULENCE PROTEINS
Why do bacterial pathogens translocate avirulence proteins into plant cells? We now know that avirulence proteins simply represent a small subset of the cocktail of proteins that are injected into host cells via the type III secretion system (Buell et al., 2003). These proteins, which are commonly referred to as TTSS effector proteins, are collectively essential to pathogenesis as mutations that block secretion abrogate disease (Alfano and Collmer, 1996). However, we are only just beginning to gain insights into the functions of specific TTSS effectors, which appear to vary widely between bacterial strains (Innes, 2003). The AvrRpt2 protein of Pseudomonas syringae has been shown to suppress defense responses in Arabidopsis and enhance the virulence of P. syringae (Chen et al., 2000). Several other TTSS effectors from P. syringae have recently been shown to suppress defense responses induced by other TTSS effectors resident in the same bacterium (i.e. mutation of a given TTSS effector can render a P. syringae strain avirulent on a normally susceptible host; Abramovitch et al., 2003; Bretz et al., 2003; Espinosa et al., 2003). One of these, HopPtoD, contains a Tyr phosphatase domain, and mutations that eliminate phosphatase activity block the HR suppression activity (Bretz et al., 2003; Espinosa et al., 2003). The targets of HopPtoD have not yet been identified. The XopD protein of Xanthomonas campestris has recently been shown to possess SUMO isopeptidase activity and to be nuclear localized, suggesting that it may target nuclear proteins and remove SUMO groups, which are ubiquitin-like peptides that modify the localization and/or stability of proteins (Hotson et al., 2003). As with HopPtoD, the specific targets of XopD, and how it contributes to virulence, are not known. The P. syringae AvrPphB protein functions as Cys protease, and furthermore, protease activity is required for induction of the HR in both Arabidopsis and tobacco (Shao et al., 2002; Shao et al., 2003; Zhu et al., 2004), suggesting that it is the enzymatic activity of AvrPphB, rather than the protein by itself, that activates resistance. Whether and how AvrPphB contributes to the virulence of P. syringae remains to be determined.
AN ALTERNATIVE MODEL FOR HOW R PROTEINS MEDIATE PATHOGEN RECOGNITION
The realization that plants have a limited repertoire of R genes, and that pathogen avirulence proteins are effectors that target host cell processes led to the development of a new model for R protein function dubbed the guard hypothesis (van der Biezen and Jones, 1998b). The authors suggested that NB-LRR genes function to guard the targets of pathogen virulence proteins. According to this model, pathogen effectors target host cell proteins in order to suppress defense responses or elicit susceptible responses (e.g. leakage of water and nutrients). NB-LRR proteins evolved as a counter-defense and function to monitor the status of the effector targets. For example, effectors could cause an allosteric change in their targets that promote binding by the NB-LRR protein, which in turn triggers the HR. Since the potential targets of pathogen virulence proteins are probably quite limited, this model would explain how a plant can detect thousands of pathogens with only few hundred R genes.
Evidence in support of the guard model has been mounting in the last 2 years, particularly for the recognition of P. syringae avirulence proteins. For example, recognition of the AvrPto protein by tomato requires the Pto gene, which encodes a protein kinase, and Prf, which encodes an NB-LRR protein. According to the guard model, Pto would represent the target and Prf the guard. Consistent with this, AvrPto physically interacts with Pto in a yeast (Saccharomyces cerevisiae) two-hybrid assay (Tang et al., 1996). More importantly, mutations have been identified in Pto that cause it to induce an HR-like response in tomato (Lycopersicon esculentum) in the absence of AvrPto, and this response is dependent on Prf (Rathjen et al., 1999). These Pto mutations may mimic the allosteric change induced by AvrPto.
A second example that supports the guard model is the interaction of the P. syringae effectors AvrB and AvrRpm1 with the Arabidopsis RIN4 protein. RIN4 interacts with these effectors in both yeast two-hybrid assays and coimmunoprecipitation assays (Mackey et al., 2002). Expression of AvrB and AvrRpm1 inside Arabidopsis cells induces phosphorylation of RIN4 (Mackey et al., 2002). RIN4 also physically interacts with RPM1, which is the NB-LRR protein that mediates recognition of both AvrB and AvrRpm1. Thus, RIN4 may represent the target of AvrB and AvrRpm1, while RPM1 guards RIN4. Intriguingly, a third P. syringae avirulence protein, AvrRpt2, induces the elimination of RIN4 (Axtell and Staskawicz, 2003; Mackey et al., 2003), suggesting that RIN4 is the target of at least three TTSS effectors. Recognition of AvrRpt2 is mediated by the NB-LRR protein RPS2, not RPM1, but RPS2 also physically interacts with RIN4 (Axtell and Staskawicz, 2003; Mackey et al., 2003), suggesting that RIN4 is guarded by at least two different R proteins. RPS2 appears to be activated by loss of RIN4, as a RIN4 knockout mutation is lethal in plants containing a functional RPS2 gene but has no obvious phenotype in plants lacking RPS2 function. It is not clear how AvrB, AvrRpm1, or AvrRpt2 induce changes in RIN4, or why RIN4 is a target. It is also not clear how RIN4 modifications activate RPM1 and RPS2.
The strongest data in support of the guard model comes from work on the P. syringae effector protein AvrPphB. Similar to the AvrPto story described above, recognition of AvrPphB by Arabidopsis requires both an NB-LRR protein, RPS5, and a protein kinase, PBS1 (Warren et al., 1999). Deletion of RPS5 or PBS1 blocks AvrPphB recognition, but these deletions have no effect on other Arabidopsis R genes, nor do they cause any morphological phenotypes. Significantly, PBS1 is cleaved by AvrPphB within the activation loop of PBS1, and such cleavage is required for activation of RPS5-mediated resistance (Shao et al., 2003). Thus, RPS5 is thought to function as a surveillance protein that senses specific damage to PBS1.
Although the above data strongly support an indirect mechanism of pathogen recognition by NB-LRR proteins, there are now two reports of physical associations between plant NB-LRR proteins and pathogen proteins. The first involves the Pi-ta protein from rice (Oryza sativa) and the AVR-Pita protein from the fungus Magnaportha grisea (Jia et al., 2000). Both yeast two-hybrid assays and an in vitro filter-binding assay demonstrated a physical interaction between the C-terminal Leu rich domain of Pi-ta and AVR-Pita. Interestingly, AVR-Pita is homologous to known metalloproteases, and mutations in the putative catalytic site of AVR-Pita obviate its physical interactions with Pi-ta, suggesting that Pi-ta might in fact be a substrate of AVR-Pita. Perhaps there is a second NB-LRR protein that guards Pi-ta.
The second example is the RRS1 protein of Arabidopsis, which has been shown to interact with the PopP2 protein of Ralstonia solanacearum using a yeast split ubiquitin yeast two-hybrid assay (Deslandes et al., 2003). In addition, RRS1 and PopP2 colocalize to the nucleus when transiently coexpressed in Arabidopsis protoplasts. Nuclear localization of RRS1 is dependent on a nuclear localization signal present in PopP2, suggesting that PopP2 drags RRS1 into the nucleus. Interpretation of these data is complicated, however, by the presence of a WRKY transcriptional activator domain on the C terminus of RRS1 and the putative SUMO-protease activity of PopP2. As with the interaction between Pi-ta and AVR-Pita, RRS1 might represent a target of PopP2, rather than being a specific receptor for PopP2.
SIMILARITIES BETWEEN PLANT NB-LRR PROTEINS AND ANIMAL PROTEINS THAT MEDIATE IMMUNE RESPONSES
Clues to how NB-LRR proteins may activate defense responses have come from comparison to animal proteins. The NB domain of plant NB-LRR proteins shares significant similarity to several animal proteins known to regulate programmed cell death and immune responses (van der Biezen and Jones, 1998a; Inohara and Nunez, 2003). Because the region of similarity extends beyond the nucleotide binding pocket, this domain has been referred to as the NB-ARC domain, for nucleotide binding and similarity to Apaf-1, R genes and Ced-4 (van der Biezen and Jones, 1998a). More recently, several mammalian NB-ARC proteins that have C-terminal LRR domains have been identified that are structurally quite similar to plant NB-LRR proteins (Inohara and Nunez, 2003). These proteins differ from plant NB-LRR domains at their N termini, with two of the best characterized members, NOD1 and NOD2, having caspase recruitment domains in this position. Significantly, NOD1 and NOD2 both function in the mammalian innate immune system and are activated by bacterial peptidoglycan fragments (Girardin et al., 2003).
Because human NOD proteins appear to function in pathogen perception and are structurally similar to plant NB-LRR proteins, an understanding of how plant NB-LRR proteins are regulated may enhance our understanding of how NOD proteins are regulated, and vice versa. Work on the human NOD1 and Apaf-1 proteins has led to development of the induced proximity model for NOD protein activation (for review, see Inohara and Nunez, 2003). In this model, the NB-ARC domains mediate homo-oligomerization (NOD stands for nucleotide oligomerization domain), but such oligomerization is inhibited by the C-terminal domains (WD-40 repeats in the case of Apaf-1 and LRRs in the case of NOD1). Oligomerization of Apaf-1 and NOD1 brings together downstream signaling proteins that are physically associated with the N-terminal caspase recruitment domains. The induced proximity of the downstream signaling proteins then leads to activation of apoptotic and inflammatory pathways.
In support of this model, deletion of the C-terminal domains of either Apaf-1 or NOD1 promotes self-oligomerization and activation of downstream responses (Inohara and Nunez, 2003). Full-length Apaf-1 can be induced to oligomerize in vitro by the addition of cytochrome C (Hu et al., 1999). Apaf-1 binds cytochrome C, and this binding requires both the WD-40 repeat domain and the NB-ARC domain, as well as ATP hydrolysis (Hu et al., 1999). Based on these data, it is hypothesized that the WD-40 repeat domain contains a ligand binding pocket that is unmasked only upon a conformational change induced by ATP hydrolysis. Binding of ligand to the WD-40 repeat domain then induces a conformational change that enables self-oligomerization via the NB-ARC domain. This general model is consistent with current data on the NOD1 and NOD2 proteins (Inohara and Nunez, 2003), although the putative NOD1 and NOD2 ligands (peptidoglycan fragments) have not been shown to bind to these proteins.
REGULATION OF PLANT NB-LRR PROTEIN FUNCTION BY INTRAMOLECULAR INTERACTIONS
It is not yet clear whether the NB-ARC domains of plant NB-LRR proteins also mediate self-oligomerization. The best insights to date come from work on the Rx protein of potato (Solanum tuberosum), which belongs to the CC-NB-LRR subclass (Moffett et al., 2002). Rx mediates resistance to potato virus X (PVX) and is specifically activated by the PVX coat protein. Although a physical interaction between the coat protein and Rx has not been shown, the coat protein somehow regulates intramolecular interactions among the CC, NB-ARC, and LRR domains of Rx; the CC-NB-ARC domain coimmunoprecipitates with the LRR domain when these domains are expressed from separate constructs, and this physical association is disrupted by coexpression of the PVX coat protein (Moffett et al., 2002). Furthermore, such coexpression reconstitutes coat protein-dependent activation of the HR. Importantly, expression of the CC-NB-ARC domain by itself does not induce an HR, unless highly overexpressed, and even then only in certain genotypes of tobacco (Bendahmane et al., 2002). Likewise, expression of the LRR domain by itself does not induce an HR. Thus, induction of the HR requires coexpression of the CC-NB-ARC, LRR, and coat proteins, but paradoxically, none of these proteins appear to be stably associated when all three are coexpressed. One plausible explanation for this result is that the coat protein does not directly interact with Rx, but in fact induces a change in a yet-to-be identified host protein, which in turn triggers Rx conformational changes.
Intramolecular interactions between the various domains of NB-LRR proteins are also indicated by work on the tomato Mi-1.1 protein. By creating domain swaps between Mi-1.1 and its close paralog Mi-1.2, Hwang and Williamson (2003) demonstrated that a mismatch between the N-terminal region and the LRR region resulted in a protein that constitutively activated an HR-like response. They further showed that a single amino acid substitution in the LRR region of Mi-1.2 could also cause a constitutive HR and that this activity could be suppressed by overexpression of the N-terminal region of Mi-1.1. These data suggest that interactions between the N-terminal domain and LRR domain may be important for regulation of HR activation.
WHAT IS THE ROLE OF NUCLEOTIDE BINDING IN NB-LRR PROTEIN FUNCTION?
The nucleotide binding domain of plant NB-LRR proteins is highly conserved and mutations in this domain invariably inactivate NB-LRR signaling (Tao et al., 2000; Tornero et al., 2002). Given its central role in NB-LRR function, surprisingly little is known about the role of nucleotide binding in NB-LRR activation. In fact, nucleotide binding has been demonstrated unequivocally for only two plant NB-LRR proteins, I-2 and Mi-1, both from tomato (Tameling et al., 2002). The NB domains of these two proteins were expressed in Escherichia coli and shown in a filter-binding assay to specifically bind ATP rather than other nucleoside triphosphates. Significantly, these recombinant proteins also hydrolyzed ATP, suggesting that nucleotide hydrolysis may represent a key step in plant NB-LRR protein activation.
This hypothesis is supported by the work on the mammalian Apaf-1 protein described above. The binding of Apaf-1 to its ligand cytochrome C and subsequent oligomerization requires dATP or ATP, and substitution of the nonhydrolyzable analog ATP-γS prevents both steps (Hu et al., 1999). This observation indicates that ATP hydrolysis rather than simple ATP binding is required for Apaf-1 to bind ligand and form oligomers. If ATP hydrolysis is similarly required for plant NB-LRR protein function, it would suggest that conformational changes associated with NB-LRR activation may only be observed in the presence of dATP/ATP.
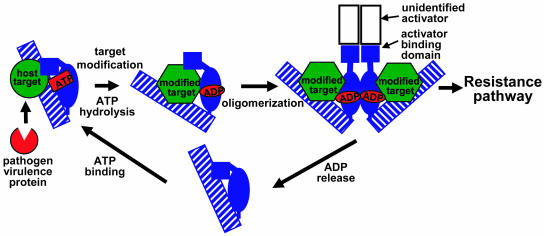
Although it is not yet clear how ATP hydrolysis regulates ligand binding by Apaf-1 or other NB-ARC containing proteins, analogies to GTP binding proteins would suggest that hydrolysis causes a conformational change, and further, that nucleotide exchange between ADP and ATP may be an important aspect of regulating signaling. Figure 1 presents a model for how ATP hydrolysis and nucleotide exchange might be incorporated into the regulation of plant NB-LRR protein function. Although speculative, this model makes several testable predictions now that potential ligands for RPM1, RPS2, and RPS5 have been identified. In particular, it predicts that modification of the ligand (a host protein) by the pathogen virulence protein induces ATP hydrolysis by the NB-LRR protein, causing a conformational change in the NB-LRR protein that allows oligomerization. Inactivation of signaling would occur when ADP dissociates. The inactive NB-LRR protein would then have to bind another molecule of ATP before it would be able to function once again.
Figure 1.
An induced proximity model based on data from the human Apaf-1 protein and consistent with current data on plant NB-LRR proteins. A pathogen virulence protein modifies a host target protein, which then causes a conformational change in the NB-LRR protein and ATP hydrolysis. This enables oligomerization of the NB-LRR protein via the NB-ARC domain (blue oval), which activates resistance via inducing proximity of an unidentified activator protein. ADP release causes disassociation of the complex. ATP must bind to restart the cycle. Specificity for the modified host protein is determined by both the N-terminal activator binding domain (blue square) and the LRR domain (hatched rectangle). These domains interact with each other in the absence of ligand. The host target protein is shown interacting with the NB-LRR protein prior to modification based on data on the Arabidopsis RIN4 protein (see text).
OTHER COMPONENTS OF THE PLANT ALARM SYSTEM
A recent flurry of papers has established that many NB-LRR proteins interact with cytosolic HSP90 proteins (Hubert et al., 2003; Lu et al., 2003; Takahashi et al., 2003). These interactions appear to be important for either the folding of NB-LRR proteins and/or the formation of a stable NB-LRR protein complex, as mutation or silencing of HSP90s causes a sharp reduction in NB-LRR protein accumulation (Hubert et al., 2003; Lu et al., 2003).
HSP90s may be assisted in the putative assembly of NB-LRR resistance protein complexes by two other conserved proteins, RAR1 and SGT1. RAR1 interacts with HSP90, and mutations in the Arabidopsis RAR1 gene cause a loss of detectable RPM1 protein (Tornero et al., 2002; Hubert et al., 2003). SGT1 interacts with both HSP90 and RAR1 (Takahashi et al., 2003), but appears to play a different role from RAR1, as silencing of SGT1 in Nicotiana benthamiana does not affect accumulation of the NB-LRR protein Rx, despite blocking Rx-mediated resistance (Lu et al., 2003).
Arabidopsis contains two SGT1 genes (Austin et al., 2002). Mutations in SGT1b compromise resistance mediated by a specific subset of NB-LRR proteins that only partially overlaps with those affected by rar1 mutations. Knockouts of both SGT1 genes in Arabidopsis appear to be lethal as homozygous double mutants have not been recovered. SGT1 has also been shown to interact with an SCF ubiquitin ligase complex and the COP9 signalosome (Azevedo et al., 2002) and thus may participate in multiple signaling pathways in plants via regulation of protein degradation. For a more detailed discussion of these papers see the recent dispatch by Schulze-Lefert (2004).
CONCLUSIONS
Although much progress has been made in the last 2 years, there still remain large gaps in our understanding of the plant alarm system. Evidence is mounting that many, and perhaps most, pathogens are detected indirectly via the enzymatic activity of their virulence proteins. However, we still lack compelling evidence that a modified host protein directly activates NB-LRR signaling. An even more glaring gap in our current understanding is how plant NB-LRR proteins activate defense responses. Presumably these proteins physically associate with one or more downstream activator proteins, but we currently have very little information as to what these activators might be. Filling these gaps will undoubtedly keep many laboratories busy for some time to come. At this point we have a partial parts list for the alarm system and have some information on how each of the parts work, but have yet to sort out the wiring.
Acknowledgments
I thank Dr. Jeff Dangl, University of North Carolina, and Dr. Jeff Ellis, CSIRO, Canberra, Australia, for sharing papers in press, and the members of my laboratory for stimulating discussions and comments on this manuscript.
This work was supported by the National Institute of General Medical Sciences (grant no. R01GM46451).
References
- Abramovitch RB, Kim YJ, Chen S, Dickman MB, Martin GB (2003) Pseudomonas type III effector AvrPtoB induces plant disease susceptibility by inhibition of host programmed cell death. EMBO J 22: 60–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfano JR, Collmer A (1996) Bacterial pathogens in plants: Life up against the wall. Plant Cell 8: 1683–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin MJ, Muskett P, Kahn K, Feys BJ, Jones JD, Parker JE (2002) Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295: 2077–2080 [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Staskawicz BJ (2003) Initiation of RPS2-Specified Disease Resistance in Arabidopsis Is Coupled to the AvrRpt2-Directed Elimination of RIN4. Cell 112: 369–377 [DOI] [PubMed] [Google Scholar]
- Azevedo C, Sadanandom A, Kitagawa K, Freialdenhoven A, Shirasu K, Schulze-Lefert P (2002) The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295: 2073–2076 [DOI] [PubMed] [Google Scholar]
- Bendahmane A, Farnham G, Moffett P, Baulcombe DC (2002) Constitutive gain-of-function mutants in a nucleotide binding site-leucine rich repeat protein encoded at the Rx locus of potato. Plant J 32: 195–204 [DOI] [PubMed] [Google Scholar]
- Bretz JR, Mock NM, Charity JC, Zeyad S, Baker CJ, Hutcheson SW (2003) A translocated protein tyrosine phosphatase of Pseudomonas syringae pv. tomato DC3000 modulates plant defence response to infection. Mol Microbiol 49: 389–400 [DOI] [PubMed] [Google Scholar]
- Buell CR, Joardar V, Lindeberg M, Selengut J, Paulsen IT, Gwinn ML, Dodson RJ, Deboy RT, Durkin AS, Kolonay JF, et al (2003) The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci USA 100: 10181–10186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Kloek AP, Boch J, Katagiri F, Kunkel BN (2000) The Pseudomonas syringae avrRpt2 gene product promotes pathogen virulence from inside plant cells. Mol Plant Microbe Interact 13: 1312–1321 [DOI] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y (2003) Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci USA 100: 8024–8029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Lawrence GJ, Catanzariti AM, Ayliffe MA, Ellis JG (2004) The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell 16: 755–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa A, Guo M, Tam VC, Fu ZQ, Alfano JR (2003) The Pseudomonas syringae type III-secreted protein HopPtoD2 possesses protein tyrosine phosphatase activity and suppresses programmed cell death in plants. Mol Microbiol 49: 377–387 [DOI] [PubMed] [Google Scholar]
- Flor HH (1956) The complementary genic systems in flax and flax rust. Adv Genet 8: 29–54 [Google Scholar]
- Gabriel DW, Rolfe BG (1990) Working models of specific recognition in plant-microbe interactions. Annu Rev Phytopathol 28: 365–391 [Google Scholar]
- Girardin SE, Boneca IG, Carneiro LA, Antignac A, Jehanno M, Viala J, Tedin K, Taha MK, Labigne A, Zahringer U, et al (2003) Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science 300: 1584–1587 [DOI] [PubMed] [Google Scholar]
- Hotson A, Chosed R, Shu H, Orth K, Mudgett MB (2003) Xanthomonas type III effector XopD targets SUMO-conjugated proteins in planta. Mol Microbiol 50: 377–389 [DOI] [PubMed] [Google Scholar]
- Hu Y, Benedict MA, Ding L, Nunez G (1999) Role of cytochrome c and dATP/ATP hydrolysis in Apaf-1-mediated caspase-9 activation and apoptosis. EMBO J 18: 3586–3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert DA, Tornero P, Belkhadir Y, Krishna P, Takahashi A, Shirasu K, Dangl JL (2003) Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J 22: 5679–5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang CF, Williamson VM (2003) Leucine-rich repeat-mediated intramolecular interactions in nematode recognition and cell death signaling by the tomato resistance protein Mi. Plant J 34: 585–593 [DOI] [PubMed] [Google Scholar]
- Innes R (2003) New effects of type III effectors. Mol Microbiol 50: 363–365 [DOI] [PubMed] [Google Scholar]
- Inohara N, Nunez G (2003) NODs: intracellular proteins involved in inflammation and apoptosis. Nat Rev Immunol 3: 371–382 [DOI] [PubMed] [Google Scholar]
- Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B (2000) Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J 19: 4004–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe B, Kajava AV (2001) The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol 11: 725–732 [DOI] [PubMed] [Google Scholar]
- Lu R, Malcuit I, Moffett P, Ruiz MT, Peart J, Wu AJ, Rathjen JP, Bendahmane A, Day L, Baulcombe DC (2003) High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J 22: 5690–5699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL (2003) Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112: 379–389 [DOI] [PubMed] [Google Scholar]
- Mackey D, Holt BF, Wiig A, Dangl JL (2002) RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108: 743–754 [DOI] [PubMed] [Google Scholar]
- Martin GB, Bogdanove AJ, Sessa G (2003) Understanding the functions of plant disease resistance proteins. Annu Rev Plant Biol 54: 23–61 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, Young ND (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J 20: 317–332 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15: 809–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Shen KA, Rohani P, Gaut BS, Michelmore RW (1998) Receptor-like genes in the major resistance locus of lettuce are subject to divergent selection. Plant Cell 10: 1833–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett P, Farnham G, Peart J, Baulcombe DC (2002) Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J 21: 4511–4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parniske M, Hammond-Kosack KE, Golstein C, Thomas CM, Jones DA, Harrison K, Wulff BB, Jones JD (1997) Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell 91: 821–832 [DOI] [PubMed] [Google Scholar]
- Rathjen JP, Chang JH, Staskawicz BJ, Michelmore RW (1999) Constitutively active Pto induces a Prf-dependent hypersensitive response in the absence of AvrPto. EMBO J 18: 3232–3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Lefert P (2004) Plant immunity: the origami of receptor activation. Curr Biol 14: R22–R24 [PubMed] [Google Scholar]
- Shao F, Golstein C, Ade J, Stoutemyer M, Dixon JE, Innes RW (2003) Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301: 1230–1233 [DOI] [PubMed] [Google Scholar]
- Shao F, Merritt PM, Bao Z, Innes RW, Dixon JE (2002) A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell 109: 575–588 [DOI] [PubMed] [Google Scholar]
- Takahashi A, Casais C, Ichimura K, Shirasu K (2003) HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc Natl Acad Sci USA 100: 11777–11782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameling WI, Elzinga SD, Darmin PS, Vossen JH, Takken FL, Haring MA, Cornelissen BJ (2002) The tomato R gene products I-2 and MI-1 are functional ATP binding proteins with ATPase activity. Plant Cell 14: 2929–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Frederick RD, Zhou J, Halterman DA, Jia Y, Martin GB (1996) Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science 274: 2060–2063 [DOI] [PubMed] [Google Scholar]
- Tao Y, Yuan F, Leister RT, Ausubel FM, Katagiri F (2000) Mutational analysis of the Arabidopsis nucleotide binding site-leucine-rich repeat resistance gene RPS2. Plant Cell 12: 2541–2554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornero P, Chao RA, Luthin WN, Goff SA, Dangl JL (2002) Large-scale structure-function analysis of the Arabidopsis RPM1 disease resistance protein. Plant Cell 14: 435–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornero P, Merritt P, Sadanandom A, Shirasu K, Innes RW, Dangl JL (2002) RAR1 and NDR1 contribute quantitatively to disease resistance in Arabidopsis, and their relative contributions are dependent on the R gene assayed. Plant Cell 14: 1005–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Biezen EA, Jones JDG (1998. a) The NB-ARC domain: a novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr Biol 8: R226–R227 [DOI] [PubMed] [Google Scholar]
- van der Biezen EA, Jones JDG (1998. b) Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem Sci 23: 454–456 [DOI] [PubMed] [Google Scholar]
- Warren RF, Merritt PM, Holub E, Innes RW (1999) Identification of three putative signal transduction genes involved in R gene-specified disease resistance in Arabidopsis. Genetics 152: 401–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Shao F, Innes RW, Dixon JE, Xu Z (2004) The crystal structure of Pseudomonas avirulence protein AvrPphB: a papain-like fold with a distinct substrate-binding site. Proc Natl Acad Sci USA 101: 302–307 [DOI] [PMC free article] [PubMed] [Google Scholar]