Abstract
We carried out a genome-wide prediction of scaffold/matrix attachment regions (S/MARs) in Arabidopsis. Results indicate no uneven distribution on the chromosomal level but a clear underrepresentation of S/MARs inside genes. In cases where S/MARs were predicted within genes, these intragenic S/MARs were preferentially located within the 5′-half, most prominently within introns 1 and 2. Using Arabidopsis whole-genome expression data generated by the massively parallel signature sequencing methodology, we found a negative correlation between S/MAR-containing genes and transcriptional abundance. Expressed sequence tag data correlated the same way with S/MAR-containing genes. Thus, intragenic S/MARs show a negative correlation with transcription level. For various genes it has been shown experimentally that S/MARs can function as transcriptional regulators and that they have an implication in stabilizing expression levels within transgenic plants. On the basis of a genome-wide in silico S/MAR analysis, we found a significant correlation between the presence of intragenic S/MARs and transcriptional down-regulation.
The Arabidopsis genome has been sequenced, and 115 Mb of sequence were allocated on five chromosomes encoding >25,000 genes. Only for the highly repetitive regions, like the core centromeres and the nucleolar organizing regions, no sequence information is available (The Arabidopsis Genome Initiative, 2000). Nevertheless, in addition to the low copy regions located on the chromosome arms, extensive information and analysis data for the complex heterochromatic and subcentromeric regions are available. Annotation of the Arabidopsis genome is far advanced with respect to genes, but little, beyond some repetitive sequences, is known about the noncoding regions.
Among the most important sequence features involved in chromatin structure as well as in transcription controls are the scaffold/matrix attachment regions (S/MARs). These regions are about 300 bp to several kb in length and are present in all higher eukaryotes, including mammals and plants (Bode et al., 1996; Allen et al., 2000). S/MARs are operationally defined as DNA elements that bind specifically to the nuclear matrix or as DNA fragments that copurify with the nuclear matrix (Michalowski et al., 1999). S/MARs often coincide with chromatin features associated with gene function, nuclease hypersensitivity, non-B-DNA structures, origins of replication, and hotspots of retroelement insertion (Avramova et al., 1998; Paul and Ferl, 1998; Tikhonov et al., 2000, 2001). S/MARs are notable for their AT richness and likely narrowing of the minor groove (Gasser et al., 1989; Bode et al., 1995, 1996). S/MARs may additionally be defined as cis-acting elements typically found outside the transcribed regions and within introns. They typically augment transcription rates in a highly context-dependent manner (Schubeler et al., 1996) but are separable from enhancer sequences on the basis of transient expression analyses (Bode et al., 1995).
In the interphase and metaphase nuclei, chromosomal DNA is tightly packaged into higher order structures. A DNA-histone protein complex forms the nucleosome core. This structure is further condensed to the chromatin fiber. According to the loop domain model, these fibers are attached at their bases to the nuclear scaffold, and the unanchored fiber loops out from the point of attachment. Such loops are not randomly attached to the matrix but involve associations with specific stretches of DNA, the S/MAR landmarks themselves. Such looping has been implicated with the functional compartmentalization of the genome. Human chromosomes 18 and 19 provide evidence for such a functional compartmentalization of the genome (Croft et al., 1999). Chromosome 19 is actively transcribed and is both tightly associated with the nuclear matrix and localized to the interior of the nucleus. Chromosome 18 is poorly transcribed, and its peripheral location is associated with heterochromatin (Croft et al., 1999). This demonstrates some importance for association with the nuclear matrix for transcription. However, thus far, this has not yet been demonstrated in plants.
Studies in Arabidopsis and maize (Zea mays; Paul and Ferl, 1998) have shown that the plant genome is not packaged by random gathering into domains of indiscriminate length, but, rather, the genome is gathered into specific domains, and a gene consistently occupies a discrete physical section of the genome. The average loop size in Arabidopsis and maize has been estimated as 25 and 45 kb, respectively (Paul and Ferl, 1998), although other studies (e.g. van Drunen et al., 1997) have suggested smaller domain sizes. Some loops may remain permanently condensed and inactive, even within the euchromatic portions of the genome. Others can be extended to produce a transcriptionally poised conformation in appropriately differentiated cells (Hassan et al., 1994). Data on the location of transcribed elements within structural loops at the supragenic level are however limited to only a few studies. These suggest that attachment to the matrix and transcription is not systematically associated (Mirkovitch et al., 1988; Surdej et al., 1990), though S/MARs are associated with the ends of some DNaseI-sensitive (transcriptionally poised) domains (Bonifer et al., 1991). There is, however, a wide variety of examples that suggest a correlation between the presence of an S/MAR and gene expression (Jarman and Higgs, 1988; Farache et al., 1990; Mielke et al., 1990). S/MARs have also been identified within introns of genes (e.g. Cockerill et al., 1987; Avramova and Paneva, 1992; Romig et al., 1994). Cockerill et al. (1987) suggested that MARs flanking enhancer sequences may act as positive and/or negative regulators of enhancer function. Romig et al. (1994) stated that nothing is known about the function of MARs that have been identified within introns of genes (for further examples of intronic MARs, see references cited therein). The size of the S/MAR anchored loops has been implicated in relative expression efficacy; single genes contained within shorter loops have higher expression levels than multiple genes existing within a longer loop (Mirkovitch et al., 1988; Mlynarova et al., 1995).
The promoter-S/MAR distance is, however, an important factor in the correct functioning of the loop (Mlynarova et al., 1995; Schubeler et al., 1996). In addition to the S/MAR-associated enhancement of gene expression, S/MARs have a proposed role in the negative regulation of gene expression. Such negative regulation is the proposed default mode of action for S/MARs both closely associated with the promoter sequence or when appearing downstream of the promoter (Schubeler et al., 1996). Such S/MARs would block progression by RNA polymerase II, so they may be either nonfunctional in vivo or have a regulated matrix-binding activity (Schubeler et al., 1996). Investigation of these intronic S/MARs in plants shows that they generally have a lower affinity for binding to the nuclear matrix and may therefore differ in their association contacts with the matrix (Tikhonov et al., 2000). Additional specific S/MAR-binding soluble factors might be involved in mediating a regulated binding of weak S/MARs to the nuclear matrix (Mielke et al., 1990; Bode et al., 1996; Schubeler et al., 1996). Strong S/MARs delimit the structural domains containing the transcriptional units in yeast (Saccharomyces cerevisiae; Brun et al., 1990). This has not been found for weak S/MARs, though the possibility remains that weak S/MARs are involved in dynamic folding of loops into subdomains (Avramova et al., 1995).
S/MARs have been further demonstrated in a variety of functional tests to act as insulators (Mlynarova et al., 2003), according to the loop domain model, by protecting a loop from the effects of the neighboring chromatin or associated enhancer sequences. However, this is not a general rule since other S/MARs do not show such insulating activity in enhancer-blocking assays, and some known insulators do not show properties typical for S/MARs (Jarman and Higgs, 1988; Kellum and Schedl, 1992; Li and Stamatoyannopoulos, 1994; Antes et al., 2001; Razin, 2001).
The association of an S/MAR with a transgene may lead to a transgene forming its own chromatin domain and allows separation from the local chromatin environment (Hall et al., 1991), thus providing the transcription machinery with better access to the gene (Phi-Van et al., 1990). This may be observed experimentally as a change in DNaseI sensitivity (Allen et al., 2000). S/MARs appear to greatly increase transgene expression in plant cell lines (Mlynarova et al., 1995; Allen et al., 1996). In mammalian cell line systems, the use of S/MARs within transgene constructs has allowed for reporter expression levels proportional to the number of transgene insertions (Phi-Van et al., 1990; McKnight et al., 1992). S/MARs may also be effective against transcriptional silencing induced by cis-interactions within repeated transgene arrays (Allen et al., 2000).
The paradox that ortholog genes from different species will show similar functions in different chromosomal settings has been proposed (Tikhonov et al., 2000). The presence of anchors that stably position genes within the spatial architecture of the nucleus seems a reasonable hypothesis (Flavell, 1994). This has been tested using large collinear chromosomal continuums between maize and sorghum (Sorghum bicolor; Tikhonov et al., 2000) and rice (Oryza sativa) and sorghum (Avramova et al., 1998). With the virtual subtraction of retrotransposon sequences, the gene coding sequences were observed to occur within comparable loops. Although the chromosomal segments are small (78 kb in sorghum), there is evidence for a more extensive localization of S/MARs within the coding (intronic) sequences. These intronic S/MARs are preserved in homologous genes and in ortholog introns in maize and sorghum (Tikhonov et al., 2000). This supports a notion of conserved function.
Plant systems are ideal for the study of S/MARs on a genome-wide scale. Gene structures within plants are typically composed of compact genes with short introns. This is in contrast to mammalian systems in which genes are expanded and short exons are interspersed by enormous introns. To date, there have been no published genome-scale investigations of S/MARs in plants. In this article, we present a genome-scale in silico analysis of putative S/MARs in Arabidopsis.
We have undertaken a comprehensive bioinformatics-based analysis of S/MAR sequences across the Arabidopsis genome. This reveals two distinct types of S/MARs, those that can be mapped to exons or more frequently to intronic sequences that we have termed intragenic S/MARs. The bulk of S/MARs map to the intergenic portion of the genome. In this article, we focus on the intragenic S/MARs, the elements that have been hypothesized to have a role within the regulation of gene expression. In particular, we carried out an exhaustive combinatorial analysis on the correlation between transcriptional abundance and presence/absence of S/MAR elements. Apart from expressed sequence tag (EST) data, expression data generated by the massively parallel signature sequencing (MPSS) method were used to investigate potential correlations.
RESULTS
Concatenated pseudomolecules representing the five sequenced Arabidopsis chromosome arms were assembled from the bacterial artificial chromosome tiling path available from MAtDB (http://mips.gsf.de/proj/thal; Schoof et al., 2002). Suitability of SMARTest for the purpose of detection of S/MAR elements in plants has been demonstrated previously (Frisch et al., 2002). Therefore, SMARTest was used to predict S/MARs within each of the pseudomolecules.
No S/MAR Clusters Appear on Chromosomal Level
A total of 21,705 S/MARs were detected throughout the genome, which is in the same order of magnitude as the number of genes (Table I). The average density of S/MARs detected was about 1 S/MAR per 5.5 kb of genomic sequence. The predicted S/MAR sequences total 16 Mb of sequence and account for approximately 13.5% of the Arabidopsis genome.
Table I.
Number and density of S/MARs in Arabidopsis chromosomes
| Chromosome | Number of Genes | Number of S/MARs | Chromosome Size | S/MAR Density |
|---|---|---|---|---|
| 1 | 6,704 | 5,521 | 30,136 kb | 5,528 |
| 2 | 4,151 | 3,945 | 19,847 kb | 5,031 |
| 3 | 5,272 | 4,018 | 23,752 kb | 5,912 |
| 4 | 3,886 | 3,245 | 17,790 kb | 5,483 |
| 5 | 6,014 | 4,976 | 26,944 kb | 5,462 |
| Total | 26,027 | 21,705 | 118,471 kb | 5,487 |
This table shows the number of genes and S/MARs found in the Arabidopsis genome ranked by chromosome and the relative S/MAR density between the different chromosomes. Density was defined as the number of nucleotides divided by the number of S/MARs, which were represented by single positions for this purpose.
The Arabidopsis chromosomes have pronounced cytogenetic and clearly defined molecular topological features such as heterochromatic and subcentromeric regions, which contain a high density of transposable elements and large-scale insertions of mitochondrial DNA (Copenhaver et al., 1999; Lin et al., 1999; Mayer et al., 1999; CSHL/WUGSC/PEB Arabidopsis Sequencing Consortium, 2000; Fransz et al., 2000; Stupar et al., 2001). No evidence was found for a significant genomic location bias (hotspots of high or low S/MAR density) with respect to chromosomal localization and the presence of subcentromeric or heterochromatic regions (data not shown). Consistent with the absence of S/MARs in organelles, no S/MAR element has been detected within the approximate 620-kb mitochondrial insertion on Arabidopsis chromosome 2.
Intragenic S/MARs within the Arabidopsis Genome
Numerous S/MARs have been reported in plant genomes (Chinn and Comai, 1996; van Drunen et al., 1997; Avramova et al., 1998). Thus, we asked the question as to whether S/MARs in Arabidopsis showed any correlation with genes on a whole-genome scale. A potential preference for the localization of S/MARs in transcribed and nontranscribed regions was investigated by comparison of the predicted S/MAR coordinates with the positions of the annotated genes.
A gene was only considered as containing an S/MAR if there was at least 50 bp of sequence overlap between the coding region of a gene and the S/MAR in question because S/MAR elements are of considerable length (by definition at least 300 bp using SMARTest), and exact S/MAR borders, thus far, cannot be defined. Using these criteria, we found 8.2% of all genes to contain at least 1 S/MAR (2,135 out of 26,027 genes). We carried out an identical analysis on 50 datasets generated by randomizing the distribution of S/MARs across the chromosomes (adapted from Frisch et al., 2002) in order to address the significance of S/MAR-gene correlations. This analysis indicates an approximate 4-fold underrepresentation of S/MARs within genes (Table II).
Table II.
S/MARs are underrepresented in exons and introns
| Observed | Expected | Underrepresentation (x-fold) | |
|---|---|---|---|
| Genes | 26,027 | ||
| Genes containing an S/MAR (excluding untranslated regions) | 2,135 (8.2%) | 9,427 ± 53.6 (37.3%) | 4.4 |
This table shows the observed and expected numbers of S/MARs found within coding regions and introns of genes. The expected numbers were obtained by averaging the results from 50 test sets in which the number of S/MARs actually found was randomly distributed throughout the Arabidopsis genome. The sd is given for the 50 control datasets.
Correlation of Intragenic S/MARs with Gene Expression
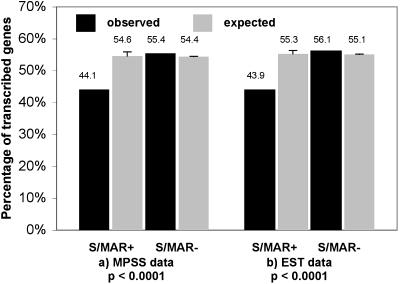
In mammalian and viral systems, S/MARs are known to be involved in chromatin remodeling and transcriptional control (Liu et al., 1997; Wang et al., 1999; Stunkel et al., 2000). S/MARs have also been found to be involved in plant gene regulation, and intragenic S/MARs were found to be different from intergenic S/MARs (Tikhonov et al., 2000). We investigated whether there is a correlation between S/MARs and gene expression on a whole-genome scale in Arabidopsis. We assessed correlation of the presence of intragenic S/MARs with the expression level of genes by using MPSS data as indication of whether a gene is expressed or not (Fig. 1A). MPSS is a powerful means for the quantitative measurement of gene expression. It enables one to identify and analyze the level of expression of all genes in a sample by counting the number of individual mRNA molecules. A 17- to 20-bp signature (or tag) is identified from the 3′ end of each transcript in a population of mRNAs; the abundance of identical tags in a library is indicative of the level of expression of the gene from which the tag was derived. Therefore, the MPSS data provide a quantitative estimate of expression as opposed to the relative estimates derived from hybridization signal intensities on microarrays. We also evaluated the same correlation using EST data instead of MPSS data, as a correlation between the abundance of ESTs and the expression of genes was already reported (Pires Martins et al., 2001). Since only 8.2% of all genes contain S/MARs, we estimated whether the sample size might cause a bias against MPSS or EST-correlated genes by randomly selecting 50 sets of 2,135 genes (i.e. the number of S/MAR-containing genes) from all genes and calculating the MPSS and EST correlation for the random sample. The small sample size did not influence the expected amount of MPSS and EST correlation, while there are clearly fewer S/MAR-containing genes associated with MPSS sequences or EST matches (Fig. 1). The average number of MPSS sequences or ESTs associated with S/MAR-containing genes is also significantly reduced, suggesting an on average lower expression level of the affected genes.
Figure 1.
S/MAR-containing genes are associated with fewer MPSS matches and ESTs than S/MAR-less genes. A, This graph shows the relative coverage of MPSS hits of genes with (S/MAR+) or without (S/MAR−) intragenic S/MARs. A gene was only considered containing an S/MAR when the predicted S/MAR overlapped at least 50 bp with an annotated gene. The number of genes containing an S/MAR (S/MAR+) or without an S/MAR (S/MAR−) for which at least 10 MPSS hits were found is shown as percentage of all genes of this class. The exact values and sd are given above the columns. P < 0.0001. Calculation of expected values is detailed in the methods. B, The graph shows results of an analogous analysis as in A using EST data. P < 0.0001. All values were calculated by InStat3 from GraphPad software (www.graphpad.com).
Annotation of higher eukaryotic genomes is an error-prone process. Thus, such a finding could potentially result from a 10% gene annotation error generating spurious genes or concatenated genes accidentally located around intergenic S/MARs, which would not correspond to ESTs. However, EST and MPSS data are only available for expressed sequences. In addition, correlation analysis as outlined above, but restricted to experimentally verified genes, was also carried out for MPSS as well as EST data. A smaller but still significant reduction of MPSS hits and ESTs associated with S/MAR-containing genes was observed (data not shown). Thus, we can rule out that the significant deviation in expression level is attributable to spurious gene models and/or annotation errors.
In summary, there is a considerable bias for genes with intragenic S/MAR to have a correspondingly lower number of MPSS hits and EST coverage than genes that are not associated with an S/MAR.
In the human genome, involvement of intragenic S/MARs in repression of gene expression has been reported. An S/MAR located within the first intron of the hepatic-specific human cholesterol 7α-hydroxylase (CYP7A1) inhibits transcription of the gene by recruiting histone deacetylase to CCAAT displacement protein binding sites within the S/MAR (Antes et al., 2000). The first intron is a favorable place for intragenic regulatory regions, bringing the S/MARs close to the promoter region, i.e. overlapping the 5′ end of the gene or within the first or second intron (exons usually do not contain S/MARs).
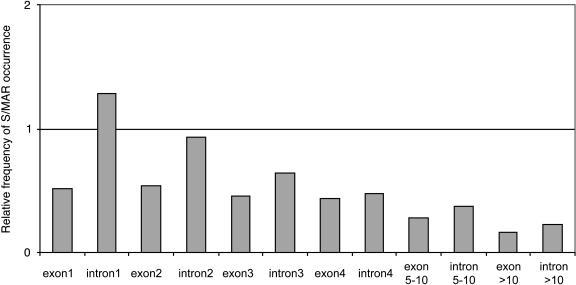
Therefore, we investigated the relative distribution of S/MAR elements within the S/MAR-containing genes of Arabidopsis. As depicted in Figure 2, there is a clear bias toward the 5′-half of the genes most pronounced in the first intron. The expected value was corrected for relative number and size of introns and exons, and we verified that the different AT content of Arabidopsis exons versus introns did not have a significant influence on the results.
Figure 2.
Intragenic S/MARs in Arabidopsis are located preferentially in the 5′-half of the genes. This graph shows the relative frequency of an S/MAR overlapping or occurring within a particular intron or exon. A value of 1.0 is indicative of the observed value equaling the expected value; a value greater than 1.0 is indicative of an overrepresentation of S/MAR occurrence with this feature; and a value of less than 1.0 is indicative of an underrepresentation of S/MAR occurrence with this feature. Numbers have been corrected for actual numbers and length of introns and exons.
DISCUSSION
The presence and conservation of S/MARs in plant genomes is well established (Chinn and Comai, 1996; Avramova et al., 1998; Fukuda, 2000; Tikhonov et al., 2000), and S/MARs have been reported for individual genes of Arabidopsis as well (Sawasaki et al., 1998). From these studies it is also well established that S/MARs fulfill important biological functions. However, a genome-wide analysis of S/MARs within plant genomes thus far has not been carried out. Here, we report, to our knowledge, the first genome-wide prediction of S/MARs in a plant genome, Arabidopsis, using the SMARTest software (Frisch et al., 2002). In this study, we predicted 21,705 S/MAR elements within the Arabidopsis genome. SMARTest was shown to predict S/MARs with a specificity of about 70% and a sensitivity of about 40% (Frisch et al., 2002). Our data represent an approximation of number and location of Arabidopsis S/MARs and allow carrying out of meaningful correlation analysis on a genome-wide scale, which had been impossible.
We observed no clustering of S/MARs along the whole genome of Arabidopsis, independent of low or high complexity regions. No potential S/MAR elements have been detected within the large-scale insertion of mitochondrial DNA in chromosome 2, which is consistent with the absence of full-length S/MARs in organelle DNA.
S/MARs have been implicated with the organization of chromatin structure and remodeling. This function is thought to be independent of any distance correlation with genes (Bode et al., 1996; Tikhonov et al., 2000). Our genome-wide association studies proved that S/MARs are predominantly located within the intergenic regions, and the number of S/MARs detected (21,705) is in the same order of magnitude as the number of genes (26,027). S/MARs within Arabidopsis are clearly associated with genes. However, due to the compact organization of the genome, it cannot be ruled out that the S/MAR-gene association might merely be a consequence of the short intergenic regions (average length approximately 2,500 bp).
One of the functions S/MARs are involved in is modulation of transcription both in mammalians and plants (Chinn and Comai, 1996; Whitelaw et al., 2000), and S/MAR-binding proteins are directly involved in transcriptional control (Nepveu, 2001). Intronic S/MARs were also previously linked functionally to gene regulation (Oancea et al., 1997), prompting us to search for a connection between intragenic S/MARs and gene transcription. Absence of ESTs for a given gene does not necessarily indicate a lack of transcription but may result from low levels of transcription causing the mRNA to be missed during EST sequencing. However, as MPSS data are more than 60 times deeper than EST data, this makes such undersampling unlikely. Using ESTs as a measure for gene transcription levels was also successfully applied in at least one other study (Pires Martins et al., 2001) and supported the negative correlation between intragenic S/MARs and transcription level of Arabidopsis genes already deduced from MPSS sequence correlation.
Recent results obtained by genome-wide expression analysis on high density oligonucleotide tiling arrays showed that the 600-Mb insertion of mitochondrial DNA on Arabidopsis chromosome 2 shows a high level of transcriptional activity (Yamada et al., 2003). One hypothesis coincident with our suggested transcriptional repressive function of S/MARs is that the absence of any full-length S/MARs in this insert, which is now under the same control mechanisms as the chromosomal DNA, might be one reason for the observed high transcriptional activity.
A mechanism for gene silencing by an S/MAR located in the first intron of the human cholesterol 7α-hydroxylase gene has been reported (Antes et al., 2000); further examples for S/MAR-mediated repressive effects are reported by Banan et al. (1997) and Kohwi-Shigematsu et al. (1997). However, to our knowledge, no large-scale correlation analysis has been carried out thus far. We analyzed all Arabidopsis S/MAR-containing genes and found S/MAR locations to be biased toward the 5′-half of those genes (most prominent was the first intron). This is consistent with findings in human and mouse (Glazko et al., 2003). The clear tendency of S/MARs to be located within the first two introns, especially in genes with no EST correlation, suggests a connection between S/MAR presence and gene expression. Further support comes from a study on function of MAR-binding filament-like protein (MFP1) in various plants (Harder et al., 2000). MFP1 has been shown to depend on organ specificity and developmental timing. This has been interpreted with a possible role of MAR-binding proteins in creating chromatin environments in tissue and/or development-dependent manner for either expression or repression of MAR-regulated genes. These data are compatible with a scenario in which intragenic S/MARs tend to fulfill a down-regulatory function. Our genome-wide S/MAR prediction led to the formulation of a correlation-supported functional hypothesis for intragenic S/MARs. Although the mechanism(s) of S/MAR induced repression in Arabidopsis remains unclear, S/MARs might play a role in negatively controlling particular groups of genes depending on particular tissue environments or environmental clues. Considering this and taking into account probable chromatin-dependant regulatory influences on particular genes of interest might well impact future functional characterization of individual genes.
Experimental S/MAR detection is usually carried out by matrix-reassociation assays and currently is not being performed on a whole-genome scale. Functional properties like the down-modulatory effect on gene transcription proposed in this study possibly would have escaped detection, since only one S/MAR property, matrix binding, is tested in most assays.
Current knowledge did not allow establishing similar correlations for the intergenic S/MARs. It remains unclear whether the observed preferred distance between intragenic and intergenic S/MARs of about 1,500 bp (data not shown) is a mere consequence of the small size of genes and intergenic regions, or the preferential location of intragenic S/MARs is there to ensure such S/MAR pairs with the observed distances. However, successful correlation of the intragenic S/MARs with functional features suggests that the intergenic S/MARs may also be functional, e.g. to provide a biologically meaningful scaffolding of genomes for further in silico as well as laboratory-based experimental research.
MATERIALS AND METHODS
Five pseudomolecules representing each of the Arabidopsis chromosomes were concatenated using genome-tiling path data from MAtDB (08/11/02 release). Gene annotation and coordinates for each of the annotated protein-coding features were anchored to the pseudomolecules, and coordinates were stored in a relational database.
S/MAR prediction was performed using the SMARTest program (http://www.genomatix.de; Genomatix Software, Munich) as published (Frisch et al., 2002). SMARTest does not use a consensus sequence approach. The approach is based on a library of S/MAR-associated, AT-rich patterns derived from comparative sequence analysis of experimentally defined S/MAR sequences. Density analysis of the matches of these S/MAR-associated weight matrices is used for the prediction of S/MARs in genomic DNA sequences. The original training set of SMARTest contained 16 plant S/MARs (7 of these were from Arabidopsis) in a total of 34 S/MARs, and plant and mammalian S/MARs can be located with the same program and settings. Sensitivity (38%) and specificity (68%) of SMARTest has been confirmed using genomic sequences with experimentally defined S/MARs for evaluation (Frisch et al., 2002). We used SMARTest default settings for the analysis of the Arabidopsis genome. The coordinates delimiting the chromosomal location of each of the candidate S/MARs was anchored to the pseudomolecules and appended to the database schema.
The technology has also been applied for a genome-wide prediction of S/MARs in mammalian genomes (human, mouse, and rat). These and also the Arabidopsis data are available in the Genomatix ElDorado system (http://www.genomatix.de). The list of Arabidopsis S/MARs is also available as Supplemental Tables I to V (available at www.plantphysiol.org).
For the random-test analysis of the results, the mapped S/MARs were randomly distributed across the chromosome on which they were originally found. The distribution procedure was invoked 50 times, so there were 50 different copies of the dataset with randomized S/MAR locations. While the S/MARs were distributed throughout the genome, size constraints were maintained so each S/MAR maintained its original size.
To assess the effect of intragenic S/MARs upon gene expression, we made use of the Arabidopsis MPSS data available at the University of Delaware (http://mpss.udel.edu). MPSS involves parallel sequencing of millions of short cDNA fragments from a specific tissue (Brenner et al., 2000a, 2000b). The 8.8 × 106 MPSS sequence reads were consolidated into a matrix describing MPSS sequences and their relative abundance in the 10 different experimental tissues and conditions measured. Each MPSS signature was anchored to corresponding Arabidopsis genes, and ambiguous MPSS sequences were removed. An ambiguous sequence was defined as a sequence that could not be clearly anchored to an annotated Arabidopsis gene or to those sequences that matched more than one Arabidopsis gene.
For each gene cognate, MPSS tag counts were summed up for all 10 experiments used. For all Arabidopsis genes containing at least 50 bp of S/MAR overlap with either intron or exon structure, the average number of MPSS sequence reads per gene was determined. As a background noise threshold, a gene was only scored if within the sum of all 10 experiments used, a total normalized abundance of 10 transcripts per million had been scored. As a control, all genes within the genome that are not associated with an S/MAR were subjected to the same analysis and classification. The number of genes that did not contain any MPSS tags above threshold was also compared between genes containing or not containing S/MARs. To exclude the possibility that any results were caused by sampling effects, 10 replicates of the experiment were repeated using randomly selected populations of Arabidopsis genes while retaining the size of both the S/MAR-containing or non-S/MAR-containing populations. The analyses were repeated using just the experimentally validated, known Arabidopsis genes.
To validate our observations of the effect of intragenic S/MARs upon gene expression, we made use of EST databases. The complete collection of Arabidopsis ESTs from dbEST (March 2002) was anchored to Arabidopsis genes on the basis of BLASTN sequence similarities. An EST was assigned only to the best Arabidopsis gene match, and matches that did not satisfy the assignment criteria of 10−10 were rejected. If at least one EST could be assigned, the gene was counted as transcribed/expressed. The March 2002 dbEST was used since the 113,000 Arabidopsis ESTs represented were from standard cDNA libraries and represented undirected clone sequencing strategies. Newer EST collections contained full-length cDNA sequences and are less representative of random gene expression.
S/MAR assignment to intron/exon features used both the S/MAR coordinates and the coordinates of the gene features. An S/MAR was assigned to a gene feature when a minimal sequence overlap of 50 bp was measured. The number of S/MARs assigned to a feature was expressed as the percentage of all S/MARs that could be assigned to the feature. Relative overrepresentation or underrepresentation of S/MARs was assessed by expressing the observed data against the expected data averaged from the 50 jumbled datasets as a ratio.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers NC_003070, NC_003071, NC_003074, NC_003075, and NC_003076.
Supplementary Material
This work was supported by the Bundesministerium für Bildung und Forschung (grant no. 0312270/4) and the National Science Foundation Plant Genome Research Program (award no. 0110528).
The online version of this article contains Web-only data.
References
- Allen GC, Hall G Jr, Michalowski S, Newman W, Spiker S, Weissinger AK, Thompson WF (1996) High-level transgene expression in plant cells: effects of a strong scaffold attachment region from tobacco. Plant Cell 8: 899–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen GC, Spiker S, Thompson WF (2000) Use of matrix attachment regions (MARs) to minimize transgene silencing. Plant Mol Biol 43: 361–376 [DOI] [PubMed] [Google Scholar]
- Antes TJ, Chen J, Cooper AD, Levy-Wilson B (2000) The nuclear matrix protein CDP represses hepatic transcription of the human cholesterol-7alpha hydroxylase gene. J Biol Chem 275: 26649–26660 [DOI] [PubMed] [Google Scholar]
- Antes TJ, Namciu SJ, Fournier RE, Levy-Wilson B (2001) The 5′ boundary of the human apolipoprotein B chromatin domain in intestinal cells. Biochemistry 40: 6731–6742 [DOI] [PubMed] [Google Scholar]
- Avramova Z, Paneva E (1992) Matrix attachment sites in the murine alpha-globin gene. Biochem Biophys Res Commun 182: 78–85 [DOI] [PubMed] [Google Scholar]
- Avramova Z, SanMiguel P, Georgieva E, Bennetzen JL (1995) Matrix attachment regions and transcribed sequences within a long chromosomal continuum containing maize Adh1. Plant Cell 7: 1667–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avramova Z, Tikhonov A, Chen M, Bennetzen JL (1998) Matrix attachment regions and structural colinearity in the genomes of two grass species. Nucleic Acids Res 26: 761–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banan M, Rojas IC, Lee WH, King HL, Harriss JV, Kobayashi R, Webb CF, Gottlieb PD (1997) Interaction of the nuclear matrix-associated region (MAR)-binding proteins, SATB1 and CDP/Cux, with a MAR element (L2a) in an upstream regulatory region of the mouse CD8a gene. J Biol Chem 272: 18440–18452 [DOI] [PubMed] [Google Scholar]
- Bode J, Schlake T, Rios-Ramirez M, Mielke C, Stengert M, Kay V, Klehr-Wirth D (1995) Scaffold/matrix-attached regions: structural properties creating transcriptionally active loci. Int Rev Cytol 162A: 389–454 [DOI] [PubMed] [Google Scholar]
- Bode J, Stengert-Iber M, Kay V, Schlake T, Dietz-Pfeilstetter A (1996) Scaffold/matrix-attached regions: topological switches with multiple regulatory functions. Crit Rev Eukaryot Gene Expr 6: 115–138 [DOI] [PubMed] [Google Scholar]
- Bonifer C, Hecht A, Saueressig H, Winter DM, Sippel AE (1991) Dynamic chromatin: the regulatory domain organization of eukaryotic gene loci. J Cell Biochem 47: 99–108 [DOI] [PubMed] [Google Scholar]
- Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, Ewan M, et al (2000. a) Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol 18: 630–634 [DOI] [PubMed] [Google Scholar]
- Brenner S, Williams SR, Vermaas EH, Storck T, Moon K, McCollum C, Mao JI, Luo S, Kirchner JJ, Eletr S, et al (2000. b) In vitro cloning of complex mixtures of DNA on microbeads: physical separation of differentially expressed cDNAs. Proc Natl Acad Sci USA 97: 1665–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun C, Dang Q, Miassod R (1990) Studies of an 800-kilobase DNA stretch of the Drosophila X chromosome: comapping of a subclass of scaffold-attached regions with sequences able to replicate autonomously in Saccharomyces cerevisiae. Mol Cell Biol 10: 5455–5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinn AM, Comai L (1996) The heat shock cognate 80 gene of tomato is flanked by matrix attachment regions. Plant Mol Biol 32: 959–968 [DOI] [PubMed] [Google Scholar]
- Cockerill PN, Yuen MH, Garrard WT (1987) The enhancer of the immunoglobulin heavy chain locus is flanked by presumptive chromosomal loop anchorage elements. J Biol Chem 262: 5394–5397 [PubMed] [Google Scholar]
- Copenhaver GP, Nickel K, Kuromori T, Benito MI, Kaul S, Lin X, Bevan M, Murphy G, Harris B, Parnell LD, et al (1999) Genetic definition and sequence analysis of Arabidopsis centromeres. Science 286: 2468–2474 [DOI] [PubMed] [Google Scholar]
- Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA (1999) Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol 145: 1119–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CSHL/WUGSC/PEB Arabidopsis Sequencing Consortium (2000) The complete sequence of a heterochromatic island from a higher eukaryote. The Cold Spring Harbor Laboratory, Washington University Genome Sequencing Center, and PE Biosystems Arabidopsis Sequencing Consortium. Cell 100: 377–386 [PubMed] [Google Scholar]
- Farache G, Razin SV, Rzeszowska-Wolny J, Moreau J, Targa FR, Scherrer K (1990) Mapping of structural and transcription-related matrix attachment sites in the alpha-globin gene domain of avian erythroblasts and erythrocytes. Mol Cell Biol 10: 5349–5358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell RB (1994) Inactivation of gene expression in plants as a consequence of specific sequence duplication. Proc Natl Acad Sci USA 91: 3490–3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz PF, Armstrong S, de Jong JH, Parnell LD, van Drunen C, Dean C, Zabel P, Bisseling T, Jones GH (2000) Integrated cytogenetic map of chromosome arm 4S of A. thaliana: structural organization of heterochromatic knob and centromere region. Cell 100: 367–376 [DOI] [PubMed] [Google Scholar]
- Frisch M, Frech K, Klingenhoff A, Cartharius K, Liebich I, Werner T (2002) In silico prediction of scaffold/matrix attachment regions in large genomic sequences. Genome Res 12: 349–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda Y (2000) Interaction of nuclear proteins with intrinsically curved DNA in a matrix attachment region of a tobacco gene. Plant Mol Biol 44: 91–98 [DOI] [PubMed] [Google Scholar]
- Gasser SM, Amati BB, Cardenas ME, Hofmann JF (1989) Studies on scaffold attachment sites and their relation to genome function. Int Rev Cytol 119: 57–96 [DOI] [PubMed] [Google Scholar]
- Glazko GV, Koonin EV, Rogozin IB, Shabalina SA (2003) A significant fraction of conserved noncoding DNA in human and mouse consists of predicted matrix attachment regions. Trends Genet 19: 119–124 [DOI] [PubMed] [Google Scholar]
- Hall G Jr, Allen GC, Loer DS, Thompson WF, Spiker S (1991) Nuclear scaffolds and scaffold-attachment regions in higher plants. Proc Natl Acad Sci USA 88: 9320–9324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder PA, Silverstein RA, Meier I (2000) Conservation of matrix attachment region-binding filament-like protein 1 among higher plants. Plant Physiol 122: 225–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan AB, Errington RJ, White NS, Jackson DA, Cook PR (1994) Replication and transcription sites are colocalized in human cells. J Cell Sci 107: 425–434 [DOI] [PubMed] [Google Scholar]
- Jarman AP, Higgs DR (1988) Nuclear scaffold attachment sites in the human globin gene complexes. EMBO J 7: 3337–3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellum R, Schedl P (1992) A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol Cell Biol 12: 2424–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi-Shigematsu T, Maass K, Bode J (1997) A thymocyte factor SATB1 suppresses transcription of stably integrated matrix-attachment region-linked reporter genes. Biochemistry 36: 12005–12010 [DOI] [PubMed] [Google Scholar]
- Li Q, Stamatoyannopoulos G (1994) Hypersensitive site 5 of the human beta locus control region functions as a chromatin insulator. Blood 84: 1399–1401 [PubMed] [Google Scholar]
- Lin X, Kaul S, Rounsley S, Shea TP, Benito MI, Town CD, Fujii CY, Mason T, Bowman CL, Barnstead M, et al (1999) Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature 402: 761–768 [DOI] [PubMed] [Google Scholar]
- Liu J, Bramblett D, Zhu Q, Lozano M, Kobayashi R, Ross SR, Dudley JP (1997) The matrix attachment region-binding protein SATB1 participates in negative regulation of tissue-specific gene expression. Mol Cell Biol 17: 5275–5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer K, Schuller C, Wambutt R, Murphy G, Volckaert G, Pohl T, Dusterhoft A, Stiekema W, Entian KD, Terryn N, et al (1999) Sequence and analysis of chromosome 4 of the plant Arabidopsis thaliana. Nature 402: 769–777 [DOI] [PubMed] [Google Scholar]
- McKnight RA, Shamay A, Sankaran L, Wall RJ, Hennighausen L (1992) Matrix-attachment regions can impart position-independent regulation of a tissue-specific gene in transgenic mice. Proc Natl Acad Sci USA 89: 6943–6947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalowski SM, Allen GC, Hall GE Jr, Thompson WF, Spiker S (1999) Characterization of randomly-obtained matrix attachment regions (MARs) from higher plants. Biochemistry 38: 12795–12804 [DOI] [PubMed] [Google Scholar]
- Mielke C, Kohwi Y, Kohwi-Shigematsu T, Bode J (1990) Hierarchical binding of DNA fragments derived from scaffold-attached regions: correlation of properties in vitro and function in vivo. Biochemistry 29: 7475–7485 [DOI] [PubMed] [Google Scholar]
- Mirkovitch J, Gasser SM, Laemmli UK (1988) Scaffold attachment of DNA loops in metaphase chromosomes. J Mol Biol 200: 101–109 [DOI] [PubMed] [Google Scholar]
- Mlynarova L, Hricova A, Loonen A, Nap JP (2003) The presence of a chromatin boundary appears to shield a transgene in tobacco from RNA silencing. Plant Cell 15: 2203–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlynarova L, Jansen RC, Conner AJ, Stiekema WJ, Nap JP (1995) The MAR-mediated reduction in position effect can be uncoupled from copy number-dependent expression in transgenic plants. Plant Cell 7: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepveu A (2001) Role of the multifunctional CDP/Cut/Cux homeodomain transcription factor in regulating differentiation, cell growth and development. Gene 270: 1–15 [DOI] [PubMed] [Google Scholar]
- Oancea AE, Berru M, Shulman MJ (1997) Expression of the (recombinant) endogenous immunoglobulin heavy-chain locus requires the intronic matrix attachment regions. Mol Cell Biol 17: 2658–2668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AL, Ferl RJ (1998) Higher order chromatin structures in maize and Arabidopsis. Plant Cell 10: 1349–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phi-Van L, von Kries JP, Ostertag W, Stratling WH (1990) The chicken lysozyme 5′ matrix attachment region increases transcription from a heterologous promoter in heterologous cells and dampens position effects on the expression of transfected genes. Mol Cell Biol 10: 2302–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires Martins R, Leach RE, Krawetz SA (2001) Whole-body gene expression by data mining. Genomics 72: 34–42 [DOI] [PubMed] [Google Scholar]
- Razin SV (2001) The nuclear matrix and chromosomal DNA loops: Is their any correlation between partitioning of the genome into loops and functional domains? Cell Mol Biol Lett 6: 59–69 [PubMed] [Google Scholar]
- Romig H, Ruff J, Fackelmayer FO, Patil MS, Richter A (1994) Characterisation of two intronic nuclear-matrix-attachment regions in the human DNA topoisomerase I gene. Eur J Biochem 221: 411–419 [DOI] [PubMed] [Google Scholar]
- Sawasaki T, Takahashi M, Goshima N, Morikawa H (1998) Structures of transgene loci in transgenic Arabidopsis plants obtained by particle bombardment: junction regions can bind to nuclear matrices. Gene 218: 27–35 [DOI] [PubMed] [Google Scholar]
- Schoof H, Zaccaria P, Gundlach H, Lemcke K, Rudd S, Kolesov G, Arnold R, Mewes HW, Mayer KF (2002) MIPS Arabidopsis thaliana Database (MAtDB): an integrated biological knowledge resource based on the first complete plant genome. Nucleic Acids Res 30: 91–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler D, Mielke C, Maass K, Bode J (1996) Scaffold/matrix-attached regions act upon transcription in a context-dependent manner. Biochemistry 35: 11160–11169 [DOI] [PubMed] [Google Scholar]
- Stunkel W, Huang Z, Tan SH, O'Connor MJ, Bernard HU (2000) Nuclear matrix attachment regions of human papillomavirus type 16 repress or activate the E6 promoter, depending on the physical state of the viral DNA. J Virol 74: 2489–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar RM, Lilly JW, Town CD, Cheng Z, Kaul S, Buell CR, Jiang J (2001) Complex mtDNA constitutes an approximate 620-kb insertion on Arabidopsis thaliana chromosome 2: implication of potential sequencing errors caused by large-unit repeats. Proc Natl Acad Sci USA 98: 5099–5103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surdej P, Got C, Miassod R (1990) Developmental expression pattern of a 800 kb DNA continuum cloned from the Drosophila X chromosome 14B-15B region. Biol Cell 68: 105–118 [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Tikhonov AP, Bennetzen JL, Avramova ZV (2000) Structural domains and matrix attachment regions along colinear chromosomal segments of maize and sorghum. Plant Cell 12: 249–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov AP, Lavie L, Tatout C, Bennetzen JL, Avramova Z, Deragon JM (2001) Target sites for SINE integration in Brassica genomes display nuclear matrix binding activity. Chromosome Res 9: 325–337 [DOI] [PubMed] [Google Scholar]
- van Drunen CM, Oosterling RW, Keultjes GM, Weisbeek PJ, van Driel R, Smeekens SC (1997) Analysis of the chromatin domain organisation around the plastocyanin gene reveals an MAR-specific sequence element in Arabidopsis thaliana. Nucleic Acids Res 25: 3904–3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Goldstein A, Zong RT, Lin D, Neufeld EJ, Scheuermann RH, Tucker PW (1999) Cux/CDP homeoprotein is a component of NF-muNR and represses the immunoglobulin heavy chain intronic enhancer by antagonizing the bright transcription activator. Mol Cell Biol 19: 284–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw CB, Grolli S, Accornero P, Donofrio G, Farini E, Webster J (2000) Matrix attachment region regulates basal beta-lactoglobulin transgene expression. Gene 244: 73–80 [DOI] [PubMed] [Google Scholar]
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, et al (2003) Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302: 842–846 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.