Abstract
Carefully curated proteomes of the inner envelope membrane, the thylakoid membrane, and the thylakoid lumen of chloroplasts from Arabidopsis were assembled based on published, well-documented localizations. These curated proteomes were evaluated for distribution of physical-chemical parameters, with the goal of extracting parameters for improved subcellular prediction and subsequent identification of additional (low abundant) components of each membrane system. The assembly of rigorously curated subcellular proteomes is in itself also important as a parts list for plant and systems biology. Transmembrane and subcellular prediction strategies were evaluated using the curated data sets. The three curated proteomes differ strongly in average isoelectric point and protein size, as well as transmembrane distribution. Removal of the cleavable, N-terminal transit peptide sequences greatly affected isoelectric point and size distribution. Unexpectedly, the Cys content was much lower for the thylakoid proteomes than for the inner envelope. This likely relates to the role of the thylakoid membrane in light-driven electron transport and helps to avoid unwanted oxidation-reduction reactions. A rule of thumb for discriminating between the predicted integral inner envelope membrane and integral thylakoid membrane proteins is suggested. Using a combination of predictors and experimentally derived parameters, four plastid subproteomes were predicted from the fully annotated Arabidopsis genome. These predicted subproteomes were analyzed for their properties and compared to the curated proteomes. The sensitivity and accuracy of the prediction strategies are discussed. Data can be extracted from the new plastid proteome database (http://ppdb.tc.cornell.edu).
Plastids are essential organelles of prokaryotic origin that are present in nearly every plant cell. Plastids are built up out of several compartments: (1) the outer and inner envelope membranes surrounding the plastids, providing a barrier for proteins and small molecules; (2) the soluble stroma within plastids, expected to contain thousands of different proteins; and (3) in the case of chloroplasts, the thylakoid membrane, an internal membrane system, holding the photosynthetic electron transport chain, as well as an unknown number of other proteins. The thylakoid membrane system forms large vesicles and its intrathylakoid space is called the thylakoid lumen, which contains an additional set of proteins.
To understand plastid function, biogenesis, and biosynthetic pathways, it is critical to characterize the plastid proteome: This includes protein expression levels, protein-protein interactions, and subplastid localization. A first step in the characterization of the plastid proteome is the careful assembly of all experimentally determined plastid proteins and their suborganellar localization. In addition to experimental data sets, prediction tools can and have been developed to predict the plastid proteome and suborganellar proteomes, with varying degrees of success (Abdallah et al., 2000; Emanuelsson, 2002; Koo and Ohlrogge, 2002; Peltier et al., 2002).
Numerous individual plastid proteins, as well as larger sets of plastid proteins, can be collected from the literature (for review, see van Wijk, 2004). Thus, it is possible to assemble high-quality experimental data sets for different chloroplast subproteomes. Since various experimental approaches have been used for identification of these proteins, a bias for a particular class of proteins is likely avoided. In addition, extensive biochemical analysis, as well as recent successes in x-ray crystallography of the higher plant photosynthetic thylakoid membrane protein complexes, allows for accurate determination of the number of transmembrane domains (TMDs) of many thylakoid proteins (Barber, 2002; Ben-Shem et al., 2003; Stroebel et al., 2003; Liu et al., 2004). Such well-curated experimental protein sets can be used to extract valuable information about the physical-chemical properties and to evaluate localization and TMD prediction strategies.
Most proteins localized in plastids are synthesized as precursor proteins in the cytosol with a cleavable chloroplast transit peptide (cTP). Features of these cTPs are discussed by von Heijne (1989); Claros et al. (1997); and Bruce (2000). The subcellular localization prediction programs, Predotar (http://www.inra.fr/Internet/Produits/Predotar/) and TargetP (http://www.cbs.dtu.dk/services/TargetP; Emanuelsson et al., 2000) predict the presence of a cTP with fairly good accuracy and sensitivity. The neural network predictor TargetP can also predict the cleavage site of the cTP, which is important when analyzing the features of mature plastid proteins, as will also become evident in the current study. TargetP has been used in combination with TMD predictors (TMHMM), signal peptides (SignalP; Nielsen et al., 1997; Krogh et al., 2001), and experimentally derived filters to predict the lumenal (Peltier et al., 2002), as well as the hydrophobic, proteome and possible candidates in the inner chloroplast envelope (Ferro et al., 2002; Koo and Ohlrogge, 2002). The proteins in the outer envelope membrane generally do not have a cTP and cannot easily be predicted. However, since many are identified as β-barrel proteins, proteins with predicted β-sheets can be evaluated as candidate members of the outer envelope membrane (Schleiff et al., 2003).
One particularly important question in plastid biogenesis is how nuclear-encoded integral membrane proteins (more than 500 are predicted) are targeted to the inner envelope and thylakoid membrane. With the exception of those proteins that carry a lumenal transit peptide (lTP) for targeting of the N terminus to the lumen (Mori and Cline, 2001; Robinson et al., 2001) or an L18 domain in the case of a subset of chlorophyll-binding thylakoid proteins (Tu et al., 2000), it is unclear how these putative integral membrane proteins are sorted within the chloroplast. Is this selection taking place early during envelope translocation, possibly involving Tic110 and/or Tic40 (Chou et al., 2003; Inaba et al., 2003), or does it occur after processing of the cTP in the stroma, following the so-called conservative sorting principle?
In this study, we carefully collected published experimental sets of integral thylakoid membrane proteins and integral inner envelope proteins. We analyzed these subproteomes, as well as the experimental thylakoid lumenal proteome, for their properties, with the objective of extracting potential predictors of subplastid localization and evaluating the number of putative chloroplast membrane proteins and their TMDs. Subsequently, we used the existing predictors, TargetP, SignalP, and TMHMM, as well as a newly developed predictor for the lumenal proteome, LumenP (Westerlund et al., 2003), in combination with different filters to predict four chloroplast subproteomes: (1) the soluble lumenal proteome; (2) the stromal proteome; (3) the combined integral membrane proteome of thylakoids and inner membrane; and (4) the thylakoid membrane proteins with lTPs. These predicted plastid subproteomes were compared to the curated experimental sets. Distributions of predicted functional domains across the subproteomes are discussed. All parameters and the curated data sets can be extracted from the new plastid proteome database (PPDB; http://ppdb.tc.cornell.edu).
RESULTS
Assembly of the Curated Experimental Integral Inner Envelope and Thylakoid Proteomes
The nonredundant accession numbers for known thylakoid and inner envelope proteins of Arabidopsis were carefully collected from the literature and public databases. All proteins were then evaluated for being an integral membrane protein; for the most part, this was based on experimental data (e.g. not extractable with urea or salts, x-ray structure, or topology mapping by proteolysis, etc.) and, in some cases, based on prediction of TMDs by TMHMM or the consensus prediction reported in the Aramemnon database (http://aramemnon.botanik.uni-koeln.de/). If available, the number of experimentally determined TMDs for each protein was recorded. Importantly, only those proteins were listed for which it was very clear that they are integral either to the thylakoid membrane or to the inner envelope membrane. In total, 65 (75 when including alternative gene models in The Arabidopsis Information Resource [TAIR]; see also below) nuclear-encoded thylakoid membrane and 24 (27 when including different gene models) nuclear-encoded inner envelope membrane proteins were obtained (see Supplemental Table I at www.plantphysiol.org).
We also assembled extended experimental thylakoid and inner envelope integral membrane sets, adding additional integral membrane proteins for each membrane system, using data from Friso et al. (2004), Ferro et al. (2003), and Froehlich et al. (2003), respectively. Proteins were manually selected based on the confirmed presence of a cTP and the convincing presence of multiple TMDs (as above). This added, respectively, 18 (20 with alternative gene models) and 15 (16 with alternative gene models) proteins to the thylakoid membrane and inner envelope membrane sets (see Supplemental Table I; see also the PPDB). In addition, 51 (53 when allowing for different gene models) thylakoid lumen proteins assembled earlier (Peltier et al., 2002; Westerlund et al., 2003) were included in the analysis (see Supplemental Table I; see also the PPDB). Finally, all 38 plastid-encoded integral thylakoid membrane proteins, as well as 39 plastid-encoded stromal localized proteins, were assembled (see Supplemental Table I).
Comparing the Experimental Proteomes
A number of parameters (length, pI, grand average of hydrophobicity [GRAVY; Kyte and Doolittle, 1982], number of TMDs, and amino acid composition) were calculated or predicted for precursor and processed proteins. Median and average values for each parameter were calculated for each of the experimental sets (Table I) and frequency distribution plots were generated. These parameters are available for each accession via the PPDB. None of the frequency distribution plots for the different parameters showed normal distributions, and distributions for some parameters were bi- or trimodal. Therefore, it is important to compare both median and average values (Table I) and examine the plotted frequency distributions.
Table I.
Median and average values for different physicochemical parameters for the curated experimental subproteomes
| Curated Experimental Setsa | Median (Average) | Median (Average) | Median (Average) | Median (Average) | |
|---|---|---|---|---|---|
| Unprocessed | Lengthb | GRAVYc | pId | Rel. Cys | ne |
| Inner envelope membrane | 447 (479) | 0.08 (0.12) | 9.19 (8.75) | 0.011 (0.013) | 27 (24) |
| Expanded envelope | 433 (455) | 0.11 (0.11) | 9.14 (8.64) | 0.012 (0.013) | 43 (39) |
| Thylakoid membrane | 261 (289) | −0.02 (0.02) | 6.89 (7.44) | 0.005 (0.008) | 76 (65) |
| Expanded thylakoid | 254 (276) | −0.02 (0.01) | 7.28 (7.51) | 0.006 (0.008) | 95 (83) |
| Lumen (soluble) | 247 (284) | −0.21 (−0.19) | 8.53 (7.78) | 0.011 (0.014) | 53 (51) |
| Stroma | 284 (345) | −0.26 (−0.25) | 7.62 (7.46) | 0.008 (0.009) | 86 (73) |
| Minus cTP | |||||
| Inner envelope membrane | 398 (427) | 0.06 (0.15) | 8.77 (8.12) | 0.010 (0.013) | 27 (24) |
| Expanded envelope | 378 (404) | 0.12 (0.14) | 8.70 (8.01) | 0.011 (0.012) | 43 (39) |
| Thylakoid membrane | 213 (244) | −0.05 (0.01) | 5.33 (6.13) | 0.005 (0.008) | 76 (65) |
| Expanded thylakoid | 204 (232) | −0.05 (0.01) | 5.22 (6.02) | 0.005 (0.008) | 95 (83) |
| Lumen (soluble) | 208 (239) | −0.22 (−0.21) | 5.82 (6.49) | 0.009 (0.011) | 53 (51) |
| Stroma | 234 (293) | −0.33 (−0.32) | 5.63 (6.42) | 0.005 (0.008) | 86 (73) |
| Minus cTP + lTP | |||||
| Lumen (soluble) | 165 (201) | −0.33 (−0.34) | 5.31 (6.26) | 0.006 (0.009) | 53 (51) |
| Plastid encoded integral thylakoid | 140 (252) | 0.66 (0.67) | 6.45 (6.85) | 0.005 (0.005) | 38 |
| Plastid encoded stromal proteins | 138 (357) | −0.41 (−0.43) | 10.32 (9.77) | 0.013 (0.015) | 39 |
The accession numbers for each curated set are listed in Supplemental Table 1.
Prediction of cTP by ChloroP;
length in number of amino acids;
measure of hydrophobicity;
bimodal distribution;
number of proteins, including all gene models in TAIR and, in parentheses, the number when only counting a gene model for each accession.
Protein Size
Very significant differences in protein size were observed between the experimentally identified lumenal, the integral thylakoid membrane, and the integral inner envelope proteomes (Table I). The envelope proteins were, on average, twice as large as the lumenal proteins and 75% bigger than the integral thylakoid proteins. The plastid-encoded integral thylakoid membrane proteins were, on average, 252 amino acids, but with a median value of only 140 amino acids (Table I).
Number of TMDs and Hydrophobicity
The number of TMDs of the curated thylakoid and inner envelope proteome was predicted either by TMHMM or by consensus prediction as reported in the Aramemnon database. In the case of TMHMM, cTPs and lTPs were excluded as potential TMDs, and, in the case of Aramemnon, the authors report that TMDs overlapping with predicted N-terminal signal peptides (by SignalP) are removed. TMDs for the plastid-encoded integral thylakoid proteins were only predicted by TMHMM, since they are not included in Aramemnon.
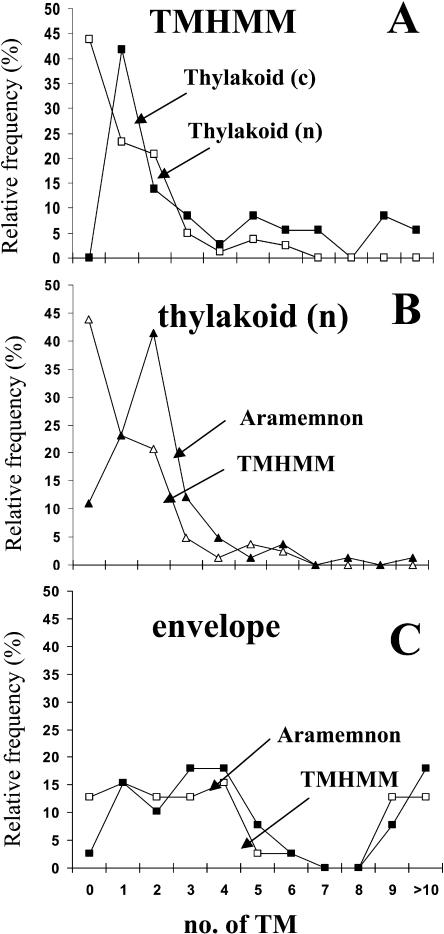
The distribution of these predicted TMDs is shown in Figure 1, A to C. Figure 1A shows the distribution of predicted TMDs by TMHMM for the chloroplast- and nuclear-encoded thylakoid membrane proteins. Clearly, the thylakoid proteome is dominated by proteins predicted to have zero to three TMDs and a small group of proteins with nine or more TMDs (Fig. 1A). These TMD predictions were compared with the experimental TMD determinations reported in the literature. In the case of the chloroplast-encoded thylakoid membrane proteins, TMHMM slightly overpredicted the TMDs, with a total of 77 predicted for 72 known TMDs.
Figure 1.
Frequency distribution of predicted TMDs of the curated membrane subproteomes. The curated sets are the extended nuclear-encoded (n) thylakoid membrane proteome with 83 proteins (A–C), the extended inner envelope membrane proteome with 39 proteins (A and B), and the chloroplast-encoded (c) thylakoid membrane proteome with 38 members (A). TMD predictions were by TMHMM (A–C) or consensus prediction as reported in Aramemnon (B and C). Predicted transit peptides were removed. All accession numbers are listed in Supplemental Table I.
In the case of the nuclear-encoded thylakoid membrane proteins, a very significant percentage was not predicted to have any TMD. It turned out that TMHMM has a specific problem with predicting the 29 nuclear-encoded chlorophyll- and carotenoid- binding thylakoid membrane proteins (light-harvesting complexes [LHCs], Elips, PsbS, and Seps or Lils, Hlips, Scps, and Ohps; Jansson, 1999; Adamska, 2001). This LHC superfamily has an average of 2.82 TMDs, but TMHMM predicted only an average of 0.31. In contrast, Aramemnon listed quite an accurate prediction (Fig. 1), with an average of 2.10 TMDs. The poor prediction by TMHMM is most likely related to the presence of (conserved) positively and negatively charged residues within the TMDs used for helix-helix interactions (Adamska, 2001). Aramemnon and TMHMM both predicted very similarly that the curated envelope proteome falls into two classes of proteins with either one to four TMDs or nine or more TMDs (Fig. 1C).
Surprisingly, approximately 50% of the curated lumenal proteome was predicted to be a membrane protein by Aramemnon. In contrast, only one out of the 53 confirmed lumenal proteins was a predicted membrane protein by TMHMM after removal of predicted cTPs. This overprediction was caused by the very long bipartite targeting sequence of lumenal proteins. The lTP, which is typically 40 to 50 amino acids down-stream of the N terminus, is identified as a TMD by many TMD predictors; a typical example, for instance, is the well-known lumenal electron transporter plastocyanin (At1g20340).
A different way to characterize the integral membrane proteomes is by hydrophobicity, as calculated by the GRAVY index. The processed integral thylakoid proteome was, on average, slightly less hydrophobic than the processed inner envelope membrane proteome (Table I). The chloroplast-encoded thylakoid membrane proteome was significantly more hydrophobic than both nuclear-encoded proteomes (Table I). Cross-correlation of the GRAVY index with the number of predicted TMDs per amino acid listed in Aramemnon showed a linear correlation for the nuclear-encoded envelope and thylakoid membrane proteomes (Fig. 2, A and B). Since the chloroplast-encoded proteins are not in the Aramemnon database, we plotted for this group only the cross-correlation of the GRAVY index with the number of TMDs predicted by TMHMM (Fig. 2C); a good linear correlation was observed, as is evident from the regression analysis (Fig. 2C). This shows that the chloroplast-encoded thylakoid proteins are more hydrophobic because they have more TMDs per amino acid length.
Figure 2.
Hydrophobicity and TMD prediction of the curated membrane proteomes. Correlation of the ratio between predicted number of TMDs and the number of amino acid residues (TMD:aa) and GRAVY index for the expanded nuclear-encoded inner membrane envelope (A) and the expanded thylakoid membrane proteomes (B), as well as the chloroplast-encoded thylakoid membrane proteome (C). These are the same sets as used for Figure 1. TMDs were obtained from Aramemnon (A and B) or predicted by TMHMM (C). The regression analysis is indicated for each plot.
pI
The distribution of pI is strongly bimodal for the processed lumenal proteome (Table I; Fig. 3A). Removal of the cleavable cTPs and lTPs has a dramatic influence on the pI distribution, with median pI values of 8.53 for the unprocessed lumenal proteome and 5.31 for the processed proteome (Table I; Fig. 3A). In an earlier study of the thylakoid lumen proteome, we showed that these predicted pI values of processed proteins matched well (typically within 0.5 pH unit deviation) to the experimental pI values determined from two-dimensional gels (Peltier et al., 2002). This emphasizes that in silico proteome analysis of physical-chemical parameters should be done after removal of cleavable signal/transit peptides. Also, the processed integral thylakoid membrane proteome is characterized by acidic median (5.22) and average (6.0) pI values (Table I; Fig. 3A). In contrast, the processed (and unprocessed) inner envelope proteome is, on average, basic, with a median pI value of 8.7 and an average pI value of 8.1 (Table I; Fig. 3A). The two membrane proteomes showed a bi- or trimodal distribution of pI values (Fig. 3A). Manual inspection of the proteins at high and low pI did not suggest particular functions for the outliers in both populations. The low pI of the thylakoid membrane and lumenal proteome can be explained by a high content of the abundant acidic residues Asp (D) (pKr = 3.9) and Glu (E) (pKr = 4.07), when compared to the envelope proteome (Supplemental Fig. 1).
Figure 3.
pI and Cys content of the expanded curated lumenal, thylakoid membrane and inner envelope membrane proteome. A, Frequency distribution of the pI for the full-length and processed curated proteomes (full-length, white squares, cTP removed, black squares, and cTP and lTP removed, black triangles). B, Frequency distribution of proteins in the three curated proteomes based on the number of Cys residues. Membrane sets are the same as used in Figures 1 and 2, and the lumenal set contained 51 proteins (see Supplemental Table I).
Cys Content
Cys residues play an important role in redox reactions, as ligands and in stabilization of proteins and protein complexes by formation of disulfide bonds (Giles et al., 2003). Surprisingly, the Cys content is much lower for thylakoid proteins in the lumen and membrane, as compared to proteins in the inner envelope or elsewhere in the chloroplast (Table I). Importantly, the plastid-encoded integral thylakoid membrane proteome also has a low relative Cys content, with a median of 0.003 and an average of 0.005, expressed as the abundance ratio between Cys over all amino acid residues. This is about 3 times less than the Cys content in the envelope membrane proteome. The chloroplast-encoded stromal proteome has a much higher average Cys content of 0.015, clearly suggesting that the reduced Cys content in the thylakoid is related to its particular function. When expressed into the number of Cys residues per protein, 35% to 40% of the thylakoid lumen and membrane proteins have no Cys at all, whereas nearly all experimentally identified inner envelope membrane proteins have one or more Cys residues, with a peak at four Cys residues per protein (Fig. 3B). This significance will be discussed below.
cTP Characteristics
An important question in plastid biogenesis is how nuclear-encoded thylakoid membrane and inner envelope proteins are directed to the correct membrane within the chloroplast. We compared, therefore, the predicted cTPs for the inner envelope proteome, the thylakoid membrane proteome, the lumenal proteome, as well as a newly assembled set of stromal and peripheral proteins (Supplemental Table III). Average and median lengths of the predicted cTPs for the inner envelope proteome were 10% to 13% longer than for the integral and lumenal thylakoid proteomes (Supplemental Table II). To evaluate if any amino acid motifs are present upstream and downstream of the predicted cTP cleavage site, we aligned the proteins within each of the four curated subproteomes around the predicted cTP, using sequence logos (a graphic representation of sequence conservation and amino acid frequency (Schneider and Stephens, 1990), but no significant differences were observed (Supplemental Fig. 2).
Functional Domains
Functional domains for all TAIR accessions were predicted by automated PFAM-HMM (with a cutoff of E = 0.1; Bateman et al., 2002) and are available via the PPDB. The top-scoring domains for the curated envelope proteome are related to biosynthesis or transport, whereas the top-scoring domains for the curated thylakoid membrane proteome are chlorophyll a/b-binding (24×), peptidase family M41 (5×), protein kinase (5×), AAA domains (4×), and bacterial DNA-binding domains (4×). We compared these distributions to the predicted functional domains of all 577 plastid-predicted integral membrane proteins. This gave 430 different domains, with the top-scoring domains being C3HC4-type Zinc finger (24×), acyltransferase (22×), branched-chain amino acid transport system (21×), and protein kinase domain (19×).
TargetP Sensitivity for the Experimental Plastid Subproteomes
The TargetP sensitivity (defined as the fraction of plastid-predicted proteins out of all true plastid proteins in our data sets) was 96%, 86%, and 78%, respectively, for the thylakoid lumen and membrane proteomes of the thylakoid and inner envelope (Supplemental Table II). These sensitivities are in the same range as the 85% reported originally (Emanuelsson et al., 2000). Investigation of the overlap between the TargetP training set and the curated set of lumenal, thylakoid, and envelope proteins showed that only 15 curated proteins were part of the original training set (details are provided under “Materials and Methods”). Removal of those 15 did not affect the sensitivity.
The experimental sets were then extended to encompass all known thylakoid proteins (lumenal, peripheral, and integral membrane), as well as proteins copurified with thylakoid and envelope proteins (stromal), totaling 283 proteins (Supplemental Table I). All are confirmed chloroplast proteins and they were initially identified by various biochemical and genetic approaches, thus avoiding experimental bias. TargetP predicted 89% of the 283 proteins correctly (Supplemental Table II) and 90% correctly after removal of 43 (15%) proteins with homology to a protein in the TargetP training set. A larger, unpublished, experimental data set with well over 400 stromal proteins from chloroplasts and non-green plastids confirmed this approximately 90% sensitivity (J.B. Peltier, Y. Cai, G. Friso, L. Giacomelli, V. Zabrouskov, Q. Sun, and K. van Wijk, unpublished data). The high sensitivity of TargetP is important for the prediction of plastid subproteomes, as detailed below.
Prediction of Plastid Subproteomes
We predicted the plastid proteome and subplastid localization by screening the latest annotation of the Arabidopsis genome (4.0 of ATH1.pep) using two prediction schemes (Fig. 4, A and B). The predictor, TargetP, was either combined with LumenP (Westerlund et al., 2003) or SignalP and, in both cases, followed by TMHMM. The predictions were improved by inclusion of several filters developed earlier (Peltier et al., 2002) and were either incorporated into the predictor LumenP (Fig. 4A) or added after prediction by SignalP (Fig. 4B). These prediction schemes resulted in four chloroplast subproteomes: (1) plastid stroma; (2) thylakoid lumen, split in two populations based on the presence or absence of a twin arginine motif (TAT; Mori and Cline, 2001); (3) integral membrane proteins, in which inner envelope membrane proteins and thylakoid membrane proteins are clustered, since no method using amino acid sequence data only exists to separate them; and (4) integral membrane proteins with an lTP.
Figure 4.
Overview of the two prediction strategies and comparisons of predictions and experimentation. A, Prediction strategy using LumenP. B, Prediction strategy using SignalP. C, Overlap of proteins predicted by the two prediction strategies with 291 and 285 proteins, respectively, and the experimental lumenal proteome with 53 proteins (using all gene models in TAIR). D, Comparison of distribution cTP + lTP length for the curated and predicted lumenal proteomes.
It was predicted that 4,255 proteins have a cTP (14.9% of the total Arabidopsis proteome; Table II). Of those, 520 have one or more TMDs and are located in the inner envelope or thylakoid membrane, 291 (LumenP) or 285 (SignalP) are predicted to be soluble in the lumen, and an additional 57 integral membrane proteins were predicted to have an lTP. The remaining 3,387 proteins are assigned stromal proteins; however, it is important to note that these can be permanently or transiently associated with the thylakoid and/or inner envelope membrane via protein-protein, electrostatic, or hydrophobic interactions, as well as lipid anchors, but this is currently not possible to predict.
Table II.
Size of the predicted subproteomes, using a combination of TargetP, TMHMM, LumenP, and SignalP, as well as filters derived from experimental data sets (as outlined in Fig. 4, A and B)
| n | % | % | |
|---|---|---|---|
| All Arabidopsis | 28,581a | 100 | n.r. |
| All membraneb | 6,320 | 22.11 | n.r. |
| All Ctp | 4,255c | 14.90 | 100 |
| Stromad | 3,387 | 11.80 | 79.60 |
| Membranede | 520 | 1.80 | 12.20 |
| Membrane + lTPd | 57 | 0.20 | 1.30 |
| Lumend (LumenP) | 291 | 1.00 | 6.80 |
| Lumen + TATd (LumenP) | 71 | 0.25 | 1.70 |
| Lumen (SignalP) | 285 | 1.00 | 6.70 |
| Lumen + TAT (SignalP) | 50 | 0.17 | 1.12 |
Represents 28,581 gene models for 27,170 genes.
TMHMM; 7,238 (25.3%) in Aramemnon.
Represents 4,013 genes with 4,255 gene models.
Using the prediction scheme with LumenP (see Fig. 4A).
1,015 (23.9% of all cTP-predicted proteins) in Aramemnon.
Thus, our prediction scheme suggests that 13.5% of the predicted plastid proteins are integral membrane proteins, compared to 22% predicted membrane proteins for the complete Arabidopsis genome (Table II). The consensus TMD prediction listed in the Aramemnon database currently reports that 24% of the 4,028 cTP proteins are predicted membrane proteins, compared to 25% of the Arabidopsis proteome (Table II; see “Discussion” for further details).
Comparing the Predicted and Experimental Lumenal Proteomes
We compared the two prediction strategies (outlined in Fig. 4, A and B) for the lumenal proteome. The total number of predicted proteins was very similar for the two approaches, and both approaches gave very similar sensitivities (56% and 60%; Supplemental Table II; Fig. 4C). Twenty-six out of 51 confirmed lumenal proteins were correctly predicted by both approaches, and both approaches predicted some additional known lumenal proteins correctly, as well as 150 proteins with no confirmed lumenal location. However, both strategies also predicted an additional, nonoverlapping set of over 100 proteins. Since the rest of the lumenal proteome is unknown, it is hard to evaluate these two nonoverlapping sets.
We then compared the features of the experimental lumenal proteome with the predicted lumenal proteomes. The average and median lengths of proteins in the two predicted lumenal proteomes were 50% to 60% longer than the experimental lumenal proteome. This suggests that either the experimental set was biased toward smaller proteins or the predicted proteome contained a number of false positives. Comparison of the length of the cTP + lTP between experimental sets and predicted sets shows higher median and average values for the predicted sets (Fig. 4D). This suggests that the cutoff for maximum cTP + lTP length was too high. Strikingly, the cTP + lTP length for the predicted TAT proteins was very similar to the experimentally determined length, suggesting a more robust prediction for this subclass, as also demonstrated earlier (Peltier et al., 2002). This seems logical since the TAT motif in a narrow window upstream of the predicted lTP cleavage site is unique.
Comparing the Predicted Integral Membrane Proteins and Curated Thylakoid and Envelope Proteomes
The properties (length, GRAVY, pI, number of TMDs, and relative Cys content) of the predicted subproteomes, before and after removal of the predicted cTPs and lTPs, were analyzed and displayed using frequency distribution plots (data not shown). Mean and average values were calculated (Supplemental Table IV). The median and average lengths of the predicted chloroplast integral membrane proteome (inner envelope and thylakoid) after removal of the predicted cTP are 327 and 382 amino acids, respectively. This is in between the values for the experimental integral thylakoid proteome (213/244; Table I) and the integral inner envelope membrane proteome (398/427). The average and median pI values of the predicted total plastid membrane proteome are trimodal, with an average pI value of 7.39 (Supplemental Table IV).
Predicted Integral Membrane Proteins with an lTP
Several proteins have been identified that are integral to the thylakoid membrane and have an N-terminal lumenal transit peptide. Known cases are CFo-II (At4g32260), psbW (At2g30570), PsbX (At2g06520), psbT1 (At3g21055), and psaF (At1g31330). Insertion of these proteins seems to occur without assistance of other proteins or energy requirements (Robinson et al., 2000). Our analysis predicted a total of 57 proteins in this class and psaF was one of them. This predicted lTP class had a lower Cys content and was smaller and less basic than the curated envelope proteome, but similar to the curated thylakoid membrane proteome, suggesting that indeed a significant fraction of these proteins are localized to the thylakoid.
Alternative Gene Models in TAIR
The TAIR database reports more than one gene model for 1,411 (5%) out of 27,170 genes of the annotated Arabidopsis genome (1,141 genes with two, 109 with three, and 17 with four or more gene models; each gene model might be biologically relevant). The 4,255 gene models predicted to have a cTP represented 4,013 genes (Table II). cTP prediction differentiated between the gene models for 36 out of those 4,013 genes. In the case of the curated subproteomes, TargetP prediction was only rarely affected by the different gene models presented in TAIR. Exceptions include carbonic anhydrase (At3g01500), for which three forms are present in TAIR (.1, .2, and .3). At3g01500.1 is not predicted to go to the plastid, whereas version .2 and .3 are correctly predicted to be plastid localized. The explanation is that At3g01500.1 is N-terminally truncated, thus lacking a proper cTP.
DISCUSSION
For a complete understanding of plant functions and biosynthetic and signaling pathways, it is important to determine and characterize the proteomes at different subcellular locations. This will also be important in long-term efforts to develop faithful, quantitative models for plant processes (for discussion, see Raikhel and Coruzzi, 2003).
Experimental proteomics using modern mass spectrometry has become a powerful tool, with continuous improvements in sensitivity, dynamic resolution, and quantification (Aebersold and Mann, 2003). However, it remains challenging to identify proteins that are expressed only under particular conditions (e.g. adverse growth conditions, particular developmental stage, etc.), or with very low expression levels. It may be possible to predict these low abundant or transient proteins using prediction strategies as discussed in this study. Focused experimental approaches can then be used to identify these candidate proteins experimentally.
Experimental proteome analysis of subcellular compartments can provide extensive protein sets to either train predictors or extract experimental parameters for design of filters, as we demonstrated earlier for the thylakoid lumen (Peltier et al., 2002). Indeed, there are now a fairly large number of localization predictors specialized for different organelle locations (Nakai, 2000; Emanuelsson and von Heijne, 2001). Accurate prediction of integral membrane proteins and their topology is equally important, since membrane proteins often fulfill critical functions. However, accurate prediction of TMDs is rather difficult because protein topology is not only determined by primary amino acid sequence, but also by membrane chaperones and post-translational modifications (Ott and Lingappa, 2002).
In this study, we focused on curation, analysis, and prediction of the chloroplast subproteomes encoded by the nuclear and plastid genomes of Arabidopsis. We carefully assembled proteins located in either the thylakoid membrane system or the plastid inner envelope membrane, with the goal of obtaining a better overview of the respective integral membrane proteomes and finding specific features for each set of proteins, possibly with predictive value for membrane localization. Significant sets of proteins could indeed be assembled and their analysis showed clear differences in the properties of each membrane proteome and associated functions. It is unlikely that these differentiating properties are due to an experimental bias, since these proteins and their corresponding genes were originally identified using various strategies, ranging from reverse and forward genetics screens, highly specific cross-link experiments, as well as more recent proteomics approaches involving gels or chromatography, followed by different types of mass spectrometry techniques.
The pI distribution was bimodal for both membrane systems (possibly trimodal for the envelope proteome), with, on average, a basic integral inner envelope membrane proteome and an acidic integral thylakoid membrane proteome. These pI distributions were strongly affected by removal of the predicted transit peptides. Currently, there is no good explanation for this pI distribution of thylakoid and envelope proteome, but it is likely related to the pH in the lumenal, stromal, and intra-envelope space. It is unclear if and how this connects to the positive-inside rule, which states that membrane proteins have, on average, a net positive charge on the loops facing the cis-side of the membrane (Gavel et al., 1991; Sipos and von Heijne, 1993). Proteins are least soluble when the pH of the medium is close to their pI. The stromal pH fluctuates between 7 and 8 and, indeed, the majority of the stromal proteome generally avoid this pI range. The curated soluble lumenal proteome shows a clear avoidance of pI values between 6.5 and 8.5, whereas the pH in the lumenal compartments fluctuates widely between 3.5 and 7. Thus the relationship between the lumenal pH and the pI of the lumenal proteome cannot simply be explained in terms of solubility. pI distribution was also bimodal for bacterial proteomes and trimodal for eukaryotic proteomes of yeast, worm, and fly. This was also related to localization, but an explanation is not known (VanBogelen et al., 1999; Schwartz et al., 2001).
Cys residues have a unique reactivity and they are involved in catalysis, redox activity, structural stabilization, and metal binding (Frand et al., 2000; Giles et al., 2003). In addition, a powerful technique has been developed for comparative proteomics, in which Cys residues are used to link small tags with different stable isotopes (ICAT; Gygi et al., 1999). This prompted us to compare the relative Cys content for the experimental proteomes. The processed thylakoid integral membrane and lumen proteomes have a low Cys content, with median values of 0.005 to 0.006, about half of that of the envelope membrane with median values of 0.012 to 0.013. To put the Cys content in perspective, the median Cys content (Cys/all amino acids) for the complete predicted Arabidopsis proteome (28,786 proteins) is 0.016 and 0.015 for the plastid predicted proteome (4,255 proteins). The primary function of the thylakoid is to carry out light-driven electron transfer reaction. It is possible that the reductive environment of the thylakoid membrane, in combination with excess of protons, is not compatible with a prominent role of disulfide bonds in protein stabilization. The low Cys content could also be a way to avoid unproductive transfer and uptake of electrons, possibly resulting in oxidative damage. We propose that the low Cys content in the lumenal and thylakoid membrane proteins (both nuclear and chloroplast-encoded) is related to the role of the thylakoid membrane in electron transport. It remains to be determined whether this is a phenomenon also observed in other electron transport systems, such as the inner membrane of mitochondria.
Prediction of Inner Envelope and Thylakoid Membrane Localization
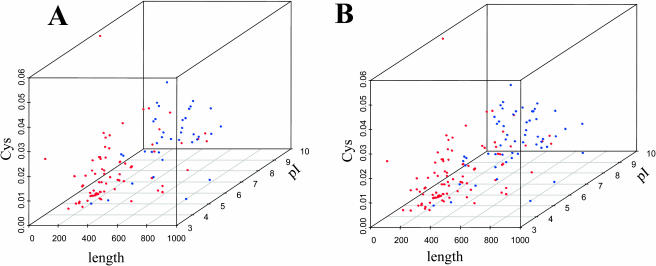
Thylakoid proteins were, on average, smaller, more acidic, and, most significantly, contained less Cys residues when compared to the inner envelope proteome. The question is if these differences contain enough predictive power in order to discriminate integral inner envelope membrane proteins from integral thylakoid membrane proteins when applied to uncharacterized proteins. A three-dimensional plot in which pI, number of Cys residues, and protein length are combined shows that the expanded curated thylakoid and inner envelope membrane proteins (83 and 39 proteins, respectively; see Supplemental Table I) are generally well separated (Fig. 5A). Recently, published experimental envelope proteome studies (Ferro et al., 2002, 2003; Froehlich et al., 2003) and a new thylakoid membrane study (Friso et al., 2004) did identify potential additional inner envelope membrane and thylakoid membrane proteins. After removing overlap between the new experimental thylakoid and envelope data sets and filtering for TargetP prediction and presence of predicted transmembrane domains using TMHMM, this added 27 and 21 putative integral thylakoid and inner envelope membrane proteins, respectively. Creation of a three-dimensional plot (using pI, number of Cys residues, and protein length) with these extended sets (110 and 60 proteins) shows that thylakoid and inner envelope proteins are still generally well separated (Supplemental Fig. 5B).
Figure 5.
Scatter plot of the number of Cys residues, protein length, and pI for the curated sets of inner envelope and thylakoid membrane proteins. Predicted cTPs are removed. A, 83 thylakoid and 39 envelope membrane proteins. B, 110 and 61 thylakoid and envelope membrane proteins, respectively. Envelope proteins are symbolized by blue dots and thylakoid proteins by red dots.
The cTP cleavage site motifs and upstream and downstream amino acid regions for the different (curated and extended) subproteomes did not exhibit any striking differences and did not offer any predictive value for subplastid localization.
Prediction of Membrane Proteins
TMHMM was successful at predicting TMDs for both plastid- and nuclear-encoded thylakoid membrane proteins that were not part of the LHC superfamily (Jansson, 1999; Adamska, 2001). This LHC superfamily with chlorophyll-binding domains was generally not recognized as membrane proteins by TMHMM, but were well predicted by the consensus prediction listed in Aramemnon. This group of proteins is unusual in that some of the TMDs contain one or more conserved charged residues, as well as a conserved Pro. Apparently, TMHMM is sensitive for such unusual features, although TMHMM outperformed all other available predictors according to a recent evaluation (Moller et al., 2001).
It is interesting to note that, when using TMHMM, only 13% of the predicted plastid proteins have predicted TMDs whereas 22% of all annotated Arabidopsis genes are predicted to encode for membrane proteins. There could be two additive biological reasons for this relatively low percentage of membrane proteins in the plastid: (1) a plastid is not a complete organism and has only one internal membrane compartment (the thylakoid), whereas the complete cell has many types of organelles and corresponding membrane systems; and (2) the cTP set includes proteins in both non-green plastids without internal thylakoid membrane system and chloroplasts. The soluble, stromal proteome is expected to be quite different between non-green plastids and chloroplasts, whereas the integral membrane composition of the inner envelope might not be significantly different and an internal membrane system is absent in non-green plastids. The diversity of the plastid would thus add a lot of extra soluble proteins, but possibly few integral membrane proteins.
The consensus prediction listed in the Aramemnon database suggests that the number of membrane proteins in the plastid is 24%, similar to the 25% prediction of membrane proteins in the total Arabidopsis proteome. Manual evaluation of this discrepancy between TMHMM and the accessions listed in Aramemnon for the plastid predicted proteome suggests that many of these discrepancies represent soluble proteins. This is likely resulting from assignment of a significant percentage of lumenal transit peptides and cTPs as TMDs by a number of predictors used in Aramemnon.
Prediction of Proteins with an lTP
Two strategies were used to predict lTPs in both the predicted membrane and soluble proteins. Interestingly, the predicted membrane proteins with lTP have a low Cys content relative to the other predicted subproteomes, suggesting that a significant population was indeed located in the thylakoid. Otherwise, the prediction of the lumenal proteome still seems to be difficult, even when including the experimentally derived filters and considering the fairly large training set (>200 proteins) used for developing LumenP. As mentioned before, only the lumenal proteins with a TAT motif could be fairly well predicted.
CONCLUSIONS
We conclude that analysis of curated plastid subproteomes from existing literature suggests a striking difference in Cys content between integral inner envelope membrane proteins and integral and lumenal thylakoid proteins, in addition to significant differences in protein length, pI, and TMD distribution. Analysis of these assembled thylakoid and inner envelope membrane proteomes did not reveal an obvious sorting signal for either membrane system. Despite partial success, suborganellar location prediction is still in its infancy. More large-scale experimental identifications of subproteomes from different subcellular locations are needed to improve subcellular localization scheme predictions.
MATERIALS AND METHODS
Collection and Curation of Experimental Subproteomes
The literature and public databases were carefully screened for plastid proteins for which the plastid sublocalization was determined. All proteins were then evaluated as being an integral membrane protein. This was mostly based on experimental data (e.g. not extractable with urea or salts; x-ray structure, or topology mapping by proteolysis, etc.) and, in some cases, based on prediction of TMDs by TMHMM or the consensus prediction reported in the Aramemnon database.
Prediction of the Subproteomes
Proteins believed to possess a cTP were extracted using TargetP (accepting all reliability classes; Emanuelsson et al., 2000), and this subset was further processed through (1) TMHMM 2.0 (Krogh et al., 2001) to discover proteins with potential transmembrane regions (helices), and (2) a new lTP prediction program, LumenP (Westerlund et al., 2003), or using SignalP to find proteins with a potential thylakoid lumen location. The predictions of cleavage sites were taken from ChloroP for the cTP cleavage sites and from LumenP or SignalP (neural network version 2.0; Nielsen et al., 1997, 1998) for the lTP cleavage sites. The SignalP results were obtained following the outline in Peltier et al., 2002, running both gram-negative and gram-positive versions of the predictor and submitting N-terminally truncated versions of the full-length proteins to simulate the cTP removal. In total, 26 predictions were obtained for each protein (13 different truncation variants, processed through two versions, gram-negative and gram-positive, of the SignalP predictor) and the one with highest cleavage site score (as determined by the SignalP Y-score) was chosen to represent the protein. In the case of LumenP predictions, a scoring matrix developed specifically to recognize the lTP cleavage site was used to decide on the cleavage site (Westerlund et al., 2003). For both LumenP and SignalP prediction strategies, the maximum length of the cTP + lTP localization signals was 150 amino acids. In the case of predicted lumenal proteins that did not have a TAT motif within their lTP (positions −32 to −18 relative to the lTP cleavage site), the −3, −1 motif (positions −3 and −1 relative to the cleavage site) was required to be AxA (Peltier et al., 2002). All proteins with at least one TMD were assigned as membrane proteins. Some proteins contained both a TMD (or several) and an lTP. If the predicted lTP overlapped with a TMD, the TM prediction was deemed as stronger and the protein was predicted to be located in the membrane. However, if the predicted lTP did not overlap with a TMD, the protein was put into the group membrane proteins with lTP. The prediction results are also listed for each accession in PPDB (http://ppdb.tc.cornell.edu/).
Calculation of Physical-Chemical Parameters
Molecular weight, pI, GRAVY, and amino acid composition were calculated using the Emboss software suite (Rice et al., 2000). pI values were obtained from (Bjellqvist et al., 1994). These prediction parameters are also listed for each accession in the PPDB.
BLAST Analysis of the Original TargetP Training Set
The positive training set for TargetP consisted of a set of 141 confirmed chloroplast proteins from different higher plant species (data set available at http://www.cbs.dtu.dk/services/TargetP/datasets/datasets.php). A BLAST search was carried out to find the Arabidopsis orthologues for each of these 141 proteins; E-values were between 9.10−9 and 0, with 90% below 10−40, indicating that homologs were found for all 141 proteins. Due to one-to-many or many-to-many homology relationships, the actual number of Arabidopsis homologs to these 141 proteins was 113 (see Supplemental Table I). Only 15 of these 113 proteins were part of the most conservative curated lumenal, thylakoid membrane, and inner envelop set (1 in the lumenal set, 13 in thylakoid membrane set, and 1 in the inner envelope).
Construction of the Plastid Proteome Database
The database engine for the PPDB (http://ppdb.tc.cornell.edu/) is an MS SQL Server. The Web interface for the PPDB is developed on ASP.NET platform using C# language. The functional domain prediction was based on PFAM analysis, with a cutoff E-value at 0.1 (Bateman et al., 2002). The experimental data in the PPDB (including curated gene information) are provided by members of the van Wijk lab.
Supplementary Material
Acknowledgments
We thank members of the van Wijk lab for helpful discussions and corrections and Prof. Gunnar von Heijne for comments and support. Rainer Schwacke is gratefully acknowledged for sending an Aramemnon data set and for helpful discussions.
This work was supported by the National Science Foundation (MCB no. 0090942) and NYSTAR (grant to K.J.v.W.). All large-scale data collection at Cornell was conducted using the resources of the Cornell Theory Center, which receives funding from Cornell University, New York State, federal agencies, foundations, and corporate partners.
The online version of this article contains Web-only data.
References
- Abdallah F, Salamini F, Leister D (2000) A prediction of the size and evolutionary origin of the proteome of chloroplasts of Arabidopsis. Trends Plant Sci 5: 141–142 [DOI] [PubMed] [Google Scholar]
- Adamska I (2001) The Elip family of stress proteins in the thylakoid membrane of pro- and ukaryota. In E-M Aro, B Anderson, eds, Regulation of Photosynthesis, Vol 11. Kluwer Academic Publishers, London, UK
- Aebersold R, Mann M (2003) Mass spectrometry-based proteomics. Nature 422: 198–207 [DOI] [PubMed] [Google Scholar]
- Barber J (2002) Photosystem II: a multisubunit membrane protein that oxidises water. Curr Opin Struct Biol 12: 523–530 [DOI] [PubMed] [Google Scholar]
- Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer EL (2002) The Pfam protein families database. Nucleic Acids Res 30: 276–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Frolow F, Nelson N (2003) Crystal structure of plant photosystem I. Nature 426: 630–635 [DOI] [PubMed] [Google Scholar]
- Bjellqvist B, Basse B, Olsen E, Celis JE (1994) Reference points for comparisons of two-dimensional maps of proteins from different human cell types defined in a pH scale where isoelectric points correlate with polypeptide compositions. Electrophoresis 15: 529–539 [DOI] [PubMed] [Google Scholar]
- Bruce BD (2000) Chloroplast transit peptides: structure, function and evolution. Trends Cell Biol 10: 440–447 [DOI] [PubMed] [Google Scholar]
- Chou ML, Fitzpatrick LM, Tu SL, Budziszewski G, Potter-Lewis S, Akita M, Levin JZ, Keegstra K, Li HM (2003) Tic40, a membrane-anchored co-chaperone homolog in the chloroplast protein translocon. EMBO J 22: 2970–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros MG, Brunak S, vonHeijne G (1997) Prediction of N-terminal protein sorting signals. Curr Opin Struct Biol 7: 394–398 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O (2002) Predicting protein subcellular localisation from amino acid sequence information. Brief Bioinform 3: 361–376 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, von Heijne G (2001) Prediction of organellar targeting signals. Biochim Biophys Acta 1541: 114–119 [DOI] [PubMed] [Google Scholar]
- Ferro M, Salvi D, Brugiere S, Miras S, Kowalski S, Louwagie M, Garin J, Joyard J, Rolland N (2003) Proteomics of the chloroplast envelope membranes from Arabidopsis thaliana. Mol Cell Proteomics 2: 325–345 [DOI] [PubMed] [Google Scholar]
- Ferro M, Salvi D, Riviere-Rolland H, Vermat T, Seigneurin-Berny D, Grunwald D, Garin J, Joyard J, Rolland N (2002) Integral membrane proteins of the chloroplast envelope: identification and subcellular localization of new transporters. Proc Natl Acad Sci USA 99: 11487–11492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frand AR, Cuozzo JW, Kaiser CA (2000) Pathways for protein disulphide bond formation. Trends Cell Biol 10: 203–210 [DOI] [PubMed] [Google Scholar]
- Friso G, Giacomelli L, Ytterberg AJ, Peltier JB, Rudella A, Sun Q, Wijk KJ (2004) In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. Plant Cell 16: 478–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froehlich JE, Wilkerson CG, Ray WK, McAndrew RS, Osteryoung KW, Gage DA, Phinney BS (2003) Proteomic study of the Arabidopsis thaliana chloroplastic envelope membrane utilizing alternatives to traditional two-dimensional electrophoresis. J Proteome Res 2: 413–425 [DOI] [PubMed] [Google Scholar]
- Gavel Y, Steppuhn J, Herrmann R, von Heijne G (1991) The ‘positive-inside rule’ applies to thylakoid membrane proteins. FEBS Lett 282: 41–46 [DOI] [PubMed] [Google Scholar]
- Giles NM, Giles GI, Jacob C (2003) Multiple roles of cysteine in biocatalysis. Biochem Biophys Res Commun 300: 1–4 [DOI] [PubMed] [Google Scholar]
- Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R (1999) Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol 17: 994–999 [DOI] [PubMed] [Google Scholar]
- Inaba T, Li M, Alvarez-Huerta M, Kessler F, Schnell DJ (2003) atTic110 functions as a scaffold for coordinating the stromal events of protein import into chloroplasts. J Biol Chem [DOI] [PubMed]
- Jansson S (1999) A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci 4: 236–240 [DOI] [PubMed] [Google Scholar]
- Koo AJ, Ohlrogge JB (2002) The predicted candidates of Arabidopsis plastid inner envelope membrane proteins and their expression profiles. Plant Physiol 130: 823–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305: 567–580 [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157: 105–132 [DOI] [PubMed] [Google Scholar]
- Liu Z, Yan H, Wang K, Kuang T, Zhang J, Gui L, An X, Chang W (2004) Crystal structure of spinach major light-harvesting complex at 2.72 A resolution. Nature 428: 287–292 [DOI] [PubMed] [Google Scholar]
- Moller S, Croning MD, Apweiler R (2001) Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 17: 646–653 [DOI] [PubMed] [Google Scholar]
- Mori H, Cline K (2001) Post-translational protein translocation into thylakoids by the Sec and DeltapH-dependent pathways. Biochim Biophys Acta 1541: 80–90 [DOI] [PubMed] [Google Scholar]
- Nakai K (2000) Protein sorting signals and prediction of subcellular localization. Adv Protein Chem 54: 277–344 [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G (1997) A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int J Neural Syst 8: 581–599 [DOI] [PubMed] [Google Scholar]
- Nielsen H, Krogh A (1998) Prediction of signal peptides and signal anchors by a hidden Markov model. Proc Int Conf Intell Syst Mol Biol 6: 122–130 [PubMed] [Google Scholar]
- Ott CM, Lingappa VR (2002) Integral membrane protein biosynthesis: why topology is hard to predict. J Cell Sci 115: 2003–2009 [DOI] [PubMed] [Google Scholar]
- Peltier JB, Emanuelsson O, Kalume DE, Ytterberg J, Friso G, Rudella A, Liberles DA, Soderberg L, Roepstorff P, von Heijne G, et al (2002) Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell 14: 211–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raikhel NV, Coruzzi GM (2003) Achieving the in silico plant. Systems biology and the future of plant biological research. Plant Physiol 132: 404–409 [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16: 276–277 [DOI] [PubMed] [Google Scholar]
- Robinson C, Thompson SJ, Woolhead C (2001) Multiple pathways used for the targeting of thylakoid proteins in chloroplasts. Traffic 2: 245–251 [DOI] [PubMed] [Google Scholar]
- Robinson C, Woolhead C, Edwards W (2000) Transport of proteins into and across the thylakoid membrane. J Exp Bot 51 Spec No: 369–374 [DOI] [PubMed] [Google Scholar]
- Schleiff E, Eichacker LA, Eckart K, Becker T, Mirus O, Stahl T, Soll J (2003) Prediction of the plant beta-barrel proteome: a case study of the chloroplast outer envelope. Protein Sci 12: 748–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T, Stephens RM (1990) Sequence logos: a new way to display consensus sequences. Nucleic Acids Res 18: 6097–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz R, Ting CS, King J (2001) Whole proteome pI values correlate with subcellular localizations of proteins for organisms within the three domains of life. Genome Res 11: 703–709 [DOI] [PubMed] [Google Scholar]
- Sipos L, von Heijne G (1993) Predicting the topology of eukaryotic membrane proteins. Eur J Biochem 213: 1333–1340 [DOI] [PubMed] [Google Scholar]
- Stroebel D, Choquet Y, Popot JL, Picot D (2003) An atypical haem in the cytochrome b(6)f complex. Nature 426: 413–418 [DOI] [PubMed] [Google Scholar]
- Tu CJ, Peterson EC, Henry R, Hoffman NE (2000) The L18 domain of light-harvesting chlorophyll proteins binds to cpSRP43. J Biol Chem 275: 13187–13190 [DOI] [PubMed] [Google Scholar]
- van Wijk KJ (2004) Chloroplast proteomics. In D Leister, ed, Plant Functional Genomics. The Haworth Press, Inc., Book Division, Binghamton, NY
- VanBogelen RA, Schiller EE, Thomas JD, Neidhardt FC (1999) Diagnosis of cellular states of microbial organisms using proteomics. In Process Citation. Electrophoresis 20: 2149–2159 [DOI] [PubMed] [Google Scholar]
- von Heijne G, Steppuhn J Hermann SG (1989) Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem 80: 535–545 [DOI] [PubMed] [Google Scholar]
- Westerlund I, Von Heijne G, Emanuelsson O (2003) LumenP–a neural network predictor for protein localization in the thylakoid lumen. Protein Sci 12: 2360–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.