Abstract
Controlled vocabularies are increasingly used by databases to describe genes and gene products because they facilitate identification of similar genes within an organism or among different organisms. One of The Arabidopsis Information Resource's goals is to associate all Arabidopsis genes with terms developed by the Gene Ontology Consortium that describe the molecular function, biological process, and subcellular location of a gene product. We have also developed terms describing Arabidopsis anatomy and developmental stages and use these to annotate published gene expression data. As of March 2004, we used computational and manual annotation methods to make 85,666 annotations representing 26,624 unique loci. We focus on associating genes to controlled vocabulary terms based on experimental data from the literature and use The Arabidopsis Information Resource-developed PubSearch software to facilitate this process. Each annotation is tagged with a combination of evidence codes, evidence descriptions, and references that provide a robust means to assess data quality. Annotation of all Arabidopsis genes will allow quantitative comparisons between sets of genes derived from sources such as microarray experiments. The Arabidopsis annotation data will also facilitate annotation of newly sequenced plant genomes by using sequence similarity to transfer annotations to homologous genes. In addition, complete and up-to-date annotations will make unknown genes easy to identify and target for experimentation. Here, we describe the process of Arabidopsis functional annotation using a variety of data sources and illustrate several ways in which this information can be accessed and used to infer knowledge about Arabidopsis and other plant species.
Genome Overview
Arabidopsis is an annual plant of the Brassicaceae family and is commonly found in temperate regions of the world. Its suitability for molecular and genetic experiments has made it one of the most widely studied plants today. It was the first plant genome to be completely sequenced and remains the most completely sequenced eukaryotic genome to date (Arabidopsis Genome Initiative, 2000). Approximately 13,000 researchers around the world are currently engaged in unraveling the functions of this genome and applying the knowledge gained to other plants. When the sequence of the Arabidopsis genome was first reported (Arabidopsis Genome Initiative, 2000), the annotation included a total of 25,498 predicted protein-coding genes. Of these, 69% were classified into nine functional categories using the PEDANT analysis system (Frishman et al., 2001): cellular metabolism, transcription, plant defense, signaling, growth, protein fate, intracellular transport, transport, and protein synthesis. The remaining 30% of gene products could not be assigned to any of these categories. The most recent version of the Arabidopsis genome annotation (The Institute for Genome Research [TIGR] release 5.0) includes 26,207 protein-coding genes and 3,786 pseudogenes (ftp://ftp.tigr.org/pub/data/a_thaliana/ath1/). While computational methods can shed some light on the general categories to which many of these genes belong, experimental approaches are essential to confirm computational predictions and supply the function of genes in cases where no computational prediction is currently possible. Given the large number of uncharacterized genes, experimental characterization of groups of genes, rather than single genes, is essential if significant progress is to be made in the near future. To meet this challenge, the projects initiated under the National Science Foundation 2010 initiative, as well as those supported by other funding agencies such as Deutsche Forschungsgemeinschaft (German Research Foundation), aim to decipher the function of every Arabidopsis gene by the year 2010 (MASC Committee, 2003) by combining high-throughput approaches with domain expertise. About 20,500 unique genes are currently being studied by various functional genomics project investigators (http://www.arabidopsis.org/info/2010_projects/index.jsp). The results of this massive experimental effort need to be summarized, stored in an easily accessible manner, and combined with information available from studies of individual genes.
A central goal of The Arabidopsis Information Resource (TAIR) project is to integrate information from various data sources and present the research community with a comprehensive view of each Arabidopsis gene. Functional annotation is defined as the process of collecting information about and describing a gene's biological identity—its various aliases, molecular function, biological role(s), subcellular location, and its expression domains within the plant. At TAIR, we obtain this information from reading the published literature and by soliciting contributions from the research community as well as from computational analyses of the genome sequence. We present the collated information in two ways: (1) in a short summary for each gene that contains its essential attributes and (2) as multiple gene-term associations (or annotations) between a controlled vocabulary term and the gene product. Each annotation is associated with an evidence code, an evidence description, and a reference on which the association is based. We use the Gene Ontology (GO) vocabularies (www.geneontology.org; GO Consortium, 2001) as well as TAIR's Arabidopsis anatomy and developmental stage ontologies as the sources for the controlled vocabulary terms.
Controlled Vocabularies
A controlled vocabulary is a standardized, restricted set of defined terms designed to reduce ambiguity in describing a concept. For example, one publication might refer to enzyme A as having phytochromobilin synthase activity, while another says that enzyme B has phytochromobilin:ferredoxin oxidoreductase activity. Both enzyme A and enzyme B perform identical functions; the terms describing them are synonymous. Without an explicitly defined standard term, searching for all gene products with this function is difficult and requires knowledge of all possible synonyms.
The GO vocabularies are gaining widespread acceptance within the scientific community as the standard set of terms to use for functional annotation (Dwight et al., 2002; Camon et al., 2003; Hazbun et al., 2003; Hennig et al., 2003; Kanapin et al., 2003; King et al., 2003; Sprague et al., 2003). The terms are organized into three categories that represent molecular functions, biological processes, and subcellular compartments (GO Consortium, 2001). Molecular function terms describe the biochemical activity performed by a gene product (e.g. kinase activity). Biological process terms describe the ordered assembly of more than one molecular function (e.g. flower development). Cellular component terms describe the subcellular compartments of a cell (e.g. nucleus). The terms are used to describe these separate aspects of a gene product's biological identity. The vocabularies are developed and maintained by a consortium of model organism databases (MODs). Curators from the MODs work together to ensure that the terms are uniformly agreed upon, clearly defined, and broadly applicable to a wide taxonomic range of species. As a GO consortium member since 2000, TAIR has been instrumental in modifying and expanding the vocabularies so that they can be used to accurately describe plant genes. The consortium maintains a central database (http://www.godatabase.org/cgi-bin/go.cgi) that stores the gene-term associations contributed by its member MODs. Having a central repository for all annotation information allows one to retrieve groups of genes from multiple species that are associated with a single term. There are currently 16,808 terms: 8,181 for biological processes, 7,278 for molecular functions, and 1,379 for cellular components (http://www.geneontology.org/index.shtml#downloads).
Most of the terms have explicit definitions, and all of them are arranged in an ontology, a structured hierarchy with defined relationships between terms (GO Consortium, 2001). The term definitions and relationships between terms are intended to reflect the current state of knowledge about a particular term. The terms are organized such that the broader concepts, or parent terms, appear on the top level on the tree structure and are composed of more specific concepts, or child terms. Broader concepts, for example, the term plastid, are used to group more specific concepts, such as amyloplast, chloroplast, chromoplast, and etioplast together. Parent-child relationships are structured such that a child term can be either an instance of or part of a parent term. Thus, a chloroplast is an instance of a plastid, while a plastid is a part of the cytoplasm. Additionally, a child term may have more than one parent term and inherits the characteristics of each parent term. To accommodate instances of multiple parentages, parent-child relationships between terms are represented using a directed acyclic graph (DAG) rather than a simple hierarchy (Fig. 1). In such cases, each parent-child relationship reflects a different aspect of this term's definition. Terms and their relationships with one another are added to, evaluated, and updated on a regular basis to keep pace with the knowledge in that field.
Figure 1.
Visualizing controlled vocabularies and DAGs. TAIR's Keyword Browser (http://www.arabidopsis.org/servlets/Search?action=new_search&type=keyword) allows users to navigate through the parent-child relationships of the ontologies, look up definitions, and view associated data. Hyperlinks are underlined, and clicking on them will open data pages that list the associated information in greater detail. Section A offers an option to view various data type associated with the term. Section B provides the term name, its identification, and an explicit definition of the term. Section C is a legend for interpreting the icons within the tree structure. Section D allows one to browse any listed ontology other than the one being viewed. Section E illustrates the multiple parentage concept in a DAG using the biological process term germination. In this example, germination is an instance of three different parent terms: cell differentiation, post-embryonic development, and physiological process.
Since the scope of GO does not extend to terms describing supracellular structures and developmental stages, we used the principles underlying the GO ontologies to develop two additional sets of controlled vocabulary terms describing Arabidopsis anatomy and developmental stages that can be used to describe gene expression patterns and mutant phenotypes. Under the auspices of the Plant Ontology Consortium (www.plantontology.org), we are collaborating with Gramene, Maize Genetics and Genomics Database (MaizeGDB), the Missouri Botanical Garden, and the University of Missouri (St. Louis) to merge these terms into a common vocabulary that will be used to annotate gene expression and phenotypes of major groups of agriculturally and economically important plants.
RESULTS
Current State of Functional Annotation of the Arabidopsis Genome
Annotation of all Arabidopsis genes to controlled vocabulary terms that describe their biological identity is an ongoing process begun by TAIR in 2002. As of March 2004, we associated a total of 26,624 loci to 1,095 biological process terms, 1,146 molecular function terms, 260 cellular component terms, 120 anatomy terms, and 33 developmental stage terms for a total of 85,666 annotations. Of these, 33,733 annotations to 20,260 loci were manual annotations done by a curator. One gene may have multiple process, function, and/or component annotations, depending on the amount of information available in its associated literature. We have identified approximately 3,600 Arabidopsis genes that have been described in about 6,500 publications obtained from PubMed, Agricola, BIOSIS, and the meeting abstracts of the International Conference on Arabidopsis Research and have assigned at least one GO term to nearly all of these genes. Annotations are made not only to sequenced protein-coding genes and pseudogenes but also to approximately 570 mapped genetic loci where the molecular sequence has not been identified and the only information available pertains to their mutant phenotypes.
We have also used computational methods to generate annotations to a large number of genes, many of which have not been described in the literature. There are currently about 42,500 annotations from INTERPRO2GO mapping, about 11,500 annotations based on TargetP predictions, about 600 from Metacyc2GO mapping, and about 350 from a string matching algorithm. Taking these annotations into account, 20,818 genes (69% of the genome) have at least one GO annotation from TAIR. Upon integration of TIGR's GO annotations, the total number of Arabidopsis genes with at least one GO assignment to a known term increases to 22,570 genes, including protein coding genes, pseudogenes, and genetic loci, covering approximately 75% of the genome (Table I). This results in a 6% increase in functional classification since the initial Arabidopsis Genome Initiative genome analysis in 2000, which covered 69% of the genome.
Table I.
Arabidopsis genome functional annotation statistics as of March 4, 2004
| Number of Annotations | Number of Genes Annotateda | |
|---|---|---|
| Functional annotations made by TAIR and TIGRb | 121,933 | 28,331 |
| Biological process annotations: | ||
| Known | 25,955 | 14,621 |
| Unknown | 13,241 | 12,853 |
| Total annotated | 39,196 | 27,469 |
| Unannotated | n/a | 3,713 |
| Molecular function annotations: | ||
| Known | 36,686 | 16,432 |
| Unknown | 11,657 | 11,588 |
| Total | 48,343 | 27,959 |
| Unannotated | n/a | 3,223 |
| Cellular component annotations: | ||
| Known | 22,115 | 15,752 |
| Unknown | 11,323 | 10,951 |
| Total annotated | 33,438 | 26,703 |
| Unannotated | n/a | 4,479 |
| Functional annotations made by TAIRb | 85,666 | 26,624 |
| TAIR GO annotationsb | 84,708 | 30,063 |
| TAIR computational annotations to GO | 50,975 | 19,218 |
| TAIR manual annotations to GOb | 33,733 | 20,260 |
| TAIR annotations to anatomy and temporal ontology | 958 | 443 |
| TAIR annotations to anatomy ontology | 867 | 423 |
| TAIR annotations to temporal ontology | 91 | 76 |
Numbers include annotations to genetic loci that have not been sequenced. This leads to a higher total number of genes than that predicted from the genome sequence.
Numbers include annotations to unknown terms.
n/a, Not applicable.
Since not all genes, even those that have been described in the literature, have been characterized in detail, curators assign the terms molecular function unknown, cellular component unknown, or biological process unknown to any gene that has been manually inspected and does not have any evidence (in the literature or by computational prediction) to support a known process, function, or subcellular component annotation. For example, a gene that has been shown in expression studies to be involved in the biological process response to pathogen may have an undetermined molecular function or subcellular localization. Annotations to the unknown GO terms are useful for delineating what is unknown about a gene and informs the user that the literature for these genes has been inspected and no information on a known function, process, or location was available at the time of annotation. Including associations to unknown terms, 28,331 genes (94% of the genome) have at least one GO annotation. Unannotated genes, which have not yet been assigned a term by computational methods or by a curator, reflect the ongoing nature of this annotation project.
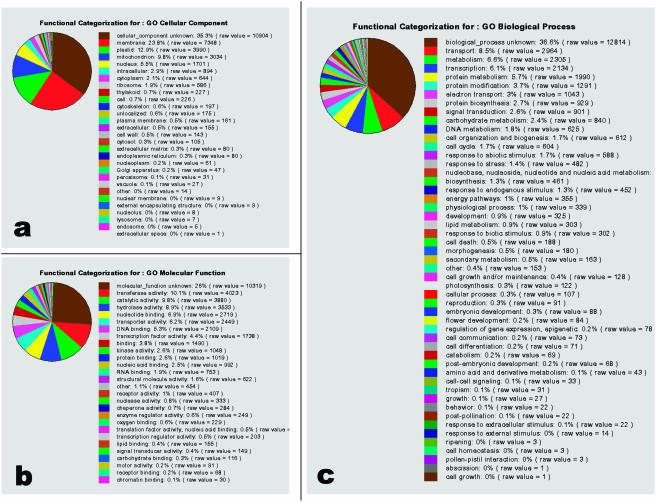
To get an overview of the distribution of the annotations within each ontology, we have chosen some of the high-level terms from each GO hierarchy that are useful for grouping genes into broad categories. Taking the earlier example of plastids, the more specific terms chromoplast, etioplast, chloroplast, and amyloplast can be represented by the single parent term plastid. These high-level terms, called GOslims, are a simplified version of the full ontologies composed of about 40, as opposed to several thousand, terms per ontology. There are several GOslims in use by the GO Consortium; TAIR uses one developed with plant annotations in mind (ftp://ftp.geneontology.org/go/GO_slims/). Using the plant GOslim terms, we have classified the genome into an array of broad functional categories that aid in assessing the distribution of genes among different functions, processes, and subcellular locations (Fig. 2). The resulting distribution shows that most cellular component annotations are to unknown (35%), membrane (24%), and plastid (13%). Molecular function annotations are largely to unknown (26%), followed by transferase activity and catalytic activity (both 10%). The most common biological processes are unknown (37%), transport (8%), and metabolism (7%). The current distribution of genes in known GOslim categories may not accurately reflect biological reality because of the large proportion of computationally derived annotations. As the number of unknown genes is decreased by further experimentation and refinement of computational methods, the number of genes within each category will more accurately reflect the actual distributions of functions, processes, and subcellular locations.
Figure 2.
Functional classification of the whole Arabidopsis genome representing the distribution of genes based on their annotations to terms in the GO cellular component (a), GO molecular function (b), and GO biological process vocabularies (c).
Annotation of Temporal and Spatial Gene Expression Data
In addition to making GO annotations, we have also been using controlled vocabularies to describe the anatomical parts and developmental stages in which a gene is expressed. As part of this effort, we have annotated the protein and/or mRNA expression patterns of more than 400 genes (about 900 annotations; Table I). Combining the GO annotations with the anatomy and temporal annotations for a given gene provides a comprehensive view of the role of a gene in the cell.
Accessing Arabidopsis Controlled Vocabulary Annotations
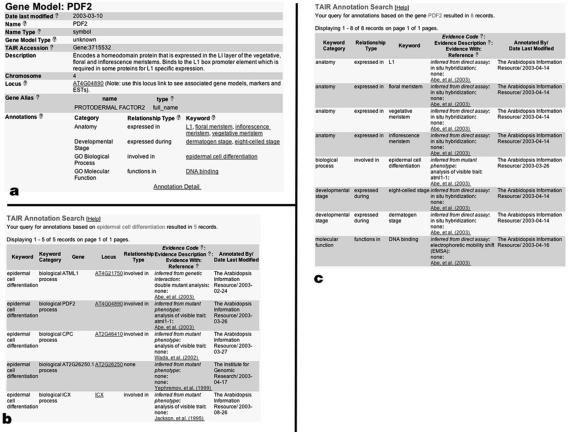
To enable the research community to effectively use these controlled vocabulary annotations, we have developed several tools to search, browse, and download them from TAIR's Web site. Table II provides a complete set of URLs where tools to access the vocabularies and annotations at TAIR and related Web sites can be found. The main search tools for finding genes and associated terms include TAIR's Gene Search and Keyword Browser. The Gene Search allows users to specify the vocabulary type, term name, and many gene-related attributes. Search results are displayed on the Gene detail page (Fig. 3a), which links to the Term Annotation detail (Fig. 3b) and Gene Annotation detail pages (Fig. 3c). Browsing of all the controlled vocabularies and their associated genes can be done using the TAIR Keyword Browser (Fig. 1). One can retrieve GO annotations and plant GOslim mappings for a list of Arabidopsis Genome Initiative locus codes (i.e. AT1G01010) by entering or uploading a locus list into the TAIR GO annotation search, functional categorization, and download tool (http://www.arabidopsis.org/tools/bulk/go/index.jsp). The complete annotation set can be downloaded by ftp (file transfer protocol).
Table II.
Useful Web site links to aid the searching with controlled vocabularies
| Page Names | URL | Usage |
|---|---|---|
| TAIR Gene search | http://www.arabidopsis.org/servlets/Search?action=new_search&type=gene | Search for genes using controlled vocabularies |
| TAIR Keyword Browser | http://www.arabidopsis.org/servlets/Search?action=new_search&type=keyword | Search for or browse controlled vocabulary terms; view term details and term relationships |
| TAIR GO bulk download | http://www.arabidopsis.org/tools/bulk/go/index.jsp | Download GO annotations and functionally categorize a set of genes |
| TAIR and TIGR GO annotations | ftp://ftp.arabidopsis.org/home/tair/Genes/Gene_Ontology/ | Download GO annotations for the whole Arabidopsis genome |
| TAIR anatomy and temporal ontologies | ftp://ftp.arabidopsis.org/home/tair/Ontologies/ | Download Arabidopsis anatomy and temporal ontologies |
| TAIR anatomy annotations | ftp://ftp.arabidopsis.org/home/tair/Genes/Gene_Anatomy/ | Download anatomy annotations for the whole genome |
| TAIR temporal annotations | ftp://ftp.arabidopsis.org/home/tair/Genes/Gene_Developmentalstage/ | Download temporal annotations for the whole genome |
| GO consortium | http://www.geneontology.org/ | Gene Ontology Web site |
| GO database browser | http://www.godatabase.org/cgi-bin/go.cgi | Search for terms and annotations in the GO database |
| OBO | http://obo.sourceforge.net/ | Open Biological Ontologies Web site, which hosts most of the controlled vocabularies |
| Plant Ontology consortium | http://plantontology.org/ | Plant Ontology Web site |
Figure 3.
Display of controlled vocabulary association on the TAIR Gene detail page (a), which summarizes information relevant to gene, the Term Annotation detail page (b), which displays all annotations made to the term in question, and the Gene Annotation detail page (c), which displays all controlled vocabulary annotations made to that gene. These pages are interlinked so that one can get from one page to the next by clicking on the appropriate hyperlink.
Components of Controlled Vocabulary Annotations
A controlled vocabulary association has several parts as defined by the GO Consortium: gene name, associated term and ID, evidence code, reference, annotation date, and annotating database/person (GO Consortium, 2001). To these standard GO annotation components, we have added two fields to present a complete picture of the annotation to the users: evidence description and relationship type. These fields are not submitted to the GO database and are displayed only on TAIR Web site pages.
The combination of evidence code, evidence description, and reference defines the basis for annotation and provides the information necessary for a user to interpret an annotation correctly. The evidence code indicates how the association between the gene and the term is supported. There are 11 evidence codes in use by TAIR and TIGR (see Table III). Annotations derived from computational predictions that have not been reviewed by a curator are given the evidence code IEA (inferred from electronic annotation). Annotations that have been reviewed by a curator are given one of the other evidence codes depending on the type of experimental evidence that was used to make the association. The evidence description provides additional information on the evidence used to support the annotation. In the example shown in Figure 3c, the association between the gene PDF2 and the term epidermal cell differentiation is supported by an IMP (inferred from mutant phenotype) evidence code with an evidence description of analysis of visible trait. Here, the phrase analysis of visible trait provides information about the type of method used to support the association between the gene and the GO term. Evidence descriptions used by TAIR are also a controlled vocabulary currently composed of 107 descriptions. Table IV shows an example of the evidence descriptions used in conjunction with the IPI (inferred from physical interaction) evidence code. Finally, the reference linked to each association gives users a concrete source where the experimental evidence can be found and read about in greater depth. We strive to capture all relevant data, including conflicting views, permitting users to evaluate the supporting evidence themselves.
Table III.
Evidence codes used in functional annotations
| Evidence Code Abbreviation | Evidence Code Definition |
|---|---|
| Computational: | |
| IEA | Inferred from electronic annotation |
| Manual: | |
| IDA | Inferred from direct assay |
| IMP | Inferred from mutant phenotype |
| IEP | Inferred from expression pattern |
| ISS | Inferred from sequence similarity |
| IGI | Inferred from genetic interaction |
| IPI | Inferred from physical interaction |
| TAS | Traceable author statement |
| NAS | Nontraceable author statement |
| ND | No biological data available |
| IC | Inferred by curator |
Table IV.
Unique fields used in TAIR functional annotations
| Unique Field Name | Description | Number of Annotations |
|---|---|---|
| Relationship type | Has | 23,400 |
| Located in | 29,701 | |
| Involved in | 29,087 | |
| Functions as | 760 | |
| Expressed in | 857 | |
| Related to | 639 | |
| Functions in | 194 | |
| Is subunit of | 111 | |
| Constituent of | 75 | |
| Expressed during | 60 | |
| Required for | 31 | |
| Not involved in | 31 | |
| Regulates | 23 | |
| Not expressed in | 24 | |
| Is down-regulated by | 20 | |
| Expressed only in | 20 | |
| Not functions as | 6 | |
| Not located in | 6 | |
| Represses | 3 | |
| Expressed only during | 5 | |
| Not required for | 2 | |
| None | 38,067a | |
| Evidence Description | Yeast two-hybrid assay | 56 |
| (For IPI evidence code) | Coimmunoprecipitation | 28 |
| Copurification | 4 | |
| Yeast one-hybrid | 5 | |
| Cosedimentation | 3 | |
| Sos-recruitment assay | 2 | |
| Far-western analysis | 2 | |
| Split-ubiquitin assay | 1 | |
| None | 36,729a |
Most of the annotations with none as the relationship type or evidence description are those made by TIGR, who have not implemented these extra fields
Relationship type refers to terms that define the association between the gene and the controlled vocabulary term. For example, Figure 3a displays several annotations, one of which states that PDF2 is involved in epidermal cell differentiation. Here, involved in is the relationship type that links the gene PDF2 with the controlled vocabulary term epidermal cell differentiation. The relationship type provides a specific context for the association between the term and the gene that can be used for searching and data mining purposes. It also allows the annotation to be read in a more logical, sentence-like format, helping users understand the functional annotation more intuitively. There are 21 relationship types currently in use by TAIR (Table IV). The relationship types that include the word not allow curators to capture specific negative results that have been described in the literature, which may be contrary to previously known data. The GO consortium has recognized the utility of TAIR's relationship types and may move toward adding them to the current standard for consortium-wide GO annotations.
DISCUSSION AND CONCLUSION
Advantages to Using Controlled Vocabularies
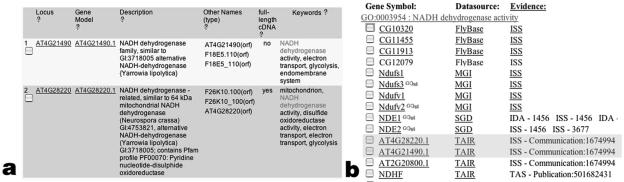
There are several advantages to using controlled vocabularies for functional annotation of a genome. First, it allows one to perform powerful intraspecies and cross-species genome queries. For example, one can identify all of the genes in Arabidopsis that are associated to the term NADH dehydrogenase activity using the TAIR gene search (Fig. 4a), or one can identify all of the genes in the central GO database that are associated to the same term using the AmiGO browser (Fig. 4b).
Figure 4.
Searching with controlled vocabulary terms within one species and across multiple species. a, Screenshot from a TAIR Web page showing a partial list of all Arabidopsis genes associated to the GO term NADH dehydrogenase activity. This page can be retrieved by entering the GO term on the TAIR gene search page (http://www.arabidopsis.org/servlets/Search?action=new_search&type=gene). b, Screenshot from a GO Web page showing a partial list of genes from multiple organisms associated to the term NADH dehydrogenase activity. This page can be reached by entering the GO term on the GO database/ontology browser (http://www.godatabase.org/cgi-bin/go.cgi) or by clicking on the GO database hyperlink from the TAIR keyword detail page.
Second, one can quantitatively assess the similarity/dissimilarity of any two sets of genes or genomes by comparing the distribution of their annotations among GOslim categories. Functional categorization of the whole genome using GOslim terms provides researchers the ability to view the distribution of the entire genome into categories describing cellular location, molecular function, and biological process. This large-scale view may assist in directing future research to areas that are in need of more attention. Exploring these areas of biology may reduce the number of unknown genes and lead to better understanding of the overall nature of the genome. The plant GOslim terms are also useful in classifying and comparing smaller sets of genes, such as those identified by common expression patterns in a microarray experiment. In a previous section, we described the retrieval of annotations for lists of genes. In addition to getting the association counts in a tabular format, users can also draw pie charts (such as those in Fig. 2) based on the GOslim mapping for analysis and presentation purposes. A researcher can group the genes in one data set and compare their distribution among GOslim categories to a second set of genes or the genome as a whole to determine which categories are overrepresented or underrepresented.
Third, one can use the annotated genome of any one species to transfer knowledge to another genome. Since Arabidopsis has the most comprehensive functional annotation of any plant genome, its annotation can serve as a foundation upon which the functional annotation of other plant genomes such as rice (Oryza sativa), tomato (Lycopersicon esculentum), cotton (Gossypium hirsutum), maize (Zea mays), and related Brassica species can be built. For example, there are approximately 22,000 tentative tomato consensus sequences (TIGR Tomato Gene Index version 9.0, April 2003) that have been generated from approximately 182,000 tomato expressed sequence tags in several sequencing projects. Many of these tentative consensus sequences have >50% amino acid sequence similarity to an Arabidopsis protein over the entire sequence length (http://aztec.stanford.edu/cold/cgi-bin/analysis.cgi). Transferring at least the molecular function annotations of the Arabidopsis genes to the homologous tomato sequences with an IEA evidence code would be a reasonable first step in annotating the tomato genome. Expanding this example to a large-scale transfer of annotations makes the construction of a scaffold functional annotation of a new plant genome possible. This approach is also valid for smaller sets of genes. Researchers focusing on other plant species can find Arabidopsis genes similar to their genes of interest using sequence similarity methods. This gene list can be used to obtain functional annotation from the Arabidopsis genome (see above), which can be used to infer information and suggest experiments for these other systems.
Finally, complete functional annotation of a genome allows detailed evaluation of known versus unknown genes in that genome. For example, one can easily assess the number of genes with unknown molecular function, biological process, or cellular component. The lack of information in the literature, which is reflected by the unknown annotation, could guide researchers to a set of genes in need of further research. In addition, evidence codes can be used to determine to what extent a gene has been characterized. For example, a gene whose sequence is similar to known glycosyl transferases but has no experimental evidence for the activity may be annotated to glycosyl transferase activity with an ISS (inferred from sequence similarity) evidence code indicating that no experimental evidence supporting this prediction exists. By using a combination of GO terms and evidence codes, a researcher looking for a new project can get an up-to-date view of genes still requiring experimental characterization.
TAIR's annotations using controlled vocabularies are based on clearly defined sources of evidence, either experimental or computational. Both methods have their advantages—computational data can supply hypotheses that suggest experimental approaches and supply a basic level of annotation for genes not yet characterized experimentally. Experimental data, on the other hand, provides confirmation of a gene's biological role and also provides the basis for future computational analysis. When it is available, experimental data must take precedence over computational data, but both kinds of information are useful in combination to examine relationships between structure and function and answer evolutionary questions.
Continuing and Expanding Functional Annotation of the Arabidopsis Genome
Once we have captured the basic information for each published gene, we will be faced with the task of keeping the functional annotations up to date, including adding new genes as they are described and capturing new information about existing genes. Keeping the annotations current is essential to reflecting the most recent state of knowledge about the genome. The most efficient way to accomplish both of these tasks will be to switch from our current gene-based curation approach to a paper-based approach in which we will extract all relevant information from new papers (approximately 100 per month) as they are incorporated into TAIR's PubSearch database. New genes will be annotated with GO terms describing their identity or with unknown terms to indicate missing information. For existing genes, we will use new information to replace existing unknown annotations with the appropriate GO terms, add GO and TAIR terms for newly described phenomena, and update existing known annotations based upon the latest experimental data. We also regularly update annotations based on comments from the research community. Since our user community is ultimately the best judge of the annotation quality, we strongly encourage them to contact us if we have made erroneous annotations or incorrectly captured data from the literature. Researchers can give their feedback by (1) adding comments to genes by clicking on the Add My Comments button on each gene detail page, (2) e-mailing us directly at curator@arabidopsis.org, or (3) giving us comments in person when at scientific meetings such as the International Conference on Arabidopsis Research or the Annual Meeting of the American Society of Plant Biologists.
Building on our experience in extracting gene-related information from the literature, we are in the early stages of the next large task of annotation of mutant and natural variant alleles and their associated germ plasms and phenotypes. Incorporation of data into TAIR will capture what processes and/or expression patterns are disrupted or modified as a result of allelic variance. From a survey of almost 8,400 full-text Arabidopsis articles held in-house at TAIR, there are about 5,000 unique alleles described to varying degrees in the literature. Allele-related data is extremely complex and challenging to curate, and we anticipate that this project will last several years. Initially, we will describe the phenotypes using text summaries similar to gene descriptions. We will then move to using controlled vocabularies for describing basic phenotypes as well. Along with many other model organism databases, we have participated in a series of Phenotype Ontology meetings that discussed the need for a controlled vocabulary to describe phenotypes (http://obo.sourceforge.net/pheno/). Such a vocabulary would facilitate querying and comparison of phenotypes between different species. The common desire for a phenotype annotation standard has led to the development of a prototype controlled vocabulary (available from http://obo.sourceforge.net/) that will be modified and updated by TAIR and the other databases in much the same way as the GO vocabularies.
We have begun capturing information in the literature pertaining to genetic interactions and will expand this effort to cover signal transduction and transcriptional regulation pathways. Finally, we will begin making more complex associations by including environmental condition or genotype information in our annotations as well as by tying annotations to two separate controlled vocabularies to each other. Examples of this kind of information include: gene X is expressed in the radicle during germination or gene Y is expressed in the nucleus in the ecotype Columbia-0 but in the cytoplasm in the ecotype Landsberg erecta. Other types of composite annotations could capture conditional subcellular localization depending on phosphorylation status of the protein or association of a signal molecule. A combination of these types of annotations with the existing controlled vocabulary annotations will provide the researcher with a more complete summary of a gene's identity in a computationally accessible format.
MATERIALS AND METHODS
Computational Annotation Methods
The following methods were used to computationally generate GO assignments: (1) INTERPRO2GO transfer, a mapping between all Arabidopsis proteins containing INTERPRO domains (Mulder et al., 2003) and the corresponding GO identification assigned to the individual INTERPRO domain using the INTERPRO2GO mapping file (http://www.geneontology.org/external2go/interpro2go). (2) TargetP analysis (Emanuelsson et al., 2000), which uses a pattern recognition program that detects consensus targeting sequences within the entire predicted Arabidopsis proteome. The subcellular locations determined by this analysis were mapped to the corresponding GO term. (3) Metacyc2go transfer. The metacyc2go mapping file (http://www.geneontology.org/external2go/metacyc2go) is used to generate GO annotations in a manner similar to the INTERPRO2GO mapping, in which GO identifications for particular metabolic processes and functions were assigned to genes that had been annotated to Metacyc biochemical pathways and reactions (Krieger et al., 2004). (4) String matching, an algorithm in which gene descriptions obtained from TIGR were matched to a corresponding GO term. All annotations derived using these methods are given the IEA evidence code and associated to a reference describing the analysis in detail. Our computational analyses are repeated on each successive genome release to ensure that they remain up to date.
Manually Reviewed Annotation Methods
We also associate genes with controlled vocabulary terms based on evidence found in the published literature. This entails obtaining appropriate papers that describe Arabidopsis genes, reading the papers, and associating the controlled vocabulary terms to the genes along with the evidence supporting the association. To facilitate literature-based annotation, we developed PubSearch, a literature curation software package that stores gene, paper, and controlled vocabulary data, automatically indexes the literature against genes and controlled vocabulary terms, and provides a user-friendly Web interface for manual verification of matches and curation (http://pubsearch.org/). PubSearch is maintained by TAIR and is one of the literature curation tools for the Generic Model Organism Database project. Its source code is available under the General Public License from Sourceforge (http://www.gmod.org). PubSearch is both extensible, allowing new types of biological objects to be added, and flexible, allowing programmatic implementation of different curation strategies. The software automatically assigns new genes each day to individual curators and displays the number of genes completed and in progress. The criteria used by PubSearch for selecting genes to be curated are modified according to the priorities of the curation team. All curation at TAIR is stored in the PubSearch database, and updates are sent to the production database on a weekly basis. Table V gives an overview of the data types that are stored in the TAIR installation of the PubSearch database.
Table V.
PubSearch data types and statistics
| Data Types | Numbers | |
|---|---|---|
| All literature records | 21,532 | |
| Research papers | 16,427 | |
| Research papers with abstracts | 11,888 | |
| Articles with full text | 8,633 | |
| Gene names (including aliases) | 118,484 | |
| Controlled vocabulary terms | 17,178 | |
| Anatomy terms | 268 | |
| Developmental stage terms | 102 | |
| GO molecular function terms | 7,278 | |
| GO biological process terms | 8,181 | |
| GO cellular component terms | 1,379 | |
| Hits between terms and articles | 177,210 | |
| Curator-reviewed hits between genes and articles | 19,974 | |
| Valid hits | 15,604 | |
| Invalid hits | 4,301 | |
| Maybe hits | 69 | |
The stored titles and abstracts of publications are first indexed against the gene names and aliases to generate hits, or associations, between papers and genes. For example, a paper that mentions the gene HST in its abstract will be associated with the gene HST. Because gene symbols are often not unique (for example, there are two GPX genes, two PUP1 genes, etc.), each match of a gene to an abstract is verified by a curator if the association is correct. Thus, several gene entries may exist with the same gene symbol but with different associated publications. After verification, the set of articles associated to a gene serves as the reading material for the curator who is updating a specific gene's annotations. The automated association of genes to papers frees curators from the need to search the literature for gene-related articles each time a gene record is updated or revisited.
We use the following procedure in extracting information from each gene's associated body of literature. First, the most recent paper or review about the gene is read to determine whether the process, function, and/or cellular location are known. If some or all of these aspects are known, the original paper describing the details of the experiments leading to that conclusion is located and the relevant information (i.e. subcellular localization method) is translated into a GO term, evidence code, and description. Each gene and annotation is stamped with the date it was last modified and the name of the annotating curator.
We select the most specific GO term that is appropriate for describing that aspect of the gene's identity. For example, we would select Ser/Thr kinase activity rather than enzyme activity to describe a Ser/Thr kinase. If the appropriate term is not present in the ontologies, curators propose a new term together with a definition and parentage and enter it as a temporary term through the PubSearch user interface. Annotations made to the new terms are not released to the public until the term has been accepted and added to the GO/TAIR vocabularies. Two members of our curation team periodically go through the list of proposed terms and, after review and consultation with the GO consortium and/or the rest of the TAIR curation team, add it to the appropriate vocabulary, at which point the term becomes available for the entire community to use.
Finally, we incorporate annotations made by external groups such as individual researchers sending corrections by mail, gene family experts sending annotations in spreadsheet files, and major database groups such as TIGR. TIGR has been annotating genes based on their membership in paralogous gene families. This has resulted in annotation of 21,893 genes. TIGR's paralogous family groupings are based on sequence similarity, identification of Pfam and TIGRFAM domain signatures, and potential novel domains in the Arabidopsis proteome (Wortman et al., 2003). GO terms that are associated with certain protein domains are then ascribed to all gene products that are members of paralogous families, if they are deemed appropriate. In cases where some members of the paralogous family had been described in the literature, annotations for biological process and/or cellular component were added as well.
Quality Control Methods
We employ several methods to assure a consistent and accurate standard of annotation. First, to minimize variability in annotation between curators, individual annotations are randomly selected and checked by verifying the association between the gene and the controlled vocabulary term. Rules for making associations are clarified when necessary. Second, at the level of data input, the curation software checks ensure that all the necessary fields are filled in to complete an annotation. User interfaces for editing information are designed to minimize human error. Third, at the level of data exchange between the PubSearch database and the TAIR production and GO databases, a number of software checks ensure data integrity (e.g. that annotations made to temporary terms are not sent out and that all references used in the annotation are present in the TAIR database). Fourth, we have implemented a method of computationally updating annotations based on a combination of evidence code and whether the association is made to an unknown term or not. Annotations of a gene to unknown terms are updated when an annotation of the same gene to a known term in the same ontology is made. Annotations with an IEA evidence code are replaced when a curator adds a non-IEA based annotation to the gene using a term in the same ontology. Finally, we incorporate feedback from the scientific community who provide corrections to the annotations or point out papers that were missing from our database.
This work was supported by the National Science Foundation (grant no. DBI–9978564) and the National Institutes of Health (grant no. HG02273–03).
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Camon E, Magrane M, Barrell D, Binns D, Fleischmann W, Kersey P, Mulder N, Oinn T, Maslen J, Cox A, Apweiler R (2003) The Gene Ontology Annotation (GOA) project: implementation of GO in SWISS-PROT, TrEMBL, and InterPro. Genome Res 13: 662–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium GO (2001) Creating the gene ontology resource: design and implementation. Genome Res 11: 1425–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwight SS, Harris MA, Dolinski K, Ball CA, Binkley G, Christie KR, Fisk DG, Issel-Tarver L, Schroeder M, Sherlock G, et al (2002) Saccharomyces Genome Database (SGD) provides secondary gene annotation using the Gene Ontology (GO). Nucleic Acids Res 30: 69–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, Svon Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Frishman D, Albermann K, Hani J, Heumann K, Metanomski A, Zollner A, Mewes HW (2001) Functional and structural genomics using PEDANT. Bioinformatics 17: 44–57 [DOI] [PubMed] [Google Scholar]
- Hazbun TR, Malmstrom L, Anderson S, Graczyk BJ, Fox B, Riffle M, Sundin BA, Aranda JD, McDonald WH, Chiu CH, et al (2003) Assigning function to yeast proteins by integration of technologies. Mol Cell 12: 1353–1365 [DOI] [PubMed] [Google Scholar]
- Hennig S, Groth D, Lehrach H (2003) Automated Gene Ontology annotation for anonymous sequence data. Nucleic Acids Res 31: 3712–3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanapin A, Batalov S, Davis MJ, Gough J, Grimmond S, Kawaji H, Magrane M, Matsuda H, Schonbach C, Teasdale RD, Yuan Z (2003) Mouse proteome analysis. Genome Res 13: 1335–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King OD, Lee JC, Dudley AM, Janse DM, Church GM, Roth FP (2003) Predicting phenotype from patterns of annotation. Bioinformatics 19 (suppl. 1): I183–I189 [DOI] [PubMed] [Google Scholar]
- Krieger CJ, Zhang P, Mueller LA, Wang A, Paley S, Arnaud M, Pick J, Rhee SY, Karp PD (2004) MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res 32: D438–D442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MASC Committee (2003) The Multinational Coordinated Arabidopsis thaliana Functional Genomics Project: Annual Report 2003. MASC Committee, Madison, WI
- Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Barrell D, Bateman A, Binns D, Biswas M, Bradley P, Bork P, et al (2003) The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res 31: 315–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague J, Clements D, Conlin T, Edwards P, Frazer K, Schaper K, Segerdell E, Song P, Sprunger B, Westerfield M (2003) The Zebrafish Information Network (ZFIN): the zebrafish model organism database. Nucleic Acids Res 31: 241–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortman JR, Haas BJ, Hannick LI, Smith RK Jr, Maiti R, Ronning CM, Chan AP, Yu C, Ayele M, Whitelaw CA, et al (2003) Annotation of the Arabidopsis genome. Plant Physiol 132: 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]