Abstract
Data mining methods have been used to identify 356 Cyt P450 genes and 99 related pseudogenes in the rice (Oryza sativa) genome using sequence information available from both the indica and japonica strains. Because neither of these genomes is completely available, some genes have been identified in only one strain, and 28 genes remain incomplete. Comparison of these rice genes with the 246 P450 genes and 26 pseudogenes in the Arabidopsis genome has indicated that most of the known plant P450 families existed before the monocot-dicot divergence that occurred approximately 200 million years ago. Comparative analysis of P450s in the Pinus expressed sequence tag collections has identified P450 families that predated the separation of gymnosperms and flowering plants. Complete mapping of all available plant P450s onto the Deep Green consensus plant phylogeny highlights certain lineage-specific families maintained (CYP80 in Ranunculales) and lineage-specific families lost (CYP92 in Arabidopsis) in the course of evolution.
The publication of the Arabidopsis genome in December of 2000 offered the world its first glimpse of a complete plant genome (Arabidopsis Genome Initiative, 2000). The inventorying of gene families and pathway components in this plant began a process of annotation that may take decades to complete. One of the notable gene families characterized in Arabidopsis is that encoding the very large set of Cyt P450 monooxygenases (P450s). In the superfamily of divergent genes that encode these proteins, 246 full-length, putatively functional P450 coding sequences represent approximately 1% of the Arabidopsis gene complement (Paquette et al., 2000; Werck-Reichhart et al., 2002; Schuler and Werck-Reichhart, 2003). Contrasting with this, between 54 and 105 P450s exist in the human, mouse, Takifugu rubripes, Anopheles gambiae, Drosophila melanogaster, Caenorhabditis elegans, Ciona intestinalis, and Ciona savignyi genomes (Nelson et al., 2004), representing approximately 0.1% to 0.5% of each of these gene complements.
Not surprisingly, sequencing of large eukaryotic genomes has placed early emphasis on the evolution of gene families in animals and, especially, in mammals. Comparisons between the mouse and human genomes have confirmed the impression that mammals are generally very similar at the genomic level and suggested that few surprises will be encountered in future characterizations of dog, cat, rat, or bat genomes. Complexities that exist in the biochemical reactions and genomes of more diverse phyla, as in the insecta, which includes 900,000 named species, and in angiosperms, which includes more than 230,000 named species (Margulis and Schwartz, 1998), suggest there is still much to be discovered. This is especially apparent in the proliferation of P450-mediated reactions already characterized in plants (Kahn and Durst, 2000; Werck-Reichhart et al., 2002) that hint at a biochemical repertoire, well beyond that known to exist in animals. This is also apparent in the large contingents of Arabidopsis P450 sequences that have become available through earlier sequencing efforts and rice (Oryza sativa) P450 sequences that have recently become available. Although these two species can hardly be representative of their 170,000 dicot and 65,000 monocot relatives, comparison of the more than 600 P450 genes present in these two species opens a door into the evolution of this large superfamily in plants and allows one to begin sampling the diversity of these sequences in monocots.
RESULTS AND DISCUSSION
Because of redundancies in much of the genomic information, the rice genome (430 Mb) is thought to contain substantially more genes (32,000–56,000) than the smaller Arabidopsis genome (129 Mb, 26,439 genes; http://mips.gsf.de/proj/thal/db/tables/chrall_tables/exons.html). Cyt P450 genes provide clear evidence of this level of reiteration. Compared to 246 full-length P450s in Arabidopsis and 26 identifiable pseudogenes, the rice genome contains 328 full-length P450 genes and 99 designated pseudogenes with 28 partial P450 sequences that may represent 24 additional genes and/or pseudogenes. From the summary of P450 families in rice and Arabidopsis presented in Table I, it is clear that there are families unique to each species and plant P450 families absent from both species. The rice expressed sequence tags (ESTs) were searched for plant P450 families not found in the available genomic sequence and for rice sequences that were only partial in the genome. A complete side-by-side list of all genes and pseudogenes in rice and Arabidopsis is given in the supplemental material (Supplemental Table I; see www.plantphysiol.org) and on the Cyt P450 Homepage (http://drnelson.utmem.edu/rice.arab.list.htm).
Table I.
Cyt P450 genes and pseudogenes by family in Arabidopsis and rice (Jan. 1, 2004)
| CYP | Arabidopsis | Rice |
|---|---|---|
| 51 | 2 | 10 + 2P |
| 71 | 52 + 2P | 90 + 28P |
| 72 | 9 + 1P | 13 + 4P |
| 73 | 1 | 3 + 1P |
| 74 | 2 | 5 + 1P |
| 75 | 1 | 3 |
| 76 | 8 + 1P | 30 + 12P |
| 77 | 5 + 2P | 2 |
| 78 | 6 | 8 |
| 79 | 7 + 5P | 4 |
| 80 | 0 | 0 |
| 81 | 18 | 12 + 1P |
| 82 | 5 | 0 |
| 83 | 2 | 0 |
| 84 | 2 | 3 + 1P |
| 85 | 2 | 1 |
| 86 | 11 | 5 + 6P |
| 87 | 1 + 1P | 11 + 2P |
| 88 | 2 | 1 |
| 89 | 7 | 15 + 6P |
| 90 | 4 | 5 + 1P |
| 91 | No longer used | |
| 92 | 0 | 9 + 5P |
| 93 | 1 | 3 + 4P |
| 94 | 6 + 1P | 18 + 8P |
| 95 | No longer used | |
| 96 | 13 + 2P | 12 |
| 97 | 3 | 3 |
| 98 | 3 | 3 + 2P |
| 99 | 0 | 2 |
| 701 | 1 | 4 |
| 702 | 5 + 3P | 0 |
| 703 | 1 | 1 |
| 704 | 3 | 7 |
| 705 | 25 + 8P | 0 |
| 706 | 7 | 4 |
| 707 | 4 | 2 |
| 708 | 4 | 0 |
| 709 | 3 | 11 + 3P |
| 710 | 4 | 4 + 2P |
| 711 | 1 | 5 |
| 712 | 2 | 0 |
| 713 | No longer used | |
| 714 | 2 | 6 + 1P |
| 715 | 1 | 1 + 3P |
| 716 | 2 | 0 |
| 717 | No longer used | |
| 718 | 1 | 0 |
| 719 | 0 | 0 |
| 720 | 1 | 0 |
| 721 | 1 | 2 |
| 722 | 1 | 1 |
| 723 | 0 | 2 + 1P |
| 724 | 1 | 1 |
| 725 | 0 | 0 |
| 726 | 0 | 0 |
| 727 | 0 | 1 |
| 728 | 0 | 11 + 4P |
| 729 | 0 | 2 |
| 730 | 0 | 11* |
| 731 | 0 | 1 |
| 732 | 0 | 1 |
| 733 | 0 | 1 |
| 734 | 1 | 4 + 1P |
| 735 | 2 | 2 |
| 736 | 0 | 0 |
The data were obtained from the complete Arabidopsis genome and the Beijing Genomics Institute (rice indica strain) genome assembly and the publicly available japonica rice strain sequences in GenBank from the IRGSP and other sources (approximately 90% genome coverage at approximately 71% finished quality; IRGSP Working Group Meeting Feb. 4, 2004; http://demeter.bio.bnl.gov/Tsukuba04.html).
All CYP730 sequences are incomplete.
Organization of P450 Clans in Plants
These genome-wide comparisons of P450s have allowed us to further develop the relationships between clans, or recognizable clusters, of related P450 families. With the inclusion of the new rice P450 sequences, it is apparent that there are 10 clans in plants that are designated by their lowest-numbered family members, CYP71, CYP72, CYP85, and CYP86, or their only family, as in CYP51, CYP74, CYP97, CYP710, CYP711, and CYP727 (Fig. 1). Grouping the families from each clan together, one sees that only four plant P450 clans contain multiple P450 families (Figs. 2–6) and that six clans contain only single families. The clustering of plant P450s into these larger groups, which was first observed in 1995 (Durst and Nelson, 1995), has been supported by a large number of sequences collected from dicot as well as monocot species. The exact number of clans differs slightly between dicots, which contain nine, and monocots, which have a tenth, designated as the CYP727 clan. More distant comparisons with Pinus ESTs and other gymnosperm sequences currently available indicate that all clans except CYP711 and CYP727 exist in gymnosperms. The presence of CYP711-related sequences in Chlamydomonas reinhardtii implies it will also be found in gymnosperms. It is thus likely that CYP727 will be the only plant P450 clan found in a limited taxonomic range.
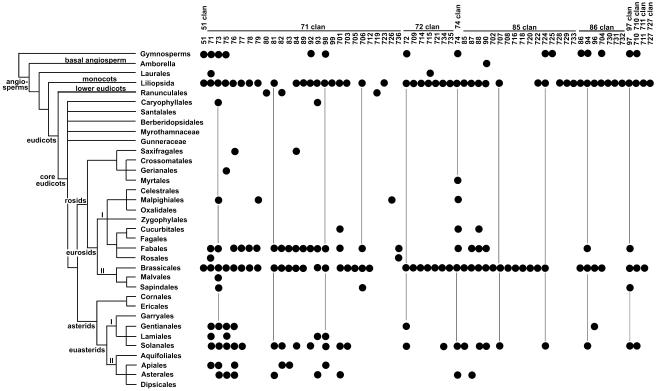
Figure 1.
Distribution of P450 families across plant phylogeny. A total of 1,098 named plant P450s were sorted by family and taxonomic position in the phylogeny of plants. Each dot may represent many genes, as in the rice CYP71s in the monocot branch (117 named P450s). P450 families are grouped in the 10 plant P450 clans. Vertical lines are placed to help visualize the P450 family position. Arabidopsis is in the Brassicales branch. Only rice and Arabidopsis had complete genomes available at the time of this compilation.
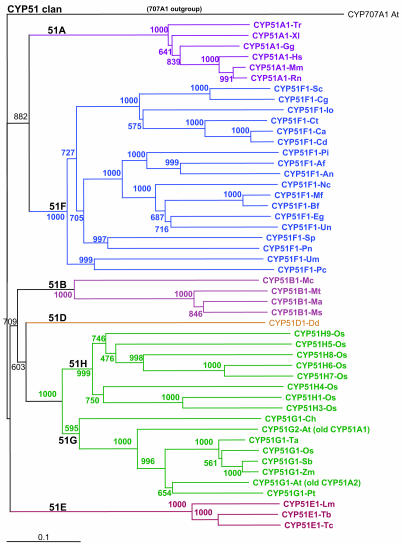
Figure 2.
The CYP51 clan. A total of 48 sequences in the CYP51 family across all taxa are shown as a Neighbor-Joining tree computed with a PAM350 scoring matrix. In this new nomenclature, subfamilies in CYP51 represent major taxonomic divisions, not the usual ≥55% sequence identity. CYP51H sequences in rice are an exception, apparently diverging from the usual CYP51 function. Abbreviations for the 75 species used in Figures 2 to 6 are as follows: Af, Aspergillus fumigatus; An, Aspergillus nidulans; Ao, Asparagus officinalis; At, Arabidopsis (thale cress); Bf, Botryotinia fuckeliana; Bs, Berberis stolonifera; Ca, Candida albicans; Cd, Candida dubliniensis; Cg, Candida glabrata; Ch, C. reinhardtii (green algae); Ci, Cicer arietinum (chickpea); Cj, Coptis japonica; Cm, Cucurbita maxima (pumpkin); Cr, Catharanthus roseus (Madagascar periwinkle); Ct, Candida tropicalis; Dd, Dictyostelium discoideum (cellular slime mold); Ec, Eschscholzia californica (California poppy); Eg, Erysiphe graminis; El, Euphorbia lagascae; Ga, Gossypium arboreum (cotton); Ge, Glycyrrhiza echinata (licorice); Gg, Gallus gallus (chicken); Gm, soybean; Hs, Homo sapiens (human); Ht, Helianthus tuberosus (Jerusalem artichoke); Io, Issatchenkia orientalis (fungi); Le, tomato; Lj, Lotus japonicus (lotus); Lm, Leishmania major (Leishmaniasis parasite); Lr, Lolium rigidum (ryegrass); Ls, Liquidambar styraciflua (sweetgum); Ma, Mycobacterium avium; Mc, Methylococcus capsulatus; Me, Manihot esculenta (cassava); Mf, Monilinia fructicola; Mm, Mus musculus (mouse); Ms, Medicago sativa (alfalfa); Mt, Mycobacterium tuberculosis; Mu, Musa acuminata (banana); Mys, Mycobacterium smegmatis; Nc, Neurospora crassa; Np, Nicotiana plumbaginifolia; Nt, tobacco; Os, rice; Pa, Persea americana (avocado); Pas, Pastinaca sativa (wild parsnip); Pc, Pyrus communis (pear); Ph, Petunia hybrida; Phc, Phanerochaete chrysosporium (white rot fungus); Pi, Penicillium italicum; Pnc, Pneumocystis carinii; Pr, Pinus radiata (Monterey pine); Ps, pea; Pso, Papaver somniferum (opium poppy); Pt, Pinus taeda (loblolly pine); Rn, Rattus norvegicus (rat); Sa, Sinapis alba (white mustard); Sb, Sorghum bicolor; Sc, yeast; Sm, Solanum melongena cv Sinsadoharanasu (eggplant); Sp, Schizosaccharomyces pombe (fission yeast); St, potato; Ta, wheat; Tc, Taxus cuspidata (Japanese yew); Tm, Triglochin maritima; Tr, T. rubripes (Japanese pufferfish); Trb, Trypanosoma brucei (African sleeping sickness parasite); Trc, Trypanosoma cruzi (Chagas disease parasite); Um, Ustilago maydis; Un, Uncinula necator (grape powdery mildew fungus); Vr, Vigna radiata (mung bean); Vs, Vicia sativa; Vv, Vitis vinifera (grape); Xl, Xenopus laevis (African clawed frog); and Zm, Z. mays (maize).
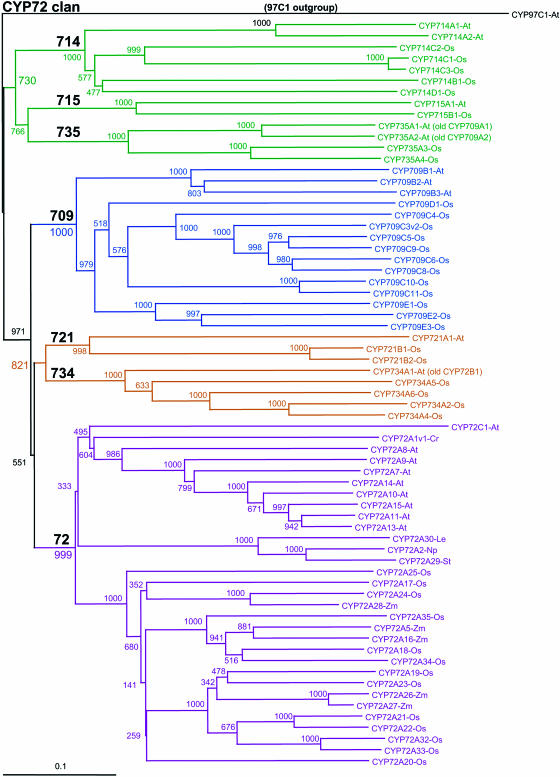
Figure 6.
The CYP72 clan. This tree illustrates the frustration of nomenclature efforts, which must act on sequences as they are found. New data can cause relocation of branches as in the CYP709A1 and CYP709A2 sequences. These have moved to a new family as CYP735A1 and CYP735A1. CYP72B1 has also moved to a new location as CYP734A1. This tree of 67 sequences with CYP97C1 as the outgroup is a Neighbor-Joining tree using a Gonnett scoring matrix.
Inspection of these clans indicates that each has diversified over time to the expanded sets that we see today. At least eight of the 10 clans, and probably CYP711, are present in gymnosperms and angiosperms and, therefore, predate the divergence of gymnosperms and angiosperms estimated at 340 million years ago (Wolfe et al., 1989; Troitsky et al., 1991). It is tempting to speculate that these clans mirror early plant P450 evolution with one or only a few members in very early plants mediating fundamental processes such as synthesis of sterols (CYP51, Fig. 2; Yoshida et al., 1997; Lamb et al., 1998), modification of sterols and cyclic terpenes in the brassinosteroid, abscisic acid, and GA pathways (CYP85 clan, Fig. 3; Helliwell et al., 2001; Bishop and Koncz, 2002; Kushiro et al., 2004), synthesis of allene oxide and oxylipin derivatives in the jasmonate and octadecanoid pathways (CYP74; Laudert et al., 1996; Creelman and Mullet, 1997; Agrawal et al., 2004), hydroxylations of fatty acids (CYP86 clan, Fig. 4; Benveniste et al., 1998; Kahn et al., 2001; Wellesen et al., 2001), hydroxylations of carotenoids (CYP97 clan; Tian et al., 2004), modifications of shikimate products and intermediates (CYP71 clan, Fig. 5), and catabolism of isoprenoid hormones (CYP72 clan, Fig. 6; Turk et al., 2003). For a broad survey of plant P450 biochemistry, see Werck-Reichhart et al. (2002).
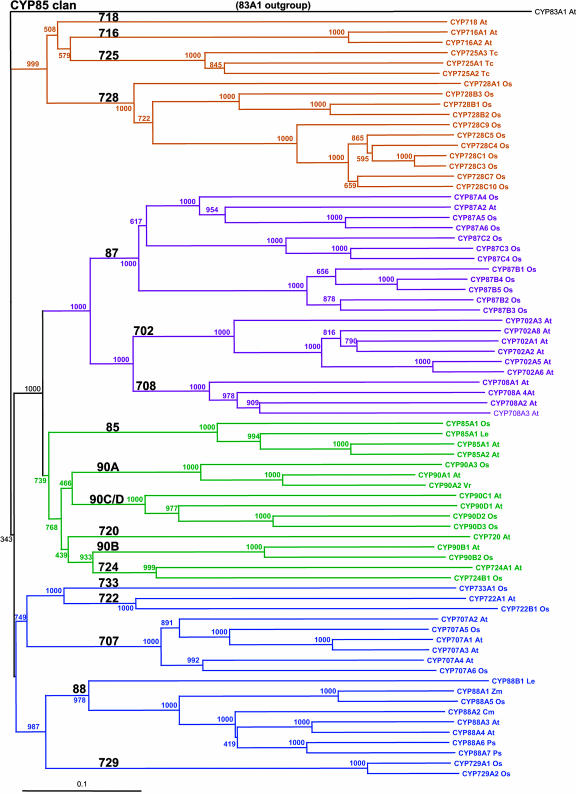
Figure 3.
The CYP85 clan. The 85 clan is one of four clans with multiple P450 families. Here, 74 sequences from 16 families are shown with CYP83A1 as an outgroup. This tree was made using the Neighbor-Joining algorithm and a Gonnett scoring matrix. CYP720 and CYP724 cluster with the CYP90s. They are not named as CYP90 subfamilies because CYP720 is only 33% identical to CYP90C1, and CYP724 is only 34% identical to CYP720. The CYP90 subfamilies could probably stand as separate families.
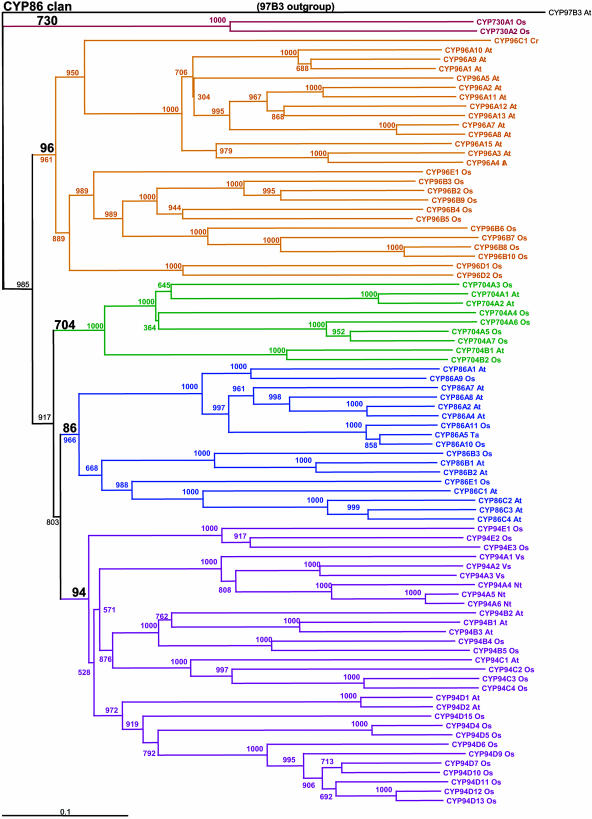
Figure 4.
The CYP86 clan. This clan contains only five P450 families. The tree has 84 sequences with CYP97B3 as the outgroup. This tree was made using the Neighbor-Joining algorithm and a Gonnett scoring matrix.
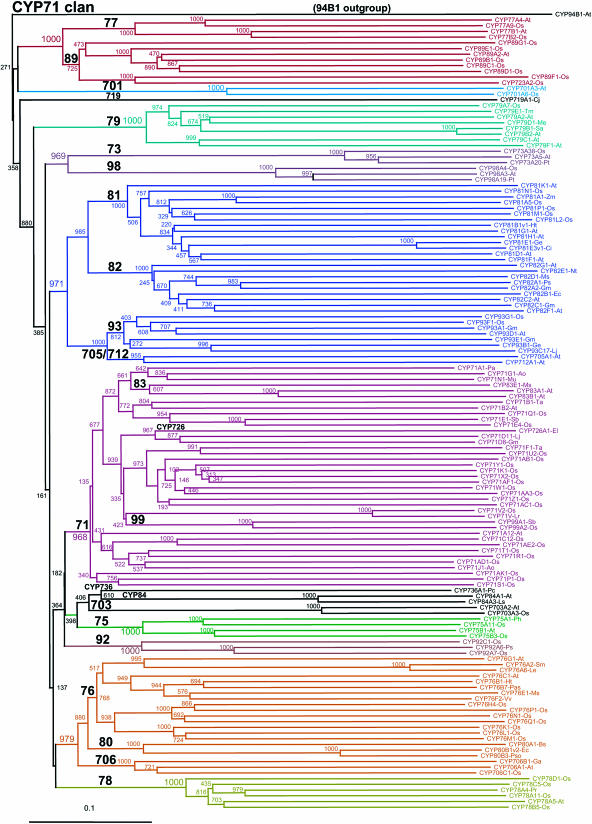
Figure 5.
The CYP71 clan. This is the largest set of P450s in plants. The extreme diversification of families and subfamilies in the CYP71 clan indicates great utility for plant evolution, yet there are no CYP71 clan members detected in C. reinhardtii. This tree has 139 sequences in 26 families. It is a Neighbor-Joining tree using a Gonnett scoring matrix. Notice that CYP83, CYP99, and CYP726 are inside the CYP71 family.
In the unicellular green algae, C. reinhardtii (Chlorophyta), which is considered a valid representative of the ancestor of terrestrial plants (Willis and McElwain, 2002), orthologs of CYP51, CYP97, CYP710, and CYP711 exist in currently available EST and genomic DNA sequences. The known functions of several members of these families in sterol (CYP51; Werck-Reichhart et al., 2002) and carotenoid (CYP97; Tian et al., 2004) modifications emphasize the significance of these orthologous P450 families in plants. Although a specific homolog of CYP97C1, which has a role in the carotenoid conversions of Arabidopsis (Tian et al., 2004), has not been identified in Chlamydomonas, orthologs of both CYP97A and CYP97B exist in this genome. Based on our phylogenetic comparisons, CYP97C represents a CYP97 subfamily that very recently split off from the CYP97B subfamily (http://www.biobase.dk/P450/cyp_allsubfam_NJ_102103.pdf; Galbraith and Bak, 2004). The conservation of two other families in Chlamydomonas whose functions have not yet been defined (CYP710 and CYP711) suggests that they mediate essential and conserved functions needed in all plant taxa. A listing of the relative identity and similarity between CYP51, CYP97, CYP710, and CYP711 across Chlamydomonas, rice, and Arabidopsis can be seen in Table II. It may be significant that these four families belong to plant P450 clans containing only one family, even though they must be several hundred million years old. In the moss Physcomitrella patens, an EST project (http://moss.nibb.ac.jp/) has revealed the presence of the CYP71 clan in vascular plants. Orthologs of CYP73, the cinnamate 4-hydroxylase, as well as of Phe ammonia lyase and other genes involved in phenylpropanoid synthesis, are found in P. patens. The CYP71 clan thus seems to appear in terrestrial vascular plants. The absence of CYP71 orthologs from Chlamydomonas probably highlights the importance of the CYP71 clan in the production of the vast array of secondary metabolites that have proliferated in terrestrial vascular plants (Durst and Nelson, 1995; Paquette et al., 2003). In addition to the recently completed Chlamydomonas genome, the genome sequences of a moss, a fern, and a gymnosperm will inevitably lead to a deeper understanding of evolution of higher plant P450s.
Table II.
Comparisons of the four P450 families conserved between C. reinhardtii, rice, and Arabidopsis
| 51G1-At | 51G2-At | 51G1-Ch | |||
| CYP51G1-Ch | 55 | 52 | 100 | ||
| CYP51G1-Os | 76 | 65 | 56 | ||
| CYP51G3-Os | 55 | 50 | 46 | ||
| CYP51G4P-Os | 53 | 47 | 43 | ||
| CYP51H1-Os | 48 | 47 | 40 | ||
| CYP51H3-Os | 46 | 46 | 40 | ||
| CYP51H4-Os | 49 | 47 | 39 | ||
| CYP51H5-Os | 49 | 50 | 44 | ||
| CYP51H6-Os | 51 | 43 | 42 | ||
| CYP51H7-Os | 48 | 46 | 40 | ||
| CYP51H8-Os | 50 | 47 | 42 | ||
| CYP51H9-Os | 53 | 50 | 43 | ||
| 97A3-At | 97B3-At | 97C1-At | 97A-Ch | 97B-Ch | |
| CYP97A3-At | 100 | 45 | 52 | 60 | 48 |
| CYP97B3-At | 45 | 100 | 41 | 46 | 52 |
| CYP97C1-At | 52 | 41 | 100 | 50 | 45 |
| CYP97A4-Os | 70 | 47 | 51 | 59 | 63 |
| CYP97B4-Os | 46 | 76 | 43 | 45 | 57 |
| CYP97C2-Os | 49 | 45 | 78 | 48 | 56 |
| 710A1-At | 710A2-At | 710A3-At | 710A4-At | 710-Ch | |
| CYP710-Ch | 43 | 40 | 41 | 42 | 100 |
| CYP710A5-Os | 60 | 58 | 54 | 52 | 40 |
| CYP710A6-Os | 60 | 58 | 55 | 53 | 41 |
| CYP710A7-Os | 61 | 60 | 57 | 56 | 42 |
| CYP710A8-Os | 65 | 62 | 58 | 56 | 43 |
| 711A1-At | 711-Ch | ||||
| CYP711A1-At | 100 | 34 | |||
| CYP711A2-Os | 63 | 32 | |||
| CYP711A3-Os | 68 | 34 | |||
| CYP711A4-Os | 64 | 34 | |||
| CYP711A5-Os | 52 | 34 | |||
| CYP711A6-Os | 60 | 32 |
Abbreviations are as in Figure 1. Values are in percent identity.
A General Comparison of Rice and Arabidopsis P450s
One of the first interesting features of these comparisons is that the CYP82, CYP83, CYP702, CYP705, CYP708, CYP712, CYP716, CYP718, and CYP720 families are absent from the rice genome but present in the Arabidopsis genome. Phylogenetic comparisons indicate that, of these, the CYP702, CYP708, CYP716, CYP718, and CYP720 families are related families within the CYP85 clan (Fig. 3). This clan is notable in that it includes Arabidopsis CYP85A1, CYP85A2, CYP90A1, and CYP90B1 genes and the rice CYP90D2 gene involved in brassinosteroid biosynthesis (Szekeres et al., 1996; Azpiroz et al., 1998; Choe et al., 1998; Clouse, 2001; Shimada et al., 2001, 2003; Hong et al., 2002, 2003; Mori et al., 2002; Fujioka and Yokota, 2003), as well as the Arabidopsis CYP88A3 and CYP88A4 genes involved in GA biosynthesis (Helliwell et al., 2001). The fact that 14 Arabidopsis P450s clustered in five families of the same P450 clan are absent in rice suggests that biochemical differences exist between rice and Arabidopsis in their metabolism of steroids and/or isoprenoids. Whether these particular gene differences exist in other dicots and not in monocots awaits many of the sequencing efforts that are now under way.
Another of the missing rice families, CYP705, is represented by a large family of 25 full-length genes and eight pseudogenes in Arabidopsis. To date, this CYP705 family has only been identified in the Brassicaceae, represented by Arabidopsis and Brassica napus. Sequences in this family are closely related to the small Arabidopsis CYP712 family, and the complete absence of both families from the rice genome suggests that they represent families of lineage-specific P450 genes. The appearance of the CYP705 family in the Arabidopsis genome seems to be a recent event as this family contains a relatively large number of pseudogenes, suggesting that it is has recently undergone multiple duplication events in the process of evolving new functions (Moore and Purugganan, 2003). The related CYP712 sequences branch outside the CYP705 cluster; however, the CYP712 sequences share 40% to 43% sequence identity with some CYP705s, thus including them in the same family. Historically, CYP712 was given a separate family name, but it should be treated as a subfamily of CYP705 (Fig. 5).
Phylogenetic comparisons also indicate that the CYP82 and CYP83 families, which are also missing from the rice genome, are most closely related to the CYP81 (37%–38% identical to some CYP81D sequences) and CYP71 families (40%–43% to CYP71B sequences) that exist in both Arabidopsis and rice. The absence of the CYP83 family from the rice genome is particularly interesting since CYP83B1 was shown to play a key role in auxin homeostasis and plant development in Arabidopsis by CYP83B1 regulating the flux of indole-3-acetaldoxime into indoleglucosinolates and auxin (Barlier et al., 2000; Bak et al., 2001). This absence suggests that different metabolic processes ensure indole-3-acetic acid homeostasis in different plant phyla. The relationships of CYP83A1 and CYP83B1, which are well known to mediate the synthesis of Met and Trp-derived glucosinolates in Arabidopsis (Bak and Feyereisen, 2001; Bak et al., 2001; Hemm et al., 2003; Naur et al., 2003) and to play a role in defense and stress-responses within the massive CYP71 family, are shown in Figure 5. Similar to the CYP99 family, which sorts within the CYP71 family, the CYP83 family has been absorbed within the CYP71 family as it has grown. When CYP83 is viewed as just another CYP71 subfamily, then its absence from the rice genome is less notable. Similarly, the CYP82 family sorts evolutionarily very close to the CYP81 family (bootstrap value of 985/1,000 iterations), and it is envisioned that the ancestor of the CYP81 and the CYP82 families split into a CYP82 family that resides only in dicots and a CYP81 family that resides in both monocots and dicots. Interestingly, members of the CYP82 family are reported to be highly stress responsive in pea (Pisum sativum), tobacco (Nicotiana tabacum), or soybean (Glycine max; Frank et al., 1996; Schopfer and Ebel, 1998; Ralston et al., 2001), but the function of these enzymes remains unknown.
Conversely, the CYP92, CYP99, CYP723, and CYP727 to CYP733 gene families exist in rice but not in Arabidopsis. Of these, the CYP92 family exists in many other plant species, including tobacco, pea, and Pinus. The fact that CYP92A6, which has been proposed to mediate the 2-hydroxylation of brassinosteroids in pea (Kang et al., 2001), is absent from Arabidopsis indicates that this conserved function has been assumed by a more divergent P450 or another class of enzymes. CYP99 sorts inside the CYP71 cluster and can be viewed as a CYP71 subfamily. The remaining families CYP723 and CYP727 to CYP733 are unique to rice and/or other grasses, possibly reflecting grass-specific pathways not previously sampled in the sequenced set of P450 genes.
Distributions of P450 Families within the Plant Kingdom
Sorting this collection of rice and Arabidopsis P450 sequences with those in other plants against the best available plant phylogeny from the Deep Green Project, a worldwide effort to determine plant evolutionary relationships (http://ucjeps.berkeley.edu/bryolab/GPphylo/), has indicated that 39 of 59 currently identified P450 families are shared between monocots and dicots (Fig. 1). This numbering includes treating CYP99 and CYP726 as subfamilies within the CYP71 family and CYP712 as a subfamily within the CYP705 family and including the CYP92 family present in many dicots but not Arabidopsis. By this estimate, two-thirds of known plant P450 families predate the divergence of monocots and dicots at 200 million years ago. This number may increase as the genomes of additional plant species become available.
At this level of comparison, one interesting feature is that subfamilies are often not conserved over the time frame of the monocot-dicot divergence. Whereas 31 subfamilies are present in both species, 121 subfamilies are present in only one of these two genomic prototypes. Not unexpectedly, several subfamilies within the set of conserved subfamilies are already known to be involved in carrying out fundamental reactions necessary for growth and development in all plants. These include CYP51G, CYP73A, CYP85A, CYP88A, CYP90A, CYP90B, CYP90D, CYP97C, and CYP98A, which, as previously mentioned, are involved in sterol, brassinosteroid, GA, phenylpropanoid, and carotenoid syntheses. Several of these and other conserved subfamilies are represented by single gene subfamilies that have maintained relatively high degrees of sequence identity between Arabidopsis and rice. Examples of these clearly orthologous P450s include CYP77B1 and CYP77B2 (60% identity); CYP90B1 and CYP90B2 (70% identity); CYP97C1 and CYP97C2 (77% identity); CYP703A3 and CYP703A4 (72% identity); and CYP704B1 and CYP704B2 (68% identity). Other conserved subfamilies contain variable numbers of paralogous genes. The remaining nonconserved subfamilies have been drifting apart in sequence, so they now retain only 40% to 50% identity. While the family relationships among these are still easily detectable, subfamily relationships have been lost, with <55% identity retained between sets of genes in rice and Arabidopsis. Based on our knowledge at the present time, these subfamilies may have arisen from one or a small number of precursors in the common ancestor of monocots and dicots.
Another interesting feature is that some families have greatly expanded in one species but not both (Table I). The most interesting example of such a family expansion is in the CYP51 family. Owing to its involvement in sterol biosynthesis, CYP51 is the only P450 family present in all kingdoms, and, in most phyla that have been compared, only a single CYP51 gene exists. Quite atypically, two CYP51 genes exist in Arabidopsis and 10 CYP51 genes exist in rice (Fig. 2). Among these many rice CYP51 genes, one (CYP51G1 in the recently revised CYP51 nomenclature) is clearly orthologous to the other plant CYP51 sequences and the Arabidopsis CYP51G1 (formerly CYP51A2). This rice ortholog is 76% identical to Arabidopsis CYP51G1 and less than 56% identical to the other nine rice CYP51 sequences (Table II). The sterol synthetic function of Arabidopsis CYP51G1, which has been demonstrated in yeast (Saccharomyces cerevisiae) complementation experiments (Kushiro et al., 2001) is expressed in many tissues (S. Ali, H. Duan, Y. Ferhatoglu, J. Thimmapuram, A. Hehn, S. Goepfer, C. Asnaghi, M. Band, D. Werck-Reichhart, and M.A. Schuler, unpublished data). The more divergent rice CYP51 sequences, which share lower degrees of identity with this sequence, are probably not involved in demethylating obtusifoliol but rather are evolving to mediate new functions. This degree of gene duplication and evolution in plant CYP51 sequences is especially interesting because the CYP51 genes have long been treated as unique among P450s because they exist in all kingdoms of life. Owing to the preservation of orthologous obtusifoliol demethylation functions in bacteria and plants (Bellamine et al., 1999; Jackson et al., 2002, 2003), the family definition (>40% sequence identity) has been broken in naming CYP51 sequences. As a result, this family contains substantially lower identity (34%–37% identity between rice CYP51G1 and bacterial CYP51 sequences) than other P450 families described here.
The existence of 10 CYP51 genes in rice has now forced the issue of naming CYP51 subfamilies. Based on this need, Figure 2 shows a cladogram of CYP51 genes from all taxa, including bacteria and protists with subfamily designations indicating membership in particular taxa. Again, owing to high diversity within this family, the 55% identity rule for subfamily membership has not been enforced here since it would generate too many subfamilies (especially in fungi) and the value of the nomenclature would be considerably reduced. In the nomenclature system presented in Figure 2, the single copy CYP51 genes existing in animal genomes are referred to merely as CYP51 or CYP51A1. Species identifiers are added for the requirements of this figure, but these name extensions are not part of the official P450 nomenclature. CYP51 is absent in nematodes and insects, as they are sterol heterotrophs and CYP51 has become superfluous and extinct over time in these organisms. The CYP51 genes existing in bacteria, which number only four at the present time, are designated as CYP51B. The suggestion that bacterial CYP51 genes represent a horizontal gene transfer from plants (Debeljak et al., 2000, 2003) is supported by the CYP51 tree (Fig. 2) with a bootstrap value of 709/1,000. Although no CYP51 sequences have yet been identified in protist groups in the Chromista (Stramenopiles and heterokonts), the CYP51C designation has been reserved for future designations in these groups. The sequence searches that have been completed to date indicate that the Plasmodium falciparum genome, present in an intracellular parasite not requiring its own sterols, does not contain any P450 genes. By contrast, Tetrahymena and Paramecium genomes, present in free-living ciliates requiring sterols, are expected to contain CYP51 sequences. Continuing with this nomenclature system, CYP51D is for Dictyostelium and other Mycetozoa sequences. CYP51E is for Euglenozoa (Leishmania and Trypanosoma), CYP51F is for fungi, and CYP51G is for green plants and related photosynthetic species such as red algae and Cyanophora. Notably, the green algal Chlamydomonas CYP51 sequence clusters with the higher plant CYP51G sequences, reflecting the conserved and orthologous function of CYP51G1 in these photosynthetic species. Also noteworthy is the fact that the additional CYP51G sequences present in rice and Arabidopsis are not orthologs of one another. Accordingly, these genes have been named CYP51G2 (previously CYP51A1) in Arabidopsis and CYP51G3 in rice. The reason why Arabidopsis and rice contain two paralogous CYP51G genes is currently not understood. Based on their apparently independent derivation from a sterol 14α-demethylase parent at a position in the CYP51 tree outside the monocot-dicot separation, it is likely that they are acquiring or have acquired a new function. The CYP51G3 sequence is not included in Figure 2 because it clusters with either other CYP51G sequences or with the CYP51H sequences, making its location in the tree variable. It has the highest sequence similarity to CYP51G sequences (55% identical to CYP51G1-Os).
Within the plant CYP51 sequences, but branching outside Chlamydomonas CYP51G1, are eight rice sequences. The CYP51H subfamily designation is reserved for these CYP51 sequences that clearly have diverged away from the parent sterol 14α-demethylase function. In agreement with the potential for neofunctionalization in these CYP51H sequences, the CYP51H branch is deeper than the monocot-dicot split as a consequence of these sequences evolving with a much higher evolutionary rate than the conserved CYP51G orthologs. Although the CYP51H9 sequence sometimes branches with the other CYP51H sequences and sometimes not, it has been named as part of this gene subfamily. In contrast to gene families that are highly conserved, stable over time (e.g. the CYP97 and CYP710 families), and rarely have pseudogenes, the existence of two CYP51 pseudogenes in the rice genome (CYP51G4P and CYP51H2P) suggests that divergent offshoots of the CYP51 family are not stable. In conclusion, this new CYP51 nomenclature recognizes deep divisions in the CYP51 family across phyla while preserving the unity of this most conserved P450 family.
Other expanded P450 families include the CYP87 family that contains 1 gene in Arabidopsis compared with 11 counterparts in rice, the CYP709 family that contains 3 genes in Arabidopsis compared with 11 in rice, the CYP711 family that contains 1 gene in Arabidopsis with 5 counterparts in rice, and the CYP94 family that contains 6 genes in Arabidopsis compared with 18 in rice. By contrast, another fatty acid metabolizing family (CYP86) decreases from 11 genes in Arabidopsis to 5 genes in rice. In another contrast, the sets of 11 CYP728 and 11 CYP730 genes that exist in rice have no counterparts in Arabidopsis. These enumerations of the CYP86 family and others that occur in the CYP77, CYP79, and CYP81 families make it clear that family expansion is not necessarily correlated to genome size. Furthermore, apparent conservations of gene numbers within families do not imply conservations in functions. This is exemplified best in the CYP98A8 and CYP98A9 genes that have functions different from CYP98A3 in Arabidopsis (Schoch et al., 2001) and no apparent orthologs in the 3 CYP98 genes in rice.
Nomenclature Revisions
The present nomenclature system for the P450s has been in use since 1987 (Nebert et al., 1987). From time to time it has been necessary to change a P450 name due to revisions in incomplete or inaccurate sequences, accidental assignment of the same name to two different genes, improper naming of sequences by authors without consulting the nomenclature committee, and the effects of adding new sequences on the location of branches on the trees used in assigning names. This last process has resulted in three names that require changing due to addition of 455 rice genes to the nomenclature. Not unexpectedly, the nomenclature system as well as publications, grants, microarrays, and patents are impacted by these changes. In the case of minor changes, such as shifting a sequence to a new subfamily within the same family, it is sometimes best to retain the original name. This is the case with Arabidopsis CYP83B1, which was originally sorted into its own subfamily rather than being designated CYP83A2 (Bak and Feyereisen, 2001). More complete and accurate sequencing has indicated that CYP83B1 is 58% identical to CYP83A1 and it should be listed in the same subfamily. However, the nomenclature committee decided to retain its original designation.
In the case of major changes, such as when a sequence shifts from within a family to another distinct location on a tree, it is not appropriate to retain the original name. In these cases, the confusion caused by having one family with two locations on the phylogenetic tree outweighs the cost of retaining the original name. The need for reordering of these names is especially apparent for CYP709A1 and CYP709A2 sequences in Arabidopsis, which exist within the CYP72 clan in a region of the phylogenetic tree that has separated from the CYP709 family (Fig. 6, CYP72 clan). With the availability of new rice sequences, it has become clear that these sequences join to form a new CYP735 family with their new numbers being designated as Arabidopsis CYP735A1 and CYP735A2, respectively. Since no publications exist with allusion to the CYP709A subfamily, this name change will have little impact on existing literature. More problematically, the Arabidopsis CYP72B1 (alias BAS-1; Neff et al., 1999; Turk et al., 2003) in the CYP72 clan has joined five rice and two tomato (Lycopersicon esculentum) sequences in a newly designated CYP734 family. Nomenclature pages for Arabidopsis P450s retain an entry for the three old names, noting that they have been renamed. With the availability of nearly 1,100 plant P450 sequences in the databases, the plant P450 nomenclature committee expects that future name changes will occur only at a low frequency and only be made for compelling reasons.
P450 Orthologs in the Two Rice Strains
The two rice strains (indica and japonica) sequenced for the compilation of the rice genome are incomplete in public access files. As a result, some P450 sequences are only represented from one strain. The International Rice Genome Sequencing Project (IRGSP) summarized their progress in the japonica sequence as of Nov. 16, 2003 (http://demeter.bio.bnl.gov/Shanghai_summary.html), and indicated that there are 1,277 clones yet to finish, covering 24.5 Mb of sequence in 56 physical gaps with another 12 Mb of sequence remaining unanchored. The P450 sequences from the indica strain that have no orthologous genes in japonica probably fall in one or more of these gaps. One clear example of this is the set of indica CYP730 sequences that currently contain two complete and nine incomplete P450 genes. None of these sequences are present in the available japonica sequence. There are currently 28 P450s that are incomplete.
Comparisons between the completed indica and japonica sequences indicate that orthologous genes are very similar. In 100 orthologous pairs taken in order of their indica accession numbers, 30 pairs are 100% identical at the protein level and another 49 are >99% identical, 6 more are 98% to 99% identical, 15 are 94% to 98% identical, and 5 of the lowest percent orthologs are pseudogenes. The phylogenetic trees presented in this article were of necessity composed of both japonica and indica sequences, but, in the absence of complete sequence information, the high degree of amino acid identity between orthologs clearly justifies this mixture of sequences.
Anomalies in the Definition of P450 Transcription Units/Genes
While the comparisons described above detail the identities and divergences existing in the coding sequences of individual P450 loci, there are a number of lines of evidence suggesting that a number of Arabidopsis P450 loci produce more than one type of transcript and, therefore, more than one P450 protein. Drawn from alignments of available EST and full-length cDNA sequences, variations in the transcriptional start site in the CYP708A2 locus generate a longer version (518 amino acids) and a shorter version (477 amino acids) of the CYP708A2 protein differing only in their N-terminal amino acids (J. Thimmapuram, H. Duan, and S. Schuler, unpublished data). Variations in the 3′ splice site selection process within the first intron generate a full-length version (522 amino acids) and an N-terminally truncated version (439 amino acids) of the CYP711A1 protein, again differing only in their N-terminal amino acids. More unusually, transcription read-through of the tandem CYP96A9 and CYP96A10 loci and in-frame splicing of the CYP96A9 coding sequence to the CYP96A10 coding sequence generate a dimeric P450 protein containing two P450 signature motifs (J. Thimmapuram, H. Duan, and S. Schuler, unpublished data). These unusual transcripts coexist with transcripts terminating downstream from these individual CYP96A9 and CYP96A10 loci, indicating that this set of adjacent P450 genes is technically capable of coding for three P450 proteins. Similarly, transcription read-through followed by in-frame splicing events fuse the CYP71B10 open reading frame to the reading frame of a non-P450 protein. Along a slightly different vein, transcription read-throughs of the tandem CYP705A15 and CYP705A16 loci and the tandem CYP71B34 and CYP71B35 loci without locus-spanning splicing events generate long, apparently bicistronic transcripts containing two P450 open reading frames and the entire intergenic region separating these adjacent loci. As is the case with the coding region fusions described above, these unusual loci coexist with transcripts terminating downstream from individual loci. But, unlike the coding region fusions described above, these transcripts are not predicted to result in alternative proteins, even if they were capable of initiating translation of the downstream open reading frame and are, therefore, not relevant to the protein comparisons that are the subject of this article. These additional truncated and fused P450s add another level of complexity to the P450 nomenclatures in Arabidopsis. But, until cloning of full-length rice cDNAs makes it clearer whether alternative P450 transcripts exist in rice, comparisons with these unusual Arabidopsis P450s are not possible.
Evolution of Some Specific Pathways in Plants
The sequencing of multiple genomes allows for a deeper understanding at the molecular level of the presence of biochemical pathways and biochemical features. Except for the CYP71 clan, the other nine clans seem primarily involved in conserved functions that relate to sterol and isoprenoid biosynthesis (85 and 51 clans), fatty acid metabolism (86 clan), carotenoids (97 clan), biosynthesis of oxylipids (74 clan), and plant hormone homeostasis (72 clan). The proliferation of the CYP71 clan coincided with the explosion of terrestrial vascular plants approximately 425 million years ago. Although the CYP71 clan is absent in the green algae, a few CYP71 clan members can be found in mosses, such as CYP73 (phenylpropanoid biosynthesis; Teutsch et al., 1993; Werck-Reichhart et al., 2002), CYP98 like (early phenylpropanoid biosynthesis; Schoch et al., 2001; Franke et al., 2002a, 2002b), and CYP78 (function unknown). Ring-B flavonoid hydroxylases (CYP75; Holton et al., 1993; Werck-Reichhart et al., 2002) are present in gymnosperms, while late enzymes in the lignin and flavonoid synthesis (CYP84 [Humphreys et al., 1999; Werck-Reichhart et al., 2002] and CYP93 [Akashi et al., 1999; Steele et al., 1999]) pathways are found only in angiosperms. This is in agreement with the different lignin compositions between gymnosperms (G-lignin) and angiosperms (G/S lignin; Boerjan et al., 2003) and increased accompanying increases in the complexity of flavonoid structures. Many of the other 71 clan families and, in particular, subfamilies appear to be species specific and represent the success in recruiting P450s for evolutionary novelty. Among these, the CYP79 family is particularly interesting because several of its members are amino acid N-hydroxylases involved in synthesis of cyanogenic glucosides and glucosinolates, two defense-related secondary metabolites (Wittstock and Halkier, 2002). Cyanogenic glucosides are widespread in nature and are evolutionary older than glucosinolates, as they are present in ferns as well as gymnosperms and angiosperms. Glucosinolates reside only in the Brassicaceae. Accordingly, members of the CYP79 family exist in both rice, which synthesizes cyanogenic glucosides, and Arabidopsis, which synthesizes glucosinolates. Interestingly, rice has only four CYP79 genes and no pseudogenes, while Arabidopsis has seven full-length genes and five pseudogenes. The high number of pseudogenes in Arabidopsis probably reflects the fact that the acquisition of the capacity to synthesize glucosinolates is an evolutionarily recent event that has not stabilized in the Arabidopsis genome (Kroymann et al., 2003).
CONCLUSIONS
The open space of Figure 1 reveals the great holes in our knowledge about plant P450s. There are two completed rows of data points crossing the page, one for rice and one for Arabidopsis. Beyond that, sequences for soybean (Fabales) and tomato/potato (Solanum tuberosum; Solanales) are filling in, and the ongoing analysis of their ESTs will continue this process. Current counts for ESTs at GenBank show 549,926 ESTs for wheat (Triticum aestivum), 391,145 for Zea mays, 352,924 for barley (Hordeum vulgare), and 345,723 for soybean. Of the top 32 species listed (with more than 100,000 ESTs), 14 are plants. Many of these have a page at The Institute for Genomic Research Gene Indices (http://www.tigr.org/tdb/tgi/plant.shtml), where the ESTs have been sorted into contigs and partially identified as to their protein families. Careful examination of this data will eventually allow us to fill in many more data points in the P450 phylogenetic trees that we are currently extending.
The numbers are daunting. As of March 31, 2004, 1,098 plant P450s have been named, with most in the two species that are the subject of this review. Calculating that there are 35 lines in Figure 1, each containing approximately 300 P450s per line, projections suggest 10,500 P450 sequences are needed to completely fill in this chart of higher plant P450 proteins. Extensions of this analysis into the genomes of Chlamydomonas, ferns, liverworts, stoneworts, mosses, and cycads begin to open a path into this nearly unexplored territory. But, it is now clear that we have a P450 framework that these new sequences can be mapped onto. With their addition to this matrix and a definition of critical P450 functions, we will have a significantly better understanding of the role of P450 diversity in the acquisition of novel biochemical functions. While the numbers of P450 families (about 60 so far) and clans, or clusters, of families (10 so far) may increase to some extent, we at least will know geographical (and biochemical) constraints on this enormous superfamily of proteins.
From the data we have now, two thirds of rice and Arabidopsis families are shared between these monocot and dicot prototypes. While sequencing of more species (and not just more grass species) will increase this count, it is clear that much of plant P450 diversity existed prior to the divergence of monocots and dicots, estimated to have occurred 200 million years ago. Except for the CYP727 clan, it is now obvious that all of the plant P450 clans existed before gymnosperms branched from angiosperms (360 million years ago). This fact is comparable to the conclusions drawn from an examination of P450 evolution in chordates (Nelson, 2003). In this, fish (Takifugu, Danio, and Tetraodon), which diverged from tetrapods about 420 million years ago, contain all the same P450 families as mammals except for one (CYP39). Based on our current analysis of the Arabidopsis and rice sequences, it appears that once P450 families become established in plants or animals, they tend to persist in a recognizable and, presumably, functional form over hundreds of millions of years. Our evidence indicates that plants have created at least three times more P450 families than chordates in a 400 million-year time frame. One remaining question that will be decided in future genomic comparisons is whether the number and types of P450s existing in early plants gave them an advantage in the colonization of land, possibly by allowing adaptive chemistries to evolve water-tight barriers and structural components for life outside the water. It is likely that the multigenome history of plants, depending as it did on symbioses with cyanobacteria, supplied the primordial plants with additional P450s to mediate functions other than the obligatory CYP51 sequence needed for sterol synthesis. Some of the P450s found in cyanobacteria today are obviously eukaryotic-type P450s, such as the CYP110 sequences from Anabaena and CYP120 from Synechocystis. These or related P450s could have given the primordial plant some extra P450s beyond the obligatory CYP51 needed for sterol synthesis. P450s are found in cyanobacteria today, and they are eukaryotic forms. CYP110 sequences from Anabaena, CYP120 sequences from Synechocystis, or related P450 sequences could have existed in that first land plant 425 million years ago. The present day 3:1 ratio of P450 families seen in the plants as compared to the chordates could be due to unequal numbers of progenitor genes in each of these lineages and not to more rapid evolution in the plant lineage. With the large number of P450 genes available in each genome for examination of these evolutionary events, the tale will rapidly unfold as we move from analysis of the Chlamydomonas genome to mosses and ferns.
Having nearly completed comparisons of the P450 superfamilies in Arabidopsis and rice, one of the big challenges for the future is to define the range of functions for individual P450 proteins within each of these diversified gene families. Because of potential redundancies in protein functions, this, of necessity, is best accomplished by expression of individual P450 cDNAs in heterologous systems that provide the interactive P450 reductases needed for catalytic function (Schuler and Werck-Reichhart, 2003). With these functions defined, it will become possible to examine the degree of evolution within the catalytic sites of P450s mediating similar catalytic reactions and to define that range of amino acid changes dictating the evolution of new enzyme activities.
MATERIALS AND METHODS
The Arabidopsis P450 sequences have been collected over many years and annotated by various groups. Complete collections of the Arabidopsis P450 genes are available at two Arabidopsis P450 sites (http://www.biobase.dk/P450/ and http://arabidopsis-P450.biotec.uiuc.edu) and a Cyt P450 Homepage (http://drnelson.utmem.edu/cytochromeP450.html), as well as P450 entries at The Arabidopsis Information Resource (http://tair.stanford.edu/info/genefamily/p450.html). Based on comparisons with all available full-length cDNAs and ESTs, the Arabidopsis Cyt P450s have recently been reannotated and corrected for problems in gene models with the National Science Foundation 2010 Project at http://arabidopsis-P450.biotec.uiuc.edu, providing a searchable Arabidopsis P450 database at http://arabidopsis-P450.biotec.uiuc.edu/cgi-bin/p450.pl.
The rice (Oryza sativa) P450s have been identified by systematic BLAST searches against the public project sequences from the japonica strain at GenBank (http://www.ncbi.nlm.nih.gov/BLAST/, nr division, limit to Oryza sativa) and the indica strain from the Beijing Genomics Institute (http://www.ncbi.nlm.nih.gov/BLAST/, WGS division, limit to Oryza sativa). The strategy for finding all members of the P450 family in a genome has been described (Nelson, 2002). Briefly, in this evaluation one member from each plant P450 clan was used in the search process conducted at the protein sequence level. The hits were collected and placed in a BLAST searchable file on a local server. Accession numbers were sorted in alphanumeric order, and new searches were compared against this list to identify novel accession numbers. The indica and japonica strains were treated separately to avoid creating hybrid genes. Sequences of the indica strain were substantially easier to search, since they had been assembled and duplicate sequences had been removed from the database. Sequences of the japonica strain had to be assembled into intact genes from multiple overlapping sequences. Annotation is a self-correcting endeavor. Occasionally, this process caused revisions in the nomenclature, as incorrectly assembled genes were detected and split into individual genes. For example, Arabidopsis CYP705A7 became CYP705A30 and CYP705A32 and the CYP705A7 name was retired. Communication between the P450 database sites listed above has resulted in an agreed set of annotated Arabidopsis P450 genes and pseudogenes.
The collection of rice sequences obtained above was BLAST searched against one another to find overlapping or identical sequences, which were then placed in unique gene bins. All of these genes were extended to their complete length if possible, by examining translations of EST, cDNA, or genomic DNA sequences. The process of finding BLAST hits was continued using members from individual families within each P450 clan to better cover the sequence space. As new rice sequences were generated and assembled, these were used to find more closely related sequences. Sequences unique to one strain were searched against the other strain to identify orthologs. Representative members of P450 families not immediately evident in rice were also used in searches for missing genes.
Phylogenetic trees were generated using the Neighbor-Join (ClustalX 1.83) or Unweighted Pair Group Method with Arithmetic Mean (UPGMA) with BioNJ (PAUP version 4.0) methods, and underlying alignments used the Gonnett or PAM matrices (ClustalX 1.83). Sequences used were taken from the rice and Arabidopsis FASTA sequence files of the Cyt P450 Web page or from GenBank using accession numbers given on the nomenclature file Biblio D (Plant P450s) at http://drnelson.utmem.edu/bibliod.html. Choices of algorithms, scoring matrices, and gap penalties for making alignments that produced the trees in Figures 2 to 6 were driven by a desire to keep known or suspected orthologous gene clusters in the same branch on the trees. The CYP51 clan is an example of the variations occurring within a family when using different algorithms, with the Neighbor-Join tree often splitting the fungal CYP51 sequences into multiple branches and the UPGMA tree maintaining all fungal CYP51 sequences in a single branch. Trees were selected that kept presumed orthologous clusters together (as in the CYP72 sequences) and made the best evolutionary sense (kept all fungal CYP51 sequences united) and fit closely to the established P450 nomenclature.
Supplementary Material
Acknowledgments
The efforts of Dr. Jyothi Thimmapuram in annotation and reannotation of the Arabidopsis P450s is greatly appreciated.
This work is supported by the National Science Foundation (2010 Project grant no. MCB 0115068 to M.A.S.), by the Danish National Research Foundation (grant to Center for Molecular Plant Physiology [PlaCe]), and by the Danish Agricultural and Veterinary Research Council (grant no. 23–02–0095 to S.B.).
The online version of this article contains Web-only data.
References
- Agrawal GK, Tamogami S, Han O, Iwahashi H, Rakwal R (2004) Rice octadecanoid pathway. Biochem Biophys Res Commun 317: 1–15 [DOI] [PubMed] [Google Scholar]
- Akashi T, Fukuchi-Mizutani M, Aoki T, Ueyama Y, Yonekura-Sakakibara K, Tanaka Y, Kusumi T, Ayabe S (1999) Molecular cloning and biochemical characterization of a novel cytochrome P450, flavone synthase II, that catalyzes direct conversion of flavanones to flavones. Plant Cell Physiol 40: 1182–1186 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Azpiroz R, Wu Y, LoCascio JC, Feldmann KA (1998) An Arabidopsis brassinosteroid-dependent mutant is blocked in cell elongation. Plant Cell 10: 219–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak S, Feyereisen R (2001) The involvement of two P450 enzymes, CYP83B1 and CYP83A1, in auxin homeostasis and glucosinolate biosynthesis. Plant Physiol 127: 108–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak S, Tax FE, Feldmann KA, Galbraith DW, Feyereisen R (2001) CYP83B1, a cytochrome P450 at the metabolic branch point in auxin and indole glucosinolate biosynthesis in Arabidopsis. Plant Cell 13: 101–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlier I, Kowalczyk M, Marchant A, Ljung K, Bhalerao R, Bennett M, Sandberg G, Bellini C (2000) The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc Natl Acad Sci USA 97: 14819–14824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellamine A, Mangla AT, Nes WD, Waterman MR (1999) Characterization and catalytic properties of the sterol 14alpha-demethylase from Mycobacterium tuberculosis. Proc Natl Acad Sci USA 96: 8937–8942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste I, Tijet N, Adas F, Philipps G, Salaun JP, Durst F (1998) CYP86A1 from Arabidopsis thaliana encodes a cytochrome P450-dependent fatty acid omega-hydroxylase. Biochem Biophys Res Commun 243: 688–693 [DOI] [PubMed] [Google Scholar]
- Bishop GJ, Koncz C (2002) Brassinosteroids and plant steroid hormone signaling. Plant Cell 14 (suppl.): S97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Choe S, Dilkes BP, Fujioka S, Takatsuto S, Sakurai A, Feldmann KA (1998) The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22α-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10: 231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD (2001) Brassinosteroids. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/10.1199/tab.0009, http://www.aspb.org/publications/arabidopsis/
- Creelman RA, Mullet JE (1997) Biosynthesis and action of jasmonates in plants. Annu Rev Plant Physiol Plant Mol Biol 48: 355–381 [DOI] [PubMed] [Google Scholar]
- Debeljak N, Fink M, Rozman D (2003) Many facets of mammalian lanosterol 14 α-demethylase from the evolutionarily conserved cytochrome P450 family CYP51. Arch Biochem Biophys 409: 159–171 [DOI] [PubMed] [Google Scholar]
- Debeljak N, Horvat S, Vouk K, Lee M, Rozman D (2000) Characterization of the mouse lanosterol 14α-demethylase (CYP51), a new member of the evolutionarily most conserved cytochrome P450 family. Arch Biochem Biophys 379: 37–45 [DOI] [PubMed] [Google Scholar]
- Durst F, Nelson DR (1995) Diversity and evolution of plant P450 and P450 reductases. Drug Metabol Drug Interact 12: 189–206 [DOI] [PubMed] [Google Scholar]
- Frank MR, Deyneka JM, Schuler MA (1996) Cloning of wound-induced cytochrome P450 monooxygenases expressed in pea. Plant Physiol 110: 1035–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke R, Hemm MR, Denault JW, Ruegger MO, Humphreys JM, Chapple C (2002. a) Changes in secondary metabolism and deposition of an unusual lignin in the ref8 mutant of Arabidopsis. Plant J 30: 47–59 [DOI] [PubMed] [Google Scholar]
- Franke R, Humphreys JM, Hemm MR, Denault JW, Ruegger MO, Cusumano JC, Chapple C (2002. b) The Arabidopsis REF8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J 30: 33–45 [DOI] [PubMed] [Google Scholar]
- Fujioka S, Yokota T (2003) Biosynthesis and metabolism of brassinosteroids. Annu Rev Plant Physiol Plant Mol Biol 54: 137–164 [DOI] [PubMed] [Google Scholar]
- Galbraith DW, Bak S (2004) Functional genomics of the cytochrome P450 gene superfamily in Arabidopsis thaliana. In D Leister, ed, Plant Functional Genomics. Haworth Press, Binghamton, NY, in press
- Helliwell CA, Chandler PM, Poole A, Dennis ES, Peacock WJ (2001) The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc Natl Acad Sci USA 98: 2065–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemm MR, Ruegger MO, Chapple C (2003) The Arabidopsis ref2 mutant is defective in the gene encoding CYP83A1 and shows both phenylpropanoid and glucosinolate phenotypes. Plant Cell 15: 179–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holton TA, Brugliera F, Lester DR, Tanaka Y, Hyland CD, Menting JG, Lu CY, Farcy E, Stevenson TW, Cornish EC (1993) Cloning and expression of cytochrome P450 genes controlling flower colour. Nature 366: 276–279 [DOI] [PubMed] [Google Scholar]
- Hong Z, Ueguchi-Tanaka M, Shimizu-Sato S, Inukai Y, Fujioka S, Shimada Y, Takatsuto S, Agetsuma M, Yoshida S, Watanabe Y, et al (2002) Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J 32: 495–508 [DOI] [PubMed] [Google Scholar]
- Hong Z, Ueguchi-Tanaka M, Umemura K, Uozu S, Fujioka S, Takatsuto S, Yoshida S, Ashikari M, Kitano H, Matsuoka M (2003) A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15: 2900–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys JM, Hemm MR, Chapple C (1999) New routes for lignin biosynthesis defined by biochemical characterization of recombinant ferulate 5-hydroxylase, a multifunctional cytochrome P450-dependent monooxygenase. Proc Natl Acad Sci USA 96: 10045–10050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CJ, Lamb DC, Marczylo TH, Parker JE, Manning NL, Kelly DE, Kelly SL (2003) Conservation and cloning of CYP51: a sterol 14 alpha-demethylase from Mycobacterium smegmatis. Biochem Biophys Res Commun 301: 558–563 [DOI] [PubMed] [Google Scholar]
- Jackson CJ, Lamb DC, Marczylo TH, Warrilow AG, Manning NJ, Lowe DJ, Kelly DE, Kelly SL (2002) A novel sterol 14alpha-demethylase/ferredoxin fusion protein (MCCYP51FX) from Methylococcus capsulatus represents a new class of the cytochrome P450 superfamily. J Biol Chem 277: 46959–46965 [DOI] [PubMed] [Google Scholar]
- Kahn R, Durst F (2000) Function and evolution of plant cytochrome P450. Recent Adv Phytochem 34: 151–189 [Google Scholar]
- Kahn RA, Le Bouquin R, Pinot F, Benveniste I, Durst F (2001) A conservative amino acid substitution alters the regiospecificity of CYP94A2, a fatty acid hydroxylase from the plant Vicia sativa. Arch Biochem Biophys 39: 180–187 [DOI] [PubMed] [Google Scholar]
- Kang J-G, Yun J, Kim DH, Chung KS, Fujioka S, Kim JI, Dae HW, Yoshida S, Takatsuto S, Song PS, et al (2001) Light and brassinosteroid signals are integrated via a dark-induced small G protein in etiolated seedling growth. Cell 105: 625–636 [DOI] [PubMed] [Google Scholar]
- Kroymann J, Donnerhacke S, Schnabelrauch D, Mitchell-Olds T (2003) Evolutionary dynamics of an Arabidopsis insect resistance quantitative trait locus. Proc Natl Acad Sci USA 100: 14587–14592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushiro M, Nakano T, Sato K, Yamagishi K, Asami T, Nakano A, Takatsuto S, Fujioka S, Ebizuka Y, Yoshida S (2001) Obtusifoliol 14alpha-demethylase (CYP51) antisense Arabidopsis shows slow growth and long life. Biochem Biophys Res Commun 285: 98–104 [DOI] [PubMed] [Google Scholar]
- Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, Asami T, Hirai N, Koshiba T, Kamiya Y, Nambara E (2004) The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: key enzymes in ABA catabolism. EMBO J 23: 1647–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb DC, Kelly DE, Kelly SL (1998) Molecular diversity of sterol 14alpha-demethylase substrates in plants, fungi and humans. FEBS Lett 425: 263–265 [DOI] [PubMed] [Google Scholar]
- Laudert D, Pfannschmidt U, Lottspeich F, Hollander-Czytko H, Weiler EW (1996) Cloning, molecular and functional characterization of Arabidopsis thaliana allene oxide synthase (CYP 74), the first enzyme of the octadecanoid pathway to jasmonates. Plant Mol Biol 31: 323–335 [DOI] [PubMed] [Google Scholar]
- Margulis L, Schwartz KV (1998) Five Kingdoms. An illustrated Guide to the Phyla of Life on Earth, Ed 3. W.H. Freeman, New York
- Moore RC, Purugganan MD (2003) The early stages of duplicate gene evolution. Proc Natl Acad Sci USA 100: 15682–15687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Nomura T, Ooka H, Ishizaka M, Yokota T, Sugimoto K, Okabe K, Kajiwara H, Satoh K, Yamamoto K, et al (2002) Isolation and characterization of a rice dwarf mutant with a defect in brassinosteroid biosynthesis. Plant Physiol 130: 1152–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naur P, Petersen BL, Mikkelsen MD, Bak S, Rasmussen H, Olsen CE, Halkier BA (2003) CYP83A1 and CYP83B1, two nonredundant cytochrome P450 enzymes metabolizing oximes in the biosynthesis of glucosinolates in Arabidopsis. Plant Physiol 133: 63–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebert DW, Adesnik M, Coon MJ, Estabrook RW, Gonzalez FJ, Guengerich FP, Gunsalus IC, Johnson EF, Kemper B, Levin W, et al (1987) The P450 gene superfamily: recommended nomenclature. DNA 6: 1–11 [DOI] [PubMed] [Google Scholar]
- Neff MM, Nguyen SM, Malancharuvil EJ, Fujioka S, Noguchi T, Seto H, Tsubuki M, Honda T, Takatsuto S, Yoshida S, et al (1999) BAS1: A gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc Natl Acad Sci USA 96: 15316–15323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DR (2002) Mining databases for cytochrome P450 genes. Methods Enzymol 357: 1–15 [DOI] [PubMed] [Google Scholar]
- Nelson DR (2003) Comparison of P450s from human and fugu: 420 million years of vertebrate P450 evolution. Arch Biochem Biophys 409: 18–24 [DOI] [PubMed] [Google Scholar]
- Nelson DR, Zeldin DC, Hoffman SMG, Maltais LJ, Wain HM, Nebert DW (2004) Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes, and alternative-splice variants. Pharmacogenetics 14: 1–18 [DOI] [PubMed] [Google Scholar]
- Paquette SM, Bak S, Feyereisen R (2000) Intron-exon organization and phylogeny in a large superfamily, the paralogous cytochrome P450 genes of Arabidopsis thaliana. DNA Cell Biol 19: 307–317 [DOI] [PubMed] [Google Scholar]
- Paquette SM, Møller BL, Bak S (2003) On the origin of family 1 plant glycosyltransferases. Phytochemistry 62: 399–413 [DOI] [PubMed] [Google Scholar]
- Ralston L, Kwon ST, Schoenbeck M, Ralston J, Schenk DJ, Coates RM, Chappell J (2001) Cloning, heterologous expression, and functional characterization of 5-epi-aristolochene-1,3-dihydroxylase from tobacco (Nicotiana tabacum). Arch Biochem Biophys 393: 222–235 [DOI] [PubMed] [Google Scholar]
- Schoch G, Goepfert S, Morant M, Hehn A, Meyer D, Ullmann P, Werck-Reichhart D (2001) CYP98A3 from Arabidopsis thaliana is a 3′-hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. J Biol Chem 276: 36566–36574 [DOI] [PubMed] [Google Scholar]
- Schopfer CR, Ebel J (1998) Identification of elicitor-induced cytochrome P450s of soybean (Glycine max L.) using differential display of mRNA. Mol Gen Genet 258: 315–322 [DOI] [PubMed] [Google Scholar]
- Schuler MA, Werck-Reichhart D (2003) Functional genomics of P450s. Annu Rev Plant Biol 54: 629–667 [DOI] [PubMed] [Google Scholar]
- Shimada Y, Fujioka S, Miyauchi N, Kushiro M, Takatsuto S, Nomura T, Yokota T, Kamiya Y, Bishop GJ, Yoshida S (2001) Brassinosteroid-6-oxidases from Arabidopsis and tomato catalyze multiple C-6 oxidations in brassinosteroid biosynthesis. Plant Physiol 126: 770–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y, Goda H, Nakamura A, Takatsuto S, Fujioka S, Yoshida S (2003) Organ-specific expression of brassinosteroid-biosynthetic genes and distribution of endogenous brassinosteroids in Arabidopsis. Plant Physiol 131: 287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele CL, Gijzen M, Qutob D, Dixon RA (1999) Molecular characterization of the enzyme catalyzing the aryl migration reaction of isoflavonoid biosynthesis in soybean. Arch Biochem Biophys 367: 146–150 [DOI] [PubMed] [Google Scholar]
- Szekeres M, Németh K, Koncz-Kálmán Z, Mathur J, Kauschmann A, Altmann T, Rédei GP, Nagy F, Schell J, Koncz C (1996) Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85: 171–182 [DOI] [PubMed] [Google Scholar]
- Teutsch HG, Hasenfratz MP, Lesot A, Stoltz C, Garnier JM, Jeltsch JM, Durst F, Werck-Reichhart D (1993) Isolation and sequence of a cDNA encoding the Jerusalem artichoke cinnamate 4-hydroxylase, a major plant cytochrome P450 involved in the general phenylpropanoid pathway. Proc Natl Acad Sci USA 90: 4102–4106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Musetti V, Kim J, Magallanes-Lundback M, DellaPenna D (2004) The Arabidopsis LUT1 locus encodes a member of the cytochrome p450 family that is required for carotenoid epsilon-ring hydroxylation activity. Proc Natl Acad Sci USA 101: 402–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troitsky AV, Melekhovets YuF, Rakhimova GM, Bobrova VK, Valiejo-Roman KM, Antonov AS (1991) Angiosperm origin and early stages of seed plant evolution deduced from rRNA sequence comparisons. J Mol Evol 32: 253–261 [DOI] [PubMed] [Google Scholar]
- Turk EM, Fujioka S, Seto H, Shimada Y, Takatsuto S, Yoshida S, Denzel MA, Torres QI, Neff MM (2003) CYP72B1 inactivates brassinosteroid hormones: an intersection between photomorphogenesis and plant steroid signal transduction. Plant Physiol 133: 1643–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellesen K, Durst F, Pinot F, Benveniste I, Nettesheim K, Wisman E, Steiner-Lange S, Saedler H, Yephremov A (2001) Functional analysis of the LACERATA gene of Arabidopsis provides evidence for different roles of fatty acid omega-hydroxylation in development. Proc Natl Acad Sci USA 98: 9694–9699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werck-Reichhart D, Bak S, Paquette S (2002) Cytochrome P450. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/10.1199/tab.0028, http://www.aspb.org/publications/arabidopsis
- Willis HJ, McElwain JC (2002) The Evolution of Plants. Oxford University Press, Oxford
- Wittstock U, Halkier BA (2002) Glucosinolate research in the Arabidopsis era. Trends Plant Sci 7: 263–270 [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Gouy M, Yang YW, Sharp PM, Li WH (1989) Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc Natl Acad Sci USA 86: 6201–62052762323 [Google Scholar]
- Yoshida Y, Noshiro M, Aoyama Y, Kawamoto T, Horiuchi T, Gotoh O (1997) Structural and evolutionary studies on sterol 14-demethylase P450 (CYP51), the most conserved P450 monooxygenase: II. Evolutionary analysis of protein and gene structures. J Biochem (Tokyo) 122: 1122–1128 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.