Abstract
Here, we report our effort in generating an ORFeome collection for the Arabidopsis transcription factor (TF) genes. In total, ORFeome clones representing 1,282 Arabidopsis TF genes have been obtained in the Gateway high throughput cloning pENTR vector, including 411 genes whose annotation lack cDNA support. All the ORFeome inserts have also been mobilized into a yeast expression destination vector, with an estimated 85% rate of expressing the respective proteins. Sequence analysis of these clones revealed that 34 of them did not match with either the reported cDNAs or current predicted open-reading-frame sequences. Among those, novel alternative splicing of TF gene transcripts is responsible for the observed differences in at least five genes. However, those alternative splicing events do not appear to be differentially regulated among distinct Arabidopsis tissues examined. Lastly, expression of those TF genes in 17 distinct Arabidopsis organ types and the cultured cells was profiled using a 70-mer oligo microarray.
Transcription factors (TFs) play critical roles in all aspects of a higher plant's life cycle. It is the programmed and regulated interactions between TFs and genomic DNA that bring a genome to its life and define many of its functional features (Grandori et al., 2000; Dimova et al., 2003; Kohler et al., 2003). An initial analysis indicated that Arabidopsis has at least 1,533 TF genes (approximately 6% of the coding capacity of its genome) that belong to more than 30 different families, each possessing a highly conserved and characteristic region recognized as the DNA-binding domain (Riechmann et al., 2000, 2002). Besides the DNA-binding domain in different TF families, there are conserved sequence motifs that help to further classify the TF genes into subgroups (Hosoda et al., 2002; Heim et al., 2003; Parenicova et al., 2003; Toledo-Ortiz et al., 2003). Among the Arabidopsis TFs, about 45% are plant-specific, whereas the rest share DNA-binding domains common to other eukaryotes (Riechmann et al., 2000, 2002). Arabidopsis has several large TF families, each with more than 100 members. Those include the MYB, bHLH, MADS, and AP2/EREBP family of TFs (Riechmann and Ratcliffe, 2000; Hosoda et al., 2002; Heim et al., 2003; Parenicova et al., 2003; Toledo-Ortiz et al., 2003).
Although extensive studies have been carried out for functional analysis of individual TFs, the function of only a small fraction of these TFs has been revealed so far (Riechmann, 2002). The complete sequence of the Arabidopsis genome (The Arabidopsis Genome Initiative, 2000) made it possible not only to approach the function of TFs on a genomic scale, but to examine the transcriptional network and cascade involved. Transcriptional networks or cascades are common features in controlling Arabidopsis development and its response to various environmental challenges (Shinozaki and Yamaguchi-Shinozaki, 2000; Riechmann, 2002). In these regards, microarray profiling has become an important approach to analyze TF genes at the genome level. For example, profiling of 402 Arabidopsis TF genes under various environmental conditions revealed that 74 of the bacterial pathogen-responsive TF genes were also involved in salicylic acid, jasmonic acid, and ethylene signaling (Chen et al., 2002). Among the 43 genes that were activated during senescence, 28 were also induced by other stress treatments, indicating the possibility of extensive overlaps of cellular events downstream of the same TFs (Chen et al., 2002). Recently, a PCR-amplified fragment-based microarray containing approximately 95% of all TF genes from Arabidopsis was generated and used to reveal genome-wide differential expression of TF genes between white light- and dark-grown seedlings (Jiao et al., 2003; Ma et al., 2003). Although microarray profiling is a power tool in revealing TF gene expression patterns and in some instances the temporal and functional interdependency among TF gene expression, it alone often cannot provide sufficient knowledge of TF function.
Further functional analysis of TF genes requires a careful examination of the encoded proteins and their interaction in the cells. A prerequisite for genome-wide analysis of TF genes at the protein level is a collection of cDNA clones with intact open-reading-frames (ORFs). Unfortunately, the initial identification of TF genes in the Arabidopsis genome sequence was carried out mainly by ab initio gene predictions, sequence homology comparisons, motif analysis, and other nonexperimental methods (The Arabidopsis Genome Initiative, 2000). Dramatic progress in Arabidopsis genome annotation was achieved by expressed sequence tag and full-length cDNA analysis (Seki et al., 2002) and tiling-path oligo micorarray studies (Yamada et al., 2003). However, transcriptional activity was demonstrated for only about 70% of the Arabidopsis genes by microarray analyses and currently available full-length cDNA clones cover only about 41% of Arabidopsis genes (Yamada et al., 2003). Thus there is an urgent need for higher coverage of ORFeome clones (cDNA clones containing full-length ORFs) of TF genes for the Arabidopsis community.
Here we report our genome wide effort in generating Arabidopsis TF ORFeome clones, which succeeded in covering 1,282 unique Arabidopsis TF genes. In the process, sequence analysis of our ORFeome clones allowed us to correct a number of errors in the annotation of these genes. Further, comprehensive expression profiles of those TF genes in the Arabidopsis life cycle were conducted. This ORFeome clone collection has been deposited in the Arabidopsis stock center and is available to the research community for in-depth functional analysis of Arabidopsis TF genes.
GENERATION OF ORFeome cDNA CLONES FOR 1282 ARABIDOPSIS TF GENES
Through searching MIPS database using previously described InterPro and GenBank accessions as family identifier (Riechmann et al., 2000), we obtained a collection of 1,581 TF genes from the Arabidopsis genome at the start of this study (see “Materials and Methods”). Table I summarizes TF gene numbers in each family as reported by a prior study (Riechmann et al., 2000) and the number of TF genes for which we were able to retrieve sequence information at the time. Note that a total of about 20 TF genes that could not be placed in any known families were reported as “others.” Further, about 30 TF genes in a newly defined AS family were included as well. Based on the retrieved sequence and annotation information, primer pairs were designed for all TF genes based on a recombinant cloning strategy and used to amplify ORF regions of mRNAs through reverse transcription (RT) and PCR (see Fig. 1 and “Materials and Methods”). A variety of Arabidopsis tissue types and treatments (see “Materials and Methods”) were employed to isolate total RNA to serve as template for RT-PCR. In total, we succeeded in obtaining ORFeome clones for 1,282 TF genes in the pENTR TOPO vector system. Of the 1,282 TF genes that had ORFeome clones, 411 had no matches in current cDNA collections (data not shown).
Table I.
The number of genes encoding putative TFs in the Arabidopsis genome
| TF Families | InterPro or GenBank Acc. Access | A | B | C |
|---|---|---|---|---|
| ABI3/VP1 | CAA48241 | 14 | 17 | 4 |
| ALFIN-like | AAA20093 | 7 | 7 | 7 |
| AP2/EREBP | IPR001471 | 144 | 118 | 142 |
| ARF | AAC49751 | 23 | 23 | 11 |
| ARID | IPR001606 | 4 | 5 | 3 |
| AS2 | 0 | 0 | 30 | |
| AUX/IAA | AAC39440 | 26 | 28 | 26 |
| bHLH | IPR001092 | 139 | 108 | 81 |
| BZIP | IPR001871 | 81 | 82 | 54 |
| C2C2 (Zn)-co-like | A56133 | 33 | 32 | 24 |
| C2C2 (Zn)-dof | CAA66600 | 37 | 35 | 32 |
| C2C2 (Zn)-gata | IPR000679 | 28 | 22 | 33 |
| C2C2 (Zn)-yabby | AAD30526 | 6 | 5 | 3 |
| C2H2 (Zn) | IPR000822 | 105 | 152 | 92 |
| C3H-type1 (Zn) | IPR000232 | 17 | 46 | 32 |
| C3H-type (Zn) | CAA65242 | 16 | 26 | 10 |
| CCAAT | A26771/P13434/Q02516 AAB51375 | 36 | 36 | 34 |
| CPP (Zn) | CAA09028 | 8 | 8 | 5 |
| E2F/DP | O00716/Q64163 | 8 | 8 | 1 |
| EIL | AAC49750 | 6 | 6 | 5 |
| GARP | AAD55941/BAA74528 | 56 | 53 | 24 |
| GRAS | AAB06318 | 32 | 32 | 26 |
| HB | IPR001356 | 89 | 83 | 56 |
| HMG-box | IPR000910 | 10 | 13 | 10 |
| HSF | IPR000232 | 26 | 16 | 12 |
| JUMONJI | T30254 | 9 | 8 | 4 |
| LFY | AAA32826 | 1 | 1 | 0 |
| MADS | IPR002100 | 82 | 91 | 70 |
| MYB | IPR001005/IPR000818 | 190 | 258 | 243 |
| NAC | BAB10725 | 109 | 106 | 93 |
| Nin-like | CAB61243 | 15 | 14 | 2 |
| PCG | 4 | 4 | 2 | |
| SBP | CAB56581 | 16 | 16 | 14 |
| TCP | AAC26786 | 25 | 21 | 19 |
| Trihelix | S39484 | 28 | 21 | 1 |
| TUB | IPR000007 | 11 | 9 | 7 |
| WRKY (Zn) | S72443 | 72 | 70 | 54 |
| Others | 20 | 20 | 16 | |
| Totals | 1,533 | 1,581 | 1,282 |
Column A, number of putative TFs reported by Riechmann et al. (2000); Column B, number of TFs whose sequence and annotation can be retrieved from MIPS database at the time we started this effort (December, 2000); Column C, number of TFs whose ORFs were cloned in the current work. All genes are counted only once, even in some genes that belong to more than one family based on their multiple signature motifs.
Figure 1.
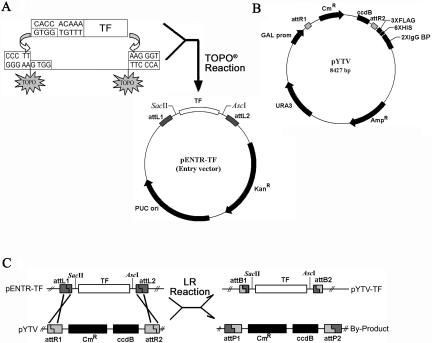
Schematic diagrams showing strategies of high-throughput cloning of blunt-ended PCR amplified TF gene ORFeome products. A, Cloning of ORFeome product into the pENTR-TOPO vector. The PCR products containing individual TF gene ORFs were directionally cloned into pENTR-TOPO vectors using the TOPO DNA recombination reaction facilitated by topoisomerase I attached to one of the vector strands. B, Map of pYTV. This yeast expression vector was modified based on vector pDEST 52 (Invitrogen). The fused tags at the C-terminal of TF ORF include three copies of Flag, six His amino acids, and two copies of IgG-binding motif from protein A. Those tags should enable tandem affinity purification (TAP) of the TF proteins (Rigaut et al., 1999). C, Generation of yeast expression cassette using pENTR clones and pYTV vector. Recombination between different pENTR-TFs and pYTV vectors were carried out in the presence of Gateway LR Clonase enzyme mix. AscI and SacII were designed for determination of the insert size. For further details about this gateway system, please refer to the Invitrogen manual (www.invitrogen.com).
As expected, most of the ORFeome clones matched to the existing cDNA sequences or gene annotation based on single read sequencing from both ends of each clone. Our sequence analysis also revealed differences in 39 clones. Among those, 34 ORFeome clones were completely sequenced and their differences confirmed from either reported cDNA sequences or annotation in public databases (Tables II and III). Table II summarizes those 15 TF genes that have prior deposited cDNA sequences but nevertheless show clear sequence discrepancy between our ORFeome clones and the cDNA. Table III summarizes those 19 Arabidopsis TF genes that had predicted annotation without experimental support. In these cases, we were able to correct inaccuracy in the gene annotation using our ORFeome clones. All those 34 ORFeome clone sequences were submitted to GenBank and their corresponding accession numbers are listed in Table II and III.
Table II.
Fifteen of our TF genes showed different in ORF sequences from previously reported cDNA submission
| Families | Locus ID | Old Acc. | New Acc. | ORF in bp
|
Differences (New VS old) | |
|---|---|---|---|---|---|---|
| Old | New | |||||
| C2H2 (Zn) | At1g03840 | BT006209 | AJ630476 | 1,521 | 1,515 | 6 bp deleted at 1511 |
| bHLH | At1g26260 | RAFL 16–92-I23 | AJ630483 | 1,173 | 1,020 | 153 bp deleted at 178 |
| C2H2 (Zn) | At1g34790 | ATH318491 | AJ630477 | 912 | 909 | 3 bp deleted at 219 |
| C2H2 (Zn) | At1g72050 | RAFL 09–22-M23 | AJ630478 | 975 | 1,239 | 9 bp omitted and 273 bp added from codon 1 |
| SBP | At1g76580 | RAFL 08–16-H24 | AJ630503 | 1,467 | 1,510 | 43 bp added at 1,440 |
| SBP | At2g42200 | RAFL 04–13-I11 | AJ628864 | 1,128 | 1,137 | 9 bp added at 824 |
| C2H2 (Zn) | At3g13810 | RAFL 09–12-L04 | AJ630504 | 1,542 | 1,551 | 9 bp added at 1752 |
| MYB | At3g46590 | RAFL 07–12-A13 | AJ630475 | 939 | 1,659 | 3 bp deleted at 81, 6 bp deleted and 729 bp added at 933 |
| bHLH | At3g61950 | RAFL 19–75-M14 | At3g61950 | 924 | 1,077 | 153 bp added from codon 1 |
| HMG | At4g11080 | AY133687 | AJ630485 | 1,341 | 1,353 | 12 bp added at 90 |
| WRKY (Zn) | At4g26640 | RAFL 07–10-M14 | AJ630479 | 1,458 | 1,674 | 12 bp omitted and 228 bp added from codon 1 |
| bHLH | At4g29930 | RAFL 17–41-A21 | AJ630482 | 765 | 792 | 89 bp deleted and 116 bp added at 676 |
| WRKY (Zn) | At5g22570 | RAFL 21–21-O16 | AJ630480 | 870 | 867 | 3 bp deleted at 677 |
| C3H-type2 (Zn) | At5g46730 | RAFL 15–50-L08 | AJ630502 | 873 | 813 | 60 bp deleted at 586 |
| C2H2 (Zn) | At5g54630 | RAFL 09–11-B05 | AJ630501 | 1,419 | 593 | 826 bp deleted at 568 |
Nucleotide positions counted from the A of translation initiation codon.
This 9-bp sequence was not found in the Arabidopsis genome database.
Table III.
Nineteen clones that were different in their ORF from those predicted annotations in the genome database
| Families | Locus ID | New GenBank Accessions | ORF in bp
|
Differences (Ours VS MIPS)1 | |
|---|---|---|---|---|---|
| MIPS | Ours | ||||
| MYB | At1g14600 | AJ630486 | 252 | 768 | 33 bp deleted and 549 bp added at 219 |
| ABI3/VP1 | At1g28300 | AJ630496 | 1,092 | 1,089 | 3 bp deleted at 1,011 |
| WRKY (Zn) | At1g29280 | AJ630494 | 780 | 762 | 18 bp omitted from codon 1 |
| AP2/EREBP | At1g79700 | AJ580379 | 927 | 912 | 6 bp deleted at 171, 9 bp deleted at 250 |
| C2H2 (Zn) | At3g01030 | AJ630491 | 1,062 | 1,116 | 54 bp added at 633 |
| MYB | At3g10000 | AJ630487 | 1,491 | 1,446 | 13 bp added and 58 bp deleted at 368 |
| CPP (Zn) | At3g16160 | AJ630495 | 1,083 | 1,107 | 3 bp deleted at 137, 6 bp added at 405, 16 bp added and 4 bp deleted at 630, 9 bp added at 716 |
| AP2-EREBP | At3g23230 | AJ580377 | 369 | 420 | 51 bp added at 293 |
| ABI3/VP1 | At3g26790 | AJ630497 | 942 | 933 | 9 bp deleted at 715 |
| C2H2 (Zn) | At3g27970 | AJ630492 | 1,065 | 1,074 | 9 bp added at 315 |
| MYB | At4g12670 | AJ630488 | 1,563 | 1,500 | 63 bp deleted at 1,232 |
| MYB | At4g39160 | AJ630489 | 1,638 | 1,806 | 168 bp added at 49 |
| C2H2 (Zn) | At5g03150 | AJ630493 | 1,506 | 1,512 | 6 bp added at 187 |
| MYB | At5g17780 | AJ630490 | 1,260 | 1,254 | 6 bp deleted at 418 |
| bHLH | At5g38860 | AJ630499 | 1,062 | 897 | 165 bp deleted at 665 |
| AP2/EREBP | At5g50080 | AJ580378 | 663 | 663 | 27 bp deleted at 75, 27 bp added at 202 |
| bHLH | At5g53210 | AJ630498 | 885 | 1,095 | 210 bp added at 583. |
| C2H2 (Zn) | At5g54360 | AJ630500 | 813 | 763 | 9 bp deleted at 681, 24 bp deleted at 697, 17 bp deleted at 750 |
| bHLH | At5g61270 | AJ630484 | 804 | 837 | 19 bp added at 159, 14 bp added at 245 |
All nucleotide positions started from A of the translation initiation codon.
EXPRESSION OF REPRESENTATIVE TF ORFeome CLONES IN YEAST
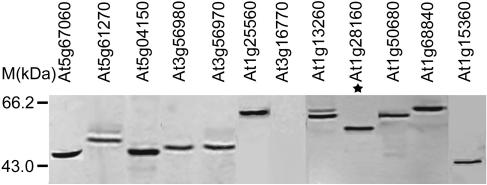
As a way to confirm the intactness of ORFeome clones and to test the feasibility of high-throughput expression of TF proteins, all the ORFeome clone inserts in our collection were transferred into a yeast expression vector (pYTV; see Fig. 1) for expression analysis in yeast. About 300 representative ORFeome clones in pYTV vector were selected from different TF gene families and their expression in yeast was examined via protein-blotting analyses. Using antibodies against a His-tag fused to ORFeome inserts in the pYTV vector, our results indicated that up to 85% of ORFeome clones expressed TF proteins above the detection limit. Examination of the protein size in SDS-PAGE indicated that about 90% of the expressed proteins were of expected Mr (data not shown). For example, as illustrated in Figure 2, of the yeast protein extracts from 12 distinct TF ORFeomes, 10 of them produced strong protein blot signals that match with the calculated protein sizes, while 1 protein migrated significantly slower than predicted (Fig. 2). Protein extract from 1 clone failed to produce a detectable amount of protein (Fig. 2). These results largely confirmed the intactness of our ORFeomes clones. The small amount of clones (<10%) that failed to produce proteins with the expected size (with most migrating slower) suggest that the proteins encoded by these ORFeome clones may have unusual conformations or promiscuous interactions with other proteins in yeast such that they migrated in larger size than expected. For those failed to be detected, the proteins might be unstable or expressed at very low levels in yeast.
Figure 2.
Protein-blot analysis of Arabidopsis TF protein expression in Saccharomyces cerevisiae. Total proteins were fractionated by SDS-PAGE, probed with 1 ug/mL monoclonal antipolyhistidine antibody, and visualized after incubating with goat anti-mouse AP-conjugated secondary antibody. * marks the protein that migrated significantly slower than its predicted Mr.
EXPRESSION PROFILING OF ARABIDOPSIS TF GENES
An Arabidopsis 70-mer oligo microarray covering more than 25,000 Arabidopsis genes were used for organ-specific expression analysis (L.G. Ma, N. Sun, X.G. Liu, Y.L. Jiao, H.Y. Zhao, and X.W. Deng, unpublished data). In this array, 1,222 of the 1,282 ORFeome genes were present. The profiling data of those 1,222 TF genes from the 17 representative Arabidopsis organs and suspension cultured cells were extracted from the above-mentioned data set and allow us to estimate the relative expression abundance for each transcript in different organs. As the detailed characterization of the AP2/EREBP and MYB families of TF genes will be reported separately, the expression patterns for the remaining 858 cloned TF genes are summarized in this report. The expression patterns of MADS family of TF genes among the 17 organs and cultured cells were illustrated in Figure 3, while the expression patterns for the entire 858 TF genes are shown in an appendix figure available at www.plantphysiol.org. One notable feature of the TF gene expression profile is that a vast majority of the TF genes exhibited organ specific expression patterns. On the other hand, some notable exceptions are present. For example, four genes (At5g65670, At2g22430, At2g18160, and At1g30970) were found to express at quite higher levels in all organs and cultured cells tested in the current work. The relative expression levels of the 858 TF genes follow similar distribution in most organ types (Fig. 4) with a general pattern not much different from the total gene expression level distribution in each organ type (data not shown).
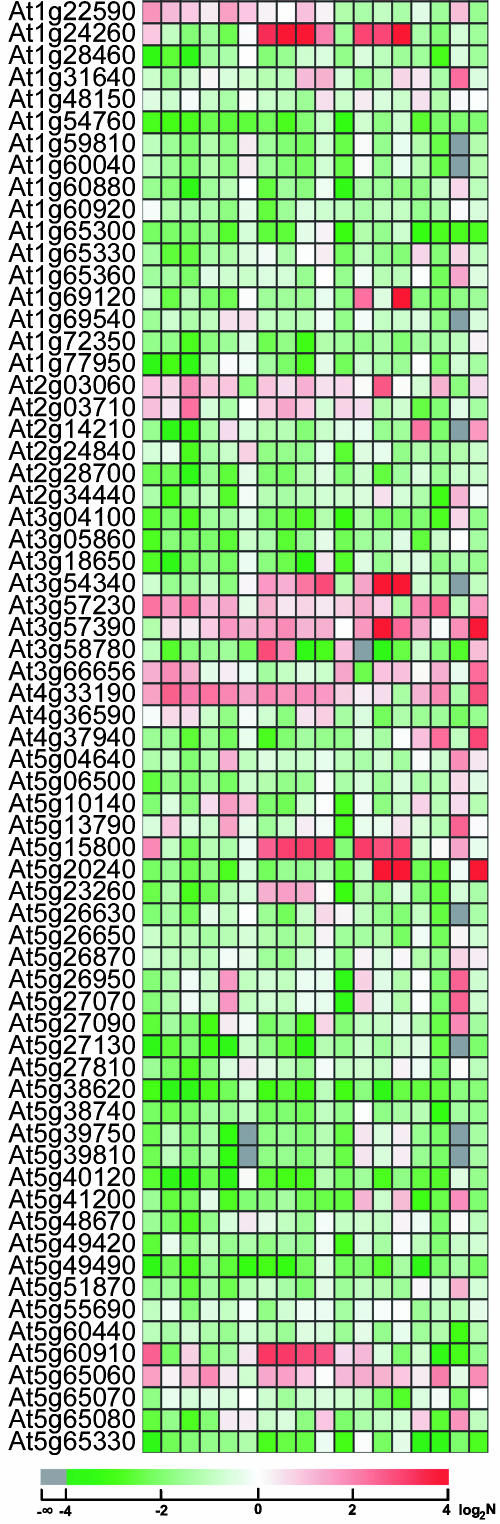
Figure 3.
Expression profiles of 66 Arabidopsis MADS family TF genes in 17 different Arabidopsis organs and cultured cells. Locus ID is listed at the left of the column, and only those MADS box genes whose ORFeome clones are presented in our collection were analyzed. All comparisons were done against the absolute median point obtained for the respective organ. The ratio value of the normalized signal intensity in each organ for a given gene (see “Materials and Methods”) relative to the median value from that organ was first calculated. Then this ratio was subjected to logarithmic (log2) transformation, with the resulting value as indicator of relative expression level among organ type. Therefore, a value of zero indicates an expression level equal to the median of all gene expression in that organ, a positive value indicates an expression level higher than the media, and a negative value indicates an expression level lower than the median. The 18 lanes are as follows: a, cauline leaf; b, light cotyledon; c, rosette leaf; d, dark cotyledon; e, dark hypocotyls; f, light hypocotyl; g, pistil 1 d after pollination; h, pistil 1 d before pollination; i, Silique 3 d after pollination; j, silique 8 d after pollination; k, stem; l, sepal; m, stamen; n, petal; o, dark root; p, light root; q, germinating seed; and r, cultured cells.
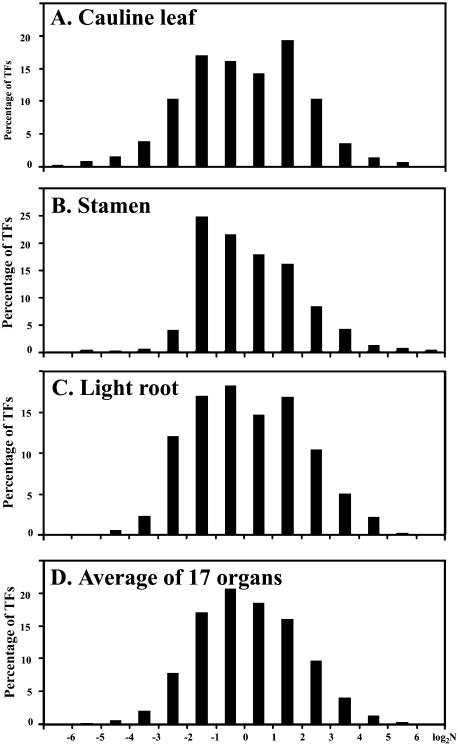
Figure 4.
Distribution of expression abundance for the 858 TF genes. A, Distribution of TF gene expression levels in cauline leaf. B, Distribution of TF gene levels in stamen. C, Distribution of TF gene levels in light root. D, An averaged distribution of TF gene expression levels in all 17 organs. A similar method was used to calculate relative expression as described in the legend for Figure 3 and the log (2) values for the ratio for the normalized expression levels with the median are shown in the x axis.
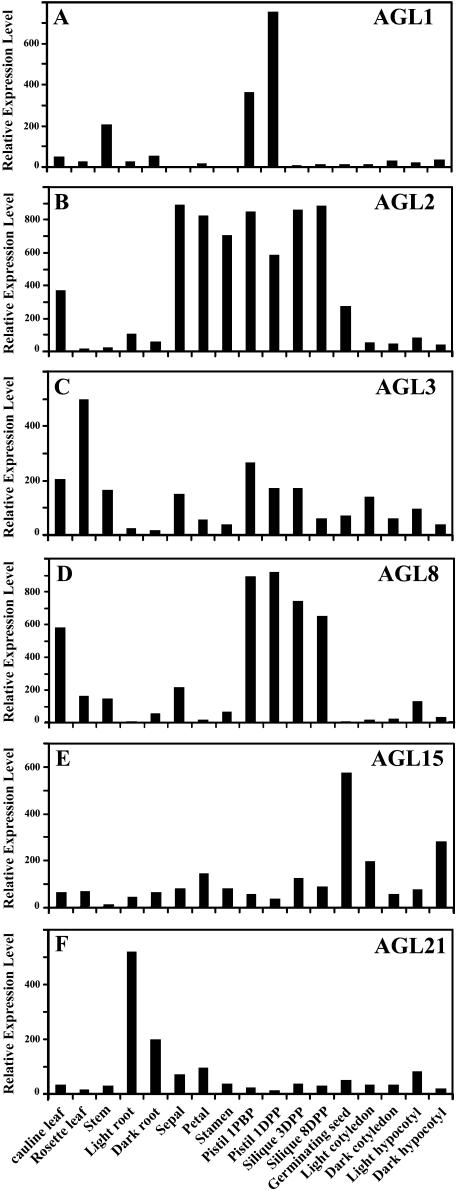
As described in a previous analysis (L.G. Ma, N. Sun, X.G. Liu, Y.L. Jiao, H.Y. Zhao, and X.W. Deng, unpublished data), close comparisons between the known expression patterns of several well-characterized TF genes and the microarray result is a valuable mean to validate our microarray data. The genes examined in that work (L.G. Ma, N. Sun, X.G. Liu, Y.L. Jiao, H.Y. Zhao, and X.W. Deng, unpublished data) included the PISTILLATA (Goto and Meyerowitz, 1994; Honma and Goto, 2000), APETALA1 (Honma and Goto, 2001; Ng and Yanofsky, 2001), and LATERAL ORGAN BOUNDARIES genes (Shuai et al., 2002). The data from the microarray analysis all exhibited organ or tissue expression patterns that are consistent with prior studies (L.G. Ma, N. Sun, X.G. Liu, Y.L. Jiao, H.Y. Zhao, and X.W. Deng, unpublished data). In this report, we extend this comparative analysis to 14 known MADS box TF genes that have expression data derived from in situ or northern-blot analysis. We found that all 14 TF genes exhibited largely similar expression patterns between our microarray analysis and previous reported nonmicroarray data (Table IV). The detailed expression patterns from six of those MADS box genes are illustrated in Figure 5. For example, AGL1 is specifically expressed in particular regions of the gynoecium and ovule (Fig. 5A). AGL2 transcript is very abundant in the primordia of all four floral organs: sepals, petals, stamens, and carpels. The AGL2 transcript remains abundant in each organ during morphological differentiation but diminishes as each organ undergoes the final maturation phase of development (Fig. 5B). AGL3 is expressed in all aerial organs but roots (Fig. 5C). AGL8 RNA does not accumulate during vegetative growth; it accumulates to high levels in the inflorescence apical meristem as well as in the inflorescence stem and cauline leaves (Fig. 5D). AGL15 is preferentially expressed in embryos and accumulates significantly in germinating seedlings (Fig. 5E). AGL21 is highly expressed in roots and in developing embryos (Fig. 5F). The only possibly minor exception is the FLC gene, which exhibited a small difference in relative expression level in inflorescence organs from our microarray analysis compared to prior studies. In a prior northern analysis, it was shown that FLC expression was not detectable in inflorescence organs (Michaels and Amasino, 1999), while our microarray result indicated that FLC expressed at levels slightly below the median expression level in floral organs. This minor discrepancy could be due to the not exactly same organ types used or cross-hybridization in the oligo microarray. Overall, the results suggest that whole genome oligo microarray is a valid approach to determine specific gene expression patterns. As knowledge of expression patterns of a TF gene often offers the initial clues needed in dissecting its biological function, these results should provide useful information for further functional studies.
Table IV.
Comparison of our microarray results with prior studies for representative MADS box genes
| Locus ID | Gene name | Reference | Comment |
|---|---|---|---|
| At3g58780 | AGL1 | Flanagan et al. (1996) | |
| Ma et al. (1991) | Microarray data are consistent with reported results | ||
| Kofuji et al. (2003) | |||
| At5g15800 | AGL2 SEP1 | Flanagan and Ma (1994) | Microarray data are consistent with reported results |
| Ma et al. (1991) | |||
| Kofuji et al. (2003) | |||
| At2g03710 | AGL3 | Huang et al. (1995) | Microarray data are consistent with reported results |
| Ma et al. (1991) | |||
| Kofuji et al. (2003) | |||
| At5g60910 | AGL8 FUL | Mandel and Yanofsky (1995) | Microarray data are consistent with reported results |
| Kofuji et al. (2003) | |||
| At5g13790 | AGL15 | Rounsley et al. (1995) | Microarray data are consistent with reported results |
| Kofuji et al. (2003) | |||
| At4g37940 | AGL21 | Burgeff et al. (2002) | Microarray data are consistent with reported results |
| Kofuji et al. (2003) | |||
| At1g24260 | AGL9 SEP3 | Kofuji et al. (2003) | Microarray data are consistent with reported results |
| At2g03060 | AGL30 | Kofuji et al. (2003) | Microarray data are consistent with reported results |
| At3g57230 | AGL16 | Kofuji et al. (2003) | Microarray data are consistent with reported results |
| At5g20240 | PI | Goto and Meyerowitz (1994) | Microarray data are consistent with reported results |
| Kofuji et al. (2003) | |||
| At5g23260 | AGL32 | Kofuji et al. (2003) | Microarray data are consistent with reported results |
| At1g69120 | AGL7/AP1 | Mandel et al. (1992) | Microarray data are consistent with reported results |
| Kofuji et al. (2003) | |||
| At3g54340 | AP3 | Jack et al. (1992) | Microarray data are consistent with reported results |
| Kofuji et al. (2003) | |||
| At5g10140 | AGL25 FLC | Michaels et al. (1999) | Possible minor discrepancy in inflorescence or floral organs |
| Kofuji et al. (2003) |
Figure 5.
The relative expression levels of the 6 well-characterized MADS box TF genes in 17 organ types as determined by our microarray analysis. A, AGL1 (At3g58780); B, AGL2 (At5g15800); C, AGL3 (At2g03710); D, AGL8 (At5g60910); E, AGL15 (At5g13790); and F, AGL21 (At4g37940). The observed expression patterns from our microarray studies are consistent with the previously reported results (see Table IV).
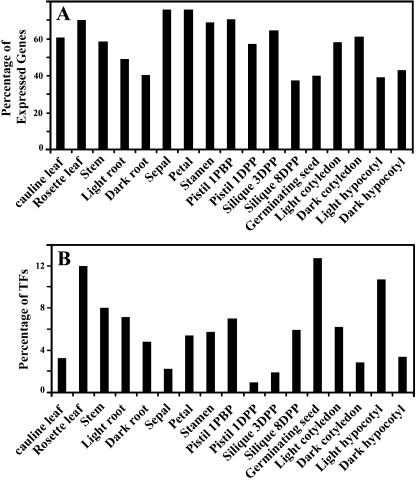
We also examined the number (and percentage) of the 858 TF genes whose expression can be detected experimentally in each organ type and in any of the organs examined. This analysis revealed that the expression for 831 (97%) out of the 858 genes can be detected in at least one of the 17 organs or cultured cells examined. This result confirms that the vast majority of known and predicted TFs are expressed during Arabidopsis development; while the percentage of TF genes expressed in each organ types varied from 37.4% (silique 8 d-post-pollination) to 75.8% (petal; see Fig. 6A). We also calculated the numbers of genes exhibiting highest relative expression levels in each organ types. As shown in Figure 6B, the percentage of the highest expressed TF genes in each organ type varies from organ to organ. Vegetative organs have large numbers of TF genes that have the highest expression level. This may be consistent with the fact that vegetative organs are where most of metabolism activities reside. About 20% TF genes exhibit highest expression levels in flower organs, which may hint at their special roles in flower development and reproduction. The germinating seed has the highest number of TFs, with highest expression among all organs examined here. It is interesting to note that the germinating seed has a very low percentage of the total TF genes with detectable expression (Fig. 6A). This result indicated that during seed germination, a relatively large fraction of genes turned on highly are TFs. This is consistent with the fact that those early expressed genes will initiate the developmental and metabolic processes to follow.
Figure 6.
Summary of TF gene expression characteristics among Arabidopsis organ types. A, The percentage of TF genes expressed in different organs. Abbreviations: DPP, day post pollination; DBP, day before pollination. A total 858 TF genes (see text) are analyzed, and 61% (cauline leaf), 70% (rosette leaf), 58% (stem), 49% (light root), 41% (dark root), 76% (sepal), 76% (petal), 79% (stamem), 71% (pistil 1DBP), 57% (pistil 1DPP), 65% (silique 3DPP), 37% (silique 8DPP), 40% (germinating seed), 58% (light cotyledon), 61% (dark cotyledon), 39% (light hypocotyl), 43% (dark hypocotyl) were detected expression respectively. B, Distribution of TF genes in their highest expression level among 17 different issues. Among the 858 TF genes analyzed, 3% (cauline leaf), 12% (rosette leaf), 8% (stem), 7% (light root), 5% (dark root), 2% (sepal), 5% (petal), 6% (stamem), 7% (pistil 1DBP), 1% (pistil 1DPP), 2% (silique 3DPP), 6% (silique 8DPP), 13% (germinating seed), 6% (light cotyledon), 3% (dark cotyledon), 11% (light hypocotyl), and 3% (dark hypocotyl) are expressed at their highest level in the indicated organ types, respectively.
ALTERNATIVE SPLICING OF FIVE TF GENES
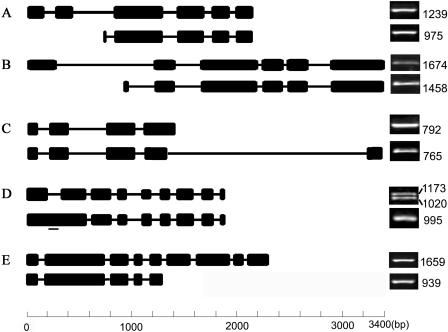
For the 34 TF genes where our ORFeome sequences differ from prior cDNA sequence or predicted gene annotation (Tables II and III), we examined whether alternative splicing contributes to the observed differences. Indeed, we were able to confirm that alternative splicing variants were present for five TF genes (At1g26260, At1g72050, At3g46590, At4g26640, and At4g29930) by RT-PCR (Fig. 7) and were responsible for the observed differences in their ORFeome clone sequences. Different mRNA forms of At1g72050 and At4g26640 appear to be generated by using extra or different exons located at the very 5′ end, while At4g29930 and At3g46590 include alternative exons at the 3′ of the mature RNAs. In At1g26260, one form of the mRNAs simply contains the first intron that is spliced out in the other form of transcript. In the case of At1g72050, the previously reported mature transcript contains 5 exons with a 975 bp ORF (GenBank accession nos. AY054225 and AY066042), and its encoded protein possesses one C2H2-type Zinc-finger domain (PSSIM-Id: 20248) plus a partial domain (PSSIM-Id: 21389) that is incomplete in both ends. By using two extra exons at the 5′ end of the transcript, the new ORF predicted from our cloned mRNA species is 1,239 bp in length and has two C2H2-type Zinc-finger domains, a scenario resembling a previously reported alternative splicing event for the rice Myb7 gene (Magaraggia et al., 1997). In that case, one of the two transcripts that encodes a partial DNA-binding domain was known to act as a repressor to switch off the expression of its target genes, while the other served as a transcription activator (Magaraggia et al., 1997). However, in the other four genes, alternative splicing variants do not affect the DNA-binding domains.
Figure 7.
Alternative splicing of five Arabidopsis TF genes. A diagram of the exon/intron structures of the alternative spliced transcripts of At1g72050 (A), At4g26640 (B), At4g29930 (C), At1g26260 (D), and At3g46590 (E) is shown on the left. Sequence-specific primers were synthesized for each form of the mature RNA as outlined on the left side. For each gene, our version of gene structure and PCR product is placed on top, while the gene structure and PCR products based on prior information is placed on bottom. The levels of the two mature RNA molecules from Arabidopsis plant samples were analyzed by RT-PCR, and only one representative result is shown as ethidium bromide-stained DNA band, with the length of full-length ORF marked to the right. At the bottom is a scale that indicates the lengths of introns (represented by thin lines) and exons (thick black boxes) in all the genes compared. The small bar to the lower transcript shown in D designates the position of the forward primer used to amplify the RAFL reported version of cDNA since the 5′ end of these two mature RNA molecules were identical and hence only the 995 bp band represented a partial ORF. However, the PCR primer pair for the 1,173 bp cDNA (based on the full-length ORF of RAFL cDNA) can amplify both the 1,020 bp cDNA band (our new version) and the 1,173 bp cDNA, so that doublet bands were observed.
In all five cases, the typical GT-AG binucleotide splicing junctions were observed in the alternatively spliced transcripts. To test if alternative splicing of these genes is developmentally regulated, we designed specific PCR primer pairs for the two alternative spliced transcripts of each of those five genes and used semiquantitative RT-PCR to examine the presence and abundance of those alternative mRNAs in selected Arabidopsis tissue samples (Fig. 7). The alternative spliced transcripts for each of those five genes were present at similar abundance in all tissue types tested (data not shown), suggesting that alternative splicing of these genes is constitutive and is not regulated by developmental or environmental conditions tested.
MATERIALS AND METHODS
Plant Materials
Arabidopsis (ecotype Columbia) plants were grown in fully automated growth chambers (Conviron, Canada) under 16 h light illumination each 24 h period. Plants were maintained at 23°C during the light period and 21°C during the dark period.
To provide additional RNA samples to cover those TF genes that may not be expressed under normal growth conditions, Arabidopsis plants at 6 to 8 rosette stages were subjected to the following 8 specific treatments and were used for total RNA isolation: (1) NaCl treatment, whole pots were submerged in 300 mm NaCl for 8 h. (2) Heat shock (heat), plants were preconditioned at 37°C for 2 h before being transferred to 45°C for another 2 h. (3) UV treatment, plants were radiated with UV light (100 J m−2) for 6 h. (4) Water depletion treatment, entire plants were uprooted, placed on filter papers, and allowed to dry for 6 h. (5) Ethylene treatment, plants were placed in a closed jar containing 100 ppm C2H4 for 24 h. (6) Cold treatment, plants were placed in a 4°C cold room for 8 h. (7) Wound treatment, rosette leaves were cut into approximately 5 mm strips and were left in the growth chamber for 8 h before being used for RNA isolation. (8) Dark adaptation, plants were placed in darkness for 48 h before being harvested for RNA isolation.
Identification of Arabidopsis Transcription Factor Genes
All known and predicted TF genes were selected from the MIPS Arabidopsis genome database (http://www.mips.biochem.mpg.de/proj/thal/db/index.html) as of December 21, 2000. Each gene was identified by its chromosome locus ID (e.g. At5g61270). The MIPS Arabidopsis database August 17, 2003 update was used as our final reference for gene annotation.
Isolation and Cloning of ORFeome into pENTR and Expression Vectors
Total RNA was isolated from pooled Arabidopsis plant samples harvested from the 6 to 8 rosette leaves before bolting and plants a week after flowering using the RNeasy plant mini kit (QIAGEN, Germany) and was quantified at 260 nm with a spectrophotometer. This RNA sample was used as a generic initial template for RT-PCR. For any TF genes that were not able to be cloned from those generic RNA samples, RNA samples from specific treated Arabidopsis seedlings (see “Plant Materials” section) were used as an alternative template for RT-PCR amplification.
Three μg of total RNA sample was reverse transcribed using SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA) in a total volume of 20 μL. Primers for ubiquitin amplification (forward: 5′-GGTGCTAAGAAGAGGAAGAAT-3′ and reverse: 5′-CTCCTTCTTTCTGGTAAACGT-3′) were added as the internal control together with gene-specific primers. PCR was performed using Pfu polymerases (Sangon, China). For tissue-specific expression analyses, different plant materials harvested at indicated stages were used. PCR products were purified with a gel extraction kit (CLONTECH Laboratories, Palo Alto, CA), cloned into pENTR/D/TOPO vector (Invitrogen), and verified by sequencing using M13 primers. Primers for different TF genes were designed using information obtained from the Arabidopsis genome. The forward primer contained the sequence 5′-CACCACAAA-3′ at the 5′end. The CACC base paired with the overhang sequence, GTGG, in pENTR TOPO vector (Fig. 1A). The yeast expression vector, pYTV, was a modified version of the pDEST 52 (Invitrogen). The original tag was removed and was replaced by 3XFLAG, 6Xhis, and a 3C cleavage and 2XIgG binding protein added as C-terminal tags to facilitate purification of the fusion protein (Fig. 1B). To clone the gene of interest in frame with C-terminal tags present in the pYTV, the reverse primer was designed in such a way that the stop codon in the target gene was deleted in the final PCR product for ORF amplification for initial cloning into pENTR TOPO vector (Fig. 1C).
Protein-Blot Analysis of TF Proteins in Yeast
Total proteins extracted from 50 to 75 μL saturated yeast cells expressing target genes were fractionated by SDS-PAGE. Each gel was probed with 1 ug/mL monoclonal antipolyhistidine antibody (R&D Systems, Minneapolis) and visualized after incubating with goat anti-mouse AP-conjugated secondary antibody (Promega, Madison, WI).
Expression Profile Analysis using Microarray
Gene-specific 70-mer oligos were designed based on Arabidopsis genome annotation data available on February 20, 2002 by Qiagen (http://omad.qiagen.com/download/genelist/arabidopsis_V1_384.prn), and the microarray slide was printed at Yale University as described (L.G. Ma, N. Sun, X.G. Liu, Y.L. Jiao, H.Y. Zhao, and X.W. Deng, unpublished data). The signal was scanned at 532 nm (Cy3) and 635 nm (Cy5) wavelengths with an Axon GenePix 4000B scanner (Axon, Foster City, CA) at 5-nm resolution and quantified with Axon GenePix Pro 3.0 image analysis. The intensity of different organs was normalized by equalizing the median value of all gene intensities from each organ. The normalized intensity value for each gene was considered its relative expression level (L.G. Ma, N. Sun, X.G. Liu, Y.L. Jiao, H.Y. Zhao, and X.W. Deng, unpublished data).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers listed in Tables II and III.
Supplementary Material
Acknowledgments
We thank Dr. Lei Li for commenting on this manuscript.
This work was supported by a grant from the Chinese National Natural Science Foundation (grant no. 30221120261).
The online version of this article contains Web-only data.
References
- The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Burgeff C, Liljegren SJ, Tapia-López R, Yanofsky MF, Alvarez-Buylla ER (2002) MADS-box gene expression in lateral primordia, meristems and differentiated tissues of Arabidopsis thaliana roots. Planta 214: 365–372 [DOI] [PubMed] [Google Scholar]
- Chen W, Provart NJ, Glazebrook J, Katagiri F, Chang H-S, Eulgem T, Mauch F, Luan S, Zou G, Whitham SA, et al (2002) Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14: 559–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova DK, Stevaux O, Frolov MV, Dyson NJ (2003) Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev 17: 2308–2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan CA, Hu Y, Ma H (1996) Specific expression of the AGL1 MADS-box gene suggests regulatory functions in Arabidopsis gynoecium and ovule development. Plant J 10: 343–353 [DOI] [PubMed] [Google Scholar]
- Flanagan CA, Ma H (1994) Spatially and temporally regulated expression of the MADS-box gene AGL2 in wild-type and mutant Arabidopsis flowers. Plant Mol Biol 26: 581–595 [DOI] [PubMed] [Google Scholar]
- Goto K, Meyerowitz EM (1994) Function and regulation of the Arabidopsis f homeotic gene PISTILLATA. Genes Dev 8: 1548–1560 [DOI] [PubMed] [Google Scholar]
- Grandori C, Cowley SM, James LP, Eisenman RN (2000) The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol 16: 653–699 [DOI] [PubMed] [Google Scholar]
- Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC (2003) The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol 20: 735–747 [DOI] [PubMed] [Google Scholar]
- Honma T, Goto K (2000) The Arabidopsis floral homeotic gene PISTILLATA is regulated by discrete cis-elements responsive to induction and maintenance signals. Development 127: 2021–2030 [DOI] [PubMed] [Google Scholar]
- Honma T, Goto K (2001) Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409: 525–528 [DOI] [PubMed] [Google Scholar]
- Hosoda K, Imamura A, Katoh E, Hatta T, Tachiki M, Yamada H, Mizuno T, Yamazaki T (2002) Molecular structure of the GARP family of plant Myb-related DNA binding motifs of the Arabidopsis response regulators. Plant Cell 14: 2015–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Tudor M, Weiss CA, Hu Y, Ma H (1995) The Arabidopsis MADS-box gene AGL3 is widely expressed and encodes a sequence-specific DNA-binding protein. Plant Mol Biol 28: 549–567 [DOI] [PubMed] [Google Scholar]
- Jack T, Brockman LL, Meyerowitz EM (1992) The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68: 683–697 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Yang H, Ma L, Sun N, Yu H, Liu T, Gao Y, Gu H, Chen Z, Wada M, et al (2003) A genome-wide analysis of blue-light regulation of Arabidopsis transcription factor gene expression during seedling development. Plant Physiol 133: 1480–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofuji R, Sumikawa N, Yamasaki M, Kondo K, Ueda K, Ito M, Hasebe M (2003) Evolution and divergence of the MADS-box gene family based on genome-wide expression analysis. Mol Biol Evol 20: 1963–1977 [DOI] [PubMed] [Google Scholar]
- Kohler C, Hennig L, Spillane C, Pien S, Gruissem W, Grossniklaus U (2003) The polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev 17: 1540–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Yanofsky MF, Meyerowitz EM (1991) AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev 5: 484–495 [DOI] [PubMed] [Google Scholar]
- Ma LG, Zhao HY, Deng XW (2003) Analysis of the mutational effects of the COP/DET/FUS loci on genome expression profiles reveals their overlapping yet not identical roles in regulating Arabidopsis seedling development. Development 130: 969–981 [DOI] [PubMed] [Google Scholar]
- Magaraggia F, Solinas G, Valle G, Giovinazzo G, Coraggio I (1997) Maturation and translation mechanisms involved in the expression of a myb gene of rice. Plant Mol Biol 35: 1003–1008 [DOI] [PubMed] [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF (1992) Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360: 273–277 [DOI] [PubMed] [Google Scholar]
- Mandel MA, Yanofsky MF (1995) The Arabidopsis AGL18 MADS box gene is expressed in inflorescence meristems and is negatively regulated by APETALA1. Plant Cell 7: 1763–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M, Yanofsky MF (2001) Activation of the Arabidopsis B class homeotic genes by APETALA1. Plant Cell 13: 739–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenicova L, de Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Cook HE, Ingram RM, Kater MM, Davies B, et al (2003) Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15: 1538–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL (2002) Transcriptional regulation: a genomic overview. In CR Somerville, EM Meyerwitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/10.1199/tab.0085, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang CZ, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110 [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Ratcliffe OJ (2000) A genomic perspective on plant transcription factors. Curr Opin Plant Biol 3: 423–434 [DOI] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 17: 1030–1035 [DOI] [PubMed] [Google Scholar]
- Rounsley SD, Ditta GS, Yanofsky MF (1995) Diverse roles for MADS box genes in Arabidopsis development. Plant Cell 7: 1259–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Kamiya A, Ishida J, Satou M, Sakurai T, Nakajima M, Enju A, Akiyama K, Oono Y, et al (2002) Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141–147 [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K (2000) Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol 3: 217–223 [PubMed] [Google Scholar]
- Shuai B, Reynaga CG, Springer PS (2002) The LATERAL ORGAN BOUNDARIES gene defines a novel, plant-specific gene family. Plant Physiol 129: 747–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Hug E, Quail PH (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, et al (2003) Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302: 842–846 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.