Abstract
A significant fraction (approximately 17%) of Arabidopsis genes are members of tandemly repeated families and pose a particular challenge for functional studies. We have used the Ac-Ds transposition system to generate single- and double-knockout mutants of two tandemly duplicated cytochrome P450 genes, SPS/BUS/CYP79F1 and CYP79F2. We have previously described the Arabidopsis supershoot mutants in CYP79F1 that exhibit massive overproliferation of shoots. Here we use a cytokinin-responsive reporter ARR5::uidA and an auxin-responsive reporter DR5::uidA in the sps/cyp79F1 mutant to show that increased levels of cytokinin, but not auxin, correlate well with the expression pattern of the SPS/CYP79F1 gene, supporting the involvement of this gene in cytokinin homeostasis. Further, we isolated Ds gene trap insertions in the CYP79F2 gene, and find these mutants to be defective mainly in the root system, consistent with a root-specific expression pattern. Finally, we generated double mutants in CYP79F1 and CYP79F2 using secondary transpositions, and demonstrate that the phenotypes are additive. Previous biochemical studies have suggested partially redundant functions for SPS/CYP79F1 and CYP79F2 in aliphatic glucosinolate synthesis. Our analysis shows that aliphatic glucosinolate biosynthesis is completely abolished in the double-knockout plants, providing genetic proof for the proposed biochemical functions of these genes. This study also provides further demonstration of how gluconisolate biosynthesis, regarded as secondary metabolism, is intricately linked with hormone homeostatis and hence with plant growth and development.
Plant growth and development requires coordination of networks of biological processes within the plant, as well as with responses to external environments. The control of shoot branching by auxin and cytokinin is a well-known example of hormone interactions in controlling plant development. Cytokinin plays a key role in promoting bud growth, whereas auxin has an inhibitory effect. Therefore, the outcome appears to depend on the ratio of the two hormones (for review, see Tamas, 1995; Li and Bangerth, 1992). The two plant hormones not only play opposite roles in controlling plant growth and development, but also influence each other hormone homeostasis (Binns et al., 1987; Palni et al., 1988; Bangerth, 1994; Zhang et al., 1995; Makarova et al., 1996). We and others have described Arabidopsis mutants called supershoot (sps) or bushy (bus), which are disrupted in the gene encoding the cytochrome P450 CYP79F1 (Reintanz et al., 2001; Tantikanjana et al., 2001). These mutants exhibit massive proliferation of shoots, together with other developmental defects. The quantification of hormone levels in sps/cyp79F1 mutant plants indicates that both auxin and cytokinins are higher in the mutants, but the phenotypes of the sps/cyp79F1 plants are consistent with higher levels of cytokinins rather than auxins as the key factor responsible for the change in branching pattern (Tantikanjana et al., 2001).
Despite the fact that sps/cyp79F1 plants resemble hormone mutants, biochemical studies have shown that SPS/CYP79F1 gene encodes an enzyme catalyzing metabolism of both short-chain and long-chain elongated Met-derivatives in the biosynthesis of aliphatic glucosinolates (Hansen et al., 2001; Chen et al., 2003). Glucosinolates are a group of secondary plant metabolites known to play a role in plant defense and are sources of flavor compounds, cancer-preventing agents, and bioherbicides. It has recently been shown that blockage of plant secondary metabolic pathways can result in severe repercussion for hormone homeostasis. The high levels of auxin in the superroot1 (sur1) and superroot2 (sur2) are caused by indirect effects due to blockage of the C-S lyase (Mikkelsen et al., 2004) and the cytochrome P450 CYP83B1, respectively (Barlier et al., 2000; Bak et al., 2001), in the biosynthetic pathway of the natural plant products glucosinolates. The high-auxin phenotypes of the sur1 and sur2 are derived from accumulation of indole-3-acetaldoxime that is channelled into indole-3-acetic acid (IAA), which shows that indole-3-acetaldoxime plays a critical role in auxin homeostasis and functions as a key branching point between primary and secondary metabolism.
In order to elucidate the potential roles of cytokinin and auxin in controlling branching pattern in the sps/cyp79F1 mutant plants, we have used cytokinin and auxin-responsive reporters to study the changes of hormone levels with greater spatial resolution. For comparison, we have investigated the effect of disruption of the CYP79F2 gene, which is a tandem duplication of SPS/CYP79F1, by the isolation and characterization of cyp79F2 mutants with transposon insertions. Furthermore, we have generated double mutants of sps/cyp79F1 and cyp79F2 to analyze redundant functions of these two closely linked genes using an approach based on multiple transpositions of a Ds element. This method could be utilized to generate double-knockout lines of other tandemly repeated genes in the genome. Characterization of the phenotypic changes and glucosinolate profiles in these double-knockout mutants are presented.
RESULTS
Expression Patterns of the Cytokinin and Auxin-Responsive Reporters in the sps/cyp79F1 Mutants
As previously reported, several physiological changes in the aerial part of the sps/cyp79F1 mutant plants are strikingly similar to the effects of increased cytokinins levels. These physiological changes include the release of lateral buds from apical dominance and an increase in bud initiation, as well as the delay of senescence. Quantification of the cytokinin levels in the sps/cyp79F1 mutants yielded results consistent with this prediction, as several types of cytokinins in the sps/cyp79F1 mutants are present at higher levels than in wild-type plants (Tantikanjana et al., 2001). These mutants also have elevated auxin levels (Reintanz et al., 2001; Tantikanjana et al., 2001), however, the proliferation of shoots observed in the sps/cyp79F1 mutant is inconsistent with increased auxin levels, which is expected to produce the opposite effect. It has been shown that when cytokinin homeostasis is perturbed, the level of auxin can be affected (Binns et al., 1987; Makarova et al., 1996). Therefore we suggested that the phenotypic changes in sps/cyp79F1 mutants result primarily from increased cytokinin levels, and that the changes in auxin levels are a consequence of feedback mechanisms involved in hormone interactions (Tantikanjana et al., 2001).
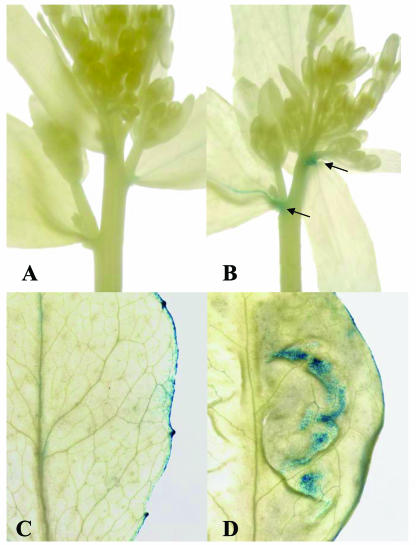
To further investigate the relationship between hormone levels and the sps/cyp79F1 mutant, we have used a cytokinin-responsive reporter ARR5::uidA (D'Agostino et al., 2000) and a synthetic auxin-responsive reporter DR5::uidA (Ulmasov et al., 1997) to study developmental changes affected by cytokinin and auxin levels. These reporters are useful to investigate changes in hormone levels within defined regions of wild-type and mutant plants (e.g. Casson et al., 2002; Harrar et al., 2003; Werner et al., 2003 for the cytokinin reporter ARR5; and Blilou et al., 2002; Long et al., 2002; Willemsen et al., 2003 for the auxin reporter DR5), because the hormone measurements on entire aerial parts of the plants may not reflect hormone levels at specific developmental sites, which may be masked by the changes elsewhere in the plant. We selected the sps1-1 allele for the investigation, as this allele does not exhibit β-glucuronidase (GUS) expression derived from the Ds gene trap insertion. The analysis showed that expression levels of both cytokinin and auxin-responsive reporters in the sps/cyp79f1 mutants are indeed higher than those of wild-type plants, in agreement with the hormone measurements. Importantly, the reporters revealed that the increases in the two hormone levels occur at different sites in the plant. The level of the cytokinin-responsive reporter is increased the most at the axil of the mutant leaf (Fig. 1B), as compared to that of the wild type (Fig. 1A). On the other hand, we could not detect any expression of the auxin-responsive reporter at the leaf axil in neither the mutant nor wild-type plants. Instead, expression of the auxin-responsive reporter was found at higher level in the leaf blade of the mutant (Fig. 1D) than in the wild type (Fig. 1C). These data further support the possible involvement of the SPS/CYP79F1 gene in modulating cytokinin level at the site of bud initiation and that changes in levels of cytokinin at these sites, rather than auxin, are responsible for the altered shoot branching pattern.
Figure 1.
Expression patterns of the cytokinin-responsive reporter ARR5::uidA and the synthetic auxin-responsive reporter DR5::uidA in sps/cyp79F1 mutants. A, Expression patterns of the cytokinin-responsive reporter ARR5::uidA in young inflorescences of wild type and B, sps/cyp79F1 mutant plant. Arrows indicate expression of the GUS reporter at the sites of bud initiation in the sps/cyp79F1 mutant. C, Expression patterns of the synthetic auxin-responsive reporter DR5::uidA in the wild type leaf, and D, in the sps/cyp79F1 mutant leaf.
Distinct Roles of SPS/CYP79F1 and CYP79F2 in Plant Growth and Development
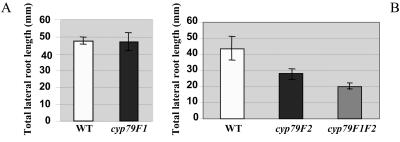
We have shown that disruption of the SPS/CYP79F1 gene leads to severe developmental defects in the aerial architecture of the plants. The expression pattern of the SPS/CYP79F1 gene and analysis of mosaic plants has prompted the suggestion that this gene acts locally in its effects on plant growth and development (Tantikanjana et al., 2001). Immediately upstream of the SPS/CYP79F1 gene, there is a second closely related gene that has been designated CYP79F2 (Nelson, 1999; http://biobase.dk/P450), which has also been shown to function in aliphatic glucosinolate metabolism (Chen et al., 2003). The two genes share 89% sequence identity at both nucleotide and amino acid levels. In order to reveal possible physiological functions of the CYP79F2 gene, a collection of Ds insertion lines were used to screen for cyp79F2 knockout mutants by reverse-genetics approach. A total of three cyp79F2 alleles were isolated. Locations of the Ds elements in different cyp79F2 alleles are shown in Figure 2. In all three cyp79F2 alleles, the mutant plants have slight reduction of aerial growth but otherwise normal aerial structure. Unlike the sps/cyp79F1 mutant that has severe defects in the aerial parts of the plant, cyp79F2 mutants are predominantly defective in the root system. When lateral root length of 8-d-old seedlings were measured, cyp79F2 mutants exhibit only 65% root growth compared to the wild-type starter line that was used to generate cyp79F2 mutants (see “Materials and Methods”; Fig. 3), whereas the number of lateral roots in the mutant is unaffected (data not shown). There were no obvious differences in cell size between cyp79F2 and wild-type (starter line) plants, suggesting that the primary effect of the mutant might be on cell division. In contrast, the lateral root length, as well as the lateral root number, of the sps/cyp79F1 mutant does not differ from the wild-type starter line (Fig. 3). The observation of root defects only in cyp79F2 mutants, but not in cyp79F1 mutants, both of which were derived from the same starter line, strongly suggests that only the CYP79F2 gene has a role in root growth. The reduction in lateral root length of the cyp79F2 mutants is observed only when seedlings are grown in soil. This defect is not obvious in mutant seedlings growing on agar plates containing Murashige and Skoog basal medium, suggesting that expression of the phenotypic changes in cyp79F2 mutants is influenced by growth conditions. We could not detect any obvious changes in the primary root length under the conditions where we observed an effect on lateral root lengths, but we note that it is more difficult to obtain intact main roots as they grow deep into the soil. We have found that the severity of the sps/cyp79F1 mutant phenotypes is also affected by growth conditions such as growing medium and light condition (data not shown).
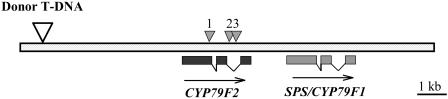
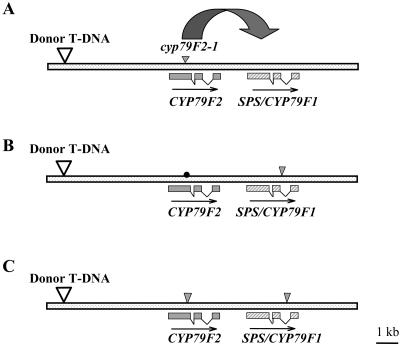
Figure 2.
Positions of the donor T-DNA and insertion sites of Ds gene trap elements in different cyp79F2 alleles. Positions of the SPS/CYP79F1 and CYP79F2 genes on the bacterial artificial chromosome clone accession number AC006341 are shown. Boxes represent exons. Insertion sites of Ds elements in the CYP79F2 gene are indicated as small arrowheads; position of the donor T-DNA is indicated as big arrowhead. Insertion site of the cyp79F2-1 is in the first exon, whereas the cyp79F2-2 and cyp79F2-3 are in the second intron.
Figure 3.
Lateral root length of sps/cyp79F1, cyp79F2 mutants and sps/cyp79F1 cyp79F2 double mutant. A, Lateral root length of sps/cyp79F1 and wild-type plants. B, Lateral root length of cyp79F2, sps/cyp79F1 cyp79F2 double-mutant and wild-type plants. The studies of lateral root development between sps/cyp79F1 and wild-type plants (A), and cyp79F2, sps/cyp79F1 cyp79F2 double-mutant and wild-type plants (B) were done in separate experiments but in the same controlled growth chamber conditions. Error bars indicate sd (n = 21–24).
To further understand its role in controlling root development, we analyzed expression pattern of the CYP79F2 gene using the Ds gene trap insertions. The Ds gene trap insertions in CYP79F2 should provide an accurate expression profile of the gene, because the GUS reporter fusion in the gene trap is in the correct chromosomal context (Sundaresan et al., 1995). Two of the cyp79F2 alleles confer GUS reporter gene expression. Both cyp79F2 alleles show GUS expression pattern primarily in the root system. Occasionally, GUS staining was detected in the vascular tissue of the cotyledons. However, no expression was observed in the leaf or other parts of the aerial structure. In the root system, CYP79F2 gene is developmentally regulated. CYP79F2 is expressed above the root elongation zone and continues for a few millimeters before fading away in the more mature region (Fig. 4). We could not detect any expression in the root primodium. The data suggest that the CYP79F2 gene also affects plant growth and development locally, as was observed with the SPS/CYP79F1 gene. The nonoverlapping expression patterns, together with the observed phenotypic changes, indicate that SPS/CYP79F1 and CYP79F2, in addition to being involved in biosynthesis of aliphatic glucosinolates, might function in different parts of the plant with the former primarily in the aerial parts and the latter primarily in the root system.
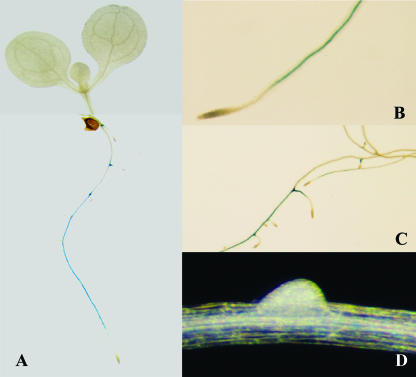
Figure 4.
CYP79F2 expression patterns monitored by GUS expression from the Ds gene trap insertion in the cyp79F2-2 allele. A, GUS-staining pattern detected in the root system of a 10-d-old seedling. The picture is the composite image of the aerial part and root system from the same seedling. B to D, Staining detected in the root of a 2-week-old seedling; B, At the region above root apical meristem; C, Staining is not detectable in more mature regions of the root. D, No GUS staining is detected in the root primodium.
Generation of sps/cyp79F1 and cyp79F2 Double Mutants by Multiple Ds Transpositions
Despite high sequence similarity between SPS/CYP79F1 and CYP79F2 genes, the analysis of single-knockout mutants and the expression patterns of the genes suggest that the two genes might play different roles, via differences in their spatial regulation. Recently SPS/CYP79F1 and CYP79F2 genes have been shown to have distinct but overlapping metabolic functions. SPS/CYP79F1 metabolizes both short-chain and long-chain aliphatic Mets, whereas CYP79F2 exclusively metabolizes the long-chain elongated aliphatic Mets (Hansen et al., 2001; Chen et al., 2003). In order to reveal any possible overlapping physiological roles, as well as to confirm metabolic roles of the SPS/CYP79F1 and CYP79F2 genes, we generated sps/cyp79F1 and cyp79F2 double mutants by multiple Ds transpositions. Transposon mutagenesis using the Ac-Ds system has features that are advantageous for generating multiple mutations within closely linked physical distances. First, the Ac/Ds elements preferentially transpose to linked genomic sites (for review, see Sundaresan, 1996). Furthermore, the mutation can be retained at the original donor site after remobilization of the transposon, either by the element that is still present at the donor site or by the footprint left behind at the empty donor site after excision of the element, as the Ac/Ds element undergoes transposition either during or after chromosome replication (for review, see Fedoroff, 1983).
An experimental procedure was designed so that the screening for double-knockout mutants could be done with ease. Because cyp79F2 mutants have normal aerial structure and are fertile, cyp79F2-1 allele containing a Ds element inserted into the first exon of the gene was used for the reactivation of Ds transposition. Although a PCR screen for the double knockouts could be performed, we preferred a phenotypic screen, reasoning that if the Ds element excises from the CYP79F2 gene and reinserts into the SPS/CYP79F1, we would observe families segregating for plants with defects in the aerial structure in the subsequent generation. After reactivation of the Ds transposition, a total of 516 families were screened for plants displaying abnormal aerial architecture. Four independent families segregating for plants resembling the sps/cyp79F1 mutant were isolated. Of these four families, two families still segregating for mutant plants in the next generation were characterized in detail. Positions of the Ds elements in the SPS/CYP79F1 genes were confirmed by PCR using a SPS/CYP79F1-specific primer and a Ds primer. The PCR products were further verified by DNA sequencing. Both independent double-knockout mutants, designated sps-7 cyp79F2-1 and sps-8 cyp79F2-1, contained Ds elements inserted into the second intron of the SPS/CYP79F1 gene but at different positions (Fig. 5). The presence of Ds elements or footprints left by germinal excisions of Ds elements in the donor CYP79F2 gene was investigated by determination of the genomic sequences at the original donor sites. Of the two independent double-knockout mutants isolated, one contained an empty donor site with a footprint that generates a frame shift mutation in the CYP79F2 gene. The other double-knockout mutant retained the Ds element at the original location in the CYP79F2 gene in addition to the new insertion in the SPS/CYP79F1 gene. Positions of the mutations in the two independent double mutants are shown in Figure 5.
Figure 5.
Generation of sps/cyp79F1 cyp79F2 double mutants. A, Position of the Ds element in the cyp79F2-1 allele used as a starter line for the reactivation of Ds transposition. B to C, Positions of the Ds elements in the SPS/CYP79F1 and CYP79F2 genes of the two independent double-knockout mutants, designated sps-7 cyp79F2-1 allele (B) and sps-8 cyp79F2-1 allele (C). Boxes represent exons. Insertion sites of Ds elements in the SPS/CYP79F1 and CYP79F2 genes are indicated as small arrowheads; positions of the donor T-DNAs are indicated as big arrowheads. Dot represents position of the frame-shift mutation in the CYP79F2 gene after excision of the Ds element.
Phenotypes and Glucosinolate Profiles of the sps/cyp79F1 and cyp79F2 Double Mutants
The sps-7 cyp79F2-1 double-mutant plants were further analyzed for their physiological and metabolic effects. Results from expression patterns of the SPS/CYP79F1 and CYP79F2, as well as phenotypic characterization of single mutants, indicate that the two genes function mainly in different parts of the plant. sps/cyp79F1 and cyp79F2 double mutants are defective in both shoot and root systems. As expected from the phenotypic screen used to initially identify the double-knockout mutants, the aerial structure of the double mutants is similar to the previously described supershoot phenotype of sps/cyp79F1 single mutants in terms of shoot branching (Tantikanjana et al., 2001), but the plants are slightly smaller in size (data not shown). In addition, the lateral root length of the double mutants is also reduced relative to wild-type plants, to a level comparable to that in the single cyp79F2 mutant, though slightly shorter (Fig. 3B). Therefore the developmental effects of the double knockout are largely additive, as may be expected from their essentially nonoverlapping patterns of expression.
Biochemical studies have previously shown that SPS/CYP79F1 and CYP79F2 have overlapping substrate specificity in the synthesis of aliphatic glucosinolates. SPS/CYP79F1 catalyzes the conversion of homomethionine, di-, tri-, tetra-, penta-, and hexahomomethionine to their aldoximes, whereas CYP79F2 catalyzes the conversion of penta- and hexahomomethionine to their aldoximes (Chen et al., 2003). The biochemical data have been supported by the presence of long-chain aliphatic glucosinolates in both the single null sps/cyp79F1 mutant and the single null cyp79F2 mutant, while short-chain glucosinolates are abolished only in sps/cyp79F1 nulls. Double-knockout mutants obtained in our studies allowed us to test the overlapping metabolic roles of the SPS/CYP79F1 and CYP79F2 directly. Glucosinolate profiles of the wild type, sps/cyp79F1 and cyp79F2 single-knockout mutants, as well as sps/cyp79F1 and cyp79F2 double-knockout mutants, are shown in Table I. The analysis of glucosinolate profiles in the double-knockout mutant plants shows that, in addition to the absence of short-chain aliphatic glucosinolates, long-chain elongated glucosinolates are completely abolished when both genes are disrupted. This result provides genetic confirmation for the partial functional redundancy of these two genes in glucosinolate metabolism and demonstrates that disruption of both genes results in the complete absence of aliphatic glucosinolates in the plant. It has previously been shown that indole glucosinolates are also increased in the sps/cyp79F1 mutant (Reintanz et al., 2001; Chen et al., 2003). However, the double knockout in both SPS/CYP79F1 and CYP79F2 genes did not significantly enhance biosynthesis of indole glucosinolates as compared to the single knockout in SPS/CYP79F1 gene (Table I).
Table I.
Glucosinolate profile of cyp79F1 and cyp79F2 mutants and cyp79F1 cyp79F2 double mutant
| Short-Chain Glucosinolates
|
Long-Chain Glucosinolates
|
Indole Glucosinolates
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 3-msp | 5-msp | 3-mtp | 7-msh | 8-mso | 8-mto | i-3ym | 4mi-3ym | Nmi-3ym | |
| Rosette Leaves | |||||||||
| Wild type | 10.67 ± 0.92 | 0.24 ± 0.02 | 0.24 ± 0.02 | 0.05 ± 0.002 | 0.61 ± 0.04 | 0.06 ± 0.008 | 1.33 ± 0.06 | 0.25 ± 0.02 | 0.29 ± 0.06 |
| cyp79F1 | ND | ND | ND | 0.05 ± 0.006 | 1.66 ± 0.12 | 0.14 ± 0.009 | 2.14 ± 0.07 | 0.24 ± 0.01 | 0.013 ± 0.001 |
| cyp79F2 | 9.88 ± 0.85 | 0.22 ± 0.03 | 0.24 ± 0.02 | 0.05 ± 0.008 | 0.47 ± 0.05 | 0.05 ± 0.007 | 1.42 ± 0.13 | 0.25 ± 0.01 | 0.27 ± 0.04 |
| cyp79F1 cyp79F2 | ND | ND | ND | ND | ND | ND | 2.2 ± 0.08 | 0.24 ± 0.02 | 0.03 ± 0.02 |
| Root | |||||||||
| Wild type | 7.38 ± 0.69 | 0.24 ± 0.05 | 0.22 ± 0.01 | 0.02 ± 0.002 | 0.36 ± 0.02 | 0.12 ± 0.01 | 0.9 ± 0.06 | 0.24 ± 0.02 | 11.3 ± 0.87 |
| cyp79F1 | ND | ND | ND | 0.03 ± 0.004 | 0.58 ± 0.04 | 0.21 ± 0.02 | 0.73 ± 0.08 | 0.15 ± 0.01 | 7.97 ± 0.35 |
| cyp79F2 | 5.02 ± 0.37 | 0.32 ± 0.05 | 0.18 ± 0.02 | 0.01 ± 0.0002 | 0.17 ± 0.03 | 0.37 ± 0.02 | 0.79 ± 0.04 | 0.16 ± 0.007 | 13.3 ± 0.58 |
| cyp79F1 cyp79F2 | ND | ND | ND | ND | ND | ND | 0.44 ± 0.07 | 0.11 ± 0.03 | 6.05 ± 1.43 |
| Stem | |||||||||
| Wild type | 8.38 ± 0.22 | 0.1 ± 0.01 | 0.07 ± 0.02 | 0.04 ± 0.01 | 0.44 ± 0.10 | 0.005 ± 0.001 | 0.33 ± 0.01 | 0.06 ± 0.007 | 0.007 ± 0.001 |
| cyp79F1 | ND | ND | ND | 0.19 ± 0.01 | 5.58 ± 0.39 | 0.03 ± 0.004 | 2.15 ± 0.16 | 0.002 ± 0.0003 | 0.02 ± 0.005 |
| cyp79F2 | 14.29 ± 0.86 | 0.14 ± 0.03 | 0.13 ± 0.01 | 0.07 ± 0.01 | 0.44 ± 0.11 | 0.02 ± 0.001 | 0.47 ± 0.02 | 0.05 ± 0.01 | 0.01 ± 0.002 |
| cyp79F1 cyp79F2 | ND | ND | ND | ND | ND | ND | 2.12 ± 0.17 | 0.002 ± 0.0002 | 0.018 ± 0.002 |
| Flower | |||||||||
| Wild type | 33.02 ± 0.99 | 0.27 ± 0.03 | 0.93 ± 0.05 | 0.57 ± 0.03 | 7.76 ± 0.46 | 0.06 ± 0.007 | 1.32 ± 0.07 | 0.03 ± 0.005 | 0.009 ± 0.002 |
| cyp79F1 | ND | ND | ND | 0.41 ± 0.007 | 11.8 ± 0.31 | 0.18 ± 0.008 | 3.02 ± 0.08 | 0.04 ± 0.003 | 0.01 ± 0.002 |
| cyp79F2 | 28.36 ± 2.11 | 0.24 ± 0.02 | 1.17 ± 0.01 | 0.47 ± 0.008 | 5.47 ± 0.11 | 0.08 ± 0.01 | 1.72 ± 0.07 | 0.03 ± 0.001 | 0.02 ± 0.007 |
| cyp79F1 cyp79F2 | ND | ND | ND | ND | ND | ND | 2.77 ± 0.05 | 0.03 ± 0.001 | 0.01 ± 0.0006 |
3-msp, 3-methylsulfinylpropyl glucosinolate; 5-msp, 5-methylsulfinylpentyl glucosinolate; 3-mtp, 3-methylthiopropyl glucosinolate; 7-msh, 7-methylsulfinylheptyl glucosinolate; 8-mso, 8-methylsulfinyloctyl glucosinolate; 8-mto, 8-methylthiooctyl glucosinolate; i-3ym, indole-3-ylmethyl glucosinolate; 4mi-3ym, 4-methoxyidol-3-ylmethyl glucosinolate; Nmi-3ym, N-methoxyindol-3-ylmethyl glucosinolate.
ND, Not determined.
DISCUSSION
Recent biochemical and genetic studies have shown that disruption of glucosinolate biosynthesis, considered to be secondary metabolism, has important effects on hormone homeostasis in Arabidopsis. The loss-of-function mutations in the CYP83B1 gene (SUR2) and the C-S lyase (SUR1) in the glucosinolate pathway result in plants with elevated IAA levels. Consequently, the sur1 and sur2 mutants confer high-auxin phenotypes, including severe apical dominance and adventitious root development from hypocotyl tissue. In addition, disruption of CYP83B1 function also up-regulates Trp biosynthesis and other stress-induced pathways (Smolen and Bender, 2002). Likewise, characterization of the bushy/sps/cyp79F1 mutants has shown that disrupting the SPS/CYP79F1 gene required for the synthesis of short-chain and long-chain aliphatic glucosinolates severely affects hormone homeostasis (Hansen et al., 2001; Reintanz et al., 2001; Tantikanjana et al., 2001). Despite the fact that sps/cyp79F1 mutants have higher level of both auxin and cytokinin, the mutants resemble plants with cytokinin overproduction. The role of SPS/CYP79F1 as a modulator for cytokinin homeostasis is unclear, and the lack of detailed understanding of cytokinin metabolism makes it difficult to identify the biochemical link between glucosinolate biosynthetic pathway and cytokinin homeostasis.
In order to relate the changes of auxin and cytokinin levels with developmental defects observed in the sps/cyp79F1 mutants, we used the cytokinin-responsive reporter ARR5::uidA and the synthetic auxin-responsive reporter DR5::uidA to detect changes of hormone levels at particular sites. The study reveals that higher levels of cytokinin, particularly at the site of bud initiation correlates well with the increase in branching in the sps/cyp79F1 mutants, whereas the dramatic increase in auxin content is in the leaf blade. The pattern of increased cytokinin levels in the mutants revealed by the reporter gene correlated well with the expression pattern of the SPS/CYP79F1 gene. These data support the hypothesis that the primary effect of disrupting SPS/CYP79F1 function is on cytokinin homeostasis rather than auxin homeostasis. It is well documented that both auxin and cytokinin can influence the hormone levels of each other. Even though the exact mechanisms that regulate these hormone interactions are not fully understood, it has been shown that auxin can stimulate oxidative breakdown of active cytokinin (Palni et al., 1988; Zhang et al., 1995). In addition, it has been proposed that auxin levels may influence cytokinin biosynthesis, because removal of the endogenous source of auxin by decapitation leads to an increase in the cytokinin content of xylem exudates (Bangerth, 1994). Unlike the effect of auxin that results in decreased cytokinin levels, the effect of cytokinin on auxin levels is less well understood. Manipulation of cytokinin levels in plants by transformation with a bacterial cytokinin biosynthesis gene results in accumulation of increased levels of auxin (Binns et al., 1987; Makarova et al., 1996). Complex interactions are also observed between different glucosinolate biosynthetic pathways. Disruption of the SPS/CYP79F1 gene in the aliphatic glucosinolate biosynthesis leads to increased levels of indole glucosinolates (Reintanz et al., 2001; Chen et al., 2003). The connection between disruptions of different glucosinolate biosynthetic pathways that can influence the levels of different hormones opens a possibility that hormone interaction can occur, in part, through a complex network of secondary metabolites and/or common intermediates. A direct connection between aliphatic and indole glucosinolate biosynthetic pathways is not well understood. It is unlikely that the higher cytokinin levels and the consequent developmental effects in these mutants is due to the higher indole glucosinolate levels, as the double mutants, despite their more extreme phenotypes, show comparable levels of indole glucosinolates to the single CYP79F1/sps mutant (Table I). A more likely possibility arises from the observation that the CYP79B2/B3 genes are up-regulated in stressed plants, resulting in increased indole glucosinolates and IAA (Mikkelsen et al., 2003). Therefore, it is also possible that sps/cyp79F1 mutants are stressed because of the perturbation of cytokinin homeostasis, which in turn up-regulates CYP79B2/B3 genes. Glucosinolates are the largest group of secondary metabolites known in Arabidopsis and other genera of Brassicaceae (Halkier, 1999). Several glucosinolate biosynthetic enzymes have been cloned and shown to be members of the cytochrome P450 superfamily. Gene duplication, tandem clustering of the duplicated genes, and gene conversion of the cytochrome P450 genes provide evidence that rapid evolution led to generation of diverse enzymatic reactions. Analysis of the highly related SPS/CYP79F1 and CYP79F2 genes provide a clear picture of a recent gene duplication undergoing diversification through the utilization of distinct but overlapping substrates, as well as through different spatial expression profiles.
Reintanz et al. (2001) and Chen et al., (2003) showed that, by using SPS/CYP79F1 and CYP79F2 promoter-GUS fusion, both promoters conferred GUS expression in aerial parts as well as in root. However, we showed by using gene trap insertion that SPS/CYP79F1 and CYP79F2 genes were expressed mainly in aerial parts and in root, respectively. These discrepancies in the expression patterns of SPS/CYP79F1 and CYP79F2 genes may derive from different types of reporter gene fusions used in the studies, i.e. gene trap fusions in this study versus ectopic transgene fusions. We also cannot rule out the possibility that the distinct expression patterns of the SPS/CYP79F1 and CYP79F2 genes observed by different groups may reflect ecotype-dependant allelic variations, as different Arabidopsis ecotypes were used in the studies, i.e. the Wassilewskija ecotype in this study, versus the Columbia ecotype. Recently, it has been reported that glucosinolates show extensive variations in both the composition and concentration among different ecotypes (Kliebenstein et al., 2001; Petersen et al., 2002; Reichelt et al., 2002; Brown et al., 2003).
Arabidopsis has an estimated 273 different cytochrome P450 genes in the genome (for review, see Nelson, 1999; Schuler and Werck-Reichhart, 2003; http:/biobase.dk/P450). With the large number of related cytochrome P450 sequences in the genome, it is not surprising that functions of some of the closely related cytochrome P450 genes are redundant or overlapping. The construction of double mutants by genetic recombination is the most direct approach to uncover redundant functions of duplicated genes. However, this approach can be very cumbersome when the duplicated genes are tightly linked as a tandem duplication, as is the case with nearly 17% of the Arabidopsis genes (The Arabidopsis Genome Initiative, 2000). We have successfully used the Ac-Ds transposition system to generate double mutations of two closely related genes in tandem, i.e. the SPS/CYP79F1 and CYP79F2 genes, to study their overlapping metabolic and physiological functions. The isolation of sps/cyp79F1 and cyp79F2 double mutants from a small set of transposed lines (approximately one in 100 lines) demonstrates that the approach is efficient. This approach of using multiple transpositions of the Ds transposon may be of general utility for elucidating functions of other clustered duplicated genes in the genome.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis mutants and wild-type plants were derived from Wassilewskija ecotype. Plants were grown on soil under controlled growth chamber conditions at 20°C, 60% to 70% relative humidity, 16/8 photoperiod, and a photosynthetic flux of 100 μmol photons m−2 s−1. Plants used for glucosinolate analysis were grown under a photosynthetic flux of 250 μmol photons m−2 s−1 for 3 weeks before the samples were collected as pooled materials.
Isolation of cyp79F2 Mutants
A collection of Ds insertion lines, generated by transpositions of a Ds gene trap element from a single T-DNA (Tantikanjana et al., 2001), was used to screen for insertion lines containing Ds element in the CYP79F2 genes. Approximately 900 insertion lines were used for the screening using CYP79F2 gene-specific primers (5′-TCTTCATCGCATCAATCACTTTAC-3′; 5′GCGTGGCTCAACAGACAA-3′) and transposon-specific primer (5′-CCGTTTACCGTTTTGTATATCCCCG-3′; 5′-CGATTACCGTATTTATCCCGT-3′). Young leaves from four independent insertion lines were pooled for DNA extraction, and pooled DNA from a total of 24 insertion lines were used for each PCR reaction. The putative lines were then identified and verified by sequencing.
Generation of sps/cyp79F1 and cyp79F2 Double-Knockout Mutants
The mutant cyp79F2-1 allele containing a Ds element inserted into the first exon of the gene was used as a starter line for the reactivation of Ds transposition. Because expression of the transposase used in this tagging system is driven by the induction of a heat shock promoter (Balcells et al., 1994; Tantikanjana et al., 2001), transactivation of the Ds element was performed by subjecting the starter line to high temperature (40°C to 42°C) as described (Balcells et al., 1994). The heat-shocked seeds were then sowed and allow to set seeds. Screening for families, segregating for plants with abnormal aerial structure resembling to the sps/cyp79F1 mutant, was done in the next generation. Approximately eight to 12 plants derived from each family were used for the screening. Positions of the Ds elements in SPS/CYP79F1 gene were verified by PCR and sequencing using SPS1/CYP79F1-specific primer (5′-GGAGGATGCAAGAACCA-3′) and 3′ Ds-specific primers (5′-CGATTACCGTATTTATCCCGT-3′ or 5′-CCGGTATATCCCGTTTTCG-3′). A footprint of the Ds element after excision from CYP79F2 gene in the sps-7 cyp79F2-1 allele was verified by PCR and sequencing using CYP79F2-specific primers (5′-TCTTCATCGCATCAATCACTTTAC-3′ and 5′-GCGTGGCTCAACAGACAA-3′).
Analysis of Lateral Root Length and Lateral Root Number
Seedlings were grown on soil for 8 d before root samples were washed gently to maintain integrity and collected for the analysis. The lateral root length of mutant and wild-type plant was derived from the analysis of 21 to 24 seedlings. For the analysis of lateral root length and lateral root number, total lateral root length and lateral root number derived from the first 1 cm of the primary roots, starting from the hypocotyls-root junction, were used in the investigation.
Staining for GUS Expression
Plant materials were stained in GUS-staining solution (100 mm Na Phosphate at pH 7.0, 10 mm EDTA, 0.1% Triton X-100, 1 mg/mL of X-Gluc [Biosynth AG]), containing 5 or 10 mm potassium ferricyanide and 5 or 10 mm potassium ferrocyanide. After being placed under vacuum for 10 min in a dessicator, the samples were incubated at 37°C for 2 h to overnight. The stain solution was removed and the tissues were cleared by incubating with several changes of 70% ethanol.
Glucosinolate Analysis by HPLC
Glucosinolates were extracted from approximately 20 mg slightly homogenized freeze-dried rosette leaves, stems, roots, or flowers, either from five to eight single plants or from three to six pools of three to eight plants, by boiling 4 min in 4 mL 70% methanol. The supernatant was collected, and the plant material was washed with 2 mL 70% methanol. The extracts were combined and applied to 200 μL DEAE Sephadex CL-6B (Amersham-Pharmacia Biotech, Uppsala) columns (Bio-Rad Polyprep; Copenhagen) equilibrated with 1 mL 0.02 m KOAc, pH 5.0 and washed with 1 mL water. The columns were washed with 2 mL 70% methanol, 2 mL water, and 0.02 m KOAc, pH 5. Following addition of 100 μL of 2.5 mg/mL Helix pomatia sulfatase (Sigma-Aldrich, St. Louis), the columns were sealed and left overnight. The resulting desulphoglucosinolates were eluted with 2 × 1 mL water. The eluate was lyophilized until dryness and resuspended in 200 μL water. Aliquots of 100 μL were applied to a Shimadzu Spectachrom HPLC system equipped with a Supelco supelcosil LC-ABZ 59142 RP-amid C-16 (25 cm × 4.6 mm, 5 mm; Supelco, Bellfonte, PA; Holm & Halby, Brendby, Denmark) and an SPD-M10AVP photodiode array detector (Shimadzu, Columbia, MD). The flow rate was 1 mL min−1. Desulphoglucosinolates were eluted with water for 2 min followed by a linear gradient from 0% to 60% methanol in water (48 min), a linear gradient from 60% to 100% methanol in water (3 min), and with 100% methanol (14 min). Detection was performed at 229 nm and 260 nm using a photodiode array. Desulphoglucosinolates were quantified based on response factors (Buchner, 1987; Haughn et al., 1991) and an internal p-hydroxybenzylglucosinolate (Bioraf, Akirkeby, Denmark) standard as previously described (Petersen et al., 2001, 2002). The standard was added at the beginning of the extraction procedure.
Sequence data used in this article are taken from GenBank accession number AC0006341.
Acknowledgments
We thank Joseph Kieber for the ARR5::uidA and Tom Gulifoyle for the DR5::uidA transgenic lines.
This work was supported by the National Science Foundation, the Danish National Research Foundation, and the University of California, Davis.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.040113.
References
- Bak S, Tax FE, Feldmann KA, Galbraith DW, Feyereisen R (2001) CYP83B1, a cytochrome P450 at the metabolic branch point in auxin and indole glucosinolate biosynthesis in Arabidopsis. Plant Cell 13: 101–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcells L, Sundberg E, Coupland G (1994) A heat-shock promoter fusion to the Ac transposase gene drives inducible transposition of a Ds element during Arabidopsis embryo development. Plant J 5: 755–764 [Google Scholar]
- Bangerth F (1994) Response of cytokinin concentration in the xylem exudates of bean (Phaseolus vulgaris L.) plants to decapitation and auxin treatment and relationship to apical dominance. Planta 194: 439–442 [Google Scholar]
- Barlier I, Kowalczyk M, Marchant A, Ljung K, Bhalerao R, Bennett M, Sandberg G, Bellini C (2000) The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc Natl Acad Sci USA 97: 14819–14824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binns AN, Labriola J, Black RC (1987) Initiation of auxin autonomy in Nicotiana glutinosa cells by the cytokinin-biosynthesis gene from Agrobacterium tumefaciens. Planta 171: 539–548 [DOI] [PubMed] [Google Scholar]
- Blilou I, Frugier F, Folmer S, Serralbo O, Willemsen V, Wolkenfelt H, Eloy NB, Ferreira PC, Weisbeek P, Scheres B (2002) The Arabidopsis HOBBIT gene encodes a CDC27 homolog that links the plant cell cycle to progression of cell differentiation. Genes Dev 16: 2566–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PD, Tokuhisa JG, Reichelt M, Gershenzon J (2003) Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry 62: 471–481 [DOI] [PubMed] [Google Scholar]
- Buchner R (1987) Glucosinolates in Rapeseeds: Analytical Aspects. In JP Wathelet, ed, World Crops: Production, Utilization, Description. Martinus Nijhoff Publishers, Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 50–58
- Casson SA, Chilley PM, Topping JF, Evans IM, Souter MA, Lindsey K (2002) The POLARIS gene of Arabidopsis encodes a predicted peptide required for correct root growth and leaf vascular patterning. Plant Cell 14: 1705–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Glawischnig E, Jorgensen K, Naur P, Jorgensen B, Olsen C-E, Hansen CH, Rasmussen H, Pickett JA, Halkier BA (2003) CYP79F1 and CYP79F2 have distinct functions in the biosynthesis of aliphatic glucosinolates in Arabidopsis. Plant J 33: 923–937 [DOI] [PubMed] [Google Scholar]
- D'Agostino I, Deruere J, Kieber JJ (2000) Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol 124: 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff NV (1983) Controlling element in maize. In J Shapiro, ed, Mobile Genetic Elements. Academic Press, New York, pp 1–63
- Halkier BA (1999) Glucosinolates. In R Ikan, ed, Naturally Occurring Glycosides: Chemistry, Distribution and Biological Properties. John Wiley and Sons, Chichester, UK, pp 193–223
- Hansen CH, Wittstock U, Olsen CE, Hick AJ, Pickett JA, Halkier BA (2001) Cytochrome P450 CYP79F1 from Arabidopsis catalyzes the conversion of dihomomethionine and trihomomethionine to the corresponding aldoximes in the biosynthesis of aliphatic glucosinolates. J Biol Chem 276: 11078–11085 [DOI] [PubMed] [Google Scholar]
- Harrar Y, Bellec Y, Bellini C, Faure JD (2003) Hormonal control of cell proliferation requires PASTICCINO genes. Plant Physiol 132: 1217–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughn GW, Davin L, Giblin M, Underhill EW (1991) Biochemical genetics of plant secondary metabolites in Arabidopsis thaliana. Plant Physiol 97: 217–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Kroymann J, Brown P, Figuth A, Pedersen D, Gershenzon J, Mitchell-Olds T (2001) Genetic control of natural variation in Arabidopsis thaliana glucosinolate accumulation. Plant Physiol 126: 811–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CJ, Bangerth F (1992) The possible role of cytokinins, ethylene and indoleacetic acid in apical dominance. In C Karssen, et al., eds, Progress in Plant Growth and Regulation. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 431–436
- Long JA, Woody S, Poethig S, Meyerowitz EM, Barton MK (2002) Transformation of shoots into roots in Arabidopsis embryos mutant at the TOPLESS locus. Development 129: 2797–2806 [DOI] [PubMed] [Google Scholar]
- Makarova RV, Borisova TA, Machackova I, Kefeli VI (1996) Effect of alien ipt gene on hormonal concentrations of plants. In AR Smith et al., eds, Plant Hormone Signal Perception and Transduction. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 171–173
- Mikkelsen MD, Naur P, Halkier BA (2004) Arabidopsis mutants in the C-S lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J 37: 770–777 [DOI] [PubMed] [Google Scholar]
- Mikkelsen MD, Petersen BL, Glawischnig E, Jensen AB, Andreasson E, Halkier BA (2003) Modulation of CYP79 genes and glucosinolate profiles in Arabidopsis by defense signaling pathways. Plant Physiol 131: 298–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DR (1999) Cytochrome P450 and the individuality of species. Arch Biochem Biophys 369: 1–10 [DOI] [PubMed] [Google Scholar]
- Palni LMS, Burch L, Horgan R (1988) The effect of auxin concentration on cytokinin stability and metabolism. Planta 194: 231–234 [DOI] [PubMed] [Google Scholar]
- Petersen BL, Andreasson E, Bak S, Agerbirk N, Halkier BA (2001) Characterization of transgenic Arabidopsis thaliana with metabolically engineered high levels of p-hydroxybenzylglucosinolate. Planta 212: 612–618 [DOI] [PubMed] [Google Scholar]
- Petersen BL, Chen S, Hansen CH, Olsen CE, Halkier BA (2002) Composition and content of glucosinolates in developing Arabidopsis thaliana. Planta 214: 562–571 [DOI] [PubMed] [Google Scholar]
- Reichelt M, Brown PD, Schneider B, Oldham NJ, Stauber E, Tokuhisa J, Kliebenstein DJ, Mitchell-Olds T, Gershenzon J (2002) Benzoic acid glucosinolate esters and other glucosinolates from Arabidopsis thaliana. Phytochemistry 59: 663–671 [DOI] [PubMed] [Google Scholar]
- Reintanz B, Lehnen M, Reichelt M, Gershenzon J, Kowalczyk M, Sandberg G, Godde M, Uhl R, Palme K (2001) bus, a Bushy Arabidopsis CYP79F1 knockout mutant with abolished synthesis of short-chain aliphatic glucosinolates. Plant Cell 13: 351–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler MA, Werck-Reichhart D (2003) Functional genomics of P450s. Annu Rev Plant Biol 54: 629–667 [DOI] [PubMed] [Google Scholar]
- Smolen G, Bender J (2002) Arabidopsis cytochrome P450 cyp83B1 mutation activate the tryptophan biosynthesis pathway. Genetics 160: 323–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan V (1996) Horizontal spread of transposon mutagenesis: new uses for old elements. Trends Plant Sci 1: 184–190 [Google Scholar]
- Sundaresan V, Springer P, Volpe T, Haward S, Jones JDG, Dean C, Ma H, Marteinssen R (1995) Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev 9: 1797–1810 [DOI] [PubMed] [Google Scholar]
- Tamas IA (1995) Hormonal regulation of apical dominance. In PJ Davis, Plant Hormone. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 572–579
- Tantikanjana T, Yong JWH, Letham DS, Griffith M, Hussain M, Ljung K, Sandberg G, Sundaresan V (2001) Control of axillary bud initiation and shoot architecture in Arabidopsis through the SUPERSHOOT gene. Genes Dev 15: 1577–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmulling T (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15: 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen V, Friml J, Grebe M, van den Toorn A, Palme K, Scheres B (2003) Cell polarity and PIN protein positioning in Arabidopsis require STEROL METHYLTRANSFERASE1 function. Plant Cell 15: 612–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Zhang X, Wang J, Letham DS, Mckinney AA, Higgins TJV (1995) The effect of auxin on cytokinin levels and metabolism in transgenic tobacco tissue expressing and ipt gene. Planta 196: 84–94 [Google Scholar]