Abstract
This work describes the purification and characterization of enzymes that exhibit β-d-xylosidase activity in stem tissues of Arabidopsis. This is the first detailed investigation that concerns the characterization of catalytic properties and sequence identity of enzymes with β-d-xylosidase activities in a dicotyledonous plant. Three different enzymes, ARAf, XYL4, and XYL1 with apparent molecular masses of 75, 67, and 64 kD, respectively, were purified to homogeneity. ARAf was identified as a putative α-l-arabinofuranosidase, and XYL4 and XYL1 as putative β-d-xylosidases using matrix-assisted laser-desorption ionization time of flight. ARAf belongs to family 51 and XYL4 and XYL1 to family 3 of glycoside hydrolases. ARAf and XYL1 have highest specificity for p-nitrophenyl-α-l-arabinofuranoside and XYL4 for p-nitrophenyl-β-d-xylopyranoside and natural substrates such as xylobiose and xylotetraose. XYL4 was shown to release mainly d-Xyl from oat spelt xylan, rye arabinoxylan, wheat arabinoxylan, and oligoarabinoxylans. ARAf and XYL1 can also release d-Xyl from these substrates but less efficiently than XYL4. Moreover, they can also release l-Ara from arabinoxylans and arabinan. Overall, the results indicate that XYL4 possesses enzymatic specificity characteristic for a β-d-xylosidase, while ARAf and XYL1 act as bifunctional α-l-arabinofuranosidase/β-d-xylosidases. Analysis of the activity of these three enzymes in stem tissues at different stages of development has shown that young stems possess the highest activities for all three enzymes in comparison to the activities of the enzymes present in stems at older stages of development. High enzyme activities are most likely related to the necessary modifications of cell wall structure occurring during plant growth.
Plant cells are surrounded by an extracellular matrix known as the cell wall. The regulation of cell wall morphology and composition is important for the determination of cell size and shape, cellular interactions with the environment, mechanical resistance, and defense against pathogen attacks (Reiter, 2002). The cell wall is also involved in plant growth and development (Stolle-Smits et al., 1999; Obel et al., 2002). In addition, although cell walls consist mainly of polysaccharides broadly classified as celluloses, hemicelluloses, and pectins, their constitution varies, not only from plant to plant, but also in different tissues of the same plant (Heredia et al., 1995; Cosgrove, 1997; Popper and Fry, 2003). A coordinated series of biochemical processes occur during plant development resulting in the biosynthesis and degradation of cell wall components. Consequently, numerous enzymes must be implicated in these processes (Heredia et al., 1995; Cosgrove, 1997; Stolle-Smits et al., 1999; Obel et al., 2002; Reiter, 2002; Popper and Fry, 2003). However, to date, little information is available concerning such enzymes, their mode of action, and their physiological role. Increased research interest has focused on enzymes involved in the metabolism of hemicelluloses in recent years because of their possible applications in biotechnology, considering the great abundance of hemicelluloses in nature.
Hemicelluloses constitute up to 30% to 35% of the dry weight of the plant cell wall and are mainly composed of xyloglucan and xylan with minor amounts of several mannose-containing polysaccharides (Bewley et al., 1997; Handford et al., 2003). Xyloglucan is coated and cross-linked with cellulose microfibrils. Xyloglucans have a β-1,4-linked d-glucosyl backbone structure substituted with xylosyl, galactosyl, and l-fucosyl residues (Renard et al., 1992). Xylan is the major hemicellulose component in wood and therefore is the second most abundant polysaccharide, next to cellulose, in nature (Prade, 1996; Kulkarni et al., 1999). It has a relatively complex structure based on a β-(1,4)-linked d-Xyl backbone substituted to varying extents with acetyl, l-arabinofuranosyl, galactosyl, glucuronyl, and 4-O-methylglucuronyl groups (Coughlan and Hazlewood, 1993; Prade, 1996; Kulkarni et al., 1999). Xylans can form cross-linkages with lignin via α-l-arabinofuranosyl residues, which can be esterified with hydroxycinnamic acids, such as ferulic or p-coumaric acids (Carpita, 1996).
It has been hypothesized that the metabolism of hemicelluloses in plant cells involves different hydrolytic enzymes that can modify polysaccharides (Coughlan and Hazlewood, 1993; Cosgrove, 1997; Prade, 1996; Kulkarni et al., 1999). However, the mechanism of regulation of hemicellulose structure is difficult to establish because the number of enzymes characterized so far is limited. Xylan, the major component of hemicelluloses, is greatly heterogeneous; therefore, its hydrolysis requires the action of a battery of enzymes (Rahman et al., 2003; Tuncer and Ball, 2003). This battery is usually composed of β-1,4-endoxylanase, β-d-xylosidase, α-l-arabinofuranosidase, α-d-glucuronidase, acetylxylan esterase, and phenolic acid esterase (Sunna and Antranikian, 1997). These enzymes act cooperatively to convert xylan into its constituting sugars (Rahman et al., 2003; Tuncer and Ball, 2003), and they have been reported to occur widely in microorganisms (Kulkarni et al., 1999; Subramaniyan and Prema, 2000; Beg et al., 2001), fungi (Coughlan and Hazlewood, 1993; Sunna and Antranikian, 1997), and animals (Yamaura et al., 1997). Side chain cleaving enzymes, such as α-l-arabinofuranosidase, α-d-glucuronidase, and acetylxylan esterase, remove substituent functional groups of heteroxylans. Removal of side chain substituents in xylan is considered to be a limiting step in achieving efficient hydrolysis of the polysaccharide polymer (Rahman et al., 2003; Tuncer and Ball, 2003), since these groups create steric obstacles for the enzyme-substrate complex formation (Gorbacheva and Rodionova, 1977a, 1977b). l-Ara, as a side chain substituent of xylan, is present in various plant structural polysaccharides such as rhamnogalacturonan-I (RG-I) and rhamnogalacturonan-II (Ridley et al., 2001; Willats et al., 2001; Glushka et al., 2003).
Endoxylanases and β-d-xylosidases are enzymes responsible for the cleavage of the xylan backbone. Endo-β-1,4-xylanases hydrolyze the insoluble xylan backbone into shorter, soluble xylo-oligosaccharides, while β-d-xylosidases hydrolyze xylo-oligosaccharides and xylobiose from their nonreducing ends to liberate d-Xyl (Sunna and Antranikian, 1997). Endoxylanases have been identified in several fruits and higher plants (Slade et al., 1989; Banik et al., 1997; Caspers et al., 2001; Suzuki et al., 2002). β-d-Xylosidase, a key enzyme for complete degradation of xylan, has been purified and characterized in a variety of bacteria and fungi (Sunna and Antranikian, 1997). One β-d-xylosidase from young barley (Hordeum vulgare) seedlings was recently purified, sequenced, and biochemically characterized (Lee et al., 2003). Other plant β-d-xylosidases have been purified and partially characterized in wheat flour (Cleemput et al., 1997), sycamore cell cultures (Tezuka et al., 1993), pea seedlings (O'Neill et al., 1989), and sugarcane (Chinen et al., 1982). Recently, a gene from Arabidopsis encoding a putative β-d-xylosidase was identified, and a role for this enzyme in secondary cell wall metabolism and plant development has been proposed (Goujon et al., 2003).
We have chosen to study enzymes with β-d-xylosidase activity from stem tissues of the model plant Arabidopsis. Xylan is very abundant in Arabidopsis stems (Gardner et al., 2002); therefore, this plant part is likely to contain enzymes that can degrade xylan. However, until now, Arabidopsis stems have not been carefully explored for the presence of such enzymes. In addition to its fundamental interest, the elucidation of the plant strategy toward xylan degradation is of interest because of possible biotech applications (Kulkarni et al., 1999; Beg et al., 2001; Subramaniyan and Prema., 2002). Xylan degradation has the potential to be a future energy resource; however, its practical exploitation demands effective and economical methods for its hydrolysis to d-Xyl, methods which are not yet available.
Therefore, the objective of this work was the purification and identification of enzymes that possess β-d-xylosidase activity from stem extracts of Arabidopsis and the determination of their substrate specificity and kinetic and molecular properties. The results obtained allow us to propose a biological role for the characterized enzymes in stem development and in the degradation of not only oligoxylan, xylan, and arabinoxylan, but also in arabinan constituting side chains of RG-I.
RESULTS
Purification of Enzymes That Exhibit β-d-Xylosidase Activity in Stem Tissues of Arabidopsis
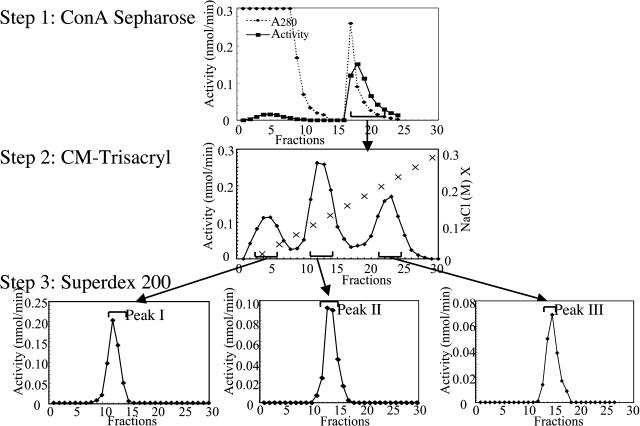
Enzymes with β-d-xylosidase activity were purified from the crude protein extract of Arabidopsis stems. p-Nitrophenyl-β-d-xylopyranoside (pNPX) was used as the substrate to monitor this enzymatic activity. Figure 1 summarizes the purification procedure, and Table I indicates the degree of purification and yield for each step. The purification protocol involves three steps of column chromatography: lectin concanavalin A (Con A), cation exchange, and gel filtration. A single peak of activity was eluted from lectin Con A Sepharose column, and only a nonsignificant proportion of this activity was not retained (Fig. 1, Step 1). Pooled fractions showing β-d-xylosidase activity after this first step were subjected to a cation-exchange chromatography on a CM-Trisacryl column. Three peaks showing β-d-xylosidase activity were resolved in this step (Fig. 1, Step 2). The enzyme present in each peak was further purified in a final step using gel filtration on Superdex 200 (Fig. 1, Step 3). These three β-d-xylosidase activities were designated as peaks I, II, and III. In this purification procedure, the lectin-affinity chromatography increased the β-d-xylosidase specific activity by approximately a factor of 7 (Table I). Since Con A Sepharose is normally used for the purification of glycoproteins (Farooqi et al., 1997; Sheldon et al., 1998), this result suggests that the three purified enzymes with β-d-xylosidase activity are glycosylated.
Figure 1.
Chromatographic purification of enzymes exhibiting β-d-xylosidase activity from stem tissues of Arabidopsis (see also Table I). Purification Step 1, A 1-mL sample of stem tissue cell-free extract (2.5 mg proteins) was loaded on a 0.5- × 3-cm column of Con A Sepharose. The column was eluted with 7 mL of 20 mm Tris-HCl (pH 7.4), 0.5 m NaCl, and 0.2 m methyl-α-glucopyranoside buffer. Fractions of 1 mL were collected, and aliquots of 50 μL were assayed for β-d-xylosidase activity. Purification Step 2, Pooled fractions from the first Con A Sepharose chromatography were dialysed, concentrated, and loaded on a CM-Trisacryl cation-exchange column (1.5 × 5 cm). The column was eluted discontinuously in 15 steps with 2-mL fractions of 25 mm Na-acetate, pH 5.0, and 0.015% Triton X-100, containing increasing concentrations of NaCl from 0.0 to 5.0 m. Fractions of 1 mL were collected, and aliquots of 100 μL were assayed for β-d-xylosidase activity. Step 3, Gel-filtration chromatography. The pooled fractions showing β-d-xylosidase activity were concentrated to 500 μL and loaded on a Superdex 200 HR10/30 column (Amersham Pharmacia Biotech). Elution was performed at room temperature with 20 mm Na-acetate buffer (pH 5.0) containing 150 mm NaCl. Fractions of 0.4 mL were collected at a flow rate of 0.5 mL/min, and 100 μL of each fraction was assayed for β-d-xylosidase activity as described in “Materials and Methods.”
Table I.
Enzyme yields and purification factors for peaks I, II, and III
| Step of Purification
|
Yield
|
Specific Activity
|
Recoverya
|
Purification Factora
|
|
|---|---|---|---|---|---|
| Protein | Activity | ||||
| mg | nmol/min | nmol min−1 mg−1 | % | fold | |
| Crude homogenate | 2.500 | 15.00 | 5.8 | 100 | 1 |
| Con A Sepharose | 0.270 | 12.00 | 44.4 | 80 | 7.7 |
| Peak I | |||||
| CM-Trisacryl | 0.027 | 1.28 | 47.3 | 8.5 | 8.2 |
| Superdex 200 | 0.008 | 0.78 | 97.0 | 5.2 | 16.7 |
| Peak II | |||||
| CM-Trisacryl | 0.028 | 1.95 | 69.8 | 13.0 | 12.0 |
| Superdex 200 | 0.006 | 1.40 | 234.0 | 9.4 | 40.3 |
| Peak III | |||||
| CM-Trisacryl | 0.031 | 1.41 | 45.5 | 9.4 | 7.8 |
| Superdex 200 | 0.003 | 0.51 | 169.0 | 3.4 | 29.1 |
Recoveries are expressed as percentage of initial activity, and purification factors are calculated on the basis of specific activities.
SDS-PAGE Analysis of the Isolated Enzymes
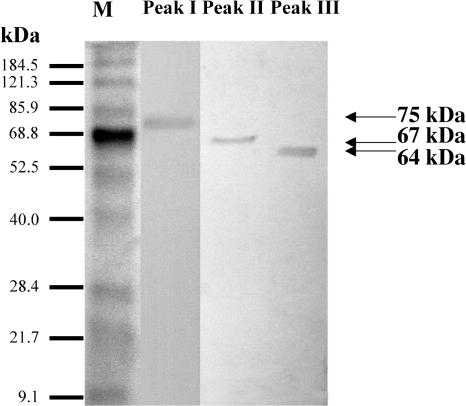
The purity of the three proteins isolated after the final gel filtration was examined by SDS-PAGE. Staining with Coomassie Brilliant Blue R-250 revealed one band for each peak (Fig. 2). The apparent molecular masses of the proteins corresponding to peaks I, II, and III were determined to be 75, 67, and 64 kD, respectively. In addition, Superdex 200 chromatography of the three isolated proteins suggests that they are native monomers since they were eluted at positions corresponding to molecular masses similar to those determined on denaturing SDS-PAGE (data not shown).
Figure 2.
SDS-PAGE of purified enzymes. The purified peak fractions from Figure 1 containing about 2 μg of protein were analyzed by SDS-PAGE (10% polyacrylamide gel), and proteins were visualized by Coomassie Brilliant Blue R-250. Lane M, Marker proteins (the sizes are indicated).
Kinetic Properties and Enzymatic Characterization
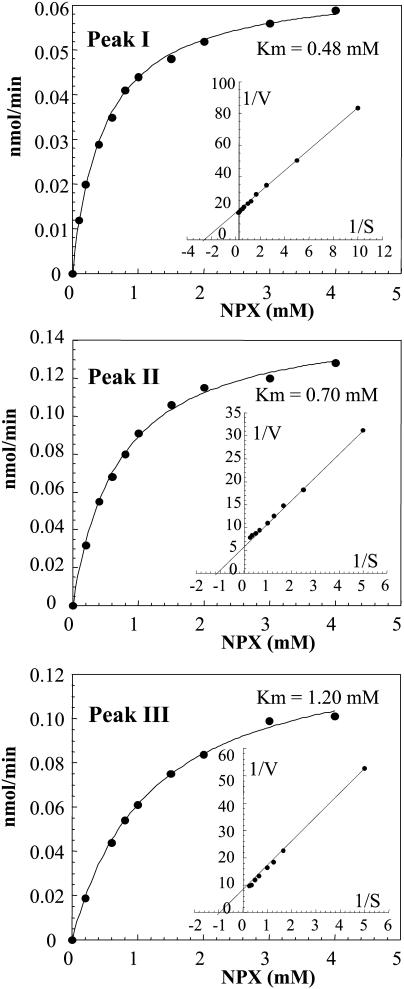
The kinetic parameters for the three peaks with β-xylosidase activity were studied using pNPX as the substrate (Fig. 3; Table II). Substrate saturation curves for peaks I, II, and III were fitted to the Michaelis-Menten equation to obtain kinetic parameters and are shown in Figure 3. Km values of 0.48, 0.70, and 1.20 mm were obtained for peaks I, II, and III, respectively. These Km values are similar to those previously reported for β-d-xylosidases from barley (Lee et al., 2003) and numerous β-d-xylosidases from fungi and bacteria (Bachmann and McCarthy, 1989; Nanmori et al., 1990; Kumar and Ramon, 1996; Saha, 2003). The corresponding kcat/Km ratios of the three enzymes are not significantly different (Table II).
Figure 3.
pNPX saturation curves of the three purified enzymes. Fractions exhibiting β-d-xylosidase activity obtained after Superdex 200 treatment were pooled (Fig. 1). Aliquots containing approximately 0.1 μg of protein were assayed for β-d-xylosidase activity in a total volume of 0.5 mL. The mixture was incubated at 37°C for 1 h in the presence of increasing concentrations of pNPX from 0.2 to 4.0 mm. The insert presents the corresponding double-reciprocal plot for the determination of Km.
Table II.
Kinetic parameters of peaks I, II, and III
| Peak of Activity | Km (37°C) | kcat | kcat/Km |
|---|---|---|---|
| mm | s−1 | mm−1 s−1 | |
| Peak I | 0.48 | 0.87 | 1.81 |
| Peak II | 0.70 | 1.65 | 2.36 |
| Peak III | 1.20 | 1.35 | 1.13 |
The influence of pH on the activity of the three enzymes was probed in the range of pH 3 to 8. The pH optima determined were 4.3, 4.7, and 4.9 for peaks I, II, and III, respectively. These values are in the same range as those previously determined for β-d-xylosidases from barley (Lee et al., 2003) and certain β-d-xylosidases from fungi and bacteria (John and Schmidt, 1988; Lachke, 1988; Matsuo and Yasui, 1988; Kumar and Ramon, 1996; Saha, 2003).
The temperature dependence of the three β-d-xylosidase activities was tested in the range of 20°C to 70°C. The apparent optimum temperatures were 67°C, 60°C, and 55°C for peaks I, II, and III, respectively. Again, these values are close to temperature optima reported for β-d-xylosidases from some fungi and bacteria (John and Schmidt, 1988; Lachke, 1988; Kumar and Ramon, 1996; Saha, 2003).
Substrate Specificities of the Purified Enzymes
The substrate specificity of the three purified enzymes was tested using various p-nitrophenyl (pNP)-glycosides and two oligoxylans (xylobiose and xylotetraose; Table III). High-performance anion-exchange chromatography (HPAEC) was used to separate xylobiose and xylotetraose from the reaction product, d-Xyl. All three enzymes hydrolyzed xylobiose to d-Xyl. Similarly, the xylotetraose was degraded to d-Xyl and xylotriose by all three enzymes. Although xylotriose can be further hydrolyzed to xylobiose, it was only detected in incubations of more than 2 h. A longer period of incubation seemed necessary because neoformed xylotetraose was present in low concentrations.
Table III.
Relative activity of peaks I, II, and III determined with different substrates
| Substrate
|
Relative Activity
|
||
|---|---|---|---|
| Peak I | Peak II | Peak III | |
| % | |||
| pNP-β-d-Xylopyranoside | 15 | 49 | 29 |
| oNP-β-d-Xylopyranoside | 3 | 45 | 8 |
| Xylobiose | 14 | 100 | 16 |
| Xylotriose | 12 | 56 | 12 |
| pNPAf | 100 | 15 | 100 |
| pNPA | 6 | 17 | 12 |
| pNP-β-d-Galactopyranoside | 19 | 3 | 15 |
| pNP-α-d-Galactopyranoside | 3 | 20 | 17 |
| pNP-β-d-Fucopyranoside | 7 | 5 | 11 |
| pNP-β-d-Glucopyranoside | 1 | 2 | 1 |
Aryl glycosides were used as substrates with purified proteins (0.1 μg) in standard assays at a final concentration of 4 mm. Activity was expressed as percent of activity against maximal obtained substrate activity. The maximal activity is indicated in bold. No activity was detected for pNP-α-d-glucopyranoside, pNP-β-l-arabinopyranoside, Me-β-d-xylopyranoside, pNP-α-d-xylopyranoside, pNP-α-d-fucopyranoside, pNP-β-d-mannopyranoside, pNP-α-d-mannopyranoside, and pNP-β-d-glucuronide.
The activities obtained using a standard substrate concentration of 4 mm, which is near saturation in the case of pNPX, showed that the three purified enzymes exhibit different substrate specificities (Table III). Although pNP-xyloside had been used for convenience in the purification procedures, the results obtained show that the three enzymes had hydrolytic activity toward oNP-xyloside, xylobiose, and xylotetraose, typical substrates for β-d-xylosidases. However, the rate of hydrolysis was higher using xylobiose than xylotetrose. Peak I showed optimal activity for pNP-α-l-arabinofuranoside (pNPAf) rather than for pNPX. This characteristic has been already observed for α-l-arabinofuranosidases isolated from other sources (Kelly et al., 1987; Beylot et al., 2001). Peak II showed an optimal efficiency with the natural substrate xylobiose suggesting that this enzyme is more specifically a β-d-xylosidase. Purified peak III showed 3-fold higher activity with pNPAf than with pNPX. Thus, the three purified enzymes can hydrolyze different substrates in addition to pNPX and pNPAf, such as pNP-β-d-galactopyranoside, pNP-α-d-galactopyranoside, pNP-β-d-fucopyranoside, and pNP-α-l-arabinopyranoside (pNPA) to various degrees (Table III).
Identification of the Purified Enzymes
In order to further identify the enzymes corresponding to the three peaks, the bands obtained after SDS-PAGE were analyzed by MALDI-TOF. The identity of the enzymes was established by the algorithms used in ProteinProspector (http://prospector.ucsf.edu/). The band corresponding to peak I was identified as a putative α-l-arabinofuranosidase, designated as ARAf encoded by gene At3g10740 in Arabidopsis (accession no. NP_187685 in the BLAST data bank). This result is in agreement with the higher specificity for pNPAf exhibited by this enzyme. Peak II was identified as a putative β-d-xylosidase, designated as XYL4 encoded by the gene At5g64570 in Arabidopsis, and named AtBXL4 by Goujon et al. (2003; accession no. BAB11424 in the BLAST data bank). This result is also in agreement with the higher specificity exhibited by this enzyme for xylobiose. Peak III was identified as a putative β-d-xylosidase, designated as XYL1 corresponding to protein BAB09906, encoded by the gene At5g49360, and named AtBXL1 in Goujon et al. (2003). On the basis of the results reported in Table III, XYL1 has the highest specificity for pNPAf. To confirm this functional specificity of XYL1, we further tested its catalytic efficiency toward pNPAf and pNPX substrates. The catalytic efficiencies obtained were 3.45 mm−1 s−1 and 1.13 mm−1 s−1, respectively. Small differences in catalytic efficiencies for these two substrates suggest that XYL1 acts as bifunctional α-l-arabinofuranosidase/β-d-xylosidase.
The bifunctional origin of XYL1 was also confirmed by comparing the profile of peak III activity upon CM-Trisacryl chromatography of crude protein extracts from the wild-type Arabidopsis and that from a T-DNA mutant for the At5g49360 gene. This null mutant completely lacks the β-d-xylosidase and α-l-arabinofuranosidase activities corresponding to peak III (data not shown).
Analysis of the Primary Structure of the Purified Enzymes
The amino acid sequences of the three proteins obtained using MALDI-TOF and the genetic analysis described above were aligned together with the known sequences of barley β-d-xylosidases in order to determine molecular properties and position of putative catalytic amino acid residues (Fig. 4). This analysis predicts the existence of signal sequences, the true NH2-terminal residue in the ARAf, XYL4, and XYL1 mature enzymes, being at positions 21, 39, and 31, respectively (Fig. 4). In the case of β-d-xylosidase from barley, the predicted NH2-terminus residue was experimentally confirmed (Lee et al., 2003).
Figure 4.
Alignment of XYL4, XYL1, and ARAf amino acid sequences. Identical residues in the three sequences are shown in bold. The XYL4, XYL1, and ARAf cDNAs encode polypeptides of 748, 774, and 678 amino acids, respectively. The start of the NH2-terminal residue, after removing signal sequences, is shown by arrowhead. A vertical line is used to indicate the likely COOH terminus, and asterisks indicate potential N-glycosylation sites. Arrows indicate the putative catalytic nucleophiles (Glu-464, Asp-308, and Asp-296 for ARAf, XYL4, and XYL1, respectively) and putative catalytic acid/bases (Glu-388, Glu-512, and Glu-500 for ARAf, XYL4, and XYL1, respectively). Overlines and underlines indicate peptide sequencing with MALDI-TOF after proteolytic cleavage by trypsin for XYL4, XYL1, and ARAf, respectively.
On this basis, the molecular masses calculated from the sequences of XYL4 and XYL1 after removal of the NH2-terminal signal sequences are 79.9 and 80.2 kD, respectively (Fig. 4). These values are significantly higher than those obtained by SDS-PAGE (67 and 64 kD, respectively; Fig. 2). This discrepancy suggests that there is additional posttranslation processing of the primary translation product, by cleavage of one part of the peptide during enzyme maturation. Such a discrepancy between the apparent molecular masses obtained for the purified α-l-arabinofuranosidase and β-d-xylosidase from barley seedlings and those predicted from the cDNAs was recently reported and attributed to COOH-terminal processing (Lee et al., 2003). Therefore, we propose that a similar processing occurs at the COOH-terminal of XYL4 and XYL1 (Fig. 4). Such a deletion would result in final molecular masses of 64.7 and 64.6 kD, values which are in good agreement with those obtained from SDS-PAGE (Fig. 2).
Based on the same multiple sequences alignments obtained for the three enzymes from Arabidopsis with homolog enzymes from barley seedlings, the putative catalytic nucleophiles are predicted to be Glu-464, Asp-308, and Asp-296 for ARAf, XYL4, and XYL1, respectively. These alignments also suggest that catalytic amino acids are at positions Glu-388, Glu-512, and Glu-500 for ARAf, XYL4, and XYL1, respectively. These catalytic amino acid residues are strictly conserved in family 3 of glycoside hydrolases in the case of XYL1 and XYL4, and in family 51 of glycoside hydrolases in the case of ARAf (Henrissat, 1998; afmb.cnrs-mrs.fr). In addition, other common features of glycoside hydrolases from the family 3 are observed in the XYL4 and XYL1 sequences, including the conserved WGR and KH motifs, beginning at residues Trp-183 and Lys-224 for XYL4, and Trp-172 and Lys-212 for XYL1, respectively. These motifs are thought to be involved in substrate binding (Hrmova et al., 2001, 2002).
Finally, ARAf, XYL4, and XYL1 possess 4, 4, and 1 potential sites for N-glycosylation, respectively (Fig. 4). Consistent with this, the prediction of enzyme cellular localization by NetNGlyc software suggests that all three enzymes are localized in the extracellular matrix.
Hydrolysis of Xylopolysaccharides and Xylooligosaccharides by ARAf, XYL4, and XYL1
To determine whether the purified enzymes can degrade native plant polysaccharides, they were incubated with xylopolysaccharides, xylooligosaccharides, and arabinan, and the neoformed products were analyzed by HPAEC. The results obtained indicate that XYL4 released mainly d-Xyl from oat spelt xylan (OSX), wheat arabinoxylan (WAX), and rye arabinoxylan (RAX; Table IV). In addition, XYL4 released d-Xyl more efficiently than ARAf and XYL1 from oligoarabinoxylans that were produced by the hydrolysis of arabinoxylans (WAX and RAX; Table IV). These results further confirm that XYL4 possess enzymatic characteristics specific for a β-xylosidase. By contrast, ARAf and XYL1 hydrolyze d-Xyl less efficiently than XYL4 from these substrates (xylan, arabinoxylans, and oligoarabinoxylans) but hydrolyze l-Ara more efficiently. The highest production of l-Ara by ARAf and XYL1 was established using sugar beet arabinan (SBA) as the substrate. These results strongly suggest that ARAf and XYL1 can be considered as bifunctional α-L arabinofuranosidase/β-d-xylosidases.
Table IV.
Concentration of l-Ara and D-Xyl released by incubation of OSX, WAX, RAX, SBA, and oligoarabinoxylans with the purified ARAf, XYL1, and XYL4
| Substrates | Released Sugar | ARAf | XYL4 | XYL1 |
|---|---|---|---|---|
| nmol/0.5 mL | ||||
| Incubation with OSX, WAX, RAX, and SBA | ||||
| OSX | d-Xyl | 0.2 | 7.2 | 0.3 |
| l-Ara | ND | ND | ND | |
| WAX | d-Xyl | 10.9 | 36.4 | 21.0 |
| l-Ara | 2.9 | 1.0 | 5.2 | |
| RAX | d-Xyl | 13.4 | 131.7 | 19.8 |
| l-Ara | 1.2 | 0.7 | 2.1 | |
| SBA | d-Xyl | ND | ND | ND |
| l-Ara | 27.7 | 3.7 | 62.5 | |
| Incubation with Oligoarabinoxylan | ||||
| Wheat oligo-AX | d-Xyl | 36.8 | 52.5 | 54.8 |
| l-Ara | 9.8 | 4.6 | 9.8 | |
| Rye oligo-AX | d-Xyl | 25.7 | 35.0 | 40.4 |
| l-Ara | 5.9 | 1.9 | 6.1 |
The enzyme activities were probed by incubating the enzymes (0.1 μg) with OSX, WAX, RAX, SBA, and oligoarabinoxylan for 72 h at pH 5.0 at 37°C. The cleavage products were fractionated on HPAEC system (Dionex X500) equipped with a CarboPac PA-1 column combined with pulsed amperometric detection. Hydrolysis products of d-Xyl and l-Ara released during the incubation were quantified by the integration of peak areas in positions corresponding to retention times for d-Xyl and l-Ara and after calculation expressed as nmol/0.5 mL of l-Ara or d-Xyl. ND, Not detected; AX, arabinoxylan.
Analysis of the β-d-Xylosidase Activities at Different Stages of Stem Development
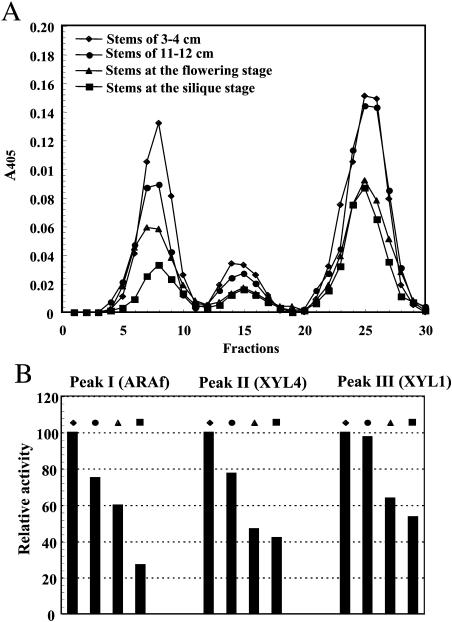
Stems at four different stages of development were analyzed to determine the potential changes in the levels of activity of the three enzymes. This analysis was performed using stems of 3 to 4 cm and 11 to 12 cm in height size and with stems at the flowering and at the silique stage of development. The protein extract from each sample was fractionated by CM-Trisacryl chromatography, and the fractions were assayed for β-d-xylosidase activity. The results obtained (Fig. 5A) demonstrated that all the tissues tested exhibited peaks of β-d-xylosidase activities corresponding to the positions of ARAf, XYL4, and XYL1 (Fig. 1, Step 2). The highest β-d-xylosidase activities were obtained in stems of 3 to 4 cm for each peak. By contrast, stems at the silique stage showed the smallest activities for all peaks. This higher β-d-xylosidase activity in young stems strongly suggests that the three enzymes might be regulated in relation to stem development.
Figure 5.
Chromatographic analysis of β-d-xylosidase activity at different stages of stem development. A, Elution profile of β-d-xylosidase activity after cation exchange chromatography on CM-Trisacryl at different stages of stem development. A 0.5-mL dialyzed cell-free stem tissue extract (1 mg of protein) was eluted as described in Figure 2. Aliquots of 100 μL were assayed for β-d-xylosidase activity in the presence of 2 mm pNPX. Analysis was performed with stems of 3 to 4 cm and of 11 to 12 cm in length, and with stems at the flowering stage and at the silique stages of development. B, Proportions of the three peaks of β-d-xylosidase activity at the different stages of stem development. Results are expressed as a percentage of the specific activity obtained in comparison with each peak in stems of 3 to 4 cm in length.
DISCUSSION
In this work, we have purified and characterized three enzymes that exhibit β-d-xylosidase activity from stem of Arabidopsis, a dicotyledonous plant. Until now, only the purification and partial characterization of one β-d-xylosidase from dicotyledonous plant has been described (Tezuka et al., 1993) for sycamore cells (Acer pseudoplantanus). In this study, we describe the purification, enzymatic properties, sequence identity, and action on various substrates of these β-d-xylosidases.
The purified enzymes have been designated as ARAf, XYL4, and XYL1. Examination of their substrate specificity and kinetic properties indicated that these three enzymes can hydrolyze pNPX and various natural substrates such as xylan, oligoxylans, arabinoxylans, and oligoarabinoxylans. In plants, only one β-d-xylosidase from young barley seedlings has been characterized and sequenced to date (Lee et al., 2003). This barley β-d-xylosidase (XYL) has pH optimum, Km, and molecular mass similar to the parameters that we determined in the case of ARAf, XYL4, and XYL1. Noteworthy, the optimum pH of these enzymes is close to that of the cell wall environment (Nari et al., 1986). XYL4 shows the highest substrate specificity characteristic of β-d-xylosidases, whereas examination of the substrate specificities and kinetic properties of ARAf and XYL1 showed that these enzymes have both α-l-arabinofuranosidase and β-d-xylosidase activities. Thus, enzymes exhibiting β-d-xylosidase activities in Arabidopsis can specifically hydrolyze substrates other than those normally acted upon by β-d-xylosidases. The three purified enzymes can hydrolyze the substrates pNPX, pNPAf, and pNPA to various degrees. Similar observations have been reported for barley β-d-xylosidases (Lee et al., 2003). This indicates that the activity of each of these enzymes is not restricted toward a single substrate but exhibits properties of glycosyl hydrolases having broad substrate specificity.
The cell wall of plant stems is composed of several heterogeneous polysaccharides, which are subjected to modifications during plant growth and development. Enzymes exhibiting hydrolytic activity toward polysaccharides are responsible for such structural changes since the degradation of polysaccharides allows the modification and restructuring of the cell wall. The broad substrate specificity of the glycosyl hydrolases studied here might correspond to a particular aptitude of the plant to degrade various saccharide residues.
As mentioned above, the identified enzymes not only exhibit β-d-xylosidase activity but also α-l-arabinofuranosidase and α-l-arabinopyranosidase activities. This suggests that they may participate in the degradation of xylan, arabinoxylan, and arabinan, which is related to the presence of these saccharides in the cell wall of Arabidopsis (Zablackis et al., 1995; Gardner et al., 2002). Here, we show that XYL4 releases Xyl from OSX, arabinoxylan (WAX and RAX), and oligoarabinoxylans (obtained from WAX and RAX) more efficiently than XYL1 and ARAf (Table IV). This observation supports the proposal that in vivo XYL4 could be involved in the hydrolysis of the xylan backbone. By contrast, ARAf and XYL1, which release both d-Xyl and l-Ara, could be involved mainly in debranching the xylan and arabinoxylan polymers, by hydrolyzing the α-l-arabinofuranoside side groups. In addition, the results obtained suggest that in vivo ARAf and XYL1 are involved in the hydrolysis of arabinan side chains of RG-I, a polysaccharide of the primary cell wall (Ridley et al., 2001; Willats et al., 2001; Glushka et al., 2003).
Arabidopsis antisense lines for XYL1 were reported to show alterations in plant development leading to a marked phenotype at the level of the leaf and silique shapes and a reduced height when compared to wild-type plants grown under the same growth conditions (Goujon et al., 2003). The cell walls of this antisense line show an increased susceptibility to hydrolysis by cellulases in vitro (data not shown). This suggested that XYL1 is an important enzyme for normal development and cell wall structure in Arabidopsis.
Two genes of Arabidopsis encoding putative l-α-arabinofuranosidases, AtASD1 and AtASD2, were identified, and expression analysis has been reported (Fulton and Cobbett, 2003). Both AtASD1 and AtASD2 show different expression patterns during Arabidopsis development suggesting that both may have multiple roles during development. High expression level of AtASD1 (encoding ARAf) is related to various stages of vegetative growth, such as morphogenesis, senescence, and abscission of floral organs.
In this work, we have observed that stems of 3 to 4 cm in height possess the highest activities for all the three enzymes in comparison to older Arabidopsis stems. In young stems, most of the cells are undergoing cell division and expansion. High enzyme activities are probably related to dynamic modifications of cell wall structure at this stage of development. Recently, it was shown that xyloglucan could determine the mechanical properties of the cell wall in azuki bean epicotyls (Kaku et al., 2002). In addition, wall maturation in wheat seedlings is characterized by a decreased degree of Ara substitution in arabinoxylan (Obel et al., 2002). Therefore, the three enzymes characterized here, which can contribute to polysaccharide modifications, could play an important role in regulating mechanical properties of the cell wall in Arabidopsis.
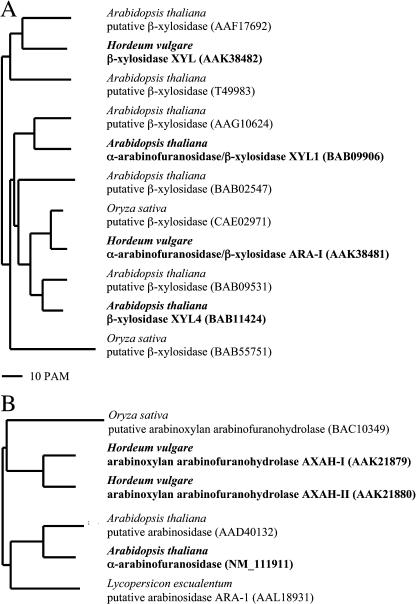
The phylogeny of proteins whose amino acid sequences are homologous to XYL4 and XYL1 is shown in Fig. 6A. Previously, biochemically characterized enzymes such as a β-d-xylosidase (XYL) and also a bifunctional α-l-arabinofuranosidase/β-d-xylosidase (ARA-I) (Lee et al., 2003) both from barley seeds were phylogenetically related to the Arabidopsis β-d-xylosidases. However, it appears that although all these enzymes share a high similarity (Fig. 6A), their substrate specificities are quite different. Five putative xylosidase genes that encode proteins that are phylogenetically related to XYL4 and XYL1 have been identified in the Arabidopsis genome, but their enzymatic properties have yet to be determined.
Figure 6.
Phylogenetic trees of plants on the basis of the purified enzymes and homologs. A, For XYL1 and XYL4. B, For ARAf. Branch lengths are drawn to scale. The GenBank accession numbers of the protein sequences are shown. Characterized enzymes are indicated in bold. The BLAST analysis were restricted to proteins from barley, rice (Oryza sativa), and Arabidopsis characterized until December 2003 and showing more than 40% sequence identities with purified enzymes.
A phylogeny of proteins homologous to ARAf is shown in Figure 6B. ARAf is related to two α-arabinoxylan arabinofuranohydrolases (AXAH-I and AXAH-II) characterized in barley seedlings (Lee et al., 2001). To date, only six genes encoding related proteins have been identified in plants. Amino acid sequence identities between barley seedling AXAH-I and AXAH-II and the Arabidopsis enzyme ARAf are 52% and 63%, respectively. It appears that ARAf and barley AXAH-I and AXAH-II belong to two phylogenetically distinct classes, characteristic of monocot and dicot plants (Fig. 6B). At present, only two genes have been identified in dicotyledonous plants, but the enzymatic properties of the corresponding proteins have not been determined and ARAf is the first α-l-arabinofuranosidase characterized in a dicotyledonous plant.
Based on amino acid sequence similarities, β-d-xylosidases from various sources are currently divided into five families, namely 3, 39, 43, 52, and 54 (Henrissat, 1998). Members of each family exhibit characteristic substrate specificities, reaction mechanisms, and three-dimensional structures (Henrissat, 1998). Plant β-d-xylosidases, XYL4, XYL1, and barley β-d-xylosidase (XYL) belong to family 3 of glycoside hydrolases (Henrissat, 1998; afmb.cnrs-mrs.fr). α-l-Arabinofuranosidases from various sources are classified in the glycoside hydrolase families 3, 43, 51, 54, and 62 (Henrissat, 1998). ARAf and the three α-l-arabinofuranosidases expressed in barley seeds belong to families 3 (AXAH-I, AXAH-II) and 51 (ARA-I). Thus, it appears that all characterized and sequenced enzymes that exhibit β-d-xylosidase activities in plants belong only to families 3 and 51. However, the small number of enzymes characterized so far does not allow extensive generalization.
In conclusion, three novel enzymes from Arabidopsis stems, which possess β-d-xylosidase activity, were purified and characterized. All three enzymes can hydrolyze various natural oligosaccharides and polysaccharides and probably play a significant role in determining plant cell wall structure since their level of activity changes during plant development. In our laboratory, Arabidopsis T-DNA knockout mutants for the genes encoding these enzymes are currently being analyzed to determine the role of these enzymes in plant physiology and cell wall structure and composition.
MATERIALS AND METHODS
Plant Material
Wild-type Arabidopsis, Wassilewskija ecotype, was grown in the greenhouse at 20°C to 22°C with a 16-h photoperiod at 150 μE m−2 s−1. Stem tissues at the flowering stage (9–13 cm in height) were used for protein purification. For analysis of β-xylosidase activities at different stages of plant development, the enzyme assays were performed on extracts from young elongating stems (3–4 cm and 11–12 cm), from stems at the flowering stage (14–15 cm), and from mature stems at the silique stage (20–24 cm).
Chemicals
pNP-glycoside substrates, d-Xyl, l-Ara, pNP, and OSX, were purchased from Sigma (St. Louis). Xylobiose, xylotetraose, WAX, RAX, and SBA were purchased from Megazyme International (Bray, Ireland).
Preparation of Protein Extracts from Stems of Arabidopsis
Stem tissues from Arabidopsis at the flowering stage were used for analysis. Approximately 2 g of stems were suspended in 2 mL of ice-cold extraction buffer and blended for 5 min. The extraction buffer consisted of 25 mm BisTris, pH 7.0, 200 mm CaCl2, 10% (v/v) glycerol, 4 μm Na-cacodylate, and 1/200 (v/v) protease inhibitor cocktail (P-9599; Sigma). The ground and suspended material was centrifuged twice at 4°C for 3 min at 10,000g, and the supernatant was additionally centrifuged for 15 min at 17,000g. The resulting supernatant was used for chromatographic analyses.
Purification of Enzymes Exhibiting β-d-Xylosidase Activity
The protein extract obtained from the stems of Arabidopsis was used for purification of β-d-xylosidases. The protein purification was performed in three steps as described below.
Step 1: Lectin chromatography. A 0.5- × 3-cm column was filled with 1 mL of Con A Sepharose (Sigma) and washed with 3 mL of 20 mm Tris-HCl and 0.5 m NaCl buffer (pH 7.4). The soluble protein extract was added and then washed with 10 mL of this buffer at a flow rate of 5 mL/h. Proteins were eluted with 0.2 m methyl-α-glucopyranoside in the same buffer. The eluates were collected, and 50-μL samples from each fraction were tested for β-d-xylosidase activity as described below.
Step 2: Cation-exchange chromatography. The pooled fractions were concentrated and equilibrated in 25 mm Na-acetate buffer (pH 5.0) containing 5% (v/v) glycerol and 0.015% (w/v) Triton X-100 (pH 5.0), and loaded on a CM-Trisacryl (Amersham, Buckinghamshire, UK) cation-exchange column (1.5 × 5 cm; Sigma). Proteins were eluted with the same buffer, followed by a 0.0 to 0.5 m NaCl discontinuous gradient at a flow rate of 20 mL/h. A loss of activity was observed in absence of detergent. Thus, to prevent enzymatic inactivation by low ionic strength of the buffer, Triton X-100 (0.015%) was added to the solution. In this way, enzyme activities were stable for approximately 3 weeks at temperature of 4°C to 8°C. One-milliliter fractions were collected and assayed for β-d-xylosidase activity. Peak fractions showing β-d-xylosidase activity were pooled and used in the third step of purification.
Step 3: Gel-filtration chromatography. The pooled and concentrated fractions showing β-d-xylosidase activities were fractionated by FPLC (Pharmacia, Piscataway, NJ) on a Superdex 200 HR10/30 column (Amersham Pharmacia Biotech, Uppsala) precalibrated with the following markers of known molecular mass: thyroglobuline (670 kD), bovine gamma globulin (158 kD), chicken ovalbumin (44 kD), equine myoglobin (17 kD), and vitamin B-12 (1.35 kD). Equilibration and elution were performed at room temperature with 20 mm Na-acetate buffer (pH 5.0), containing 150 mm NaCl and 0.015% (w/v) Triton X-100. Fractions of 0.4 mL were collected at a flow rate of 0.5 mL/min, and 100 μL of each fraction was assayed for β-d-xylosidase activity. Fractions exhibiting β-d-xylosidase activities were pooled, concentrated (150 μL), and dialyzed against 20 mm Na-acetate buffer, pH 5.0, in the presence of 5% (v/v) glycerol.
SDS-PAGE
Protein-denaturing SDS-PAGE was carried out using 10% polyacrylamide gels (Laemmli, 1970). Standard markers (BenchMark; Invitrogen, Carlsbad, CA) were used to approximately determine the molecular masses of purified proteins in gels stained with Coomassie Brilliant Blue R-250.
Identification of Proteins by Mass Spectroscopy
The individual protein bands obtained after SDS-PAGE were excised and in-gel digested with trypsin according to conditions for loading and elution described by Santoni et al. (2003). Tryptic peptides from each protein were analyzed by MALDI-TOF mass spectroscopy on a REFLEX III instrument (Bruker Instruments, Billerica, MA). Finally, proteins were identified using MS-Fit (http://prospector.ucsf.edu/ucsfhtml4.0/msfit.htm).
β-d-Xylosidase Activity
The reaction mixture contained 2 mm pNPX (Sigma), 0.1 m acetate buffer (pH 5.0), 2 mm sodium azide, and 50 to 100 μL of protein extract in a total volume of 0.5 mL. The reaction was carried out at 37°C for 60 min and stopped by the addition of 0.5 mL of 0.4 m sodium bicarbonate to the assay mixture. Concentration of the resulting pNP was determined spectrophotometrically at 405 nm, and its amount was estimated from a calibration curve. Specific activity was expressed as the amount of protein required to release 1 nmol/min of d-Xyl. The activity toward other pNP-glycosides was determined as described above for β-d-xylosidase.
Preparation of Arabinoxylan Oligosaccharides
The hydrolysis of 100 mg of RAX and WAX was performed in 10 mL of 10 mm NaOAc buffer, pH 5.5, using 200 units of endo-(1→4)-β-d-xylanase (M6 Xylanase, lot 80201; Megazyme International) at 28°C for 16 h. To remove polymeric materials, hydrolysates were precipitated with 4 volumes of cold ethanol. After centrifugation, the oligomers present in the supernatants were freeze-dried and solubilized in 1 mL of water.
Hydrolysis of Polysaccharides and Oligosaccharides by the Purified Enzymes
The assay was based on the liberation of d-Xyl and l-Ara from xylo-oligosaccharides and insoluble xylo-polysaccharides. The capacity of the purified enzymes to hydrolyze polysaccharides was determined by incubation of 0.5% (w/v) OSX (Sigma), WAX, RAX, and SBA at 37°C for 72 h. Hydrolysis of oligoarabinoxylans were performed in the same way as for the polysaccharides but using 100 μL of the prepared oligosaccharides as described above. The reaction was stopped by heating the assay mixture at 100°C for 3 min. Controls were stopped at time 0. The mixture was then centrifuged at 10,000g for 15 min and supernatants used for analysis.
The cleavage products were fractionated using a HPAEC system (Dionex X500; Sunnyvale, CA), equipped with a CarboPac PA-1 column (Dionex), and combined with pulsed amperometric detection. Degradation products were quantified by the integration of peak areas. The concentrations of released l-Ara and d-Xyl were determined from a calibration curve. Samples (20 μL) were eluted at 1 mL/min with the following NaOAc gradients in 100 mm NaOH conditions: (1) for xylobiose and xylotetraose digests: 0 to 5 min, isocratic initial condition 0 mm NaOAc; 5 to 20 min, linear gradient of 0 to 50 mm NaOAc; 20 to 22 min, linear gradient 50 to 100 mm NaOAc. The column was then washed with 0.1 m NaOAc in 100 mm NaOH for 3 min and equilibrated for 5 min with 100 mm NaOH. (2) For xylan enzymatic digests: 0 to 5 min, isocratic initial condition 0 mm NaOAc; 5 to 20 min, linear gradient of 0 to 50 mm NaOAc; 20 to 42 min, linear gradient 50 to 300 mm NaOAc, isocratic step 300 mm NaOAc; 42 to 45 min. Each elution was followed by a wash with 1 m NaOAc in 100 mm NaOH for 5 min and subsequent equilibration for 5 min with 100 mm NaOH. The retention times were 4.1, 5.5, 10, 15.9, and 19.2 for l-Ara, d-Xyl, xylobiose, xylotriose, and xylotetraose, respectively.
pH and Temperature Profiles
The determination of temperature dependence was carried at pH 5.0 as described above for the β-xylosidase assay, except that the temperature ranged from 30°C to 70°C. For the determination of pH optimum, the temperature was 37°C and the pH varied from 4.0 to 8.0 in 100 mm acetate citrate buffer.
Kinetic Analyses
Kinetic parameters of purified enzymes were determined for the substrate pNPX in a concentration range of 0.05 to 4.0 mm. Assays were performed in 100 mm sodium acetate buffer, pH 5.0, containing 0.015% (w/v) Triton X-100. Triton X-100 was added to the buffer to prevent enzymatic inactivation during kinetic analysis. sd values for assays were less than 5%. Kinetic data were processed using Kaleidagraph program (Synergy Software, Reading, PA) based on Michaelis-Menten enzyme kinetics (Atkins and Nimmo, 1980).
Bioinformatics Analyses
Sequences were aligned using the FASTA program (http://fasta.bioch.virginia.edu/fasta/align.htm). Cleavage and glycosidase sites of deduced proteins were studied using PSORT and NetNGlyc software (Nielsen et al., 1997; Nakai and Horton, 1999) at http://psort.nibb.ac.jp/form.html and http://www.cbs.dtu.dk/services/NetNGlyc/. The analysis to predict cellular enzyme localization was performed using PSORT software (http://psort.nibb.ac.jp/form.html). Multiple sequence alignments and the resulting phylogenetic tree were generated at http://prodes.toulouse.inra.fr/multalin/multalin.html (Corpet, 1988).
Protein Measurements
Protein concentrations were determined according to the Coomassie Brilliant Blue method using bovine serum albumin as a standard (Bradford, 1976).
Acknowledgments
We thank Profs. Guy Hervé, Elita Pastra Landis, and Leslie Mac Carty for reading and improving this manuscript, and Christian Malosse and Martine Gonneau for the mass spectrometric analyses.
This work was partly supported by the Genoplante Program Af2001–009. Z.M. was funded by an Institut National de la Recherche Agronomique grant.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.041269.
References
- Atkins GL, Nimmo IA (1980) Currents trends in the estimation of Michaelis-Menten parameters. Anal Biochem 104: 1–9 [DOI] [PubMed] [Google Scholar]
- Bachmann SL, McCarthy AJ (1989) Purification and characterization of a thermostable beta-xylosidase from Thermomonospora fusca. J Gen Microbiol 135: 293–299 [Google Scholar]
- Banik M, Li CD, Langridge P, Fincher GB (1997) Structure, hormonal regulation, and chromosomal location of genes encoding barley (1-4)-beta-xylan endohydrolases. Mol Gen Genet 253: 599–608 [DOI] [PubMed] [Google Scholar]
- Beg QK, Kapoor M, Mahajan L, Hoondal GS (2001) Microbiol xylanases and their industrial applications: a review. Appl Microbiol Biotechnol 56: 326–338 [DOI] [PubMed] [Google Scholar]
- Bewley JD, Burton RA, Morohashi Y, Fincher GB (1997) Molecular cloning of a cDNA encoding a (1-4)-beta-mannan endohydrolase from the seeds of germinated tomato (Lycopersicon esculentum). Planta 203: 454–459 [DOI] [PubMed] [Google Scholar]
- Beylot MH, McKie VA, Voragen AG, Doeswijk-Voragen CH, Gilbert HJ (2001) The Pseudomonas cellulosa glycoside hydrolase family 51 arabinofuranosidase exhibits wide substrate specificity. Biochem J 358: 607–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Carpita NC (1996) Structure and biogenesis of the cell walls of grasses. Annu Rev Plant Physiol Plant Mol Biol 47: 445–476 [DOI] [PubMed] [Google Scholar]
- Caspers MP, Lok F, Sinjorgo KM, van Zeijl MJ, Nielsen KA, Cameron-Mills V (2001) Synthesis, processing and export of cytoplasmic endo-beta-1,4-xylanase from barley aleurone during germination. Plant J 26: 191–204 [DOI] [PubMed] [Google Scholar]
- Chinen I, Oouchi K, Tamaki H, Fukuda N (1982) Purification and properties of thermostable beta-xylosidase from immature stalks of Saccharum officinarum L. (sugar cane). J Biochem (Tokyo) 92: 1873–1881 [DOI] [PubMed] [Google Scholar]
- Cleemput G, Hessing M, Van Oort M, Deconynck M, Delcour JA (1997) Purification and characterization of a β-d-xylosidase and endo-xylanase from wheat flour. Plant Physiol 113: 377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16: 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ (1997) Assembly and enlargement of the primary cell wall in plants. Annu Rev Cell Dev Biol 13: 171–201 [DOI] [PubMed] [Google Scholar]
- Coughlan MP, Hazlewood GP (1993) beta-1,4-D-Xylan-degrading enzyme systems: biochemistry, molecular biology and applications. Biotechnol Appl Biochem 17: 259–289 [PubMed] [Google Scholar]
- Farooqi M, Saleemuddin M, Ulber R, Sosnitza P, Scheper T (1997) Bioaffinity layering: a novel strategy for the immobilization of large quantities of glycoenzymes. J Biotechnol 55: 171–179 [DOI] [PubMed] [Google Scholar]
- Fulton LM, Cobbett CS (2003) Two alpha-L-arabinofuranosidase genes in Arabidopsis thaliana are differentially expressed during vegetative growth and flower development. J Exp Bot 54: 2467–2477 [DOI] [PubMed] [Google Scholar]
- Gardner SL, Burrell MM, Fry SC (2002) Screening of Arabidopsis thaliana stems for variation in cell wall polysaccharides. Phytochemistry 60: 241–254 [DOI] [PubMed] [Google Scholar]
- Glushka JN, Terrell M, York WS, O'Neill MA, Gucwa A, Darvill AG, Albersheim P, Prestegard JH (2003) Primary structure of the 2-O-methyl-alpha-L-fucose-containing side chain of the pectic polysaccharide, rhamnogalacturonan II. Carbohydr Res 338: 341–352 [DOI] [PubMed] [Google Scholar]
- Gorbacheva IV, Rodionova NA (1977. a) Studies on xylan degrading enzymes. I. Purification and characterization of endo-1,4-beta-xylanase from Aspergillus niger str. 14. Biochim Biophys Acta 484: 79–93 [DOI] [PubMed] [Google Scholar]
- Gorbacheva IV, Rodionova NA (1977. b) Studies on xylan-degrading enzymes. II. Action pattern of endo-1,4-beta-xylanase from Aspergillus niger str. 14 on xylan and xylooligosaccharides. Biochim Biophys Acta 484: 94–102 [DOI] [PubMed] [Google Scholar]
- Goujon T, Minic Z, El Amrani A, Lerouxel O, Aletti E, Lapierre C, Joseleau JP, Jouanin L (2003) AtBXL1, a novel higher plant (Arabidopsis thaliana) putative beta-xylosidase gene, is involved in secondary cell wall metabolism and plant development. Plant J 33: 677–690 [DOI] [PubMed] [Google Scholar]
- Handford MG, Baldwin TC, Goubet F, Prime TA, Miles J, Yu X, Dupree P (2003) Localisation and characterisation of cell wall mannan polysaccharides in Arabidopsis thaliana. Planta 218: 27–36 [DOI] [PubMed] [Google Scholar]
- Henrissat B (1998) Glycosidase families. Biochem Soc Trans 26: 153–156 [DOI] [PubMed] [Google Scholar]
- Heredia A, Jimenez A, Guillen R (1995) Composition of plant cell walls. Z Lebensm Unters Forsch 200: 24–31 [DOI] [PubMed] [Google Scholar]
- Hrmova M, De Gori R, Smith BJ, Fairweather JK, Driguez H, Varghese JN, Fincher GB (2002) Structural basis for broad substrate specificity in higher plant β-d-glucohydrolases. Plant Cell 14: 1033–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrmova M, Varghese JN, De Gori R, Smith BJ, Driguez H, Fincher GB (2001) Catalytic mechanisms and reaction intermediates along the hydrolytic pathway of a plant beta-D-glucan glucohydrolase. Structure 9: 1005–1016 [DOI] [PubMed] [Google Scholar]
- John M, Schmidt J (1988) Xylanases and beta-xylosidase of Trichoderma lignorum. Methods Enzymol 160: 662–671 [Google Scholar]
- Kaku T, Tabuchi A, Wakabayashi K, Kamisaka S, Hoson T (2002) Action of xyloglucan hydrolase within the native cell wall architecture and its effect on cell wall extensibility in Azuki bean epicotyls. Plant Cell Physiol 43: 21–26 [DOI] [PubMed] [Google Scholar]
- Kelly MA, Sinnott ML, Herrchen M (1987) Purification and mechanistic properties of an extracellular alpha-L-arabinofuranosidase from Monilinia fructigena. Biochem J 245: 843–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni N, Shendye A, Rao M (1999) Molecular and biotechnological aspect of xylanases. FEMS Microbiol Rev 23: 411–456 [DOI] [PubMed] [Google Scholar]
- Kumar S, Ramon D (1996) Purification and regulation of the synthesis of a beta-xylosidase from Aspergillus nidulans. FEMS Microbiol Lett 135: 287–293 [Google Scholar]
- Lachke AH (1988) 1,4-beta-D-xylan xylohydrolase of Sclerotium rolfstii. Methods Enzymol 160: 679–684 [Google Scholar]
- Laemmli UK (1970) Cleavage of the structural proteins during the assembly of the head of the bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lee RC, Burton RA, Hrmova M, Fincher GB (2001) Barley arabinoxylan arabinofuranosidases: purification, characterization and determination of primary structures from cDNA clones. Biochem J 356: 181–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Hrmova M, Burton RA, Lahnstein J, Fincher GB (2003) Bifunctional family 3 glycoside hydrolases from barley with α-L-arabinofuranosidase and β-D-xylosidase activity. J Biol Chem 278: 5377–5387 [DOI] [PubMed] [Google Scholar]
- Matsuo M, Yasui T (1988) beta-xylosidases of several fungi. Methods Enzymol 160: 684–694 [Google Scholar]
- Nakai K, Horton P (1999) PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci 24: 34–36 [DOI] [PubMed] [Google Scholar]
- Nanmori T, Watanabe T, Shinke R, Kohno A, Kawamura Y (1990) Purification and properties of thermostable xylanase and beta-xylosidase produced by a newly isolated Bacillus stearothermophilus strain. J Bacteriol 172: 6669–6672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nari J, Noat G, Diamantidis G, Woudstra M, Ricard J (1986) Electrostatic effects and the dynamics of enzyme reactions at surface of plant cells. 3. Interplay between limited cell-wall autolysis, pectin methyl esterase activity and electrostatic effect in soybean cell walls. Eur J Biochem 155: 199–202 [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, Heijne G (1997) A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int J Neural Syst 8: 581–599 [DOI] [PubMed] [Google Scholar]
- Obel N, Porchia AC, Scheller HV (2002) Dynamic changes in cell wall polysaccharides during wheat seedling development. Phytochemistry 60: 603–610 [DOI] [PubMed] [Google Scholar]
- O'Neill RA, Albersheim P, Darvill AG (1989) Purification and characterization of a xyloglucan oligosaccharide-specific xylosidase from pea seedlings. J Biol Chem 264: 20430–20437 [PubMed] [Google Scholar]
- Popper ZA, Fry SC (2003) Primary cell wall composition of bryophytes and charophytes. Ann Bot (Lond) 91: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prade RA (1996) Xylanases: from biology to biotechnology. Biotechnol Genet Eng Rev 13: 101–131 [DOI] [PubMed] [Google Scholar]
- Rahman AK, Sugitani N, Hatsu M, Takamizawa K (2003) A role of xylanase, α-l-arabinofuranosidase, and xylanase in xylan degradation. Can J Microbiol 49: 58–64 [DOI] [PubMed] [Google Scholar]
- Reiter WD (2002) Biosynthesis and properties of the plant cell wall. Curr Opin Plant Biol 5: 536–542 [DOI] [PubMed] [Google Scholar]
- Renard CM, Lomax JA, Boon JJ (1992) Apple-fruit xyloglucans: a comparative study of enzyme digests of whole cell walls and their alkali-extracted xyloglucans. Carbohydr Res 232: 303–320 [DOI] [PubMed] [Google Scholar]
- Ridley BL, O'Neill MA, Mohnen D (2001) Pectins: structure, biosynthesis, and oligogalacturonide related signaling. Phytochemistry 57: 929–967 [DOI] [PubMed] [Google Scholar]
- Saha BC (2003) Purification and properties of an extracellular β-xylosidase from a newly isolated Fusarium proliferatum. Bioresour Technol 90: 33–38 [DOI] [PubMed] [Google Scholar]
- Santoni V, Vinh J, Pflieger D, Sommerer N, Maurel C (2003) A proteomic study reveals novel insights into the diversity of aquaporin forms expressed in the plasma membrane of plant roots. Biochem J 373: 289–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon PS, Keen JN, Bowles DJ (1998) Purification and characterization of N-glycanase, a concanavalin-A binding protein from jackbean (Canavalia ensiformis). Biochem J 330: 13–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade AM, Hoj PB, Morrice NA, Fincher GB (1989) Purification and characterization of three (1-4)-beta-D-xylan endohydrolases from germinated barley. Eur J Biochem 185: 533–539 [DOI] [PubMed] [Google Scholar]
- Stolle-Smits T, Beekhuizen JG, Kok MT, Pijnenburg M, Recourt K, Derksen J, Voragen AG (1999) Changes in cell wall polysaccharides of green bean pods during development. Plant Physiol 121: 363–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniyan S, Prema P (2000) Cellulase-free xylanases from Bacillus and other microorganisms. FEMS Microbiol Lett 183: 1–7 [DOI] [PubMed] [Google Scholar]
- Subramaniyan S, Prema P (2002) Biotechnology of microbial xylanases: enzymology, molecular biology, and application. Crit Rev Biotechnol 22: 33–64 [DOI] [PubMed] [Google Scholar]
- Sunna A, Antranikian G (1997) Xylanolytic enzymes from fungi and bacteria. Crit Rev Biotechnol 17: 39–67 [DOI] [PubMed] [Google Scholar]
- Suzuki M, Kato A, Nagata N, Komeda Y (2002) A xylanase, AtXyn1, is predominantly expressed in vascular bundles, and four putative xylanase genes were identified in the Arabidopsis genome. Plant Cell Physiol 43: 759–767 [DOI] [PubMed] [Google Scholar]
- Tezuka K, Hayashi M, Ishihara H, Nishimura M, Onozaki K, Takahashi N (1993) Purification and substrate specificity of beta-xylosidase from sycamore cell (Acer pseudoplatanus L.): application for structural analysis of xylose-containing N-linked oligosaccharides. Anal Biochem 211: 205–209 [DOI] [PubMed] [Google Scholar]
- Tuncer M, Ball AS (2003) Co-operative actions and degradation analysis of purified xylose-degrading enzymes from Thermonospora fusca BD25 on spelt xylan. J Appl Microbiol 94: 1030–1105 [DOI] [PubMed] [Google Scholar]
- Willats WG, McCartney L, Mackie W, Knox JP (2001) Pectin: cell biology and prospects for functional analysis. Plant Mol Biol 47: 9–27 [PubMed] [Google Scholar]
- Yamaura I, Koga T, Matsumoto T, Kato T (1997) Purification and some properties of endo-1,4-beta-D-xylanase from a fresh-water mollusk, Pomacea insularus (de Ordigny). Biosci Biotechnol Biochem 61: 615–620 [DOI] [PubMed] [Google Scholar]
- Zablackis E, Huang J, Muller B, Darvill AG, Albersheim P (1995) Characterization of the cell-wall polysaccharides of Arabidopsis thaliana leaves. Plant Physiol 107: 1129–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]