Abstract
Two-component signaling systems, involving His kinases, His-containing phosphotransfer proteins, and response regulators, have been implicated in plant responses to hormones and environmental factors. Genomic analysis of Arabidopsis supports the existence of 22 response regulators (ARRs) that can be divided into at least two distinct groups designated type-A and type-B. Phylogenetic analysis indicates that the type-B family is composed of one major and two minor subfamilies. The expression of the type-B ARRs was examined by using both reverse transcription-PCR and β-glucuronidase fusion constructs. The major subfamily of type-B ARRs showed particularly high expression in regions where cytokinins play a significant role, including cells in the apical meristem region and in young leaves that would be undergoing cell division. Multiple members within this same subfamily of type-B ARRs were expressed near the root tip with highest expression in the root elongation zone. β-Glucuronidase-fusions to full-length ARR2, ARR12, and ARR19 were nuclear localized, consistent with a role in transcriptional regulation. These data suggest that differing expression levels of the type-B ARRs may play a role in modulating the cellular responses to cytokinin.
Plant two-component signaling systems have been implicated in vital cellular processes such as the responses to cytokinins, ethylene, red light, and osmosensing (Schaller, 2000; Hutchison and Kieber, 2002; Hwang et al., 2002; Schaller et al., 2002). Two-component systems were originally identified in bacteria, and in their simplest form involve a receptor kinase that autophosphorylates itself on a conserved His residue in response to an environmental stimulus (Mizuno, 1997; Stock et al., 2000). This phosphate is then transferred to a conserved Asp residue within the receiver domain of a response regulator. Phosphorylation of the response regulator modulates its ability to mediate downstream signaling in the pathway. Of particular relevance to plants is a permutation on the two-component system known as the multi-step phosphorelay (Swanson et al., 1994; Schaller, 2000). The multi-step phosphorelay makes use of three components: a hybrid receptor kinase that contains both His-kinase and receiver domains in one protein, a His-containing phosphotransfer (HPt) protein, and a separate response regulator. In these multi-step phosphorelays the phosphate is transferred from amino acid to amino acid in sequence His to Asp to His to Asp.
In Arabidopsis, proteins with homology to all elements of the two-component system have been identified, including His kinases, response regulators, and HPt proteins (Schaller, 2000, 2002; Hutchison and Kieber, 2002; Hwang et al., 2002). Phosphorylation activity has been confirmed in each case. Thus, all elements needed to establish a histidyl-aspartyl phosphorelay are represented in plants. Analysis of the Arabidopsis genome reveals the existence of 8 His kinases, 22 response regulators, and 5 HPt proteins that contain all the conserved residues required for enzymatic activity (Hutchison and Kieber, 2002; Hwang et al., 2002; Schaller et al., 2002). The Arabidopsis response regulators (ARRs) can be classified into at least two distinct groups based on domain structure and sequence: type-A and type-B (Imamura et al., 1999). The type-A response regulators are relatively small, containing the receiver domain common to response regulators along with short N- and C-terminal extensions. The type-B response regulators each have a large C-terminal extension following the receiver domain and, based on several lines of evidence, act as transcriptional regulators (Sakai et al., 2000; Lohrmann et al., 2001; Hosoda et al., 2002).
Two-component signaling elements have been clearly implicated in cytokinin signal transduction (Hwang and Sheen, 2001; Hutchison and Kieber, 2002; Heyl and Schmülling, 2003; Kakimoto, 2003). The cytokinin receptor family of Arabidopsis is composed of three hybrid His-kinases, CRE1 (also known as WOL1 and AHK4), AHK2, and AHK3 (Inoue et al., 2001; Suzuki et al., 2001; Ueguchi et al., 2001), with all members of the family reported to directly bind cytokinins (Yamada et al., 2001; Kakimoto, 2003). A knockout mutation in the type-B response regulator ARR1 results in decreased sensitivity to cytokinin in shoot regeneration and root elongation assays (Sakai et al., 2001). Overexpression of either ARR1 or ARR2 in Arabidopsis plants results in increased sensitivity to cytokinin (Hwang and Sheen, 2001; Sakai et al., 2001). The type-A response regulators are all induced, to varying levels and with varying kinetics, by cytokinin (Brandstatter and Kieber, 1998; Hutchison and Kieber, 2002). This cytokinin-dependent induction of the type-A response regulators is at least partially dependent upon transcriptional regulation by type-B response regulators (Hwang and Sheen, 2001; Sakai et al., 2001). These data are consistent with a model in which the initial steps of cytokinin signal transduction are mediated by a multi-step phosphorelay, involving cytokinin receptors, AHPs, and type-B ARRs. These relay the cytokinin signal from membrane to nucleus, where the type-B ARRs then induce transcription of the type-A genes. The type-A response regulators may mediate downstream responses to cytokinin and/or act as negative regulators of the initial signal transduction pathway (Hwang and Sheen, 2001; Hutchison and Kieber, 2002; Kakimoto, 2003).
According to this model, type-B response regulators play a pivotal role in the early response of plants to cytokinin. None, however, were previously identified in screens for mutants with altered cytokinin sensitivity. Through use of reverse genetic approaches, a T-DNA insertion mutant in ARR1 was shown to shift but not eliminate the cytokinin sensitivity of the mutant plant (Sakai et al., 2001). These observations are consistent with functional redundancy for cytokinin signal transduction among the type-B ARRs. We hypothesized that such functional redundancy might be revealed through overlapping expression patterns within this gene family. Here we take a unified approach to examine the phylogeny and the expression of all 11 type-B ARRs. Our results are consistent with a role for some members of the type-B ARRs family in cytokinin signaling. Our results also suggest that the expression level of type-B ARRs in particular cells may play an important role in modulating signal output and thus the responsiveness of these cells to cytokinin.
RESULTS
The Type-B ARR Gene Family of Arabidopsis
Analysis of the Arabidopsis genome reveals the presence of 22 response regulators with all the residues required for activity (Schaller et al., 2002). Two major families of response regulators have been distinguished: the type-A and the type-B response regulators (Imamura et al., 1999). Both type-A and type-B response regulators contain a receiver domain. However, the type-B ARRs also contain a large C-terminal extension with a Myb-like DNA binding domain referred to as the GARP domain (Fig. 1A). The GARP domain is a motif specific to plant transcription factors and is named after GOLDEN2 from maize (Zea mays), ARRs from Arabidopsis, and Psr1 from Chlamydomonas (Imamura et al., 1999; Hosoda et al., 2002). Also present in Arabidopsis are nine pseudo response regulators (APRRs), so called because they lack the conserved Asp that is phosphorylated in the receiver domain (Makino et al., 2000).
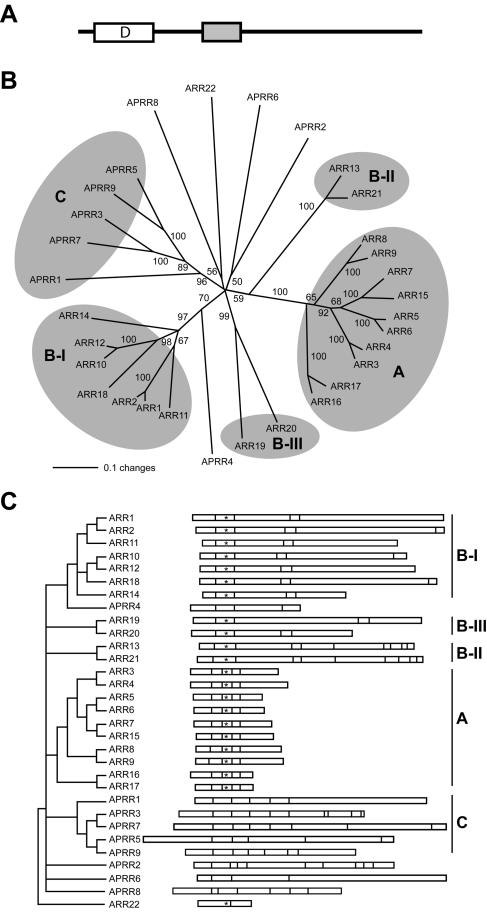
Figure 1.
Phylogenetic analysis of type-B ARRs. A, Structural features of Type-B response regulators. The receiver domain, with the conserved Asp residue that gets phosphorylated (D), is indicated by a white box. The GARP domain is indicated by a gray box. B, Phylogenetic relationship among response regulators and pseudo response regulators based upon amino acid sequence of the receiver domain. Numbers indicate percentage of bootstrapped replicates that give the same branch after 1,000 iterations. The type-A response regulators (A), subfamilies of type-B response regulators (B-I, B-II, and B-III), and circadian clock-involved pseudo response regulators (C) are highlighted. C, Distribution of introns and exons among response regulators and pseudo response regulators. Vertical lines within each box represent intron positions within the coding sequence for each ARR. The genes are aligned around the site of phosphorylation (*). A different view of the same phylogenetic tree shown in B, with branches below 60% bootstrap support collapsed, is included to show family groupings. Branch lengths of the phylogenetic tree are not to scale.
An unrooted phylogenetic tree based on the receiver domain was constructed using bootstrap analysis (Fig. 1B). The 10 type-A response regulators all fall on one branch. The type-B response regulators form three distinct subfamilies. The largest subfamily, hereafter referred to as subfamily I, contains ARR1, ARR2, ARR10, ARR11, ARR12, ARR14, and ARR18. Notably, this subfamily contains ARR1, ARR2, and ARR11 that have been previously implicated in cytokinin responses based on loss-of-function and/or overexpression studies (Hwang and Sheen, 2001; Sakai et al., 2001; Imamura et al., 2003). A second subfamily (subfamily II) of the type-B response regulators is composed of ARR13 and ARR21. A third subfamily (subfamily III) of the type-B response regulators is composed of ARR19 and ARR20. The response regulator ARR22 does not group with members of the type-A or type-B families, and previous analysis indicates that it is more closely related to the receiver domains of the hybrid His kinases than to those of the other Arabidopsis response regulators (Schaller et al., 2002). Among the pseudo response regulators, APRR1, 3, 5, 7, and 9, which are implicated in the circadian clock mechanism (Matsushika et al., 2000; Strayer et al., 2000; Makino et al., 2001), all fall on one branch of the phylogenetic tree. In contrast, APRR4, which contains a GARP domain (Schaller et al., 2002), is as closely related to subfamily I of the type-B ARRs as it is to any of the pseudo response regulators.
Analysis of intron positions (Fig. 1C) supports the phylogenetic analysis of the Arabidopsis response regulators. The type-A response regulators all contain four introns, similarly positioned, within their receiver domains. In contrast, the type-B response regulators contain only two introns, similarly positioned, within their receiver domains. Most of the type-B response regulators, including all members of subfamily I, contain an additional two introns of similar position in their C-terminal extensions. ARR13 and 21 of subfamily II share intron positions in their C-terminal extensions not found in the other type-B subfamilies. Members of the circadian subfamily of pseudo response regulators contain two shared intron positions within their receiver domains and a third conserved intron position (lacking in the type-A and type-B ARRs) just C-terminal to the receiver domain.
Expression of Type-B ARRs Based on Reverse Transcription-PCR
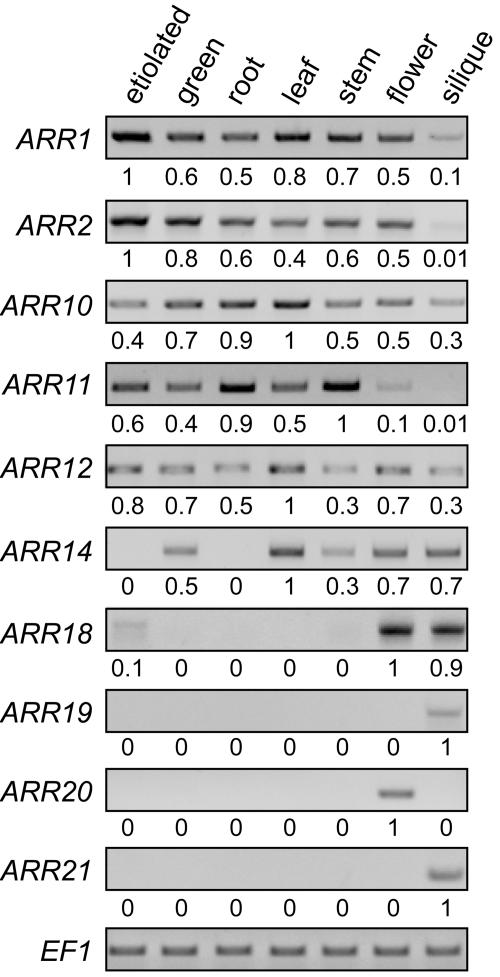
To obtain an overview of ARR expression throughout the plant, RNA was extracted from Arabidopsis, reverse transcribed, and the resultant cDNA analyzed for the relative levels of each type-B ARR by PCR (Fig. 2). Expression was examined in whole seedlings grown in the light or in the dark. This provided a means to assess whether there were any apparent effects of light upon expression of the ARR genes. In addition, one could potentially detect expression in whole seedlings that might be missed when only examining isolated tissues. This analysis supports expression for all members of subfamily I of the type-B response regulators. This analysis also suggests that the expression of ARR14 is light regulated, because ARR14 had a low level of expression in etiolated seedlings compared to light-grown seedlings of similar age (Fig. 2).
Figure 2.
Expression analysis of type-B response regulators by RT-PCR. RNA isolated from etiolated and green seedlings, as well as from roots, leaves, stems, flowers (including buds and floral meristems), and siliques was reverse transcribed and used as template for PCR amplification. Levels of template used were first standardized based on levels of EF1. PCR reactions were performed using ARR specific primers under optimal conditions for each primer set (ARR1, 34 cycles; ARR2, 35 cycles; ARR10, 29 cycles; ARR11, 31 cycles; ARR12, 30 cycles, ARR14, 34 cycles; ARR18, 36 cycles; ARR19, 30 cycles; ARR20, 45 cycles; ARR21, 34 cycles). Numbers indicate expression levels relative to the most abundant PCR product in that reaction set, with 0 indicating no detectable product. For ARR14 and ARR18, PCR product was observed for all templates when the number of PCR cycles was increased. No RT-PCR product was observed for ARR13 although several primer sets were tested.
Expression of the ARR genes was also examined in various tissues isolated from plants grown in the light, including roots, mature leaves, stems, flowers (with buds and floral meristem), and green siliques (Fig. 2). Transcripts from the members of subfamily I were detected in all tissues tested, although at differing levels. ARR1, 2, 10, 11, and 12, in particular, had broad patterns of expression consistent with previous expression studies of ARR1, ARR2, and ARR10 (Sakai et al., 1998; Imamura et al., 1999). ARR14 and ARR18 demonstrated greater specificities of expression (Fig. 2) but could be detected in all tissues when examined by reverse transcription (RT)-PCR using increased numbers of cycles (results not shown). The presence of ARR14 transcript at low levels in roots compared to aerial tissues provides additional evidence for light regulation of ARR14 expression. Members of subfamilies II and III were more restricted than members of subfamily I in expression, being found predominantly in the flowers and/or siliques. A transcript for ARR13 was not detected even when different primer combinations were tested.
Expression of Subfamily I Type-B ARRs Based on Fusions to the β-Glucuronidase Reporter Gene
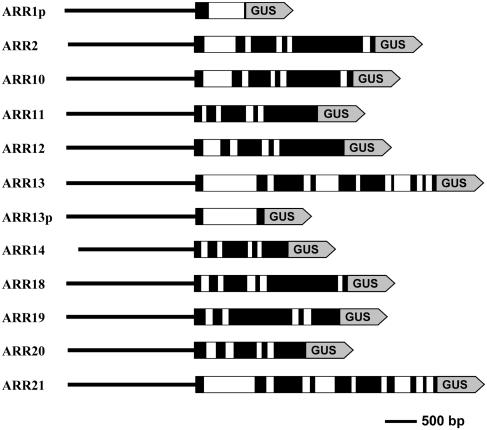
Expression of the type-B ARR proteins was also investigated by use of fusions with the β-glucuronidase (GUS) reporter gene. Whenever possible, the fusions contained full-length genomic copies of each ARR with approximately 2 kb upstream of the predicted start codon (Fig. 3). This approach was taken so that GUS expression levels would (1) reflect potential regulatory sequences for transcription in the introns as well as in the promoter (Jeon et al., 2000; Chang and Sun, 2002); and (2) reflect potential posttranscriptional regulation of the protein (Colon-Carmona et al., 1999; Gray et al., 2001). For each ARR, at least 15 transformed lines were analyzed with a minimum of five T2 plants per line examined. To improve data consistency, histochemical analysis was conducted on at least two independently grown sets of plants per transgenic line. The GUS staining patterns common to multiple lines for each transgene are described below.
Figure 3.
Schematic diagram of chimeric GUS constructs used for expression analysis. Black and white boxes represent exons and introns, respectively, within the coding region. A straight line represents the region upstream of the coding region and includes the 5′-UTR and the promoter. The shaded box represents the GUS reporter gene (not to scale). ARR1p and ARR13p are promoter fusions that translationally fused to GUS via their second exon. The scale bar indicates 500 bp.
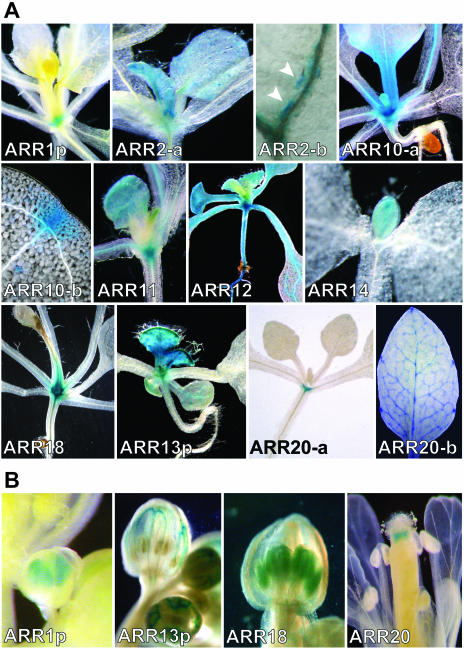
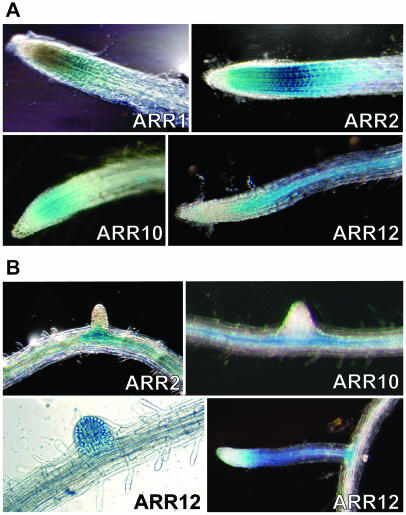
RT-PCR data shows that the members of subfamily I are broadly expressed throughout the plant (Fig. 2). In particular, transcripts for ARR1, ARR2, ARR10, ARR11, and ARR12 were readily detectable in most tissues tested. Consistent with this, GUS analysis also revealed overlapping expression of these ARRs, most notably within the shoot apical meristem region and in young rosette leaves (Fig. 4A). As leaf development progressed, GUS activity of the ARR fusions was predominantly observed at the leaf base adjacent to the petiole. Activity of ARR10::GUS was detected more readily than that of the other four ARR::GUS fusions in multiple transgenic lines, potentially indicating a higher level of expression for ARR10 under these growth conditions.
Figure 4.
Histochemical localization of type-B ARRs in aerial portions of Arabidopsis based on GUS reporter gene expression. A, GUS activity within vegetative tissues. Images show the GUS activity of each ARR fusion in the region around the shoot apical meristem and/or young developing leaves. Additional images of ARR2, ARR10, and ARR20 GUS-fusions show localization in mature leaves. ARR2-b shows the punctate staining pattern (arrows) observed along the leaf vascular tissue. ARR10-b shows GUS activity at a cotyledon hydathode. ARR20-b shows the GUS activity in the rosette leaf vascular tissue and hydathodes. Images are organized so that all subfamily I members are together. B, GUS activity within floral tissues for ARR1, ARR13, ARR18, and ARR20 GUS-fusions.
In contrast to what was observed in young leaves, expression of the GUS fusions for ARR1, ARR2, ARR10, and ARR12 was localized to specific cell types and regions within the mature leaf. Expression for ARR11::GUS dropped to below detectable limits in mature leaves. Expression of the ARR1, ARR2, ARR10, and ARR12 GUS-fusions was predominantly in the vascular tissue and the hydathodes (Fig. 4A). ARR10::GUS and ARR12::GUS showed consistent staining along the length of the vasculature. In contrast, ARR2::GUS staining appeared intermittently along the leaf vascular system (Fig. 4A, ARR2-b). A similar staining pattern was occasionally seen in ARR1p::GUS lines (data not shown). These patches of expression appear randomly along the vascular tissue and do not appear to be associated with vascular junctions. In addition to vascular staining, ARR2::GUS and ARR12::GUS were found in leaf and floral stem trichomes (Fig. 4, ARR2a; Fig. 6). ARR12::GUS was only found in the trichomes at the base of young rosette leaves, whereas ARR2::GUS was observed in trichomes on leaves of all ages.
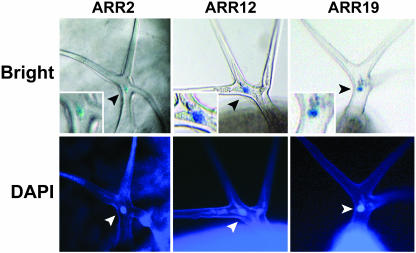
Figure 6.
Nuclear localization of type-B ARRs. Trichomes from leaves showing nuclear localized ARR::GUS activity were counterstained with DAPI. Upper sections show nuclear localized GUS expression, with insets showing close-ups of nuclei. Lower sections show DAPI fluorescence of the same tissue.
We also observed overlapping patterns of expression for subfamily I members within the roots. GUS activity for the ARR1, ARR2, ARR10, and ARR12 fusions was observed in the zone of elongation near the root tip (Fig. 5). Within lines showing higher levels of GUS expression (Fig. 5, ARR2), it appears that a gradient of increasing ARR::GUS activity begins at the root meristem, reaches a peak in the zone of elongation, and then decreases to below detectable limits in mature roots. In addition to being found at the root tip, GUS fusions of ARR2, ARR10, and ARR12 were found associated with developing lateral roots. Expression was initially observed in the vascular tissue of the main root adjacent to the site of lateral root formation. As lateral roots developed, the ARR::GUS fusions took on the expression patterns observed with the primary root tip. Occasionally ARR11::GUS was detected in the root-shoot junction but was not observed near the root tip or lateral root junctions (data not shown).
Figure 5.
Histochemical localization of type-B ARRs in roots based on GUS reporter gene expression. A, Expression of ARR1, ARR2, ARR10, and ARR12 GUS-fusions in the root tip region. B, Expression of ARR2, ARR10, and ARR12 GUS-fusions at lateral root junctions.
Within the floral structures, GUS expression for the ARR1 construct was observed in young developing anthers (Fig. 4B), consistent with the ready detection of ARR1 in flowers by RT-PCR (Fig. 2). However, as the anthers matured, ARR1p::GUS activity decreased to below the limit of detection.
RT-PCR reveals that ARR14 and ARR18 are unique members of subfamily I in that they display pronounced expression in a subset of tissues (Fig. 2). RT-PCR revealed a higher level of ARR14 expression in leaves than in other tissues examined. Likewise, GUS analysis revealed ARR14 expressed predominantly in young leaf tissue, although its expression decreased as the leaves matured (Fig. 4A). ARR18::GUS was observed in developing anthers (Fig. 4B), consistent with transcript being detected in flowers by RT-PCR (Fig. 2). Like other members of subfamily I, ARR18::GUS was also found in young leaf tissue, its expression decreasing to below detection levels in mature leaves (Fig. 4A).
Expression of Subfamily II Type-B ARRs Based on Fusions to the GUS Reporter Gene
Comparison of the intron-exon structure and sequence of ARR13 with its close relative, ARR21, suggested that the original ARR13 genomic annotation had incorrectly predicted the C terminus of ARR13. GUS constructs were made to ARR13 and ARR21 based on shared features that were consistent with the genomic annotation for the C terminus of ARR21. Although we were not able to observe any tissue staining using the full-length ARR13::GUS fusion, we were able to detect GUS activity using a promoter fusion construct (ARR13p::GUS). ARR13p::GUS showed relatively strong GUS expression only in the aerial portions of the plants, particularly in young leaves (Fig. 4A). Like many of the subfamily I genes, ARR13p::GUS expression became predominantly localized to the vascular tissue as the leaves matured. ARR13p::GUS was also seen in the vascular tissue of the sepals (Fig. 4B). For ARR21::GUS, we observed weak GUS activity in germinating seedlings (data not shown), but did not observe consistent GUS staining patterns in mature plants despite the detection of transcripts in siliques by RT-PCR.
Expression of Subfamily III Type-B ARRs Based on Fusions to the GUS Reporter Gene
ARR19::GUS expression was observed in the trichomes at the base of the youngest rosette leaves (Fig. 6) similar to that of subfamily I member, ARR12. However, unlike ARR12, ARR19 was not observed in the roots, shoot apical meristem, or mature leaves. Additionally, we did not detect the expression of the ARR19::GUS fusion in the silique, even though RT-PCR supports its expression in this tissue. GUS activity for the second member of subfamily III, ARR20, was observed in the mature pistil tip (Fig. 4B), consistent with expression in the flower as determined by RT-PCR. ARR20::GUS was also observed in the shoot apical meristem region as well as the vascular tissue and hydathodes of leaves (Fig. 4A).
Nuclear Localization of Type-B ARRs
Previous work has shown that ARR1, ARR2, and ARR10 under the control of the 35S promoter will localize to the nucleus of transiently transformed onion (Allium cepa) or parsley (Petroselinum crispum) cells (Sakai et al., 2000; Imamura et al., 2001; Lohrmann et al., 2001; Hosoda et al., 2002). We examined the subcellular localization of the full-length ARR fusions to GUS described here (Fig. 3), focusing in particular on the trichome with its clearly resolved nucleus. The GUS fusions to ARR2, ARR12, and ARR19 all appeared to localize to the nucleus of trichomes on rosette leaves (Fig. 6). Counterstaining of the leaves with 4′,6-diamino-phenylindole (DAPI) confirmed nuclear localization based on the overlapping staining profiles of GUS and the DAPI-stained trichome nucleus (Fig. 6). Subcellular localization of ARR2, ARR10, and ARR12 GUS-fusions consistent with the nucleus was also observed in root tips but could not be confirmed by DAPI staining due to the high levels of background fluorescence.
DISCUSSION
The type-B response regulators function in the final step of Arabidopsis histidyl-aspartyl phosphorelays and are thought to be transcription factors based on several lines of evidence. First, all type-B ARRs contain a GARP domain shown capable of directly binding DNA in studies of ARR10 (Hosoda et al., 2002). Second, several members of subfamily I (ARR1, ARR2, ARR10, and ARR11) can bind specific DNA sequences and enhance transcription (Sakai et al., 2000, 2001; Hosoda et al., 2002; Imamura et al., 2003). Third, type-B ARRs contain putative nuclear localization signals and transiently expressed subfamily I members (ARR1, ARR2, and ARR10) are localized to the nucleus (Sakai et al., 2000; Imamura et al., 2001; Lohrmann et al., 2001; Hosoda et al., 2002). Consistent with a role in transcription, we have localized ARR2, ARR12, and ARR19 GUS-fusions under control of their native promoters to the nucleus. Notably, whereas previous analyses were confined to members of subfamily I, ARR19 is a member of subfamily III. Our results thus support a general role for the type-B ARRs in transcription that is not limited to specific subfamilies.
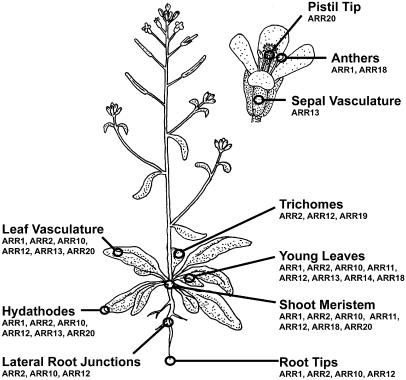
Both RT-PCR and GUS analysis reveals that type-B ARRs have overlapping expression patterns in Arabidopsis. These results are consistent with those recently reported by Tajima et al. (2004), where they also noted a broader expression profile for subfamily I members compared to other type-B ARRs based on analysis by RT-PCR. Results based on our GUS analysis are summarized in Figure 7. Overlapping expression based on GUS analysis is particularly evident in the shoot apical meristem and young developing leaves, a region of proliferating tissue where all members of subfamily I are expressed. The presence of subfamily I members in proliferating tissue is consistent with their proposed role in mediating cytokinin signal transduction (Hwang and Sheen, 2001; Sakai et al., 2001; Hutchison and Kieber, 2002; Kakimoto, 2003). Two recent studies in particular illustrate the role of cytokinins in promoting cell division in these tissues. Expression of the cyclin D3, which is induced by cytokinins and mediates the effect of cytokinins on cell division, is particularly high in the shoot meristem and in young leaf primordia (Riou-Khamlichi et al., 1999). In addition, reduction of endogenous cytokinin levels in tobacco or Arabidopsis by overexpression of cytokinin oxidase results in smaller shoot meristems and leaves due to the reduced rate of cell division (Werner et al., 2001, 2003). Thus, members of subfamily I contain regulatory elements that induce expression in cells destined to undergo division in response to endogenous levels of cytokinin.
Figure 7.
Overview of Arabidopsis type-B ARR localization based on GUS analysis. (Drawing by G.E.S.)
A subset of subfamily I members are also expressed in mature leaf tissue. The expression of the ARR1, ARR2, ARR10, and ARR12 GUS fusions appeared ubiquitous throughout the young leaves. However, as the leaves matured, these proteins became predominantly localized to the vascular tissue and the hydathodes (Fig. 4A). Two out of these four subfamily I members, ARR2 and ARR12, were also found to be expressed in trichomes (Fig. 6). This suggests that the type-B ARRs have different functions within leaves of different developmental stages and/or that their activity is mainly required by particular cell types as the leaves mature. ARR2::GUS fusions produced patchy leaf vascular staining in which GUS activity appears randomly along the vascular tissue (Fig. 4A, ARR2-b). A similar staining pattern was occasionally observed in ARR1p::GUS plants (data not shown). The expression pattern of type-B ARRs in mature leaves may relate to the regulation of leaf vascular differentiation, because cytokinin-deficient plants had reduced leaf vasculature (Werner et al., 2003). The expression pattern may also explain the seemingly random emergence of adventitious buds along the vascular tissue of plants overexpressing the ipt gene for cytokinin biosynthesis (Estruch et al., 1991). The results of this previous study had suggested that different cells in the leaf have different cytokinin sensitivities and thus different capacities to produce new meristemoids (Estruch et al., 1991), a capacity that could potentially be mediated by the varying cellular levels of type-B ARRs.
Overlapping expression of subfamily I members ARR1, ARR2, ARR10, and ARR12 was also observed in the lateral root junctions and root tips, with each ARR forming a gradient of increasing ARR::GUS activity beginning in the root meristem, reaching a peak in the zone of elongation and then decreasing below detectable limits in the mature root (Fig. 5). This localization is interesting in light of the role of cytokinins in inhibiting both root growth rate and the formation of lateral roots, a role that contrasts with the stimulatory effect of cytokinin upon shoot growth (Werner et al., 2001, 2003). The expression pattern of these four type-B ARRs could relate to several possible functions in root development that are not mutually exclusive. First, the expression pattern is consistent with a role in modulating postembryonic vascular differentiation. Cytokinins, in conjuntion with auxin, are implicated in regulating vascular differentiation (Aloni, 1993; Demura et al., 2002), and the analysis of cytokinin-deficient plants revealed a decrease in the leaf vasculature (Werner et al., 2001, 2003). Second, higher levels of expression in the region adjacent to the root meristem could play a role in restricting the size of the meristem, based on the finding that a decrease in cytokinin levels results in enlarged root meristems with increased cell number (Werner et al., 2001, 2003). Third, the localization of type-B ARRs in the elongation zone may relate to the interconnection between cytokinin and ethylene signaling. Cytokinin stimulates ethylene production in roots, with ethylene then inhibiting cell elongation (Woeste et al., 1999).
RT-PCR and GUS analysis reveals that the members of subfamilies II and III have unique expression patterns but that these still overlap to some extent with those of subfamily I members. For example, ARR19 (subfamily III) is expressed in young trichomes, similar to ARR2 and ARR12 (Fig. 6). However, unlike these subfamily I ARRs, ARR19 was not detected in the shoot apical meristem, the vascular tissue of mature leaves, or the roots. ARR13::GUS expression (subfamily II) overlaps with several subfamily I members in aerial tissues but, unlike them, is not detected in root tissues.
From the expression analysis, it is clear that there is overlapping expression among the type-B family members. ARR1, ARR2, ARR10, and ARR12 from subfamily I in particular are expressed in many of the same cell types. However, the overall pattern of expression for each ARR is distinct, the differences being most apparent when examined in mature differentiated cell types. For example, ARR::GUS fusions were expressed in three different structures of the flower: ARR1 and ARR18 were found in the anthers, ARR13 was found in the sepal vasculature, and ARR20 was found at the pistil tip (Fig. 4B). Although not a focus of this study, the potential light regulation of ARR14 expression suggests that the patterns of expression for each ARR may also differ based on environmental signaling factors.
Based on the finding that multiple type-B ARRs are expressed in regions of the shoot undergoing rapid cell division, it will be of interest to determine the extent of the correlation between type-B ARR expression and cell division in proliferating tissues. In preliminary experiments, we did not observe GUS activity in developing embryos, although several lines of ARR1p::GUS and ARR10::GUS did display GUS activity in the funiculus (data not shown). The inability to detect type-B ARR expression in embryos may indicate that the level of type-B ARR::GUS activity was below detectable limits and/or that this ARR-mediated signaling pathway has a more restricted role in embryogenesis. The wol1 mutation, for example, is within a gene encoding a cytokinin receptor, but its effect is not until the torpedo stage of embryogenesis and is restricted to cell division of the vascular initials (Mahonen et al., 2000). In contrast, we did observe GUS activity in callus tissues generated from several ARR10::GUS and ARR12::GUS lines (data not shown). The expression of type-B ARRs in callus tissue is consistent with the well-documented role of cytokinins in regulation of callus growth and differentiation (Skoog and Miller, 1957).
Analysis of ARR gene expression provides limited functional relevance by itself although, based on previous studies, potential functional roles of type-B ARRs can be inferred. Because a correlation has been made between type-B ARRs and cytokinin signal transduction (Hwang and Sheen, 2001; Sakai et al., 2001; Hutchison and Kieber, 2002; Kakimoto, 2003), it is pertinent to compare the type-B ARR expression patterns with those of other genes involved in cytokinin signaling. The isopentyltransferase (IPT) and cytokinin oxidase (CKX) gene products catalyze cytokinin synthesis and degradation, respectively (Kakimoto, 2001; Mok and Mok, 2001). Unlike the type-B ARRs, the Arabidopsis IPT and CKX genes display limited expression overlap within their gene families (Werner et al., 2003; Miyawaki et al., 2004). Some members from both the IPT and the CKX families are expressed in areas where type-B ARRs are found (e.g. lateral root junctions, trichomes, and leaf vasculature), but overall, expression of the IPT and CKX genes does not closely correlate with that of the type-B ARRs. Notably, none of the IPT genes known to be involved in cytokinin synthesis are expressed in the shoot meristem based on GUS reporter analysis (Miyawaki et al., 2004). This is not entirely unexpected as the type-B ARRs appear to function at the site of cytokinin action, which can differ from the site of its synthesis and degradation. In contrast, expression of type-B ARRs, especially subfamily I members, overlaps with the expression of the type-A ARRs that have been established as cytokinin primary response genes (D'Agostino et al., 2000; Kiba et al., 2002). These data are consistent with the proposed role for type-B ARRs in regulating the expression of these primary response genes (Hwang and Sheen, 2001; Sakai et al., 2001; Hutchison and Kieber, 2002; Kakimoto, 2003).
In conclusion, type-B response regulators display distinct, yet overlapping expression patterns in Arabidopsis, with subfamily I members showing the broadest expression throughout the plant. The more specific localization of subfamily II and III members suggests that these proteins may play more specialized roles in plant growth and development. Expression of type-B ARRs, in particular subfamily I members, overlaps with the expression of cytokinin primary response genes such as those encoding type-A ARRs (D'Agostino et al., 2000; Kiba et al., 2002) and cyclin D3 (Riou-Khamlichi et al., 1999). These data are consistent with a role for type-B ARRs in mediating the induction of these primary response genes and thus modulating cytokinin responses. The differing expression levels of type-B ARRs observed in specific cell types would result in vastly different abilities of the cells to respond to cytokinin or other potential upstream signals. This is likely to be an important component in modulating the cellular response to different hormone doses.
MATERIALS AND METHODS
Phylogenetic Analysis
The receiver domain for each response regulator was determined by analysis using the Simple Modular Architecture Research Tool search algorithms (http://smart.embl-heidelberg.de; Schultz et al., 1998; Letunic et al., 2002), then aligned using ClustalW from Megalign 5.03 (DNASTAR, Madison, WI). The unrooted phylogenetic tree was constructed with PAUP 4.0b10 (Swofford, 2002) by using a distance method based on comparison of 1,000 bootstrap replications. The divergent ARR22 was designated as the outgroup for construction of the rectangular cladogram. Intron positions for the response regulators were determined based on the unspliced sequence representation at the MIPS Web site and by comparison between related family members to identify regions of sequence conservation.
Plant Materials and Growth Conditions
Wild-type Arabidopsis (ecotype Columbia) was used for all experiments. Tissues used for RT-PCR were obtained from plants grown on soil, liquid media, and agar plates. Growth on soil involved stratifying the sown seeds at 4°C for 3 d prior to their incubation at 22°C under a 16-h light (fluorescent illumination)/8-h dark cycle. Seeds germinated on plates for use in RT-PCR experiments were surface-sterilized and then sown on 0.8% (w/v) agar plates of one-half-strength Murashige and Skoog (MS) basal medium (pH 5.7) containing Gamborg's vitamins (MS media, Sigma-Aldrich, St.Louis). Seeds were stratified at 4°C for 3 d and then placed under constant light at 22°C for 8 d. Plants grown for GUS histochemical analysis were grown similarly to plants used for RT-PCR except the seeds were sown onto full-strength MS media containing 0.1% (w/v) Suc and grown for approximately 3 weeks under constant light. For growth of etiolated seedlings, sown seeds were exposed to light for 12 h and then grown in the dark for 3 d. For growth of plants in liquid media, seeds were surface sterilized and placed in 50-mL one-half-strength MS media containing 1% (w/v) Suc under a 16-h light/8-h dark cycle for 16 d.
Semiquantitative RT-PCR
Total RNA was isolated using the RNeasy plant mini kit (Qiagen, Valencia, CA) from 3.5-d-old dark grown seedlings, 8-d-old green seedlings, 16-d-old roots grown in liquid MS media, and rosette leaves, stems, flowers (including buds and floral meristem), and green siliques from 5-week-old plants. cDNA was synthesized from 1 μg total RNA using the Reverse Transcription System (Promega, Madison, WI) after pretreatment with RNase Free DNase I (Qiagen).
PCR reactions were performed using HotMaster Taq (Eppendorf, Westbury, NY). Primers specific for the sequences of interest were designed so that for each primer set at least one primer spans a region that contains an intron in the genomic sequence. The following primers were used for the PCR reactions: EF1, 5′-TAGGGCTGGTATCTCTAAG-3′ and 5′-CGAAGGGGCTTGTCTGATG-3′; ARR1, 5′-CTTTTCTTTTTTTGTTTCTTGGGTT-3′ and 5′-CGGTATTTCTGGAGGTGACTTG-3′; ARR2, 5′-TTCGGGTACTGCTGCTGGTG-3′ and 5′-CTACTGGCAACATCATTCCGCT-3′; ARR10, 5′-CGTTGCTCTGAAGAAGGTGT-3′ and 5′-GATTGGCTCTGTTCCTGTGT-3′; ARR11, 5′-CCTGTAATAATGATGTCGGT-3′ and 5′-CATAGGAACTTTGACTTGGC-3′; ARR12, 5′-CTGTCATAATGTTGTCTGCG-3′ and 5′-TAGAATGCGGTAATGGAGAG-3′; ARR13, 5′-ATGGCTTTTGCTCAATCTGTCT-3′ and 5′-TTGGGCACCACCTTATCATAAC-3′, 5′-AAAGGACGCAAATGTTAGTGT-3′ and 5′-CTATCCGAAGAAAGCATTATC-3′, 5′-ATGTACGGATTCGGAATAGAA-3′ and 5′-GCCATCATCACTAGGACCAC-3′; ARR14, 5′-ATTATGATGTCTGTTGATGG-3′ and 5′-TGTCTGATTCTGTTGTTGTT-3′; ARR18, 5′-AGGCTGTTCCCAAAAAAATA-3′ and 5′-TGGTTGTCATTCTCTGGCTT-3′; ARR19, 5′-TTTTACGGTGCTTGTGACTATG-3′ and 5′-TGATACGGTGTTTCTGAAGATG-3′; ARR20, 5′-TGACACAGTATTCCTATCAAGTAACGA-3′ and 5′-TTGACGGTACTTCTGGAGATGACTG-3′; ARR21, 5′-CCAAATACAGAGATCCATCAG-3′ and 5′-ATACGGTGCGGCTCATAG-3′. EF1 was used as an RT-PCR control and the amount PCR template used for the different samples was normalized to the expression level of EF1. The number of PCR cycles used for each ARR was determined such that the level of product from each tissue was in the linear range of the reaction. The amplified fragments were separated by agarose gel electrophoresis, stained with ethidium bromide, digitally scanned under UV light, and the relative intensities of the bands quantified using the software GelExpert 3.5 (Nucleotech, San Carlos, CA).
Plasmid Construction and Plant Transformation
PCR primers were designed to amplify the genomic clone of each type-B response regulator including approximately 2 kb of DNA upstream of the start of translation and the entire coding sequence up to but not including the translational stop codon. ARR1 was an exception to this, in that 2 kb of the ARR1 promoter and the coding sequence up to and including part its second exon was amplified by PCR. A similar PCR was performed to amplify the ARR13 promoter region in addition to a full-length clone. The PCR products were cloned into a pCAMBIA-3381 vector (Cambia, Canberra, Australia) modified to include the GATEWAY system (Invitrogen Life Technologies, Carlsbad, CA). Constructs were introduced into Agrobacterium tumefaciens strain GV3101 and used to transform Arabidopsis by the floral dipping method (Bent and Clough, 1998).
GUS Analysis
Histochemical analysis of GUS activity in stably transformed lines of Arabidopsis was performed as described (Jefferson et al., 1987) with a few alterations. Briefly, tissues were immersed in GUS staining solution (2 mm 5-bromo-4-chloro-3-indolyl β-d-glucuronide [X-gluc, Gold BioTechnology, St. Louis], 0.5% [v/v] Triton X-100, 0.1% [v/v] Tween 20, 0.5 mm potassium ferricyanide, 0.5 mm potassium ferrocyanide, 10 mm EDTA, and 50 mm sodium phosphate buffer [pH 7]) and subjected to 3 5-min vacuum infiltrations. The plants were then left overnight in the dark at 37°C before being cleared of chlorophyll by incubating in 70% (v/v) ethanol for several hours. The GUS stained tissues were visualized and photographed using a stereo microscope with a dark field base.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
This work was supported by the National Science Foundation (grant no. 0114965 to J.J.K. and G.E.S.) and by the USDA-NRICGP (grant no. 2001–35311–10988 to D.E.M.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.038109.
References
- Aloni R (1993) The role of cytokinin in organized differentiation of vascular tissues. Aust J Plant Physiol 20: 601–608 [Google Scholar]
- Bent AF, Clough SJ (1998) Agrobacterium germ-line transformation: transformation of Arabidopsis without tissue culture. In SB Gelvin, RA Schilperout, eds, Plant Molecular Biology Manual, Ed 2. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 1–14
- Brandstatter I, Kieber JJ (1998) Two genes with similarity to bacterial response regulators are rapidly and specifically induced by cytokinin in Arabidopsis. Plant Cell 10: 1009–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CW, Sun TP (2002) Characterization of cis-regulatory regions responsible for developmental regulation of the gibberellin biosynthetic gene GA1 in Arabidopsis thaliana. Plant Mol Biol 49: 579–589 [DOI] [PubMed] [Google Scholar]
- Colon-Carmona A, You R, Haimovitch-Gal T, Doerner P (1999) Technical advance: spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J 20: 503–508 [DOI] [PubMed] [Google Scholar]
- D'Agostino IB, Deruere J, Kieber JJ (2000) Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol 124: 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demura T, Tashiro G, Horiguchi G, Kishimoto N, Kubo M, Matsuoka N, Minami A, Nagata-Hiwatashi M, Nakamura K, Okamura Y, et al (2002) Visualization by comprehensive microarray analysis of gene expression programs during transdifferentiation of mesophyll cells into xylem cells. Proc Natl Acad Sci USA 99: 15794–15799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estruch JJ, Prinsen E, Onckelen HV, Schell J, Spena A (1991) Viviparous leaves produced by somatic activation of an inactive cytokinin-synthesizing gene. Science 254: 1364–1366 [DOI] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Heyl A, Schmülling T (2003) Cytokinin signal perception and transduction. Curr Opin Plant Biol 6: 480–488 [DOI] [PubMed] [Google Scholar]
- Hosoda K, Imamura A, Katoh E, Hatta T, Tachiki M, Yamada H, Mizuno T, Yamazaki T (2002) Molecular structure of the GARP family of plant Myb-related DNA binding motifs of the Arabidopsis response regulators. Plant Cell 14: 2015–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CE, Kieber JJ (2002) Cytokinin signaling in Arabidopsis. Plant Cell Suppl 14: S47–S59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Chen HC, Sheen J (2002) Two-component signal transduction pathways in Arabidopsis. Plant Physiol 129: 500–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J (2001) Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413: 383–389 [DOI] [PubMed] [Google Scholar]
- Imamura A, Hanaki N, Nakamura A, Suzuki T, Taniguchi M, Kiba T, Ueguchi C, Sugiyama T, Mizuno T (1999) Compilation and characterization of Arabidopsis thaliana response regulators implicated in His-Asp phosphorelay signal transduction. Plant Cell Physiol 40: 733–742 [DOI] [PubMed] [Google Scholar]
- Imamura A, Kiba T, Tajima Y, Yamashino T, Mizuno T (2003) In vivo and in vitro characterization of the ARR11 response regulator implicated in the His-to-Asp phosphorelay signal transduction in Arabidopsis thaliana. Plant Cell Physiol 44: 122–131 [DOI] [PubMed] [Google Scholar]
- Imamura A, Yoshino Y, Mizuno T (2001) Cellular localization of the signaling components of Arabidopsis His-to-Asp phosphorelay. Biosci Biotechnol Biochem 65: 2113–2117 [DOI] [PubMed] [Google Scholar]
- Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T (2001) Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409: 1060–1063 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: ß-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon JS, Lee S, Jung KH, Jun SH, Kim C, An G (2000) Tissue-preferential expression of a rice alpha-tubulin gene, OsTubA1, mediated by the first intron. Plant Physiol 123: 1005–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakimoto T (2001) Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate: ATP/ADP isopentenyltransferases. Plant Cell Physiol 42: 677–685 [DOI] [PubMed] [Google Scholar]
- Kakimoto T (2003) Perception and signal transduction of cytokinins. Annu Rev Plant Biol 54: 605–627 [DOI] [PubMed] [Google Scholar]
- Kiba T, Yamada H, Mizuno T (2002) Characterization of the ARR15 and ARR16 response regulators with special reference to the cytokinin signaling pathway mediated by the AHK4 histidine kinase in roots of Arabidopsis thaliana. Plant Cell Physiol 43: 1059–1066 [DOI] [PubMed] [Google Scholar]
- Letunic I, Goodstadt L, Dickens NJ, Doerks T, Schultz J, Mott R, Ciccarelli F, Copley RR, Ponting CP, Bork P (2002) Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res 30: 242–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohrmann J, Sweere U, Zabaleta E, Baurle I, Keitel C, Kozma-Bognar L, Brennicke A, Schafer E, Kudla J, Harter K (2001) The response regulator ARR2: a pollen-specific transcription factor involved in the expression of nuclear genes for components of mitochondrial complex I in Arabidopsis. Mol Genet Genomics 265: 2–13 [DOI] [PubMed] [Google Scholar]
- Mahonen AP, Bonke M, Kauppinen L, Riikonen M, Benfey PN, Helariutta Y (2000) A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev 14: 2938–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S, Kiba T, Imamura A, Hanaki N, Nakamura A, Suzuki T, Taniguchi M, Ueguchi C, Sugiyama T, Mizuno T (2000) Genes encoding pseudo-response regulators: insight into His-to-Asp phosphorelay and circadian rhythm in Arabidopsis thaliana. Plant Cell Physiol 41: 791–803 [DOI] [PubMed] [Google Scholar]
- Makino S, Matsushika A, Kojima M, Oda Y, Mizuno T (2001) Light response of the circadian waves of the APRR1/TOC1 quintet: when does the quintet start singing rhythmically in Arabidopsis? Plant Cell Physiol 42: 334–339 [DOI] [PubMed] [Google Scholar]
- Matsushika A, Makino S, Kojima M, Mizuno T (2000) Circadian waves of expression of the APRR1/TOC1 family of pseudo- response regulators in Arabidopsis thaliana: insight into the plant circadian clock. Plant Cell Physiol 41: 1002–1012 [DOI] [PubMed] [Google Scholar]
- Miyawaki K, Matsumoto-Kitano M, Kakimoto T (2004) Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J 37: 128–138 [DOI] [PubMed] [Google Scholar]
- Mizuno T (1997) Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of Escherichia coli. DNA Res 4: 161–168 [DOI] [PubMed] [Google Scholar]
- Mok DW, Mok MC (2001) Cytokinin metabolism and action. Annu Rev Plant Physiol Plant Mol Biol 52: 89–118 [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi C, Huntley R, Jacqmard A, Murray JA (1999) Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283: 1541–1544 [DOI] [PubMed] [Google Scholar]
- Sakai H, Aoyama T, Bono H, Oka A (1998) Two-component response regulators from Arabidopsis thaliana contain a putative DNA-binding motif. Plant Cell Physiol 39: 1232–1239 [DOI] [PubMed] [Google Scholar]
- Sakai H, Aoyama T, Oka A (2000) Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J 24: 703–711 [DOI] [PubMed] [Google Scholar]
- Sakai H, Honma T, Aoyama T, Sato S, Kato T, Tabata S, Oka A (2001) ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294: 1519–1521 [DOI] [PubMed] [Google Scholar]
- Schaller GE (2000) Histidine kinases and the role of two-component systems in plants. Adv Bot Res 32: 109–148 [Google Scholar]
- Schaller GE, Mathews DE, Gribskov M, Walker JC (2002) Two-component signaling elements and histidyl-aspartyl phosphorelays. In C Somerville, E Meyerowitz, eds, The Arabidopsis Book, DOI/10.1199/tab.0086, http://www.aspb.org/publications/arabidopsis/. American Society of Plant Biologists, Rockville, MD [DOI] [PMC free article] [PubMed]
- Schultz J, Milpetz F, Bork P, Ponting CP (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA 95: 5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoog F, Miller CO (1957) Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp Soc Exp Biol 11: 118–131 [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN (2000) Two-component signal transduction. Annu Rev Biochem 69: 183–215 [DOI] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Mas P, Panda S, Kreps JA, Kay SA (2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Miwa K, Ishikawa K, Yamada H, Aiba H, Mizuno T (2001) The arabidopsis sensor his-kinase, ahk4, can respond to cytokinins. Plant Cell Physiol 42: 107–113 [DOI] [PubMed] [Google Scholar]
- Swanson RV, Alex LA, Simon MI (1994) Histidine and aspartate phosphorylation: two-component systems and the limits of homology. Trends Biochem Sci 19: 485–490 [DOI] [PubMed] [Google Scholar]
- Swofford DL (2002) PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sinauer Associates, Sunderland, MA
- Tajima Y, Imamura A, Kiba T, Amano Y, Yamashino T, Mizuno T (2004) Comparative studies on the type-B response regulators revealing their distinctive properties in the His-to-Asp phosphorelay signal transduction of Arabidopsis thaliana. Plant Cell Physiol 45: 28–39 [DOI] [PubMed] [Google Scholar]
- Ueguchi C, Sato S, Kato T, Tabata S (2001) The AHK4 gene involved in the cytokinin-signaling pathway as a direct receptor molecule in Arabidopsis thaliana. Plant Cell Physiol 42: 751–755 [DOI] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmülling T (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15: 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Motyka V, Strnad M, Schmülling T (2001) Regulation of plant growth by cytokinin. Proc Natl Acad Sci USA 98: 10487–10492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woeste KE, Vogel JP, Kieber JJ (1999) Factors regulating ethylene biosynthesis in etiolated Arabidopsis thaliana seedlings. Physiol Plant 105: 478–484 [Google Scholar]
- Yamada H, Suzuki T, Terada K, Takei K, Ishikawa K, Miwa K, Mizuno T (2001) The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol 42: 1017–1023 [DOI] [PubMed] [Google Scholar]