Abstract
Cellular proteins are constantly damaged by reactive oxygen species generated by cellular respiration. Because of its metal-chelating property, the histidine residue is easily oxidized in the presence of Cu/Fe ions and H2O2 via metal-catalyzed oxidation, usually converted to 2-oxohistidine. We hypothesized that cells may have evolved antioxidant defenses against the generation of 2-oxohistidine residues on proteins, and therefore there would be cellular proteins which specifically interact with this oxidized side chain. Using two chemically synthesized peptide probes containing 2-oxohistidine, high-throughput interactome screening was conducted using the E. coli K12 proteome microarray containing >4200 proteins. Ten interacting proteins were identified, and successfully validated using a third peptide probe, fluorescence polarization assays, as well as binding constant measurements. We discovered that 9 out of 10 identified proteins seemed to be involved in redox-related cellular functions. We also built the functional interaction network to reveal their interacting proteins. The network showed that our interacting proteins were enriched in oxido-reduction processes, ion binding, and carbon metabolism. A consensus motif was identified among these 10 bacterial interacting proteins based on bioinformatic analysis, which also appeared to be present on human S100A1 protein. Besides, we found that the consensus binding motif among our identified proteins, including bacteria and human, were located within α-helices and faced the outside of proteins. The combination of chemically engineered peptide probes with proteome microarrays proves to be an efficient discovery platform for protein interactomes of unusual post-translational modifications, and sensitive enough to detect even the insertion of a single oxygen atom in this case.
The complexity of the proteome arises in a large part because of the hundreds of post-translational modifications (PTMs)1 already discovered. Many PTMs are enzyme-catalyzed, such as phosphorylation, glycosylation, or ubiquitination (1, 2), but there are also numerous nonenzymatic PTMs caused by chemical reactions between reactive molecules and protein side chains, such as glycation, nitrosylation, and oxidation by reactive oxygen species (ROS) (3, 4). When protein side chains are enzymatically modified, there are generally specialized factors in the cell to recognize such changes. For instance, 14-3-3 family protein can recognize protein phosphorylation motifs (5) and various lectins can recognize protein glycosylation (6). However, recognition factors may also exist for nonenzymatic PTMs, such as receptor for advanced glycation end-products (RAGE) (7). In this study we seek to uncover cellular binding factors for 2-oxohistidine, the oxidized product of histidine, which is an important but little-understood nonenzymatic PTM.
The generation of ROS is an unavoidable consequence of cellular respiration, which leads to the oxidation of proteins, lipids, and nucleic acids (4, 8). ROS play regulatory roles in cellular signaling pathways under low levels (9), but high levels of ROS are cytotoxic and lead to the accumulation of damaged cellular components (10, 11). The reactions of proteins with ROS may lead to almost 100 types of side chain modifications (12, 13). Histidine is highly susceptible to ROS damage, because it has strong metal chelation affinity and often constitutes the binding site for metal ions (14, 15). The presence of H2O2 and redox-active metals (Cu and Fe) can lead to metal-catalyzed oxidation (MCO, also called Fenton-type chemistry), which converts histidine side chain to 2-oxohistidine (16, 17).
The conversion of histidine to 2-oxohistidine alters its charge state, hydrogen bonding property, and metal chelation affinity, and hence may have serious impacts on protein structure and function. The net reaction is oxygen insertion (+16 Da), which makes it an irreversible PTM. It is unclear if cells simply tolerate such damages on histidines or employ active mechanisms to recognize them and use them as redox sensors or as damage markers for promoting protein degradation. The only known biological function of 2-oxohistidine is to serve as a redox sensor on bacterial transcription factor PerR (18), whereas other studies have used 2-oxohistidine as a stable marker of protein damage during oxidative stress (12, 19).
Judging by the potential biological significance of 2-oxohistidine modification, we hypothesized that there may be cellular factors to recognize it. Previous research on 2-oxohistidine had been impeded by the difficulty in generating this side chain with reasonable yields. Recently, we managed to greatly improve the yield of 2-oxohistidine conversion by optimizing MCO reaction conditions using the copper/ascorbate system (20), allowing us to synthesize and purify peptide probes containing 100% 2-oxohistidine for this study.
Here, we used 2-oxohistidine-containing peptides to mimic the oxidative conversion of histidine residues on native proteins. Then, we utilized the Escherichia coli (E. coli) K12 proteome chip to identify 2-oxohistidine-interacting proteins via high-throughput screening, and the interactors turned out to be largely involved redox-related metabolism. From the bacterial interactors we predicted a consensus binding motif, which could be validated across different species and correctly predicted S100A1 as a human binding factor for 2-oxohistidine. Thus, recognition of 2-oxohistidine appears to be an evolutionarily conserved capacity from bacteria to human.
EXPERIMENTAL PROCEDURES
Fabrication of E. coli K12 Proteome Chip
The high throughput protein expression, protein purification, and protein printing were modified from the previous study (21). Briefly, we expressed and purified E. coli K12 proteins in 96-well plate format and subsequently printed the proteome microarray. All purified proteins were spotted in duplicate on each aldehyde slide (BaiO, Shanghai, China) by SmartArrayer 136 (CapitalBio, Beijing, China) at 4 °C. After printing proteins, the proteome microarray chips were kept at 4 °C for protein immobilization on the slides for 12 h. The chips were stored at −80 °C before probing with samples.
Peptide Oxidation
Solutions containing 1 mm peptide, 5 mm Cu2+ and 200 mm sodium ascorbate were exposed to air with gentle shaking at 37 °C for 24 h (AG and SE peptide) or 6 h (IA peptide). The oxidation reaction was quenched with 20 mm EDTA and analyzed by reverse-phase high-performance liquid chromatography (HPLC) (10–30% acetonitrile and 0.1% TFA in water, C18 column from Dr. Maisch, Ammerbuch, Germany) to determine the reaction yield. For liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis of crude reaction mixtures and HPLC fractions, 10 μl samples were acidified with 2 μl 10% TFA and desalted with ZipTip (Millipore, Billerica, MA) following manufacturer's protocols. Oxidized peptides were purified by semi-preparative HPLC (C18 column, Dr. Maisch). LC-MS/MS experiments were conducted under previously reported conditions (20).
Peptide Labeling
Oxidized and nonoxidized peptides were dissolved in 50 mm sodium borate buffer at pH 7.5 and analyzed by HPLC to determine peptide concentration by 210 nm absorbance. DyLight-conjugated NHS esters (Thermo, Waltham, MA) were dissolved in anhydrous DMF to 10 mg/ml and added to peptide solutions for 1 h incubation at room temperature, at the following fluorophore/peptide ratios: DyLight 650/AG = 3:1, DyLight 650/SE = 5:1, DyLight 650/oxo-IA = 1.5:1; DyLight 550/oxo-AG = 5:1, DyLight 550/oxo-SE = 7:1, DyLight 550/IA = 3:1. Labeled peptides were analyzed and purified by HPLC as described above. Labeled products were verified by LC-MS/MS, and quantified by absorbance measurements based on known fluorophore properties.
E. coli K12 Proteome Chip Assays with 2-oxohistidine Peptides
The chips were first blocked with 3% bovine serum albumin (BSA) (Sigma, St. Louis, MO) for 5 min. DyLight 550-conjugated 2-oxohistidine peptide and DyLight 650-conjugated nonoxidized peptide (10 μm each) were probed together onto the chip with LifterSlips (Thermo) at room temperature for 45 min. Finally, the chips were washed by Tris-buffered saline-Tween 20 (TBST) on an orbital shaker three times and 5 min each time. The chip was dried by centrifugation and then scanned with a LuxScan microarray scanner (CapitalBio). Signal intensities, defined as foreground median subtracted by background median, were acquired and analyzed using GenePix Pro 6.0 software. Then, we used quantile normalization to normalize the signal intensity from both 2-oxohistidine containing probes and nonoxidized probes. To identify positive 2-oxohistidine interacting proteins, four cutoff criteria were set: (1) The signal from experimental group was >1.5 standard deviations above the mean for all experimental groups. (2) To identify large signal differences between experimental groups and negative controls, the delta, defined as signal difference between experimental group and control group, was >1.5 standard deviations above the mean for all deltas. (3) To exclude the nonspecific binding to 2-oxohistidine residue, the signal from the negative control was >1.5 standard deviations below the mean for all control groups. (4) To remove irreproducible hits among triplicate chip assays, the student's t test p values between experimental groups and negative controls were less than 0.05.
Heat Map
The R programming language (22) was used to display heat map. The data was presented by the signal intensity of foreground subtracted by background. The gplots package (23) was used for classifying 2-oxohistidine containing peptides and nonoxidized peptides in hierarchy.
Functional Interaction Analysis
The identified proteins were used for functional interaction analyses by using EcID (24) and Cytoscape (25). Briefly, the files of EcID entities and EcID pairs were downloaded from EcID database. Before mapping identified proteins to their EcID entities and EcID pairs, we removed the pairs that were based on the prediction mode, such as phylogenetic profiles, gene neighborhood, mirror tree, in silico two-hybrid, or context mirror. After mapping, we used Cytoscape to generate the functional interaction network, and visualized the identified proteins and their interacting proteins. Subsequently, we used AmiGO 2 (26) and KOBAS 2.0 (27) to generate gene ontology (GO) (28) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (29) results, respectively.
Fluorescence Polarization Assay
After blocking the 96-well black plate (Thermo) with 1% BSA at room temperature for 1 h, the identified proteins were added to the plate. The concentrations of 10 identified proteins (ThrS, YqjG, YajL, HemE, IlvA, PrpD, Zwf, Eda, Gor, and PqqL) were 12.0, 25.7, 10.7, 15.6, 3.4, 18.6, 19.5, 11.8, 26.1, and 5.9 μm, respectively. The concentrations of BSA, as a negative control, were identical to the protein being compared with. A 10 nm solution of DyLight 550-conjugated 2-oxohistidine peptide was incubated with target protein or BSA in a Micromixer MX4 (FINEPCR, Gunpo, Korea) at room temperature for 1 h. After incubation, the degree of polarization of each well was detected by a Synergy 2 (BioTek, Winooski, VT), using an excitation wavelength of 540 nm and an emission wavelength of 590 nm with a dichroic mirror of 570 nm.
Measurement of Dissociation Constant
Identified proteins and S100A1 (Abnova, Taipei, Taiwan) were printed on aldehyde chips in a multiple-well format. After printing, the chips were immobilized at 4 °C for 12 h and then stored at −80 °C. The printed chips were blocked at room temperature for 5 min with 3% BSA. Two-fold serial-diluted DyLight 550-conjugated 2-oxohistidine peptides, DyLight 650-conjugated nonoxidized peptides, and quenched fluorescent dyes were added into the chip wells individually with Multi-Well Microarray Hybridization Cassettes (Arrayit, Sunnyvale, CA), and incubated at room temperature for 45 min. The fluorescent dyes, NHS esters of DyLight 550 and DyLight 650, were already quenched by Tris-HCl (Bionovas, Toronto, Canada). To check whether calcium affects interaction between S100A1 and 2-oxohistidine, 1 mm CaCl2 was added in the assay buffer. After several washes with TBST, the chips were dried by centrifugation and then scanned with a microarray scanner. The Kd value was calculated by double-reciprocal plot analysis, in which y is one divided by fluorescence intensity, and x is one divided by peptide concentration. We set the regression line formula in the form of y = ax, where “a” is the slope of regression line. The Kd value will be “a” multiplied by the concentration of identified protein.
Motif Search with GLAM2
All identified proteins were converted to FASTA format and analyzed by Gapped Local Alignment of Motifs (GLAM2) (30) for surveying consensus motif. The parameters of GLAM2 were set as default. The resultant motif was then searched in entire E. coli K12 proteome and human proteome by GLAM2SCAN (30).
Protein 3D Structure and Secondary Structure Prediction
All protein 3D structures were obtained from published studies (31–38) and RCSB PDB (39), and visualized by RasMol software (40). We used the EcoGene 3.0 (41), which contained the QUARK prediction method (42), to predict the secondary structure of proteins without reported 3D structures.
RESULTS
Many studies have reported that 2-oxohistidine residue had been found in various peptides and proteins (16, 43–51). To identify proteins which may bind specifically to 2-oxohistidine residue, we devised an experimental strategy illustrated in Fig. 1. First, we fabricated the E. coli K12 proteome chip, generated the 2-oxohistidine containing peptides, and probed these peptides with E. coli K12 proteome chips. After identifying the positive hits, we used fluorescence polarization assays to validate the interactions and measured the binding affinity by dose-response measurements. Then, we surveyed the consensus motif among these identified proteins and applied to human proteome to look for possible human 2-oxohistidine interacting proteins. Finally, we used the functional interaction network to find out the possible interacting proteins and used GO and KEGG to figure out related processes and pathways (Fig. 1).
Fig. 1.
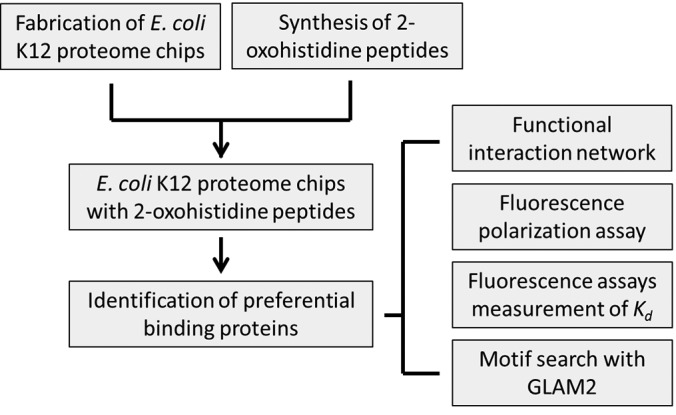
Overall strategy for the identification of 2-oxohistidine interacting proteins using E. coli K12 proteome chip. We expressed and purified ∼4,300 E. coli proteins in high-throughput manner to fabricate the E. coli K12 proteome chip. We used an improved condition to obtain 2-oxohistidine peptides in high yield and purity. 2-Oxohistidine peptides were then probed to E. coli K12 proteome chip and preferential binding proteins were identified. We also built their functional interaction network to investigate their biological roles. Fluorescence polarization assays were used to validate the identified binding proteins. We conducted dose-response fluorescence assays to measure the Kd of these proteins. Furthermore, we used GLAM2 to search consensus motif among these identified proteins and also applied this motif to the entire E. coli K12 proteome and human proteome by GLAM2SCAN.
Oxidation of Peptide Histidine Residue
To synthesize peptide probes, histidine residues were placed in the middle of 12-mer or 13-mer peptides to eliminate possible charge effects at N terminus and C terminus, creating a context similar to proteins. Easily oxidized amino acids, such as methionine, cysteine, tyrosine, tryptophan, phenylalanine, lysine, and arginine, were avoided. Three peptides containing a single histidine residue and random selections of other residues, namely AGAQVAHGNEVAG (AG), SEAGVNHGSAGQA (SE), and IAVENVHGGLA (IA), were used for chip assays. We carried out MCO reactions using the copper/ascorbate/air system shown in Fig. 2 to convert them to 2-oxohistidine containing peptides (Oxo-AG, Oxo-SE, Oxo-IA). The HPLC yield of peptides Oxo-AG and Oxo-SE were around 10%, and for Oxo-IA peptide around 20% (Fig. 2). The site of oxidative modification was confirmed by LC-MS/MS for all peptides (supplemental Fig. S1).
Fig. 2.
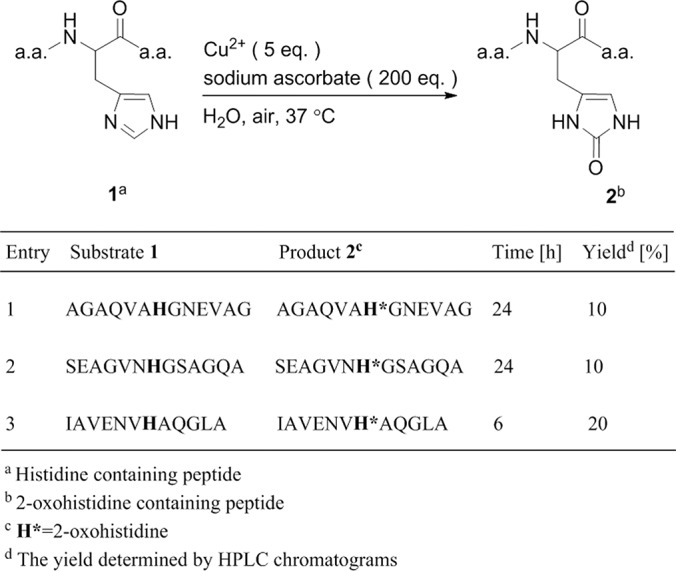
Summary scheme for the synthesis of 2-oxohistidine-containing peptides. Using metal-catalyzed oxidation, the histidine side chain on three different peptides (AG, SE, IA) was converted to 2-oxohistidine.
E. coli K12 Proteome Chip Assays
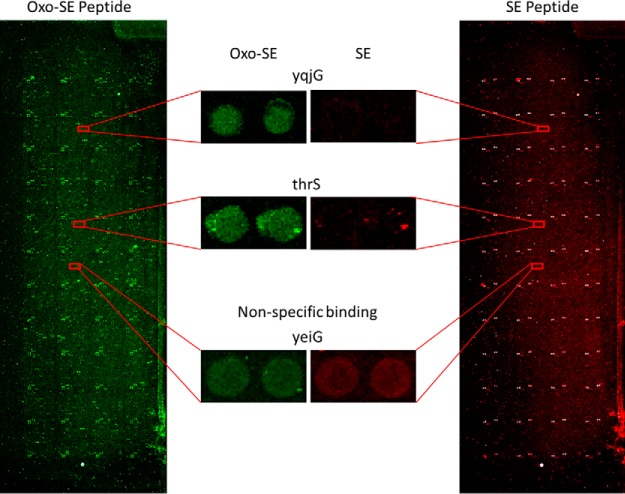
To investigate 2-oxohistidine interacting proteins, AGAQVAH*GNEVAG (Oxo-AG peptide) and SEAGVNH*GSAGQA (Oxo-SE peptide) were conjugated to DyLight 550 fluorophore, where H* denotes 2-oxohistidine. Nonoxidized AG and SE peptides were conjugated to DyLight 650 as negative controls. In the chip assay, 2-oxohistidine containing peptide and its negative control were probed with E. coli K12 proteome chips in triplicate (Fig. 3). Examples of 2-oxohistidine interacting proteins compared with nonspecific binding proteins were shown in Fig. 4.
Fig. 3.
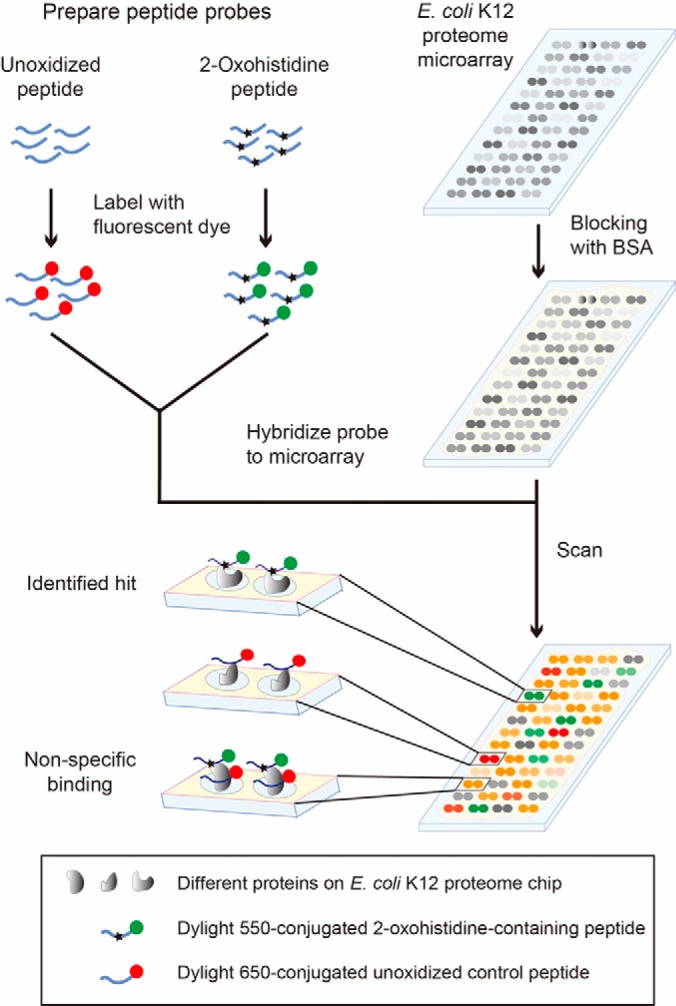
Schematic of E. coli K12 proteome chip assays with 2-oxohistidine peptide probes. To detect 2-oxohistidine interacting proteins, E. coli K12 proteome chips were probed with 2-oxohistidine-containing peptides and unoxidized control peptides labeled with different fluorophores. Each protein was printed in duplicate spots on the chip.
Fig. 4.
Representative images of the E. coli K12 proteome chips probed with 2-oxohistidine containing peptide (Oxo-SE peptide) and unoxidized peptide (SE peptide). The representative positive hits (yqjG and thrS) and nonspecific binding protein (yeiG) on the chip are enlarged from sample images of Oxo-SE peptide and SE peptide, respectively. The contrast and brightness of images have been adjusted equally using the same parameters.
To identify specific hits to 2-oxohistidine peptides, we set several cutoffs. First, we chose the hits which had strong intensities in experimental groups. Second, we selected for the hits which had high signals in experimental groups and low signals in negative controls. Thus, we chose the hits which had large differences between experimental groups and negative controls. Third, although we chose the hits which had large differences between two groups, there were still some strong signals in negative controls. To exclude this kind of nonspecific binding to 2-oxohistidine residue, we removed the hits >1.5 standard deviations above the mean for all negative controls. Fourth, in order to have reproducible results among triplicate chip assays, we excluded the hits which had large variances as described in the section of experimental procedures. Under such stringent criteria, 38 and 20 protein hits were found to bind Oxo-SE peptide and Oxo-AG peptide, respectively (supplemental Table S1–S2). To avoid the nonspecific binding because of the different peptide sequences, we chose the hits shared by both 2-oxohistidine peptides among those proteins. Only 10 proteins (ThrS, YqjG, YajL, HemE, IlvA, PrpD, Zwf, Eda, Gor, and PqqL) were identified by both 2-oxohistidine containing peptides (Table I).
Table I. 2-Oxohistidine interacting proteins identified by E. coli K12 proteome chips. There were 38 and 20 proteins are identified by Oxo-AG peptide and Oxo-SE peptide chip assays, respectively. There were 10 proteins which showed affinity toward both peptide probes, as shown below.
| Accession ID | Protein Symbol | Protein Name | Protein Function |
|---|---|---|---|
| EG11001 | thrS | Threonyl-tRNA synthetase | An enzyme involved in protein synthesis, which is regulated by aerobic and anaerobic metabolisms |
| EG12746 | yqjG | Glutathionyl-hydroquinone reductase | Reduction of organic small molecules |
| EG13272 | yajL | Anti-oxidative stress chaperone | A covalent chaperone for thiol-containing proteome, also promoting disulfide formation |
| EG11543 | hemE | Uroporphyrinogen decarboxylase | Involved in the synthesis of heme group, which is a critical cofactor for antioxidant enzymes |
| EG10493 | ilvA | Threonine dehydratase | A metabolic enzyme that converts threonine to 2-oxobutanoate, regulated by an oxygen-responsive promoter |
| EG13603 | prpD | 2-Methylcitrate dehydratase | A metabolic enzyme in the methylcitrate cycle that converts propionyl-CoA to pyruvate |
| EG11221 | zwf | Glucose-6-phosphate dehydrogenase | A metabolic enzyme in the pentose-phosphate pathway that supplies reducing power to cells by generating NADPH |
| EG10256 | eda | KDPG aldolase | An enzyme in the Entner-Doudoroff pathway, also a multi-function aldolase to detoxify aldehydes generated by oxidative stress |
| EG10412 | gor | Glutathione reductase | An enzyme that generates glutathione to maintain a reducing environment in the cell |
| EG11744 | pqqL | Putative periplasmic M16 family zinc metalloendopeptidase | An enzyme involved in pyrroloquinoline quinone biosynthesis, which is a redox cofactor that supplies reducing power |
We used heat map to visualize the intensity of these 10 identified proteins among 2-oxohistidine and nonoxidized probing results (Fig. 5). The heat map shows that our 10 identified proteins clearly classified the 2-oxohistidine peptides from nonoxidized peptides.
Fig. 5.
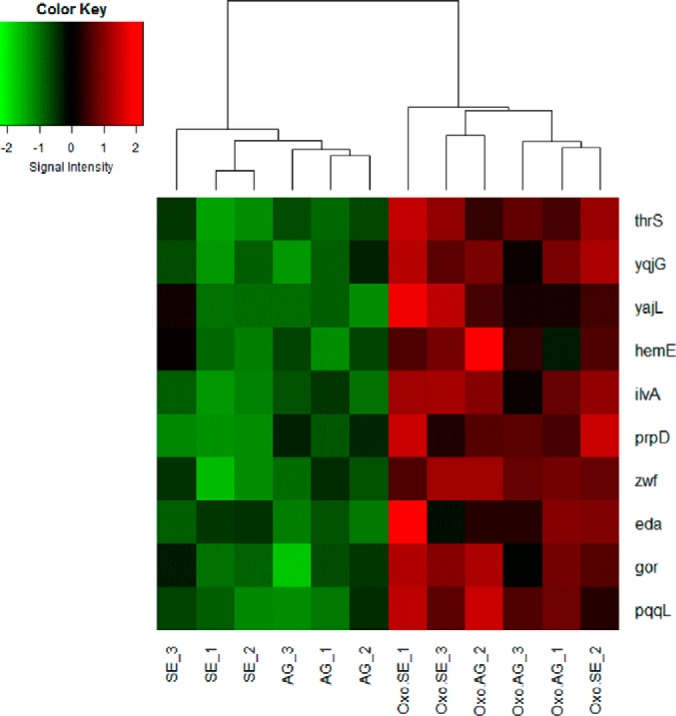
The heat map of 10 identified proteins. The heat map shows the classification of 10 identified proteins in Oxo-AG, Oxo-SE, AG, and SE chip assay probing results. Each peptide probe were analyzed in triplicate. The R programming language and gplots package were used to display the heat map.
Functional Interaction Analysis
We exploited EcID to find the interacting partners of 2-oxohistidine-binding proteins. The EcID database (Escherichia coli Interaction Database) (24) provided a framework for the integration of several protein interaction data sources, including EcoCyc (metabolic pathways, protein complexes, and regulatory information), KEGG (metabolic pathways), MINT and IntAct (protein interactions), high-throughput experiments (protein complexes), and iHOP (text mining).
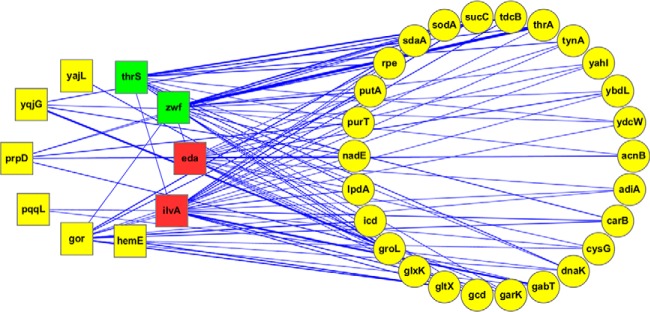
In this study, we only selected interactions supported by experimental evidence from more than one database to ensure reliability and confidence. Through functional network analysis, we identified 26 additional “secondary interacting proteins” which are known to interact with 3 or more of the 10 proteins that bind 2-oxohistidine. As shown in Fig. 6, four of these 2-oxohistidine binding proteins (ThrS, Zwf, Eda, IlvA) appeared to be “hubs” that connected many interacting proteins in the network. We further analyzed this functional interaction network, including 10 identified 2-oxohistidine binding proteins and 26 secondary interacting proteins, using AmiGO 2 (26) and KOBAS 2.0 (27) to provide the GO (28) and KEGG (29) results, respectively (supplemental Table S3–S5). Interestingly, 15 out of the 36 proteins (∼40%) were related to oxidation-reduction process, which showed significant enrichment (p < 0.05). Table II summarizes the related GO terms and KEGG pathways. Oxidation-reduction processes are metabolic activities involving the transfer of electrons between chemical species (52). Because 2-oxohisitidine is a product of oxidative modification, it is reasonable to find its binding partners and secondary interactors to be associated with redox pathways. Moreover, 21 of the 36 proteins are related to oxoacid metabolism, which regulates energy metabolism, reducing power, and ROS levels in the cell (53).
Fig. 6.
The functional interaction network of the 10 identified 2-oxohistidine binding proteins and 26 secondary interacting proteins. The interaction pairs for 10 identified proteins were downloaded from EcID database, and functional interaction network was visualized by Cytoscape. We found 26 secondary interacting proteins that interacted with at least three out of the 10 proteins which bind 2-oxohistidine. Four out of ten identified proteins, (eda, ilvA, zwf, and thrS) have many interactions and were considered to be hubs. Square shapes represent the 10 identified proteins, and round shapes represent the 26 secondary interacting proteins. The node colors reflect the number of interactions: the red for >10 interactions, the green for 5–10 interactions, and the yellow for <5 interactions. The thickness of connecting lines reflects the strength of protein-protein interactions, determined by the number of reported interactions from database searches.
Table II. Summary for functional analysis of 36 proteins from functional interaction network. A total of 36 proteins, including 10 identified 2-oxohistidine binding proteins and 26 secondary interacting proteins, are used to conduct the functional analysis. The GO and KEGG results are generated by AmiGO 2 and KOBAS 2.0, respectively. We summarize the related GO terms and KEGG pathways in this table. Detailed information on GO and KEGG results are given in supplementary Table S3–S5.
| ID | Proteins involved | p value | |
|---|---|---|---|
| GO Term (Biological process) | |||
| Oxoacid metabolic process | GO:0043436 | 21 | 5.29E-08 |
| Oxidation-reduction process | GO:0043436 | 15 | 5.03E-03 |
| GO Term (Molecular function) | |||
| Ion binding | GO:0043167 | 27 | 3.43E-05 |
| Cofactor binding | GO:0048037 | 15 | 2.38E-06 |
| KEGG | |||
| Carbon metabolism | eco01200 | 11 | 2.89E-03 |
| Biosynthesis of secondary metabolites | eco01110 | 19 | 1.25E-02 |
In terms of molecular functions, ion binding and cofactor binding were enriched in our network, which were indicative of important enzymes in the cell. In terms of KEGG results, carbon metabolism and biosynthesis of secondary metabolites were significantly involved. Redox changes in molecules are frequently associated with the biosynthesis and modification of secondary metabolites (54), which may explain this result. These data suggest that 2-oxohistidine binding proteins and their interacting proteins may be involved in redox processes, oxygen-sensitive environmental responses, and central metabolic pathways, and that 2-oxohistidine may be a sensor or marker for oxidative stress.
Fluorescence Polarization Assays
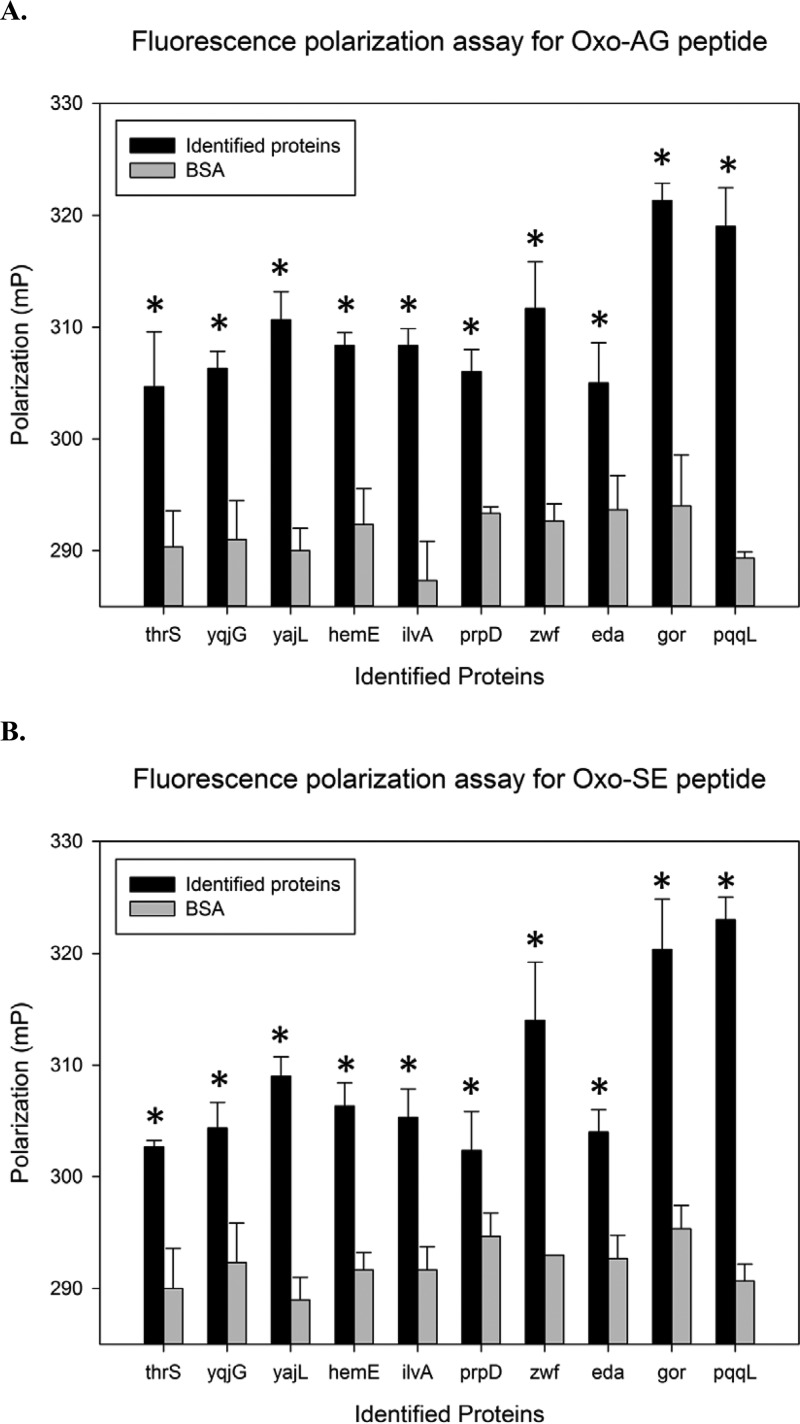
Although we found positive hits in the proteome chip assay, we could not exclude the possible bias of this type of heterogeneous binding approach. On the other hand, fluorescence polarization assay is a homogeneous binding detection method to mimic the interaction between two compounds in the cellular solution environment (55–59). We used fluorescence polarization assays to investigate the binding between fluorescent peptide probes and the 10 identified interacting proteins. Once the protein binds to the fluorescent 2-oxohistidine containing peptide, an increase in the degree of polarization is expected. As shown in Fig. 7, all the 10 identified proteins displayed higher polarization than the negative control, BSA. Besides, the polarization distribution of two oxidized peptides was similar to each other. It indicated that interaction between proteins and 2-oxohistidine was little affected by different peptide sequences. The result confirmed that the 10 identified proteins can preferentially bind to 2-oxohistidine in both AG and SE peptides.
Fig. 7.
Validation of the interactions between 2-oxohistidine peptides and identified proteins using fluorescence polarization assays. Tested proteins were compared with identical concentrations of BSA, which served as the negative control. A, The fluorescence polarization assays for Oxo-AG peptide and identified proteins. B, The fluorescence polarization assays for Oxo-SE peptide and identified proteins. The error bars represent S.E. The asterisks indicate p < 0.05 by unpaired t test against the BSA control.
Measurement of Binding Affinity
Dissociation constant (Kd) describes the propensity of a ligand-protein complex to dissociate reversibly into its components. We measured the Kd of these identified proteins to oxidized peptides, normal peptides, and quenched fluorescent dyes by dose-response measurements. Fluorescent 2-oxohistidine-containing peptides at different concentrations were probed onto the slide, where the identified proteins were immobilized (supplemental Fig. S2A). Using double-reciprocal plot analysis, we calculated the Kd values for all identified proteins (supplemental Fig. S2B). The same procedures were also repeated for normal peptides and fluorescent dyes. The result showed that our 10 identified proteins had strong affinities to 2-oxohistidine, ranging from 10−8 to 10−10 m, especially the HemE protein, which had the strongest Kd (∼10−10 m) for both 2-oxohistidine containing peptides (Table III). We also found that our proteins slightly preferred Oxo-SE peptide than Oxo-AG peptide, but the difference in Kd was within one order of magnitude. Importantly, the Kd values for oxidized peptides were significantly higher than nonoxidized peptides and quenched fluorescent dyes (p < 0.05). To further validate this interaction, we synthesized a third peptide probe, IAVENVH*QGLA (Oxo-IA peptide) and its negative control (IAVENVHQGLA, IA peptide), and we also swapped their fluorescent dyes to avoid the influence of fluorescent dyes. The result also showed statistically significant difference between Oxo-IA and IA peptides (p < 0.05). This indicated that our 10 identified proteins had strong binding affinities to 2-oxohistidine, which were not affected by different peptide sequences and different fluorescent dyes.
Table III. Kd values for 2-oxohistidine peptides binding to identified proteins. All Kd values were determined by dose-response measurements. Three pairs (oxidized vs. nonoxidized) of peptide probes with different sequences were investigated.
| Name | Oxidized peptides |
Unoxidized peptides |
Fluorescent dyes |
|||||
|---|---|---|---|---|---|---|---|---|
| DyLight 550 Oxo-AG | DyLight 550 Oxo-SE | DyLight 650 Oxo-IA | DyLight 650 AG | DyLight 650 SE | DyLight 550 IA | DyLight 550 | DyLight 650 | |
| thrS | 1.2E-8 ± 9.6E-10a | 6.7E-9 ± 1.9E-9a | 3.9E-8 ± 4.7E-9a | 1.4E-7 ± 3.5E-8 | 2.5E-7 ± 8.3E-8 | 1.0E-7 ± 6.8E-8 | 8.5E-8 ± 1.1E-8 | 1.1E-7 ± 1.6E-8 |
| yqjG | 1.1E-8 ± 9.2E-10a | 3.4E-9 ± 2.4E-10a | 1.4E-8 ± 1.7E-9a | 4.9E-7 ± 2.1E-7 | 1.0E-7 ± 4.5E-9 | 1.2E-7 ± 7.0E-8 | 2.9E-7 ± 9.7E-8 | 1.4E-7 ± 2.7E-8 |
| yajL | 5.6E-8 ± 2.8E-8a | 1.4E-8 ± 7.5E-9a | 1.0E-7 ± 3.1E-8a | 3.7E-7 ± 1.9E-7 | 8.6E-7 ± 2.8E-7 | 2.2E-7 ± 1.2E-7 | 2.5E-7 ± 1.3E-7 | 4.4E-7 ± 2.9E-7 |
| hemE | 8.7E-10 ± 6.1E-11a | 6.9E-10 ± 2.3E-11a | 5.6E-9 ± 5.3E-10a | 1.9E-7 ± 3.4E-8 | 1.2E-8 ± 7.7E-10 | 1.2E-7 ± 3.5E-8 | 1.4E-7 ± 5.4E-8 | 4.8E-8 ± 5.7E-9 |
| ilvA | 1.6E-8 ± 4.1E-9a | 2.8E-8 ± 3.6E-8a | 5.9E-7 ± 1.7E-7a | 2.9E-7 ± 4.1E-8 | 2.9E-7 ± 5.9E-8 | 1.5E-6 ± 6.8E-7 | 8.5E-8 ± 5.4E-8 | 1.3E-6 ± 5.3E-7 |
| prpD | 1.6E-7 ± 1.7E-7a | 9.0E-8 ± 1.8E-7a | 1.3E-7 ± 2.1E-8a | 7.3E-7 ± 1.0E-7 | 6.2E-7 ± 4.2E-7 | 7.9E-7 ± 1.7E-7 | 1.3E-6 ± 8.7E-7 | 2.4E-6 ± 5.7E-7 |
| zwf | 1.4E-8 ± 1.5E-9a | 2.3E-8 ± 2.7E-8a | 7.2E-8 ± 1.7E-8a | 1.2E-6 ± 5.3E-7 | 5.8E-7 ± 2.2E-7 | 3.7E-7 ± 2.7E-7 | 5.6E-7 ± 3.6E-7 | 6.4E-7 ± 2.7E-7 |
| eda | 3.9E-9 ± 2.7E-10a | 2.0E-9 ± 2.6E-10a | 9.2E-8 ± 1.6E-8a | 3.3E-7 ± 1.1E-7 | 2.6E-7 ± 9.4E-8 | 2.6E-7 ± 1.1E-7 | 2.2E-7 ± 2.4E-7 | 4.6E-7 ± 2.1E-7 |
| gor | 2.4E-8 ± 3.5E-9a | 1.2E-8 ± 1.8E-9a | 8.3E-8 ± 3.4E-8a | 1.1E-6 ± 5.5E-7 | 8.6E-7 ± 5.0E-7 | 2.2E-7 ± 8.2E-8 | 9.0E-7 ± 5.0E-7 | 6.1E-7 ± 3.7E-7 |
| pqqL | 5.4E-9 ± 4.7E-10a | 1.8E-9 ± 2.3E-10a | 6.7E-8 ± 1.1E8a | 3.0E-7 ± 2.0E-7 | 3.0E-7 ± 1.1E-7 | 3.7E-7 ± 2.7E-7 | 1.5E-7 ± 9.2E-8 | 6.3E-7 ± 3.8E-8 |
a Significant difference to its normal peptide control and fluorescent dye control (p < 0.05, unpaired t-test).
Motif Searching in E. coli Proteome and Human Proteome
Based on fluorescence polarization and binding affinity results, we used GLAM2 (Gapped Local Alignment of Motifs) (30) to search for a consensus motif among these identified proteins. In this study, we found the consensus motif among these identified proteins to be [DS][VQ][DTE]A[YIL]X[AK][ARL][MV][ETK][LV][AYLF]E, where X indicates any amino acid (Fig. 8). In addition, we used this motif to query the entire E. coli K12 proteome by GLAM2SCAN (30). The result showed that the top ten ranking proteins containing this motif were identical to our identified proteins (Table IV). This indicated that the motif was significantly unique in the entire E. coli K12 proteome (p < 0.05). We also applied this motif to the entire human proteome, and found the top-ranking protein to be S100 Calcium Binding Protein A1 (S100A1), which is a member of the S100 family (supplemental Table S6).
Fig. 8.
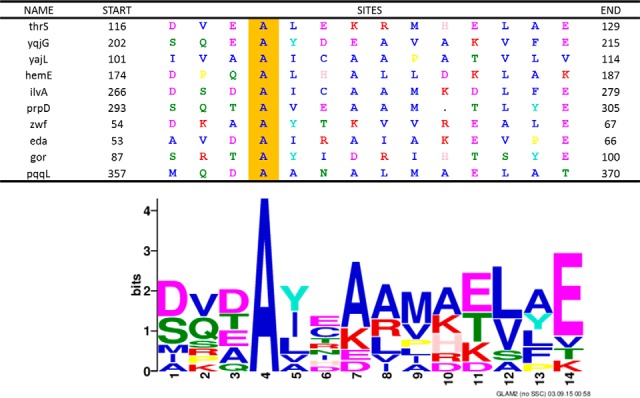
Consensus motif among the 10 validated proteins. The logo shows the consensus motif [DS][VQ][DTE]A[YIL]X[AK][ARL][MV][ETK][LV][AYLF]E identified by GLAM2. The table shows the protein sequences of 10 validated 2-oxohistidine binding proteins aligned with the consensus motif.
Table IV. Top 10 protein hits from [DS][VQ][DTE]A[YIL]X[AK][ARL][MV] [ETK][LV][AYLF]E motif-scan for the entire E. coli K12 proteome. The motif was searched in entire E. coli K12 proteome by GLAM2SCAN.
| Rank | Name | EcoGene accession | Start | Sequence | End | Score |
|---|---|---|---|---|---|---|
| 1 | ilvA | EG10493 | 266 | DSDAICAAMKDLFE | 279 | 29.2 |
| 2 | thrS | EG11001 | 116 | DVEALEKRMHELAE | 129 | 28.1 |
| 3 | yqjG | EG12746 | 202 | SQEAYDEAVAKVFE | 215 | 26 |
| 4 | pqqL | EG11744 | 357 | MQDAANALMAELAT | 370 | 24.3 |
| 5 | prpD | EG13603 | 293 | SQTAVEAAM.TLYE | 305 | 23 |
| 6 | eda | EG10256 | 53 | AVDAIRAIAKEVPE | 66 | 22.6 |
| 7 | gor | EG10412 | 87 | SRTAYIDRIHTSYE | 100 | 22.5 |
| 8 | zwf | EG11221 | 54 | DKAAYTKVVREALE | 67 | 21.8 |
| 9 | yajL | EG13272 | 101 | IVAAICAAPATVLV | 114 | 21.1 |
| 10 | hemE | EG11543 | 174 | DPQALHALLDKLAK | 187 | 20 |
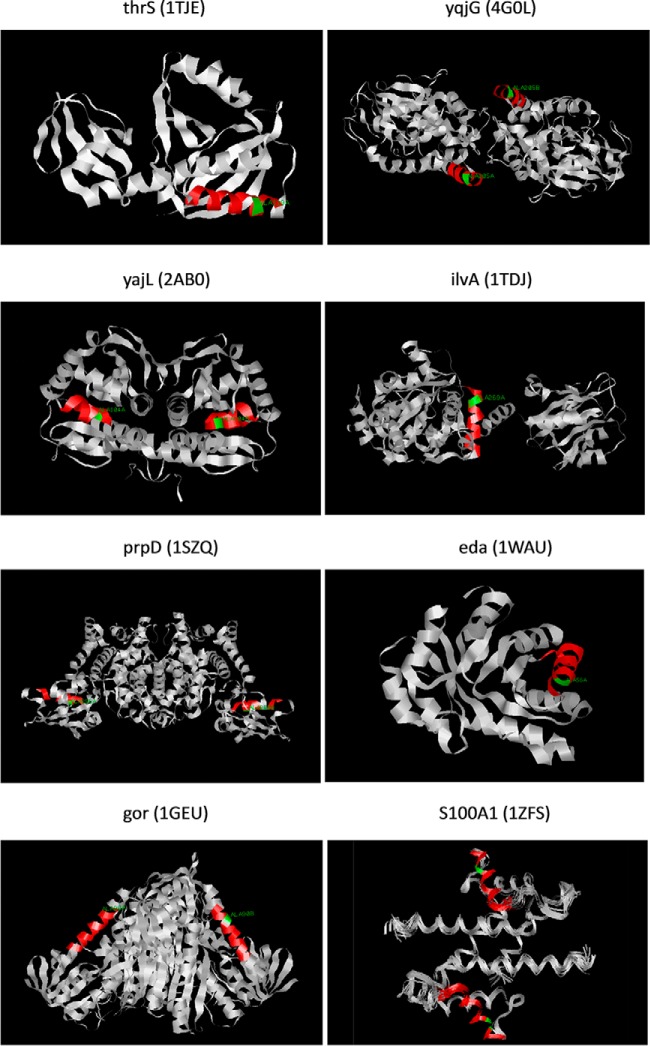
After motif screening in E. coli and human proteomes, we further investigated the secondary structure of the motif region in our identified proteins and S100A1 using protein 3D structures (Fig. 9). However, there were 3 proteins (HemE, Zwf, and PqqL) lacking published 3D structures. For these three proteins, we used the QUARK prediction method to predict their secondary structures. By 3D structure analysis and QUARK prediction, the result showed that this consensus motif usually formed alpha-helices in these proteins except for YajL, which contained 36% beta-sheet and 64% alpha-helix in the motif (Table V). Besides, we found that these helical motifs generally faced the outside of the protein, which meant they had the chance to interact with other molecules. Our finding suggested that 2-oxohistidine recognition motif was an alpha-helical structure and conserved between E. coli and humans.
Fig. 9.
Protein 3D structures of E. coli identified proteins and human S100A1. Seven of the E. coli identified proteins (thrS, yqjG, yajL, ilvA, prpD, eda, gor) and human S100A1 have published protein 3D structures, which are visualized by RasMol software. The red region represents the consensus motif found by GLAM2, and the green region indicates the conserved alanine at the fourth position in the consensus motif. YqjG, yajL, prpD, gor and S100A1 were deposited in RCSB PDB as homodimer structures, whereas the others were monomers.
Table V. Secondary structure of motifs from 10 E. coli K12 identified proteins and human S100A1. The secondary structure of the consensus motif in each protein is determined from published crystal structures. However, hemE, zwf and pqqL do not have available crystal structures, and we used the QUARK prediction in EcoGene 3.0 to predict their secondary structures.
| Name | Secondary structure of motif | Source | PDB ID | QUARK ID | Reference |
|---|---|---|---|---|---|
| thrS | α-helix | RCSB PDB | 1TJE | (34, 39) | |
| yqjG | α-helix | RCSB PDB | 4G0L | (38, 39) | |
| yajL | β-sheet + α-helix | RCSB PDB | 2AB0 | (36, 39) | |
| hemE | α-helix | EcoGene 3.0 | E11780 | (41, 42) | |
| ilvA | α-helix | RCSB PDB | 1TDJ | (33, 39) | |
| prpD | α-helix | RCSB PDB | 1SZQ | (31, 39) | |
| zwf | α-helix | EcoGene 3.0 | E14278 | (41, 42) | |
| eda | α-helix | RCSB PDB | 1WAU | (37, 39) | |
| gor | α-helix | RCSB PDB | 1GEU | (32, 39) | |
| pqqL | α-helix | EcoGene 3.0 | E12551 | (41, 42) | |
| S100A1 | α-helix | RCSB PDB | 1ZFS | (35, 39) |
Kd Measurement Between Human S100A1 Protein and the Oxidized Peptides
To validate the interaction of human S100A1 protein that we found by GLAM2SCAN for the entire human proteome, we calculated the Kd values according to dose-response measurements for all oxidized peptides, including Oxo-AG, Oxo-SE, and Oxo-IA peptide. The result showed that S100A1 protein had a strong affinity to all 2-oxohistidine-containing peptides, significantly stronger than nonoxidized peptides and fluorescent dyes (p < 0.05) (Table VI). The binding affinity of S100A1 to 2-oxohistidine peptides was 10-fold to 100-fold higher than the negative controls, indicating that S100A1 actually had the ability to recognize 2-oxohistidine. Because we knew that S100A1 was a calcium binding protein, we wondered whether calcium would affect the interaction or not. The result showed that calcium was not involved in the interaction of S100A1 to 2-oxohistidine peptides or the other groups (p > 0.1). This suggested the E. coli K12 proteome chip was a suitable platform for motif screening in cross-species studies.
Table VI. Kd values for 2-oxohistidine peptides binding to S100A1.
| S100A1 | Oxidized peptides |
Unoxidized peptides |
Fluorescent dyes |
|||||
|---|---|---|---|---|---|---|---|---|
| DyLight 550 Oxo-AG | DyLight 550 Oxo-SE | DyLight 650 Oxo-IA | DyLight 650 AG | DyLight 650 SE | DyLight 550 IA | DyLight 550 | DyLight 650 | |
| no calcium | 5.3E-9 ± 4.9E-9a,b | 5.2E-9 ± 2.1E-9a,b | 1.2E-8 ± 4.2E-9a,b | 3.9E-8 ± 4.0E-8 | 8.2E-8 ± 5.7E-8 | 3.1E-8 ± 2.0E-8 | 3.4E-7 ± 2.4E-7 | 3.6E-7 ± 3.6E-7 |
| w/calcium | 7.3E-9 ± 4.8E-9a | 2.9E-9 ± 6.4E-10a | 2.8E-8 ± 9.8E-9a | 1.3E-7 ± 9.1E-8 | 8.5E-8 ± 5.5E-8 | 6.6E-8 ± 3.7E-8 | 5.6E-8 ± 3.8E-8 | 1.2E-7 ± 8.4E-8 |
a Significant difference to its normal peptide control and fluorescent dye control (p < 0.05, unpaired t-test).
b No significant difference compared to calcium group (p > 0.1, unpaired t-test).
DISCUSSION
Enzymatic and nonenzymatic PTMs are comparable in their diversity and chemical complexity, but past research efforts have mostly focused on the former, leaving a huge gap in our understanding of biological phenomena associated with nonenzymatic PTMs. Even though nonenzymatic PTMs are not generated by enzyme actions, there may still be specific enzymes to chemically reverse such modifications, or specific receptors to detect such modification. For example, the chemical oxidation of methionine to methionine sulfoxide can be reduced back to methionine by specific reductases MsrA and MsrB (60); RAGE can recognize protein glycation and lead to inflammatory responses (7). However, there are still many nonenzymatic PTMs with elusive biological functions.
Among nonenzymatic PTMs, 2-oxohistidine is particularly interesting because of its minimal size, involving the insertion of just one oxygen atom. It probably represents the smallest atomic-scale alteration associated with a known PTM, and we investigated if cells have evolved the ability to monitor such a small change on the surface of proteins. Because the histidine residue often plays critical roles in protein function, both structurally and catalytically, we hypothesized there would be cellular factors that specifically recognize 2-oxohistidine side chains, and this hypothesis was tested with specially synthesized peptide probes and E. coli proteome chips.
Using three peptide probes with homogeneous 2-oxohistidine modifications, we were able to identify 10 proteins that show preferential binding for 2-oxohistidine-containing peptides over nonoxidized control peptides (Table I). Because these three probes have very different flanking sequences, it is very likely that we have identified proteins which specifically recognize side-chain differences between 2-oxohistidine and histidine, and we will refer to them as 2-oxohistidine recognition factors. Before this study, the recognition factors of 2-oxohistidine had never been proposed or identified.
In theory, the recognition of 2-oxohistidine could play several different biological roles. First, it may act as a redox sensor, similar to S-nitrosylation (61). Secondly, it may identify oxidatively damaged proteins and mark it for degradation. Third, it may trigger cellular stress responses and antioxidant pathways. Although there is no known involvement of 2-oxohistidine in various E. coli physiological pathways, several of the recognition factors in E. coli appear to be related to redox pathways and antioxidant pathways.
Among the 10 putative recognition factors identified via the proteome microarray, 9 seemed to be involved in redox-related cellular functions. Gor is a glutathione reductase, involved in the generation of glutathione, which maintains the reducing environment of the cell (62). YqjG is glutathionyl hydroquinone reductase, which utilizes glutathione to reduce a wide range of organic molecules (38). HemE is a uroporphyrinogen decarboxylase involved in the biosynthesis of the heme group, which is an important cofactor for antioxidant enzymes like catalase and peroxidase (63). Zwf is a glucose-6-phosphate dehydrogenase, which helps supply dihydronicotinamide-adenine dinucleotide phosphate (NADPH) through the pentose phosphate pathway (64), and NADPH is a cofactor used as a reducing agent by many metabolic enzymes (65, 66). PqqL in E. coli is a putative zinc metalloprotease, but functionally it may be similar to PqqF in Klebsiella pneumoniae, which has a supportive role in pyrroloquinoline quinone biosynthesis (67). Pyrroloquinoline quinone is a redox cofactor that provides reducing power for the cell, and also a ROS scavenger (68).
YajL is an anti-oxidative-stress chaperone, which promotes disulfide formation to help maintain order in the thiol proteome (69). Interestingly, the human homolog of YajL, DJ-1, is also an antioxidative stress protein, and its mutations are known to cause familial Parkinsonism (70). On the other hand, IlvA and ThrS are both involved in threonine metabolism, and known to be regulated by oxygen levels in the cell. IlvA, a threonine dehydratase, converts threonine to 2-oxobutanoate, and its promoter is activated by oxygen (71). ThrS is a threonyl-tRNA synthetase, and potentially also an oxygen sensor in the cell through Cys182 oxidation (72). Eda, Entner-Doudoroff aldolase (also called KDPG aldolase), is involved in the Entner-Doudoroff pathway that generates pyruvate and NADPH by consuming glucose. Eda is a multi-functional aldolase which also catalyzes the addition of pyruvate to electrophilic aldehydes to detoxify harmful byproducts generated by oxidative stress (73).
PrpD, a 2-methylcitrate dehydratase, does not appear to be directly involved in redox functions, but it converts propionyl-CoA into pyruvate through the methylcitrate cycle (74), and pyruvate can be utilized by the aforementioned Eda enzyme to detoxify oxidized organic molecules with aldehydes. Therefore, all 10 putative recognition factors for 2-oxohistidine identified here appear to be involved in supplying reducing power to the cell or involved in oxygen-sensitive regulation of carbon metabolism. This strongly implies that recognition of 2-oxohistidine in E. coli may play certain roles in redox sensing and metabolic regulation, but further experiments are required to elucidate the actual biological function.
Using motif analysis by GLAM2 and GLAM2SCAN, we identified a putative 2-oxohistidine binding motif from these 10 recognition factors, which turned out to be: [DS][VQ][DTE]A[YIL]X[AK][ARL][MV][ETK][LV][AYLF]E. It is plausible that the binding pocket is constituted by the highly conserved Ala at the 4th position and the hydrophobic [YIL] at the fifth position. This pocket may accommodate the uncharged 2-oxohistidine side chain by hydrophobic interaction. In contrast, histidine is positively charged at pH 7 and cannot stably bind to a hydrophobic pocket. Histidine side chain is also likely repelled by the [AK][ARL] residues downstream of the conserved Ala, and attracted to the [DS][VQ][DTE] residues upstream. This may offer a rudimentary explanation for the basis for selective binding for 2-oxohistidine, but further structural and mutagenesis data will be required to support our prediction.
We further validated this binding motif by searching for the highest-scoring match in the human proteome, which turned out to be DVDAVDKVMKELDE on S100A1 protein, and we verified that S100A1 indeed exhibited 2-oxohistidine binding affinity. S100A1 is a calcium binding protein highly expressed in the brain and heart, and its calcium binding affinity is greatly enhanced by the oxidative nitrosylation of Cys86 (75). It is believed to regulate calcium and nitric oxide signaling in neuronal cells, affecting neurotransmitter release as well as inflammation (76). Interestingly, because S100A1 is also secreted extracellularly (77), it may bind to oxidized amyloid beta (Aβ) with 2-oxohisitidine side chains, which are released from extracellular senile plaques that trap metals and generate ROS (45, 78, 79). Because misfolded Aβ is known to cause calcium misregulation (80), oxidative stress (81), and inflammatory response (82) in the brain, the interaction between S100A1 and oxidized Aβ through 2-oxohistidine recognition may play a role in Alzheimer's disease (AD) pathogenesis, which warrants future investigation.
Our preliminary evidence suggests that both bacteria and humans have cellular factors which can recognize 2-oxohistidine side chains, and a conserved binding motif has been putatively identified. Through the course of evolution, the recognition of 2-oxohistidine may carry important cellular functions related to redox signaling. We have also shown that E. coli K12 proteome microarray is capable of being exploited as a motif library for screening small molecule binding, and that even a single-atom modification on the molecule may be recognized. We expect a wide application of this approach for studying the interaction of other post-translational modifications, such as phosphorylation, methylation, acetylation, amidation, thiolation, sulfation, nitrosylation, as well as many nonenzymatic PTMs. With regard to 2-oxohistidine, future work is required to elucidate how single-oxygen insertion can be recognized on the protein surface, and how recognizing this modification regulates biological functions.
Supplementary Material
Footnotes
Author contributions: H.T. and C.C. designed research; J.L., Y.T., Y. Liu, and Y. Lin performed research; J.L., H.T., and C.C. analyzed data; J.L., H.T., and C.C. wrote the paper.
* CSC was supported by the Ministry of Science and Technology, R.O.C. (MOST 104-2320-B-008-002-MY3 and MOST 104-2627-M-008-001), the Aim for the Top University Project (104G705-4), the Cathay General Hospital (104CGH-NCU-A1) and the Landseed Hospital, Taiwan (NCU-LSH-104-A-001). HCT was supported by grant MOST 103-2113-M-002-007-MY2.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- PTM
- post-translational modification
- MCO
- metal-catalyzed oxidation
- RAGE
- receptors for advanced glycation end-products
- Aβ
- amyloid beta
- AD
- Alzheimer's disease
- GO
- Gene Ontology
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- BSA
- bovine serum albumin
- TBST
- Tris-buffered saline with Tween 20
- Kd
- dissociation constant
- AG peptide
- AGAQVAHGNEVAG, SE peptide
- SEAGVNHGSAGQA
- IA peptide
- IAVENVHAQGLA
- Oxo-AG peptide, AG peptide with 2-oxohistidine residue
- Oxo-SE peptide
- SE peptide with 2-oxohistidine residue
- Oxo-IA peptide
- IA peptide with 2-oxohistidine residue
- NADPH
- dihydronicotinamide-adenine dinucleotide phosphate.
REFERENCES
- 1. Wold F. (1981) In vivo chemical modification of proteins (post-translational modification). Annu. Rev. Biochem. 50, 783–814 [DOI] [PubMed] [Google Scholar]
- 2. Walsh C. T., Garneau-Tsodikova S., and Gatto G. J. Jr. (2005) Protein posttranslational modifications: the chemistry of proteome diversifications. Angew Chem. Int. Ed. 44, 7342–7372 [DOI] [PubMed] [Google Scholar]
- 3. Harding J. J. (1985) Nonenzymatic covalent posttranslational modification of proteins in vivo. Adv. Protein Chem. 37, 247–334 [DOI] [PubMed] [Google Scholar]
- 4. Davies M. J. (2005) The oxidative environment and protein damage. Biochim. Biophys. Acta 1703, 93–109 [DOI] [PubMed] [Google Scholar]
- 5. Morrison D. K. (2009) The 14-3-3 proteins: integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol. 19, 16–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kilpatrick D. C. (2002) Animal lectins: a historical introduction and overview. Biochim. Biophys. Acta 1572, 187–197 [DOI] [PubMed] [Google Scholar]
- 7. Sparvero L. J., Asafu-Adjei D., Kang R., Tang D., Amin N., Im J., Rutledge R., Lin B., Amoscato A. A., Zeh H. J., and Lotze M. T. (2009) RAGE (Receptor for Advanced Glycation Endproducts), RAGE ligands, and their role in cancer and inflammation. J. Transl. Med. 7, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Muller F. L., Lustgarten M. S., Jang Y., Richardson A., and Van Remmen H. (2007) Trends in oxidative aging theories. Free Radic. Biol. Med. 43, 477–503 [DOI] [PubMed] [Google Scholar]
- 9. Ray P. D., Huang B. W., and Tsuji Y. (2012) Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal 24, 981–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Martin K. R., and Barrett J. C. (2002) Reactive oxygen species as double-edged swords in cellular processes: low-dose cell signaling versus high-dose toxicity. Hum. Exp. Toxicol. 21, 71–75 [DOI] [PubMed] [Google Scholar]
- 11. Jang Y. Y., and Sharkis S. J. (2007) A low level of reactive oxygen species selects for primitive hematopoietic stem cells that may reside in the low-oxygenic niche. Blood 110, 3056–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shacter E. (2000) Quantification and significance of protein oxidation in biological samples. Drug Metab. Rev. 32, 307–326 [DOI] [PubMed] [Google Scholar]
- 13. Xu G., and Chance M. R. (2007) Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chem. Rev. 107, 3514–3543 [DOI] [PubMed] [Google Scholar]
- 14. Tainer J. A., Roberts V. A., and Getzoff E. D. (1991) Metal-binding sites in proteins. Curr. Opin. Biotechnol. 2, 582–591 [DOI] [PubMed] [Google Scholar]
- 15. Regan L. (1993) The design of metal-binding sites in proteins. Annu. Rev. Biophys. Biomol. Struct. 22, 257–287 [DOI] [PubMed] [Google Scholar]
- 16. Uchida K., and Kawakishi S. (1994) Identification of oxidized histidine generated at the active site of Cu, Zn-superoxide dismutase exposed to H2O2. Selective generation of 2-oxo-histidine at the histidine 118. J. Biol. Chem. 269, 2405–2410 [PubMed] [Google Scholar]
- 17. Lewisch S. A., and Levine R. L. (1995) Determination of 2-oxohistidine by amino acid analysis. Anal. Biochem. 231, 440–446 [DOI] [PubMed] [Google Scholar]
- 18. Traore D. A., El Ghazouani A., Jacquamet L., Borel F., Ferrer J. L., Lascoux D., Ravanat J. L., Jaquinod M., Blondin G., Caux-Thang C., Duarte V., and Latour J. M. (2009) Structural and functional characterization of 2-oxo-histidine in oxidized PerR protein. Nat. Chem. Biol. 5, 53–59 [DOI] [PubMed] [Google Scholar]
- 19. Davies M. J., Fu S., Wang H., and Dean R. T. (1999) Stable markers of oxidant damage to proteins and their application in the study of human disease. Free Radic. Biol. Med. 27, 1151–1163 [DOI] [PubMed] [Google Scholar]
- 20. Huang C. F., Liu Y. H., and Tai H. C. (2015) Synthesis of peptides containing 2-oxohistidine residues and their characterization by liquid chromatography-tandem mass spectrometry. J. Pept. Sci. 21, 114–119 [DOI] [PubMed] [Google Scholar]
- 21. Chen C. S., Korobkova E., Chen H., Zhu J., Jian X., Tao S. C., He C., and Zhu H. (2008) A proteome chip approach reveals new DNA damage recognition activities in Escherichia coli. Nat. Methods 5, 69–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Team R. C. (2015) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing [Google Scholar]
- 23. Warnes G. R., Bolker B., Bonebakker L., Gentleman R., Huber W., Liaw A., Lumley T., Maechler M., Magnusson A., and Moeller S. (2009) gplots: Various R programming tools for plotting data. R package Version 2 [Google Scholar]
- 24. Andres Leon E., Ezkurdia I., Garcia B., Valencia A., and Juan D. (2009) EcID. A database for the inference of functional interactions in E. coli. Nucleic Acids Res. 37, D629–D635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Carbon S., Ireland A., Mungall C. J., Shu S., Marshall B., Lewis S., Ami G. O. H., and Web Presence Working, G. (2009) AmiGO: online access to ontology and annotation data. Bioinformatics 25, 288–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xie C., Mao X., Huang J., Ding Y., Wu J., Dong S., Kong L., Gao G., Li C. Y., and Wei L. (2011) KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316–W322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., and Sherlock G. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kanehisa M., and Goto S. (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Frith M. C., Saunders N. F., Kobe B., and Bailey T. L. (2008) Discovering sequence motifs with arbitrary insertions and deletions. PLoS Comput. Biol. 4, e1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. PDB ID: 1SZQ Rajashankar K. R., Kniewel R., Solorzano V., Lima C. D., Burley S. K., New York SGXResearch Center for Structural Genomics. Crystal structure of 2-methylcitrate dehydratase.
- 32. Mittl P. R., Berry A., Scrutton N. S., Perham R. N., and Schulz G. E. (1994) Anatomy of an engineered NAD-binding site. Protein Sci. 3, 1504–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gallagher D. T., Gilliland G. L., Xiao G., Zondlo J., Fisher K. E., Chinchilla D., and Eisenstein E. (1998) Structure and control of pyridoxal phosphate dependent allosteric threonine deaminase. Structure 6, 465–475 [DOI] [PubMed] [Google Scholar]
- 34. Dock-Bregeon A. C., Rees B., Torres-Larios A., Bey G., Caillet J., and Moras D. (2004) Achieving error-free translation; the mechanism of proofreading of threonyl-tRNA synthetase at atomic resolution. Mol. Cell 16, 375–386 [DOI] [PubMed] [Google Scholar]
- 35. Wright N. T., Varney K. M., Ellis K. C., Markowitz J., Gitti R. K., Zimmer D. B., and Weber D. J. (2005) The three-dimensional solution structure of Ca(2+)-bound S100A1 as determined by NMR spectroscopy. J. Mol. Biol. 353, 410–426 [DOI] [PubMed] [Google Scholar]
- 36. Wilson M. A., Ringe D., and Petsko G. A. (2005) The atomic resolution crystal structure of the YajL (ThiJ) protein from Escherichia coli: a close prokaryotic homologue of the Parkinsonism-associated protein DJ-1. J. Mol. Biol. 353, 678–691 [DOI] [PubMed] [Google Scholar]
- 37. Fullerton S. W., Griffiths J. S., Merkel A. B., Cheriyan M., Wymer N. J., Hutchins M. J., Fierke C. A., Toone E. J., and Naismith J. H. (2006) Mechanism of the Class I KDPG aldolase. Bioorg. Med. Chem. 14, 3002–3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Green A. R., Hayes R. P., Xun L., and Kang C. (2012) Structural understanding of the glutathione-dependent reduction mechanism of glutathionyl-hydroquinone reductases. J. Biol. Chem. 287, 35838–35848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., and Bourne P. E. (2000) The Protein Data Bank. Nucleic Acids Res. 28, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sayle R. A., and Milner-White E. J. (1995) RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 20, 374. [DOI] [PubMed] [Google Scholar]
- 41. Zhou J., and Rudd K. E. (2013) EcoGene 3.0. Nucleic Acids Res. 41, D613–D624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu D., and Zhang Y. (2012) Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins 80, 1715–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Uchida K., and Kawakishi S. (1993) 2-Oxo-histidine as a novel biological marker for oxidatively modified proteins. FEBS Lett. 332, 208–210 [DOI] [PubMed] [Google Scholar]
- 44. Lewisch S. A., and Levine R. L. (1999) Determination of 2-oxohistidine by amino acid analysis. Methods Enzymol. 300, 120–124 [DOI] [PubMed] [Google Scholar]
- 45. Atwood C. S., Huang X., Khatri A., Scarpa R. C., Kim Y. S., Moir R. D., Tanzi R. E., Roher A. E., and Bush A. I. (2000) Copper catalyzed oxidation of Alzheimer Abeta. Cell Mol Biol 46, 777–783 [PubMed] [Google Scholar]
- 46. Schoneich C. (2000) Mechanisms of metal-catalyzed oxidation of histidine to 2-oxo-histidine in peptides and proteins. J. Pharm. Biomed. Anal. 21, 1093–1097 [DOI] [PubMed] [Google Scholar]
- 47. Gunther M. R., Peters J. A., and Sivaneri M. K. (2002) Histidinyl radical formation in the self-peroxidation reaction of bovine copper-zinc superoxide dismutase. J. Biol. Chem. 277, 9160–9166 [DOI] [PubMed] [Google Scholar]
- 48. Hovorka S. W., Biesiada H., Williams T. D., Huhmer A., and Schoneich C. (2002) High sensitivity of Zn2+ insulin to metal-catalyzed oxidation: detection of 2-oxo-histidine by tandem mass spectrometry. Pharm. Res. 19, 530–537 [DOI] [PubMed] [Google Scholar]
- 49. Schoneich C., and Williams T. D. (2002) Cu(II)-catalyzed oxidation of beta-amyloid peptide targets His13 and His14 over His6: Detection of 2-Oxo-histidine by HPLC-MS/MS. Chem. Res. Toxicol. 15, 717–722 [DOI] [PubMed] [Google Scholar]
- 50. Schiewe A. J., Margol L., Soreghan B. A., Thomas S. N., and Yang A. J. (2004) Rapid characterization of amyloid-beta side-chain oxidation by tandem mass spectrometry and the scoring algorithm for spectral analysis. Pharm. Res. 21, 1094–1102 [DOI] [PubMed] [Google Scholar]
- 51. Inoue K., Garner C., Ackermann B. L., Oe T., and Blair I. A. (2006) Liquid chromatography/tandem mass spectrometry characterization of oxidized amyloid beta peptides as potential biomarkers of Alzheimer's disease. Rapid Commun. Mass Spectrom. 20, 911–918 [DOI] [PubMed] [Google Scholar]
- 52. Boyer R. F. (2005) Concepts in Biochemistry, 3 edition Ed., Wiley [Google Scholar]
- 53. Jensen P. R., and Michelsen O. (1992) Carbon and energy metabolism of atp mutants of Escherichia coli. J. Bacteriol. 174, 7635–7641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dewick P. M. (2009) Secondary Metabolism: The Building Blocks and Construction Mechanisms. Medicinal Natural Products, pp. 7–38, John Wiley & Sons, New York, NY. [Google Scholar]
- 55. Lundblad J. R., Laurance M., and Goodman R. H. (1996) Fluorescence polarization analysis of protein-DNA and protein-protein interactions. Mol. Endocrinol. 10, 607–612 [DOI] [PubMed] [Google Scholar]
- 56. Jameson D. M., and Seifried S. E. (1999) Quantification of protein-protein interactions using fluorescence polarization. Methods 19, 222–233 [DOI] [PubMed] [Google Scholar]
- 57. Allen M., Reeves J., and Mellor G. (2000) High throughput fluorescence polarization: a homogeneous alternative to radioligand binding for cell surface receptors. J. Biomol. Screen 5, 63–69 [DOI] [PubMed] [Google Scholar]
- 58. Parker G. J., Law T. L., Lenoch F. J., and Bolger R. E. (2000) Development of high throughput screening assays using fluorescence polarization: nuclear receptor-ligand-binding and kinase/phosphatase assays. J. Biomol. Screen 5, 77–88 [DOI] [PubMed] [Google Scholar]
- 59. Moerke N. J. (2009) Fluorescence Polarization (FP) Assays for Monitoring Peptide-Protein or Nucleic Acid-Protein Binding. Curr. Protoc. Chem. Biol. 1, 1–15 [DOI] [PubMed] [Google Scholar]
- 60. Kim H. Y., and Gladyshev V. N. (2007) Methionine sulfoxide reductases: selenoprotein forms and roles in antioxidant protein repair in mammals. Biochem. J. 407, 321–329 [DOI] [PubMed] [Google Scholar]
- 61. Martinez-Ruiz A., Araujo I. M., Izquierdo-Alvarez A., Hernansanz-Agustin P., Lamas S., and Serrador J. M. (2013) Specificity in S-nitrosylation: a short-range mechanism for NO signaling? Antioxid. Redox. Signal. 19, 1220–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mittl P. R., and Schulz G. E. (1994) Structure of glutathione reductase from Escherichia coli at 1.86 A resolution: comparison with the enzyme from human erythrocytes. Protein Sci. 3, 799–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nishimura K., Nakayashiki T., and Inokuchi H. (1993) Cloning and sequencing of the hemE gene encoding uroporphyrinogen III decarboxylase (UPD) from Escherichia coli K-12. Gene 133, 109–113 [DOI] [PubMed] [Google Scholar]
- 64. Henard C. A., Bourret T. J., Song M., and Vazquez-Torres A. (2010) Control of redox balance by the stringent response regulatory protein promotes antioxidant defenses of Salmonella. J. Biol. Chem. 285, 36785–36793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lim S. J., Jung Y. M., Shin H. D., and Lee Y. H. (2002) Amplification of the NADPH-related genes zwf and gnd for the oddball biosynthesis of PHB in an E. coli transformant harboring a cloned phbCAB operon. J. Biosci. Bioeng. 93, 543–549 [DOI] [PubMed] [Google Scholar]
- 66. Shi F., Li K., Huan X., and Wang X. (2013) Expression of NAD(H) kinase and glucose-6-phosphate dehydrogenase improve NADPH supply and L-isoleucine biosynthesis in Corynebacterium glutamicum ssp. lactofermentum. Appl. Biochem. Biotechnol. 171, 504–521 [DOI] [PubMed] [Google Scholar]
- 67. Xiong X., Yang L., Han X., Wang J., and Zhang W. (2010) Knockout and function analysis of pqqL gene in Escherichia coli. Wei Sheng Wu Xue Bao 50, 1380–1384 [PubMed] [Google Scholar]
- 68. Misra H. S., Khairnar N. P., Barik A., Indira Priyadarsini K., Mohan H., and Apte S. K. (2004) Pyrroloquinoline-quinone: a reactive oxygen species scavenger in bacteria. FEBS Lett. 578, 26–30 [DOI] [PubMed] [Google Scholar]
- 69. Le H. T., Gautier V., Kthiri F., Malki A., Messaoudi N., Mihoub M., Landoulsi A., An Y. J., Cha S. S., and Richarme G. (2012) YajL, prokaryotic homolog of parkinsonism-associated protein DJ-1, functions as a covalent chaperone for thiol proteome. J. Biol. Chem. 287, 5861–5870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bonifati V., Rizzu P., van Baren M. J., Schaap O., Breedveld G. J., Krieger E., Dekker M. C., Squitieri F., Ibanez P., Joosse M., van Dongen J. W., Vanacore N., van Swieten J. C., Brice A., Meco G., van Duijn C. M., Oostra B. A., and Heutink P. (2003) Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science 299, 256–259 [DOI] [PubMed] [Google Scholar]
- 71. Lopes J. M., and Lawther R. P. (1989) Physical identification of an internal promoter, ilvAp, in the distal portion of the ilvGMEDA operon. Gene 76, 255–269 [DOI] [PubMed] [Google Scholar]
- 72. Wu J., Fan Y., and Ling J. (2014) Mechanism of oxidant-induced mistranslation by threonyl-tRNA synthetase. Nucleic Acids Res. 42, 6523–6531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Murray E. L., and Conway T. (2005) Multiple regulators control expression of the Entner-Doudoroff aldolase (Eda) of Escherichia coli. J. Bacteriol. 187, 991–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Brock M., Maerker C., Schutz A., Volker U., and Buckel W. (2002) Oxidation of propionate to pyruvate in Escherichia coli. Involvement of methylcitrate dehydratase and aconitase. Eur. J. Biochem. 269, 6184–6194 [DOI] [PubMed] [Google Scholar]
- 75. Lenarcic Zivkovic M., Zareba-Koziol M., Zhukova L., Poznanski J., Zhukov I., and Wyslouch-Cieszynska A. (2012) Post-translational S-nitrosylation is an endogenous factor fine tuning the properties of human S100A1 protein. J. Biol. Chem. 287, 40457–40470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wright N. T., Cannon B. R., Zimmer D. B., and Weber D. J. (2009) S100A1: Structure, function, and therapeutic potential. Curr. Chem. Biol. 3, 138–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Perrin R. J., Craig-Schapiro R., Malone J. P., Shah A. R., Gilmore P., Davis A. E., Roe C. M., Peskind E. R., Li G., Galasko D. R., Clark C. M., Quinn J. F., Kaye J. A., Morris J. C., Holtzman D. M., Townsend R. R., and Fagan A. M. (2011) Identification and validation of novel cerebrospinal fluid biomarkers for staging early Alzheimer's disease. PLoS ONE 6, e16032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Curtain C. C., Ali F., Volitakis I., Cherny R. A., Norton R. S., Beyreuther K., Barrow C. J., Masters C. L., Bush A. I., and Barnham K. J. (2001) Alzheimer's disease amyloid-beta binds copper and zinc to generate an allosterically ordered membrane-penetrating structure containing superoxide dismutase-like subunits. J. Biol. Chem. 276, 20466–20473 [DOI] [PubMed] [Google Scholar]
- 79. Schoneich C., and Williams T. D. (2003) CU(II)-catalyzed oxidation of Alzheimer's disease beta-amyloid peptide and related sequences: remarkably different selectivities of neurotoxic betaAP1–40 and non-toxic betaAP40–1. Cell Mol. Biol. 49, 753–761 [PubMed] [Google Scholar]
- 80. Kuchibhotla K. V., Goldman S. T., Lattarulo C. R., Wu H. Y., Hyman B. T., and Bacskai B. J. (2008) Abeta plaques lead to aberrant regulation of calcium homeostasis in vivo resulting in structural and functional disruption of neuronal networks. Neuron 59, 214–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Garcia-Alloza M., Dodwell S. A., Meyer-Luehmann M., Hyman B. T., and Bacskai B. J. (2006) Plaque-derived oxidative stress mediates distorted neurite trajectories in the Alzheimer mouse model. J. Neuropathol. Exp. Neurol. 65, 1082–1089 [DOI] [PubMed] [Google Scholar]
- 82. Du Yan S., Zhu H., Fu J., Yan S. F., Roher A., Tourtellotte W. W., Rajavashisth T., Chen X., Godman G. C., Stern D., and Schmidt A. M. (1997) Amyloid-beta peptide-receptor for advanced glycation endproduct interaction elicits neuronal expression of macrophage-colony stimulating factor: a proinflammatory pathway in Alzheimer disease. Proc. Natl. Acad. Sci. U.S.A. 94, 5296–5301 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.