Abstract
The two-spotted spider mite Tetranychus urticae is an extremely polyphagous crop pest. Alongside an unparalleled detoxification potential for plant secondary metabolites, it has recently been shown that spider mites can attenuate or even suppress plant defenses. Salivary constituents, notably effectors, have been proposed to play an important role in manipulating plant defenses and might determine the outcome of plant-mite interactions. Here, the proteomic composition of saliva from T. urticae lines adapted to various host plants—bean, maize, soy, and tomato—was analyzed using a custom-developed feeding assay coupled with nano-LC tandem mass spectrometry. About 90 putative T. urticae salivary proteins were identified. Many are of unknown function, and in numerous cases belonging to multimembered gene families. RNAseq expression analysis revealed that many genes coding for these salivary proteins were highly expressed in the proterosoma, the mite body region that includes the salivary glands. A subset of genes encoding putative salivary proteins was selected for whole-mount in situ hybridization, and were found to be expressed in the anterior and dorsal podocephalic glands. Strikingly, host plant dependent expression was evident for putative salivary proteins, and was further studied in detail by micro-array based genome-wide expression profiling. This meta-analysis revealed for the first time the salivary protein repertoire of a phytophagous chelicerate. The availability of this salivary proteome will assist in unraveling the molecular interface between phytophagous mites and their host plants, and may ultimately facilitate the development of mite-resistant crops. Furthermore, the technique used in this study is a time- and resource-efficient method to examine the salivary protein composition of other small arthropods for which saliva or salivary glands cannot be isolated easily.
The family of spider mites (Chelicerata: Acari: Tetranychidae) comprises well over 1000 species, including several that are important pests on crops, and about 0.9 billion euro is being spent annually for their control worldwide (1, 2). These minute herbivores—about 0.5 mm in size—use their stylets to pierce leaf mesophyll cells and to inject saliva, after which they suck out the cytoplasm. This results in cell death visible as chlorotic spots sometimes accompanied by necrosis, and ultimately in leaf abscission (3, 4). Among the spider mites, the two-spotted spider mite, Tetranychus urticae, is the most polyphagous, having been reported on more than 1000 host plant species in more than 140 different families (1). However, not all these host plants are equally suitable to T. urticae, and host plant acceptance can even differ across mite populations (5–7).
Important factors determining host plant acceptance by the herbivore are plant defenses, including physical and molecular-chemical barriers that hamper herbivore feeding (8). Different herbivores can induce a different repertoire of defenses and these differential plant responses are set in motion via herbivore-specific signals, predominantly emanating from their saliva (9). Plant defenses are regulated by a set of phytohormones, primarily jasmonates (such as jasmonic acid (JA)1) (10), salicylic acid (SA) (11, 12), and ethylene (13). Hormonal interactions are believed to enable the plant to regulate and customize responses under variable biotic and abiotic stress conditions (14). Most spider mites induce a mixture of JA- and SA-defenses (15–19) while a role for ethylene remains elusive (20).
It is conceivable that some spider mites have evolved traits that enable them to resist (5, 21, 22), attenuate (23), or suppress JA- (5) and SA-related defenses (24) to maintain a high fitness (19). Although it is largely unknown which terminal plant defenses determine resistance or susceptibility to mites, negative correlations were found between mite fitness and several plant secondary metabolites (18, 25–27). How plants detect spider mite feeding is poorly understood, but analyses of transcriptional networks have suggested the involvement of receptor-like kinases reminiscent of other plant-herbivore interactions (28). These receptors may be involved in the recognition of molecules (elicitors) released during the onset of the plant-pathogen or plant-herbivore interaction (29).
Many herbivore elicitors emanate from saliva or regurgitation fluids released on or in the plant during feeding (30). Reminiscent of phytopathogens (9, 31, 32), herbivores evolved additional salivary molecules to counter the induction of defenses (8, 33, 34). Such molecules, enhancing herbivore performance, were originally referred to as “effectors.” Some plant varieties have however evolved the means to recognize these effectors, effectively turning them into elicitors which activate plant defense responses (32, 35, 36). Because of this context-dependence (37), a broader inclusive definition of the term effector was suggested (35). Effectors are defined as pathogen- or herbivore-secreted proteins and small molecules that alter host-cell structure and function. Effectors are of high interest to the plant breeding industry because they can lead to the identification of resistance genes (R genes) (38) and susceptibility genes (S genes) (39). R genes code for immune receptors, which confer recognition of pathogen- or herbivore-derived effectors or their modification inflicted on a host protein, eventually resulting in the activation of host defenses (39). S genes, on the other hand, can considered to be all plant genes that facilitate infection and support compatibility (39).
The vast majority of herbivore effectors emanate from saliva. Silencing salivary effectors in non-arthropod herbivores like nematodes has been shown to reduce their performance (40). Likewise, silencing salivary effectors in insects like aphids reduced their reproduction (41). These studies indicate that salivary components are key players in the plant-herbivore molecular battlefield, and hence their identification is a high priority. Salivary proteins can be inferred from genomic, transcriptomic and/or proteomic data using a combination of criteria. For example, combining temporal and spatial gene expression data with the predicted presence of an N-terminal signal peptide (SP) in the corresponding proteins results in lists of putative salivary proteins (e.g. (42, 43)). For T. urticae, an annotated genome is available (44), but no salivary gland-specific transcriptome and/or proteome has been obtained yet. It is known that spider mites inject salivary substances into host plant leaves (45–47). However, the proteomic composition of these substances has yet to be elucidated. The generation of gland specific transcriptomes and proteomes is hampered by the extremely small size of spider mites and the complex morphology of the glands (48) (T. urticae adults have a body length of 400–500 μm with an approximate salivary gland length of 50 μm). Salivation of several eriophyid mite species has been achieved by soaking adult mites into immersion oil (49), and of Varroa destructor mites by topical application of cholinomimetic agents (50). Protein sequences were not obtained in these studies, however. A successful approach for obtaining sufficient amounts of salivary secretions suitable for protein analysis from nonmite arthropods has been to collect secretions from artificial diets encapsulated by a membrane on which feeding has taken place. For example, using this approach, multiple proteins, in a range from 10 to 100, have been identified in the secreted saliva of aphids (51, 52) and true bugs (53).
We developed a set-up for collecting salivary secretions of T. urticae from artificial diet and analyzed the proteomic composition of these secretions. Our approach involved T. urticae lines that were reared on distinct economically important host plants for more than five generations, a period during which adaptation usually occurs (54). By including lines adapted to different hosts, we aimed to discover a broader spectrum of salivary proteins. Mite salivary secretions were harvested using a custom-developed mite feeding assay and subsequently investigated by nano-LC-MS/MS analysis. Additionally, a transcriptome of the proterosoma—harboring the salivary glands—was constructed to validate proteomic data. Evidence for the salivary origin of a selection of identified proteins was obtained by whole-mount in situ hybridizations (ISHs). Furthermore, to assess host-specificity of salivary gland productions, we investigated the host-dependence of expression of genes coding for the identified putative salivary proteins. The results from this study lay the groundwork for an improved understanding of the molecular machinery behind induction or suppression of resistance during plant-mite interactions, and may open new opportunities for mite-resistance plant breeding.
EXPERIMENTAL PROCEDURES
Establishment of T. urticae Lines on Different Host Plants
The T. urticae London strain has been maintained under laboratory conditions on bean plants (Phaseolus vulgaris cv. “Prelude,” Fabaceae) for many years. The genome of this London strain has been sequenced (44). Lines on alternative host plants were established by transferring ∼250 adult female mites from the London strain on bean to new hosts. These new host plants were cotton (Gossypium hirsutum, Malvaceae), maize (Zea mays, cv. “Ronaldinio,” Poaceae), soy (Glycine max, cv. “Merlin,” Fabaceae), and tomato (Solanum lycopersicum, cv “Moneymaker,” Solanaceae). Three independent lines were generated for cotton and tomato, whereas four independent lines were obtained for maize and soy. The mite lines were maintained in a climatically controlled environment at 26 °C with 60% RH, and a light/dark (L:D) photoperiod of 16:8 h. Mites were offered fresh plants as needed, and were used in experiments after 5 generations for all hosts, except tomato, where replicate lines derived from London were adapted and maintained on tomato for over 30 generations (23).
Collection of Artificial Diet Enriched with T. urticae Saliva
To collect saliva, spider mites were allowed to feed on an artificial diet. Briefly, a pocket-like invagination was made in stretched Parafilm® M (Sigma, Bornem, Belgium) using a custom built vacuum device (see supplemental Fig. S1), consisting of a 96-well plate (plate thickness 4.2 mm, hole diameter 4.5 mm) fitting on a vacuum manifold plate (Analytical Research Systems, Micanopy, FL) connected to a vacuum pump (model N 035.1.2 A_.18, KNF Neuberger, Freiburg, Germany). Next, 70 μl sterile holidic artificial diet (1/30 diluted aphid diet, (55)) supplemented with the antibiotic rifampicin (0.05 mg/ml) was added, after which the pocket was sealed with packaging tape (Scotch Packaging Tape, Extra Strong, 3M, Diegem, Belgium). The parafilm, with the diet-filled hemisphere side directed upwards, was cut to size (∼4 × 4 cm), mounted on the back of a small Petri dish (90 mm diameter), and placed in a large Petri dish (135 mm diameter) filled with water. Using cotton wool and paper tissue, a water barrier was created, confining the spider mites in close proximity of the diet hemisphere (Fig. 1). Thirty to 40 adult female mites were transferred to each feeding arena. These mites originated from replicate lines adapted to bean, maize, soy or tomato (mites adapted to cotton were not used for the collection of saliva). For each host plant-specific sample, mites originating from each replicated host plant line were pooled. Addition of a blue colorant (0.05 mg/ml erioglaucine, Sigma) to control diet hemispheres was used to verify spider mite feeding as assessed by staining of gut contents (Fig. 1). Petri dishes with feeding hemispheres were placed in an incubator (MLR-352H, Sanyo/Panasonic, Osaka, Japan) at 26 °C with 60% RH and 16:8 L:D. After 24 h, the remaining content of the feeding hemispheres was collected using a Hamilton microsyringe under sterile conditions (Laminar Flow cabinet, ESCO, Singapore). Samples were stored at −80 °C until enough sample (about 0.3 ml, 10–15 diet hemispheres/sample) was collected for nano-LC-MS/MS analysis. Feeding hemispheres that did not receive spider mites were treated identically and served as reference sample during nano-LC-MS/MS analysis.
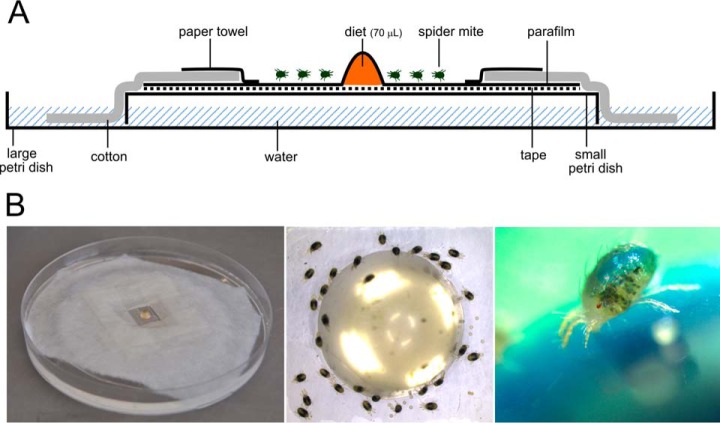
Fig. 1.
Feeding arena used to collect spider mite saliva. A, Schematic drawing of the feeding arena. B, Left panel: overview of the feeding arena; middle panel: top view of the diet hemisphere infested with spider mites; right panel: adult female spider mite feeding on a control hemisphere with a blue colorant added to the diet (note the blue color visible in the gut of the spider mite).
Nano-LC-MS/MS Analysis
Twenty micrograms of total protein (Pierce BCA Protein Assay Kit, Thermo Scientific, Aalst, Belgium) of the sample was reduced using 1.25 μl of 500 mm TRIS (2-carboxyethyl) phosphine in a volume of 100 μl 100 mm TEAB, and incubated for 1 h at 55 °C. Next, the samples were processed using the filter-aided sample preparation (FASP) procedure (FASP Protein digestion kit, Protein discovery, Knoxville, TN) according to manufacturer's instructions. In short, the samples were diluted in a urea buffer and processed on a FASP filter, alkylated with iodoacetamide and digested with trypsine (enzyme/protein ratio = 1:50) overnight. Liquid chromatography mass spectrometric analysis was performed on a Waters nanoAquity LC-Ultra system connected to a Thermo Scientific LTQ Velos Orbitrap mass spectrometer. The equivalent of 2 μg of total protein of the digested sample was dissolved in 20 μl of 2% acetonitrile in HPLC-grade water. 10 μl of the sample (1 μg) was loaded on the trapping column (Pepmap C18 300 μm × 20 mm) (Dionex, Sunnyvale, California) with an isocratic flow of 2% acetonitrile in water with 0.1% formic acid at a flow rate of 5 μl/min. After 2 min, the column-switching valve was switched, placing the pre-column online with the analytical capillary column, a Pepmap C18, 3 μm 75 μm × 150 mm nano column (Dionex). Separation was conducted using a linear gradient from 2% acetonitrile in water, 0.1% formic acid to 40% acetonitrile in water, 0.1% formic acid in 100 min. The flow rate was set at 400 nl/min. The LTQ Orbitrap Velos (Thermo Scientific) was set up in a data dependent MS/MS mode where a full scan spectrum (350–5000 m/z, resolution 60,000) was followed by a maximum of ten CID tandem mass spectra (100 to 2000 m/z). Peptide ions were selected as the 20 most intense peaks of the MS1 scan. CID scans were acquired in the LTQ iontrap part of the mass spectrometer. The normalized collision energy used was 35% in CID. We applied a dynamic exclusion list of 45 s.
Protein identification
Prior to protein identification, calibration of the data was performed using the methods described by (56), correcting the systematic bias in mass measurement in the second replicate. Peak lists obtained from MS/MS spectra were identified using OMSSA version 2.1.9 (57), X!Tandem version X! Tandem Vengeance (2015.12.15.2) (58), and MS-GF+ version Beta (v10282) (59). The search was conducted using SearchGUI version 2.8.5 (60). Protein identification was conducted against a concatenated target/decoy version of the T. urticae protein database holding 17,907 target sequences (version of December 16th 2014, see supplemental Data S1) and the common Repository of Adventitious Proteins (cRAP) database (available at http://www.thegpm.org/crap/). Reversing the target sequences in SearchGUI created the decoy sequences. The identification settings were as follows: Trypsin with a maximum of 2 missed cleavages; 10.0 ppm as MS1 and 0.5 Da as MS2 tolerances; variable modifications: carbamidomethyl c (+57.021464 Da), oxidation of m (+15.994915 Da), pyro-glu from n-term q (-17.026549 Da), acetylation of protein n-term (+42.010565 Da), pyro-cmc (-17.026549 Da) and pyro-glu from n-term e (-18.010565 Da). Peptides and proteins were inferred from the spectrum identification results using PeptideShaker version 1.10.2 (61) (see supplemental Table S1 and S2 for a detailed list of all features of the LC-MS/MS identifications at the protein and peptide level in the treatment samples (artificial diet with feeding mites), respectively, and supplemental Table S3 for a list of all features of the LC-MS/MS identifications at the protein level in the reference samples (artificial diet without feeding mites)). Peptide Spectrum Matches (PSMs), peptides and proteins were validated at a 1.0% False Discovery Rate (FDR) estimated using the decoy/hit distribution.
The mass spectrometry data along with the identification results have been deposited to the ProteomeXchange Consortium (62) via the PRIDE partner repository (63) (www.ebi.ac.uk/pride) with the data set identifiers PXD003022 and 10.6019/PXD003022.
Experimental Design and Statistical Rationale
Two technical replicates of artificial diets enriched with saliva of mites maintained on four different host plant species were analyzed in this nano-LC-MS/MS study. The different host plants to which the mite lines have been adapted were bean, maize, soy and tomato, as described in a previous section on “Establishment of T. urticae Lines on Different Host Plants.” Eight samples were analyzed in total, yielding a list of putative T. urticae salivary proteins. Care should be taken when comparing the proteomics data of the different host plant lines, as each host-specific data set is based on one pooled biological replicate (each analyzed in two technical replicates). However, we believe that our approach was sufficiently rigorous as host plant specific production of T. urticae salivary proteins was complemented by a gene expression analysis of mites maintained on the four different host plants (see “The Expression Profiles of Salivary Protein Genes Show Signs of Host Adaptation”).
Proteomics Data Filtering and Protein Quantification
Proteins identified in the artificial diet were pooled into one data set, while retaining host plant specific information. In order to retain confident proteins identifications, only proteins identified in at least one of the samples with a mean PSM value of at least two, calculated over both technical replicates per host plant, were retained in the final putative salivary protein list. A MS1-intensity based Top3 analysis was performed to derive protein abundance (64). First, the moFF (modest Feature Finder, https://github.com/compomics/moFF) tool was used to extract the MS1 intensities from the Thermo raw files for the validated PSMs matching to non-decoy (true) sequences (See supplemental Table S4). Afterward, the intensities of the three (or fewer) peptides with the highest intensities were averaged for every protein detected. Next, a normalized abundance factor (rTop3) was calculated by dividing the Top3 value by the sum of all Top3 values of the quantified proteins in each experiment (65), excluding contaminants. Additionally, the proteins were ranked according to their maximal rTop3 value, as calculated across each host-plant specific sample.
X-ray Submicron Computed Tomography
Adult female T. urticae specimens were fixed as described previously (66). Briefly, live mites were collected and then incubated in a 6:3:1 mixture of 80% ethanol, 38% formaldehyde and 100% acetic acid for 72 h. Specimens were washed with 70% ethanol and dehydrated through graded concentrations of 70 to 100% ethanol (5% steps) for 10 min per step. The 100% ethanol step was done overnight at room temperature (RT). The dehydrated specimens were critical point dried with CO2 (CPD 020, Bal-Tec, Balzers, Liechtenstein) and glued with their opisthosoma to a 0.5 mm carbon pencil lead (Staedtler, Bornem, Belgium). Spider mite specimens were scanned with Nanowood (67), an x-ray sub-micron computed tomography (CT) system developed at the Ghent University Centre for x-ray Tomography (UGCT). The sample was scanned with an open-type nanofocus x-ray tube, reaching a focal spot size < 1 μm. Samples were scanned for 1 h, resulting in scans with an isotropic voxel pitch of ∼0.5 μm. Reconstructions were performed using Octopus Reconstruction, a tomography reconstruction package for parallel, cone-beam and helical geometry (68), licensed by InsideMatters (www.insidematters.eu). Filtering was performed using the single step phase-retrieval Paganin algorithm (69). After reconstruction, a noise removal anisotropic diffusion filter was applied using Octopus Analysis, formerly known as Morpho+ (70), also licensed by InsideMatters. All visualization was performed with Fiji (71).
RNAseq Expression Analysis of the Proterosoma of T. urticae Adult Females
RNA was extracted from intact adult T. urticae females and from dissected proterosomas of adult females using the Qiagen RNeasy RNA extraction kit (Qiagen, Venlo, The Netherlands). Three replicates of 100 intact adult females from the London strain were collected from bean plants. For dissection of the proterosoma, a dissection chamber was prepared by affixing a square piece of 12.7 mm width double sided Scotch tape (3M, Maplewood, MN) on the bottom of a 35 mm Petri plate and allowing tape to cure for 24 h. Adult female mites were carefully attached to tape with their ventral side downwards and covered with 1x phosphate buffered saline solution (PBS). The dorsal side of the hysterosoma was opened using micro-dissecting needles and contents (gut, ovaries, eggs, dorsal parts of exoskeleton) were removed with dissecting needles and micropipette. Remaining tissue (proterosoma, first and second pair of legs, nervous mass, salivary glands) was gently lifted from tape and transferred to Buffer RLT (RNeasy RNA extraction kit, Qiagen) using a micropipette. In total, 250 mites were dissected and dissected tissue was pooled into one sample. RNA from this sample, as well as three samples collected from whole adult females, was used for Illumina library construction and subsequent sequencing on a HiSeq instrument (Fasteris, Plan-Les-Ouates, Switzerland). Briefly, 100 bp strand-specific paired-end reads were generated for all samples. The paired-end strand-specific Illumina RNA-seq reads were aligned to the T. urticae reference genome (44) using the two-pass alignment mode of STAR 2.5.0b (72) with a maximum intron size of 20 kb (the results were splice-aware alignments made independently of the reference genome annotation). The resulting BAM files were subsequently sorted by read name using Samtools 1.2 (73). Read counts per gene, based on the reference annotation (version October 29, 2015), were then obtained using the default settings of HTSeq 0.6.0 (74) with the “STRANDED” flag set to “yes” and the “FEATURE” flag set to “exon.” Differentially expressed genes between the proterosoma of T. urticae females and intact T. urticae females were determined using the DESeq2 (version 1.6.3, (75)) and Bioconductor (http://bioconductor.org/) R-packages. The “unfiltered DESeq2 results” settings (dds <- DESeq(dds, minReplicatesForReplace = Inf) and res <- results(dds, cooksCutoff = FALSE, independent Filtering = FALSE)) were used for differential expression analysis. Genes with a fold change (FC) higher than or equal to eight and a Benjamini-Hochberg adjusted p value less than or equal to 0.05 were considered differentially expressed (DE). All gene expression data have been uploaded to the Gene Expression Omnibus with accession number GSE81128.
Validation of Salivary Proteins by Whole-mount In-situ Hybridization
In situ hybridization in T. urticae was based on Dearden et al 2000 and 2002 (76, 77). Briefly, RNA was extracted from T. urticae mites (London strain) feeding on bean or tomato plants (Total RNA Isolation Mini Kit, Agilent), treated with TURBO DNA-freeTM Kit (ThermoScientific) to remove contaminating genomic DNA and used for cDNA synthesis using Maxima First Strand cDNA Synthesis Kit (ThermoScientific). Primers were designed using Primer3 (http://bioinfo.ut.ee/primer3/). A fragment with preferred length of about 300 bp was amplified (primers used are in supplemental Table S5). PCR products were cloned into pGEM-T plasmids (Promega, Madison, WI) and transformed into E. coli. Plasmids from liquid cultures were purified after which insert orientation and nucleotide sequence were determined by sequencing (LGC Genomics, Germany). A PCR was performed on the plasmids using pUC/M13 primers (supplemental Table S5). PCR product, containing insert flanked by T7 and SP6 promoter sites from the plasmid backbone, was checked by agarose gel electrophoresis and purified using E.Z.N.A. Cycle Pure Kit (Omega Biotek, Norcross, GA). Depending on orientation, sense or antisense digoxigenin-labeled (DIG-labeled) probes were generated using T7 or SP6 RNA polymerase (Roche, Anderlecht, Belgium), using the pUC/M13 PCR product and DIG-UTPs (Roche) in the in vitro labeling reaction. Probes were then purified using SigmaSpinTM Sequencing Reaction Clean-Up Columns (Sigma), supplemented with hybridization buffer (50% formamide (Sigma), 4× SSC (Sigma), 1× Denhardt's solution (Sigma), 250 μg/ml tRNA (wheat germ type V, Sigma), 250 μg/ml ssDNA (boiled salmon sperm DNA, Sigma), 50 μg/ml heparin (sodium salt, Sigma), 0.1% Tween-20 (Sigma), 5% dextran sulfate (sodium salt, Sigma), and stored at −20 °C until used.
T. urticae nymphs and adults (London strain) of both sexes were collected from bean and tomato plants and fixed overnight in a 1:1 mixture of heptane and PTw (PBS with 0.1% Tween-20) containing 4% formaldehyde. The mites were then washed in methanol and gradually rehydrated in PTw, followed by sonication in a sonic cleansing bath and treatment with 5 mg/ml Proteinase K during 10 min. The mites were then refixed with 4% formaldehyde in PTw. After washing in PTw, mites were prehybridized in hybridization buffer for 1 h at 52 °C. Hybridization buffer was refreshed and probe was added. The mites were then incubated overnight at 52 °C. Washing occurred at 53 °C with wash buffer composed of 50% formamide, 2× SSC and 0.1% tween-20. After washing at room temperature with PBTw (PTw with 0.1% BSA, Sigma), the mites were incubated at RT for 2 h with a 1:1000 dilution of anti-digoxigenin-AP (Fab fragments, Roche) in PBTw. The mites were then washed with PTw and alkaline phosphatase (AP) buffer (100 mm Tris pH 9.5, 100 mm NaCl, 1 m MgCl2, 0.1% Tween-20). AP buffer containing the FastRed substrate (SIGMAFASTTM Fast Red TR/Naphthol AS-MX tablets, Sigma) was added and mites were incubated at RT in the dark, until red staining was visible. Methanol was used to reduce background staining and the mites were eventually cleared in 70% glycerol in PTw (pH 8.5) after washing with pure PTw. The mites were then mounted on a microscopy glass for further microscopic investigation (Nikon A1R fluorescence confocal microscope; emission at 500–530 nm and acquisition at 488 nm for spider mite auto-fluorescence and emission at 570–620 nm and acquisition at 561.7 nm for FastRed signal). Z-stacks were created using 15 slices with 2–3 μm distance between slices. All images were processed with Fiji (71).
Transcriptome Analysis of T. urticae Maintained on Different Host Plants
For each host plant adapted mite line, three (tomato and cotton) or four (bean, maize and soy) biologically replicated RNA samples were obtained. Each RNA sample was extracted from 100–150 pooled female adult mites using the RNeasy extraction kit (Qiagen) and was subsequently treated with DNase (Turbo DNA-free kit, ThermoScientific). RNA quantity and integrity was measured using an Agilent TapeStation system. RNA samples were labeled with cyanine dyes following the Low Input Quick Amp Labeling Kit (Agilent Technologies, Diegem, Belgium), with 100 ng of total RNA as starting material. RNA samples from mites feeding on the reference bean host plant were labeled with cy3, whereas cy5-labeling was performed on all other samples. Samples were pooled per host plant transfer and hybridized to a custom-made Sureprint G3 8 × 60K array (Agilent Technologies, with a GEO platform number of GPL16890) following the standard procedure of the Gene Expression Hybridization Kit (Agilent Technologies). After washing procedures (Gene Expression Wash Buffer kit (Agilent Technologies)), raw data was extracted from the 8 × 60 k slides using the GE2_107_Sep09 protocol of the Agilent Feature Extraction Software. The intraspot correlation coefficient per array and the metrics from the arrayQualityMetrics package per host plant line were assessed for optimal background correction and normalization procedures (78). Data was background corrected using the “normexp”-method and normalized by loess and Aquantile (79). Cyanine intensities were extracted from the processed RG-object and averaged per host plant. Using the normalized MA-object, differential expression was assessed for mites on cotton, maize, soy and tomato against the corresponding ancestral mite population living on bean by an empirical Bayes approach. T. urticae gene expression data have been uploaded to the Gene Expression Omnibus with accession number GSE80337.
T. urticae Protein Family Analyses
T. urticae proteins were assigned to OrthoMCL groups using the online OrthoMCL software tool (http://www.orthomcl.org/orthomcl/proteomeUpload.do) and the T. urticae proteome (version July 29, 2015, available via the Online Resource for Community Annotation of Eukaryotes (ORCAE) database) as query (80). Signal peptides were predicted with SignalP 4.1 (81) and protein subcellular localization was predicted using WoLF PSORT (organism type: “Animal”) (82) at http://www.genscript.com/wolf-psort.html. Proteins with a sequence length less than 30 AA or not containing a start methionine (pseudogenes) were excluded from WoLF PSORT analysis. Transmembrane domains were predicted using TMHMM server 2.0 (http://www.cbs.dtu.dk/services/TMHMM/). A phylogenetic analysis was performed for proteins belonging to OrthoMCL group cluster Tu_MCL_35 and Tu_MCL_36. Except for tetur55g00110 (Tu_MCL_36) which is encoded by a pseudogene, proteins from each cluster were aligned using MUSCLE (83). Model selection was done with ProtTest 2.4 (84) and according to the Akaike information criterion WAG+G and WAG+G+F were optimal for the phylogenetic reconstruction of Tu_MCL_35 and Tu_MCL_36 proteins, respectively. Finally, for each alignment a maximum likelihood analysis was performed using Treefinder (version 2011) (85) bootstrapping with 1000 pseudoreplicates (LR-ELW). The resulting trees were midpoint rooted and edited with MEGA 6.0 software (86).
Evaluation of T. urticae Salivary Proteins Using Available Databases
T. urticae salivary proteins identified by nano-LC-MS/MS were used as query in a BLASTp search (E-value threshold 1.0 E-5) against the proteome of the non-phytophagous American house dust mite, Dermatophagoides farinae (87). Furthermore, expression levels of genes coding for putative T. urticae salivary proteins were compared between feeding (mobile) T. urticae stages (larva, nymph and adult) and the embryo stage (44), while a nano-LC-nano-ESI-QTOF MS/MS proteomic analysis of mite faeces (88) was screened for the presence of putative salivary proteins of T. urticae. A set of differentially expressed genes in diapausing T. urticae females (89) was also investigated for genes coding for putative T. urticae salivary proteins. Finally, an expression data set of T. urticae genes across different time points of host plant transfer to tomato (23) was mined for putative T. urticae salivary genes.
RESULTS
T. urticae Secretes Proteins in Artificial Diet Which Can Reliably Be Identified Through Mass Spectrometry Analysis
Spider mite lines propagated for at least 5 generations on different host plants (i.e. bean, maize, soy and tomato) were allowed to feed on an artificial diet encapsulated in parafilm (Fig. 1). Mites fed on the artificial diet within hours, as assessed by blue staining of control mites being fed an erioglaucine-supplemented diet. After 24 h, the artificial diet was collected for each plant-adapted mite line and for reference samples (artificial diet without feeding mites) and the protein content was analyzed using nano-LC-MS/MS. The detected salivary proteins from all host plant adapted mites were pooled into one data set. Ninety-five proteins (some belonging to 12 “protein inference groups” containing proteins identified by shared peptides) had a mean PSM of at least 2 and were retained as putative T. urticae salivary proteins (Table I, supplemental Table S6). Subsequently we used the normalized (relative) abundance factor rTop3, based on MS1 intensity, for abundance ranking of these putative salivary proteins. This rTop3 value has been shown to correlate with the mole fraction of the protein of interest (65, 90). Proteins with a high rTop3 factor are therefore assumed to be more prominent in T. urticae saliva. The majority (81%) of the putative T. urticae salivary proteins had a SP for secretion as predicted by SignalP. Only four (tetur03g08030, tetur10g00090, tetur10g00100, tetur22g00260) out of the 95 putative salivary proteins were predicted to have a transmembrane domain (Table I), whereas 76 (80%) were predicted to have an extracellular localization (Table I). The OrthoMCL analysis grouped 13,558 T. urticae proteins into 6397 ortholog groups. The majority of these groups already existed in the OrthoMCL database (http://www.orthomcl.org/orthomcl/), whereas 401 ortholog groups (group names starting with “Tu_MCL”) were specific for T. urticae (supplemental Table S7). From the 95 putative T. urticae salivary proteins detected by nano-LC-MS/MS, 63 proteins could be assigned to an existing OrthoMCL group, 22 proteins were grouped into a T. urticae specific OrthoMCL group and 10 proteins could not be included into any group (Table I, supplemental Table S6).
Table I. List of putative T. urticae salivary proteins identified in artificial diet using nano-LC- MS/MS. Proteins are ranked based on normalized abundance factor (rTop3). See supplemental Table S6 for a detailed overview.
| T. urticae protein IDa | Functional description | Max. rTop3b | SPc | WPd | ISHe | Dff | OrthoMCL (#)g | Feeding FCh | Proterosoma FCi |
|---|---|---|---|---|---|---|---|---|---|
| tetur01g01850 | Projectin | 0.1484 | N | C | * | OG5_126738 (10) | 0.4 | 2.9* | |
| tetur20g00560 | Hypothetical protein | 0.1137 | Y | E | A | Tu_MCL_12 (35) | 6.0 | 33.9* | |
| tetur07g00150 | Serine protease | 0.0877 | Y | E | A | * | OG5_135950 (7) | 8.0 | 28.6* |
| tetur28g01330, tetur55g00040 | Wannes-Thomas Secreted Protein (WTSP) 1 | 0.0855 | Y | E | Tu_MCL_36 (13) | 3.7 | 37.1* | ||
| tetur03g03700 | Hypothetical protein/PE-PGRS family protein | 0.0817 | Y | E | OG5_144177 (6) | 30.4 | 30.9* | ||
| tetur12g03940, tetur12g03950 | Serine protease | 0.0816 | Y | E | * | OG5_168371 (5) | emb(0) | 10.4* | |
| tetur03g03680 | Hypothetical protein/PE-PGRS family protein | 0.0738 | Y | E | OG5_144177 (6) | 17.9 | 38.9* | ||
| tetur32g00050 | Small Secreted Protein, Family F | 0.0691 | Y | E | A | Tu_MCL_35 (14) | 2.3 | 10* | |
| tetur03g03670, tetur03g03690, tetur03g03730, tetur03g10093 | Hypothetical protein/PE-PGRS family protein | 0.0628 | Y | E | OG5_144177 (6) | 15.3 | 40* | ||
| tetur06g06630 | Cystatin | 0.0530 | Y | E | A | Tu_MCL_45 (11) | emb(0) | (−) | |
| tetur06g02580 | Cathepsin B | 0.0469 | Y | E | * | OG5_127800 (28) | 1.5 | 5.9 | |
| tetur05g04580 | Wannes-Thomas Secreted Protein (WTSP) 10 | 0.0429 | Y | E | D | Tu_MCL_36 (13) | 11.4 | 9.5* | |
| tetur16g03470 | Serine protease homologue | 0.0387 | Y | E | * | OG5_152237 (2) | 3.7 | 12.6* | |
| tetur06g03320 | Wannes-Thomas Secreted Protein (WTSP) 3 | 0.0328 | Y | E | Tu_MCL_36 (13) | (−) | −3.3 | ||
| tetur16g03420 | Beta-mannosidase | 0.0313 | Y | E | * | OG5_129300 (4) | 2.5 | 18.4* | |
| tetur29g01360* | Hypothetical protein | 0.0233 | Y | E | D | 3.8 | 29.5* | ||
| tetur03g10083 | Hypothetical protein/PE-PGRS family protein | 0.0219 | Y | E | Tu_MCL_74 (6) | 9.4 | 36.1* | ||
| tetur01g02670 | MD-2-related lipid-recognition domain protein | 0.0217 | Y | E | * | OG5_158831 (2) | 4.6 | −2* | |
| tetur04g05980* | Apolipoprotein D precursor | 0.0215 | Y | E | * | OG5_130527 (50) | 6.4 | −1.3 | |
| tetur05g04560 | Wannes-Thomas Secreted Protein (WTSP) 8 | 0.0209 | Y | E | Tu_MCL_36 (13) | 10.3 | 10.5* | ||
| tetur07g00160* | Hypothetical protein | 0.0200 | Y | E | D | 4.4 | 14.6* | ||
| tetur03g03680, tetur03g03700, tetur03g10083 | Hypothetical protein/PE-PGRS family protein | 0.0196 | Y | E | OG5_144177, Tu_MCL_74 (6) | 17.9 | 38.9* | ||
| tetur40g00392, tetur95g00080 | Trypsin Inhibitor-like, cysteine rich domain protein | 0.0167 | Y | E | D | OG5_207753 (2) | 3.3 | 45.3* | |
| tetur28g00360 | Beta-mannosidase | 0.0167 | Y | E | A | * | OG5_129300 (4) | 5.5 | 44* |
| tetur06g00510 | Hypothetical protein | 0.0162 | Y | E | 3.9 | 41.1* | |||
| tetur55g00040 | Wannes-Thomas Secreted Protein (WTSP) 1b | 0.0131 | Y | E | Tu_MCL_36 (13) | 1.8 | 16.4* | ||
| tetur14g02090 | Immunoglobulin E-set | 0.0128 | Y | E | * | OG5_133467 (14) | 9.1 | −2.7* | |
| tetur07g01660 | Hypothetical protein | 0.0122 | Y | E | 5.1 | 10.5* | |||
| tetur28g01330 | Wannes-Thomas Secreted Protein (WTSP) 1a | 0.0119 | Y | E | Tu_MCL_36 (13) | 3.7 | 37.1* | ||
| tetur31g00630 | Hypothetical protein with Armadillo-type fold | 0.0118 | Y | E | Tu_MCL_25 (17) | 4.7 | 27.3* | ||
| tetur03g08710 | Hypothetical protein | 0.0111 | Y | E | D | 3.0 | 36* | ||
| tetur14g02080 | MD-2-related lipid-recognition domain protein | 0.0108 | Y | E | * | OG5_133467 (14) | 8.9 | −22.6* | |
| tetur02g04310 | Glycoside hydrolase, catalytic core | 0.0100 | Y | E | * | OG5_127620 (2) | 7.9 | −1.8* | |
| tetur516g00020 | Vitellogenin 1, partial | 0.0098 | N | C | * | OG5_132251 (7) | 336.7 | −3.9* | |
| tetur01g05230, tetur01g16473, tetur06g02570, tetur06g02580 | Cathepsin B | 0.0097 | Y | E | * | OG5_127800 (28) | 6.5 | −6.4* | |
| tetur30g01440 | Serine protease | 0.0096 | Y | E | * | OG5_152337 (1) | 5.1 | 46.4* | |
| tetur32g01880 | Hypothetical protein | 0.0094 | Y | E | Tu_MCL_43 (12) | 2.6 | 62.3* | ||
| tetur28g01720 | Short-chain dehydrogenase/reductase SDR | 0.0091 | N | C | * | OG5_128170 (22) | 4.9 | −46.1* | |
| tetur14g02070 | MD-2-related lipid-recognition domain protein | 0.0091 | Y | E | * | OG5_133467 (14) | 7.8 | −16.7* | |
| tetur10g00090† | Glycine-rich secreted protein | 0.0083 | Y | E | D | 5.0 | 31.5* | ||
| tetur09g04400* | Cathepsin L | 0.0083 | Y | E | * | OG5_126607 (22) | 8.0 | −1.1 | |
| tetur43g00010* | Vitellogenin 5 | 0.0082 | Y | E | * | OG5_132251 (7) | 536.3 | −7.5* | |
| tetur01g01010 | Orphan Secreted protein | 0.0078 | Y | E | Tu_MCL_212 (2) | 8.9 | 21.9* | ||
| tetur07g07380* | Beta-galactosidase | 0.0074 | Y | M | * | OG5_128163 (4) | 5.5 | −2.3* | |
| tetur10g00100† | Hypothetical protein | 0.0072 | Y | E | 4.2 | 24.4* | |||
| tetur04g09479 | Small Secreted Protein, Family F | 0.0068 | Y | E | Tu_MCL_35 (14) | 5.9 | 65.8* | ||
| tetur01g00950 | Orphan secreted protein | 0.0068 | Y | E | A | Tu_MCL_211 (2) | 7.5 | 28.6* | |
| tetur06g00230 | Hypothetical protein | 0.0061 | Y | E | A | Tu_MCL_25 (17) | 5.1 | 29.3* | |
| tetur12g01860* | Cathepsin L | 0.0057 | Y | E | * | OG5_126607 (22) | 8.2 | −1.5* | |
| tetur31g00830 | Hypothetical protein | 0.0056 | Y | E | Tu_MCL_25 (17) | emb(0) | 14.4 | ||
| tetur03g07920* | Glutathione S-transferase, delta class | 0.0053 | N | C | * | OG5_126942 (17) | 5.7 | −2.2* | |
| tetur31g01040, tetur31g01060 | Hypothetical protein with Armadillo-type fold | 0.0051 | Y | E | Tu_MCL_25 (17) | 8.6 | 47.7* | ||
| tetur03g08030† | Cathepsin B | 0.0051 | Y | E | * | OG5_127800 (28) | 13.3 | −15.8* | |
| tetur16g03190 | Serine protease homologue | 0.0050 | Y | E | * | OG5_141111 (2) | 5.3 | 48.7* | |
| tetur01g02510* | Glutathione S-transferase, class delta | 0.0048 | N | C | * | OG5_126942 (17) | 1.2 | 10.1* | |
| tetur39g00810* | Vitellogenin 2 | 0.0045 | Y | E | * | OG5_132251 (7) | 574.8 | 1.3* | |
| tetur23g01640 | Flavin reductase | 0.0043 | N | C | OG5_131746 (2) | 2.7 | 1.3 | ||
| tetur26g02320 | Superoxide dismutase, copper/zinc binding | 0.0043 | N | C | * | OG5_127584 (8) | 1.8 | 3.2* | |
| tetur19g01900 | Prolylcarboxypeptidase | 0.0043 | Y | E | * | OG5_130246 (1) | 3.5 | −1.8* | |
| tetur18g03030* | Alpha-2-macroglobulin | 0.0042 | N | C | * | 890.1 | −12.3* | ||
| tetur07g08034 | Serine proteases, trypsin family | 0.0042 | Y | E | * | OG5_149533 (13) | 1.0 | 4 | |
| tetur09g03880 | Serine protease | 0.0040 | Y | E | * | OG5_168371 (5) | 6.7 | 48.3* | |
| tetur06g01060 | Cystatin | 0.0039 | Y | E | * | OG5_147492 (6) | 10.1 | −2.9* | |
| tetur14g03160 | Hypothetical protein | 0.0039 | N | E | Tu_MCL_63 (8) | emb(0) | 3.1 | ||
| tetur06g01640 | Outer membrane lipoprotein Blc/Lipocalin | 0.0031 | Y | E | OG5_130527 (50) | 3.8 | −2.3* | ||
| tetur20g01290 | Hypothetical protein | 0.0030 | Y | E | OG5_126560 (8) | 9.3 | −12.2* | ||
| tetur13g03820 | Proteinase inhibitor I2, Kunitz metazoa | 0.0030 | Y | E | * | OG5_134456 (1) | 4.1 | 2.5* | |
| tetur25g00650* | Cathepsin L | 0.0030 | Y | E | * | OG5_126607 (22) | 16.4 | −1.4 | |
| tetur13g00600 | Hypothetical protein | 0.0029 | Y | E | 3.8 | 25.4* | |||
| tetur147g00020, tetur33g01260 | Glycoside hydrolase, subgroup, catalytic core | 0.0029 | Y | E | * | OG5_129423 (34) | 17.5 | −1 | |
| tetur01g00940 | Orphan secreted protein | 0.0029 | Y | E | A | Tu_MCL_212 (2) | 7.0 | 23.4* | |
| tetur09g03620, tetur09g03650, tetur09g03670 | Cystatin | 0.0028 | Y | E | OG5_147492 (6) | 7.0 | −1.8* | ||
| tetur22g00260† | Trypsin Inhibitor-like, cysteine rich domain protein | 0.0027 | Y | E | OG5_176862 (1) | 11.3 | 2.6* | ||
| tetur02g12930 | Glycoside hydrolase, subgroup, catalytic core | 0.0027 | Y | E.R. | * | OG5_129423 (34) | 8.2 | −3.9* | |
| tetur02g11340, tetur02g11390 | Aldo-keto reductase family B, member 1 | 0.0026 | N | C | * | OG5_126583 (9) | 5.0 | 8.1* | |
| tetur32g02327 | Short-chain dehydrogenase/reductase | 0.0022 | N | C | * | OG5_128075 (3) | emb(0) | 2.7* | |
| tetur01g11910 | Chitinase | 0.0022 | Y | E | * | OG5_152454 (3) | 0.7 | 28.7* | |
| tetur09g00900 | Hypothetical protein | 0.0022 | Y | E | 2.1 | 21.3* | |||
| tetur07g03440*, tetur109g00010 | Fructose-bisphosphate aldolase, class-I | 0.0018 | N | C | * | OG5_127143 (3) | 2.0 | 2.8* | |
| tetur03g09480*, tetur09g05350*, tetur09g05360 | Actin | 0.0017 | N | CS | * | OG5_126595 (3) | 0.5 | 14.1* | |
| tetur11g01500 | Carboxyl/cholinesterase | 0.0015 | Y | E | * | OG5_136350 (8) | 1.6 | −1.1 | |
| tetur03g04470 | Fibronectin, type III-like fold | 0.0012 | Y | E | Tu_MCL_153 (3) | 6.7 | 1.3* |
a accession number in the ORCAE database (http://bioinformatics.psb.ugent.be/orcae/overview/Tetur); an asterisk (*) indicates that the T. urticae protein has been found in faeces of mites (88) while a cross (†) indicates that the T. urticae protein was predicted with a transmembrane domain by TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM/).
b maximal rTop3 value across the four T. urticae host plant adapted lines (bean, soy, maize, tomato).
c presence (Y = yes, N = no) of a signal peptide as predicted by SignalP 4.0 (81).
d subcellular localization prediction by WoLF PSORT (CytoSkeleton/Cytoplast/Endoplasmic Reticulum/Extracellular/Mitochondrial) (82).
e gland expression localization determined by whole-mount in situ hybridization (ISH): Anterior or Dorsal podocephalic glands.
f homologues (BLASTp E-value threshold 1E-5) identified in the American house dust mite D. farinae (Df) (*).
g OrthoMCL group with the number of OrthoMCL members present in the T. urticae genome shown between brackets (see Table S7 for OrthoMCL details).
h overall average expression across all the feeding stages (larva, nymph, adult) relative to expression in the embryo stage (44); in case of gene groups the expression ratio of the first gene is shown; emb(0): no reads could be mapped to this gene in the embryo stage; (−) T. urticae gene was not annotated at time of experiment.
i fold change in gene expression between T. urticae proterosomas and intact T. urticae females (* = Benjamini-Hochberg corrected p value ≤ 0.05); in case of gene groups the FC of the first gene is shown; (−) no reads could be mapped to this gene.
RNAseq Analysis of T. urticae Proterosomas Supports the Salivary Origin of Many Identified Proteins
RNA was extracted from 100 intact adult female T. urticae mites with 3-fold biological replication and from 250 dissected T. urticae proterosomas (see Experimental Procedures). For all T. urticae genes the number of mapped RNAseq reads can be found in supplemental Table S8. Using the RNAseq data and the DESeq2 software, we performed a differential expression analysis (Benjamini-Hochberg adjusted p value ≤ 0.05 and |FC| ≥ 8) (supplemental Fig. S2, supplemental Table S9) between dissected T. urticae proterosomas and intact females: 1800 T. urticae genes showed a significantly higher expression in the proterosoma compared with intact females, whereas 1268 had a significantly lower expression. About 20% (591 genes) of the genes with a significantly different expression in the proterosoma coded for extracellular proteins, with 455 out of 591 genes having a significantly higher expression and 136 having a significantly lower expression in the proterosoma (Fig. 2, supplemental Table S9). Fifty-three percent of the top 100 (ranked based on decreasing log2FC values) of the genes coding for extracellular proteins with significantly higher expression in the proterosoma coded for proteins with unknown function, whereas genes coding for serine proteases (8 genes), neuropeptides (6 genes) or homologues of “salivary gland peptides” of ticks (5 genes) were also observed (supplemental Table S9).
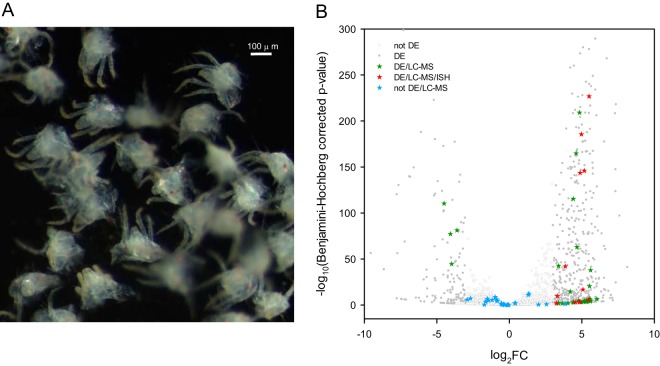
Fig. 2.
T. urticae genes that code for extracellular proteins and are differentially expressed between proterosomas and intact females. A, Dissected T. urticae proterosomas used for RNA extraction (see Fig. 3 for dissection position). B, The negative log10 of Benjamini-Hochberg adjusted p values was plotted against the log2FC in gene expression for all T. urticae genes (see supplemental Fig. S2) and subsequently filtered for genes coding for extracellular proteins (predicted to be extracellular by WoLF PSORT (82) and predicted with a SP by SignalP 4.1(81), see supplemental Table S9). Differentially expressed genes (|FC| ≥ 8, Benjamini-Hochberg adjusted p value ≤ 0.05) are shown as gray dots (DE), whereas genes that were not differentially expressed are shown as gray circles (not DE). Genes coding for extracellular proteins that were identified by nano-LC-MS/MS (“extracellular LC-MS salivary protein genes”) are depicted as stars. Extracellular LC-MS salivary protein genes that were differentially expressed are depicted as green stars (DE/LC-MS). Extracellular LC-MS salivary protein genes that were differentially expressed and for which ISH confirmed expression in salivary glands are depicted as red stars (DE/LC-MS/ISH) whereas extracellular LC-MS salivary protein genes that are not differentially expressed are shown as blue stars (not DE/LC-MS).
About half of the genes (54%) coding for T. urticae putative salivary proteins detected by nano-LC-MS/MS had significantly different expression levels (Benjamini-Hochberg adjusted p value ≤ 0.05 and |FC| ≥ 8) in the proterosoma (Fig. 2, supplemental Fig. S2, supplemental Table S9)). The majority (88%) of these genes showed a higher expression in the proterosoma whereas only 6 had a lower expression. Among those genes with a higher expression, we identified actin (tetur03g09480), an aldo-keto reductase (tetur02g11340), a glutathione S-transferase (tetur01g02510), a chitinase (tetur01g11910), a protein with a trypsin inhibitor-like, cysteine rich domain (tetur40g00392), 2 beta-mannosidases (tetur16g03420, tetur28g00360), 7 serine-proteases (tetur07g00150, tetur09g03880, tetur12g03940, tetur12g03950, tetur16g03190, tetur16g03470, tetur30g01440), and 31 genes coding for proteins with a yet unknown function. Among the 6 genes with a lower expression we found genes coding for a cathepsin (tetur03g08030), a short-chain dehydrogenase (tetur28g01720), an alpha-2-macroglobulin (tetur18g03030), conserved secreted proteins with MD-2-related lipid recognition domain (tetur14g02070 and tetur14g02080), and a gene coding for a protein with an unknown function (tetur20g01290) (Table I, supplemental Table S9, Fig. 2, supplemental Fig. S2).
Evaluation of the Salivary Protein Repertoire With Previously Published Data Sets
Fifty of the 95 proteins (53%) identified by nano-LC-MS/MS shared homology (BLASTp with E-value threshold 1.0 E-5) with proteins of the most closely related nonphytophagous acariform mite for which proteomic resources were available, the American house dust mite D. farinae (Table I, supplemental Table S6). These were mainly proteins with a predicted function in digestion (26 out of 50), like glycoside hydrolases, beta-galactosidase/mannosidase, propylcarboxypeptidase, proteinase inhibitors, and serine proteases. T. urticae proteins with an unknown function did not share homology with D. farinae proteins. A previously published data set of the T. urticae transcriptome across different developmental stages (44) was mined for the gene expression-levels of T. urticae putative salivary genes. The overall average expression for all T. urticae genes (calculated as “reads per kb of transcript per million mapped reads”: RPKM) across all the feeding stages (larva, nymph, adult) relative to the overall average expression in the embryo stage was 2.21-fold higher (44). Seventy-five out of 95 T. urticae putative salivary genes had a ratio higher than this average, whereas 28 (30%) had an expression ratio more than 8 (Table I). Moreover, although only three putative T. urticae salivary protein genes were expressed at lower levels in nondiapausing as compared with diapausing adults, we found that 26 genes were more strongly expressed in nondiapausing adults that are actively feeding ((89), Table I, supplemental Table S6). Finally, 15 putative T. urticae salivary proteins were also detected in a proteome analysis of T. urticae faeces, (Table I, (88)). These included cathepsins (tetur09g04400, tetur25g00650, and tetur12g01860), actins (tetur03g09480, tetur09g05350), a lipocalin (tetur04g05980), beta-galactosidase (tetur07g07380), fructose-biphosphate aldolase (tetur07g03440), glutathione S-transferases (tetur01g02510, tetur03g07920), vitellogenins (tetur39g00810, tetur43g00010), alpha-2-macro-globulin (tetur18g03030), and two proteins with unknown function (tetur07g00160 and tetur29g01360).
ISH of Salivary Protein Genes Combined with Morphological Analysis Distinguishes Anterior and Dorsal Podocephalic Glands
The spatial expression pattern of 15 putative T. urticae salivary protein genes was evaluated using whole-mount in situ hybridization (ISH). These genes were selected out of the nano-LC-MS/MS data set based on the possession of a SP for secretion and a relatively high expression in the proterosoma (Table I). Furthermore, we endeavored to include genes from as many different gene families as possible. To ensure that the observed ISH signal is linked to the genuine morphological structure, T. urticae internal anatomy was investigated by means of x-ray submicron computed tomography. This imaging technique resulted in high-resolution three-dimensional pictures, providing additional insights into spider mite internal anatomy from those reported previously (48, 91, 92), and allowed more accurate interpretation of the ISH signal observed by bright field and fluorescence confocal microscopy. Of the 15 genes, 14 were expressed specifically in either the anterior or dorsal podocephalic glands (Fig. 3). None of the genes we tested showed expression in both the anterior and dorsal glands, and staining was not observed when using sense control probes. We did not detect gene expression in a third pair of glands annotated previously as being salivary, and known as the coxal organs (48). Tetur13g00600 was also tested, yet no clear specific staining was visible in any gland or tissue.
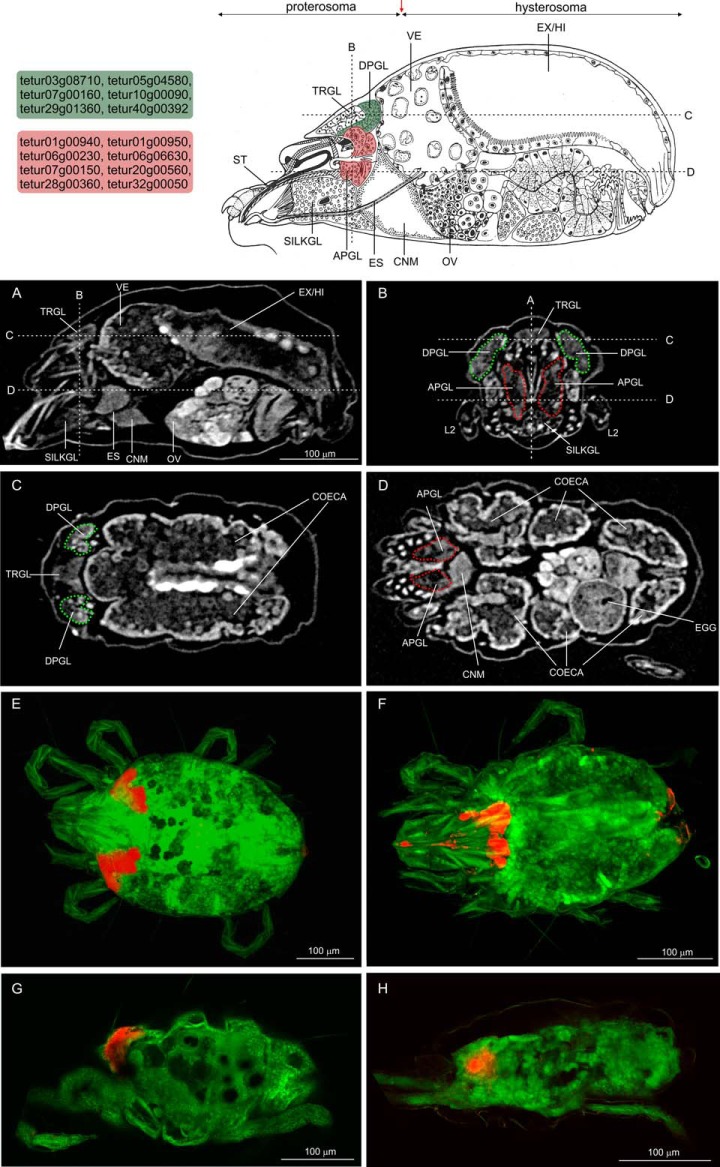
Fig. 3.
Localization of expression of T. urticae genes coding for putative salivary proteins identified by nano-LC-MS/MS. A schematic representation of a mid-sagittal section of a T. urticae female is shown on top (redrafted from (92) with the permission from Elsevier). Dorsal podocephalic glands (DPGL) are shaded green whereas the anterior podocephalic glands (APGL) are shadedred. T. urticae genes for which ISH confirmed expression in one of the podocephalic salivary glands are framed by a box shaded red (APGL) or green (DPGL). A red arrow indicates the dissection position for collection of proterosomas. A–D, virtual sections obtained by a sub-micron CT scan of a T. urticae adult female confirmed the internal morphology as described by (92). A, virtual mid-sagittal section, dashed straight lines represent the other virtual sections that were created and are shown in panels B, C, and D. B, Virtual cross section at the tracheal glands (TRGL): DPGL and APGL are indicated with a green and red dashed line, respectively. C, Virtual frontal section at TRGL, DPGL are indicated with a dashed green line. D, Virtual frontal section at the central nervous mass (CNM), APGL are indicated with a dashed red line. E–H, Confocal images of whole-mount in situ hybridization of putative T. urticae salivary protein genes. A DIG-labeled antisense probe was used for hybridization and the signal was developed using anti-DIG-AP and FastRed as substrate. The reaction product is visible as a red signal whereas the spider mite body shows green autofluorescence. Signal development corresponded to the localization of the podocephalic glands as shown in panels B, C, and D. E, G, Dorsal (Z-stack maximum intensity projection) and lateral (sagittal stack) view of signal development in the DPGL (tetur07g00160). F, H, Dorsal (Z-stack maximum intensity projection) and lateral (sagittal stack) view of signal development in the APGL (tetur28g01360, panel F, and tetur01g00950, panel H). Other abbreviations: SILKGL, silk glands; ES, esophagus; OV, ovaria; ST, stylet; VE, ventriculus; EX/HI, excretory organ/hindgut; and L2, second pair of legs.
The Expression Profiles of Salivary Protein Genes Show Signs of Host Adaptation
To single out putative T. urticae salivary proteins with a host plant dependent production, the list of all nano-LC-MS/MS identified salivary proteins was split up over the separate hosts lines (i.e. reference line on bean, 5 generations on maize, 5 generations on soy, 30 generations on tomato). Thirty-eight proteins were found in all samples. Furthermore, host-specific salivary proteins could be identified as well, 7 were found only when feeding on maize, 5 only when feeding on soy and 11 only when feeding on tomato. No proteins were unique for the reference line on bean (Fig. 4A, supplemental Table S6). Host plant specificity of putative T. urticae salivary proteins was further illustrated by means of a heat map depicting the mean rTop3 factor of a selection of candidate salivary proteins (maximum rTop3 value higher than the 30th percentile of all maximum rTop3 values) for the different investigated host plants (Fig. 4B). The most apparent case of host-specific salivary proteins was tetur55g00040/tetur28g01330 (WTSP1, belonging to OrthoMCL group TuMCL_36), proteins with an unknown function, which were uniquely and abundantly identified from saliva of the tomato-adapted mites. Based on our clustering analysis the salivary proteomic repertoire of T. urticae feeding on bean plants seemed most similar to the repertoire of soy-adapted mites (both plant species belong to the Fabaceae).
Fig. 4.
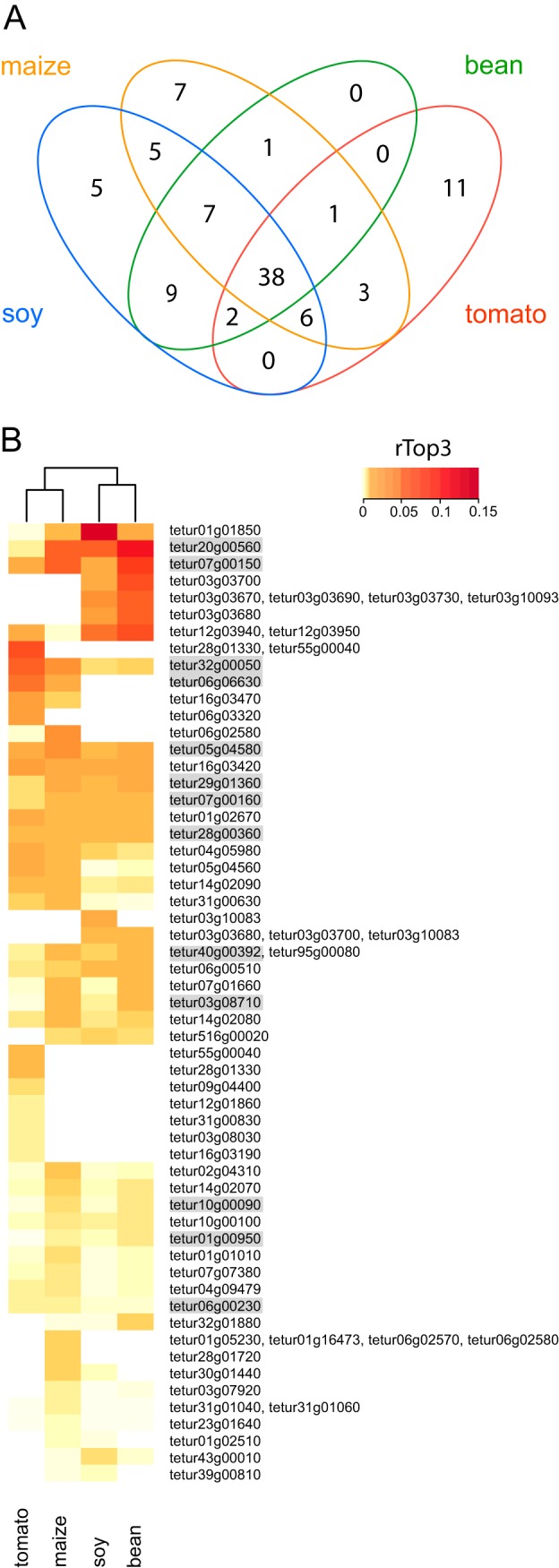
Overview of nano-LC-MS/MS identified putative T. urticae salivary proteins. A, Venn diagram depicting overlap between putative T. urticae salivary proteins secreted by mites adapted to different host plants (bean, maize, soy, tomato). Only those salivary proteins with a mean PSM of at least two in at least one of the T. urticae host plant adapted lines were used for comparison (see Table I and supplemental Table S6). B, Heat map of mean rTop3 values of putative T. urticae salivary proteins secreted by mites adapted to different host plants (bean, maize, soy, tomato). Only those salivary proteins (and “protein inference groups”) with a mean PSM of at least two in at least one of the T. urticae host plant adapted lines and with a maximum rTop3 value higher than the 30th percentile of maximum rTop3 values were used for comparison (see Table I and supplemental Table S6). The Euclidean distance metric and Ward's method were used for clustering of both rows and columns. All putative salivary proteins for which the corresponding genes were shown to be expressed in the salivary glands by ISH (Fig. 3) are shaded gray.
To validate these host-specific findings based on individual biological replicates, as well as to compare in further detail the expression level of genes coding for T. urticae putative salivary proteins across host plant species, we performed a genome-wide expression analysis of the mite lines that were used in proteomic experiments (i.e. T. urticae adapted to maize, soy or tomato relative to the reference line on bean) using an Agilent gene expression microarray. Additionally, a line adapted to cotton during 5 generations was also investigated. We determined the absolute expression levels using normalized cyanine 3 (cy3) intensity values but also calculated the expression levels relative to the mites from bean as a benchmark (see Experimental Procedures for details, Fig. 5A and 5B). We confirmed what we detected previously when comparing the rTop3 values of the proteomics data across different host plant lines (Fig. 4): the salivary composition of T. urticae is host plant dependent. For example, in the tomato-adapted mite lines several salivary protein genes were highly expressed relative to bean-adapted mites: tetur32g00050 (Tu_MCL_35), tetur28g01330 and tetur55g00040 (Tu_MCL_36) and tetur31g00830 (Tu_MCL_25). Alternatively, tetur03g03670, tetur03g10093, tetur03g03700, tetur03g03680, tetur03g03690, and tetur03g03730 (OrthoMCL group OG5_144177) were expressed at much lower levels in mites feeding from all host plants except for soy, compared with bean. The absolute expression level of the latter six genes was very high after feeding on bean or soy (cy3 intensity levels were within the top 1% of highly expressed genes in mites on either bean or soy) and relatively low after feeding on the nonleguminous host plants under study (fold changes between mites on bean and mites on tomato, maize or cotton varied between 7 and 289).
Fig. 5.
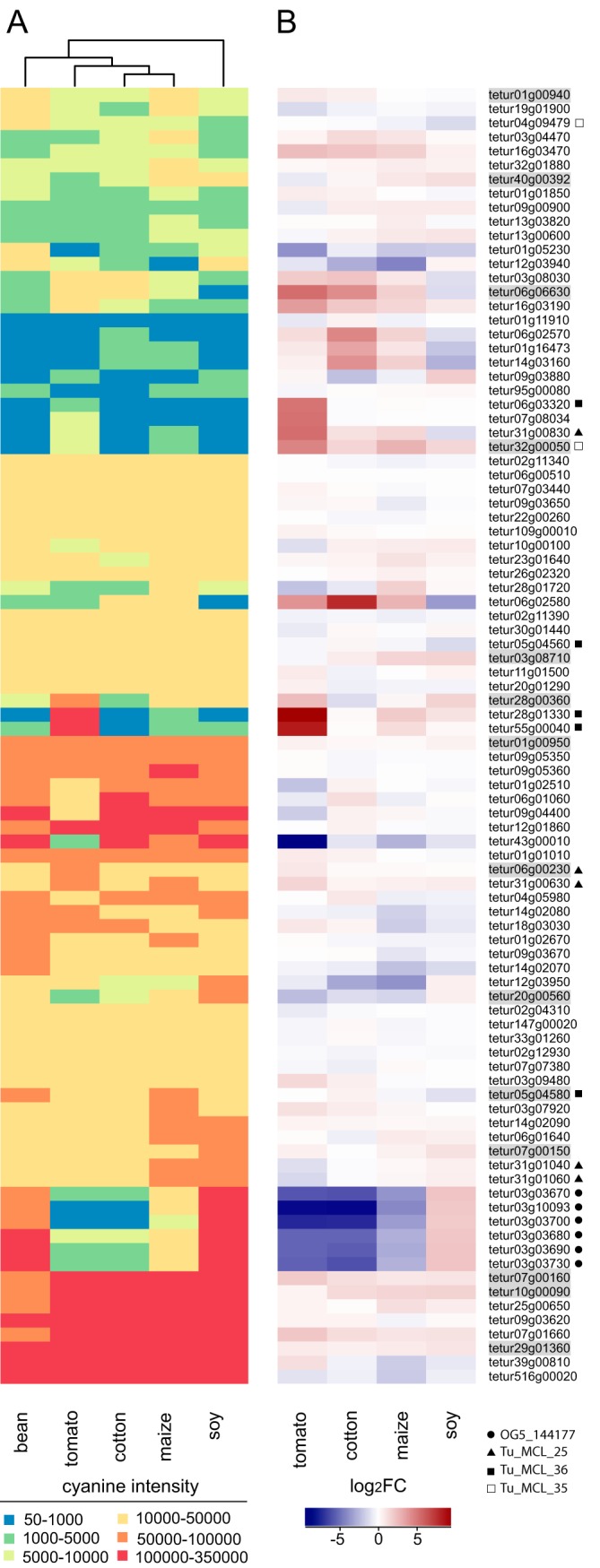
Heatmap of expression levels of putative T. urticae salivary protein encoding genes and their up- or down-regulation in mites adapted to different host plants. A, Heatmap of cyanine intensities of putative T. urticae salivary protein encoding genes. The Euclidean distance metric and Ward's method were used for clustering of both rows and columns. For 92 out of 95 putative T. urticae salivary protein genes expression data was available. B, Heatmap of log2FCs of putative salivary protein genes in mites adapted to soy, maize, cotton or tomato as compared with mites adapted to bean. Genes are sorted based on their order in panel A. Genes that were shown to be expressed in the salivary glands by ISH (Fig. 3) are shaded gray. A circle, triangle, filled square or empty square indicates whether a T. urticae gene belongs to either OrthoMCL group OG5_144177, Tu_MCL_25, Tu_MCL_36 or Tu_MCL_35, respectively.
DISCUSSION
Identification of Salivary Gland Proteins Secreted by the Spider Mite T. urticae
The spider mite T. urticae is well known for its ability to feed on an extraordinary wide range of different plant species even though their feeding activities induce plant defenses that can negatively affect mite fitness (5, 16, 93). However, arthropod populations have adapted to suppress these defenses, including T. urticae populations (19, 23, 24, 94), suggesting that effectors in their saliva are crucial determinants for success in colonizing host plants. To gain insight into the salivary proteome of T. urticae, spider mites were allowed to feed on small hemispheres filled with artificial diet. Using this setup, mites can secrete saliva into the diet, which in turn can be used for nano-LC-MS/MS analysis. By combining this approach with extensive follow-up analyses, we aimed to obtain an as accurate as possible set of proteins that are actually injected by T. urticae into host plants. In the past, this kind of approach was also successful to identify aphid salivary proteins (95–97). T. urticae gene expression profiles are known to be highly host plant dependent (23, 98). By using spider mites reared on four different host plant species for a minimum of five generations, we maximized chances of capturing a higher variety of salivary proteins. In addition, this allowed to study the influence of the host plant on spider mite salivary protein composition.
By design, our method is suitable to collect saliva from small numbers of potentially tiny arthropods. In recent studies on aphid saliva (96, 99, 100), tens of thousands of aphids were needed to collect saliva in sufficient quantities for protein identification. Despite the smaller size of T. urticae (0.4–0.5 mm) compared with aphids (> 1 mm), we managed to collect sufficient amounts of saliva for nano-LC-MS/MS analysis using only about 500 mites per sample. Limited volumes of diet were sufficient for the analysis via state-of-the-art mass spectrometry, making the analysis of salivary proteomes of small herbivorous arthropods time- and resource-efficient where feeding on an artificial diet is feasible.
Salivary proteins need to be secreted by the salivary glands into the salivary duct before they can be delivered into a host plant via the saliva. The majority of the 95 putative T. urticae salivary proteins identified by nano-LC-MS/MS possess a SP and were predicted to be localized extracellularly, consistent with secretion. However, the lack of a SP for 18 out of the 95 proteins does not exclude that these proteins are secreted, as secretory pathways independent of the canonical endoplasmic-reticulum-Golgi network may exist (101). Additionally, SPs can be missed by gene prediction programs during genome annotation. In fact, both reasons have been proposed to explain why many putative arthropod salivary proteins lack a predicted SP (e.g. (96)).
Although detection by LC-MS is one of the most robust methods to identify secreted salivary proteins, the list in Table I should not be considered exhaustive. A number of factors determine detection by nano-LC-MS/MS, including the adhesion to the material of the collection device during sample preparation, the presence of trypsin cleavage sites (102), the quality of MS/MS spectra, the type of chemical or post-translational modifications that were accounted for in the database search, and the protein hydrophobicity influencing the fractionation (103). We validated proteomic findings with additional expression data sets. Previously, transcriptome analyses of salivary glands of insects have been reported (e.g. (104–106)). Current technology, however, does not allow straightforward generation of specific spider mite salivary transcriptomes as spider mite glands are merely 50 μm in diameter. We therefore isolated spider mite proterosomas, the anterior body region that includes the salivary glands next to other tissues such as the nervous mass and the silk glands (Fig. 3). In this study, the salivary glands were confirmed to be localized in the proterosoma using x-ray tomography. In a comparison between transcriptomes of T. urticae intact females and T. urticae proterosomas, candidate genes for salivary gland expression are expected to be more highly expressed in the latter. Our analysis revealed more than 400 genes with a significantly higher expression in the proterosoma. Reassuringly, (Table I, supplemental Table S6). The majority of the identified putative T. urticae salivary proteins are predicted to be secreted, are encoded by genes of which the expression is higher in the body region containing the salivary glands, and are expressed specifically in feeding spider mites.
Genes Encoding Putative Salivary Proteins Are Expressed in the Salivary Glands
Despite multiple lines of evidence for salivary origins, the proteins we identified could still originate from another source. Of particular note, the catalogue of putative salivary proteins did show some overlap (15 out of 95), with proteins identified from spider mite fecal extracts (88). However, with ISH we showed that for genes encoding two of these proteins (tetur07g00160 and tetur29g01360) expression is specific to the salivary glands (Table I, Fig. 3). Hence, the proteins present in the faeces probably reflect a salivary origin. This indicates that secreted salivary proteins may end up in the digestive system during feeding, either directly or indirectly after ingesting cytoplasm from cells in which saliva had been injected. Interestingly, many putative T. urticae salivary proteins shared homology with D. farinae proteins that were predicted to have a function in digestion. ISH in T. urticae showed that some of the genes that code for presumptive gut proteins, for example tetur07g00150 (a serine protease) and tetur28g00360 (a beta-mannosidase), are actually expressed in the salivary glands (Table I). Hence, they may have a digestive function in the saliva instead of, or in addition to, a potential role in the gut. Nevertheless, some proteins we found in the artificial diet could be fecal contaminants. This is also suggested by the proterosomal transcriptome, as some of the proteins detected in the diet are not differentially expressed, whereas the digestive enzymes confirmed by ISH are highly expressed in proterosomas (Fig. 2, supplemental Table S9). Adult female spider mites were found to deposit eggs and silk on the diet membrane and together with the mite's cuticle these could be other potential sources of contamination, e.g. for vitellogenin (tetur39g00810, tetur43g00010 and tetur516g00020). Among the 15 genes selected for ISH, we included some coding for proteins with either high or low rTop3 values, but all were more highly expressed in the proterosoma and belonged to the most prominent proteins or protein families in the data set (Table I). Of the 15 selected genes, 14 were found to be expressed in either the paired anterior or paired dorsal podocephalic glands. Both of these podocephalic salivary glands are predicted to be rich in proteins as assessed by ribosomes presence (107) and staining for proteins with methylene blue (48). Intriguingly, our data show that the anterior and dorsal podocephalic glands are responsible for the production of a discrete subset of the salivary proteome. Such “division of labor” has been reported for the different secretory cell types in the salivary glands of aphids (108–110) and has been suggested for thrips as well (111). The regulatory mechanism by which these glands or cell types are “assigned” to produce particular proteins in spider mites, aphids, or thrips is not known. A functional distinction between anterior and dorsal podocephalic glands in spider mites was already predicted by Mothes and Seitz (48). The production of a serous secretion was attributed to dorsal glands, whereas the anterior glands were predicted to produce a mucous secretion (48). However, supporting evidence for these specific predictions is not provided by our study.
For one of the 15 genes selected for ISH, expression in the salivary glands could not be confirmed, nor did we observe staining in other tissues. This does not exclude that this gene encodes a salivary protein, as ISH “failure” may have several causes including poor probe design or expression below the detection limit.
The Saliva of T. urticae Comprises a Mixture of Different Protein Families
Among the different putative T. urticae salivary proteins we identified, several were carbohydrate and protein degrading enzymes including cathepsins, serine proteases, glycoside hydrolases, beta-galactosidases, and beta-mannosidases. Genes coding for serine proteases and beta-mannosidases also showed a higher expression in the proterosoma (Table I, supplemental Table S9). These catabolic proteins could have a digestive function during the feeding process before ingestion. For example, polysaccharide-digesting enzymes present in the saliva of the hemipteran herbivore Homalodisca vitripennis have been proposed to play a role in the degradation of cell wall material, hereby facilitating stylet penetration (112). Predigestion of plant material has been suggested to be a property of spider mite saliva (47–49) but there is no empirical evidence to support this hypothesis. Recently, the spider mite consumption rate was estimated at a single mesophyll cell per 10 min (113), which is much lower than reported previously (3). Hence, such a low consumption rate might allow predigestion of plant cells with spider mite salivary enzymes. However, secreted digestive enzymes could have a function in the mite itself, e.g. in the esophagus, after ingestion. Like caterpillar feeding, also spider mite feeding induces the plant to produce defensive proteins like serine protease inhibitors (PI-I and PI-II) (5, 15, 29, 93). The production of salivary serine proteases may be a means to compensate for inactivation of gut serine proteases. The mite itself also produces salivary proteinase inhibitors as we detected several cystatins (cysteine protease inhibitors: tetur06g01060, tetur06g06630, tetur09g03620, tetur09g03650, and tetur09g03670) secreted in the diet. Cystatins can play important roles in plant-pathogen interactions. The maize pathogen Ustilago maydis manipulates expression of a plant cystatin via its effector protein pep1, causing inactivation of defensive Cys proteases thereby making the plant susceptible to infection (114). In addition, Phytophthora infestans produces secreted serine protease inhibitors and cystatin-like effectors (115) to target key extracellular defensive proteases of its hosts and a host plant shift of this pathogen was attributed to a single amino acid change in one of these cystatin effectors (116). This illustrates how decisive effectors can be for determining host plant compatibility of a pest and it may suggest that unrelated organisms might have evolved effector proteins with similar functions (i.e. similar plant targets).
We identified a chitinase (tetur01g11910) among the T. urticae salivary proteins as well. Chitinase was also found in saliva of an aphid species (Diuraphis noxia) feeding on wheat (Triticum aestivum) and it was suggested that this enzyme might inhibit secondary fungal infections at the feeding site by hydrolyzing fungal chitin-rich cell walls (51). We found the chitinase only in the saliva of mites-lines adapted to both bean and maize (Table I, supplemental Table S6, Fig. 2) suggesting there might be a host plant specific role for these proteins. However, salivary chitinases are not uncommon and were found, for example, in saliva of humans (117), remipedes (118), octopuses (119), nematodes (120, 121), and insects (51) and were suggested to play a defensive role against chitinous pathogens. Finally, among characterized proteins, a superoxide dismutase (SOD, tetur26g02320), which targets reactive oxygen species that are often induced in plants upon damage, was also detected. Noteworthy, SODs have also been identified in the saliva of a potato leafhopper (Empoasca fabae) and two aphids (D. noxia and Ropalosiphum padi) (122).
Intriguingly, several of the most abundant proteins in the saliva are coded by multi-membered gene families of unknown function. Most of these proteins were encoded by genes with proterosoma-enriched expression, and lacked homologs in D. farinae, a nonplant feeding mite. One possibility, although speculative, is that the proliferation of these families facilitated host-range expansion in T. urticae. Polyphagous species are proposed to harbor a larger collection of salivary proteins because they are exposed to a greater diversity of selection pressures, i.e. host plants that vary in morphology and (defensive) physiology (52). Analogous gene family proliferations have been described for other proteins relevant for plant-spider mite interactions, such as detoxification enzymes (cytochrome P450s, carboxyl/choline esterases, and glutathione S-transferases), transporters (21, 123, 124), and digestive cysteine peptidases (125).
The Expression Level of Some Salivary Protein Genes is Host Plant Dependent
Secreting the full salivary protein repertoire, independent of the host, might maximize the possibility that some salivary proteins are perceived as elicitors by the plant. In addition, it is unlikely that all proteins will function optimally in all hosts as targets may differ across plants or may be absent. Therefore, it would be beneficial for polyphagous herbivores to alter the composition of their saliva according to the host plant identity. Differences between the salivary gland transcriptomes of two populations of Nilaparvata lugens, a rice specialist, maintained on either a resistant or susceptible rice variety, were proposed to be related to different virulence traits of these brown planthoppers (126, 127). Furthermore, drastically different salivary protein profiles were found when the western tarnished plant bug Lygus hesperus was fed artificial diet, cotton or pinto bean (128). Hence, we investigated host plant specific secretion and expression of the mite's salivary genes. As suggested by the clustering of proteomics data (Fig. 4), transcriptome analysis revealed that the expression of T. urticae salivary protein genes is strongly influenced by the host plant species on which the mites had been feeding (Fig. 5B). For example, proteins tetur28g01330 and tetur55g00040 were uniquely and abundantly identified from diet fed upon by tomato-adapted mites, whereas expression of the corresponding gene was also extremely high when feeding on tomato, relative to mites feeding on the other tested plants (Fig. 5). Next to individual genes, we also studied OrthoMCL gene family groups. Tu_MCL_25, Tu_MCL_35, and Tu_MCL_36 (Table I, supplemental Table S7) all have members that are highly expressed when feeding on tomato, whereas the expression of other members of these groups was not influenced by the host plant (Fig. 5, Fig. 6). Future experiments should point out whether this is because of the fact that mites from the tomato-adapted lines had been maintained on tomato for many more generations than mites on the other hosts or whether this is because of the specific allelochemicals of tomato posing digestive or defensive challenges. Members of the OrthoMCL group OG5_144177 (Table I, supplemental Table S7) were, relative to mites on bean, expressed at lower levels in mites feeding from maize, cotton, and tomato (Fig 5B). When feeding on soy however, these genes were expressed at slightly higher levels. A similar pattern can also be deduced from the proteomics data of mites adapted to bean and soy versus mites adapted to maize and tomato. Bean and soy both are legume species (Fabaceae) and mites feeding on these plants probably encounter analogous plant secondary compounds that select for or induce a similar repertoire of salivary proteins. The observation that different host plant species can differentially affect expression levels of genes coding for salivary proteins—such as effectors—has been reported for aphids as well (108, 110, 129). Elzinga et al. (129) suggested that the differential expression of salivary effector genes represents a strategy to avoid activation of defenses and to facilitate feeding.
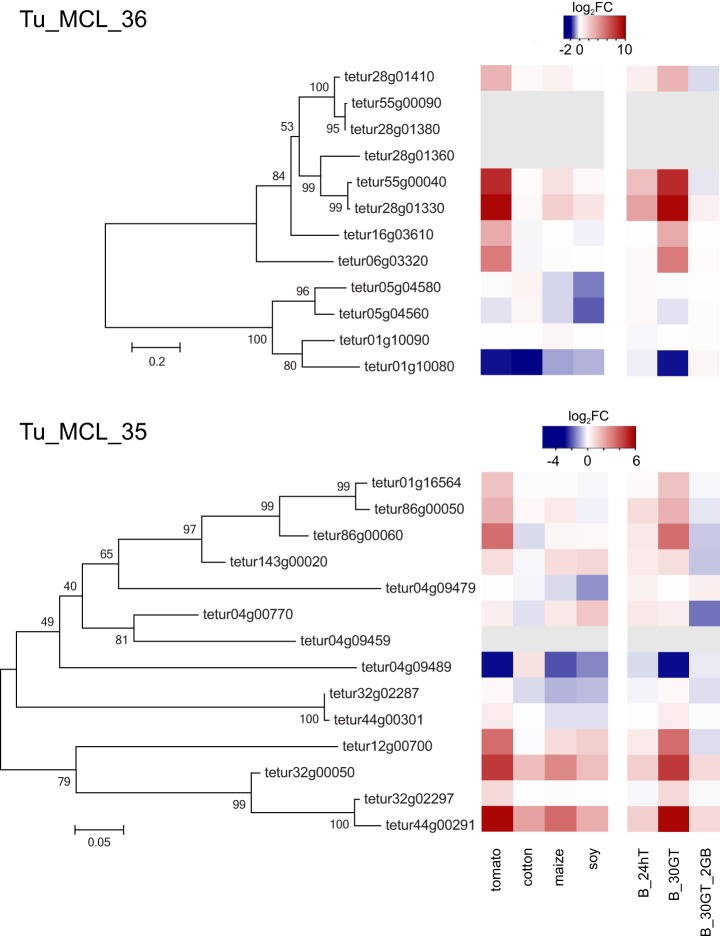
Fig. 6.
Expression profiles of genes coding for members of the expanded Tu_MCL_36 and Tu_MCL_35 protein families in T. urticae. Phylogenetic analyses of Tu_MCL_36 and Tu_MCL_35 proteins are shown next to heatmaps depicting relative gene expression of Tu_MCL_35 and Tu_MCL_36 genes in T. urticae subjected to different host plant regimes (log2FCs, relative to the expression level on bean). The heatmap on the left represents log2FCs of Tu_MCL_35 and Tu_MCL_36 genes in mites adapted to soy, maize, cotton or tomato compared with bean, whereas the heatmap on the right represents log2FCs of Tu_MCL_35 and Tu_MCL_36 genes in T. urticae after host shifts between bean and tomato (B_24hT, mites from the London reference strain on bean transferred to tomato for 24h; B_30GT, mites from the bean strain grown on tomato for 30 generations; B_30GT_2GB, mites from the bean strain grown on tomato for 30 generations and transferred back to bean for 2 generations (23)). Gray boxes indicate that for a specific gene no probes were included in the T. urticae microarray design, and hence expression could not be assayed.
In the scope of this discussion it is important to realize that only one T.urticae strain (London) was used. Although mites from this strain had been maintained on bean for many years, previous studies have shown that this population is both not fully inbred (130) and capable of extensive transcriptional plasticity upon transfer to new hosts (18, 21, 22, 44), the latter of which has been further confirmed in this study. We believe therefore that this study has captured much of the repertoire of T. urticae's biologically relevant salivary proteins. However, as marked variation of genotypes exists between T. urticae populations and across spider mite species (5, 19), the use of additional T. urticae strains or different spider mite species may deliver additional salivary proteins. Identification via shotgun proteomics will however be less straightforward without reference genomes for these strains or species.
Prediction of Effector Proteins
Confidently identifying salivary proteins as effectors is not straightforward. Arthropod effector proteins are characterized by the presence of a N-terminal SP, directing them to the secretory pathway, and their encoding genes show signatures of rapid evolution (poor sequence similarity with other genera, multiple gene copies, and high rates of non-synonymous nucleotide mutations) (131–134). Because of the highly specific function of effectors, variable expression levels depending on the host plant could also be indicative of a context-dependent function (108). The presence of R genes, which turn effectors effectively into elicitors, can differ among plant species or varieties (135–137), as does the presence of S genes (39), the target of the effector. As such, the R and S gene composition of a host may determine the specific transcriptional response of a herbivore's secretome and thus its effector repertoire. Proteins of the OrthoMCL groups OG5_144177, Tu_MCL_25, Tu_MCL_35, and Tu_MCL_36 have characteristics attributed to (arthropod) effectors: (a) they are targeted to the saliva and are secreted during feeding, (b) they share no homology with proteins of non-phytophagous mite species, (c) they belong to multigene families (> 10 members) (Table I, supplemental Table S6, supplemental Table S7), and (d) the expression level of some of the gene family members has been shown to differ across host plants (Fig. 6). Intriguingly, salivary proteins from the OrthoMCL groups Tu_MCL_25 and Tu_MCL_211 (tetur31g01040 and tetur01g01000, respectively, see Table S6), were recently shown to act as effector proteins by Villarroel et al. (138). These proteins were identified using an in silico approach and were shown to enhance T. urticae reproductive performance when transiently expressed in N. benthamiana leaves (138). Future research is needed to unravel the function of these and other T. urticae salivary constituents documented here.
CONCLUSIONS
Despite the minute size of T. urticae, we managed to collect saliva for proteome analysis through an artificial diet system. Using nano-LC-MS/MS, 95 putative T. urticae salivary proteins were identified, indicating that these mites employ a complex protein mixture in their interaction with host plants. A proterosoma specific transcriptome is presented as a second discovery pipeline, and a considerable overlap with nano-LC-MS/MS data was observed. A selection of genes coding for putative salivary proteins was confirmed to be expressed in the salivary glands by whole-mount in situ hybridizations. The expression levels of a subset of genes were strongly dependent on feeding on different host plant species. Several proteins belong to protein families with as yet unknown functions, with some having structural and gene family features suggestive of roles as effector proteins. The differential expression levels of some of these putative salivary effector genes across host plants may be an indication of a host plant specific mode of action. The search for effectors and their possible targets is essential to our understanding of polyphagy and the evolution of the plant-mite interactions, but it is also of practical importance, as it would enable plant breeders to discover new R and S genes in order to develop mite-resistant crops.
Supplementary Material
Acknowledgments
We thank Wim Reubens for help in spider mite saliva collection and in situ hybridizations. The manuscript reflects the opinions of the authors, and not necessarily those of the funding agencies.
Footnotes
Author contributions: W.J., W.D., and T.V. designed research; W.J., W.D., V.Z., J.V., and G.B. performed research; C.A.V., M.G., R.C.S., L.T., R.M.C., M.R.K., and B.V. contributed new reagents or analytic tools; W.J., W.D., N.W., R.G., G.M., and T.V. analyzed data; W.J., W.D., and T.V. wrote the paper; L.T., R.M.C., M.R.K., and B.V. revised the article.
* This project was supported by the European Commission (EC contract 618105) via FACCE ERA-NET Plus and FACCE-JP (Genomite, project ID 137), the Fund for Scientific Research Flanders (FWO) (grant G009312N to L.T. and T.V.L. and grant G053815N to L.T., T.V.L. and W.D.), The Special Research Fund (DOZA 01J131711 to W.J.) and NSF award DEB 1457346 (to R.M.C). R.G. was supported by National Institutes of Health Genetics Training Grant T32 GM07464. C.A.V. was supported by CONICYT BECAS (Chile) and M.R.K. by NWO (The Netherlands) (STW-VIDI 13492). N.W. was supported by a Marie Sklodowska-Curie Action (MSCA) Individual fellowship (658795-DOGMITE) of Horizon 2020. M.G. acknowledges funding by the Government of Canada through Genome Canada and the Ontario Genomics Institute (OGI-046) and Ontario Research Fund-Global Leadership in Genomics and Life Sciences GL2–01-035. W.D. and G.M. are postdoctoral fellows of the Fund for Scientific Research Flanders (FWO). Some of the computational resources (Stevin Supercomputer Infrastructure) and services used in this work were provided by the VSC (Flemish Supercomputer Center), funded by Ghent University, the Hercules Foundation and the Flemish Government - department EWI.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- JA
- Jasmonic Acid
- DE
- Differentially Expressed
- FC
- Fold Change
- FDR
- False Discovery Rate
- ISH
- In Situ Hybridization
- PSM
- Peptide Spectrum Match
- SA
- Salicylic Acid
- SP
- Signal Peptide
- SpC
- Spectral Counts.
REFERENCES
- 1. Migeon A., and Dorkeld F. (2006–2016) Spider Mites Web: a comprehensive database for the Tetranychidae. [Google Scholar]
- 2. Van Leeuwen T., Tirry L., Yamamoto A., Nauen R., and Dermauw W. (2015) The economic importance of acaricides in the control of phytophagous mites and an update on recent acaricide mode of action research. Pestic. Biochem. Physiol. 121, 12–21 [DOI] [PubMed] [Google Scholar]
- 3. Liesering R. (1960) Beitrag zum phytopathologischen Wirkungsmechanismus von Tetranychus urticae Koch (Tetranychidae, Acari). Z. Pflanzenkr. Pflanzenschutz 67, 524–542 [Google Scholar]
- 4. Tomczyk A., and Kropczynska D. (1985) Effect on the host plant. In: Helle W., and Sabelis M. W., eds. Spider mites. their biology, natural enemies and control. Vol. 1A, pp. 317–329, Elsevier Science Publishers B.V., Amsterdam, The Netherlands [Google Scholar]
- 5. Kant M. R., Sabelis M. W., Haring M. A., and Schuurink R. C. (2008) Intraspecific variation in a generalist herbivore accounts for differential induction and impact of host plant defences. Proc. R. Soc. Lond. B Biol. Sci. 275, 443–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. van den Boom C. E. M., van Beek T. A., and Dicke M. (2003) Differences among plant species in acceptance by the spider mite Tetranychus urticae Koch. J. Appl. Entomol. 127, 177–183 [Google Scholar]
- 7. Yano S., Wakabayashi M., Takabayashi J., and Takafuji A. (1998) Factors determining the host plant range of the phytophagous mite, Tetranychus urticae (Acari: Tetranychidae): a method for quantifying host plant acceptance. Exp. Appl. Acarol. 22, 595–601 [Google Scholar]
- 8. Kant M. R., Jonckheere W., Knegt B., Lemos F., Liu J., Schimmel B. C., Villarroel C. A., Ataide L. M., Dermauw W., Glas J. J., Egas M., Janssen A., Van Leeuwen T., Schuurink R. C., Sabelis M. W., and Alba J. M. (2015) Mechanisms and ecological consequences of plant defence induction and suppression in herbivore communities. Annals of Botany 115, 1015–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bonaventure G., VanDoorn A., and Baldwin I. T. (2011) Herbivore-associated elicitors: FAC signaling and metabolism. Trends Plant Sci. 16, 294–299 [DOI] [PubMed] [Google Scholar]
- 10. Wasternack C., and Hause B. (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Botany 111, 1021–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Caarls L., Pieterse C. M., and Van Wees S. C. (2015) How salicylic acid takes transcriptional control over jasmonic acid signaling. Front. Plant Sci. 6, 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mur L. A., Kenton P., Atzorn R., Miersch O., and Wasternack C. (2006) The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 140, 249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pieterse C. M., Leon-Reyes A., Van der Ent S., and Van Wees S. C. (2009) Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316 [DOI] [PubMed] [Google Scholar]
- 14. Erb M., Meldau S., and Howe G. A. (2012) Role of phytohormones in insect-specific plant reactions. Trends Plant Sci. 17, 250–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kant M. R., Ament K., Sabelis M. W., Haring M. A., and Schuurink R. C. (2004) Differential timing of spider mite-induced direct and indirect defenses in tomato plants. Plant Physiol. 135, 483–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Glas J. J., Alba J. M., Simoni S., Villarroel C. A., Stoops M., Schimmel B. C., Schuurink R. C., Sabelis M. W., and Kant M. R. (2014) Defense suppression benefits herbivores that have a monopoly on their feeding site but can backfire within natural communities. BMC Biol. 12, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Matsushima R., Ozawa R., Uefune M., Gotoh T., and Takabayashi J. (2006) Intraspecies variation in the Kanzawa spider mite differentially affects induced defensive response in lima bean plants. J. Chem. Ecol. 32, 2501–2512 [DOI] [PubMed] [Google Scholar]
- 18. Zhurov V., Navarro M., Bruinsma K. A., Arbona V., Santamaria M. E., Cazaux M., Wybouw N., Osborne E. J., Ens C., Rioja C., Vermeirssen V, Rubio-Somoza I., Krishna P., Diaz I., Schmid M., Gómez-Cadenas A., Van de Peer Y., Grbic M., Clark R. M., Van Leeuwen T., and Grbic V. (2014) Reciprocal responses in the interaction between Arabidopsis and the cell-content-feeding chelicerate herbivore spider mite. Plant Physiol. 164, 384–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alba J. M., Schimmel B. C., Glas J. J., Ataide L. M., Pappas M. L., Villarroel C. A., Schuurink R. C., Sabelis M. W., and Kant M. R. (2015) Spider mites suppress tomato defenses downstream of jasmonate and salicylate independently of hormonal crosstalk. New Phytol. 205, 828–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kielkiewicz M. (2002) Influence of carmine spider mite Tetranychus cinnabarinus Boisd. (Acarida: Tetranychidae) feeding on ethylene production and the activity of oxidative enzymes in damaged tomato plants (Bernini F., Nannelli R., Nuzzaci G., and de Lillo E., Eds.) Acarid Phylogeny and Evolution: Adaptation in Mites and Ticks, pp. 389–392, Springer, Dordrecht, The Netherlands [Google Scholar]
- 21. Dermauw W., Wybouw N., Rombauts S., Menten B., Vontas J., Grbić M., Clark R. M., Feyereisen R., and Van Leeuwen T. (2013) A link between host plant adaptation and pesticide resistance in the polyphagous spider mite Tetranychus urticae. Proc. Natl. Acad. Sci. U.S.A. 110, E113–E122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wybouw N., Dermauw W., Tirry L., Stevens C., Grbić M., Feyereisen R., and Van Leeuwen T. (2014) A gene horizontally transferred from bacteria protects arthropods from host plant cyanide poisoning. eLife 3, e02365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wybouw N., Zhurov V., Martel C., Bruinsma K. A., Hendrickx F., Grbić V., and Van Leeuwen T. (2015) Adaptation of a polyphagous herbivore to a novel host plant extensively shapes the transcriptome of herbivore and host. Mol. Ecol. 24, 4647–4663 [DOI] [PubMed] [Google Scholar]
- 24. Sarmento R. A., Lemos F., Bleeker P. M., Schuurink R. C., Pallini A., Oliveira M. G., Lima E. R., Kant M., Sabelis M. W., and Janssen A. (2011) A herbivore that manipulates plant defence. Ecol. Lett. 14, 229–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bleeker P. M., Mirabella R., Diergaarde P. J., VanDoorn A., Tissier A., Kant M. R., Prins M., de Vos M., Haring M. A., and Schuurink R. C. (2012) Improved herbivore resistance in cultivated tomato with the sesquiterpene biosynthetic pathway from a wild relative. Proc. Natl. Acad. Sci. U.S.A. 109, 20124–20129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chatzivasileiadis E. A., and Sabelis M. W. (1997) Toxicity of methyl ketones from tomato trichomes to Tetranychus urticae Koch. Exp. Appl. Acarol. 21, 473–484 [Google Scholar]
- 27. Jared J. J., Murungi L. K., Wesonga J., and Torto B. (2015) Steroidal glycoalkaloids: chemical defence of edible African nightshades against the tomato red spider mite, Tetranychus evansi (Acari: Tetranychidae). Pest Manag. Sci., DOI 10.1002/ps.4100 [DOI] [PubMed] [Google Scholar]
- 28. Vandoorn A., and de Vos M. (2013) Resistance to sap-sucking insects in modern-day agriculture. Front. Plant Sci. 4, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Martel C., Zhurov V., Navarro M., Martinez M., Cazaux M., Auger P., Migeon A., Santamaria M. E., Wybouw N., Diaz I., Van Leeuwen T., Navajas M., Grbic M., and Grbic V. (2015) Tomato whole genome transcriptional response to Tetranychus urticae identifies divergence of spider mite-induced responses between tomato and Arabidopsis. Mol. Plant. Microbe Interact. 28, 343–361 [DOI] [PubMed] [Google Scholar]
- 30. Schmelz E. A., Engelberth J., Alborn H. T., Tumlinson J. H. 3rd, and Teal P. E. (2009) Phytohormone-based activity mapping of insect herbivore-produced elicitors. Proc. Natl. Acad. Sci. U.S.A. 106, 653–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Boller T., and Felix G. (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406 [DOI] [PubMed] [Google Scholar]
- 32. Jones J. D., and Dangl J. L. (2006) The plant immune system. Nature 444, 323–329 [DOI] [PubMed] [Google Scholar]
- 33. Hogenhout S. A., and Bos J. I. (2011) Effector proteins that modulate plant–insect interactions. Curr. Opin. Plant Biol. 14, 422–428 [DOI] [PubMed] [Google Scholar]
- 34. Acevedo F. E., Rivera-Vega L. J., Chung S. H., Ray S., and Felton G. W. (2015) Cues from chewing insects—the intersection of DAMPs, HAMPs, MAMPs and effectors. Curr. Opin. Plant Biol. 26, 80–86 [DOI] [PubMed] [Google Scholar]
- 35. Hogenhout S. A., Van der Hoorn R. A., Terauchi R., and Kamoun S. (2009) Emerging concepts in effector biology of plant-associated organisms. Mol. Plant. Microbe Interact. 22, 115–122 [DOI] [PubMed] [Google Scholar]
- 36. Lapin D., and Van den Ackerveken G. (2013) Susceptibility to plant disease: more than a failure of host immunity. Trends Plant Sci. 18, 546–554 [DOI] [PubMed] [Google Scholar]
- 37. Schmelz E. A., Huffaker A., Carroll M. J., Alborn H. T., Ali J. G., and Teal P. E. (2012) An amino acid substitution inhibits specialist herbivore production of an antagonist effector and recovers insect-induced plant defenses. Plant Physiol. 160, 1468–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bent A. F., and Mackey D. (2007) Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 45, 399–436 [DOI] [PubMed] [Google Scholar]
- 39. van Schie C. C., and Takken F. L. (2014) Susceptibility genes 101: how to be a good host. Annu. Rev. Phytopathol. 52, 551–581 [DOI] [PubMed] [Google Scholar]
- 40. Elling A. A., and Jones J. T. (2014) Functional characterization of nematode effectors in plants. Methods Mol. Biol. 1127, 113–124 [DOI] [PubMed] [Google Scholar]
- 41. Coleman A. D., Wouters R. H., Mugford S. T., and Hogenhout S. A. (2015) Persistence and transgenerational effect of plant-mediated RNAi in aphids. J. Exp. Bot. 66, 541–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bos J. I., Prince D., Pitino M., Maffei M. E., Win J., and Hogenhout S. A. (2010) A functional genomics approach identifies candidate effectors from the aphid species Myzus persicae (green peach aphid). PLoS Genet. 6, e1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Carolan J. C., Caragea D., Reardon K. T., Mutti N. S., Dittmer N., Pappan K., Cui F., Castaneto M., Poulain J., Dossat C., Tagu D., Reese J. C., Reeck G. R., Wilkinson T. L., and Edwards O. R. (2011) Predicted effector molecules in the salivary secretome of the pea aphid (Acyrthosiphon pisum): a dual transcriptomic/proteomic approach. J. Proteome Res. 10, 1505–1518 [DOI] [PubMed] [Google Scholar]
- 44. Grbić M., Van Leeuwen T., Clark R. M., Rombauts S., Rouzé P., Grbić V., Osborne E. J., Dermauw W., Ngoc P. C., Ortego F., Hernández-Crespo P., Diaz I., Martinez M., Navajas M., Sucena É., Magalhães S., Nagy L., Pace R. M., Djuranović S., Smagghe G., Iga M., Christiaens O., Veenstra J. A., Ewer J., Villalobos R. M., Hutter J. L., Hudson S. D., Velez M., Yi S. V., Zeng J., Pires-daSilva A., Roch F., Cazaux M., Navarro M., Zhurov V., Acevedo G., Bjelica A., Fawcett J. A., Bonnet E., Martens C., Baele G., Wissler L., Sanchez-Rodriguez A., Tirry L., Blais C., Demeestere K., Henz S. R., Gregory T. R., Mathieu J., Verdon L., and Farinelli L. (2011) The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 479, 487–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Avery D. J., and Briggs J. B. (1968) The aetiology and development of damage in young fruit trees infested with fruit tree red spider mite, Panonychus ulmi (Koch). Ann. Appl. Biol. 61, 277–288 [Google Scholar]
- 46. Rodriguez J. G. (1954) Radiophosphorus in Metabolism Studies in the Two-spotted Spider Mite. J. Econ. Entomol. 47, 514–517 [Google Scholar]
- 47. Storms J. J. H. (1971) Some physiological effects of spider mite infestation on bean plants. Neth. J. Plant Path. 77, 154–167 [Google Scholar]
- 48. Mothes U., and Seitz K. (1981) Fine structure and function of the prosomal glands of the two-spotted spider mite, Tetranychus urticae (Acari, Tetranychidae). Cell Tissue Res. 221, 339–349 [DOI] [PubMed] [Google Scholar]
- 49. De Lillo E., and Monfreda R. (2004) ‘Salivary secretions’ of eriophyoids (Acari: Eriophyoidea): first results of an experimental model. Exp. Appl. Acarol. 34, 291–306 [DOI] [PubMed] [Google Scholar]
- 50. Richards E. H., Jones B., and Bowman A. (2011) Salivary secretions from the honeybee mite, Varroa destructor: effects on insect haemocytes and preliminary biochemical characterization. Parasitology 138, 602–608 [DOI] [PubMed] [Google Scholar]
- 51. Nicholson S. J., Hartson S. D., and Puterka G. J. (2012) Proteomic analysis of secreted saliva from Russian Wheat Aphid (Diuraphis noxia Kurd.) biotypes that differ in virulence to wheat. J. Proteomics 75, 2252–2268 [DOI] [PubMed] [Google Scholar]
- 52. Vandermoten S., Harmel N., Mazzucchelli G., De Pauw E., Haubruge E., and Francis F. (2014) Comparative analyses of salivary proteins from three aphid species. Insect Mol. Biol. 23, 67–77 [DOI] [PubMed] [Google Scholar]
- 53. Cooper W. R., Nicholson S. J., and Puterka G. J. (2013) Salivary Proteins of Lygus hesperus (Hemiptera: Miridae). Ann. Entomol. Soc. Am. 106, 86–92 [Google Scholar]
- 54. Fry J. (1989) Evolutionary adaptation to host plants in a laboratory population of the phytophagous mite Tetranychus urticae Koch. Oecologia 81, 559–565 [DOI] [PubMed] [Google Scholar]
- 55. Febvay G., Delobel B., and Rahbé Y. (1988) Influence of the amino acid balance on the improvement of an artificial diet for a biotype of Acyrthosiphon pisum (Homoptera: Aphididae). Can. J. Zool. 66, 2449–2453 [Google Scholar]
- 56. Gibbons B. C., Chambers M. C., Monroe M. E., Tabb D. L., and Payne S. H. (2015) Correcting systematic bias and instrument measurement drift with mzRefinery. Bioinformatics 31, 3838–3840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Geer L. Y., Markey S. P., Kowalak J. A., Wagner L., Xu M., Maynard D. M., Yang X., Shi W., and Bryant S. H. (2004) Open mass spectrometry search algorithm. J. Proteome Res. 3, 958–964 [DOI] [PubMed] [Google Scholar]
- 58. Craig R., and Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 59. Kim S., and Pevzner P. A. (2014) MS-GF+ makes progress towards a universal database search tool for proteomics. Nat. Commun. 5, 5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vaudel M., Barsnes H., Berven F. S., Sickmann A., and Martens L. (2011) SearchGUI: An open-source graphical user interface for simultaneous OMSSA and X!Tandem searches. Proteomics 11, 996–999 [DOI] [PubMed] [Google Scholar]
- 61. Vaudel M., Burkhart J. M., Zahedi R. P., Oveland E., Berven F. S., Sickmann A., Martens L., and Barsnes H. (2015) PeptideShaker enables reanalysis of MS-derived proteomics data sets. Nat. Biotechnol. 33, 22–24 [DOI] [PubMed] [Google Scholar]
- 62. Vizcaino J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolome S., Apweiler R., Omenn G. S., Martens L., Jones A. R., and Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Martens L., Hermjakob H., Jones P., Adamski M., Taylor C., States D., Gevaert K., Vandekerckhove J., and Apweiler R. (2005) PRIDE: the proteomics identifications database. Proteomics 5, 3537–3545 [DOI] [PubMed] [Google Scholar]
- 64. Silva J. C., Gorenstein M. V., Li G. Z., Vissers J. P., and Geromanos S. J. (2006) Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol. Cell. Proteomics 5, 144–156 [DOI] [PubMed] [Google Scholar]
- 65. Krey J. F., Wilmarth P. A., Shin J. B., Klimek J., Sherman N. E., Jeffery E. D., Choi D., David L. L., and Barr-Gillespie P. G. (2014) Accurate Label-Free Protein Quantitation with High- and Low-Resolution Mass Spectrometers. J. Proteome Res. 13, 1034–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Laumann M., Bergmann P., and Heethoff M. (2008) Some remarks on the cytogenetics of oribatid mites. Soil Org. 80, 223–232 [Google Scholar]
- 67. Dierick M., Van Loo D., Masschaele B., Van den Bulcke J., Van Acker J., Cnudde V., and Van Hoorebeke L. (2014) Recent micro-CT scanner developments at UGCT. Nucl. Instrum. Methods Phys. Res., Sect. B 324, 35–40 [Google Scholar]
- 68. Vlassenbroeck J., Dierick M., Masschaele B., Cnudde V., Van Hoorebeke L., and Jacobs P. (2007) Software tools for quantification of X-ray microtomography at the UGCT. Nucl. Instrum. Methods Phys. Res., Sect. A 580, 442–445 [Google Scholar]
- 69. Paganin D., Mayo S. C., Gureyev T. E., Miller P. R., and Wilkins S. W. (2002) Simultaneous phase and amplitude extraction from a single defocused image of a homogeneous object. J. Microsc. 206, 33–40 [DOI] [PubMed] [Google Scholar]
- 70. Brabant L., Vlassenbroeck J., De Witte Y., Cnudde V., Boone M. N., Dewanckele J., and Van Hoorebeke L. (2011) Three-dimensional analysis of high-resolution X-ray computed tomography data with Morpho+. Microsc. Microanal. 17, 252–263 [DOI] [PubMed] [Google Scholar]
- 71. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., and Schmid B. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Meth. 9, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T. R. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Anders S., Pyl P. T., and Huber W. (2015) HTSeq – A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Anders S., McCarthy D. J., Chen Y., Okoniewski M., Smyth G. K., Huber W., and Robinson M. D. (2013) Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Protoc. 8, 1765–1786 [DOI] [PubMed] [Google Scholar]
- 76. Dearden P., and Akam M. (2000) A role for Fringe in segment morphogenesis but not segment formation in the grasshopper, Schistocerca gregaria. Dev. Genes Evol. 210, 329–336 [DOI] [PubMed] [Google Scholar]
- 77. Dearden P. K., Donly C., and Grbić M. (2002) Expression of pair-rule gene homologues in a chelicerate: early patterning of the two-spotted spider mite Tetranychus urticae. Development 129, 5461–5472 [DOI] [PubMed] [Google Scholar]
- 78. Kauffmann A., Gentleman R., and Huber W. (2009) arrayQualityMetrics—a bioconductor package for quality assessment of microarray data. Bioinformatics 25, 415–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ritchie M. E., Silver J., Oshlack A., Holmes M., Diyagama D., Holloway A., and Smyth G. K. (2007) A comparison of background correction methods for two-colour microarrays. Bioinformatics 23, 2700–2707 [DOI] [PubMed] [Google Scholar]
- 80. Chen F., Mackey A. J., Stoeckert C. J. Jr, and Roos D. S. (2006) OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 34, D363–D368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Petersen T., Brunak S., von H, and Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Meth. 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 82. Horton P., Park K. J., Obayashi T., Fujita N., Harada H., Adams-Collier C. J., and Nakai K. (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Edgar R. C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Abascal F., Zardoya R., and Posada D. (2005) ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105 [DOI] [PubMed] [Google Scholar]
- 85. Jobb G., von Haeseler A., and Strimmer K. (2004) TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol. Biol. 4, 18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 86. Tamura K., Stecher G., Peterson D., Filipski A., and Kumar S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Chan T. F., Ji K. M., Yim A. K., Liu X. Y., Zhou J. W., Li R. Q., Yang K. Y., Li J., Li M., Law P. T., Wu Y. L., Cai Z. L., Qin H., Bao Y., Leung R. K., Ng P. K., Zou J., Zhong X. J., Ran P. X., Zhong N. S., Liu Z. G., and Tsui S. K. (2015) The draft genome, transcriptome, and microbiome of Dermatophagoides farinae reveal a broad spectrum of dust mite allergens. J. Allergy Clin. Immunol. 135, 539–548 [DOI] [PubMed] [Google Scholar]
- 88. Santamaría M. E., González-Cabrera J., Martínez M., Grbic V., Castañera P., Diaz L., and Ortego F. (2015) Digestive proteases in bodies and faeces of the two-spotted spider mite, Tetranychus urticae. J. Insect Physiol. 78, 69–77 [DOI] [PubMed] [Google Scholar]
- 89. Bryon A., Wybouw N., Dermauw W., Tirry L., and Van Leeuwen T. (2013) Genome wide gene-expression analysis of facultative reproductive diapause in the two-spotted spider mite Tetranychus urticae. BMC Genomics 14, 815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Grossmann J., Roschitzki B., Panse C., Fortes C., Barkow-Oesterreicher S., Rutishauser D., and Schlapbach R. (2010) Implementation and evaluation of relative and absolute quantification in shotgun proteomics with label-free methods. J. Proteomics 73, 1740–1746 [DOI] [PubMed] [Google Scholar]
- 91. Blauvelt W. E. (1945) The internal morphology of the common red spider mite (Tetranychus telarius Linn.). Cornell University Agricultural Experiment Station Memoir 270, Cornell University, Ithaca, N. Y. [Google Scholar]
- 92. Alberti G., and Crooker A. (1985) Internal anatomy. In: Helle W., and Sabelis M. W., eds. Spider mites. their biology, natural enemies and control. Vol. 1A, pp. 29–62, Elsevier Science Publishers B.V., Amsterdam, The Netherlands [Google Scholar]
- 93. Li C., Williams M. M., Loh Y. T., Lee G. I., and Howe G. A. (2002) Resistance of cultivated tomato to cell content-feeding herbivores is regulated by the octadecanoid-signaling pathway. Plant Physiol. 130, 494–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Godinho D. P., Janssen A., Dias T., Cruz C., and Magalhães S. (2015) Down-regulation of plant defence in a resident spider mite species and its effect upon con-and heterospecifics. Oecologia 180, 1–7 [DOI] [PubMed] [Google Scholar]
- 95. Rao S. A., Carolan J. C., and Wilkinson T. L. (2013) Proteomic Profiling of Cereal Aphid Saliva Reveals Both Ubiquitous and Adaptive Secreted Proteins. PLoS ONE 8, e57413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Chaudhary R., Atamian H. S., Shen Z., Briggs S. P., and Kaloshian I. (2015) Potato aphid salivary proteome: enhanced salivation using resorcinol and identification of aphid phosphoproteins. J. Proteome Res. 14, 1762–1778 [DOI] [PubMed] [Google Scholar]
- 97. Zheng C., and Sankoff D. (2016) Comparative transcriptomics and proteomics of three different aphid species identifies core and diverse effector sets. BMC Genomics 17, 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Díaz-Riquelme J., Zhurov V., Rioja C., Pérez-Moreno I., Torres-Pérez R., Grimplet J., Carbonell-Bejerano P., Bajda S., Van Leeuwen T., and Martínez-Zapater J. M. (2016) Comparative genome-wide transcriptome analysis of Vitis vinifera responses to adapted and non-adapted strains of two-spotted spider mite, Tetranyhus urticae. BMC Genomics 17, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Rao S. A., Carolan J. C., and Wilkinson T. L. (2013) Proteomic profiling of cereal aphid saliva reveals both ubiquitous and adaptive secreted proteins. PLoS ONE 8, e57413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Thorpe P., Cock P. J. A., and Bos J. (2016) Comparative transcriptomics and proteomics of three different aphid species identifies core and diverse effector sets. BMC Genomics 17, 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Haegeman A., Mantelin S., Jones J. T., and Gheysen G. (2012) Functional roles of effectors of plant-parasitic nematodes. Gene 492, 19–31 [DOI] [PubMed] [Google Scholar]
- 102. Vandermarliere E., Mueller M., and Martens L. (2013) Getting intimate with trypsin, the leading protease in proteomics. Mass Spectrom. Rev. 32, 453–465 [DOI] [PubMed] [Google Scholar]
- 103. Sheng S., Chen D., and Van Eyk J. E. (2006) Multidimensional Liquid Chromatography Separation of Intact Proteins by Chromatographic Focusing and Reversed Phase of the Human Serum Proteome: Optimization and Protein Database. Mol. Cell. Proteomics 5, 26–34 [DOI] [PubMed] [Google Scholar]
- 104. Stafford-Banks C. A., Rotenberg D., Johnson B. R., Whitfield A. E., and Ullman D. E. (2014) Analysis of the salivary gland transcriptome of Frankliniella occidentalis. PLoS ONE 9, e94447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Su Y. L., Li J. M., Li M., Luan J. B., Ye X. D., Wang X. W., and Liu S. S. (2012) Transcriptomic Analysis of the Salivary Glands of an Invasive Whitefly. PLoS ONE 7, e39303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Celorio-Mancera Mde l, Courtiade J., Muck A., Heckel D. G., Musser R. O., and and Vogel H. (2011) Sialome of a Generalist Lepidopteran Herbivore: Identification of Transcripts and Proteins from Helicoverpa armigera Labial Salivary Glands. PLoS ONE 6, e26676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Mills L. (1973) Morphology of glands and ducts in the two-spotted spider-mite, Tetranychus urticae Koch, 1836. Acarologia 15, 218–236 [PubMed] [Google Scholar]
- 108. Pan Y., Zhu J., Luo L., Kang L., and Cui F. (2015) High expression of a unique aphid protein in the salivary glands of Acyrthosiphon pisum. Physiol. Mol. Plant Pathol. 92, 175–180 [Google Scholar]
- 109. Mutti N. S., Louis J., Pappan L. K., Pappan K., Begum K., Chen M. S., Park Y., Dittmer N., Marshall J., Reese J. C., and Reeck G. R. (2008) A protein from the salivary glands of the pea aphid, Acyrthosiphon pisum, is essential in feeding on a host plant. Proc. Natl. Acad. Sci. U.S.A. 105, 9965–9969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Wang W., Dai H., Zhang Y., Chandrasekar R., Luo L., Hiromasa Y., Sheng C., Peng G., Chen S., Tomich J. M., Wu Y. L., Cai Z. L., Qin H., Bao Y., Leung R. K., Ng P. K., Zou J., Zhong X. J., Ran P. X., Zhong N. S., Liu Z. G., and Tsui S. K. (2015) Armet is an effector protein mediating aphid–plant interactions. The FASEB J. 29, 2032–2045 [DOI] [PubMed] [Google Scholar]
- 111. Stafford-Banks C. A., Rotenberg D., Johnson B. R., Whitfield A. E., and Ullman D. E. (2014) Analysis of the salivary gland transcriptome of Frankliniella occidentalis. PloS one 9, e94447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Backus E. A., Andrews K. B., Shugart H. J., Greve L. C., Labavitch J. M., and Alhaddad H. (2012) Salivary enzymes are injected into xylem by the glassy-winged sharpshooter, a vector of Xylella fastidiosa. J. Insect Physiol. 58, 949–959 [DOI] [PubMed] [Google Scholar]
- 113. Bensoussan N., Santamaria M. E., Zhurov V., Diaz I., Grbic M., and Grbic V. (2016) Plant-herbivore interaction: dissection of the cellular pattern of Tetranychus urticae feeding on the host plant. Front. Plant Sci. 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. van der Linde K., Hemetsberger C., Kastner C., Kaschani F., van der Hoorn R. A., Kumlehn J., and Doehlemann G. (2012) A maize cystatin suppresses host immunity by inhibiting apoplastic cysteine proteases. Plant Cell 24, 1285–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Tian M., Win J., Song J., van der Hoorn R., van der Knaap E., and Kamoun S. (2007) A Phytophthora infestans Cystatin-Like Protein Targets a Novel Tomato Papain-Like Apoplastic Protease. Plant Physiol. 143, 364–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Dong S., Stam R., Cano L. M., Song J., Sklenar J., Yoshida K., Bozkurt T. O., Oliva R., Liu Z., Tian M., Win J., Banfield M. J., Jones A. M., van der Hoorn R. A., and Kamoun S.) (2014) Effector specialization in a lineage of the Irish potato famine pathogen. Science 343, 552–555 [DOI] [PubMed] [Google Scholar]
- 117. Van Steijn G. J., Amerongen A. V., Veerman E. C., Kasanmoentalib S., and Overdijk B. (1999) Chitinase in whole and glandular human salivas and in whole saliva of patients with periodontal inflammation. Eur. J. Oral Sci. 107, 328–337 [DOI] [PubMed] [Google Scholar]
- 118. von Reumont B. M., Blanke A., Richter S., Alvarez F., Bleidorn C., and Jenner R. A. (2014) The First Venomous Crustacean Revealed by Transcriptomics and Functional Morphology: Remipede Venom Glands Express a Unique Toxin Cocktail Dominated by Enzymes and a Neurotoxin. Mol. Biol. Evol. 31, 48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Grisley M., and Boyle P. (1990) Chitinase, a new enzyme in octopus saliva. Comp. Biochem. Physiol. B 95, 311–316 [Google Scholar]
- 120. Gao B., Allen R., Maier T., McDermott J. P., Davis E. L., Baum T. J., and Hussey R. S. (2002) Characterisation and developmental expression of a chitinase gene in Heterodera glycines. Int. J. Parasitol. 32, 1293–1300 [DOI] [PubMed] [Google Scholar]
- 121. Niblack T. L., Lambert K. N., and Tylka G. L. (2006) A model plant pathogen from the kingdom Animalia: Heterodera glycines, the soybean cyst nematode. Annu. Rev. Phytopathol. 44, 283–303 [DOI] [PubMed] [Google Scholar]
- 122. Sharma A., Khan A. N., Subrahmanyam S., Raman A., Taylor G. S., and Fletcher M. J. (2014) Salivary proteins of plant-feeding hemipteroids–implication in phytophagy. Bull. Entomol. Res. 104, 117–136 [DOI] [PubMed] [Google Scholar]
- 123. Dermauw W., Osborne E. J., Clark R. M., Grbić M., Tirry L., and Van Leeuwen T. (2013) A burst of ABC genes in the genome of the polyphagous spider mite Tetranychus urticae. BMC Genomics 14, 1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Van Leeuwen T., and Dermauw W. (2016) The molecular evolution of xenobiotic metabolism and resistance in chelicerate mites. Annu. Rev. Entomol. 61, 475–498 [DOI] [PubMed] [Google Scholar]
- 125. Santamaría M. E., Hernández-Crespo P., Ortego F., Grbic V., Grbic M., Diaz I., and Martinez M. (2012) Cysteine peptidases and their inhibitors in Tetranychus urticae: a comparative genomic approach. BMC Genomics 13, 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Ji R., Yu H., Fu Q., Chen H., Ye W., Li S., and Lou Y. (2013) Comparative transcriptome analysis of salivary glands of two populations of rice brown planthopper, Nilaparvata lugens, that differ in virulence. PLoS ONE 8, e79612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Wang X., Zhang M., Feng F., and He R. (2015) Differentially regulated genes in the salivary glands of brown planthopper after feeding in resistant versus susceptible rice varieties. Arch. Insect Biochem. Physiol. 89, 69–86 [DOI] [PubMed] [Google Scholar]
- 128. Habibi J., Backus E. A., Coudron T. A., and Brandt S. L. (2001) Effect of different host substrates on hemipteran salivary protein profiles. Entomol. Exp. Appl. 98, 369–375 [Google Scholar]
- 129. Elzinga D. A., De Vos M., and Jander G. (2014) Suppression of plant defenses by a Myzus persicae (green peach aphid) salivary effector protein. Mol. Plant. Microbe Interact. 27, 747–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Van Leeuwen T., Demaeght P., Osborne E. J., Dermauw W., Gohlke S., Nauen R., Grbić M., Tirry L., Merzendorfer H., and Clark R. M. (2012) Population bulk segregant mapping uncovers resistance mutations and the mode of action of a chitin synthesis inhibitor in arthropods. Proc. Natl. Acad. Sci. U.S.A. 109, 4407–4412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Zhao C., Escalante L. N., Chen H., Benatti T. R., Qu J., Chellapilla S., Waterhouse R. M., Wheeler D., Andersson M. N., and Bao R. (2015) A massive expansion of effector genes underlies gall-formation in the wheat pest Mayetiola destructor. Curr. Biol. 25, 613–620 [DOI] [PubMed] [Google Scholar]
- 132. Aggarwal R., Subramanyam S., Zhao C., Chen M. S., Harris M. O., and Stuart J. J. (2014) Avirulence effector discovery in a plant galling and plant parasitic arthropod, the Hessian fly (Mayetiola destructor). PLos One 9:e100958, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Mitchum M. G., Hussey R. S., Baum T. J., Wang X., Elling A. A., Wubben M., and Davis E. L. (2013) Nematode effector proteins: an emerging paradigm of parasitism. New Phytol. 199, 879–894 [DOI] [PubMed] [Google Scholar]
- 134. Pitino M., and Hogenhout S. A. (2013) Aphid protein effectors promote aphid colonization in a plant species-specific manner. Mol. Plant. Microbe Interact. 26, 130–139 [DOI] [PubMed] [Google Scholar]
- 135. Stuart J. (2015) Insect effectors and gene-for-gene interactions with host plants. Curr. Opin. Insect. Sci. 9, 56–61 [DOI] [PubMed] [Google Scholar]
- 136. Stuart J. J., Chen M. S., Shukle R., and Harris M. O. (2012) Gall midges (Hessian flies) as plant pathogens. Annu. Rev. Phytopathol. 50, 339–357 [DOI] [PubMed] [Google Scholar]
- 137. Kanvil S., Powell G., and Turnbull C. (2014) Pea aphid biotype performance on diverse Medicago host genotypes indicates highly specific virulence and resistance functions. Bull. Entomol. Res. 104, 689–701 [DOI] [PubMed] [Google Scholar]
- 138. Villarroel C. A., Jonckheere W., Alba J. M., Glas J. J., Dermauw W., Haring M. A., Van Leeuwen T., Schuurink R. C., and Kant M. R. (2016) Salivary proteins of spider mites suppress defenses in Nicotiana benthamiana and promote mite reproduction. The Plant Journal 86, 119–131 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.