Abstract
Because proteins are the main mediators of most cellular processes they are also prime therapeutic targets. Identifying physical links among proteins and between drugs and their protein targets is essential in order to understand the mechanisms through which both proteins themselves and the molecules they are targeted with act. Thus, there is a strong need for sensitive methods that enable mapping out these biomolecular interactions. Here we present a robust and sensitive approach to screen proteome-scale collections of proteins for binding to proteins or small molecules using the well validated MAPPIT (Mammalian Protein-Protein Interaction Trap) and MASPIT (Mammalian Small Molecule-Protein Interaction Trap) assays. Using high-density reverse transfected cell microarrays, a close to proteome-wide collection of human ORF clones can be screened for interactors at high throughput. The versatility of the platform is demonstrated through several examples. With MAPPIT, we screened a 15k ORF library for binding partners of RNF41, an E3 ubiquitin protein ligase implicated in receptor sorting, identifying known and novel interacting proteins. The potential related to the fact that MAPPIT operates in living human cells is illustrated in a screen where the protein collection is scanned for interactions with the glucocorticoid receptor (GR) in its unliganded versus dexamethasone-induced activated state. Several proteins were identified the interaction of which is modulated upon ligand binding to the GR, including a number of previously reported GR interactors. Finally, the screening technology also enables detecting small molecule target proteins, which in many drug discovery programs represents an important hurdle. We show the efficiency of MASPIT-based target profiling through screening with tamoxifen, a first-line breast cancer drug, and reversine, an investigational drug with interesting dedifferentiation and antitumor activity. In both cases, cell microarray screens yielded known and new potential drug targets highlighting the utility of the technology beyond fundamental biology.
Every cellular function is governed by complex and dynamic networks of interactions between the proteins that are encoded by that cell. Therefore, mapping protein-protein interactions (PPIs)1, and their modulation in a changing cellular context, is an essential step in the characterization of a protein. Because of their central function in the cell, proteins are also the main targets for therapeutic intervention. The majority of marketed drugs act by binding to proteins and modulating their function. In the case of target-based small molecule drug discovery approaches, the target protein is a priori known, but many discovery efforts apply a phenotypic readout, requiring target identification once hits are identified. In addition, identification of off-target protein binding is often required to understand and to alleviate unwanted side-effects of small molecule leads. Thus, protein interaction analysis is a key aspect of both fundamental and applied biomolecular research.
Protein interactomics is a very active discipline and there is quite a wealth of PPI methodologies available (reviewed in (1, 2)). The classical yeast two-hybrid and affinity purification-based technologies still stand out as high-throughput approaches, but there are many other valuable approaches exhibiting particular features making them attractive tools to complement the basic PPI network scaffolds the former produce. To a lesser extent, also for small molecules there is a range of methods that can be applied to define the protein target spectrum (reviewed in (3)).
Our group developed the MAPPIT and MASPIT assays for PPI and small molecule-protein interaction analysis, respectively (4, 5). MAPPIT and MASPIT are complementation assays that act through reconstitution of a signaling deficient cytokine receptor (Fig. 1A, 1B). One of the main assets of these technologies is that they operate in living mammalian (human) cells, providing a highly relevant physiological environment for analyzing biomolecular interactions. Furthermore, the native background enables evaluating modulation of protein interactions triggered by exogenously induced changes in the cellular environment. Importantly, extensive benchmarking studies have indicated that the assays perform at a similar or better level than other well established interaction assays (6–8). The MAPPIT/MASPIT technology platform has been used extensively for testing interactions with individual proteins or small-scale focused target protein panels (e.g. (9, 10)) and as an orthogonal validation tool in large-scale interactome mapping efforts (e.g. (8, 11, 12)). In addition, we devised a screening platform for de novo identification of protein interactions from large proteome-wide target protein collections, using reverse transfected cell microarrays (13). An initial microtiter plate-based setup has been successfully applied to the identification of protein interaction partners of both proteins and small molecules (14–16). To render this platform compatible with high-throughput screening, we downscaled it to a microarray format and further expanded the target protein collection to near proteome-wide scale. In order to evaluate the efficiency and versatility of the screening approach, we applied the microarray platform to reveal novel (modulated) PPIs and to uncover target profiles of approved and investigational drugs.
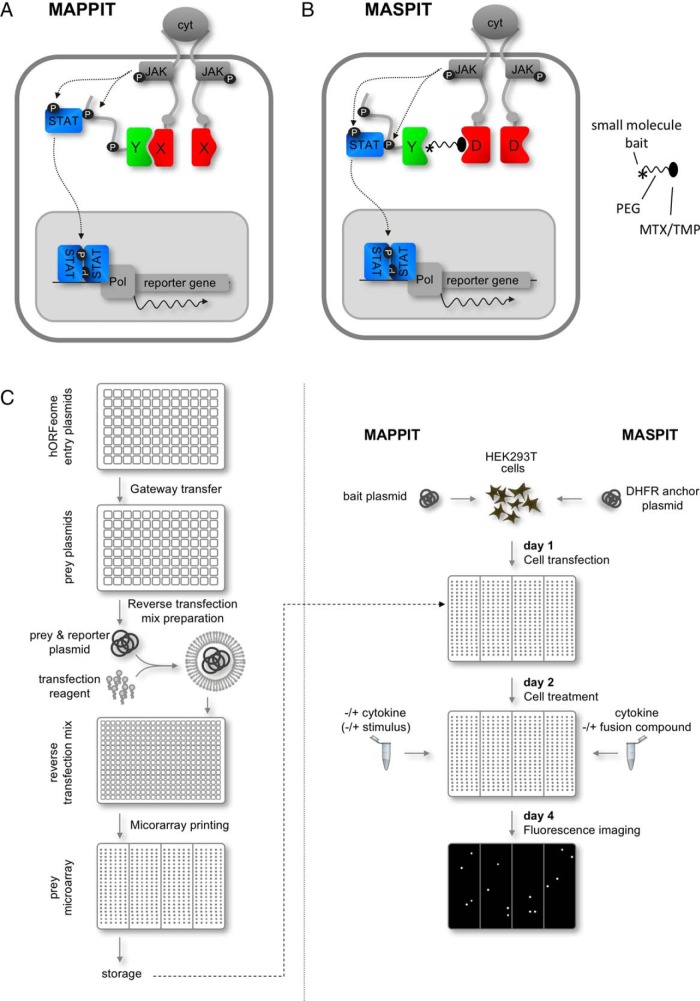
Fig. 1.
MAPPIT and MASPIT assay concept and workflow of the cell microarray screening platform. A, In MAPPIT, the bait protein of interest (X) is genetically fused to a cytokine receptor which is made signaling-deficient by mutation of signal transducer and activator of transcription (STAT) recruitment sites (gray dots), whereas the prey protein (Y) is tethered to a portion of a receptor containing intact STAT docking sites (black dots). Binding of the appropriate cytokine ligand to the chimeric bait receptor induces cross-phosphorylation (P) of the constitutively associated Janus kinases (JAKs), this way activating these. Interaction between bait and prey brings both cytokine receptor fragments into proximity, reconstituting a functional receptor complex. The activated JAKs phosphorylate the tyrosine motifs of the prey chimaera, enabling recruitment of STAT molecules. These transcription factors are in turn phosphorylated by the JAK kinases, resulting in their activation and subsequent dissociation and translocation to the nucleus. In the nucleus, STAT dimers activate transcription of a STAT-dependent reporter gene. B, The MASPIT assay is largely similar to MAPPIT, but here the bait is a small organic molecule (asterisk). A fusion compound consisting of this small molecule linked to either methotrexate (MTX) or trimethoprim (TMP) through a polyethylene glycol spacer (PEG) is added to and taken up by the cells. By virtue of the high affinity of MTX and TMP for E. coli dihydrofolate reductase (DHFR), the fusion compound is recruited to the anchor fusion which consists of the DHFR protein (D) being tethered to the same signaling-deficient cytokine receptor as in the case of the bait in MAPPIT. When a prey protein (Y) interacts with the small molecule bait, the reporter gene is activated as in MAPPIT. C, Microarray production (left panel): The ORFeome plasmid collection, which is available in Gateway entry vectors, is transferred to prey destination vectors through recombinatorial cloning. The resulting prey plasmids are individually mixed with a fluorescence reporter plasmid, transfection reagent and a number of additional agents. These reverse transfection mixes are then printed on polystyrene plates, dried and stored until needed for screening. Microarray screening (right panel): Cells are transfected in bulk with the desired bait fusion plasmid (MAPPIT) or the DHFR anchor plasmid (MASPIT) and added to the microarray plates. and the cognate cytokine is added to the appropriate wells, depending on the stimulation schedule. One day later, cells are treated with the appropriate stimuli: depending on the assay (MAPPIT or MASPIT) and the experimental setup (screen in standard conditions or comparing two physiological states), wells are either left untreated or treated with cytokine, fusion compound or additional stimuli. Next, cells are grown for another 2 days until the plates are scanned using a fluorescence imager.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
The human ORFeome v8.1 (17) and ORFeome Collaboration (18) prey collections were cloned in the pMG1 vector as described (13). Also the control prey plasmid expressing unfused gp130 (13), the plasmid encoding the C-terminal HMGCR prey (19), the STAT3-dependent firefly luciferase reporter pXP2d2-rPAPI-luciferase (4), pMet7-RNF41-Etag (20), pMet7-Flag-RNF41 (21), pMet7-Flag-SV40 large T (20), pCLG-DHFR (15), pCLG-GR (22), and the p(GRE)250hu.IL6P-luc+ GRE-luciferase reporter plasmid (22) were as described before. The pXP2d2-rPAPI-mKate2 reporter gene plasmid was generated by replacing the firefly luciferase sequence in pXP2d2-rPAPI-luciferase (4) with the mKate2 encoding sequence (23). The pMet7-Flag-VPS52, pMet7-VPS52-Etag, pMet7-Flag-ASB6 and pMet7-Etag-ASB6 constructs were generated through an LR reaction (Invitrogen, Carlsbad, CA) to transfer the VPS52 or ASB6 ORF from a Gateway entry clone of the human ORFeome collection to a previously described pMet7-Flag or pMet7-Etag destination vector, respectively (21). pMet7-RNF41-Flag was generated by replacing the Etag coding sequence in pMet7-RNF41-Etag (20) by the Flag-tag sequence. The pCLG-RNF41(Cys34Ser/His36Gln) bait expressing plasmid was generated by cloning the human RNF41 ORF into the previously described pCLG vector backbone (24) and the Cys34Ser/His36Gln double mutation was introduced using site directed mutagenesis.
Fusion Compound Synthesis
The chemical synthesis of the TMP fusion compounds has been reported elsewhere (10).
Cell Microarray Generation and Screening
Production of the microarray screening plates was based on a previously described protocol for screening in 384-well microtiter plates with a number of modifications. A fluorescent reporter gene (pXP2d2-rPAPI-mKate2; ex/em 588/633) replaced the luciferase reporter. Attractene (Qiagen, Hilden, Germany) was used as a transfection agent instead of Effectene and fibronectin (Sigma-Aldrich, St. Louis, MO) was added to the reverse transfection mixtures in order to enhance cell adherence. Printing was done in tissue culture-treated polystyrene plates with four rectangular wells (Thermo Scientific, Nunc, Waltham, MA) using a 2470 arrayer (Aushon Biosystems, Billerica, MA) equipped with a printhead holding 48 solid pins with 500 μm diameter print surface.
For screening, HEK293T cells, cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and incubated at 37 °C, 5.5% CO2, were transfected in bulk with the bait or DHFR anchor plasmid using polyethyleneimine and the cell suspension was added to the screening plates at 2 × 106 cells/well. Twenty-four hours later, depending on the screening setup, appropriate wells were stimulated with 100 ng/ml mouse leptin (Peprotech, Rocky Hill, NJ), 1 μm dexamethasone (GR MAPPIT screen; Sigma-Aldrich), 0.1 μm TMP-tamoxifen or 0.1 μm TMP-reversine (MASPIT screens). Another 48 h later, plates were imaged and fluorescence intensity of each individual spot was determined using an CellCelector microarray scanner (Automated Lab Solutions, Jena, Germany).
Fluorescence intensity data were preprocessed prior to their analysis. Raw microarray data were log-transformed (base 2) after which an “ANOVA normalization model” was fit that includes terms accounting for plate and within-plate effects (25). Subsequently, the residuals, which represent normalized values, were used as input data to calculate the rank product statistic for each prey in the screen (26). Having ranked the preys by their rank product, the corresponding p values were obtained using an algorithm developed by Huskes et al. (27). In order to cope with the multiple testing problem, the final selection was based on FDR-adjusted p values (q-values) (28).
MAPPIT and MASPIT Assays
HEK293T cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, incubated at 37 °C, 8% CO2. Cells were transfected with the indicated bait and prey plasmids and the pXP2d2-rPAPI-luciferase reporter plasmids using a standard calcium phosphate transfection method, as described earlier (13), and luciferase activity was measured 48 h after transfection using the Luciferase Assay System kit (Promega, Madison, WI) on an Envision luminometer (PerkinElmer Life Sciences, Waltham, MA). Stimulations with murine leptin (Peprotech; 100 ng/ml final concentration), dexamethasone (Sigma-Aldrich) and the TMP fusion compounds were done 24 h after transfection. In Fig. 3B, scoring was performed as described in (8), yielding detection and false positive rates of 20 and 2%, respectively: the interaction of RNF41 with a particular ORF prey was scored positive when the fold change (average stimulated cells divided by average nonstimulated cells) was at least nine times higher than that of both control bait-ORF prey and RNF41 bait-control prey control transfections; when the fold change of control bait-ORF prey was at least nine times higher than that of the control bait-control prey transfection, the interaction was scored aspecific positive; in all other cases the interaction was scored negative.
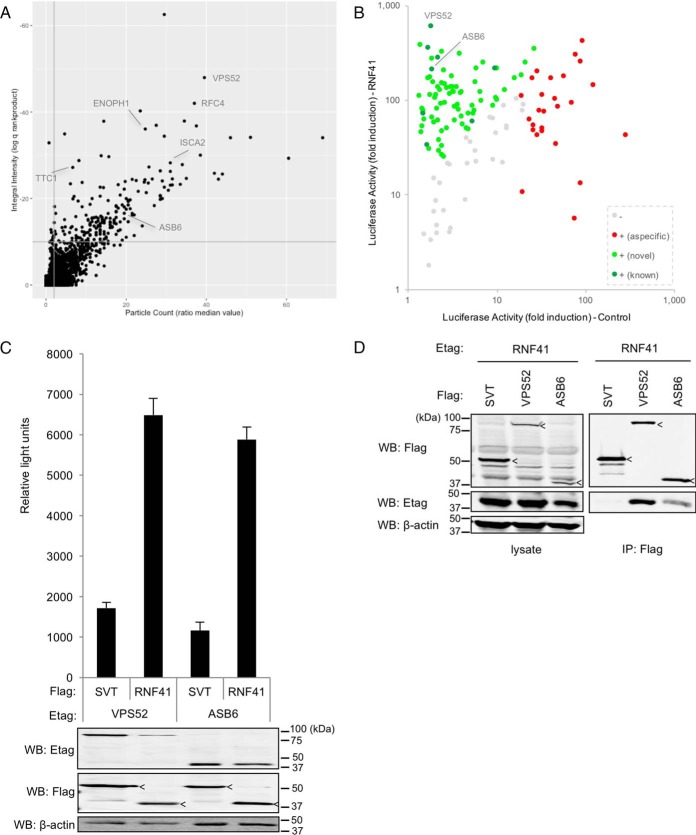
Fig. 3.
A MAPPIT cell microarray screen identifies known and novel RNF41 interaction partners. A, The 15k ORF prey collection was screened with the RNF41(Cys34Ser/His36Gln) bait chimera, for each ORF yielding quadruplicate samples for both the stimulated (+leptin) and the unstimulated (-leptin) condition. The Y-axis represents the log transformed q-value calculated using the rank product statistical test on the ratio of the fluorescence integral intensity of normalized stimulated versus unstimulated samples (see Experimental Procedures for details). On the x-axis, the ratio of the median value of the fluorescence particle count of stimulated versus unstimulated samples is depicted. Thresholds of 10−10 and 2 were applied to the integral intensity and particle count parameters, respectively, as these result in high specificity (low false positive rate) based on previous screening experiments (gray lines). Previously reported RNF41 binding proteins are indicated. See supplemental Table S2 for raw data. B, ORF preys that scored positive in the primary screen (138) were re-evaluated by testing their interaction with either the RNF41(Cys34Ser/His36Gln) or a negative control bait (E. coli DHFR) in MAPPIT. The fold induction of the average luciferase activity of triplicate leptin-stimulated versus unstimulated samples for either bait is shown. Applying a set of previously defined criteria for sensitive and specific MAPPIT analysis (see text for more details), signals were scored negative (-; gray) or positive (+). Positive signals were further categorized as corresponding with an aspecific interaction (red), where the ORF prey binds to a component of the MAPPIT bait chimera rather than the RNF41 bait (as deduced from the fact that it also generates a signal when combined with the control bait), or an RNF41-specific interaction (green). Signals corresponding with known RNF41 binders are indicated in dark green, those from potentially novel RNF41 interacting proteins in light green. C, The interaction between RNF41 and both VPS52 and ASB6 was confirmed in an AlphaScreen assay. HEK293T cells were transiently cotransfected with plasmids encoding E-tagged VPS52 or ASB6 and FLAG-tagged RNF41 or FLAG-tagged SV40 large T (SVT) as a negative control and signals were detected using the AlphaScreen FLAG detection kit. The average of triplicate samples is shown; error bars represent S.D. Expression of E-and FLAG-tagged proteins was confirmed by Western blotting (bands corresponding with SV40 Large T and RNF41 are indicated by arrowheads), and β-actin was stained as a loading control. D, Coimmunoprecipitation analysis confirms binding of VPS52 and ASB6 to RNF41. HEK293T cells were transiently cotransfected with E-tagged RNF41 and Flag-tagged VPS52, ASB6 or SV40 Large T (SVT) as a negative control. Anti-Flag immunoprecipitates and total lysates were visualized with anti-Etag and anti-Flag antibodies (bands corresponding with SV40 Large T, VPS52 and ASB6 are indicated by arrowheads). β-actin was stained as a loading control.
Coimmunoprecipitation
HEK293T cells (1.8 × 106) were seeded in 90 mm Petri dishes in DMEM with 10% FCS and transfected the next day with 3 μg of each of the indicated expression constructs. Two days after transfection, cells were washed with cold PBS and lysed in lysis buffer containing 50 mm Tris-HCl pH 7.5, 125 mm NaCl, 1.5 mm MgCl2, 0.2% Nonidet P-40, 5% glycerol and Complete Protease Inhibitor Mixture without EDTA (Roche, Basel, Switzerland) followed by two freezing rounds of 10 min at −80 °C. Lysates were incubated overnight at 4 °C with anti-Flag M2 beads (Sigma-Aldrich) after being pre-cleared with Sepharose 4B beads (GE Healthcare, Chicago, IL). The beads were then washed 3 times and eluted with 200 μg/ml of 1× flag peptide (Sigma-Aldrich). Loading buffer (5×: 155 mm Tris-HCl pH 6.8, 5% SDS, 20% glycerol, 0.025% Bromphenol blue, 1.8 m β-mercaptoethanol) was added to each sample. After boiling, samples were separated on SDS-polyacrylamide gels and transferred to nitrocellulose membranes (GE Healthcare) which were probed with monoclonal mouse anti-Etag (GE Healthcare) and rabbit anti-Flag antibodies (Sigma-Aldrich). Rabbit anti-β actin antibody (Sigma-Aldrich) was used as a loading control. Secondary goat-anti-mouse Dylight 680 antibody (Thermo Scientific, Waltham, MA) and secondary goat anti-rabbit Dylight 800 antibody (Thermo Scientific) were used for detection. Western blot analysis was performed using the Odyssey Infrared Imaging System (Li-Cor Biosciences, Lincoln, NE).
AlphaScreen
HEK293T cells were seeded in six-well plates at a density of 2 × 106 cells per well in DMEM with 10% FCS. Cells were cotransfected with 1 μg of the appropriate E- and Flag-tagged expression vectors and lysed 48 h post transfection in TAP lysis buffer (50 mm Tris-HCl ph7.5, 125 mm NaCl, 5% glycerol, 0.2% Nonidet P-40, 1.5 mm MgCl2, 25 mm NaF, 1 mm Na3VO4, and Complete Protease Inhibitor Mixture without EDTA (Roche)). Alpha screen experiments were performed according to the manufacturer's protocol (PerkinElmer). Briefly, lysates were incubated for two hours at 4 °C with 0.7 μg/ml anti-Etag antibody biotinylated with sulfo-NHS-Biotin (Thermo Scientific). Equal concentrations (20 ng/ml) of anti-FLAG M2 acceptor beads and Streptavidin donor beads were subsequently added for another incubation period of 1 h (at 4 °C) and 30 min (at room temperature) respectively. The samples were transferred in triplicate into 384-well plates and measured using the EnVision plate reader (PerkinElmer). Part of the lysate was used to verify expression of the constructs using mouse anti-Flag antibody (Sigma-Aldrich), monoclonal mouse anti-Etag antibody (GE Healthcare) and rabbit anti-β actin antibody (Sigma-Aldrich) as a loading control. SDS-PAGE, and Western blotting were performed as described above (Coimmunoprecipitation).
GR Reporter Assay
A549 cells stably transfected with the p(GRE)250hu.IL6P-luc+ GRE-luciferase reporter plasmid were seeded in 96-well microtiter plates and transfected with 50 nm of each siRNA using the calcium phosphate precipitation technique as described before (22). Three days later, the cells were treated either with vehicle or with 1 μm Dex for 6 h and luciferase activity was analyzed using the Luciferase Assay System kit (Promega) on an Envision luminometer (PerkinElmer). Z-scores were determined as follows: for each candidate the fold change of the average of the Dex-stimulated versus vehicle-stimulated values of three technical triplicates was calculated; for each candidate, this value was normalized against the one of the nontargeting control siRNA; the z-score of each candidate was calculated as (x-m)/S.D., where m and S.D. are the average and the standard deviation of the normalized values of all candidates in a single experiment, respectively.
RESULTS
Development of a High-density Cell Microarray Screening Platform for MAPPIT and MASPIT
The MAPPIT and MASPIT assays require the coexpression of three plasmids: a STAT-dependent reporter gene plasmid, a plasmid encoding the protein bait (MAPPIT) or dihydrofolate reductase (DHFR) anchor (MASPIT) fusion, and a prey fusion encoding plasmid (Fig. 1A, 1B). In the cell microarray screening setup, a large collection of prey fusion proteins is tested in parallel. This collection is derived from the combined human ORFeome version 8.1 (17) and ORFeome collaboration (18) collections, in total covering 14,816 single-colony, fully sequenced, full size human open reading frames (ORFs). Each prey expression vector, together with the reporter plasmid, is complexed with a lipid transfection reagent, cell adhesion molecules and a number of stabilizing components, and printed on a polystyrene plate (Fig. 1C left panel). Microtiter-size plates consisting of four large rectangular wells are used, each well containing 1728 individual spots, covering 6912 spots in total per plate. Individual spots are about 500 μm in diameter, printed at a center-to-center spacing of 750 μm. Importantly, these arrays can be produced batch-wise off-line, and stored for several weeks prior to screening.
In order to perform a screen using either the MAPPIT or MASPIT assay, HEK293T adherent cells are bulk transfected with the chimeric bait protein (MAPPIT) or the DHFR anchor fusion (MASPIT), and this cell suspension is spread over the array and allowed to adhere (Fig. 1C right panel). The cells are treated with the appropriate stimuli (the cognate cytokine to activate the bait or DHFR anchor cytokine receptor fusion; a methotrexate (MTX) or trimethoprim (TMP) fusion compound in the case of MASPIT; potentially an additional stimulus is added, the effect of which on the bait interactome is being evaluated) and left on the array for 3 days. During that time the cells growing on top of the spots are being reverse transfected with prey and reporter plasmid. In cells coexpressing an interacting bait-prey pair, cytokine stimulation results in activation of the fluorescent reporter, which is detected by automated microscopic imaging. The fluorescence intensity of each spot is extracted using imaging analysis software applying a superimposed grid on each well of the array (Fig. 2A). Typically, quadruplicate wells of each tested condition (e.g. with or without cytokine in the case of MAPPIT, or cytokine with or without MTX/TMP fusion compound in the case of MASPIT) are measured. The fluorescence intensity ratio between the differentially treated spots is calculated and analyzed using the Rank Product approach in order to generate a q-value used to sort the interactions (26).
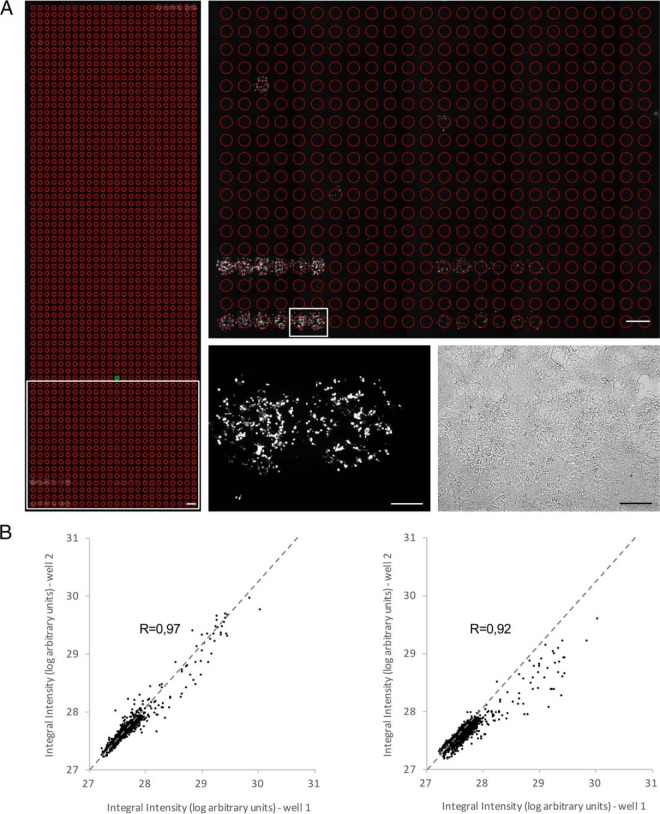
Fig. 2.
Quality and performance of the cell microarray screening approach. A, Microscopic image of a microarray sample. Left panel: example of the imaging output of a whole well containing 1728 spots screened with the GR bait and stimulated with leptin. The individual regions of interest (ROI) for which the fluorescence signal is measured are indicated as red circles. Constitutive positive controls are present at the lower left (2 series of 6 replicates) and upper right (1 series of 6 replicates). Scale bar: 1 mm. Upper right panel: magnification of the region indicated by the white rectangle in the lower left panel. Scale bar: 1 mm. Lower right panels: fluorescence (left) and bright field (right) image of the area indicated by the white rectangle in the upper right panel. Individual spots are covered by ∼500 cells. Scale bar: 250 μm. B, Dot plot showing the correlation between technical and biological replicates of a microarray well. The integral fluorescence intensity of the 1728 spots from a well corresponding to a microarray plate screened with the DHFR anchor bait and stimulated with leptin was compared with the fluorescence signal of the spots from a replicate well in the same (within screen variation, left panel) or a separate independent (between screen variation, right panel) screen that had been similarly stimulated. Pearson's correlation coefficient (R) is indicated. See supplemental Table S1 for raw data.
Fig. 2A shows a representative image of a scan of an entire well. The image magnification centered on individual positive spots illustrates that the fluorescent signal is confined within the boundaries of the printed area and that neighboring signals are spatially well separated. The robustness and reproducibility of the microarray printing and screening procedure is indicated by the limited within and between screen variation (Fig. 2B).
A MAPPIT Screen Identifies Novel RNF41 Binding Proteins
We first evaluated the microarray screening platform as a tool for identifying novel PPIs. As bait protein we selected the RNF41 (also called Nrdp1 or FLRF) RING finger-type E3 ubiquitin ligase. This enzyme coordinates ubiquitin transfer to specific substrate proteins, this way determining their ubiquitination status. Through this regulatory mechanism it has been implicated in various signaling pathways, including the EGFR and TLR signal transduction cascades (29). Our group previously established that RNF41 also controls the sorting and processing of a number of JAK2-associated cytokine receptors, including the leptin, LIF and IL-6 receptors (20, 21). It does so by modulating the ubiquitination status of USP8, which in turn regulates ubiquitination and stability of the ESCRT-0 complex involved in transport of internalized cytokine receptors toward multivesicular bodies for subsequent degradation in lysosomes. RNF41-dependent ubiquitination and ensuing destabilization of USP8 results in ESCRT-0 ubiquitination and degradation, abolishing receptor sorting to the lysosomes. Instead, receptors are recycled to the plasma membrane leading to enhanced ectodomain shedding by ADAM proteases (21).
In an ongoing effort to further elucidate this pathway, we performed a MAPPIT cell microarray screen with the RNF41 bait protein against the current prey collection. Because it was shown previously that RNF41 is subject to auto-ubiquitination, we used an RNF41 mutant that carries two mutations (Cys34Ser/His36Gln) in the RING finger domain, preventing recruitment of the E2 conjugating enzyme (29). Using the RNF41 (Cys34Ser/His36Gln) bait, we screened the 15k ORF prey set, ranked the ORFs according to normalized fluorescence signal intensity and applied a combination of predetermined selection thresholds, yielding 189 hits (Fig. 3A). Recurrent candidates which were previously identified in MAPPIT screens with unrelated bait proteins, suggesting that these bind aspecifically to the MAPPIT bait receptor backbone rather than to the fused bait protein itself, were filtered out and the 138 remaining hits were retested in a small scale MAPPIT experiment (Fig. 3B). Each hit was evaluated in parallel for interaction with the RNF41 bait and a negative control bait (E. coli DHFR), in order to allow discrimination of potential novel RNF41 interaction partners (binding only to the RNF41 bait) from aspecific MAPPIT bait receptor binders (binding to the DHFR-fused chimera as well). Extensive benchmarking studies, which involved testing large reference protein-protein interactions sets (up to 500 literature curated positive and 700 random negative interactions; (7, 8)), allowed establishing scoring criteria correlated with highly specific and sensitive MAPPIT analyses. We applied selection thresholds that were shown to yield a detection rate of around 20% at a false positive rate of 2%. Under these conditions, 80 (58%) of the 138 primary screening hits could be confirmed as candidate RNF41 binding proteins and 26 (19%) were found to also interact with the control bait and were therefore classified as aspecific hits, potentially interacting with a component of the MAPPIT bait receptor chimera. Taken together, 77% of the selected hits could be confirmed, validating the robustness of the microarray screening setup. Among the confirmed hits were a number of proteins that had been previously reported to bind to RNF41 (VPS52, RFC4, ENOPH1, ISCA2, TTC1, ASB6, HOMER2, and ARL6IP4). Because they had been implicated in intracellular protein trafficking, two of these in particular attracted our attention, VPS52 and ASB6.
VPS52 (Vesicular Sorting Protein 52), the most prominent hit in the screen, is a subunit of the GARP (Golgi-Associated Retrograde Protein) retrograde transport complex and the recently discovered EARP (Endosome-Associated Recycling Protein) endocytic recycling complex (30, 31). ASB6 (Ankyrin repeat and Socs Box containing 6) is an E3 ubiquitin ligase implicated in insulin-dependent degradation of SH2B (also known as APS) (32), a protein that facilitates Cbl-dependent ubiquitination and subsequent internalization of the insulin receptor (IR) (33). Binding of both proteins to RNF41 had not been formally established, but the interactions had been identified in a large-scale yeast two-hybrid screen (8). We therefore set out to confirm binding of VPS52 and ASB6 with RNF41 applying two orthogonal interaction methods. Using AlphaScreen (Amplified Luminescent Proximity Homogeneous Assay; PerkinElmer), a bead-based proximity assay, we obtained a strong and specific signal for both interactions (Fig. 3C). The interactions were corroborated using classic coimmunoprecipitation (Fig. 3D). These data suggest that RNF41 can engage in a genuine interaction with VPS52 and ASB6. Further research on the functional implications of these interactions is currently ongoing (Masschaele et al., in preparation; De Ceuninck et al., in preparation).
MAPPIT Microarray Screening Uncovers Dexamethasone-modulated Glucocorticoid Receptor Interactions
One of the main advantages of MAPPIT compared with yeast-based two-hybrid assays or classic in vitro interaction assays is that it operates in intact mammalian (human) cells, which allows evaluating biological interactions under different physiological conditions. For example, a bait protein of interest can be scanned for binding partners in different activation states, induced by addition of its cognate ligand.
To explore this opportunity, we performed a MAPPIT microarray screen to identify glucocorticoid receptor (GR) binding proteins for which the interaction was up- or downregulated upon addition of dexamethasone, a commonly used synthetic ligand of this steroid receptor. In unliganded state, the GR resides in the cytoplasm, complexed with various chaperone proteins such as the heat shock proteins Hsp70 and Hsp90, which maintain the receptor in its proper inactive conformation. Upon binding of a steroid ligand, these chaperones are released and the activated GR travels to the nucleus, where it engages with coactivator or -repressor proteins or other transcription factors to regulate gene transcription which will result in an anti-inflammatory outcome (34).
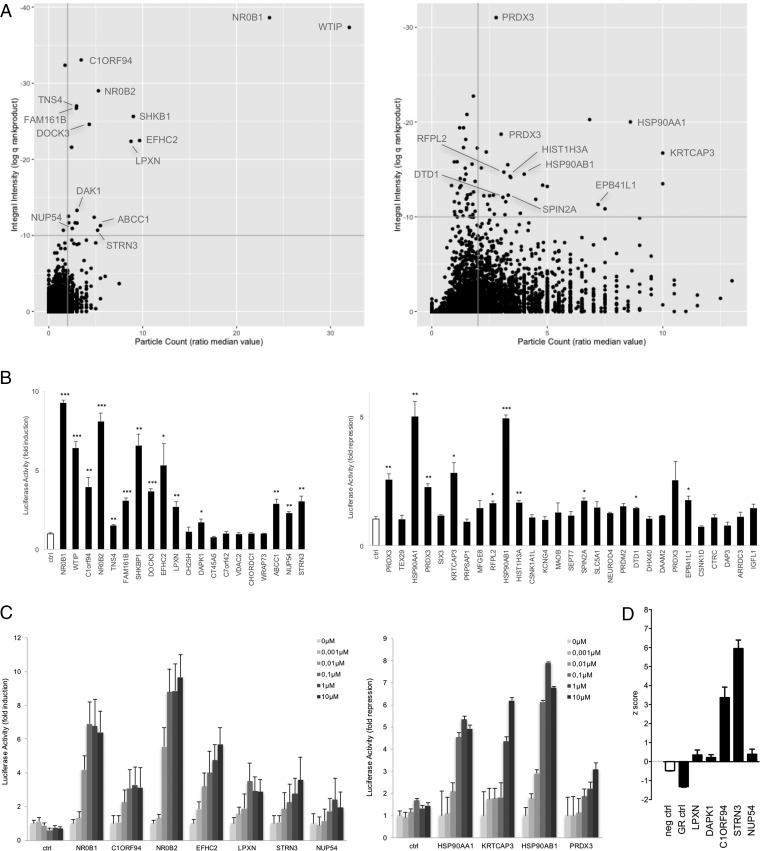
The ORF prey collection was screened in cells expressing the GR bait and differentially stimulated with dexamethasone. For 20 ORFs the interaction with GR was found to be up-regulated, 29 proteins were identified as candidate downregulated GR interaction partners (Fig. 4A). Evaluation in MAPPIT retests confirmed 14 and 10 up- and downregulated hits, respectively (Fig. 4B). To further corroborate these findings, selected interactions were subject to a dexamethasone concentration gradient, confirming a dose-dependent response for these ORF preys in MAPPIT interaction tests (Fig. 4C). Interestingly, among the confirmed hits were a number of known GR binding proteins, including HSP90AA1, HSP90AB1, NR0B1, and NR0B2. As expected for the chaperones, interaction with the Hsp90 proteins was lost upon treatment with dexamethasone. Consistent with previous reports, the interaction of GR with the NR0B1 and NR0B2 transcriptional corepressors is strongly induced upon dexamethasone stimulation of the GR bait expressing cells (35, 36). Next, selected GR interaction partners were validated in a functional GR transactivation assay. The effect of siRNA-mediated knock-down for a number of the GR binders was evaluated in an A549 cell line that endogenously expressed GR and had been engineered with a GRE-luciferase reporter consisting of the GR response element (GRE) fused to a firefly luciferase gene (Fig. 4D). Luciferase signals were normalized against the effect of a nontargeted control siRNA. As a positive control, an siRNA directed against the GR was included. As expected, GR knock-down resulted in a negative z-score, indicative of reduced dexamethasone-induced GRE reporter activity. Silencing of the selected candidates resulted in positive scores, suggesting that these proteins act as corepressors. The most significant effect on GR transactivation activity was observed for STRN3 (Striatin-3), a scaffold protein involved in organizing a variety of signaling complexes across different cellular processes (37). Detailed analysis of the implications of STRN3 binding on GR signaling pointed out that this protein recruits a phosphatase to the GR complex, resulting in GR dephosphorylation, this way suppressing GR mediated transactivation (Petta et al., in preparation).
Fig. 4.
Identification of dexamethasone-modulated interactions with GR in a MAPPIT cell microarray screen. A, The ORF prey collection was screened with the GR bait, with quadruplicate samples of both cell treatments: leptin and 1 μm dexamethasone costimulated and leptin only stimulated. Data was analyzed as described in Fig. 3A for up- (left panel) and downregulated signals (right panel), the upper right quadrant containing the candidate hits (20 up- and 29 downregulated candidates). Interacting proteins, the modulated binding to GR of which was confirmed in MAPPIT confirmation experiments (Fig. 4B), are indicated. See supplemental Table S3 for raw data. B, ORF preys selected upon application of the preset thresholds (q<10−10; ratio median value particle count>2) were re-evaluated for dexamethasone-modulated binding in MAPPIT. Cells coexpressing the GR bait and the indicated ORF preys were stimulated with leptin alone or leptin combined with 1 μm dexamethasone. Three independent experiments were performed and the fold induction (for up-regulated GR binders; left panel) or fold repression (for downregulated GR binders; right panel) was calculated as the average luciferase activity of leptin+dexamethasone treated versus leptin only treated (for up-regulated GR binders) or leptin only treated versus leptin+dexamethasone treated (for downregulated GR binders) samples. Error bars represent S.E. An empty prey (unfused gp130) was taken along as a control and used to compare data with applying a two-tailed Student's t test (*: p < 0.05; **: p < 0.01; ***: p < 0.001). C, Dose-dependent modulation of the GR interaction of selected ORF preys was tested in MAPPIT. Cells coexpressing the GR bait and the indicated ORF preys were stimulated either with leptin alone or with leptin and dexamethasone in a concentration gradient from 0.001 to 10 μm. The bars indicate the fold induction (for up-regulated GR binders; left graph) or fold repression (for downregulated binders; right graph) as in B, calculated using the average of triplicate samples. Error bars represent S.D. D, Functional effect of silencing of selected GR interaction partners in A549 cells. A549 cells harboring the GRE-luc GR-responsive reporter were transfected with siRNA specific for the indicated proteins and treated either with vehicle or with 1 μm dexamethasone before measuring luciferase activity. Controls include a nontargeting siRNA (neg ctrl) and an siRNA for GR (GR ctrl). Each condition was performed in triplicate. The graph represents the z-score [(x-m)/S.D.)] of three independent experiments.
Known and Novel Targets of Tamoxifen and Reversine Are Detected Through MASPIT Cell Microarray Screening
The utility of the screening platform extends beyond protein-protein interactions, as it also enables identification of new target proteins of small molecules through MASPIT. To evaluate the efficiency of the new cell microarray setup toward this appealing application, we screened two molecules that had previously been validated in MASPIT using known targets, tamoxifen and reversine (10, 15).
Tamoxifen is a selective estrogen receptor (ER) antagonist that has been in the clinic as part of the standard therapy for treatment of ER-positive breast cancer since the seventies (38). The ER is overexpressed in the majority of breast carcinomas, driving transcription of cell cycle-related genes and downregulating anti-proliferative and pro-apoptotic genes (39). Tamoxifen acts as a competitive antagonist of binding of endogenous estrogen hormone ligands, this way preventing pro-tumorigenic gene transcription. Interestingly, tamoxifen also exhibits cytotoxic effects in ER-negative cells by directly inducing apoptosis (40, 41). To date, the mechanism underlying tamoxifen's pro-apoptotic effects in these cells and the target proteins involved have not been identified, making it an attractive case to evaluate our target profiling approach.
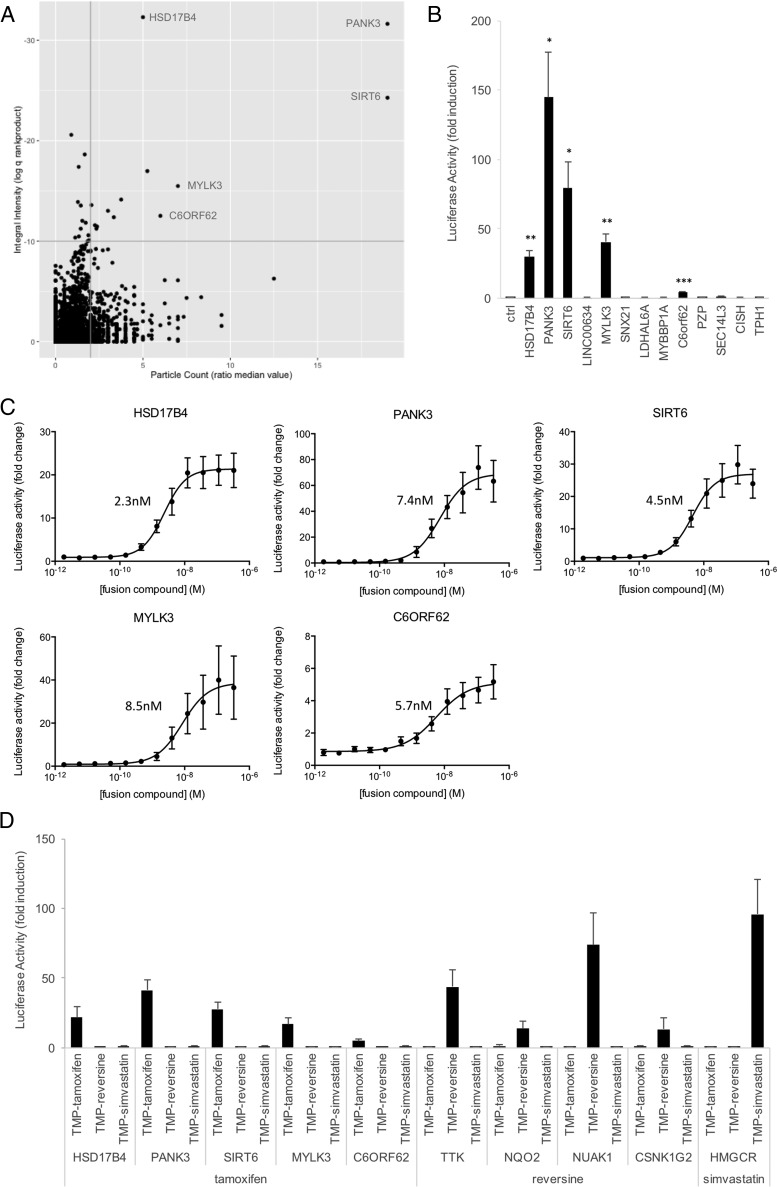
Based on the well-defined structure-activity relationship (SAR) described in the literature, we previously synthesized a tamoxifen TMP fusion and confirmed its binding in MASPIT to ER1 (or ERα), the ER isoform associated with the majority of breast cancer cases (10). The 15k ORF collection was screened with 0.1 μm TMP-tamoxifen, yielding 13 hits (Fig. 5A). Hits were cherry-picked and evaluated in small-scale MASPIT experiments, validating 5 of these (Fig. 5B). Next, we tested the interactions with a concentration gradient of the tamoxifen fusion compound, resulting in a dose-dependent MASPIT response for each of the targets and EC50 values in the nanomolar range (Fig. 5C). In order to evaluate specificity of target binding, we tested combinations of the ORF preys with fusion compounds corresponding to unrelated small molecule baits (reversine and simvastatin), confirming that only tamoxifen binds with the identified targets (Fig. 5D).
Fig. 5.
A MASPIT cell microarray screen detects tamoxifen target proteins. A, A MASPIT ORF screen was performed in which quadruplicate wells of each screening plate were differentially treated with either leptin alone or leptin combined with 0.1 μm TMP-tamoxifen fusion compound. Data was analyzed as described in Fig. 3A. Interacting proteins confirmed in MASPIT retests are indicated. See supplemental Table S4 for raw data. B, MASPIT retest of the hits selected using the standard selection thresholds (q<10−10; ratio median value particle count>2). Cells coexpressing the DHFR anchor fusion and the indicated ORF preys were stimulated with leptin alone or leptin combined with 0.1 μm TMP-tamoxifen. Fold induction of the average luciferase activity of triplicate samples in three independent experiments is shown. Error bars represent S.E. An empty prey (unfused gp130) was taken along as a control and used to compare data with applying a two-tailed Student's t test (*: p < 0.05; **: p < 0.01; ***: p < 0.001). C, MASPIT dose-response curves for the confirmed hits. Cells coexpressing the DHFR anchor protein and the indicated ORF preys were stimulated either with leptin alone or with leptin and the indicated concentration of TMP-tamoxifen. Data points depict fold induction of the average luciferase activity of triplicate samples. Error bars represent S.D. Curves were fit using 4-parameter nonlinear regression in GraphPad Prism and calculated EC50 values are indicated. D, MASPIT specificity test. Cells coexpressing the DHFR anchor fusion and the indicated ORF preys were stimulated with leptin or leptin combined with 0.1 μm of the indicated TMP fusion compounds. Bars represent fold induction of the average luciferase activity of triplicate samples, error bars represent S.D.
Among the validated hits was panthotenate kinase 3 (PANK3), which had been previously identified as a target of tamoxifen in in vitro high-throughput screens for PANK3 inhibitors and activators (42). PANK3 catalyzes the rate-limiting step in Coenzyme A (CoA) biosynthesis, and tamoxifen binding was found to interfere with binding of acetyl-CoA, this way preventing acetyl-CoA-mediated feedback inhibition of PANK3 enzymatic activity. HSD17B4, another confirmed hit, is a 17β hydroxysteroid dehydrogenase involved in the metabolism of estradiol, the endogenous ER ligand. Earlier reports have described tamoxifen binding to this enzyme, albeit with very low affinity, in the high micromolar range (43). Interestingly, HSD17B4 was also identified in a tamoxifen screen using Virotrap, a novel mammalian protein complex trapping method in which protein or small molecule target complexes are captured in virus-like particles prior to analysis by mass spectrometry (44).
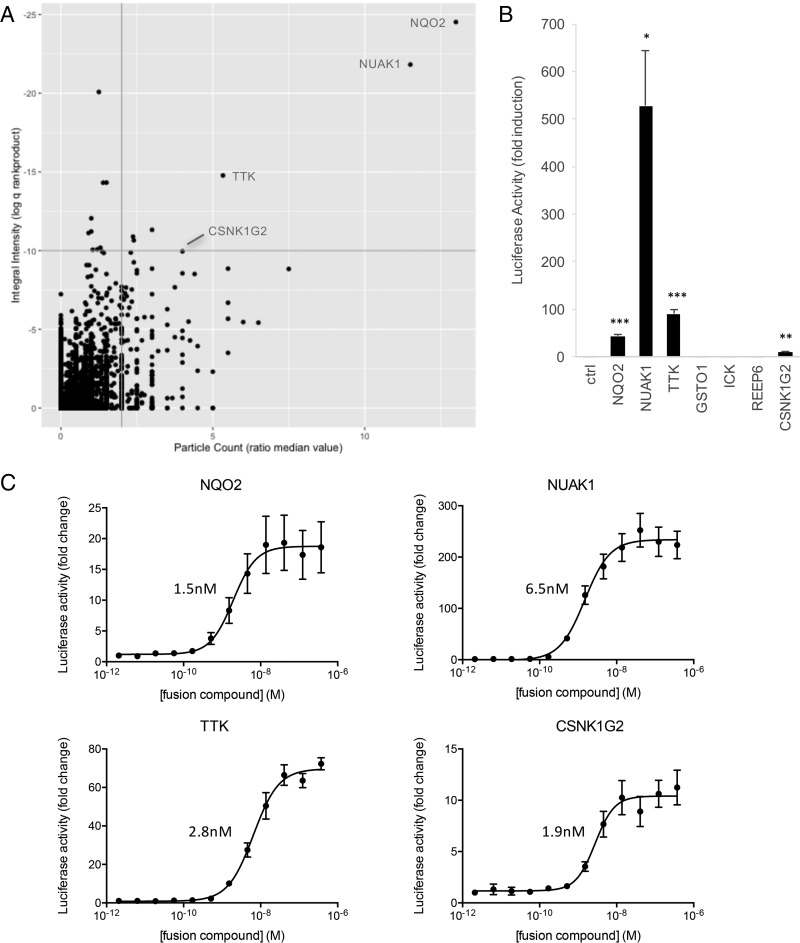
A second small molecule we evaluated in the screening platform is reversine, a purine analog identified from a compound library built around kinase-directed scaffolds in a phenotypic screen for dedifferentiation of mouse myoblasts (45). There is a high therapeutic potential associated with molecules that induce dedifferentiation of somatic cells, as this would allow generating reprogrammed adult cells to use for repair of damaged tissues. Several studies have been published on the proteins that mediate the dedifferentiating effect of reversine, and—as anticipated in view of the nature of the screened library—most of these are kinases (46–49). In addition to its dedifferentiating properties, reversine has also been shown to exhibit potent antitumor activity in a number of cancer models (47, 50–52). As for both effects of the molecule the exact mechanism of action remains to be established, we decided to screen for novel target proteins. As in the case of tamoxifen, we had previously generated a TMP-reversine fusion and MASPIT analysis had confirmed its binding to TTK, one of the reported kinase targets (10). We screened the 15k ORF collection with 0.1 μm TMP-reversine and obtained seven hits (Fig. 6A), and four of these could be confirmed in follow-up MASPIT experiments (Fig. 6B). Dose-response analysis enabled assigning EC50 values to the interactions, which—as in the case of tamoxifen—were in the nanomolar range (Fig. 6C). Cross-testing with unrelated fusion compounds (TMP-tamoxifen and TMP-simvastatin) established that these interactions were specific for the reversine fusion compound (Fig. 5D). TTK, which is considered the primary target kinase of the molecule (48), was among the confirmed hits. Interestingly, the EC50 value obtained in the MASPIT dose-response assay (2.8 nm) matches the IC50 reported based on an in vitro kinase assay using the full-size protein (36). The top hit corresponds to NQO2, a cytoplasmic quinone oxidoreductase which was also identified as a reversine target in a recent Virotrap screen (44). Notably, NQO2 was also found to be targeted by imatinib and nilotinib, two marketed drugs for the treatment of chronic myelogenous leukemia (53). Several lines of evidence indicate that inhibition of NQO2 might contribute to the antitumoral effect of these drugs. It will be interesting to see whether the same is true for reversine. The two remaining hits obtained in the reversine cell microarray screen, NUAK1 and CSNK1G2, are both kinases, consistent with the origin and nature of the compound.
Fig. 6.
Known and new reversine targets identified in a MASPIT cell microarray screen. A, A MASPIT microarray screen was performed in which quadruplicate wells of each screening plate were treated with either leptin alone or leptin combined with 0.1 μm TMP-reversine. Data was analyzed as described in Fig. 3A. Interacting proteins confirmed in MASPIT retests are indicated. See supplemental Table S5 for raw data. B, MASPIT retest of the hits selected using the standard selection thresholds (q<10−10; ratio median value particle count >2). Cells coexpressing the DHFR anchor fusion and the indicated ORF preys were stimulated with leptin alone or leptin combined with 0.1 μm TMP-reversine. Fold induction of the average luciferase activity of triplicate samples in three independent experiments is shown, error bars represent S.E. An empty prey (unfused gp130) was taken along as a control and used to compare data with applying a two-tailed Student's t test (*: p < 0.05; **: p < 0.01; ***: p < 0.001). C, MASPIT dose-response curves for the confirmed hits. Cells coexpressing the DHFR anchor protein and the indicated ORF preys were stimulated either with leptin alone or with leptin and the indicated concentration of TMP-reversine. Data points depict fold induction of the average luciferase activity of triplicate samples. Error bars represent S.D. Curves were fit using 4-parameter nonlinear regression in GraphPad Prism and calculated EC50 values are indicated.
DISCUSSION
In this study we describe the development and validation of a highly miniaturized screening platform for the identification of novel protein interactors of both proteins and small molecules using MAPPIT and MASPIT. Even though these are mammalian cell-based assays, which are typically less amenable to high-throughput applications than most in vitro or Y2H assays, the approach allows testing thousands of target proteins in parallel, with short overall timelines and requiring little hands-on time. Despite the high density of the arrays, signals are robust and reproducible, resulting in a reliable primary screening output and high confirmation rates. The identification of previously described interacting proteins and the confirmation of selected interactions with orthogonal methods validated the screening results.
The different applications described here illustrate the versatility of the screening platform. The interactome of a designated protein of interest can be determined efficiently, as such enabling “pathway walking” where serial identification of novel binding proteins supports signal transduction pathway mapping. The identification of RNF41 interaction partners is an example of such an approach: RNF41 itself was found an interaction partner of the leptin receptor complex in a MAPPIT screen (20), we here report the identification VPS52 and ASB6 as RNF41 binding proteins, and the analysis of additional MAPPIT screens to identify downstream interactors is currently ongoing (Masschaele et al., in preparation; De Ceuninck et al., in preparation). Apart from mapping the basic interactome of a protein, we showed that also dynamic aspects of the interaction pattern of a particular bait can be captured at this scale. As this type of information is in most cases not possible to obtain using in vitro methods or cellular assays that operate in lower eukaryotes (e.g. yeast two-hybrid), this represents one of the important assets of the technology. The cost efficiency of the method allows performing such studies at a scale that has been hitherto unattainable. For example, a network of proteins involved in endocytosis is currently being compiled by screening close to a hundred bait proteins that have been implicated in this trafficking process, both in standard conditions and under conditions where autophagy is induced by addition of rapamycin (Lemmens, Vauthier et al., in preparation).
A major application of the cell microarray screening approach is small molecule target profiling using MASPIT. Target deconvolution is a significant bottleneck in the drug discovery industry, both for detection of on-target binding in phenotypic screening and for identification of off-target binding sites in efforts to minimize toxicity-related attrition during drug development (54). To date, chemoproteomics approaches are often being applied, where small molecule targets are identified from cell lysates using mass spectrometry. Although state-of-the art mass spectrometers are exceptionally sensitive, low endogenous expression of the target protein or disruption of target binding because of cell lysis are common issues related to failure to recover relevant candidate target proteins. Expression-based trapping methods, such as yeast three-hybrid and MASPIT as a mammalian variant, have shown to be valuable alternatives. A modified yeast three-hybrid approach which was used to profile a number of clinically approved drugs was developed by the Johnsson group (55). We previously established the utility of MASPIT to identify known and novel targets of a number of kinase inhibitors, using a cDNA library screening platform (5). By combining MASPIT with the current cell microarray format, we now have access to a platform at a scale and throughput that enables application at industry level. The featured screens for targets of tamoxifen and reversine illustrate the efficiency of the approach. The excellent separation between background signals and candidate hits in the primary screen obviates the need for extensive hit retesting and warrants high confirmation rates. Beyond screening, the MASPIT assay allows for simple dose-response analysis and affinity determination. Although screening and confirmation of small molecule bait fusions is rapid and straightforward, the rate-limiting step is the synthesis of the TMP (or MTX) fusions. Although we streamlined fusion compound synthesis as much as possible through the development of an efficient click chemistry-based route (10, 15), in the absence of a generic methodology for small molecule derivatization this essentially requires a case-by-case approach.
A more general limitation of the technology, which applies to both small molecule and protein interaction screening, relates to the bait configuration. The fact that the bait fusion protein (or DHFR anchor fusion protein in the case of MASPIT) is inserted in the plasma membrane precludes detection of interactions with proteins that are located exclusively in the nucleus or other cellular compartments, or with integral membrane proteins. In order to extend the technology platform to cover also transmembrane proteins, which constitute a significant portion of the proteome, we recently devised an alternative assay, KISS for Kinase Substrate Sensor, that is compatible with this protein class (19). Implementing this assay in the microarray setup will be a next step in expanding the screening platform.
By combining different assays, we expect to broaden the coverage of interactions that can be detected. It is important to keep in mind that every method can only detect a subset of potential interactions. This was clearly illustrated in the benchmarking study of Braun et al. (7), where a fixed reference set of positive and negative PPIs was evaluated with a panel of interaction assays, including MAPPIT and yeast two-hybrid. Each method was found to detect around 30% of the positive reference set, and the detected subset overlapped only partially between the different methods. Probably steric or topological requirements related to the configuration of the assay components determine whether a specific interaction can or cannot be detected using a particular assay. We know that this is also the case for MAPPIT and MASPIT. For example, the MASPIT screen for tamoxifen targets failed to detect the primary target ER1, although it is present in the 15k ORF collection. We have found out that tamoxifen binding to ER1 can be detected readily with MASPIT only in the configuration where the gp130 cytokine receptor portion is fused at the C terminus of the protein (10). Tethering of gp130 at the N terminus, which is the standard configuration applied in the 15k ORF prey collection, yields a weak signal for the tamoxifen-ER1 interaction (15), below the detection threshold applied in the microarray screening setup. This observation indicates that for this particular interaction, detection in MASPIT is hampered by a fusion at the N terminus of the target protein. The scale of the microarray setup now allows expanding coverage by including both N- and C-terminal ORF fusions.
In conclusion, through the cell microarray technology we have turned the well validated MAPPIT and MASPIT assays into a versatile high-throughput platform for protein interaction analysis. As the platform will further grow by integrating newer versions of the expanding ORFeome collection as well as collections focused on alternative protein isoforms and protein subdomains, its coverage of the proteome will continue to extend. Through its sensitivity and robustness, we believe the platform will prove to be a valuable tool for both basic and applied biomedical research.
Supplementary Material
Acknowledgments
We thank Peter Van Den Hemel for the continued IT support.
Footnotes
Author contributions: S.L., J.V., D.M., L.D., I.P., S.G., L.M., K.D., C.L., and J.T. designed research; S.L., J.V., D.M., L.D., I.P., S.G., D. Defever, N.V., and A.D. performed research; V.D., V.V., I.L., D. De Clercq, S.V., L.M., D.H., and M.V. contributed new reagents or analytic tools; S.L., J.V., D.M., L.D., I.P., S.G., V.D., V.V., S.E., K.D., C.L., and J.T. analyzed data; S.L., J.V., D.M., L.D., V.D., I.L., and J.T. wrote the paper.
* This work was supported by grants from the Belgian government (Interuniversity Attraction Poles Project P6/36), the Fund for Scientific Research - Flanders (FWO-V Project G.0864.10) and the NIH (U01 HG001715). J.T. is the recipient of an ERC Advanced Grant (# 340941).
 This article contains supplemental material.
This article contains supplemental material.
Financial interests: J.T. is scientific cofounder of and has financial interests in Orionis Biosciences, which practices technologies described in this publication. S.L. is an employee of Orionis Biosciences.
1 The abbreviations used are:
- PPI
- protein-protein interaction
- GRE
- GR response element
- MAPPIT
- mammalian protein-protein interaction trap
- MASPIT
- mammalian small molecule-protein interaction trap
- MTX
- methotrexate
- ORF
- open reading frame
- SAR
- structure-activity relationship
- TMP
- trimethoprim.
REFERENCES
- 1. Snider J., Kotlyar M., Saraon P., Yao Z., Jurisica I., and Stagljar I. (2015) Fundamentals of protein interaction network mapping. Mol. Syst. Biol. 11, 848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhou M., Li Q., and Wang R. (2016) Current Experimental Methods for Characterizing Protein-Protein Interactions. ChemMedChem 11, 738–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ziegler S., Pries V., Hedberg C., and Waldmann H. (2013) Target identification for small bioactive molecules: finding the needle in the haystack. Angew Chem. Int. Ed. Engl. 52, 2744–2792 [DOI] [PubMed] [Google Scholar]
- 4. Eyckerman S., Verhee A., Van der Heyden J., Lemmens I., Ostade X. V., Vandekerckhove J., and Tavernier J. (2001) Design and application of a cytokine-receptor-based interaction trap. Nat. Cell Biol. 3, 1114–1119 [DOI] [PubMed] [Google Scholar]
- 5. Caligiuri M., Molz L., Liu Q., Kaplan F., Xu J. P., Majeti J. Z., Ramos-Kelsey R., Murthi K., Lievens S., Tavernier J., and Kley N. (2006) MASPIT: three-hybrid trap for quantitative proteome fingerprinting of small molecule-protein interactions in mammalian cells. Chem. Biol. 13, 711–722 [DOI] [PubMed] [Google Scholar]
- 6. Yu H., Braun P., Yildirim M. A., Lemmens I., Venkatesan K., Sahalie J., Hirozane-Kishikawa T., Gebreab F., Li N., Simonis N., Hao T., Rual J. F., Dricot A., Vazquez A., Murray R. R., Simon C., Tardivo L., Tam S., Svrzikapa N., Fan C., de Smet A. S., Motyl A., Hudson M. E., Park J., Xin X., Cusick M. E., Moore T., Boone C., Snyder M., Roth F. P., Barabasi A. L., Tavernier J., Hill D. E., and Vidal M. (2008) High-quality binary protein interaction map of the yeast interactome network. Science 322, 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Braun P., Tasan M., Dreze M., Barrios-Rodiles M., Lemmens I., Yu H., Sahalie J. M., Murray R. R., Roncari L., de Smet A. S., Venkatesan K., Rual J. F., Vandenhaute J., Cusick M. E., Pawson T., Hill D. E., Tavernier J., Wrana J. L., Roth F. P., and Vidal M. (2009) An experimentally derived confidence score for binary protein-protein interactions. Nat. Methods 6, 91–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rolland T., Tasan M., Charloteaux B., Pevzner S. J., Zhong Q., Sahni N., Yi S., Lemmens I., Fontanillo C., Mosca R., Kamburov A., Ghiassian S. D., Yang X., Ghamsari L., Balcha D., Begg B. E., Braun P., Brehme M., Broly M. P., Carvunis A. R., Convery-Zupan D., Corominas R., Coulombe-Huntington J., Dann E., Dreze M., Dricot A., Fan C., Franzosa E., Gebreab F., Gutierrez B. J., Hardy M. F., Jin M., Kang S., Kiros R., Lin G. N., Luck K., MacWilliams A., Menche J., Murray R. R., Palagi A., Poulin M. M., Rambout X., Rasla J., Reichert P., Romero V., Ruyssinck E., Sahalie J. M., Scholz A., Shah A. A., Sharma A., Shen Y., Spirohn K., Tam S., Tejeda A. O., Trigg S. A., Twizere J. C., Vega K., Walsh J., Cusick M. E., Xia Y., Barabasi A. L., Iakoucheva L. M., Aloy P., De Las Rivas J., Tavernier J., Calderwood M. A., Hill D. E., Hao T., Roth F. P., and Vidal M. (2014) A proteome-scale map of the human interactome network. Cell 159, 1212–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baietti M. F., Simicek M., Abbasi Asbagh L., Radaelli E., Lievens S., Crowther J., Steklov M., Aushev V. N., Martinez Garcia D., Tavernier J., and Sablina A. A. (2016) OTUB1 triggers lung cancer development by inhibiting RAS monoubiquitination. EMBO Mol. Med. 8, 288–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. De Clercq D. J., Risseeuw M. D., Karalic I., De Smet A. S., Defever D., Tavernier J., Lievens S., and Van Calenbergh S. (2015) Alternative reagents for methotrexate as immobilizing anchor moieties in the optimization of MASPIT: synthesis and biological evaluation. Chembiochem 16, 834–843 [DOI] [PubMed] [Google Scholar]
- 11. Koorman T., Klompstra D., van der Voet M., Lemmens I., Ramalho J. J., Nieuwenhuize S., van den Heuvel S., Tavernier J., Nance J., and Boxem M. (2016) A combined binary interaction and phenotypic map of C. elegans cell polarity proteins. Nat. Cell Biol. 18, 337–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Corominas R., Yang X., Lin G. N., Kang S., Shen Y., Ghamsari L., Broly M., Rodriguez M., Tam S., Trigg S. A., Fan C., Yi S., Tasan M., Lemmens I., Kuang X., Zhao N., Malhotra D., Michaelson J. J., Vacic V., Calderwood M. A., Roth F. P., Tavernier J., Horvath S., Salehi-Ashtiani K., Korkin D., Sebat J., Hill D. E., Hao T., Vidal M., and Iakoucheva L. M. (2014) Protein interaction network of alternatively spliced isoforms from brain links genetic risk factors for autism. Nat. Commun. 5, 3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lievens S., Vanderroost N., Van der Heyden J., Gesellchen V., Vidal M., and Tavernier J. (2009) Array MAPPIT: high-throughput interactome analysis in mammalian cells. J. Proteome Res. 8, 877–886 [DOI] [PubMed] [Google Scholar]
- 14. Simicek M., Lievens S., Laga M., Guzenko D., Aushev V. N., Kalev P., Baietti M. F., Strelkov S. V., Gevaert K., Tavernier J., and Sablina A. A. (2013) The deubiquitylase USP33 discriminates between RALB functions in autophagy and innate immune response. Nat. Cell Biol. 15, 1220–1230 [DOI] [PubMed] [Google Scholar]
- 15. Risseeuw M. D., De Clercq D. J., Lievens S., Hillaert U., Sinnaeve D., Van den Broeck F., Martins J. C., Tavernier J., and Van Calenbergh S. (2013) A “Clickable” MTX Reagent as a Practical Tool for Profiling Small-Molecule-Intracellular Target Interactions via MASPIT. ChemMedChem 8, 521–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brozzi F., Gerlo S., Grieco F. A., Nardelli T. R., Lievens S., Gysemans C., Marselli L., Marchetti P., Mathieu C., Tavernier J., and Eizirik D. L. (2014) A combined “omics” approach identifies N-Myc interactor as a novel cytokine-induced regulator of IRE1 protein and c-Jun N-terminal kinase in pancreatic beta cells. J. Biol. Chem. 289, 20677–20693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang X., Boehm J. S., Salehi-Ashtiani K., Hao T., Shen Y., Lubonja R., Thomas S. R., Alkan O., Bhimdi T., Green T. M., Johannessen C. M., Silver S. J., Nguyen C., Murray R. R., Hieronymus H., Balcha D., Fan C., Lin C., Ghamsari L., Vidal M., Hahn W. C., Hill D. E., and Root D. E. (2011) A public genome-scale lentiviral expression library of human ORFs. Nat. Methods 8, 659–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Collaboration O. R. (2016) The ORFeome Collaboration: a genome-scale human ORF-clone resource. Nat. Methods 13, 191–192 [DOI] [PubMed] [Google Scholar]
- 19. Lievens S., Gerlo S., Lemmens I., De Clercq D. J., Risseeuw M. D., Vanderroost N., De Smet A. S., Ruyssinck E., Chevet E., Van Calenbergh S., and Tavernier J. (2014) Kinase Substrate Sensor (KISS), a mammalian in situ protein interaction sensor. Mol. Cell. Proteomics 13, 3332–3342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wauman J., De Ceuninck L., Vanderroost N., Lievens S., and Tavernier J. (2011) RNF41 (Nrdp1) controls type 1 cytokine receptor degradation and ectodomain shedding. J. Cell Sci. 124, 921–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. De Ceuninck L., Wauman J., Masschaele D., Peelman F., and Tavernier J. (2013) Reciprocal cross-regulation between RNF41 and USP8 controls cytokine receptor sorting and processing. J. Cell Sci. 126, 3770–3781 [DOI] [PubMed] [Google Scholar]
- 22. Beck I. M., Drebert Z. J., Hoya-Arias R., Bahar A. A., Devos M., Clarisse D., Desmet S., Bougarne N., Ruttens B., Gossye V., Denecker G., Lievens S., Bracke M., Tavernier J., Declercq W., Gevaert K., Vanden Berghe W., Haegeman G., and De Bosscher K. (2013) Compound A, a selective glucocorticoid receptor modulator, enhances heat shock protein Hsp70 gene promoter activation. PLoS ONE 8, e69115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shcherbo D., Murphy C. S., Ermakova G. V., Solovieva E. A., Chepurnykh T. V., Shcheglov A. S., Verkhusha V. V., Pletnev V. Z., Hazelwood K. L., Roche P. M., Lukyanov S., Zaraisky A. G., Davidson M. W., and Chudakov D. M. (2009) Far-red fluorescent tags for protein imaging in living tissues. Biochem. J. 418, 567–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yan J., Li Q., Lievens S., Tavernier J., and You J. (2010) Abrogation of the Brd4-positive transcription elongation factor B complex by papillomavirus E2 protein contributes to viral oncogene repression. J. Virol. 84, 76–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wolfinger R. D., Gibson G., Wolfinger E. D., Bennett L., Hamadeh H., Bushel P., Afshari C., and Paules R. S. (2001) Assessing gene significance from cDNA microarray expression data via mixed models. J. Comput. Biol. 8, 625–637 [DOI] [PubMed] [Google Scholar]
- 26. Breitling R., Armengaud P., Amtmann A., and Herzyk P. (2004) Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 573, 83–92 [DOI] [PubMed] [Google Scholar]
- 27. Heskes T., Eisinga R., and Breitling R. (2014) A fast algorithm for determining bounds and accurate approximate p-values of the rank product statistic for replicate experiments. BMC Bioinformatics 15, 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Storey J. D., and Tibshirani R. (2003) Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U.S.A. 100, 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qiu X. B., and Goldberg A. L. (2002) Nrdpl/FLRF is a ubiquitin ligase promoting ubiquitination and degradation of the epidermal growth factor receptor family member, ErbB3. Proc. Natl. Acad. Sci. U.S.A. 99, 14843–14848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liewen H., Meinhold-Heerlein I., Oliveira V., Schwarzenbacher R., Luo G., Wadle A., Jung M., Pfreundschuh M., and Stenner-Liewen F. (2005) Characterization of the human GARP (Golgi associated retrograde protein) complex. Exp. Cell Res. 306, 24–34 [DOI] [PubMed] [Google Scholar]
- 31. Schindler C., Chen Y., Pu J., Guo X., and Bonifacino J. S. (2015) EARP is a multisubunit tethering complex involved in endocytic recycling. Nat. Cell Biol. 17, 639–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wilcox A., Katsanakis K. D., Bheda F., and Pillay T. S. (2004) Asb6, an adipocyte-specific ankyrin and SOCS box protein, interacts with APS to enable recruitment of elongins B and C to the insulin receptor signaling complex. J. Biol. Chem. 279, 38881–38888 [DOI] [PubMed] [Google Scholar]
- 33. Kishi K., Mawatari K., Sakai-Wakamatsu K., Yuasa T., Wang M., Ogura-Sawa M., Nakaya Y., Hatakeyama S., and Ebina Y. (2007) APS-mediated ubiquitination of the insulin receptor enhances its internalization, but does not induce its degradation. Endocr. J. 54, 77–88 [DOI] [PubMed] [Google Scholar]
- 34. Zhou J., and Cidlowski J. A. (2005) The human glucocorticoid receptor: one gene, multiple proteins and diverse responses. Steroids 70, 407–417 [DOI] [PubMed] [Google Scholar]
- 35. Borgius L. J., Steffensen K. R., Gustafsson J. A., and Treuter E. (2002) Glucocorticoid signaling is perturbed by the atypical orphan receptor and corepressor SHP. J. Biol. Chem. 277, 49761–49766 [DOI] [PubMed] [Google Scholar]
- 36. Zhou J., Oakley R. H., and Cidlowski J. A. (2008) DAX-1 (dosage-sensitive sex reversal-adrenal hypoplasia congenita critical region on the X-chromosome, gene 1) selectively inhibits transactivation but not transrepression mediated by the glucocorticoid receptor in a LXXLL-dependent manner. Mol. Endocrinol. 22, 1521–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hwang J., and Pallas D. C. (2014) STRIPAK complexes: structure, biological function, and involvement in human diseases. Int. J. Biochem. Cell Biol. 47, 118–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jordan V. C. (2003) Tamoxifen: a most unlikely pioneering medicine. Nat. Rev. Drug Discov. 2, 205–213 [DOI] [PubMed] [Google Scholar]
- 39. Frasor J., Danes J. M., Komm B., Chang K. C., Lyttle C. R., and Katzenellenbogen B. S. (2003) Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology 144, 4562–4574 [DOI] [PubMed] [Google Scholar]
- 40. Gelmann E. P. (1996) Tamoxifen induction of apoptosis in estrogen receptor-negative cancers: new tricks for an old dog? J. Natl. Cancer Inst. 88, 224–226 [DOI] [PubMed] [Google Scholar]
- 41. Mandlekar S., and Kong A. N. (2001) Mechanisms of tamoxifen-induced apoptosis. Apoptosis 6, 469–477 [DOI] [PubMed] [Google Scholar]
- 42. Leonardi R., Zhang Y. M., Yun M. K., Zhou R., Zeng F. Y., Lin W., Cui J., Chen T., Rock C. O., White S. W., and Jackowski S. (2010) Modulation of pantothenate kinase 3 activity by small molecules that interact with the substrate/allosteric regulatory domain. Chem. Biol. 17, 892–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Santner S. J., and Santen R. J. (1993) Inhibition of estrone sulfatase and 17 beta-hydroxysteroid dehydrogenase by antiestrogens. J. Steroid. Biochem. Mol. Biol. 45, 383–390 [DOI] [PubMed] [Google Scholar]
- 44. Eyckerman S., Titeca K., Van Quickelberghe E., Cloots E., Verhee A., Samyn N., De Ceuninck L., Timmerman E., De Sutter D., Lievens S., Van Calenbergh S., Gevaert K., and Tavernier J. (2016) Trapping mammalian protein complexes in viral particles. Nat. Commun. 7, 11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen S., Zhang Q., Wu X., Schultz P. G., and Ding S. (2004) Dedifferentiation of lineage-committed cells by a small molecule. J. Am. Chem. Soc. 126, 410–411 [DOI] [PubMed] [Google Scholar]
- 46. Chen S., Takanashi S., Zhang Q., Xiong W., Zhu S., Peters E. C., Ding S., and Schultz P. G. (2007) Reversine increases the plasticity of lineage-committed mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 104, 10482–10487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. D'Alise A. M., Amabile G., Iovino M., Di Giorgio F. P., Bartiromo M., Sessa F., Villa F., Musacchio A., and Cortese R. (2008) Reversine, a novel Aurora kinases inhibitor, inhibits colony formation of human acute myeloid leukemia cells. Mol. Cancer Ther. 7, 1140–1149 [DOI] [PubMed] [Google Scholar]
- 48. Santaguida S., Tighe A., D'Alise A. M., Taylor S. S., and Musacchio A. (2010) Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J. Cell Biol. 190, 73–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McMillin D. W., Delmore J., Weisberg E., Negri J. M., Geer D. C., Klippel S., Mitsiades N., Schlossman R. L., Munshi N. C., Kung A. L., Griffin J. D., Richardson P. G., Anderson K. C., and Mitsiades C. S. (2010) Tumor cell-specific bioluminescence platform to identify stroma-induced changes to anticancer drug activity. Nat. Med. 16, 483–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Keen N., and Taylor S. (2004) Aurora-kinase inhibitors as anticancer agents. Nat. Rev. Cancer 4, 927–936 [DOI] [PubMed] [Google Scholar]
- 51. Girdler F., Gascoigne K. E., Eyers P. A., Hartmuth S., Crafter C., Foote K. M., Keen N. J., and Taylor S. S. (2006) Validating Aurora B as an anti-cancer drug target. J. Cell Sci. 119, 3664–3675 [DOI] [PubMed] [Google Scholar]
- 52. Hua S. C., Chang T. C., Chen H. R., Lu C. H., Liu Y. W., Chen S. H., Yu H. I., Chang Y. P., and Lee Y. R. (2012) Reversine, a 2,6-disubstituted purine, as an anti-cancer agent in differentiated and undifferentiated thyroid cancer cells. Pharm. Res. 29, 1990–2005 [DOI] [PubMed] [Google Scholar]
- 53. Winger J. A., Hantschel O., Superti-Furga G., and Kuriyan J. (2009) The structure of the leukemia drug imatinib bound to human quinone reductase 2 (NQO2). BMC Struct. Biol. 9, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lee J., and Bogyo M. (2013) Target deconvolution techniques in modern phenotypic profiling. Curr. Opin. Chem. Biol. 17, 118–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chidley C., Haruki H., Pedersen M. G., Muller E., and Johnsson K. (2011) A yeast-based screen reveals that sulfasalazine inhibits tetrahydrobiopterin biosynthesis. Nat. Chem. Biol. 7, 375–383 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.