Abstract
Neurotransmitter ligand-gated ion channels (LGICs) are widespread and pivotal in brain functions. Unveiling their structure-function mechanisms is crucial to drive drug discovery, and demands robust proteomic quantitation of expression, post-translational modifications (PTMs) and dynamic structures. Yet unbiased digestion of these modified transmembrane proteins—at high efficiency and peptide reproducibility—poses the obstacle. Targeting both enzyme-substrate contacts and PTMs for peptide formation and detection, we devised flow-and-detergent-facilitated protease and de-PTM digestions for deep sequencing (FDD) method that combined omni-compatible detergent, tandem immobilized protease/PNGase columns, and Cys-selective reduction/alkylation, to achieve streamlined ultradeep peptide preparation within minutes not days, at high peptide reproducibility and low abundance-bias. FDD transformed enzyme-protein contacts into equal catalytic travel paths through enzyme-excessive columns regardless of protein abundance, removed products instantly preventing inhibition, tackled intricate structures via sequential multiple micro-digestions along the flow, and precisely controlled peptide formation by flow rate. Peptide-stage reactions reduced steric bias; low contamination deepened MS/MS scan; distinguishing disulfide from M oxidation and avoiding gain/loss artifacts unmasked protein-endogenous oxidation states. Using a recent interactome of 285-kDa human GABA type A receptor, this pilot study validated FDD platform's applicability to deep sequencing (up to 99% coverage), H/D-exchange and TMT-based structural mapping. FDD discovered novel subunit-specific PTM signatures, including unusual nontop-surface N-glycosylations, that may drive subunit biases in human Cys-loop LGIC assembly and pharmacology, by redefining subunit/ligand interfaces and connecting function domains.
Cell surface transmembrane (TM)1 proteins such as neurotransmitter ligand-gated ion channels (LGICs) and G protein-coupled receptors (GPCRs) are abundant in human nervous system, pivotal in signaling, and dominate drug target priority (1). Cys-loop receptor LGICs are synaptic ion channels formed by various pentamers of homologous TM subunits—each featuring a conserved disulfide Cys-loop in extracellular domain (ECD) and a vastly diverse intracellular domain (ICD). Cys-loop LGICs include Cl̄ channels: gamma-aminobutyric acid type A receptor (GABAAR) and glycine receptor (GlyR), and cation channels: nicotinic acetylcholine receptor (nAChR) and serotonin type 3 receptor (5HT3R) (2). Disrupting the balance between excitatory (cationic Na+, K+ or Ca2+) and inhibitory (Cl̄) ion channels may cause many neurological disorders, including epilepsy (3), addiction (4), and cognitive dysfunctions (5), which have limited therapeutics with serious side effects. Revealing Cys-loop LGICs structure-function mechanisms is coveted to drive drug discovery, but hydrophobic with complex modifications, their atomic structures are difficult to achieve (6), and genomic codes are insufficient to account for function complexity.
Recent breakthroughs in crystallography resolved the structures of homomeric bacterial and C. elegans analogs (and a truncated human (β3)5 GABAAR during earlier review of this manuscript) lacking ICD (7–9), yet genomic sequence alignments with them emphasize subunit similarities (2, 10, 11), but offer little clue on how small genomic differences cause large diversity in subunit-biased assembly, ligand recognition, and channel function response. Intriguingly, 19 different subunits for GABAAR in human genome—all homologous and mostly expressed in brain (2, 10)—would lead to about 500,000 unique pentameric assemblies without counting splicing variants, though only fewer than 30 were found (2, 10), suggesting unknown constraints for subunit biases in assembly. Post-translational modifications (PTMs) are emerging as a key to decipher the structural mechanisms of neurotransmitter receptors, highlighted by recent discoveries of phosphorylation-activity patterns in GPCRs (12), tetrameric Na+ and K+ channels (13, 14), and multiple phosphorylation sites on various LGIC subunits in mouse brain (15), mostly in ICD.
Dissecting LGIC mechanisms thus demands robust quantitative proteomic measurements of dynamic changes in protein levels, PTMs and conformations induced by stimuli. Peptide-centric quantitations of modifications and labels rely on correlating ion abundances, and require high peptide reproducibility and minimal contamination, because only those same peptides generated across sample states are useful, whereas irrelevant peaks may skew quantitation. Bottom-up HDX (H/D exchange labeling followed by digestion, MS and MS/MS) is emerging as a powerful tool to comprehensively map protein dynamic structures, and is highly complementary to crystallography (16–18), especially for membrane proteins (16, 17). HDX visualizes protein's natural backbone amide hydrogen exchange with solvent using D2O, and reveals a residue's architectural context by summarizing the extent and rate of %D from its peptide isotopic peak envelope. Thus HDX requires reproducibility at least in peptide forms throughout H/D-incubation time points and protein states. Label-free or tandem mass tag (TMT)-based quantitation of peptides, modifications or oxidation structural labels further demands peptide reproducibility in both form and abundance.
Additional tool of top-down ESI MS/MS grew rapidly and may distinguish modification-varied proteoforms, shown by 50 kDa membrane protein chloride ion channels using collisional dissociation MS/MS (19, 20), and by helical muscle proteins using electron capture MS/MS as well (21). Nonetheless, compared with bottom-up platform, top-down remains limited in method access and robustness, and in providing the structural detail, precision, and coverage required by charting clear atomic structure-function roadmaps for targeted drug discovery. Intact protein HDX MS provides mere overall shifts in total labels of whole proteins, and is confounded by solution-to-gas-phase-ion transition. Subprotein resolution relies on MS/MS, but collisional dissociation MS/MS scrambles H/D labels; ETD MS/MS, though able to preserve residues' labels and rapidly growing (22, 23), is complicated by H/D-envelope and incomplete sequence coverage, and currently proved practical on just the few selected H/d-peptides of interest in bottom-up pipeline. Thus bottom-up HDX MS provides the most useful robust approach readily adoptable to widely-accessible bottom-up proteomic instruments, it can routinely reach structural resolution at peptide level (3 or more residues) preserving MS/MS's sequence coverage, and may pinpoint resolution closer to the residue level when coupled with targeted H/D-peptide ETD MS/MS.
However, unbiased digestion of these modified TM proteins at high efficiency and reproducibility poses a long-standing challenge (24, 25). Nearly all plasma membrane proteins made and folded in the endoplasmic reticulum (ER) contain glycosylations at high site occupancy in matured forms (24), which prevents local sequence identification if unaddressed. Conventional digestion paradigm subjects membrane proteins to harsh denaturants, repeated separations via precipitations or dial-filtrations, and multiple proteases for day(s). Yet it suffers: low (sub-50%) digestion efficiency (25, 26), bias in protein abundance (27), bias in protease specificity sites (28), low peptide reproducibility (27, 29), low (sub-30%) sequence coverage (30), high oxidation artifacts, and high contamination (31). Current gel-free digestion methods of purified GABAAR take 2 days combining 6 (4 overnight) digestions (29, 32), only able to identify two glycosylation sites (29). Representative peptide reproducibility in global shotgun analysis is 25 ± 2%, and 15 ± 2% after DigDeAPr treatment (27), whereas membrane proteins are considered mission impossible. These limitations severely undermine quantitation and preclude HDX or oxidation label-based structural studies. Protein abundance bias arises when abundant proteins occupy most protease catalytic sites leaving few available for low-abundance proteins (27); intricate structures and bulky modifications—such as disulfide bonds, glycosylations, and associated lipids, detergents or other proteins—obstruct protease access (25), together pinpointing inadequate enzyme-protein substrate contact as the cause.
Here we target the enzyme-protein substrate contact as the limiting factor for both protease and de-PTM enzyme reactions, and present a modular proteomic workflow featuring a new, ultradeep digestion paradigm for glycosylated membrane proteins. We devised an abundance-unbiased flow-and-detergent-facilitated protease and de-PTM digestions for deep sequencing (FDD) method, that integrated detergent, tandem pepsin- and PNGase F-immobilized columns (operated as plug-flow reactors) and Cys-PTM-selective reduction, to streamline instant peptide preparation at high coverage. This platform transformed enzyme-protein contact into an equal catalytic travel path through the immobilized reactor column regardless of molecule abundance, removed products instantly preventing inhibition, tackled intricate structures via sequential multiple micro-digestions, and controlled precisely peptide formation via flow rate. Incorporating amine-free detergent n-dodecyl-β-d-maltopyranoside (DDM) as a tool for digestion ensured unbiased flow and effective catalytic contact of membrane proteins and peptides, complied fully with upstream protein structure, protease activity, direct TMT labeling, HDX HPLC MS and MS/MS analysis—without requiring removal, and allowed a fast enclosed pipeline immune from artifacts. Immobilized pepsin column emerged for fast digestion in HDX (33), but without DDM met limited success with membrane proteins (16, 18). FDD is flexible to support various enzymes and new detergents for specific studies.
Using the recently immuno-enriched interactome of full-length human (FLAG-α1)2(β3)2(γ2L)1 GABAAR and associated proteins from HEK293 in DDM/cholesteryl hemisuccinate (DDM/CHS) as a model (34), we demonstrated the FDD peptide preparation method enabled complete digestion of GABAAR within 10 s of contact at high peptide reproducibility (75–77% over basal 77–81%) and achieved unprecedented deep coverage of PTMs for hLGICs. We validated the superior metrics of this versatile membrane proteomic platform in three applications: (1) deep sequencing (88–99% subunit coverage from one run), (2) comprehensive PTM mapping with label-free quantitation of % site occupancy, which discovered novel subunit-bias signatures for human Cys-loop receptors, and (3) compatibility with quantitative structural mapping using HDX, or TMT-tagging following oxidation labeling.
EXPERIMENTAL PROCEDURES
Materials
Tetracycline-inducible human embryonic kidney cells (HEK293-TetR) were from the late Dr. H. G. Khorana's laboratory at Massachusetts Institute of Technology. Dulbecco's Modified Eagle Medium with F-12 (DMEM/F-12) was from ATCC (Manassas, VA). Fetal bovine serum was from Atlanta Biologicals (Lawrenceville, GA). Glycerol-free flavobacterium meningosepticum-secreted PNGase F solution was from New England Biolabs (Ipswich, MA). M2 anti-FLAG antibody-coupled agarose beads (average binding capacity 1.0 mg/ml FLAG-BAP), FLAG peptide bromide, γ-aminobutyric acid (GABA), protease inhibitors leupeptin, aprotinin, chymostatin, pepstatin (LACP), and phenylmethylsulfonyl fluoride (PMSF), porcine pepsin, bovine catalase-agarose beads, 99.9% (m/m) D2O, 30% (m/v) hydrogen peroxide (H2O2), N-acetyl methionine, 50% (m/v, 15 m) hydroxylamine (NH2OH), 1 m triethylammonium bicarbonate (TEAB), Tris-2-carboxyethylphosphine (TCEP), dithiothreitol (DTT), iodoacetamide (iodoaa), (2-chloro)-N,N-dimethylethylamine (ClCH2CH2N(CH3)2, Cl-DMEA), (2-bromoethyl)-N,N,N-trimethylammonium bromide (BrCH2CH2N(CH3)3+Br̄, BrE-TMAB), formic acid (FA), and anhydrous acetonitrile were from Sigma-Aldrich (St. Louis, MO). Glass column cartridge was from GE Healthcare Bio-Sciences (Piscataway, NJ). N-dodecyl-β-d-maltopyranoside (DDM, Anagrade) and cholesteryl hemisuccinate Trizma salt (CHS, Anagrade) were from Anatrace-Affymetrix (Santa Clara, CA). TMT2 126/127 labels, Applied Biosystems POROS AL beads (bead size 20 μm), phosphate buffered saline (PBS, 10×, final pH 7.4), BCATM protein assay kit, precast NuPAGE 4–12% (m/v) Bis-Tris gels with MES-SDS running buffer and EZ-RUN BP3603 (10–170 kDa) protein molecular weight markers were from Thermo Fisher Scientific (Rockford, IL).
GABAAR Protein Purification from HEK293 Cells
HEK293-TetR cells stably over-expressing full-length human (N)-FLAG-α1β3γ2L-(GGS)3GK-1D4 GABAAR at α1 (bovine): β3 isoform 2 (human): γ2L (human) subunit plasmid copy ratio of 2:2:1 were provided by Dr. Keith W. Miller (35), and final active GABAAR (at typical concentration of 25 μm in the 0.05% DDM/0.0125% CHS/10% glycerol protein buffer) was obtained by affinity purification on a low-flow-rate anti-FLAG column, as described earlier (34). Briefly, following 72-hour growth and 24-hour tetracycline-induced GABAAR expression in 5% CO2 at 37 °C, cells at 90% confluence on 15-cm plates were PBS-washed, harvested into hypotonic lysis buffer (10 mm HEPES pH 7.5, protease inhibitors LACP and PMSF), and frozen slowly at −80 °C to promote lysis. Thawed cells were lysed on ice with an ultrasonic probe, membranes were isolated, repeatedly homogenized and washed, and flash-frozen in liquid nitrogen. The membranes were solubilized by gradually adding 20 mm (1%, m/v) DDM/4 mm (0.25%, m/v, Trizma salt, equivalent to 0.2% free acid form) CHS over 2 mg/ml protein; solubilized supernatant was loaded to an anti-FLAG column operated at constant 0.3 ml/min for efficiency and consistency; GABAAR was eluted with FLAG peptide, cleaned and concentrated using dial-filtration with a 100 kDa molecular weight cutoff (MWCO) filter to 25 μm (BCA assay, using pentamer molecular weight 285 kDa), in the protein buffer composed of 1 mm (0.05% m/v) DDM, 0.2 mm (0.0125% m/v) CHS, 10% (v/v) glycerol, 50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 2 mm CaCl2, 5 mm KCl, 5 mm MgCl2, and 4 mm EDTA. Purified protein's receptor activity, state of dispersion (nonaggregation), and protein composition were validated as described earlier (34). Biological variances were covered by pooling 120 cell plates from two batches of growth at different passage number, and technical variances were addressed by a repeated split-pool strategy (34).
Pepsin-column Digestion
The pepsin-coupled POROS AL beads were prepared by covalently conjugating the N terminus (or accessible side-chain amines) of porcine pepsin to the aldehyde functional group on bead surface through a Schiff base reduction reaction, similar to previous descriptions (33, 36), but with modifications in pH to improve coupling efficiency and enzyme activity. Specifically, 40 mg porcine pepsin was dissolved in 1 ml of 50 mm Na3Citrate (Na3Cit) pH 5.0 buffer and added to 10 mg NaCNBH3 powder using a 20 ml glass vial. A 0.5 ml aliquot of 1.5 m Na2SO4 pH 5.0 buffer was then added dropwise, and the mixture was incubated for another 10 min on a shaker/rocker. Three hundred milligrams POROS AL-20 beads were then added to the solution, and 1.06 ml of 1.5 m Na2SO4 pH 5.0 buffer was added dropwise over 30 min. We incubated the mixture at 24 °C for 16 h, and quenched the reaction by adding 0.5 ml of 0.1 m ethanolamine prepared in 50 mm Na3Cit pH 4.4 buffer followed by incubation for another 2 h. We then transferred the mixture into a 15 ml centrifuge tube, and washed the beads sequentially with: 12.5 ml 50 mm Na3Cit pH 4.4 buffer twice, 12.5 ml 1 m NaCl solution prepared in 50 mm Na3Cit pH 4.4 once, and 12.5 ml 50 mm Na3Cit pH 4.4 buffer twice. Pepsin beads were stored as 1:1 (v/v) slurry in 50 mm Na3Cit pH 4.4 at 4 °C. Typical yield was 1.5 ml neat pepsin beads. To prepare columns, we rinsed and suspended the beads in 0.1% (v/v) FA/H2O pH 2.5 buffer as a 1:3 (beads/buffer, v/v) slurry, and packed into 1 mm i.d. x 20 mm column cartridges under Ar pressure. Columns were stored in 0.1% FA/H2O pH 2.5 at 4 °C; prior to use for digestion, columns were incubated at room temperature and flushed with 0.1% FA/H2O pH 2.5.
Protein samples were all adjusted to pH 2.4–2.5 using a digestion buffer composed of 0.02% (0.4 mm, ∼2 cmc) DDM, 15 mm TCEP and 0.1 m NaH2PO4-HCl (pH 2.4 for mixing protein: buffer at 1:1, v/v; pH 2.3 for 2:1; pH 2.0 for 4:1), and digested by directly passing through the pepsin column (1 mm × 20 mm) placed at room temperature (20 °C or 24 °C) at flow rate of 25 μl/min. We discarded the first 5 μl flow-through to avoid sample dilution based on a 26% porosity of tightly packed spheres in the column, and collected only the digest generated at constant flow rate. Between runs, we flushed the pepsin column sequentially with column priming solution (protein buffer and 0.1 m NaH2PO4-HCl/DDM/TCEP buffer mixed at 1:1, v/v, pH 2.5), 0.1% (v/v) FA in H2O pH 2.5, and priming solution. The pepsin-column digest at pH 2.5 was either directly injected for HPLC MS analysis, or further processed. We measured the digestion efficiency with SDS-PAGE, and with 85-min HPLC at high %B.
PNGase F-column Digestion
PNGase F was obtained in Tris buffer (20 mm Tris pH 7.5, 50 mm NaCl and 5 mm EDTA), thus before immobilization, we exchanged buffer into one composed of 10 mm NaH2PO4-NaOH, pH 7.5, 50 mm NaCl and 2.5 mm EDTA, using a dial-filtration cartridge with 30 kDa MWCO at 4 °C. PNGase F-coupled POROS AL beads were prepared similarly to pepsin-POROS AL beads described above, except for using pH 7.5. The PNGase F beads showed enzyme activity, although the coupling efficiency was subideal using such a high-salt method. The PNGase F-coupled POROS AL beads were packed into 2 mm i.d. × 20 mm column cartridges under Ar pressure.
PNGase F column was applied after pepsin column unless specified otherwise. We adjusted the pepsin-column digest from pH 2.5 to pH 7.5–8 using 100 mm NH4HCO3, and passed it through PNGase F column (2 mm × 20 mm) at flow rate of 2 μl/min at 24 °C. We discarded the first 15 μl flow-through based on a 26% column porosity, but rinsed the column with 15 μl protein buffer and combined it with the collected elute.
Peptide Reduction and Alkylation
Part of the pepsin-column digest at pH 2.5 (with basal ∼7 mm TCEP) was directly injected for HPLC MS analysis; part was adjusted to pH 7.5–8.0, deglycosylated by passing through PNGase F column, further reduced—with 10 mm DTT, 0.7 m NH2OH or nothing more than basal TCEP—at 24 °C for 30 min, alkylated with 25 mm iodoacetamide at 24 °C for 15 min in the dark, and quenched with 5% (v/v) FA back to pH 2.5 prior to injection. Reagents were typically prepared as 20× or 10× stocks. At equal concentration to iodoacetamide, alternative alkylation with Cl-DMEA or BrE-TMAB (24 °C for 30 min) was also tested but didn't improve Cys-peptide coverage in CID and HCD MS/MS (not tested for electron capture or electron transfer dissociation ETD, because unavailable).
Structural Labeling of GABAAR with HDX
Purified GABAAR in the apo state was labeled by HDX using D2O buffer. The protein buffers used in structural studies were the same as the final purified protein buffer described above, except for only 2% glycerol. The D2O buffer for this proof-of-concept HDX experiment contained a final D2O content of 90% (v/v). In each HDX labeling experiment, 4 μl of 25 μm purified GABAAR was mixed with 16 μl of either H2O or D2O version of protein buffer (fivefold dilution), incubated on ice for 100 min, quenched by mixing with 20 μl ice-cold quench solution composed of 0.1 m NaH2PO4-HCl pH 2.4, 0.02% DDM and 15 mm TCEP, and immediately digested by passing through a pepsin column (1 mm i.d. × 20 mm) at 25 μl/min placed at 20 °C. The first 5 μl flow-through was discarded, the middle section was collected into ice-cold sample vials, sealed, and injected for HPLC MS analysis within minutes.
Structural Labeling of GABAAR with H2O2
Purified GABAAR in the apo or ligand (GABA)-bound state was labeled by covalent oxidation using high concentration of H2O2 (3%, m/v). A 4.4 μm purified GABAAR, diluted from 25 μm using the 2% glycerol protein buffer, served as the starting protein sample. The sample was divided into two parts, 60 μl and 40 μl, and incubated with either 1.8 μl protein buffer (apo) or 1.2 μl of 50 mm GABA stock (GABA-bound) for one hour at 4 °C. The final concentration ratio of GABA ligand/protein was about 30:1 (150 μm/4.4 μm). Following ligand incubation, the samples were oxidation-labeled by adding 30% (m/v) H2O2 to a final concentration of 3%. After oxidation reaction time of 1 min (apo only), 4 min or 7 min at 24 °C, a 20 μl aliquot of the sample was retrieved, quenched by mixing with precleaned catalase-agarose beads (drained from 10 μl 1:1 slurry) for 10–15 min at 24 °C, and finally flash-frozen in liquid nitrogen, to remove bubbles and to provide an equal ground for sample comparison. Upon thawing, the sample was mixed with 10 μl of a buffer of 0.1 m NaH2PO4-HCl pH 2.4, 0.02% DDM, 15 mm TCEP and 100 μm N-acetyl methionine, and liquid was separated from beads with a gel-loading tip, without needing centrifuge; the beads were then rinsed with another 10 μl of the same buffer, solutions were combined and applied to pepsin-column digestion at 24–30 °C.
TMT2 Tagging of Peptides from H2O2-labeled GABAAR
TMT2 126/127 tags were applied to the H2O2 oxidation-labeled apo or GABA-bound GABAAR, after pepsin-column digestion. Unless specified otherwise, TMT2–126 was applied to digests of apo with 1 min, 4 min and 7 min oxidation labeling, respectively, and TMT2–127 was applied to digests of the GABA-bound version with 4 min and 7 min oxidation labeling. Aliquots from various labeled peptides were then paired as TMT2 mixtures. Specifically, 0.8 mg TMT2–126 or 127 reagent was dissolved in 44 μl anhydrous acetonitrile right prior to use. Each 10 μl aliquot of digest at pH 2.5 was adjusted to pH 7.5–8.0 by adding 2 μl of 1 m TEAB, mixed with 10 μl of TMT2–126 or 127 reagent solution, and incubated for 60 min at 24 °C. The reaction was quenched by adding 15 m NH2OH to a final concentration of 0.7 m followed by incubation for another 30 min at 24 °C. The samples (with basal ∼7 mm TCEP) were pooled as TMT2 126/127 pairs at volume ratio of 1:1, alkylated with 25 mm iodoacetamide for 15 min in the dark at 24 °C, and quenched by adding neat FA to reach pH 2.5. After acetonitrile removal by Ar at 25 kPa for 5 min on ice, samples were readjusted to pH 2.5 with neat FA, Ar-sealed, flash-frozen in liquid nitrogen, and stored at −80 °C until HPLC injection. TMT was applied in over twofold excess to the total primary amines in peptide solution. Volatile buffers were used to avoid desalting.
Mass Spectrometry
Peptide digests at pH 2–2.5 were analyzed by reversed-phase (RP) HPLC MS and higher energy collisional dissociation (HCD) MS/MS on a Q-Exactive orbitrap mass spectrometer equipped with nanoelectrospray ionization (nESI) source and an EASY-nLC 1000 system (Thermo Scientific, San Jose, CA); HDX HPLC high-resolution MS spectra were acquired on an LTQ-Orbitrap XL mass spectrometer equipped with nESI, Agilent 1100 binary pump (Agilent Technologies, Palo Alto, CA) with nanoliter flow splitter, and Famos autosampler (LC Packings, San Francisco, CA) operated in direct injection mode, at the Taplin Mass Spectrometry Facility. About 4 μl peptide sample was loaded in each run. HPLC buffer A was 0.1% (v/v) FA, and buffer B contained 0.1% (v/v) FA and 60% (v/v) acetonitrile. Peptides were loaded at 5 μl/min and separated on a C18 fused silica microcapillary column (3 μm 200 Å Maccel AQ C18 beads, 125 μm i.d. × 18 cm, hand-pulled tip i.d. 5 μm) at a flow rate of sub-0.5 μl/min over a 75-min 0–40% linear gradient of buffer B. The total HPLC analysis time (including loading, separation, and column equilibration) was 85 min. For HPLC MS in HDX experiments, a short 7 min linear gradient of buffer B was applied and the total HPLC analysis time was 15 min. A high-high (H-H) top15 data-dependent method was used for MS and HCD MS/MS acquisition: one high-resolution full MS scan in the orbitrap (m/z 390–1600) was followed by high-resolution HCD MS/MS scans of product ions from 15 most abundant precursor ions analyzed in the orbitrap. HCD settings were: precursor ion isolation width 2.0 Da for the quadrupole (Q) mass filter and normalized collision energy 25% for the high-pressure collision cell. Dynamic exclusion of precursor ions was applied with a 0.2 Da width for durations of 20 s. For the few high-low (H-L) HPLC MS and CID MS/MS experiments on LTQ-Orbitrap XL using top10 method, data acquisition settings were the same as described earlier (34). MS/MS acquisition included singly charged precursors to strengthen peptide redundancy desired in HDX and pan-PTM mapping, to cover nonpeptide endogenous components (16), and to aid method evaluation and diagnose in this pilot platform development.
Proteomic Data Processing
High-high HCD MS/MS spectra were assigned by searching against the GABAAR protein sequences supplied for overexpression (to use for subsequent quantitative and structural studies), or the human proteome (downloaded May 23, 2013, Ensembl, GRCh37.71; to check contamination and associated proteins), using the SEQUEST or SEQUEST-HT algorithm embedded in Proteome Discoverer (PD, version 1.3 or 1.4 respectively). All peptide assignments were further filtered to a peptide false discovery rate (FDR) below 1%, using q-value-based Percolator embedded in PD1.3 or PD1.4. Data were not filtered further to refine protein FDR. A protein was considered identified only when three or more unique peptides were identified. SEQUEST-HT was used for peak-area and TMT quantitations and for human proteome search. Other results showed SEQUEST unless specified otherwise, because it was more conservative for multi-K sequences.
For method validation, all modification searches against GABAAR sequence were performed in the dynamic mode, using default conditions (unless specified otherwise), including: cysteine (Cys, C) carbamidomethylation (+57.02146 Da), methionine (M) oxidation (+15.99492 Da), and asparagine (N) deamidation (+0.98402 Da, taken as N-deglycosylation when PNGase F-specific), at mass variation tolerance of 10 ppm for MS and 0.02 Da for MS/MS, charge state 1 to 7, and peptide length 3–40 (PD1.3) or 4–40 (PD1.4). Search against the human proteome was performed with fixed Cys carbamidomethylation (+57.02146 Da) in PD1.4. Pepsin digests were searched without enzyme specification, and porcine pepsin was added to the sequence database for test. Earlier high-low CID MS/MS data were searched with mass tolerance 10 ppm and 0.8 Da (or 0.2 Da when specified) for MS and MS/MS respectively, same as described (34).
Further, HCD MS/MS spectra were screened extensively for potential modifications (against GABAAR sequence), including: carbamidomethylation (Cb) +57.02146 Da (C, K, R), deamidation +0.98402 Da (N, Q), oxidation (Ox) +15.99492 Da (C, M, D, F, H, K, N, P, R, W, Y), di-oxidation (diOx) +31.98983 Da (C, M, F, K, P, R, W, Y), tri-oxidation (triOx) +47.98474 Da (C), phosphorylation +79.96633 Da (S, T, Y), ubiquitination (Ub) on K (K-GG, +114.0429 Da; also K-GGR for pepsin, +270.14404 Da), methylation (Me1) +14.01565 Da (K, R, C), di-methylation (Me2) +28.03130 Da (K, R), tri-methylation (Me3) +42.04695 Da (K, R), formylation (Fm) +27.99491 Da (K, R), acetylation (Ac) +42.01056 Da (K, C); C-palmitoylation (16:0, palm) +238.22967 Da, C-palmitoleylation (16:1, palme) +236.21402 Da, C-farnesyl (farn) +204.18780 Da, C-myristoylation (myri) +210.19836 Da, C to S −15.97716 Da, C to dehydroalaline (DHA) −33.98772 Da, N-terminal Q to pyroE −17.02655 Da. TMT2-labeled peptides were searched with additional modifications of +225.15583 Da in MS (any N terminus and K; dynamic, because performing TMT reaction and data-dependent MS/MS in the presence of detergents is a new method). PTMs were searched iteratively by including up to 6 types of modifications in each search, and maximum 4 dynamic modifications (maximum 3 identical types) were allowed per peptide. O-glycosylation was not examined here.
Although high-high HCD MS/MS offered high accuracy in N-deamidation identification, N-glycosylation sites were still validated by decoy deamidation (N, Q) search on PNGase F nontreated peptide samples as a control for false positive (such as spontaneous N, Q deamidation) as described earlier (34). Only those sites identified as deamidated in PNGase F-treated sample and not as deamidated in nontreated were taken as real N-glycosylation sites.
Label-free quantitation of PTM site occupancy for de-N-glycosylation (PNGase F-specific N-deamidation) and for oxidation was performed with precursor ion isotopic peak area summed from each isotope at 2 ppm mass precision, at PSM's retention time using the integrated function module in PD1.4. Modification may shift peptide elution time. All N residues, and most M residues, were covered by overlapping redundant peptide forms (backbone sequences). We calculated the % occupancy by 100% × (modified version)/(modified + unmodified) for each peptide form that contains the site of interest, and averaged all forms for the final % occupancy of each site. For confirmation, deglycosylation occupancy was additionally estimated by peptide PSMs (PD1.3). N-terminal TMT2-tagged peptides of Ox-labeled GABAAR were quantified in MS/MS by the peak height ratio of reporter ions at m/z 126 and m/z 127 (monoisotopic peak) using integrated module in PD1.4. Peptide coverage maps over protein sequence were prepared using the HDX workbench software (37), and other graphics were prepared using Excel.
We arbitrarily defined “comprehensive coverage” as: no undigested protein remains visible to gel or eye postdigestion, no visible sample loss to reaggregation pre-HPLC, and >90% overall sequence coverage by peptide MS/MS; “comprehensive PTM inventory” additionally required covering >90% of total target residues. By protein “denaturation,” we meant decrease of activity (such as deactivation of protease), and de-functioning distortion of original conformation, which for membrane proteins without sufficient or proper detergents often leads to destabilization and aggregation, not solubilization. By “enzyme column,” we specifically referred to a tightly-packed, HPLC pressure-compatible, sub-2 mm-diameter column whose performance can be controlled reproducibly with flow rate, column diameter and length in HPLC integration; similar to the theoretical number of plates for separation columns, we proposed the theoretical number of digestion events (n), to characterize enzyme column operation (supplemental Note S2).
Because a multispecific protease (pepsin) was used, to distinguish from protease's theoretical specificity of residue preference, we defined “protease digestion reliability” empirically as the combined abilities to: (1) reproducibly form largely superimposable HPLC-MS chromatographs and (as identified by MS/MS) a similar set of peptide sequences from independent digestions, (2) reproducibly tune reaction conditions to obtain similar peptide length distribution under the same settings, and (3) correctly predict the direction of shift in peptide length distribution upon varied reaction conditions, such as flow rate and temperature.
Details of peptide and protein identification and annotated PTM MS/MS spectra were included in Supplemental Materials. Raw HPLC MS and MS/MS data were deposited at UCSD MassIVE and ProteomeXchange under accession MSV000079379 and PXD003203.
Protein Sequence Alignments
To estimate the 3D location of key residues and domains, protein sequences for Cys-loop LGIC subunits were retrieved from Uniprot, and aligned to that of crystallized C. elegans GluCl (PDB: 3RHW) lacking ICL2 (8), using Clustal O (1.1.1 or 1.2.0) in Uniprot. When controversial, such as the starting point of TM4, alignment shown in Ref (8) was used. Results of protein sequence coverage and PTM mapping were also illustrated on GABAAR 3D structure prepared as such. To estimate N-glycan 3D location, resolved structures of C. elegans GluCl (3RHW)/Sf9 and mouse α1 subunit of nAChR ECD domain expressed in yeast (2QC1) (38) were compared by using the alignment function in PyMOL. Glycan structure resolved in 2QC1 was trimmed to (GlcNAc)2(Man)5, a minimal mammalian form, to illustrate mid-ECD N-glycans in GABAAR/HEK293 (GlcNAc, N-acetylglucosamine; Man, mannose).
RESULTS
Modular Membrane Proteomics Workflow: No Filter, No Centrifuge, Full Automation
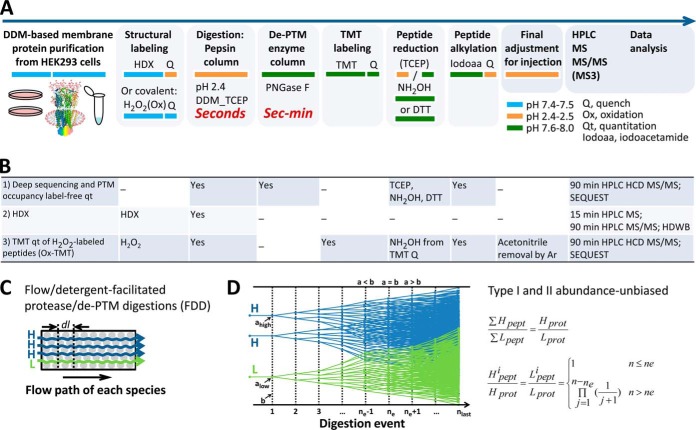
Targeting enzyme-protein substrate contacts and PTMs to solve efficient peptide formation and detection, we devised a novel digestion workflow termed flow-and-detergent-facilitated protease and de-PTM digestions for deep sequencing (FDD). FDD featured three keys to achieve streamlined abundance-unbiased peptide sample preparation within minutes: (1) DDM as a tool for digestion, (2) pepsin and PNGase F columns as tandem plug-flow reactors, and (3) Cys-selective reduction/alkylation. This approach allowed a flexible, fully-automatable modular proteomic platform that supports TMT and can instantly analyze active membrane proteins in solution, for deep sequencing, PTM occupancy quantitation, and HDX or Ox-TMT structural mapping (Fig. 1A, 1B).
Fig. 1.
Modular proteomic streamline for instant ultradeep membrane protein analysis, supporting full automation. A, Modular method workflow featuring direct FDD digestion and HPLC MS. Boxes represent modules for selection; color band shifts indicate pH switching with volatile buffers (FA, NH4HCO3 or TEAB). B, Module selection in three applications for quantitation and structural mapping. C, Flow model for FDD's flow-and-detergent-facilitated immobilized enzyme column reactors. D, Digestion model for FDD based on effective catalytic site-occupancy (detailed in supplemental Note S2). n, theoretical number of digestion; a, accessible cleavage sites on all substrates: ahigh, alow, for high- and low- abundance species H and L, respectively; b, catalytic sites on enzymes available for each digestion event; ne, nequal, the digestion event when a = b.
We discovered that detergent DDM is omni-compatible with membrane protein structures, digestion, TMT, RP HPLC MS and data-dependent MS/MS (supplemental Fig. S1D and S2D), and pivotally enables automation throughout the protein-(HDX or Ox labeling)-peptide-(TMT)-HPLC-MS/MS process. Including DDM for digestion ensured effective catalytic contacts by solubilizing hydrophobic substrates, and eliminated steps of denaturation, precipitation, filtration, centrifugation, desalting and reconcentration. Avoiding pre-HPLC sample loss from the pipeline eased quantitation of PTM % site occupancy. Nonionic DDM didn't interfere with the peptide ionization or RP-HPLC elution, when applied at 0.4–1 mm (0.02–0.05%, m/v, 2.3–5.7 cmc) over 1 μm (0.28 mg/ml) proteins. Excessive TMT may label DDM hydroxyls at slower rate than peptides, but the byproducts assume simple forms and elute away from peptides with moderate intensity (supplemental Fig. S2D, S2E, S2F), demonstrating for the first time a TMT-compatible detergent. Other high-purity low-cmc nonionic nonamine glycoside detergents likely fit TMT as well. This compatibility may allow TMT to directly label protein structures in detergent solution and in membrane.
As a general platform for membrane protein digestion, FDD is flexible to use various enzymes and recent detergents, which should be a thoughtful choice to suit specific studies. We chose pepsin that prefers multiple hydrophobic residues (FWLMC (39)) to promote comprehensive and overlapping (ladder) peptide coverage—for HDX and PTM mapping, though other proteases fit the FDD format as well. Fast enclosed FDD reactors ensured quantitative reproducible peptide formation by preventing product inhibition and precisely controlling peptide formation via flow rate, and minimized modification artifacts (gain or loss) and contamination. FDD thereby freed those data-dependent MS/MS scans—gained from faster-scan instrument, yet otherwise wasted by contaminant peptides from proteases, de-PTM enzyme, environment and artificial oxidation—for endogenous components, benefitting deeper sequence coverage. Importantly, beyond these straightforward advantages, FDD's pepsin plug-flow reactor presented a novel approach to overcome abundance bias and enhance efficiency, primarily by increasing effective enzyme concentration (over 1 mm (33) on POROS beads) to the large excess of substrates (1 μm starting protein) (supplemental Note S2).
We defined protein abundance-biases on digestion at two levels (Fig. 1D and supplemental Note S2): Type I (∑Hpept/∑Lpept)≠(Hprot/Lprot), the abundance ratio of total peptide products (all forms) for high (H)- and low (L)-abundance species doesn't maintain that of original proteins, which concerns quantitation by PSMs, and Type II (Hpepti/Hprot)≠(Lpepti/Lprot), the transmission of a given peptide (harboring residue of interest i) from its original protein varies with protein's relative abundance, which may skew peptide-centric quantitation between different samples. According to a catalytic site-occupancy model that we proposed for digestion in pepsin plug-flow reactor (supplemental Note S2), FDD pepsin column doesn't incur either type of bias (Fig. 1D), and can operate entirely within the high-efficiency region (a ≤ b).
FDD chose not to perform the traditional reduction and alkylation on intact (or denatured aggregated) proteins for multiple reasons. (1) Protein reduction/alkylation interrupts C-C bonds—functionally crucial structure features of both Cys-loop receptors and hGPCRs—thus is incompatible with structural proteomics. HDX hardly tolerates basic-pH alkylation, because prelabeling protein reduction/alkylation may distort protein conformation, and postlabeling pre- or postdigestion basic pH causes rapid loss of H/D-labels to back-exchange; but HDX can tolerate a postlabeling fast acidic reduction step co- or postdigestion. (2) For the purpose to identify Cys-sequence: at protein level, both reduction and alkylation reagents' accesses to intricate intact hLGICs are spatially biased and inefficient, and disrupting C-C bonds may collapse protein structures further challenging reagent access, as evidenced by the missing Cys-coverage in our earlier in-gel study (34). (3) For the purpose to prevent TM protein aggregation formed by random C-C bond concatenation (such as alkylation applied in high-concentration protein purification for crystallogenesis): samples for proteomics are far more diluted, and DDM as a tool for digestion already proved effective to stabilize proteins and prevent aggregation, whereas reduction/alkylation may rather pose bigger risks to destabilize and aggregate TM proteins.
Instead, to solve the challenge to efficiently digest and identify TM protein sequences spatially hindered by intricate C-C bonds, FDD applied codigestion TCEP peptide reduction, throughout the sequential digestions along the DDM-facilitated protease column to assist peptide formation, and applied postdigestion peptide alkylation to visualize peptides in deep sequencing. We reasoned that, C-C bond reduction alone may be sufficient to dissect C-C intricacy for easier digestion access—as substrates flow through the protease column with ever-present TCEP, they become increasingly simpler and more accessible for effective reduction and digestion—whereas alkylation can be used to aid Cys-peptides ionization and identification. Thus reduction and alkylation may be separately utilized in deep-sequencing-based HDX (for iteratively constructing deep MS/MS peptide matrix, and for quantifying HDX MS labels). For non-HDX applications, in-reactor codigestion TCEP and alkylation could be combined, though this might affect enzyme reactor over repeated use.
Distinct from conventional 1–2 day, denaturation-, solution-, or filter-based methods, FDD emphasized a critical new principle for membrane proteins: using all enzymes in (1) nondenaturation (at least nonaggregation) and (2) high-efficiency, precisely controllable, abundance-unbiased flow reactor formats. Unlike denaturation, FDD allowed direct proteomic measurement of protein solutions at functional state without sample loss; FDD overcame the catalytic site deficiency and slow diffusion problems intrinsic to traditional in-solution digestion (protease: substrate at 1:50 or lower, m/m). Compared with laborious tip- or filter-based methods, FDD avoided filtration and centrifuge entirely—allowing uninterrupted streamline—shifted peptide preparation from day(s) to minutes, and minimized artifacts.
Compared with earlier 2010 GPCR HDX digestion method that used single pepsin column, desalting trap, and high flow rate ESI (50 μl/min) yet endured near-baseline peptide signals (16), by avoiding desalting and adopting capillary HPLC nanoESI, FDD increased peptide ion sensitivity by over 30,000-fold and demonstrated DDM is a superior tool widely applicable to proteomics; by novel combinations with PNGase F reactor, selective reductions, alkylation and direct TMT, FDD instantly achieved high coverage at PTM and Cys sites and immediate TMT-quantitation. Proved by DDM/CHS-purified α1β3γ2 GABAAR/HEK293 and its associated proteins, FDD cleared most prominent challenges for human synaptic membrane proteins, and may be adaptable to the protein-peptide-injection step for more complex samples such as cell lysates and biofluids, with finer HPLC separation.
Closing Coverage Gaps by Targeting PTMs
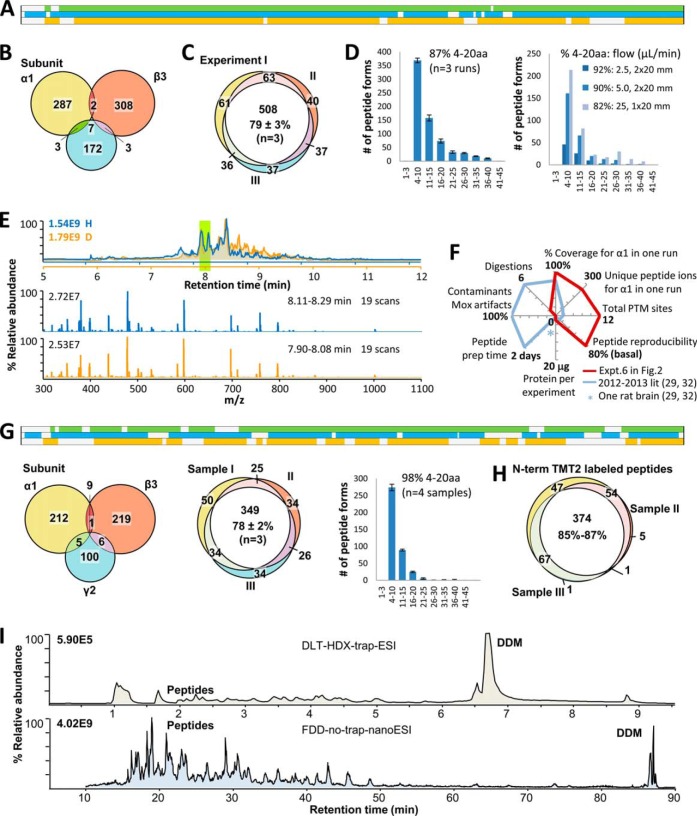
FDD with pepsin PNGase F columns and Cys-alkylation achieved high sequence coverage for all subunits of GABAAR (Fig. 2A), completely digested all proteins within sub-10-s column residence time (100%, shown by gel Fig. 2B and 85-min HPLC MS/MS supplemental Fig. S1D; supplemental Note S1), and minimized contaminations common to traditional methods: searching the HCD MS/MS data against human proteome showed GABAAR subunits were the predominant top 3 components by PSMs and unique peptides (Fig. 2C).
Fig. 2.
Direct FDD achieved ultradeep coverage of GABAAR, using module 1 in Fig. 1B. A, Closing sequence coverage gaps by targeting PTMs: N-glycosylation (N-glyco) via PNGase F column, disulfide bond (C-C) and C-palmitoylation (C-palm) via differentiating reducing reagents (TCEP, DTT and NH2OH: Expt. 4, 5 and 6 respectively). K-modifications were checked for excessive alkylation (K-Cb, Expt. 6′). Color bands represent sequence covered for each subunit (green, α1; blue, β3; orange, γ2L) in each experiment (1 to 6′); gray bands in background, genomic sequences supplied for overexpression including signaling peptides (black triangle). Key parameters for each experiment are specified in the table. Expt.3 was partly presented in Ref. 34, and is included here for comparison. TM, transmembrane helix (blue solid line); ICL, intracellular loop (red dash line); C-C loop (cyan line), used throughout figures. B, SDS-PAGE of GABAAR before (lane 1) and after (lanes 2 and 3) enzyme columns showed complete digestion within 10 s of column residence, and no intact enzymes. Each well contained one–twofold of the equivalent amount of digest injected to one HPLC run. C, Searching Expt. 6 MS/MS spectra against human proteome revealed minimal contamination from environment or protease: α1, β3, and γ2L subunits of GABAAR were the predominant top 3 by PSMs and unique peptides. A few PSMs matched to pepsin were from porcine pepsin.
Sequence coverage and number of peptides surged for all subunits upon optimized pepsin column flow rate in direct analysis (Fig. 2A Expt. 1–3). But several gaps persisted in ECD, coinciding with Asn(N) or Cys (Fig. 2A Expt. 3). Subsequent de-N-glycosylation and Cys-alkylation closed most ECD gaps (Fig. 2A Expt. 4), indicating: pepsin column and low TCEP (5–7.5 mm) effectively formed N-glycosylated peptides and reduced C-C bonds, but needed glycan removal and alkylation to visualize these residues (Cys rarely detectable as free (16)). Low TCEP was chosen from empirical screening in earlier GPCR HDX study (16), here by probing the digest with a postdigestion alkylation step, we recovered nearly all ECD Cys as Cys-Cb with many redundant partially overlapping peptide forms, which ranked among the highest-scored PSMs. Hence strong ECD Cys coverage confirmed that codigestion low TCEP in enzyme reactor was sufficient, and that peptide alkylation was necessary and effective to identify Cys. Missed TM and ICD Cys were likely caused by unknown modifications or over-digestion. Thus, FDD offered a novel solution to robustly digest spatially PTM-hindered regions including the vicinity of bulky N-glycans and intricate C-C bonds, a long-established challenge (25).
Unlike α1 and β3, γ2 intracellular loop 2 (ICL2) displayed a unique wide gap containing multiple Cys sites (Fig. 2); two of their equivalents in rat brain were identified with C-palmitoylation using NH2OH-based method (40). NH2OH, but not DTT or just TCEP, elicited multiple new peptides bearing C-Cb at these two sites, suggesting C-palmitoylation/acylation at high occupancy (Fig. 2A Expt. 6 versus Expts. 4–5; peptide MS/MS shown in supplemental Fig. S3). By now FDD covered 99%, 93 and 88% for α1, β3 and γ2, respectively (Fig. 2A Expt. 6). Searching for K-Cb didn't recover ICL2 gaps, clarifying minimal over-alkylation artifacts (Fig. 2A Expt. 6′). Broad PTM screening (see Methods) did not further increase the overall coverage.
Other few gaps dotted the TM domain (TM2-TM3 linker front, multi-F TM3 front), and had pepsin high-preference sites before/within/after them. Both gaps (such as AF, YA, LRL, and WRL) and identified TM peptides were peculiarly short (Fig. 2A Expt. 6 and supplemental Fig. S1). Rather than common under-digestion, these TM and ICL2 gaps likely stemmed from peptides escaping HPLC MS/MS capture, caused by low ionization efficiency and over-digestion because of high pepsin preference, similar to that seen in TM1 of GPCR β2AR (16), as well as from unknown modifications. This can be improved partly by longer HPLC gradient, colder or faster digestion.
Efficient digestion assisted deep PTM mapping. First, for PTM-enzyme-based site marking, such as PNGase F-specific deamidation, PTM-enzyme access to peptides is likely less confounded by spatial hindrance thus more reliable and effective than access to intact proteins. Second, forming multiple overlapping peptides for each site—rather than commonly applied single peptide sequence—increased the redundancy and power for accurate PTM measurement. Third, these glycopeptides at currently small yet tunable lengths, may further suit iterative direct ETD and HCD MS/MS to sequence both amino acids and modifiers.
However, typical to bottom-up proteomics, short discrete peptides often lose isoform-specific combinatorial PTM information that can be retained in top-down analysis of intact proteins: FDD's potential to tune for longer overlapping peptides might suit additional middle-down analysis to alleviate this problem. Unlike laborious, day-long, aggregation-plagued paradigm, FDD's instant leak-free and automatable protein-peptide pipeline may allow real-time bottom-up integration with top-down, to refine isoform-precise membrane protein PTM maps.
Digestion Coverage of Spatially Hindered N-glycosylation Sequences
Importantly, FDD's design—DDM-facilitated, flow-propelled, sequential multiple digestions through tandem high-concentration immobilized protease- and PNGase F-reactors, codigestion TCEP reduction, and peptide alkylation—resolved the challenge to robustly digest and identify spatially intertwined N-glycosylation sequences—using, remarkably, minutes, not 1–2 days. This helped refine a common speculation that PNGase F does not react at certain NXS/T sequences of N-glycopeptides.
Substrate sequence bias of PNGase F was raised using synthesized small glycopeptides (1–5 amino acids, 18–24 h): reaction with NXS/T was much faster than other sequences, though differed less between NAS/T (41). But a later proteomic study found prior (versus post) guanidine-reduction-alkylation treatment of soluble antibodies removed such sequence bias against NCG in 2-h PNGase F reactions (42), indicating minimal effect of primary sequence once higher-order spatial hindrance was solved. Consistently, Plummer et al. and Chu reported more bias at protein conformation than at peptide level. They found although PNGase F broadly recognizes N-GlcNAc-GlcNAc and immediate amino acid residues, native-folded full-length (soluble) proteins were much slower and less complete than detergent-spread versions (43–47). PNGase F didn't mandate residues before or after N-glycan though reaction could be much slower (41, 46, 47), but became nonreactive when the first N-GlcNAc was fucosylated via α1→3 (not α1→6), a link rare in mammals (48).
We noticed both human Cys-loop LGICs and hGPCRs contain NX(!P)S/T canonical N-glycosylation motifs that colocalize with C-C bonds at their most critical structural features (hLGIC Cys-loop, hGPCR ligand pocket lid), despite literature that C-C bond rigidity obstructed N-glycan occupancy (49). Our earlier in-gel study found weak evidence of two de-N-glycopeptides for the NCT on Cys-loop (common site to all GABAAR β subunits of human, mouse and rat), proving this N-glycosylation existed though occupancy was unclear (34). However, β NCT sequences were not covered in previous glyco-proteomic studies using mammalian tissue lysates or purified LGIC proteins (29, 50), which tends to suggest a PNGase F reaction bias over local sequence of glycosylated NXS/T at both protein and peptide levels despite hours of reaction. Yet direct analysis of lectin-enriched glycopeptides without PNGase F did not cover β NCT either (it found several NCS/T sites for non-GPCR non-Cys-loop-LGIC proteins) (51), suggesting the biased coverage might be caused by common upstream steps, but not the PNGase F-glycopeptide reaction per se.
Now FDD recovered multiple strong de-N-glycopeptides for NCT and other NXS/T sites. Although partly attributable to the higher efficiency of its peptide-level PNGase F column format, this suggests earlier biased weak or missing coverages likely originated collectively from ineffective upstream membrane protein reduction, alkylation and digestion, often following denaturation by urea, guanidine, boiling SDS, and protein precipitation, each of which severely damages access to hydrophobic and Cys sequences for subsequent steps. These contrasts highlighted the importance for membrane proteomics to think outside the box of denaturation (destabilization-aggregation) and to emphasize downstream compatibility in pipelines, as it zooms into the function- and therapeutic-pivotal sequences often involving intricate PTMs.
Strikingly, both N-version and d-version (PNGase F-specific deamidated original N-glyco version) of the same peptide sequences were identified for multiple N sites, permitting finer peptide peak area-based estimation of % occupancy (supplemental Table S3) and suggesting FDD is a highly efficient digestion machine despite N-glycans. We initially speculated, because N-glycans may pose spatial hindrance for protease access, the N-glyco peptides should be more difficult to form than the nonmodified N-peptides for all N sites, even though N local sequences appear regular in FWLMC density and multispecific pepsin alleviates the concern that PTMs prevent specific cleavage. If FDD pepsin column at current 25 μl/min overdigested, then N-peptides would be under-represented because of more loss to overdigestion than the N-glyco version, then N-glyco % occupancy would be over-estimated across all N-glyco sites. However, the relative peak abundance between the N and D pair varied widely across sites but remained largely consistent within the multiple peptide sequences for a given site (supplemental Table S3), suggesting N-peptides were unlikely random remains from disordered overdigestion. Nonetheless, both over- and under-digestion can be further probed by varying digestion flow rate. Because N-glycopeptides were visualized as de-N-glycopeptides, if PNGase F column reaction were incomplete, then % occupancy would be under-estimated. Because FDD solved other confounders (TM protein aggregation, ineffective Cys-reduction, digestion of intertwined regions, and Cys-alkylation), the performance of PNGase F-glycopeptide per se may be empirically tweaked apart by tuning PNGase F column flow rate and temperature.
Superior Metrics Satisfied Deep Sequencing, Peptide-centric Quantitation and Structural mapping
Using sub-10 s digestion time and sub-2 μg GABAAR for each run, FDD achieved: high sequence coverage including multiple new PTMs for all subunits (88–99%), abundant unique peptides at useful length (mostly 4–20 residues) (Fig. 3A, 3B, 3D and supplemental Fig. S1), and under HDX conditions, high peptide reproducibility (Fig. 3E), superior to traditional methods (Fig. 3F). Merits were preserved in Ox-TMT using module selection 3 in Fig. 1C (Fig. 3G; shorter peptides attributable to higher temperature). TMT labeled over 98% identified peptides (Fig. 3H). DDM, eluted in the end of HPLC, and CHS, added for human proteins, didn't interfere with peptide elution or TMT (supplemental Fig. S1D and S2D).
Fig. 3.
FDD demonstrated superior metrics for deep sequencing, quantitation and structural mapping. A, Robust 90–99% sequence coverage for all three subunits (combined search on spectra of Expt. 4, 5, and 6 in Fig. 2), using sub-2 μg GABAAR for each run. PSM distribution over sequence and HPLC-MS traces are shown in supplemental Fig. S1. B, Abundant and reasonable distribution of unique peptides for α1, β3, and γ2 subunits. C, Peptide reproducibility was 79 ± 3% between run 4, 5, and 6 (aliquots of the same deglycosylated digest treated with three different reducing regents), which served as a basal level of HPLC MS/MS run-to-run reproducibility. D, Peptide length fell mostly in the useful 4–20 residues range, and controllable by flow rate. Error bars represented standard deviations from runs 4, 5, and 6. E, HPLC and MS traces of two independent digestions under HDX conditions (module 2 in Fig. 1B), showed high reproducibility in both peptide form and abundance, suitable for HDX structural proteomics. F, Less is more: FDD metrics, critical for peptide-centric quantitative and structural proteomics, contrasting current literature. G, Superior metrics—high sequence coverage, abundant unique peptides, high peptide reproducibility (78 ± 2%) close to basal level, and desired peptide length distribution—were preserved in Ox-TMT quantitative structural mapping (module 3 in Fig. 1B), unaffected by DDM. Four aliquots of GABAAR were independently H2O2 oxidation-labeled, digested (24–30 °C), TMT2-labeled, and randomly paired as three TMT2plex samples for direct HPLC runs. PSM distribution over sequence and HPLC-MS traces are shown in supplemental Fig. S2. H, Streamlined TMT method effectively labeled over 98% of identified peptides in each sample, and 85–87% peptides were reproducibly formed, labeled, and detected between two samples. I, HPLC MS traces (total ion counts) contrasting peptide sensitivity in 2010 DLT-HDX-trap-ESI (upper, apo hGPCR β2AR) and in current FDD-no-trap-nanoESI pipeline (lower, hLGIC GABAAR), under similar concentrations of proteins and DDM.
Comprehensive PTM Inventory with % Site Occupancy: Critical for Function Insights
FDD enabled comprehensive identification and % occupancy quantitation of multitype PTMs (Fig. 4 and supplemental Table S3), a critical advantage for assessing whether PTM functions as structural hardwire or transient regulator. High-resolution MS/MS enhanced accuracy in identifying deamidation, decoy deamidation search on PNGase F nontreated sample helped eliminate non-PNGase F-specific sites (false positive), and minutes—not overnight digestion plus hours in PNGase F—of total preparation time for deglycosylated peptides likely reduced occurrence of nonenzymatic deamidation, together obviating H218O that complicates spectra. We discovered 3–5 major N-glycosylation sites and 1–3 clusters of K-Me/Me2/Me3/Ac in each subunit type, and two potential Cys-palmitoylation (acylation) sites exclusive to γ2. Without phosphatase inhibition, peptide enrichment and cell stimulation, ubiquitination, phosphorylation and R-Me/Ac were largely undetected at peptide FDR <1%. The exhaustive quantitative inventory revealed subunit-specific PTM signatures—mid-ECD high-occupancy N-glycosylation and ICD partial K-Me/Ac (Fig. 5). They likely drive subunit-biased assembly and ligand response in human Cys-loop LGICs, by redefining ECD inter-subunit and ligand interactions, and by mediating ICD ion passage and protein interactions as a feedback loop for signaling cascade, respectively. Further, complex PTMs of human TM receptors highlighted the edges of structural proteomics such as HDX to map their physiological states, because high-resolution crystallogenesis of TM receptors routinely requires truncation of flexible domains, PTM removal or control, and often resolves zero or a few residues of glycans (8, 9, 52), whereas NMR's reach for large, PTM-bearing TM receptors is limited despite mutagenesis, metabolic and chemical labeling tools.
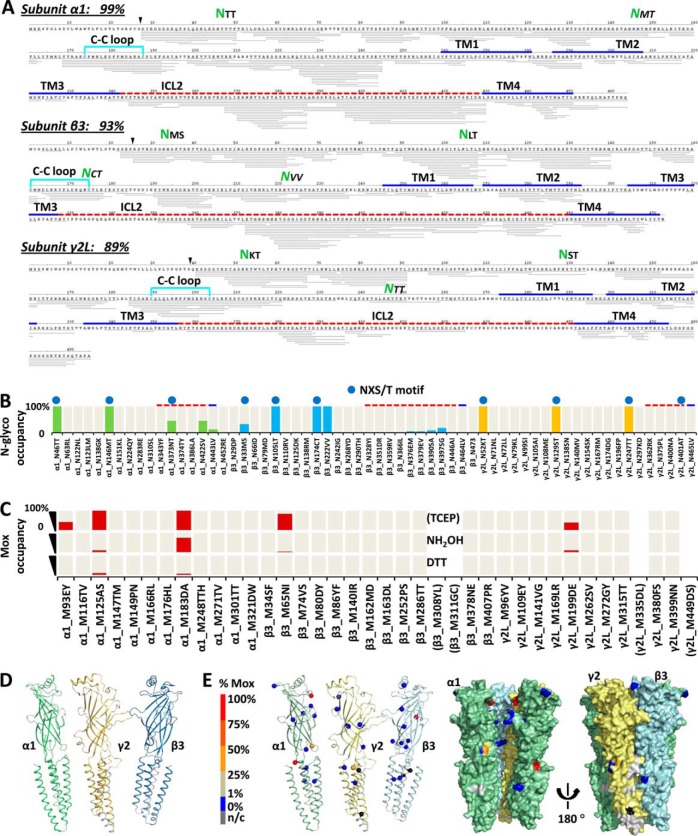
Fig. 4.
Artifact/bias-minimal FDD enabled comprehensive profiling of GABAAR PTMs with % site occupancy: N-glycosylation and endogenous M-oxidation. A, Representative peptide sequence coverage map of GABAAR acquired from one run (Expt. 6 in Fig. 2A) at peptide FDR<1% by PD 1.3 SEQUEST. Each thin line represents one unique peptide ion (modified versions combined). % coverage excluded expected signaling sequences. Green NXX highlights identified ECD N-glyco sites. Key domains were located by sequence alignment with C. elegans GluCl (3RHW) (8) (Methods and supplemental Fig. S5). B, Complete inventory of N-glyco % site occupancy. Top blue dot designates canonical NX(!P)S/T motif. Green, blue, and yellow bars, N-glyco % occupancy for α1, β3, and γ2 subunits, respectively; cream bars in background, N sites covered in PD1.4 quantitation. C, Inventory of Mox % site occupancy (red bar) using TCEP, TCEP+NH2OH, or TCEP+DTT for peptide reduction (Expt. 4, 6, and 5 in Fig. 2A respectively). Cream bars in background, covered in PD1.4 quantitation; brackets, not covered. D, Sequence coverage used for quantitation mapped to 3RHW-based 3D structures (Expt. 4–6 combined search at peptide FDR<1% by PD1.4 SEQUEST-HT). Green, α1; blue, β3; orange, γ2; silver, n/c, not covered; ICD was not shown. E, Endogenous Mox % site occupancy (sphere) by FDD-TCEP mapped to 3D models of individual α1, γ2, and β3 subunit (cartoon), and whole complex (surface). One β3 subunit was removed from the pentamer to show inner surface. N-glyco and Mox % occupancies were quantified by summing MS isotopic peak areas extracted at 2 ppm mass precision of the unmodified and modified version of the same peptide sequences: %(D/(N+D)), and %(Mox/(M+Mox)), respectively (Methods).
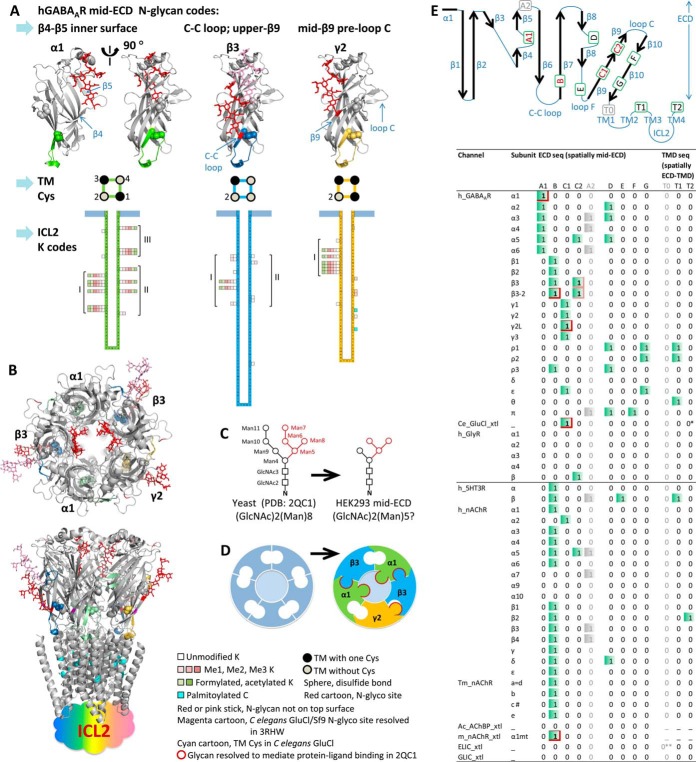
Fig. 5.
Novel subunit-distinctive PTM signatures discovered herein likely underpin high subunit-biases in hGABAAR assembly and ligand interactions. A, Novel PTM signatures for GABAAR subunits include: high-occupancy N-glycosylations in nontop-surface mid-ECD, and tentative diverse partial-occupancy K-Me/Ac in ICL2, in nonstimulated cells. TM Cys also displayed subunit-specific distribution patterns (supplemental Fig. S4C). 3D locations of mid-ECD N-glycosylations in GABAAR subunits were proposed by sequence alignment with 3RHW and 2QC1. N-glycans were superimposed to mid-ECD sites identified with high occupancy. B, Top and side views of proposed physical form of functional (α1)2(β3)2(γ2L)1 GABAAR pentamer completed with mid-ECD N-glycosylations. Top-surface N-glycans were not shown. C, Glycans shown for mid-ECD sites in hGABAAR/HEK293 were deduced by trimming that resolved in 2QC1 to (GlcNAc)2(Man)5. Immediate ligand-interacting saccharides resolved in 2QC1 were shown in red. D, Subunit interfaces in classic representation of clockwise αβαβγ GABAAR (left) can be upgraded with mid-ECD N-glycans (right, red border) to illustrate concepts of heterogeneity. E, Screening Cys-loop LGICs for NX(!P)S/T motif pinpointed 8 human mid-ECD coding regions from ECD sequence, based on alignment to 3RHW (diagram). Four mains involving αβγ-hGABAAR (red) are: A1, β4-β5 loop on inner surface (α); B, C-C loop (β); C1, preloop C mid-β9 (γ); C2, upper-β9 (β3 s site NVV). Four minors: D, β8-β8 loop; E, β8-β9 lower-loop F; F, upper-β10; G, lower-β10. TMD-originated sites for ECD-TMD interface regions: TM2/TM3 linker (T1) and TM4 tail (T2) (supplemental Fig. S4B). Red box, high-occupancy sites identified herein or by 3RHW and 2QC1; pink box, NVV. Gray, A2 is likely back to top surface (β5-β6, supplemental Fig. S4A) and T0 (TM1 front) is yet unseen though a potential site (** 0, because one NPS in ELIC). * One NAS in C. elegans GluCl β subunit but not in the 3RHW crystallized α-based construct. # One NGT on tip of loop C in fish Tm nAChR chain C (2BG9), absent in human.
A First Complete Quantitative Inventory of GABAAR N-glycosylation
FDD covered and quantified N-glycosylation % occupancy of all N residues, and found over ten mid-to-high-occupancy sites (Fig. 4A, 4B and supplemental Table S3), only two or four of whose equivalents were previously identified in rat (two in α1) or mouse (one in α1; three in γ2) (29, 50). Most N-glycosylation sites obeyed the canonical NX(!P)S/T motif and resided in ECD at high % site occupancy, but exceptions emerged (Fig. 4B). A few S/TX.NX(!P)X sites were found modified, including a high-occupancy Asn222VV in β3-ECD and multiple low-occupancy sites in α1 and β3. Besides ECD, several partial-occupancy sites were found in ICL2 of α1 (one, α1-Asn373NT at 44%) and β3 (four), yet Asn401AT in γ2-ICL2 was unmodified. Unlike all other ECD sites that showed near-full occupancy, β3-Asn33MS in ECD was modified to only 32% (two copies of the same subunits in the pentamer were undistinguished).
Noncanonical motifs NXV, NXC, and NGX were reported in mouse, using overnight urea trypsin, Glu-C or Lys-C digestion, 1-h lectin-glycopeptide enrichment and 3-h H218O-PNGase F release (50). Importantly, direct ETD MS/MS of glycopeptides, lectin-enriched from mouse brain synaptosome trypsin digest, identified the mouse version of GABAAR β3-Asn222VV bears (GlcNAc)2(Man)5 (51), directly proving a noncanonical NVV glycosylation, and explained it with the S/TX.NXX reverse motif, which was proposed earlier for human antibodies based on CID and ETD MSn of lectin-enriched endoglycosidase-trimmed glycopeptides and CID of de-N-glycopeptides from H218O-PNGase F-treated proteins (42, 53). However, most other ECD domain S/TX.NXX sites remained unmodified here (Fig. 4B), suggesting reverse motif requires further constraints. Multiple noncanonical NS/TX and NXXS/T sites (54), near-200 XX.NPS/T and XX.NXX sites (55), and NXC (56) were also observed in human breast (54), prostate (55) and lymphocyte (56) cancer tissue or cell culture respectively, using classical de-N-glycopeptide approaches via: overnight denaturation-based digestion, glycopeptide enrichment by periodate-oxidation/hydrazide-beads or lectin, and PNGase F release in H2O or H218O (54–57). However, extensive bead washing combined with sugar-specific enrichment couldn't exclude all non-PNGase-caused false positives, hours of H218O-PNGase F couldn't distinguish spontaneous deamidation that occurred during this incubation (58), and the reliance on glycopeptide enrichment and PNGase F release made a nontreated control difficult. Because FDD has proved robust to instantly form glycopeptides at small sizes likely analyzable by direct ETD to reveal both glycan and peptide sequences, FDD coupled with large-scale sampling and endoglycosidase may provide a tool to further refine these important noncanonical motifs.
Critically, upon sequence alignment with resolved crystal structures of analogs (3RHW and 2QC1), we discovered: not all ECD N-glycosylation sites resided on top surface, but each subunit contained one (α1 and γ2) or two (β3) sites intriguingly buried amid ECD (Fig. 5 and supplemental Fig. S4A and S5). Uniquely, α1-Asn146MT lined the mid-ECD inner surface of central pore, on a β4-β5 loop that is highly diverse among C. elegans GluCl, mouse α1 AChR-ECD and other Cys-loop LGIC subunits (8, 38). The high-occupancy β3-Asn174CT glycosylation, on Cys-loop arm, was unknown until FDD robustly covered Cys-loop (weak evidence in our in-gel study (34)), but its counterpart in a single-subunit mouse α1 AChR-ECD/yeast was resolved by crystallography as N-(GlcNAc)2(Man)9 to mediate protein-ligand binding and connect Cys-loop with loop C (38) (three saccharides of its counterpart in an ICD-truncated (β3)5 hGABAAR/HEK293-GnTĪ were also resolved by crystallography to interact with adjacent protein (9) during earlier editorial review of this manuscript). This reminded us that both modifications can be cotranslational and dynamic, and highlighted the impact of resolving this digestion bottleneck. A second site β3-Asn222VV (absent in other β subunits and not resolved by crystallography) was located on upper-β9 sheet, right preceding loop C—the prototypic agonist pocket in C. elegans GluCl (8) and Aplysia acetylcholine binding protein (AChBP) (52). The γ2-Asn247TT sat on mid-β9 sheet preceding loop C, lower than β3-Asn222VV but more involved in protein grooves. Mid-ECD N-glycan was absent in bacterial analogs and Aplysia AChBP/HEK293-GnTĪ (one N-hepta-saccharide on top-surface) (52), and was partially resolved at a single site on lower-β9 sheet in C. elegans GluCl/Sf9 (8) (Fig. 5B).
Mid-ECD N-glycosylations May Underpin Subunit-biased hLGIC Assembly and Ligand Response
Most of the protein/ligand-interacting saccharides in mouse α1 AChR-ECD/yeast (38) are conserved in mammalian cells, where the basic N-glycan may carry myriad further decorations (24) (Fig. 5C). Apparently by volume, ECD central pore is reluctant to fit two adjacent, or more than two, N-glycans while still maintaining stable subunit-subunit contacts (Fig. 5B). Thus we propose α's unique central-pore mid-ECD N-glycan may decide the long-enigmatic αXαXX assembly of hGABAAR pentamers (X represents non-α subunit); it further creates asymmetry on the central pore surface thus may bias ligand passage and interaction there. Sequence alignment screening across human Cys-loop LGIC subunits for NX(!P)S/T motif pinpointed 8 tentative mid-ECD coding regions (four majors for αβγ-GABAAR described above, and four minors), supporting subunit-specific mid-ECD N-glycosylation as a general coding principle (Fig. 5A, 5E and supplemental Fig. S4B). Although absent in α1β3γ2-GABAAR, the four minor regions include: β8-β8 loop (D, outer-surface counterpart of β4-β5 loop, different within α and ρ subtypes), β8-β9 lower-loop F (E, unique to 5HT3R β), upper β10 (F, GABAAR π) and lower β10 sheet (G, GABAAR ρ1, ρ2, ε). Further, single-nucleotide variation analysis of human proteome found only two gains (one in α3, one in ε) and zero loss of NXS/T motifs in GABAAR subunits (59), suggesting functional essentiality of N-glycosylation for hGABAAR. Subunit-Western blot saw N-glycosylation shifts in several GABAAR subunits (sites unknown) in human Schizophrenia brains, despite constant protein levels (60). Antibody N-glycans proved powerful to enhance antibody-target binding (61). Thus beyond conventional roles for ER-cell surface trafficking, these mid-ECD N-glycans, as receptor's architectural hardwire, create enormous diversity at subunit interfaces beyond genomic sequence, likely redefine subunit-subunit and ligand-protein interactions—via relay or disruption, and provide a PTM means to strictly yet dynamically control heteromeric assembly and ligand response. The bulky presence of these subunit-distinct, strategically-positioned, interaction-rich mid-ECD glycans suggests the concept of heterogeneity at subunit interfaces in αβαβγ GABAAR can at least be upgraded to include N-glycans (Fig. 5D). Engineering these mid-ECD N-glycosylation signatures might achieve pentameric assemblies of design, a crucial yet long-elusive tool to clarify structural mechanisms of human Cys-loop LGICs.
Intriguingly, although absent in most Cys-loop hLGIC subunits (including hGABAAR α, β and γ) and not reported by proteomic literature, besides ECD, the TM domain (TMD) sequences of a few subunits also contain the NX(!P)S/T motif, spatially likely protruding to the ECD-TMD interface: on TM2/TM3 linker (GABAAR ρ1, ρ2, θ, and 5HT3R β), or TM4 tail (nAChR β2), again all absent in the resolved bacterial, C. elegans or fish analogs (T1 and T2 in Fig. 5E). If indeed these TMD-based ECD-TMD-interface N-glycans occur at high occupancy, they may interact with receptor ECD and similarly serve as novel levers to couple TM helix (thus TM ion channel) movement with N-glycan-mediated ECD subunit-ligand and subunit-subunit interactions.
Further, several 7TM human GPCRs (UniProt sequences) also exhibit non-N-terminal NX(!P)S/T motifs on TM/TM linkers, including TM4/TM5 extracellular loop 2 the prototypic lid of TM ligand pocket (such as NET.CC in original β2AR), and TM6/TM7 linker (such as in serotonin type 2 receptor), each intertwined with C-C bonds. The motif's presence differs between species and GPCR subtypes, yet most of current hGPCR crystallography, EM and NMR structures had genetically (Asn187ET to EET (62–65)) or enzymaticly (66) removed N-glycosylation, or resolved no TM/TM modification in Sf9 (67). Indeed, nonproteomic mutagenesis-function studies of hGPCR protease-activated receptor 1 showed its extracellular loop 2 N-glycosylation existed and affected ligand-regulated activity and internalization, functionally distinct from N-terminal ones that dictated surface expression (68).
Thus we speculate, these heretofore unconsidered mid-ECD, ECD-TMD and TM/TM N-glycosylation structural hardwires, may be a widespread strategy across human cell surface TM receptors (LGICs and GPCRs), to dynamically form species-selective diversity in subtype assembly and ligand regulation; once clarified with % occupancy, they may provide an urgently needed key to precisely fine-tune pharmacotherapy for humans. FDD's potential to cover both TMD and spatially intricate N-glycosylation affords proteomics a robust platform to quantitatively map N-glycosylation across TM receptors to further decode new functions.
Subunit-specific ICL2 K-PTM May Code Ion Flux Regulation via Charge and Volume Changes
PTM screening discovered clusters of subunit-variable K-methylation/acetylation in ICL2 at partial occupancy, in nonstimulated cells (Fig. 5A and supplemental Table S4). Unlike N-glycosylation, these K-dense sequences were covered without searching for K-PTMs, suggesting low occupancy. Most modified Ks exhibited multiple versions: Me1, Me2, Me3, Ac, Fm and free (K-Me/Ac). Subunit α1 contained three K-clusters (I, II and III) each spanning 10–20 residues: one (densest) in the first half of ICL2, and two in the second half; all showed K-Me/Ac. Subunit β3 had two K-clusters, one on each half of ICL2 at similar distances from TM domain, but only the first (denser) cluster carried modifications. By contrast, γ2 had only one K-cluster, densely modified, in the first half of ICL2 close to TM3, but instead multiple Cys in remaining ICL2. Previous high-urea Lys-C and trypsin in-solution digestions of five rats identified one peptide bearing two K-Ac in GABAAR—α5 in testis, but zero in brain, other tissues, or other Cys-loop receptors (69). These preliminary K-Me/Ac sites filtered at peptide FDR <1% can be further validated in future work, as PSM b and y ions covered partial peptide sequences (partly caused by charge sequestration effect of Ks in collision-induced HCD MS/MS), despite high mass accuracy of high-high MS/MS, exclusion of artifacts-prone reagents, multiple peptides (over 20 for γ2 ICL2 K-cluster), and search criteria comparable to previous studies.
ICL2 K-Me/Ac PTMs likely regulate Cl̄ ion passage by changing ICL2 volume (K-Me) or charge state (K-Ac). Dense ICL2 K/R-clusters in large excess over D/E are common to Cl̄ channels hGABAAR and hGlyR, but rare in cationic hAChR and h5HT3R (supplemental Fig. S6). Chemically or genetically changing the charges and volumes of α1-ICL2 Ks in GABAAR/HEK293 altered Cl̄ flux functions, particularly desensitization to agonist stimulation (70). The ICL2 K-Me/Ac discovered herein suggested evidence for this regulation mechanism in vivo, and upon further examining dynamics with ligand stimulation, may afford K-Me/Ac pathway intervention as a novel strategy for GABAAR therapy. Uniformly controlling ICL2 K-PTMs, instead of ICL2 removal, may assist structural studies.
A Method to Map hLGIC Endogenous Mox Site Occupancy
Minimizing artifacts, FDD conferred the sensitivity to differentiate peptide forms between TCEP, NH2OH and DTT (Fig. 4C), and allowed using oxidation to label structures. DTT reduced both C-C and Mox to near completion; NH2OH (0.7 m) partially reversed Mox besides C-de-palmitoylation (Fig. 4C). By contrast, TCEP specifically reduced C-C at high efficiency, while leaving Mox intact. These Mox modifications identified with TCEP were likely endogenous to GABAAR, and not protein or peptide processing artifacts, because: they showed high occupancy at selected sequences, most surface-exposed Ms remained unmodified (Fig. 4E), and peptide artifacts are typically ubiquitous (34). Broad Ox screening confirmed minimal hits in other locations, except for γ2-Trp295IN in TM1 end, and γ2-Tyr46EAsp48Tyr49ASNLys53TTrp55 preceding ECD helix α1 (supplemental Table S5).
Rapidly reversible by peptide-methionine-sulfoxide reductase (Msr, abundant in selected brain regions), Mox recently emerged to modulate ion channel permeation functions (71), and to engage in aging and neurodegeneration. However, because of extensive Mox artifacts in traditional protein and peptide sample preparations (such as loss to DTT, loss to reaggregation, and gain from O2), measuring in vivo Mox has relied on tedious Mox-differential CNBr cleavage, chemical labeling, or complicated controls. In contrast, by minimizing oxidation artifacts and preserving original Mox, FDD with TCEP presented a simple robust method to directly map endogenous Mox for TM receptors, and can measure consequent conformational changes using the HDX module (Fig. 1B). Most residues covered by FDD-TCEP, particularly those on channel inner surface and ECD/TMD interface, are unlikely accessible (let alone evenly) to bulky chemical labels applied to cells, membrane pellets, or intact proteins.
Bacterial to Human LGICs: Sophistication and Precision via PTMs
Discovering extensive subunit-specific PTMs likely resolved a critical paradox: striking ECD/TMD genetic or structural similarity, against prominent subunit-biases in assembly and ligand responses, among mammalian Cys-loop LGICs and their homomeric analogs in bacteria and C. elegans lacking such PTMs. Beyond conserved ECD/TMD scaffolds that perform essential ion flux functions, subunit-specific PTMs of hLGICs afford dynamic control of channel motions, by creating myriad diversities in protein-protein/ligand interactions in ECD and ICD—absent in bacterial analogs. Further complexity may reside in the subunit-distinct TM helical Cys distribution, again missing in bacteria (supplemental Fig. S4C). These PTM-conferred diversities had remained unknown and unconsidered in modeling subunit-biased ligand interactions of hLGICs using homomeric bacterial, C. elegans or Aplysia analogs (11, 72), hence they provide crucial guidance for future modeling.
Direct Ox-TMT is Viable for Conformation Clues on GABAAR
Although vexed by reactive residues' sporadic yet often function-relevant distributions, and by slow reactions (labeling and quenching), side-chain chemical structural labeling nonetheless provides an independent means to partially cross-examine HDX study. Our platform appeared viable for direct streamlined TMT-quantitation of chemical structural labeling without detergent removal (Fig. 1B). Proof-of-principle experiments of Ox-TMT on GABAAR showed that over 98% of identified peptides contained 127/126 peak pairs in MS/MS usable for quantitation, though contributed partly by overlapping isolation at 2 Da width. This platform can readily adopt multinotch MS3 acquisition to enhance TMT quantitation accuracy.
For structural relevance, oxidized sites in Ox-TMT of apo GABAAR were first pinpointed and occupancy approximated by precursor ion MS abundance. High H2O2 oxidized multiple residues besides M, including P, F, W, C, N, and D, prominently on ECD inner surface lining central pore, and around Cys-loop at the ECD-TMD interface (supplemental Fig. S7A). Oxidation also labeled mid-ICL2. Most H2O2-oxidized sites were detected at high occupancy or exclusively—highlighting the dilemma of comparing peptides between separate analyses without TMT. TMT could allow comparing all sample states for both ox and nonox fractions. The abundance ratios of MS/MS reporter ions 127/126 in the GABA-incubated (127)/apo (126) Ox-TMT2 sample were then distributed against protein sequence, and marked for oxidized peptides (supplemental Fig. S7B). Compared with apo 7′ (127)/apo 4′ (126) control (supplemental Fig. S7C), tentative regions with increased 127/126 ratio included: ECD α1 helix and β1-β2 sheets, particularly in subunit β3. Structural validity of oxidation labeling can be improved by using faster oxidation and quenching, such as OH radicals triggered by UV laser from lower-concentration H2O2, as applied in fast photochemical oxidation of proteins (FPOP) (73–75).
DISCUSSION
This pilot study established platform methods for systematic ultradeep-sequencing-based quantitative-structural mapping of human LGIC complex and its dynamics. Targeting enzyme-substrate contact and PTMs, we presented a novel ultradeep peptide preparation method for PTM-intricate membrane proteins—FDD, flow/detergent-facilitated digestions with tandem pepsin/PNGase columns and Cys-selective reduction/alkylation. FDD achieved ultradeep coverage by maximizing the efficiency to both form and detect endogenous peptides while minimizing artifacts. FDD reduced time from day(s) to minutes, cleared most bottlenecks long hampering membrane proteomics, and enabled automatable, instant quantitative-structural integration. Using sub-2 μg DDM/CHS-purified hGABAAR interactome, FDD demonstrated fast complete digestion, high peptide reproducibility, low bias, and minimal artifact contamination. Robust deep coverage afforded a first comprehensive view of hGABAAR pan-PTM with % site occupancy—including N-glycosylation, endogenous Mox and tentative K-Me/Ac, and discovered PTMs as novel subunit signatures that likely drive the subunit-biases in assembly and ligand responses of human Cys-loop LGICs, long-standing puzzles critical to precise drug discovery. These methods could adapt to complex samples such as cell lysates and biofluids.
Conceptually unique from laborious days-long paradigms, FDD's radically but necessarily distinct procedures solved multiple conundrums accepted largely for granted yet exactly causing current impasses to membrane proteins. Contrary to common denaturation-based digestions, applying activity/TMT/HPLC/MS omni-compatible DDM avoided centrifugation and desalting, and critically enabled instant automation for both quantitative (label-free and TMT) and structural (HDX) proteomics. For PTM-intricate membrane proteins, the FDD machine systematically tackled ultradeep peptide sample preparation via a novel dual strategy: (1) first form peptides efficiently by sequential micro-digestions through the TCEP/detergent-facilitated, protease-excessive plug-flow reactor, (2) then visualize PTM- and Cys-peptides using PNGase reactor and Cys alkylation, postprotease.
Both FDD reactors iteratively digest solubilized substrates: using ever-present TCEP and detergent as tools for digestions, substrates grow progressively simpler, more accessible and less spatially biasing. FDD pepsin column lifted digestion into the high-efficiency region, unrestricted by protein relative abundance or total sample size (supplemental Note S2), and pepsin reactor can strictly control and tune peptide lengths with flow rate, column size and temperature. Crucial for PTM-intertwined membrane proteins, peptide-stage reactions for C-C reduction, alkylation and PNGase enhanced efficiency and minimized spatial bias. When these reactions treat whole TM proteins predigestion, reagent accesses to substrates are often spatially hindered and biased, which is exacerbated by protein denaturation/aggregation causing polarized nonreaction versus over-alkylation. Given the scan-limited, fixed-total, data-dependent MS/MS acquisition defined by instrument scan speed, FDD uniquely deepened sequencing further by minimizing pre-HPLC contaminant and oxidation artifact peptides—that waste MS/MS scans, obscure endogenous peptide states yet plague paradigms—thus focusing scans on meaningful peptides; this approach complements downstream HPLC MS/MS optimization, faster-scan instruments and open data search. FDD matched the ultradeep peptide sample with higher-sensitivity readout by nondesalting capillary HPLC nanoESI high-high MS/MS.
This form-and-see dual-strategy fundamentally advances proteomics toward charting precise actionable hLGIC pan-PTM maps. It enabled FDD to robustly cover Cys-loop LGIC's N-glycosylation sites intertwined with C-C bond—an intriguing feature located at the most function-critical regions in both mammalian LGICs and GPCRs—that had escaped nearly all previous proteomic reports reliant on conventional predigestion denaturation, reduction and alkylation regardless of analyzing de-N-glyco- or glyco-peptides. FDD broke this digestion barrier and suggests: the common speculation that PNGase F may not cleave N-glycans at some amino acid sequences of N-glycopeptides can be refined, as inefficient upstream digestion may mask the effects of PNGase-glycopeptide reaction per se; technically high-confidence high-coverage bottom-up N-glycosylation mapping of membrane proteins is achievable.
For N-glycosylation that applies PNGase-deamidation to mark sites, FDD pipeline enabled ultradeep N-glycosylation occupancy mapping—a first/best-in-class for human TM receptors hLGICs/hGPCRs. Coupled with upstream effective glycopeptide formation, sequential multiple digestion chances along the PNGase F column enhanced reaction efficiency, contrasting in-solution or protein-stage reaction formats; FDD's tandem fast cold reactors minimized spontaneous deamidation artifacts during both protease (seconds at low pH) and PNGase F (minutes) treatments; the modular design simplified ± PNGase F comparison and easily provided a nontreated control of otherwise equal-state peptide samples. This initial N-glycosylation sitemap provides clear leads for next in-depth analysis of glycopeptides. Further, because the entire fast cold protein-peptide-injection pipeline limits spontaneous deamidation, FDD may suggest a potential means to unveil TM receptors' cell- and protein-endogenous deamidation—a modification involved in neurodegeneration and reversible by specific enzymes, but largely untapped to proteomics because of severe artifacts in the warm day(s)-long high-pH processing.
For Ox-modification (M, C), by avoiding both artificial Mox gain and losses, FDD-TCEP instantly unmasked protein-endogenous Ox landscape, a first for full-length hLGICs/hGPCRs. Fast cold enclosed reactors without peptide cleanup or concentration minimized artificial Mox gain from O2 and loss to reaggregation; choosing TCEP for C-C reduction—not DTT that reduces both C-C and Mox—avoided Mox erase; peptide-stage reactions eliminated protein conformation bias during the TCEP/DTT/NH2OH comparison. The Mox occurrence in hGABAAR appeared intriguingly location-selective yet unrelated to outer surface exposure, suggesting potential function relevance rather than artifacts incurred at the protein- or peptide-stage. FDD-TCEP affords a simple novel alternative to elaborate Ox-specific chemical tagging/enrichment approaches that presumed overwhelming oxidation artifacts from membrane protein-peptide sample processing are unsolvable.
The pan-PTM discovery is crucial to guide the next-level species- and subtype-precise hLGIC therapeutics. FDD ultradeep sequencing enabled proteomics to unveil concrete hLGIC pan-PTM landscapes to deduce novel structure-function clues inaccessible to conventional methods. Current hLGIC efforts toward heterogeneous precision rely on genomic homology, ignore PTMs (11, 72), and permit proteomics limited if any role—hampered by low, sporadic unreliable peptide coverage/reproducibility and severe artifacts despite elaborate costly procedures.
This timely discovery of hLGIC subunit-biased PTM signatures debuts PTMs besides phosphorylation—particularly mid-ECD N-glycosylation as variable integral hardwires, endogenous Mox, and ICD K-Me/Ac that switches K volume/charge—to decipher how hLGICs dynamically assemble and function. Unconventional nontop-surface N-glycosylations likely redefine subunit and ligand interfaces, constrain assembly, and connect different function domains. Protein-endogenous Mox map suggested a first tangible structural basis to explain the long-observed oxidation effects on channel ion flux function. Original evidence of subunit-biased ICD K-methylation and K-acetylation landscape provided a lead to examine the in vivo version of emerging function modulation by K-modifications. Therefore, FDD-proteomics starts to bridge the gap between genomic similarity and function diversity among species and hLGIC subtypes—decades-long enigmas critical to drive precise therapeutic breakthroughs. This pilot pan-PTM report provides both directions and platform methods for subsequent studies to refine the dynamic structure-function roles of each PTM in hLGIC.
In summary, FDD demystified PTM-intricate membrane proteins and offered a novel paradigm of peptide sample preparation to achieve real-time integrated ultradeep quantitative-structural membrane proteomics. This study established platform methods for human LGICs to systematically examine PTM states and dynamic interactions with ligands and other proteins, at ultradeep coverage and atomic clarity to guide next-level precise drug discovery.
Supplementary Material
Acknowledgments
I thank Dr. Keith W. Miller lab for providing GABAAR/HEK293-TetR cells and other materials used in this study and for supporting publication. I thank Ross Tomaino and Dr. Steven P. Gygi at the Taplin Biological Mass Spectrometry Facility of Harvard Medical School for HPLC MS and MS/MS data acquisition, and helpful discussions on experimental design and result analysis. I thank Bruce Pascal and Dr. Patrick R. Griffin at Scripps Florida for providing the HDX workbench program. I thank Drs. Pierre Thibault, Eric Bonneil and Frederic Lamoliatte for helping with Proteome Discoverer. I thank Drs. Shelagh M. Ferguson-Miller, Pierre Thibault and Keith W. Miller for comments on the manuscript. The late Khorana lab at MIT provided the HEK293-TetR cells.
Footnotes
Author Contributions: X.Z. conceived and designed study, performed experiments, analyzed data and wrote paper.
* This work was supported in part by the National Institute of General Medical Sciences (GM 58448, K.W.M.) and the Department of Anesthesia, Critical Care and Pain Medicine, Massachusetts General Hospital. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- TM
- transmembrane
- PTM
- post-translational modification
- FDD
- flow/detergent-facilitated protease and de-PTM digestions for deep sequencing
- DDM
- n-dodecyl-β-D-maltopyranoside
- CHS
- cholesteryl hemisuccinate
- TCEP
- Tris-2-carboxyethylphosphine
- HDX
- H/D exchange
- TMT
- tandem mass tag
- DLT
- DDM-low-TCEP digestion for HDX
- TMD
- TM domain
- ICD
- intracellular domain
- ICL
- intracellular loop
- ECD
- extracellular domain
- C-C
- disulfide-bonded Cys-loop
- h
- human
- LGIC
- ligand-gated ion channel
- GPCR
- G protein-coupled receptor
- GABAAR
- gamma-aminobutyric acid type A receptor
- β2AR
- beta-2 adrenergic receptor
- nAChR
- nicotinic acetylcholine receptor
- AChBP
- acetylcholine-binding protein
- GlyR
- glycine receptor
- 5HT3R
- serotonin type 3 receptor
- ER
- endoplasmic reticulum
- HEK293
- human embryonic kidney cell line 293
- MWCO
- molecular weight cutoff
- dial-filtration
- dialysis-filtration
- cmc
- critical micelle concentration
- RP
- reversed-phase
- HCD
- higher energy collisional dissociation
- ETD
- electron-transfer dissociation
- H-L
- high-low, high resolution MS-low resolution MS/MS scans
- H-H
- high-high, high resolution MS and MS/MS scans
- H
- high-abundance
- L
- low-abundance
- PSM
- peptide spectrum match
- FDR
- false discovery rate
- PD
- Proteome Discoverer
- Mox
- Methionine oxidation
- Cb
- carbamidomethylation
- N-glyco
- N-glycosylation
- Me
- methylation
- Ac
- acetylation
- Fm
- formylation
- GlcNAc
- N-acetylglucosamine
- Man
- mannose
- FA
- formic acid
- Cl-DMEA
- (2-chloro)-N,N-dimethylethylamine
- BrE-TMAB
- (2-bromoethyl)-N,N,N-trimethylammonium bromide.
REFERENCES
- 1. Pieper U., Schlessinger A., Kloppmann E., Chang G. A., Chou J. J., Dumont M. E., Fox B. G., Fromme P., Hendrickson W. A., Malkowski M. G., Rees D. C., Stokes D. L., Stowell M. H., Wiener M. C., Rost B., Stroud R. M., Stevens R. C., and Sali A. (2013) Coordinating the impact of structural genomics on the human alpha-helical transmembrane proteome. Nat. Struct. Mol. Biol. 20, 135–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Olsen R. W., and Sieghart W. (2009) GABA A receptors: subtypes provide diversity of function and pharmacology. Neuropharmacology 56, 141–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Marini C., Harkin L. A., Wallace R. H., Mulley J. C., Scheffer I. E., and Berkovic S. F. (2003) Childhood absence epilepsy and febrile seizures: a family with a GABA(A) receptor mutation. Brain 126, 230–240 [DOI] [PubMed] [Google Scholar]
- 4. Fowler C. D., Lu Q., Johnson P. M., Marks M. J., and Kenny P. J. (2011) Habenular alpha5 nicotinic receptor subunit signalling controls nicotine intake. Nature 471, 597–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ebert D. H., and Greenberg M. E. (2013) Activity-dependent neuronal signalling and autism spectrum disorder. Nature 493, 327–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baker M. (2010) Making membrane proteins for structures: a trillion tiny tweaks. Nat. Methods 7, 429–434 [DOI] [PubMed] [Google Scholar]
- 7. Bocquet N., Nury H., Baaden M., Le Poupon C., Changeux J. P., Delarue M., and Corringer P. J. (2009) X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature 457, 111–114 [DOI] [PubMed] [Google Scholar]
- 8. Hibbs R. E., and Gouaux E. (2011) Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature 474, 54–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Miller P. S., and Aricescu A. R. (2014) Crystal structure of a human GABAA receptor. Nature 512, 270–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Luscher B., Fuchs T., and Kilpatrick C. L. (2011) GABAA receptor trafficking-mediated plasticity of inhibitory synapses. Neuron 70, 385–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Corringer P. J., Poitevin F., Prevost M. S., Sauguet L., Delarue M., and Changeux J. P. (2012) Structure and pharmacology of pentameric receptor channels: from bacteria to brain. Structure 20, 941–956 [DOI] [PubMed] [Google Scholar]
- 12. Nobles K. N., Xiao K., Ahn S., Shukla A. K., Lam C. M., Rajagopal S., Strachan R. T., Huang T. Y., Bressler E. A., Hara M. R., Shenoy S. K., Gygi S. P., and Lefkowitz R. J. (2011) Distinct phosphorylation sites on the beta(2)-adrenergic receptor establish a barcode that encodes differential functions of beta-arrestin. Sci. Signal. 4, ra51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Park K. S., Mohapatra D. P., Misonou H., and Trimmer J. S. (2006) Graded regulation of the Kv2.1 potassium channel by variable phosphorylation. Science 313, 976–979 [DOI] [PubMed] [Google Scholar]
- 14. Lin D. T., Makino Y., Sharma K., Hayashi T., Neve R., Takamiya K., and Huganir R. L. (2009) Regulation of AMPA receptor extrasynaptic insertion by 4.1N, phosphorylation and palmitoylation. Nat. Neurosci. 12, 879–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huttlin E. L., Jedrychowski M. P., Elias J. E., Goswami T., Rad R., Beausoleil S. A., Villen J., Haas W., Sowa M. E., and Gygi S. P. (2010) A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang X., Chien E. Y., Chalmers M. J., Pascal B. D., Gatchalian J., Stevens R. C., and Griffin P. R. (2010) Dynamics of the beta2-adrenergic G-protein coupled receptor revealed by hydrogen-deuterium exchange. Anal. Chem. 82, 1100–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Orban T., Jastrzebska B., Gupta S., Wang B., Miyagi M., Chance M. R., and Palczewski K. (2012) Conformational dynamics of activation for the pentameric complex of dimeric G protein-coupled receptor and heterotrimeric G protein. Structure 20, 826–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chung K. Y., Rasmussen S. G., Liu T., Li S., DeVree B. T., Chae P. S., Calinski D., Kobilka B. K., Woods V. L. Jr., and Sunahara R. K. (2011) Conformational changes in the G protein Gs induced by the beta2 adrenergic receptor. Nature 477, 611–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Howery A. E., Elvington S., Abraham S. J., Choi K. H., Dworschak-Simpson S., Phillips S., Ryan C. M., Sanford R. L., Almqvist J., Tran K., Chew T. A., Zachariae U., Andersen O. S., Whitelegge J., Matulef K., Du Bois J., and Maduke M. C. (2012) A designed inhibitor of a CLC antiporter blocks function through a unique binding mode. Chem. Biol. 19, 1460–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Whitelegge J. P. (2013) Integral membrane proteins and bilayer proteomics. Anal. Chem. 85, 2558–2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Peng Y., Chen X., Zhang H., Xu Q., Hacker T. A., and Ge Y. (2013) Top-down targeted proteomics for deep sequencing of tropomyosin isoforms. J. Proteome Res. 12, 187–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rand K. D., Zehl M., and Jorgensen T. J. (2014) Measuring the hydrogen/deuterium exchange of proteins at high spatial resolution by mass spectrometry: overcoming gas-phase hydrogen/deuterium scrambling. Acc. Chem. Res. 47, 3018–3027 [DOI] [PubMed] [Google Scholar]
- 23. Jensen P. F., Larraillet V., Schlothauer T., Kettenberger H., Hilger M., and Rand K. D. (2015) Investigating the interaction between the neonatal Fc receptor and monoclonal antibody variants by hydrogen/deuterium exchange mass spectrometry. Mol. Cell. Proteomics 14, 148–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Molinari M. (2007) N-glycan structure dictates extension of protein folding or onset of disposal. Nat. Chem. Biol. 3, 313–320 [DOI] [PubMed] [Google Scholar]
- 25. Yates J. R., 3rd (2013) The revolution and evolution of shotgun proteomics for large-scale proteome analysis. J. Am. Chem. Soc. 135, 1629–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wisniewski J. R., and Mann M. (2012) Consecutive proteolytic digestion in an enzyme reactor increases depth of proteomic and phosphoproteomic analysis. Anal. Chem. 84, 2631–2637 [DOI] [PubMed] [Google Scholar]
- 27. Fonslow B. R., Stein B. D., Webb K. J., Xu T., Choi J., Park S. K., and Yates J. R. 3rd (2013) Digestion and depletion of abundant proteins improves proteomic coverage. Nat. Methods 10, 54–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Peng M., Taouatas N., Cappadona S., van Breukelen B., Mohammed S., Scholten A., and Heck A. J. (2012) Protease bias in absolute protein quantitation. Nat. Methods 9, 524–525 [DOI] [PubMed] [Google Scholar]
- 29. Chen Z. W., Fuchs K., Sieghart W., Townsend R. R., and Evers A. S. (2012) Deep amino acid sequencing of native brain GABAA receptors using high-resolution mass spectrometry. Mol. Cell. Proteomics 11, M111 011445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen E. I., Cociorva D., Norris J. L., and Yates J. R. 3rd (2007) Optimization of mass spectrometry-compatible surfactants for shotgun proteomics. J. Proteome Res. 6, 2529–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mellacheruvu D., Wright Z., Couzens A. L., Lambert J. P., St-Denis N. A., Li T., Miteva Y. V., Hauri S., Sardiu M. E., Low T. Y., Halim V. A., Bagshaw R. D., Hubner N. C., Al-Hakim A., Bouchard A., Faubert D., Fermin D., Dunham W. H., Goudreault M., Lin Z. Y., Badillo B. G., Pawson T., Durocher D., Coulombe B., Aebersold R., Superti-Furga G., Colinge J., Heck A. J., Choi H., Gstaiger M., Mohammed S., Cristea I. M., Bennett K. L., Washburn M. P., Raught B., Ewing R. M., Gingras A. C., and Nesvizhskii A. I. (2013) The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 10, 730–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yip G. M., Chen Z. W., Edge C. J., Smith E. H., Dickinson R., Hohenester E., Townsend R. R., Fuchs K., Sieghart W., Evers A. S., and Franks N. P. (2013) A propofol binding site on mammalian GABA receptors identified by photolabeling. Nat. Chem. Biol. 9, 715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang L., Pan H., and Smith D. L. (2002) Hydrogen exchange-mass spectrometry: optimization of digestion conditions. Mol. Cell. Proteomics 1, 132–138 [DOI] [PubMed] [Google Scholar]
- 34. Zhang X., and Miller K. W. (2015) Dodecyl maltopyranoside enabled purification of active human GABA type A receptors for deep and direct proteomic sequencing. Mol. Cell. Proteomics 14, 724–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dostalova Z., Zhou X., Liu A., Zhang X., Zhang Y., Desai R., Forman S. A., and Miller K. W. (2014) Human alpha1beta3gamma2L gamma-aminobutyric acid type A receptors: High-level production and purification in a functional state. Protein Sci. 23, 157–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Busby S. A., Chalmers M. J., and Griffin P. R. (2007) Improving digestion efficiency under H/D exchange conditions with activated pepsinogen coupled columns. Int. J. Mass Spectrom. 259, 130–139 [Google Scholar]
- 37. Pascal B. D., Willis S., Lauer J. L., Landgraf R. R., West G. M., Marciano D., Novick S., Goswami D., Chalmers M. J., and Griffin P. R. (2012) HDX workbench: software for the analysis of H/D exchange MS data. J. Am. Soc. Mass Spectrom. 23, 1512–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dellisanti C. D., Yao Y., Stroud J. C., Wang Z. Z., and Chen L. (2007) Crystal structure of the extracellular domain of nAChR alpha1 bound to alpha-bungarotoxin at 1.94 A resolution. Nat. Neurosci. 10, 953–962 [DOI] [PubMed] [Google Scholar]
- 39. Hamuro Y., Coales S. J., Molnar K. S., Tuske S. J., and Morrow J. A. (2008) Specificity of immobilized porcine pepsin in H/D exchange compatible conditions. Rapid Commun. Mass Spectrom. 22, 1041–1046 [DOI] [PubMed] [Google Scholar]
- 40. Kang R., Wan J., Arstikaitis P., Takahashi H., Huang K., Bailey A. O., Thompson J. X., Roth A. F., Drisdel R. C., Mastro R., Green W. N., Yates J. R. 3rd, Davis N. G., and El-Husseini A. (2008) Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature 456, 904–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fan J. Q., and Lee Y. C. (1997) Detailed studies on substrate structure requirements of glycoamidases A and F. J. Biol. Chem. 272, 27058–27064 [DOI] [PubMed] [Google Scholar]
- 42. Valliere-Douglass J. F., Eakin C. M., Wallace A., Ketchem R. R., Wang W., Treuheit M. J., and Balland A. (2010) Glutamine-linked and non-consensus asparagine-linked oligosaccharides present in human recombinant antibodies define novel protein glycosylation motifs. J. Biol. Chem. 285, 16012–16022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Plummer T. H. Jr., Elder J. H., Alexander S., Phelan A. W., and Tarentino A. L. (1984) Demonstration of peptide:N-glycosidase F activity in endo-beta-N-acetylglucosaminidase F preparations. J. Biol. Chem. 259, 10700–10704 [PubMed] [Google Scholar]
- 44. Tarentino A. L., Gomez C. M., and Plummer T. H. Jr. (1985) Deglycosylation of asparagine-linked glycans by peptide:N-glycosidase F. Biochemistry 24, 4665–4671 [DOI] [PubMed] [Google Scholar]
- 45. Chu F. K. (1986) Requirements of cleavage of high mannose oligosaccharides in glycoproteins by peptide N-glycosidase F. J. Biol. Chem. 261, 172–177 [PubMed] [Google Scholar]
- 46. Tarentino A. L., Trimble R. B., and Plummer T. H. Jr. (1989) Enzymatic approaches for studying the structure, synthesis, and processing of glycoproteins. Methods Cell Biol. 32, 111–139 [DOI] [PubMed] [Google Scholar]
- 47. Tarentino A. L., and Plummer T. H. Jr. (1994) Enzymatic deglycosylation of asparagine-linked glycans: purification, properties, and specificity of oligosaccharide-cleaving enzymes from Flavobacterium meningosepticum. Methods Enzymol. 230, 44–57 [DOI] [PubMed] [Google Scholar]
- 48. Tretter V., Altmann F., and Marz L. (1991) Peptide-N4-(N-acetyl-beta-glucosaminyl) asparagine amidase F cannot release glycans with fucose attached alpha 1–3 to the asparagine-linked N-acetylglucosamine residue. Eur. J. Biochem. 199, 647–652 [DOI] [PubMed] [Google Scholar]
- 49. Bause E., Hettkamp H., and Legler G. (1982) Conformational aspects of N-glycosylation of proteins. Studies with linear and cyclic peptides as probes. Biochem. J. 203, 761–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zielinska D. F., Gnad F., Wisniewski J. R., and Mann M. (2010) Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell 141, 897–907 [DOI] [PubMed] [Google Scholar]
- 51. Trinidad J. C., Schoepfer R., Burlingame A. L., and Medzihradszky K. F. (2013) N- and O-glycosylation in the murine synaptosome. Mol. Cell. Proteomics 12, 3474–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hansen S. B., Sulzenbacher G., Huxford T., Marchot P., Taylor P., and Bourne Y. (2005) Structures of Aplysia AChBP complexes with nicotinic agonists and antagonists reveal distinctive binding interfaces and conformations. EMBO J. 24, 3635–3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Valliere-Douglass J. F., Kodama P., Mujacic M., Brady L. J., Wang W., Wallace A., Yan B., Reddy P., Treuheit M. J., and Balland A. (2009) Asparagine-linked oligosaccharides present on a non-consensus amino acid sequence in the CH1 domain of human antibodies. J. Biol. Chem. 284, 32493–32506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Whelan S. A., Lu M., He J., Yan W., Saxton R. E., Faull K. F., Whitelegge J. P., and Chang H. R. (2009) Mass spectrometry (LC-MS/MS) site-mapping of N-glycosylated membrane proteins for breast cancer biomarkers. J. Proteome Res. 8, 4151–4160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu Y., Chen J., Sethi A., Li Q. K., Chen L., Collins B., Gillet L. C., Wollscheid B., Zhang H., and Aebersold R. (2014) Glycoproteomic analysis of prostate cancer tissues by SWATH mass spectrometry discovers N-acylethanolamine acid amidase and protein tyrosine kinase 7 as signatures for tumor aggressiveness. Mol. Cell. Proteomics 13, 1753–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Deeb S. J., Cox J., Schmidt-Supprian M., and Mann M. (2014) N-linked glycosylation enrichment for in-depth cell surface proteomics of diffuse large B-cell lymphoma subtypes. Mol. Cell. Proteomics 13, 240–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang H., Li X. J., Martin D. B., and Aebersold R. (2003) Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol. 21, 660–666 [DOI] [PubMed] [Google Scholar]
- 58. Hao P., Ren Y., Alpert A. J., and Sze S. K. (2011) Detection, evaluation and minimization of nonenzymatic deamidation in proteomic sample preparation. Mol. Cell. Proteomics 10, O111 009381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mazumder R., Morampudi K. S., Motwani M., Vasudevan S., and Goldman R. (2012) Proteome-wide analysis of single-nucleotide variations in the N-glycosylation sequon of human genes. PLoS ONE 7, e36212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mueller T. M., Haroutunian V., and Meador-Woodruff J. H. (2014) N-Glycosylation of GABAA receptor subunits is altered in Schizophrenia. Neuropsychopharmacology 39, 528–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Song R., Oren D. A., Franco D., Seaman M. S., and Ho D. D. (2013) Strategic addition of an N-linked glycan to a monoclonal antibody improves its HIV-1-neutralizing activity. Nat. Biotechnol. 31, 1047–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cherezov V., Rosenbaum D. M., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Kuhn P., Weis W. I., Kobilka B. K., and Stevens R. C. (2007) High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science 318, 1258–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bokoch M. P., Zou Y., Rasmussen S. G., Liu C. W., Nygaard R., Rosenbaum D. M., Fung J. J., Choi H. J., Thian F. S., Kobilka T. S., Puglisi J. D., Weis W. I., Pardo L., Prosser R. S., Mueller L., and Kobilka B. K. (2010) Ligand-specific regulation of the extracellular surface of a G-protein-coupled receptor. Nature 463, 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rasmussen S. G., DeVree B. T., Zou Y., Kruse A. C., Chung K. Y., Kobilka T. S., Thian F. S., Chae P. S., Pardon E., Calinski D., Mathiesen J. M., Shah S. T., Lyons J. A., Caffrey M., Gellman S. H., Steyaert J., Skiniotis G., Weis W. I., Sunahara R. K., and Kobilka B. K. (2011) Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature 477, 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shukla A. K., Westfield G. H., Xiao K., Reis R. I., Huang L. Y., Tripathi-Shukla P., Qian J., Li S., Blanc A., Oleskie A. N., Dosey A. M., Su M., Liang C. R., Gu L. L., Shan J. M., Chen X., Hanna R., Choi M., Yao X. J., Klink B. U., Kahsai A. W., Sidhu S. S., Koide S., Penczek P. A., Kossiakoff A. A., Woods V. L. Jr., Kobilka B. K., Skiniotis G., and Lefkowitz R. J. (2014) Visualization of arrestin recruitment by a G-protein-coupled receptor. Nature 512, 218–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wu B., Chien E. Y., Mol C. D., Fenalti G., Liu W., Katritch V., Abagyan R., Brooun A., Wells P., Bi F. C., Hamel D. J., Kuhn P., Handel T. M., Cherezov V., and Stevens R. C. (2010) Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science 330, 1066–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wacker D., Wang C., Katritch V., Han G. W., Huang X. P., Vardy E., McCorvy J. D., Jiang Y., Chu M., Siu F. Y., Liu W., Xu H. E., Cherezov V., Roth B. L., and Stevens R. C. (2013) Structural features for functional selectivity at serotonin receptors. Science 340, 615–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Soto A. G., and Trejo J. (2010) N-linked glycosylation of protease-activated receptor-1 second extracellular loop: a critical determinant for ligand-induced receptor activation and internalization. J. Biol. Chem. 285, 18781–18793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lundby A., Lage K., Weinert B. T., Bekker-Jensen D. B., Secher A., Skovgaard T., Kelstrup C. D., Dmytriyev A., Choudhary C., Lundby C., and Olsen J. V. (2012) Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell. Rep. 2, 419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. O'Toole K. K., and Jenkins A. (2011) Discrete M3-M4 intracellular loop subdomains control specific aspects of gamma-aminobutyric acid type A receptor function. J. Biol. Chem. 286, 37990–37999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hoshi T., and Heinemann S. (2001) Regulation of cell function by methionine oxidation and reduction. J. Physiol. 531, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mortensen M., Iqbal F., Pandurangan A. P., Hannan S., Huckvale R., Topf M., Baker J. R., and Smart T. G. (2014) Photo-antagonism of the GABAA receptor. Nat. Commun. 5, 4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hambly D. M., and Gross M. L. (2005) Laser flash photolysis of hydrogen peroxide to oxidize protein solvent-accessible residues on the microsecond timescale. J. Am. Soc. Mass Spectrom. 16, 2057–2063 [DOI] [PubMed] [Google Scholar]
- 74. Gau B. C., Sharp J. S., Rempel D. L., and Gross M. L. (2009) Fast photochemical oxidation of protein footprints faster than protein unfolding. Anal. Chem. 81, 6563–6571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Konermann L., Stocks B. B., and Czarny T. (2010) Laminar flow effects during laser-induced oxidative labeling for protein structural studies by mass spectrometry. Anal. Chem. 82, 6667–6674 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.