Abstract
Phosphoinositides are important molecules that serve as second messengers and bind to a complex array of proteins modulating their subcellular location and activity. The enzymes that metabolize phosphoinositides can in some cases serve to terminate the signaling actions of phosphoinositides. The inositol polyphosphate 5-phosphatases (5PTases) comprise a large protein family that hydrolyzes 5-phosphates from a variety of inositol phosphate and phosphoinositide substrates. We previously reported the identification of 15 putative 5PTase genes in Arabidopsis and have shown that overexpression of the At5PTase1 gene can alter abscisic acid signaling. At5PTase1 and At5PTase2 have been shown to hydrolyze the 5-phosphate from inositol phosphate substrates. We have examined the substrate specificity of the At5PTase11 protein, which is one of the smallest predicted 5PTases found in any organism. We report here that the At5PTase11 gene encodes an active 5PTase enzyme that can only dephosphorylate phosphoinositide substrates containing a 5-phosphate. In addition to hydrolyzing known substrates of 5PTase enzymes, At5PTase11 also hydrolyzes the 5-phosphate from phosphatidylinositol (3,5) bisphosphate. We also show that the At5PTase11 gene is regulated by abscisic acid, jasmonic acid, and auxin, suggesting a role for phosphoinositide action in these signal transduction pathways.
Phosphoinositides play a role in a variety of critical eukaryotic cellular processes (Stevenson et al., 2000; Meijer and Munnik, 2003). For example, phosphatidylinositol (4,5) bisphosphate [PtdIns(4,5)P2] serves as a precursor to second messenger molecules such as inositol (1,4,5) trisphosphate [Ins(1,4,5)P3], diacylglycerol, and, in animal cells, phosphatidylinositol (3,4,5) trisphosphate [PtdIns(3,4,5)P3]. In animal cells, PtdIns(4,5)P2 is also involved in regulation of the actin cytoskeleton, stress fiber formation, and membrane trafficking (Toker, 1998). In plants, although much less is known, evidence suggests that PtdIns(4,5)P2 plays an important role in actin cytoskeleton rearrangements via its binding to profilin (Staiger et al., 1994; McCurdy et al., 2001), which has been localized to the growing ends of root hairs (Braun et al., 1999) and pollen tubes (Kost et al., 1999). In fact, either hydrolyzing PtdIns(4,5)P2 or binding it via a plextrin-homology domain disrupts tip growth in pollen tubes (Kost et al., 1999), suggesting that availability of PtdIns(4,5)P2 is crucial to growth in these cells.
The enzymes that metabolize phosphoinositides such as PtdIns(4,5)P2 can control the amount of these molecules and, in general, are considered to be important targets for understanding disease and growth control (Pendaries et al., 2003). The inositol polyphosphate 5-phosphatases (5PTases) are a large group of enzymes that have the ability to hydrolyze 5-phosphates from a variety of inositol phosphate and phosphoinositide substrates (Majerus et al., 1999). Studies on human 5PTases have classified the 5PTases into four groups according to the substrates they hydrolyze in vitro. All 5PTases characterized to date contain a conserved catalytic domain referred to as the inositol polyphosphate phosphatase catalytic (IPPc) domain. In addition, the IPPc domain is often accompanied by other domains that presumably allow for modification of 5PTases or association with specific signal transduction components (Majerus et al., 1999).
We previously reported the identification of 15 putative 5PTase genes in Arabidopsis (Berdy et al., 2001). We and others have shown that overexpression of either At5PTase1 or At5PTase2 alters abscisic acid (ABA) signaling (Sanchez and Chua, 2001; Burnette et al., 2003). The At5PTase1 and At5PTase2 gene products have been characterized biochemically and shown to hydrolyze the 5-phosphate from inositol phosphate substrates (Berdy et al., 2001; Sanchez and Chua, 2001). We have examined the substrate specificity of another At5PTase, the At5PTase11 protein, which is one of the smallest predicted 5PTases found in any organism. We report here that the At5PTase11 gene (At1g47510) encodes an active 5PTase enzyme that has a different substrate specificity as compared to other plant 5PTases. We also show that the At5PTase11 gene is regulated by ABA, jasmonic acid (JA), and auxin, suggesting a role for phosphoinositide action in these signal transduction pathways.
RESULTS
Identification of a Unique At5PTase Gene
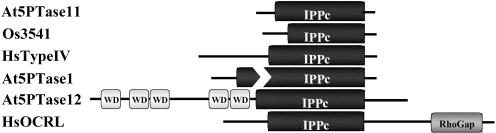
We previously identified two classes of plant 5PTases based on their phylogenetic relationships: the Group A 5PTases, such as At5PTase1 (At1g34120) and At5PTase2 (At4g18010), which contain only the IPPc domain; and the Group B 5PTases, such as At5PTase12 (At1g65580), which contain the IPPc domain and multiple N-terminal WD40 repeats (Berdy et al., 2001; Fig. 1). The At5PTase11 gene (At1g47510) is predicted to encode the smallest Group A 5PTase (331 amino acids, identified as AAD46036 in Berdy et al., 2001). At5PTase11 contains the conserved IPPc domain (amino acids 53–324; see supplemental data available at www.plantphysiol.org), and differs from the previously characterized Group A 5PTases in that it is not interrupted by a stretch of nonconserved residues (compare At5PTase1 and At5PTase11 in Fig. 1; supplemental data). By using BLASTp searches of the nonredundant protein database at the National Center for Biotechnology Information (NCBI), we determined the most closely related protein to At5PTase11 is the Os3541 protein (AAR01658) from rice (Oryza sativa). The Os3541gene is predicted to encode 301 amino acids, and has 50% identity to At5PTase11. The next most closely related protein to At5PTase11 is the yeast (Saccharomyces cerevisiae) 5PTase, Inp54 (CAA90520), which has 20.5% identity. This is similar to the 19.4% identity At5PTase11 shares with At5PTase1. Thus, At5PTase11 is more closely related to the rice Os3541 protein than it is to other Arabidopsis or yeast and animal 5PTases, indicating the possibility that At5PTase11 and Os3541 may be homologous genes.
Figure 1.
Domain structure of At5PTase11 and representative 5PTases. Amino acid sequences corresponding to At5PTase11 (AAD46036), the closely related rice gene Os3541 (ARR01658), At5PTase1 (At1g34120), At5PTase12 (At1g65580), the human Type IV 5PTase (NP_063945), and the human Type II 5PTase OCRL (Q01968) were analyzed with Pfam and SMART database tools, and the resulting domain structures are graphically presented. IPPc is the conserved 5PTase catalytic domain; WD corresponds to WD40 repeat regions.
We used reverse transcription (RT)-PCR to verify that the At5PTase11 gene is expressed in Arabidopsis tissues. Oligonucleotide primers were designed using the predicted cDNA sequence surrounding the start and stop codons. RNA from mixed Arabidopsis tissues, including seedling, roots, flowers, stems, and leaves, was used as template in an RT reaction. The products of this reaction were amplified with PCR using the At5PTase11-specific primers. The resulting At5PTase11 cDNA was sequenced and shown to be identical with the predicted cDNA (NM_103644) contained in the NCBI database. We conclude that the At5PTase11 gene is an expressed gene encoding a small and unique 5PTase.
At5PTase11 Encodes an Active 5PTase Enzyme
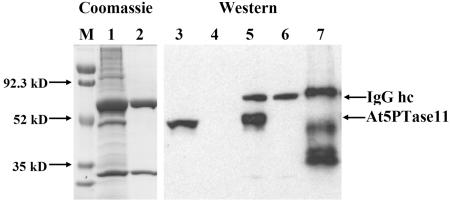
To obtain protein for 5PTase activity assays, the At5PTase11 gene was expressed as a C-terminal His fusion under control of the insect metallothionine promoter (pMT) in Drosophila melanogaster S2 tissue culture cells (S2 cells). We chose this strategy because previous efforts to obtain active, recombinant At5PTases in prokaryotic expression systems had failed, suggesting that a eukaryotic modification may be necessary for At5PTase activity. The At5PTase11 expression construct (pMTAt5PTase11) also contains a C-terminal V5 epitope tag, which we used for immunoprecipitation. We transfected S2 cells with either pMTAt5PTase11 DNA, a pMTβ-galactosidase (pMTβ-gal) control construct, or no DNA (i.e. mock transfection). After the transfection, we stimulated expression for 2 d with the addition of CuSO4. Protein extracts were made, and immunoprecipitation with an anti-V5 antibody was performed. Analysis of the soluble extracts prior to the immunoprecipitation revealed that the anti-V5 antibody did not react with any native Drosophila proteins (Fig. 2, lane 4). S2 cells transfected with the pMTAt5PTase11 construct, however, contained a recombinant protein of approximately 47 kD that reacted with the anti-V5 antibody. At5PTase11 is predicted to encode a 36.4-kD protein, and the addition of the C-terminal V5 and His tags is expected to increase the molecular mass. As is common with the addition of His tags, the apparent molecular mass of the recombinant At5PTase11 is slightly higher than expected.
Figure 2.
Expression and immunoprecipitation of recombinant At5PTase11 in Drosophila S2 cells. The pMTAt5PTase11 construct was transfected into S2 cells, and soluble extracts were prepared and immunoprecipitated with a monoclonal anti-V5 antibody as described in “Materials and Methods.” Immunoprecipitates from S2 cells transfected with pMTAt5PTase11 (lanes 1 and 5) and mock-transfected S2 cells (lanes 2 and 6) were stained with Coomassie dye after SDS-PAGE separation or were analyzed by western blotting with the monoclonal anti-V5 antibody. Lanes 3 and 4 contain a portion of the soluble protein extract from pMTAt5PTase11-transfected (lane 3) or mock-transfected (lane 4) S2 cells. Lane 7 contains purified Positope (Invitrogen) protein and was used to estimate the amount of recombinant At5PTase11 immunoprecipitated. Arrows on the left indicate the sizes of molecular mass markers (M); arrows on the right indicate the positions of At5PTase11 recombinant protein and the IgG hc.
To isolate the recombinant At5PTase11 protein, we used the anti-V5 antibody to immunoprecipitate At5PTase11 and analyzed the resulting complex with western blotting (Fig. 2). As expected, immunoprecipitated complexes from mock transfections (lane 6) or transfections with pMTβ-gal revealed the presence of the immunoglobulin heavy chain (IgG hc) from the anti-V5 antibody. Analysis of an immunoprecipitated pMTAt5PTase11 extract reveals the presence of an additional protein that is the same size as recombinant At5PTase11 protein (Fig. 2). To examine the content of our immunoprecipitations, we analyzed immunoprecipitated proteins from pMTAt5PTase11-transfected and mock-transfected S2 cells (Fig. 2, lanes 1 and 2, respectively). The immunoprecipitation from mock-transfected cells reveals the presence of the IgG hc and IgG light chain migrating at 55 and 25 kD, respectively. By contrast, an overloaded lane from an immunoprecipitation of pMTAt5PTase11-transfected cells contains one other major band, corresponding in size to recombinant At5PTase11. Since other background bands are not present in stoichiometric amounts with At5PTase11, we conclude that recombinant At5PTase can be expressed in S2 cells and immunoprecipitated in a fairly pure form.
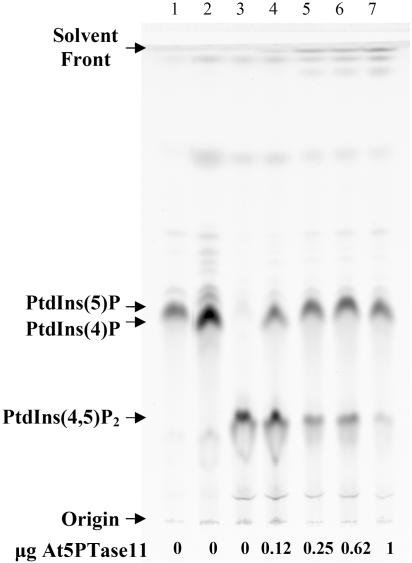
To determine whether At5PTase11 encodes an active 5PTase, we assayed for 5PTase activity directly following immunoprecipitation (Taylor and Dixon, 2001). We controlled for the amount of At5PTase11 added to activity assays by visual quantification of immunoprecipitates on SDS-PAGE gels, as shown in Figure 2, and by using a known amount of standard protein containing the V5 epitope tag (Fig. 2, lane 7). Figure 3 shows the results of incubating immunoprecipitates with a fluorescent PtdIns(4,5)P2 substrate. When PtdIns(5)P, PtdIns(4)P, or PtdIns(4,5)P2 is incubated under assay conditions in the absence of enzyme and analyzed by thin-layer chromatography (TLC), we obtain excellent separation between the mono- and bis-phosphorylated compounds. We obtained less separation of PtdIns(5)P and PtdIns(4)P. When 0.12 μg of At5PTase11 is incubated with PtdIns(4,5)P2, approximately 41% of the fluorescent PtdIns(4,5)P2 is converted to a product migrating in a similar position as PtdIns(4)P (Fig. 3, lane 4). When the amount of At5PTase11 is increased to 1 μg, the conversion of substrate to product is increased to approximately 90% (Fig. 3, lane 7). Immunoprecipitates from pMTβ-gal- or mock-transfected cells resulted in no conversion of substrate to product (Fig. 4; data not shown). This indicates that the At5PTase11 gene encodes an active 5PTase that dephosphorylates PtdIns(4,5)P2 in vitro.
Figure 3.
Effect of increasing At5PTase11 concentration on fluorescent PtdIns(4,5)P2 dephosphorylation. PtdIns(4,5)P2 fluorescent substrate (1.5 μg), or PtdIns(5)P and PtdIns(4)P standards (1.5 μg each), were incubated with 0 to 1.0 μg of At5PTase11 for 1 h at room temperature. Supernatants were processed and separated by TLC as described in “Materials and Methods.” Lane 1, PtdIns(5)P alone; lane 2, PtdIns(4)P alone; lane 3, PtdIns(4,5)P2 alone; lanes 4 to 7, PtdIns(4,5)P2 incubated with the indicated amount of At5PTase11 immunoprecipitated protein. To visualize fluorescent lipids, the TLC plate was analyzed with a Storm 860 (Molecular Dynamics).
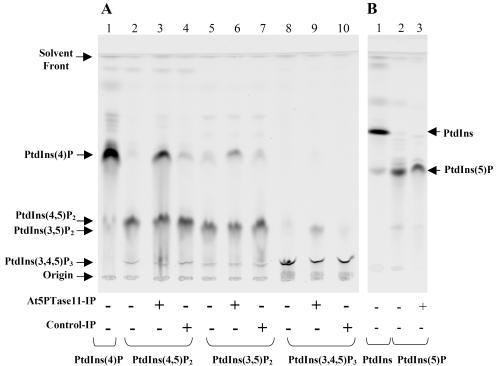
Figure 4.
Determination of At5PTase11 substrate specificity using fluorescent phosphoinositide substrates. A and B, Phosphatase reactions containing 1.5 μg of the indicated fluorescent phosphoinositide substrates were incubated in the same reaction conditions as described for Figure 3. A, Lanes 1, 2, 5, and 8 contain the substrate indicated at the bottom incubated in the presence of buffer only. Lanes 3, 6, and 9 contain the indicated substrate, buffer, and immunoprecipitated At5PTase11. Lanes 4, 7, and 10 contain the indicated substrate, buffer, and immunoprecipitates of mock-transfected S2 cells (Control-IP). B, Lanes 1 and 2 contain the PtdIns and PtdIns(5)P substrates alone. Lane 3 contains PtdIns(5)P incubated in the presence of immunoprecipitated At5PTase11. The migration of fluorescent standards and the origin are indicated (at left and right).
At5PTase11 Hydrolyzes Various 5-Phosphate-Containing Phosphoinositide Substrates
Studies of 5PTases from a variety of organisms have shown that the IPPc domain is capable of removing the 5-phosphate from both PtdIns(4,5)P2 and PtdIns(3,4,5)P3 substrates. To determine whether the At5PTase11 enzyme hydrolyzes phosphoinositide substrates other than PtdIns(4,5)P2, we examined hydrolysis of PtdIns(5)P, PtdIns(3,5)P2, and PtdIns(3,4,5)P3 by immunoprecipitates of pMTAt5PTase11- and mock-transfected S2 cells (Fig. 4). In all experiments, we incubated standards of fluorescent substrates and products in assay conditions and observed their migration on TLC plates within each experiment. As expected, the order of migration is PtdIns(5)P (fastest) > PtdIns(4)P > PtdIns(4,5)P2 > PtdIns(3,5)P2 > PtdIns(3,4,5)P3 (slowest). When immunoprecipitates from mock-transfected cells are incubated with each substrate, a small amount of product is observed, which corresponds to background hydrolysis (Fig. 4, Control-IP lanes). As expected from the results in Figure 3, when 0.25 μg of At5PTase11 is incubated with PtdIns(4,5)P2, conversion to product migrating at the same position as PtdIns(4)P is observed (Fig. 4A, lane 3). Using the same amount of At5PTase11 protein, we tested whether At5PTase11 would dephosphorylate PtdIns(3,5)P2. Figure 4A (lane 6) indicates that PtdIns(3,5)P2 is also dephosphorylated by the At5PTase11 enzyme to a greater degree than the background hydrolysis. Incubation of At5PTase11 with PtdIns(3,4,5)P3 also reveals conversion of this substrate, and the conversion is greater than what occurs in the absence of the enzyme (Fig. 4A, lane 9). The product of PtdIns(3,4,5)P3 dephosphorylation by a 5PTase is expected to be PtdIns(3,4)P2, which will migrate slower than PtdIns(4,5)P2 and PtdIns(3,5)P2. Since the observed product in lane 9 of Figure 4A migrates slower than the PtdIns(4,5)P2 and PtdIns(3,5)P2 standards, it suggests that removal of the 5-phosphate has occurred. Last, we tested whether PtdIns(5)P would be dephosphorylated by At5PTase11 and observed no increase in hydrolysis over background (Fig. 4B, compare lanes 2 and 3). From these experiments we conclude that At5PTase11 can dephosphorylate tris- and bis- but not mono-phosphorylated phosphoinositide substrates that contain a 5-phosphate.
At5PTase11 Does Not Hydrolyze Ins(1,4,5)P3 and Ins(1,3,4,5)P4 Substrates
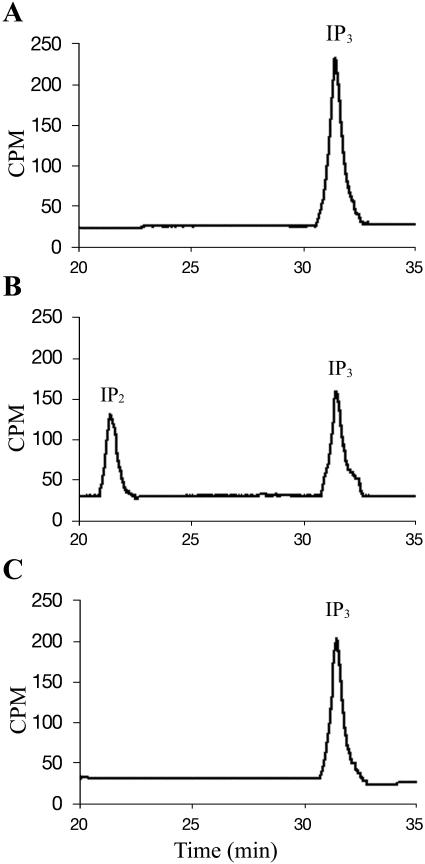
Since many 5PTase enzymes also hydrolyze inositol tris- and tetrakis-phosphate substrates containing a 5-phosphate, we tested the ability of At5PTase11 to dephosphorylate Ins(1,4,5)P3 and Ins(1,3,4,5)P4. For these experiments, we incubated immunoprecipitates from mock-transfected or pMTAt5PTase11-transfected S2 cells with [3H]myo-inositol (1,4,5)P3 or [3H]myo-inositol (1,3,4,5)P4. To analyze the products of these reactions, a small amount was injected onto a Spherisorb S5 SAX HPLC column, and separation of inositol phosphates was performed with an ammonium phosphate (AP) gradient (Stolz et al., 1998). Radiolabeled standards incubated under assay conditions were used to assign peak positions. When either At5PTase11 or no enzyme is incubated with [3H]myo-inositol (1,4,5)P3 substrate, no conversion to product [3H]myo-inositol (1,4)P2 takes place (Fig. 5, A and C, respectively). We saw a similar lack of hydrolysis when [3H]myo-inositol (1,3,4,5)P4 was incubated with At5PTase11 (data not shown). In these experiments, we used up to 10 times the amount of At5PTase11 protein as in the experiments testing phosphoinositide substrates and still found no hydrolysis. As a control for our assay conditions, we incubated 0.12 and 0.25 μg immunoprecipitated V5-tagged At5PTase1 with both [3H]myo-inositol (1,4,5)P3 and [3H]myo-inositol (1,3,4,5)P4. In each case, conversion to [3H]myo-inositol (1,4)P2 and [3H]myo-inositol (1,3,4)P3 took place as expected (Fig. 4B; data not shown). From these data, we conclude that, under our in vitro assay conditions, the substrate specificity of At5PTase11 differs from previously characterized At5PTase enzymes in that it does not hydrolyze the water-soluble inositol phosphate substrates Ins(1,4,5)P3 and Ins(1,3,4,5)P4.
Figure 5.
At5PTase11 does not hydrolyze [3H]Ins(1,4,5)P3. Immunoprecipitated At5PTase11 (0.25 μg) (A), At5PTase1 (0.25 μg) (B), or buffer (C) was incubated with [3H]Ins(1,4,5)P3. Products were separated using HPLC as described.
At5PTase11 Hydrolyzes PtdIns(4,5)P2 In Vivo in Drosophila Cells
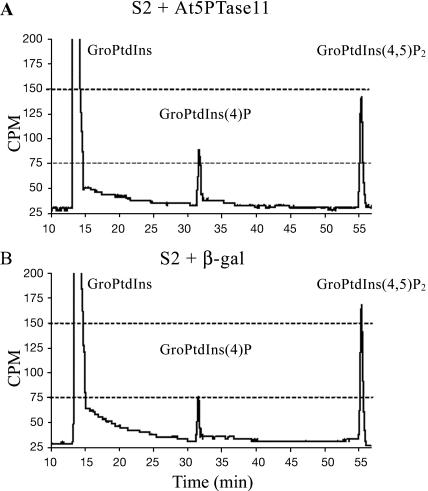
Our in vitro experiments with fluorescent phosphoinositide substrates have shown that At5PTase11 has the ability to hydrolyze PtdIns(4,5)P2, PtdIns(3,5)P2, and PtdIns(3,4,5)P3. To determine whether At5PTase11 can hydrolyze phosphoinositides produced within the cell (in vivo), we used [3H]myo-inositol labeling of S2 cells that were transiently expressing recombinant At5PTase11. S2 cells were transfected with pMTAt5PTase11 or pMTβ-gal, and 36 h postinduction, [3H]myo-inositol was added for 12 h. Deacylated phosphoinositides were then analyzed by HPLC and, in each case, data were normalized to the total [3H] counts in each run (Fig. 6). To identify the peak position of PtdIns(4,5)P2, we used a radiolabeled standard. To identify the peak position of PtdIns(4)P, we incubated extracts from labeled cells with a previously characterized, purified human 5PTase and examined the peak position of the resulting PtdIns(4)P product. Comparison of HPLC traces in Figure 6, A and B, indicates that PtdIns(4,5)P2 is reduced and PtdIns(4)P is elevated in cells transiently expressing At5PTase11. We have repeated this experiment multiple times and found that the PtdIns(4)P and PtdIns(4,5)P2 levels are always altered in cells transiently expressing At5PTase11. Specifically, levels of PtdIns(4)P are increased 15% ± 0.3% on average in our experiments, indicating that, in vivo, At5PTase11 hydrolyzes PtdIns(4,5)P2. We were not able to detect any PtdIns(3,4,5)P3 in either transfected or control cells. This is not surprising since PtdIns(3,4,5)P3 is present in very low amounts in most eukaryotic cells (Czech, 2000).
Figure 6.
PtdIns(4,5)P2 is an in vivo substrate for At5PTase11. HPLC analysis of deacylated [3H]myo-inositol-labeled lipids from S2 cells that were transfected with pMTAt5PTase11 (A) or pMTβ-gal (B). The positions of GroPtdIns standards are indicated. Equal amounts of total counts were analyzed for each sample.
Analysis of At5PTase11 Expression
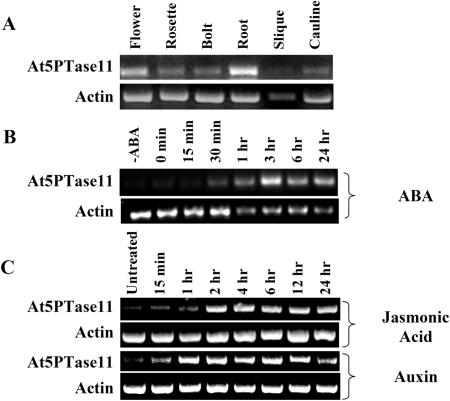
To examine expression of At5PTase11, we utilized semiquantitative RT-PCR. Oligonucleotide primers that amplify the 3′ ends of At5PTase11 and an actin gene were used to amplify cDNA products synthesized from RNAs of different tissues (Fig. 7). We have shown that the At5PTase11 primers are specific for the At5PTase11 gene by using the primer sequences as queries in a search of the BLAST short, nearly exact match database, and by comparing them to similar regions of the other At5PTase genes. The expected product from At5PTase11 cDNA using these primers is 363 bp. Contaminating At5PTase11 genomic DNA present would yield a 1,071-bp PCR product. We found that At5PTase11 is expressed in several Arabidopsis tissues, including flowers, rosette leaves, cauline leaves, roots, siliques, bolts, and seedlings (Fig. 7A).
Figure 7.
ABA, JA, and auxin treatment alters At5PTase11 expression in seedlings. Five- to 7-d seedlings were treated with either of 100 μm ABA, 10 μm auxin, or 100 μm JA and harvested at the indicated times. Total RNA was harvested and used in RT-PCR experiments with At5PTase11- and actin-specific primers as described in “Materials and Methods.”
Examination of the promoter of At5PTase11 indicated the presence of putative regulatory elements for JA and auxin. To test if At5PTase11 is regulated by JA or auxin, we measured At5PTase11 mRNA levels during JA and auxin stimulation. Since this approach was previously used to show that the At5PTase1 gene is regulated by ABA, we included seedlings stimulated by ABA in our analysis. Figure 7B shows that At5PTase11 mRNA levels increase during the first 3 h of ABA treatment and decrease slightly after this (Fig. 7B). This pattern is different from At5PTase1 regulation by ABA in which rapid induction is followed by an oscillatory behavior (Burnette et al., 2003). Seedlings treated with solvent alone (0.1% ethanol) and analyzed at various time points indicated no induction of the At5PTase11 gene (data not shown). We have also tested regulation of At5PTase11 mRNA levels in response to JA and auxin. At5PTase11 mRNA levels increased within 2 h of JA treatment and remained at induced levels for 24 h (Fig. 7C). Auxin also stimulated At5PTase11 expression (Fig. 7C). In contrast to ABA and JA treatments, auxin-stimulated At5PTase11 expression was transient, decreasing after 12 h. We conclude that ABA, JA, and auxin regulate At5PTase11 RNA expression.
DISCUSSION
We previously identified the Arabidopsis 5PTase protein family, which contains 15 putative proteins (Berdy et al., 2001). At5PTase1 and At5PTase2 have been characterized and shown to remove a 5-phosphate from the water-soluble substrates, Ins(1,4,5)P3 and Ins(1,3,4,5)P4 (Berdy et al., 2001; Sanchez and Chua, 2001). In these studies, there was no evidence that these proteins could remove a 5-phosphate from radiolabeled PtdIns(4,5)P2 as measured by TLC separation of products (Berdy et al., 2001; Sanchez and Chua, 2001). In our studies on At5PTase11, we utilized fluorescent phosphoinositide substrates that have been used by others (Giuriato et al., 2002; Laporte et al., 2002) and allows for more sensitivity in detecting reaction products (Taylor and Dixon, 2001). We show here that At5PTase11 hydrolyzes 5-phosphates from a group of phosphoinositide substrates including PtdIns(4,5)P2, PtdIns(3,5)P2, and PtdIns(3,4,5)P3. By comparing the activity of At5PTase1 and At5PTase11, we have shown that, unlike At5PTase1 and At5PTase2, At5PTase11 does not act on the water-soluble substrates Ins(1,4,5)P3 and Ins(1,3,4,5)P4 in our in vitro assay conditions. For comparison purposes, we also tested the ability of At5PTase1 and At5PTase2 to hydrolyze fluorescent PtdIns(4,5)P2 and found that, in contrast to previous reports (Berdy et al., 2001; Sanchez and Chua, 2001), these enzymes do hydrolyze phosphoinositide substrates (M.E. Ercetin and G.E. Gillaspy, unpublished data). This discrepancy is most likely due to the increased sensitivity of our assay. These data suggest that At5PTase11 has a more restricted substrate specificity than At5PTase1 and At5PTase2.
Under controlled conditions, At5PTase11 dephosphorylated more PtdIns(4,5)P2 than either of the other two substrates, PtdIns(3,5)P2 and PtdIns(3,4,5)P3, indicating a possible preference for this substrate (Fig. 4). Besides its involvement in cytoskeletal rearrangements and potential roles in vesicle trafficking and ion transport, PtdIns(4,5)P2 is known to act as a docking site for proteins via binding to plextrin-homology and Tubby protein domains (Toker, 1998). In some cases, binding to these domains can mediate the subcellular location and activity of the bound proteins. Thus, regulated PtdIns(4,5)P2 hydrolysis would lead to the loss of such docking sites and could influence several processes.
The in vitro substrate specificity of At5PTase11 is somewhat similar to that of the human Type IV 5PTase, although the Type IV enzyme has a lower Kapp for PtdIns(3,4,5)P3 than it does for PtdIns(4,5)P2 (Kisseleva et al., 2000). Because the molecular basis for 5PTase substrate specificity is of critical interest, several investigators have mutated various amino acid residues of 5PTase proteins in attempts to discern required sequence elements for specificity (Tsujishita et al., 2001). As both At5PTase11 and the human Type IV enzyme fail to act on inositol phosphate substrates in vitro, a comparison of these two protein sequences with other 5PTases that act on both phosphosinositide and inositol phosphate substrates might be expected to reveal critical residues for substrate specificity. Our sequence alignments (supplemental data) did not reveal any amino acid differences in At5PTase11 and the human Type IV enzyme as compared to other characterized 5PTases.
A unique feature of At5PTase11 is its ability to hydrolyze PtdIns(3,5)P2 in vitro. PtdIns(3,5)P2 is a recently identified phosphoinositide that was first found in hyperosmotic-stressed yeast (Dove et al., 1999) and some lower plants like Chlamydomonas moewusii (Meijer et al., 1999). However, since PtdIns(3,5)P2 levels were found to be very low in Arabidopsis and do not change after osmotic or salt stress (Pical et al., 1999; DeWald et al., 2001), the role of this phosphoinositide is unclear at this time. It is also interesting to note that, under our conditions, At5PTase11 did not hydrolyze PtdIns(5)P, which has been found to be up-regulated in response to osmotic stress in plants (Meijer et al., 2001).
We have also shown that At5PTase11 can hydrolyze PtdIns(3,4,5)P3 in vitro, and the potential role of At5PTase11 with regard to this substrate is also unclear. PtdIns(3,4,5)P3 synthesis has not been shown in any plant to date. In animal cells, PtdIns(3,4,5)P3 is present at very low levels, and synthesis is stimulated by specific signal transduction pathways. Thus, it is possible that PtdIns(3,4,5)P3 has escaped our attention in plant cells because conditions for maximal synthesis have not been examined. In support of this, it is important to note that a recombinant Arabidopsis phosphatidylinositol-4-phosphate 5-kinase (AtPK51) protein produced in insect cells can phosphorylate PtdIns(3,4)P2 producing PtdIns(3,4,5)P3 (Elge et al., 2001). In addition, an Arabidopsis homolog of a human tumor suppressor (Pten) has been shown to remove a 3-phosphate from PtdIns(3,4,5)P3 in vitro and is essential for pollen development (Gupta et al., 2002). It is possible that each of these proteins, At5PTase11, AtPten1, and AtPK51, maintains the ability to interact with PtdIns(3,4,5)P3 as a consequence of another function and that they do not encounter PtdIns(3,4,5)P3 in the plant cell. Examination of knockout mutants in these genes combined with careful labeling studies utilizing [3H]myo-inositol or [32P]orthophosphate under the appropriate signaling or growth conditions may resolve this issue.
The regulation of At5PTase11 by the three different signaling conditions tested is intriguing, and may point to the involvement of phosphoinositides in these pathways. There are many published data regarding ABA signal transduction and phosphoinositides. It is known that ABA stimulates phospholipase C activity, resulting in new Ins(1,4,5)P3 synthesis (for review, see Meijer and Munnik, 2003). Genetic studies have shown the necessity of the fiery1 gene encoding an inositol polyphosphate 1-phosphatase for ABA signaling (Xiong et al., 2001). This suggests that inositol phosphate breakdown is critical for ABA signaling. It has also been shown that ABA stimulates production of At5PTase1, which hydrolyzes Ins(1,4,5)P3, and that this terminates ABA signaling (Burnette et al., 2003). Our present data indicate that ABA-stimulated expression of At5PTase11 may also be involved in ABA signal termination. Reduction of PtdIns(4,5)P2 by At5PTase11 could reduce the pool of PtdIns(4,5)P2 available for Ins(1,4,5)P3 synthesis, imparting another level of control over Ins(1,4,5)P3 production. Reduction of PtdIns(4,5)P2 could also reduce phospholipase D activity, as some isoforms of this enzyme require PtdIns(4,5)P2 for activity (Pappan et al., 1997; Qin et al., 1997). With respect to At5PTase11 induction after auxin and JA addition, much less is known about whether these signaling pathways utilize phosphoinositides. Our data are consistent with information in the MPSS database, which reports very low levels of At5PTase11 expression except in callus tissue that was grown on auxin-containing media and in salicylic acid-treated plants (http://mpss.udel.edu/).
In summary, the cloning and identification of a novel inositol 5-phosphatase with a substrate specificity toward PtdIns(4,5)P2, PtdIns(3,5)P2, and PtdIns(3,4,5)P3 expand our knowledge of the eukaryotic 5PTase protein family and offer a unique tool to examine the importance of these phosphoinositides in plant physiological processes.
MATERIALS AND METHODS
Plant Growth and Treatment
Arabidopsis ecotype Columbia plants were used for all experiments. Growth conditions of soil-grown plants have been described previously (Berdy et al., 2001). For ABA (Sigma-Aldrich, St. Louis), JA (95% methyl jasmonate solution, Sigma-Aldrich), and auxin (α-naphthalene acetic acid, Sigma-Aldrich) experiments, seed from wild-type plants was germinated in flasks containing 1× Murashige and Skoog medium containing 1% Suc and grown for 5 d under constant light and shaking (91 rpm). Seedlings were then treated with 100 μm ABA (0.1% ethanol final concentration), 100 μm methyl jasmonate, and 10 μm 1α-naphthalene acetic acid and frozen in liquid nitrogen at the indicated times. Hormone treatment experiments were repeated three times.
Gene Cloning and Sequence Analysis
Full-length At5PTase11 cDNA was generated by RT-PCR using Arabidopsis mixed- tissue mRNA and the following oligonucleotide primers: At5PTase11-Nterm (5′ATGGGGAATAAGAATTCGATGT3′) and At5PTase11-Cterm (5′TTAACTGTTGACCCACTTCAAGCAAA3′). RT reaction (2 μL), At5PTase11-Nterm (12.5 pmol μL−1), At5PTase11-Cterm (12.5 pmol μL−1), elongase polymerase and supplied buffer (Invitrogen, Carlsbad, CA), 0.5 mm dNTPs, and 2.5 mm final MgCl2 were mixed and heated to 94°C for 3 min. PCR amplification consisted of 30 cycles (1 min 94°C, 1 min 58°C, 1.5 min 72°C). The full-length At5PTase11 cDNA product was gel purified (Qiagen gel extraction kit; Qiagen, Valencia, CA), and cloned into the T7/NT TOPO TA expression vector (Invitrogen) according to the manufacturer's instructions. The resulting prokaryotic expression construct (pNTTOPOAt5PTase11) was used as template in a PCR reaction with primers pMTAt5PTase11-Nterm (5′GCCATGGGGAATAAGAATTCGATGTGTGGGT3′) and pMTAt5PTase11-Cterm (5′ACTGTTGACCCACTTCAAGCAAA3′) to introduce a Kozak sequence for efficient translation initiation and to generate a C-terminal V5-His tag. The modified cDNA was cloned into the pMT/V5-His-TOPO vector (Invitrogen). The resulting construct (pMTAt5PTase11) was sequenced and compared to the genomic and predicted cDNA sequences present in the NCBI database (AC007519 and NM_103644). The At5PTase1 full-length cDNA (Berdy et al., 2001) was similarly expressed as a C-terminal V5-His-tagged protein in S2 cells, resulting in the production of recombinant At5PTase1 protein as judged by SDS-PAGE and western blotting. Purified human OCRL protein was a gift from Drs. Phillip Majerus and Marina Kisseleva (Washington University School of Medicine, St. Louis); its purification and activity have been described (Zhang et al., 1995). 5PTase protein alignments were created with ClustalX and Boxshade software.
Transient Expression of At5PTase11 in Drosophila melanogaster Cells
For transient expression studies, pMTAt5PTase11 was used to transfect Drosophila melanogaster S2 cells utilizing an Effectene transfection kit (Qiagen). Briefly, S2 cells were cultured in Schneider's Drosophila medium supplemented with 10% heat-inactivated (60°C for 30 min) fetal bovine serum, 50 units/mL penicillin, and 50 μg/mL streptomycin. Cells were incubated in a 28°C incubator with humidity and were routinely passaged every 3 d at a cell density of approximately 5 × 106 cells/mL. For transfections, cells were seeded into 25-cm2 flasks at a density of 1 × 106 cells/mL in 10 mL of medium; 24 h later cells were spun at 800 g for 3 min and washed with 1× phosphate-buffered saline (pH 7.2), and the resulting cell pellets were resuspended in 4 mL of fresh medium. Two micrograms of plasmid DNA, 16 μL of enhancer, and 20 μL of Effectene reagent (Qiagen) were mixed according to manufacturer's instructions and added into the cell culture. After 24 h, cells were induced with 500 μm (final concentration) CuSO4 for 2 d. Cells were harvested and washed with 1× phosphate-buffered saline (pH 7.2) and frozen in liquid N2 at −80°C. For labeling of S2 cells, we have modified a described method for human astrocytoma cells (Stephens et al., 1989). Transfections were performed as described above, except 125 μCi of [1,2 3H (N)]myo-inositol (American Radiolabeled Chemicals, St. Louis; 30 Ci/mm in sterile water) was added after 36 h of induction with CuSO4. Cells were harvested 12 h later and lipids were purified using an acidified Bligh and Dyer extraction (Bird, 1998). Deacylation of the purified lipids was performed (Bird, 1998), and the resulting deacylated lipids (GroPtdInss) were analyzed by HPLC.
Immunoprecipitation of At5PTase11 from Transiently Transfected S2 Cells
S2 cells (10 mL) transfected with pMTAt5PTase11 or pMTβ-gal were harvested and cell pellets were dissolved in 300 μL of immunoprecipitation buffer (50 mm Tris, pH 7.8, 150 mm NaCl, 5 mm MgCl2, and 0.05% Triton X-100). Cell lysis was achieved by passage through a 25-gauge syringe 10 times. The lysate was spun at 14,000 rpm for 10 min, and the resulting soluble fraction was incubated with protein A sepharose (Sigma-Aldrich) complexed with mouse anti-V5 monoclonal antibody (Invitrogen) for 2 h at 4°C with end-over-end rotating. The protein A sepharose beads were pelleted and washed three times with IP buffer followed by one wash with 5PTase activity assay buffer. A portion of this immunoprecipitate was analyzed by SDS-PAGE and western blotting to estimate the amount of immunoprecipitated protein.
Western-Blot Analysis
Protein extracts and immunoprecipitates prepared from pMTAt5PTase11-transfected, pMTβ-gal-transfected, and mock-transfected S2 cells were separated on 10% SDS-PAGE gels and either stained with Coomassie dye or transferred to nitrocellulose. Western-blot conditions have been previously described (Burnette et al., 2003). For detection of the V5 epitope tag, a 1:5,000 dilution of the mouse anti-V5 monoclonal antibody, followed by a 1:5,000 dilution of goat anti-mouse HRP conjugated antibody (Amersham, Piscataway, NJ), was used. Purified Positope V5-tag protein (175 ng; Invitrogen) was loaded on the same gel to estimate the amount of recombinant V5-tagged protein.
5PTase Activity Assays
Fluorescent substrates (Echelon Research Laboratories, Salt Lake City) were used in a modified version of a previously described phosphatase assay (Taylor and Dixon, 2001). Briefly, immunoprecipitated At5PTase11 was incubated for 1 h at room temperature with 1.5 μg of di-C6-NBD6-phosphatidylinositol 4,5-bisphosphate, di-C6-NBD6-phosphatidylinositol 3,5-bisphosphate, di-C6-NBD6-phosphatidylinositol 3,4,5-trisphosphate, or di-C6-NBD6-phosphatidylinositol 5-monophosphate, in lipid assay buffer containing 50 mm HEPES, pH 7.5, 5 mm MgCl2, and 50 mm KCl with agitation every 15 min. The reaction was stopped by addition of 100 μL acetone and was dried under low heat vacuum. After dissolving in 10 μL of methanol:2-propanol:glacial acetic acid; 5:5:2), samples were loaded on a TLC plate that had been pretreated in 1.2% potassium oxalate in 60:40 MeOH:water and dried at 100°C for 30 min. The plates were developed in 180 mL of chloroform:methanol:acetone:glacial acetic acid:water (70:50:20:20:20). To visualize fluorescence lipids, the TLC plate was dried and analyzed using a Storm 860 Blue Fluorescence Chemifluorescence and Scanner Control Software version 4.1 (Molecular Dynamics, Piscatawy, NJ). For densitometric analysis of the images produced via Storm 860, we have utilized AlphaEaseFC software version 3.1.2 (Alpha Innotech, San Leandro, CA).
For the analysis of inositol phosphate substrates, activity assays were performed using [3H]myo-inositol (1,4,5)P3 (10 μCi mL−1; NEN, Shelton, CT) and [3H]myo-inositol (1,3,4,5)P4 (10 μCi mL−1; NEN). Immunoprecipitated 5PTases were incubated with 30 nCi of [3H]myo-inositol (1,4,5)P3 and [3H]myo-inositol (1,3,4,5)P4 in reaction buffer (250 mm KCl, 3 mm MgCl2, 50 mm Tris, pH 7.5) in a total volume of 300 μL at room temperature for 1 h. The resulting products were stored at −20°C and analyzed by HPLC (Beckman System Gold; Beckman Instruments, Fullerton, CA) using a Waters Spherisorb S5 SAX 4 ×125-mm analytical column (Milford, MA) equilibrated with a 10 mm AP buffer at pH 3.8. Samples were applied to column using an autosampler, and products were eluted with a linear gradient from 10 mm AP to 340 mm AP over 30 min; 340 mm AP to 1.02 m AP over 15 min; and constant 1.02 m AP over 5 min (Stolz et al., 1998). For the analysis of deacylated phosphatidylinositols, a gradient described by Stephens et al. (1989) was used. Radioactivity was counted with an in-line Beckman 171 radioisotope detector and the data file was converted to an asci file format by 32 Karat software (Beckman Coulter, Fullerton, CA) and analyzed by Excel (Microsoft, Seattle).
RT-PCR
Conditions for semiquantitative RT-PCR have been described previously (Berdy et al., 2001). Total RNA was extracted from 100 mg of frozen tissue using the Qiagen Plant RNeasy kit according to manufacturer's specifications. RNA (1 μg) was analyzed by two-step RT-PCR utilizing a Qiagen Omniscript reverse transcriptase kit and the manufacturer's instructions. Conditions for actin amplification have been described and generate a 428-bp product (An et al., 1996; Berdy et al., 2001). For At5PTase11-specific amplification, At5PTase11 for primer (5′GAGCCGTGGCTATTCGTATT3′) and At5PTase11rev primer (5′AGTCGCTGCTTCCTACATTG3′) were used, resulting in a 363-bp product. Amplification with a cloned At5PTase11 cDNA and Arabidopsis genomic DNA was used as a control for all PCR experiments to verify the sizes of bona fide reaction products from RNA and contaminating genomic DNA. Molecular weight markers were used to determine product sizes. Each RT-PCR experiment was independently repeated three times to verify that the observed changes in expression were reproducible.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AC007519.
Supplementary Material
Acknowledgments
We thank Kevin Jones for expertise and diligence with recombinant protein expression, Phillip Majerus and Marina Kisseleva for the human 5PTase purified protein, and G. Taylor for providing advice on TLC experiments.
This work was supported by the U.S. Department of Agriculture (grant no. 2003–35318–13690 to G.E.G.) and by the Hatch project (grant no. VA–135583).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.040253.
References
- An Y, McDowell J, Huang S, McKinney E, Chambliss S, Meagher R (1996) Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass vegetative tissues. Plant J 10: 107–121 [DOI] [PubMed] [Google Scholar]
- Berdy S, Kudla J, Gruissem W, Gillaspy G (2001) Molecular characterization of At5PTase1, an inositol phosphatase capable of terminating IP3 signaling. Plant Physiol 126: 801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird I (1998) Phospholipid signaling protocols. In IM Bird, ed, Methods in Molecular Biology, Vol 105. Humana Press, Totowa, NJ
- Braun M, Baluska F, von Witsch M, Menzel D (1999) Redistribution of actin, profilin and phosphatidylinositol-4, 5-bisphosphate in growing and maturing root hairs. Planta 209: 435–443 [DOI] [PubMed] [Google Scholar]
- Burnette RN, Gunesekera BM, Gillaspy GE (2003) An Arabidopsis inositol 5-phosphatase gain-of-function alters abscisic acid signaling. Plant Physiol 132: 1011–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech MP (2000) PIP2 and PIP3: complex roles at the cell surface. Cell 100: 603–606 [DOI] [PubMed] [Google Scholar]
- DeWald DB, Torabinejad J, Jones JA, Shope JC, Cangelosi AR, Thompson JE, Prestwich GD, Hama H (2001) Rapid accumulation of phosphatidylinositol 4,5-bisphosphate and inositol 1,4,5-trisphosphate correlates with calcium mobilization in salt-stressed Arabidopsis. Plant Physiol 126: 759–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove SK, McEwen RK, Cooke FT, Parker PJ, Michell RH (1999) Phosphatidylinositol 3,5-bisphosphate: a novel lipid that links stress responses to membrane trafficking events. Biochem Soc Trans 27: 674–677 [DOI] [PubMed] [Google Scholar]
- Elge S, Brearley C, Xia HJ, Kehr J, Xue HW, Mueller-Roeber B (2001) An Arabidopsis inositol phospholipid kinase strongly expressed in procambial cells: synthesis of PtdIns(4,5)P2 and PtdIns(3,4,5)P3 in insect cells by 5-phosphorylation of precursors. Plant J 26: 561–571 [DOI] [PubMed] [Google Scholar]
- Giuriato S, Blero D, Robaye B, Bruyns C, Payrastre B, Erneux C (2002) SHIP2 overexpression strongly reduces the proliferation rate of K562 erythroleukemia cell line. Biochem Biophys Res Commun 296: 106–110 [DOI] [PubMed] [Google Scholar]
- Gupta R, Ting JT, Sokolov LN, Johnson SA, Luan S (2002) A tumor suppressor homolog, AtPTEN1, is essential for pollen development in Arabidopsis. Plant Cell 14: 2495–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisseleva MV, Wilson MP, Majerus PW (2000) The isolation and characterization of a cDNA encoding phospholipid-specific inositol polyphosphate 5-phosphatase. J Biol Chem 275: 20110–20116 [DOI] [PubMed] [Google Scholar]
- Kost B, Lemichez E, Spielhofer P, Hong Y, Tolias K, Carpenter C, Chua NH (1999) Rac homologues and compartmentalized phosphatidylinositol 4,5-bisphosphate act in a common pathway to regulate polar pollen tube growth. J Cell Biol 19: 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte J, Liaubet L, Blondeau F, Tronchere H, Mandel JL, Payrastre B (2002) Functional redundancy in the myotubularin family. Biochem Biophys Res Commun 291: 305–312 [DOI] [PubMed] [Google Scholar]
- Majerus PW, Kisseleva MV, Norris FA (1999) The role of phosphatases in inositol signaling reactions. J Biol Chem 274: 10669–10672 [DOI] [PubMed] [Google Scholar]
- McCurdy DW, Kovar DR, Staiger CJ (2001) Actin and actin-binding proteins in higher plants. Protoplasma 215: 89–104 [DOI] [PubMed] [Google Scholar]
- Meijer H, Divecha N, van den Ende H, Musgrave A, Munnik T (1999) Hyperosmostic stress induces rapid synthesis of phosphatidyl-D-inositol 3,5-bisphosphate in plant cells. Planta 208: 294–298 [Google Scholar]
- Meijer HJ, Berrie CP, Iurisci C, Divecha N, Musgrave A, Munnik T (2001) Identification of a new polyphosphoinositide in plants, phosphatidylinositol 5-monophosphate (PtdIns5P), and its accumulation upon osmotic stress. Biochem J 360: 491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer HJ, Munnik TM (2003) Phospholipid-based signaling in plants. Annu Rev Plant Biol 54: 265–306 [DOI] [PubMed] [Google Scholar]
- Pappan K, Qin W, Dyer JH, Zheng L, Wang X (1997) Molecular cloning and functional analysis of polyphosphoinositide-dependent phospholipase D, PLDbeta, from Arabidopsis. J Biol Chem 272: 7055–7061 [DOI] [PubMed] [Google Scholar]
- Pendaries C, Tronchere H, Plantavid M, Payrastre B (2003) Phosphoinositide signaling disorders in human diseases. FEBS Lett 546: 25–31 [DOI] [PubMed] [Google Scholar]
- Pical C, Westergren T, Dove SK, Larsson C, Sommarin M (1999) Salinity and hyperosmotic stress induce rapid increases in phosphatidylinositol 4,5-bisphosphate, diacylglycerol pyrophosphate, and phosphatidylcholine in Arabidopsis thaliana cells. J Biol Chem 274: 38232–38240 [DOI] [PubMed] [Google Scholar]
- Qin W, Pappan K, Wang X (1997) Molecular heterogeneity of phospholipase D (PLD). Cloning of PLDgamma and regulation of plant PLDgamma, -beta, and -alpha by polyphosphoinositides and calcium. J Biol Chem 272: 28267–28273 [DOI] [PubMed] [Google Scholar]
- Sanchez JP, Chua NH (2001) Arabidopsis plc1 is required for secondary responses to abscisic acid signals. Plant Cell 13: 1143–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger CJ, Yuan M, Valenta R, Shaw PJ, Warn RM, Lloyd CW (1994) Microinjected profilin affects cytoplasmic streaming in plant cells by rapidly depolymerizing actin microfilaments. Curr Biol 4: 215–219 [DOI] [PubMed] [Google Scholar]
- Stephens LR, Hawkins PT, Downes CP (1989) An analysis of myo-[3H]inositol trisphosphates found in myo-[3H]inositol prelabelled avian erythrocytes. Biochem J 262: 727–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson JM, Perera IY, Heilmann II, Persson S, Boss WF (2000) Inositol signaling and plant growth. Trends Plant Sci 5: 252–258 [DOI] [PubMed] [Google Scholar]
- Stolz LE, Kuo WJ, Longchamps J, Sekhon MK, York JD (1998) INP51, a yeast inositol polyphosphate 5-phosphatase required for phosphatidylinositol 4,5-bisphosphate homeostasis and whose absence confers a cold-resistant phenotype. J Biol Chem 273: 11852–11861 [DOI] [PubMed] [Google Scholar]
- Taylor GS, Dixon JE (2001) An assay for phosphoinositide phosphatases utilizing fluorescent substrates. Anal Biochem 295: 122–126 [DOI] [PubMed] [Google Scholar]
- Toker A (1998) The synthesis and cellular roles of phosphatidylinositol 4,5-bisphosphate. Curr Opin Cell Biol 10: 254–261 [DOI] [PubMed] [Google Scholar]
- Tsujishita Y, Guo S, Stolz LE, York JD, Hurley JH (2001) Specificity determinants in phosphoinositide dephosphorylation: crystal structure of an archetypal inositol polyphosphate 5-phosphatase. Cell 105: 379–389 [DOI] [PubMed] [Google Scholar]
- Xiong L, Lee B, Ishitani M, Lee H, Zhang C, Zhu JK (2001) FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev 15: 1971–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Jefferson AB, Auethavekiat V, Majerus PW (1995) The protein deficient in Lowe syndrome is a phosphatidylinositol-4,5-bisphosphate 5-phosphatase. Proc Natl Acad Sci USA 92: 4853–4856 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.