Abstract
An Arabidopsis expressed sequence tag clone, 221D24, encoding a lipase has been characterized using an antisense approach. The lipase gene is expressed during normal growth and development of Arabidopsis rosette leaves but is down-regulated as the leaves senesce. When plants are exposed to sublethal levels of UV-B radiation, expression of the lipase is strongly up-regulated. The lipase protein is localized in the cell cytosol and is present in all organs of Arabidopsis plants. Recombinant lipase protein produced in Escherichia coli preferentially hydrolyzed phospholipids, indicating that the gene encodes a phospholipase. Transgenic plants in which lipase expression is suppressed showed enhanced tolerance to UV-B stress but not osmotic stress and were unable to up-regulate PR-1 expression when irradiated with UV-B. The observations collectively indicate that the lipase is capable of deesterifying membrane phospholipids and is up-regulated in response to UV-B irradiation.
Lipases are hydrolytic enzymes that catabolize complex lipids. They include phospholipases, lipolytic acyl hydrolases, galactolipases, and triacylglycerol lipases. Certain types of lipases, those that deesterify fatty acids, contain the consensus sequence [LIV]-X-[LIVAFY]-[LIAMVST]-G-[HYWV]-S-X-G-[GSTAC] encoding the esterase motif (Derewenda and Derewenda, 1991). Lipases are important agents of cellular metabolism in that they catabolize lipids, but they have also been implicated in signal transduction, mobilization of lipids during seed germination, and catabolism of membrane lipids during senescence (Thompson et al., 1998; Wang, 2001).
Phospholipase D (PLD) is the best characterized plant phospholipase. Five isoforms of PLD (α, β, γ, δ, and ɛ) have been identified in Arabidopsis (Wang, 2001). Plant PLD contains two HXKXXXD motifs, which constitute two active-site regions necessary for activity (Xie et al., 2000). The enzyme has been implicated in a broad range of cellular functions, including the response of cells to various types of stress (Wang, 2001). For example, when PLD expression is down-regulated, stress-related senescence of detached leaves induced by the application of exogenous hormones is delayed (Fan et al., 1997). The action of phospholipase A2 generates lysophospholipids, a reaction that is stimulated by auxin (Scherer, 1995), and is also thought to be stimulated by pathogens (Munnik et al., 1998). Indeed, it is possible that linolenic acid, the precursor of jasmonic acid and other octadecanoid-derived signals that stimulate the expression of defense-related genes, is formed by the action of a specific phospholipase A2 on membrane phospholipids (Munnik et al., 1998).
Lipolytic acyl hydrolase is a nonspecific lipase that cleaves fatty acids at the Sn1 and Sn2 positions from a broad range of substrates, including phospholipids, triacylglycerols, and wax esters (Galliard, 1971). Hong et al. (2000) have isolated and characterized a cDNA clone from carnation (Dianthus caryophyllus) petals that encodes a senescence-induced lipolytic acyl hydrolase. Northern-blot analysis showed that the expression of this gene is up-regulated during natural senescence and following treatment with exogenous ethylene. The recombinant protein of this gene was shown to deesterify fatty acids from numerous substrates. A senescence-associated lipase isolated from Arabidopsis (designated SAG101) is also likely to be a lipolytic acyl hydrolase (He and Gan, 2002). Antisense suppression of SAG101 expression in Arabidopsis delayed the onset of leaf senescence, whereas overexpression resulted in precocious senescence of both attached and detached leaves (He and Gan, 2002). There is also evidence that patatin exhibits lipolytic acyl hydrolase activity (Senda et al., 1996).
Galactolipids, specifically monogalactosyldiacylglycerol (MGDG) and digalactosyldiacylglycerol (DGDG), are the dominant lipids of thylakoids, accounting for more than 60% of total polar lipids in photosynthesizing tissues. Deesterification of galactolipids is a pronounced feature of natural leaf senescence (Engelman-Silvestre et al., 1989) and is also induced in the event of drought stress (Sahsah et al., 1998) and cold stress (Kaniuga and Gemel, 1984). Proteins with galactolipid-hydrolyzing activity have been purified from leaves of Phaseolus multiflorus (Sastry and Kates, 1964), pole bean (Phaseolus vulgaris; Anderson et al., 1974), and wheat (Triticum aestivum; O'Sullivan et al., 1987). These enzymes proved capable of deesterifying fatty acids from both sn positions of MGDG and DGDG. To date, the corresponding genes for these chloroplast-associated galactolipases have not been isolated, although recently a drought-induced patatin-like gene was isolated from cowpea (Vigna unguiculata) and its cognate protein shown to have galactolipid-hydrolyzing activity (Matos et al., 2000). However, the patatin-like galactolipase is not predicted to have a chloroplast targeting sequence or transmembrane domains and is likely localized in the cytosol.
In this study, we have characterized an Arabidopsis GenBank sequence (gene accession no. At2g42690) that encodes a protein (protein accession no. AAD21737.1) exhibiting phospholipase activity. The phospholipase is localized in the cytosol and induced by treatment with sublethal levels of UV-B. Transgenic plants in which expression of the UV-B-induced lipase is suppressed exhibited enhanced tolerance to sublethal UV-B stress and were unable to up-regulate pathogenesis related protein 1 (PR-1) in response to UV-B treatment.
RESULTS
Gene Isolation and Function
Full-length cDNA corresponding to the Arabidopsis sequence, GenBank accession number At2g42690 (encoding protein AAD21737.1), was obtained by reverse transcription (RT)-PCR using RNA isolated from the rosette leaves of 4-week-old plants. A comparison of the genomic and cDNA sequences revealed that the gene contains one intron and encodes a polypeptide composed of 412 amino acid residues with an approximate molecular mass of 47 kD. Analysis of the inferred amino acid sequence (Fig. 1) using the program Predotar indicated that the protein does not contain cleavable chloroplast- or mitochondria-targeting sequences. The protein does, however, contain the lipase consensus sequence [LIV]-X-[LIVAFY]-[LIAMVST]-G-[HYWV]-S-X-G-[GSTAC] (Fig. 1), which corresponds to an esterase motif and is a characteristic feature of all known lipases that deesterify fatty acids from complex lipids. Thus, the presence of this motif indicates that the cDNA encodes a lipase. The inferred amino acid sequence also contains predicted phosphorylation, glycosylation, and myristoylation domains (Fig. 1). A BLAST search revealed that the sequence matched most closely (55% identity with an e-value of 10−136) a cytosolic lipase (GenBank accession no. AAD01804.1) isolated from carnation petals (Hong et al., 2000). The second closest match is to a putative rice (Oryza sativa) lipase (GenBank accession no. BAB39417.1), and the next six closest matches are to putative Arabidopsis lipases (GenBank accession nos. CAA16735.1, AAG52635.1, AAB63082.1, AAF63138.1, AAF63138.1, and AAG51101.1), all of which contain the lipase consensus sequence corresponding to the lipase active site.
Figure 1.
Nucleotide and inferred amino acid sequences of the cDNA corresponding to the Arabidopsis sequence, GenBank accession number AAD21737.1, illustrating the lipase consensus sequence (black) as well as predicted phosphorylation (underlined), glycosylation (dark gray), and myristoylation (light gray) motifs. *, Stop codon.
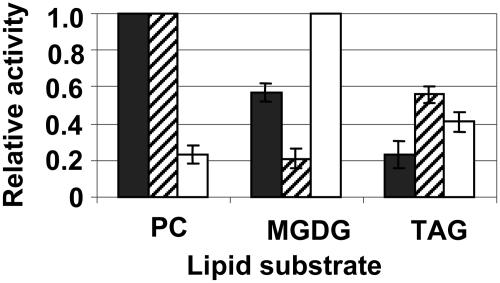
To confirm that the gene encodes a lipase, the full-length cDNA was ligated into pTrc 99a, which is isopropylthio-β-galactoside inducible, and overexpressed in Escherichia coli. SDS-PAGE and western-blot analysis indicated that the 47-kD recombinant protein was present in the pellet fraction, but not the supernatant fraction, isolated from E. coli (data not shown). This fraction also exhibited lipase activity (Fig. 2). Moreover, when the abilities of the recombinant lipase to deesterify fatty acids from soybean (Glycine max) phosphatidylcholine, monogalactolipid, and triacylglycerol were compared, the protein exhibited a strong preference for phosphatidylcholine substrate, indicating that it is a phospholipase. Specifically, its ability to hydrolyze monogalactolipid and triacylglycerol was only 58% and 22%, respectively, of its ability to hydrolyze phosphatidylcholine (Fig. 2). This distinction was further confirmed by testing commercially available fungal lipases with known substrate preferences, viz., Candida rugosa lipase, which is known to be a phospholipase, and Rhizomucor miehei lipase, which is known to be a galactolipase (Ishiguro et al., 2001; Fig. 2).
Figure 2.
Recombinant lipase catalytic activity. Activity was measured in vitro by quantifying the release of free fatty acids from phosphatidylcholine (PC), trilinolein (TAG), and MGDG. The data are expressed as relative activities compared to the maximum activity (set at a value of 1) for each lipase. The activity for the recombinant lipase is net of activity for extract from bacteria containing empty vector. Recombinant lipase (black bars); C. rugosa lipase, a known phospholipase (hatched bars); R. miehei lipase, a known galactolipase (white bars).
The possibility that the recombinant lipase exhibits specificity was examined by testing its ability to deesterify fatty acids from a range of purified molecular species of phosphatidylcholine that are available commercially. Lipid extracts of the reaction mixtures were fractionated by thin-layer chromatography (TLC), and deesterified fatty acids were quantified and identified by gas chromatography. For each of the 16:0/20:4 phosphatidylcholine, 20:4/20:4 phosphatidylcholine, 16:0/18:3 phosphatidylcholine, 16:0/18:2 phosphatidylcholine, 18:2/18:2 phosphatidylcholine, and 18:3/18:3 phosphatidylcholine substrates, only background levels of deesterified fatty acid, equivalent to those obtained for control reactions containing extract from E. coli transformed with empty pTrc 99a, were detectable over a range of substrate concentrations (data not shown). Yet the recombinant enzyme exhibited strong deesterification activity when soybean phosphatidylcholine, which contains an unknown mixture of molecular species, was used as substrate (Fig. 2). This suggests, although does not prove, that the lipase exhibits molecular species specificity and that the molecular species it is able to hydrolyze is not among those tested.
Localization and Expression
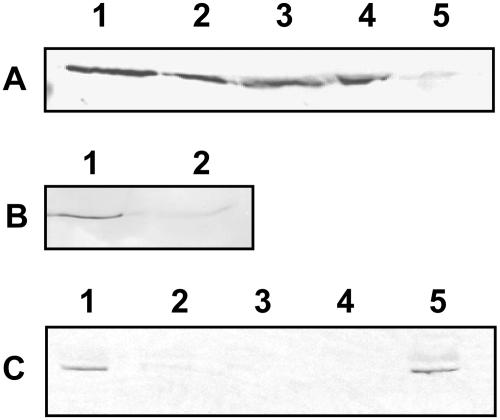
Extracts of total protein from different organs of 6-week-old Arabidopsis plants were analyzed by western blotting in order to determine the spatial localization of the native protein. An antibody (α-LIP) raised in rabbits against a synthetic peptide corresponding to amino acids 386 to 407 of the lipase was used as the primary antibody. At this stage of development, the rosette leaves are undergoing senescence, and flowers and siliques are fully developed. A 47-kD band corresponding to the native protein was evident in protein extracts from rosette leaves, stems, flowers, and siliques and at a much lower abundance in seeds (Fig. 3A). Northern analyses indicated that transcript for the native protein is also expressed in roots but at lower levels than in all the other organs tested (data not shown).
Figure 3.
Lipase subcellular localization. A, Western blot of total protein isolated from organs of 6-week-old wild-type Arabidopsis plants. Lane 1, leaf; lane 2, stem; lane 3, flower; lane 4, silique; lane 5, seed. B, Western blot of cytosolic and microsomal membrane fractions isolated from the rosette leaves of 3.5-week-old wild-type Arabidopsis plants. Lane 1, cytosol; lane 2, microsomal membranes. C, Western blot of cytosol, chloroplasts, and chloroplastic subfractions isolated from the rosette leaves of 3.5-week-old wild-type Arabidopsis plants. Lane 1, broken chloroplasts; lane 2, intact chloroplasts; lane 3, chloroplastic membranes (thylakoids and envelope); lane 4, stroma; lane 5, cytosol. The blots were probed with lipase-specific antibody.
When purified microsomal and cytosolic fractions isolated from rosette leaves of 3.5-week-old plants were analyzed by western blotting, a 47-kD band corresponding to the native protein was clearly present in the cytosolic fraction and only barely detectable in the membrane fraction (Fig. 3B). This indicated that the protein is soluble and localized in the cytosol. To confirm that it is not plastidial, chloroplasts from rosette leaves of 3.5-week-old Arabidopsis plants were subfractionated into broken chloroplasts, purified stroma, and chloroplastic membranes (thylakoids and envelope membranes), and the subfractions together with purified cytosol were analyzed by western blotting. The native protein was detectable in broken chloroplast (contaminated with cytosol) and cytosolic fractions but not in the intact chloroplasts, stroma, or chloroplastic membrane fractions, indicating that its presence in the cytosol is not attributable to leakage from broken chloroplasts (Fig. 3C).
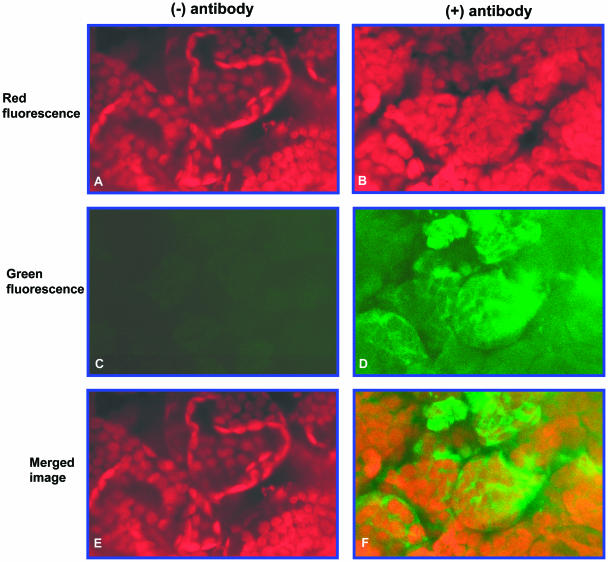
Cytosolic localization of the lipase protein was further confirmed by confocal microscopy. These experiments were performed using leaf tissue in which chloroplasts are clearly discernible by chlorophyll autofluorescence. Rosette leaf tissue from 3.5-week-old Arabidopsis plants was cut into small pieces and labeled en bloc with α-LIP antiserum and a secondary fluorescent antibody. Control tissue was not labeled with α-LIP but only with the secondary antibody. Chlorophyll autofluorescence (red) was clearly discernible in control tissue and in tissue that had been labeled with the α-LIP antibody (Fig. 4, A and B). By contrast, the lipase protein signal (green) was only detectable in tissue that had been labeled with lipase protein antiserum (Fig. 4, C and D). Moreover, when the signals were merged, the green signal corresponding to the lipase protein was evident in areas distinct from the red chloroplast signal (Fig. 4F). These observations again indicate that the lipase protein is localized in the cytosol.
Figure 4.
Confocal micrographs of rosette leaf sections from 3.5-week-old wild-type Arabidopsis plants. The sections were labeled en bloc with lipase-specific antibody. A and B, Chlorophyll autofluorescence without and with antibody labeling, respectively; C and D, fluorescein fluorescence without and with antibody labeling, respectively; E and F, merged images of A and C, and B and D, respectively.
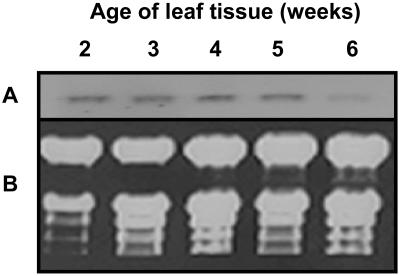
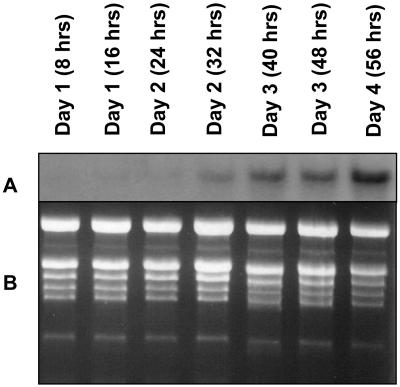
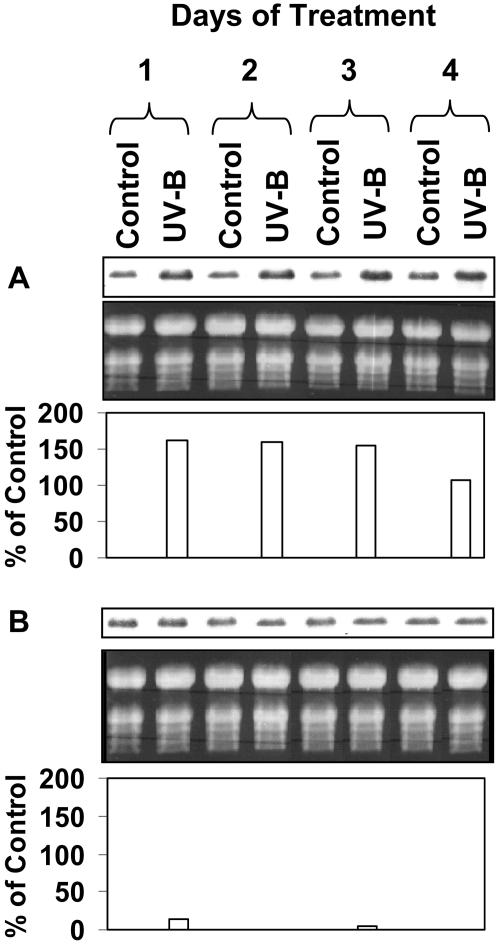
Northern-blot analysis indicated that transcript for the lipase protein is clearly present in leaves of plants 2 to 5 weeks of age but is substantially down-regulated by week 6 (Fig. 5). By week 3, the rosette leaves are fully expanded, and between weeks 4 and 5 the plants bolt and flower. By week 5, the rosette leaves have begun to senesce, and by week 6 leaf senescence is fully engaged and the leaves have turned yellow, reflecting loss of chlorophyll. Thus, expression of the gene is consistent during leaf development and begins to decline coincidently with the onset of senescence. Of particular interest, however, is the finding that its expression in leaves is strongly up-regulated when plants are subjected to sublethal UV-B stress (Fig. 6). The treatment was initiated when the plants were 3.5 weeks of age and was continued for 4 d. That the UV-B treatment was sublethal is indicated by the fact that the treated plants continued to develop and produce seed. Transcript levels began to increase within 2 d of treatment and by day 4 were pronounced (Fig. 6).
Figure 5.
Lipase expression during leaf development and senescence. A, Northern blot of total RNA isolated from the rosette leaves of wild-type Arabidopsis plants at various stages of development. The blot was probed with the 3′ untranslated region of the lipase cDNA. Lanes 1 to 5: 2-, 3-, 4-, 5-, and 6-week-old tissue, respectively. Each lane contained 10 μg of RNA. B, Corresponding ethidium bromide-stained agarose gel of the fractionated RNA.
Figure 6.
UV-B-induced lipase up-regulation. Wild-type Arabidopsis plants (3.5 weeks old) were treated for up to 4 consecutive days with UV-B. The daily treatment consisted of 16 h of exposure to photosynthetically active radiation plus UV-B followed by 8 h of darkness. Cumulative hours of actual exposure to UV-B and the days on which it occurred are indicated. A, Northern blot of total RNA isolated from rosette leaves after various periods of UV-B exposure. The blot was probed with the 3′ untranslated region of the lipase cDNA. Each lane contained 10 μg of RNA. B, Corresponding ethidium bromide-stained agarose gel of the fractionated RNA.
Antisense Transgenic Plants
Homozygous transgenic lines of Arabidopsis expressing antisense lipase cDNA were obtained by screening seeds of three successive generations on kanamycin. Three lines, 4-2A-5, 1-4C-8, and 3-2C-5, were selected based on their increased tolerance to sublethal UV-B stress.
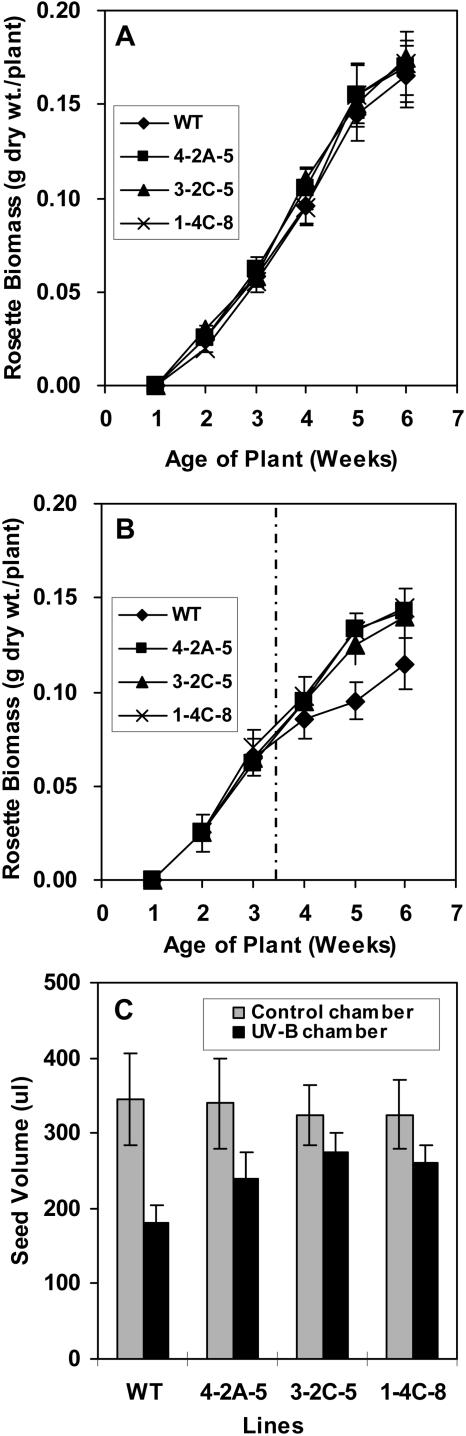
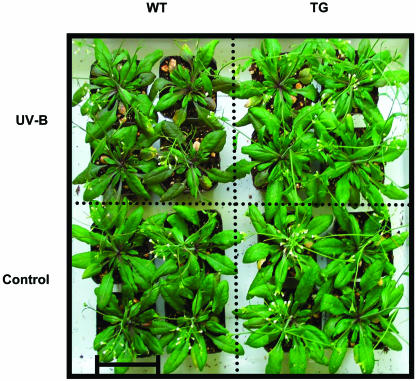
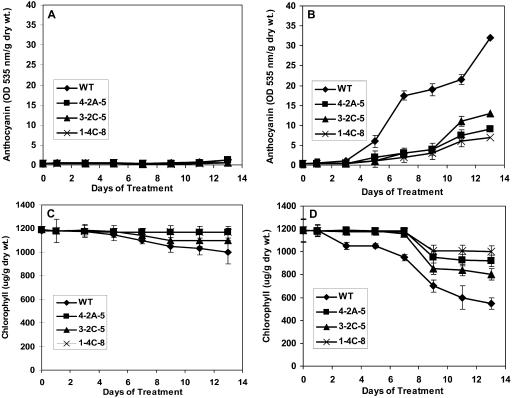
Transgenic and wild-type plants grown under normal conditions were indistinguishable at all stages of growth and development. This is evident, for example, from the fact there were no significant differences in biomass accumulation throughout growth and development (Fig. 7A). However, when wild-type and transgenic plants were maintained under conditions of sublethal UV-B stress from 3.5 weeks of age until they produced seed, the transgenic plants fared better. Within 4 d of the initiation of treatment, the transgenic plants appeared visually larger than wild-type plants, whereas wild-type and transgenic plants of the same age that were maintained under control conditions were of comparable size (Fig. 8). By 5 weeks of age, when the plants had been exposed to UV-B for 1.5 weeks, the biomass of wild-type plants was on average 30% less than that of the transgenic plants under conditions of UV-B stress (Fig. 7B). The reduction in seed yield attributable to the sublethal UV-B stress was also less for the transgenic lines 4-2A-5, 1-4C-8, and 3-2C-5 than for wild-type plants (Fig. 7C). In addition, the formation of anthocyanin was much more pronounced in the wild-type plants than in any of the transgenic lines, indicating that the transgenic plants are less susceptible to UV-B stress. Indeed, after 13 d of UV-B stress, levels of anthocyanin were only 25% of the level in wild-type plants for line 1-4C-8, 30% for line 4-2A-5, and 43% for line 3-2C-5 (Fig. 9B). By contrast, anthocyanin was barely detectable in the leaves of wild-type and transgenic lines in the absence of UV-B stress (Fig. 9A). The UV-B stress also induced premature leaf senescence as evidenced by a reduction in leaf chlorophyll content, but again the effect was more pronounced in wild-type plants than in the transgenic lines. For example, a reduction in leaf chlorophyll was evident by 3 d after initiation of the stress for wild-type plants but not until 8 d for the transgenic lines (Fig. 9D). After 13 d of treatment, leaf chlorophyll levels in wild-type plants had declined by 50%, whereas those for the transgenic plants had declined by only 15%, 23%, and 30% for lines 1-4C-8, 4-2A-5, and 3-2C-5, respectively (Fig. 9D). During the same period, there was virtually no reduction in leaf chlorophyll levels of wild-type and transgenic plants that were not subjected to UV-B stress (Fig. 9C).
Figure 7.
Effects of UV-B treatment on rosette biomass and seed yield of wild-type and transgenic lipase-suppressed Arabidopsis plants grown under sublethal UV-B stress from 3.5 weeks of age until seed maturation. A, Rosette biomass at various stages of development for plants grown under control conditions; B, rosette biomass at various stages of development for plants treated with UV-B (vertical dotted line indicates the initiation of UV-B treatment at 3.5 weeks of age); C, seed yield from 8-week-old plants grown under control conditions and treated with UV-B. WT, wild type; 4-2A-5, 3-2C-5, and 1-4C-8, transgenic lines. Means ± se for n = 8 are shown.
Figure 8.
Photograph of 3.5-week-old wild-type and transgenic lipase-suppressed Arabidopsis plants (line 4-2A-5) after 4 d of treatment with UV-B. WT, wild type; TG, transgenic. Bar = 6 cm.
Figure 9.
Effects of UV-B treatment on anthocyanin and chlorophyll levels in the rosette leaves of wild-type and transgenic lipase-suppressed Arabidopsis plants. At 3.5 weeks of age, plants were either treated with UV-B for a period of 14 d or maintained under normal growth conditions for 14 d. A and C, Control growth conditions; B and D, treated with UV-B. WT, wild type; 4-2A-5, 3-2C-5, and 1-4C-8, transgenic lipase-suppressed lines. Means ± se for n = 8 are shown.
Wild-type and antisense transgenic plants were also subjected to sublethal salt stress and drought stress. For salt stress, each plant received 50 mL of 20 mm NaCl every other day, and for drought stress, each plant received 50 mL of 15% polyethylene glycol every second day. The treatments were initiated when the plants were 3.5 weeks of age and were continued until they matured and produced seed. Unlike their response to UV-B stress, the transgenic and wild-type plants proved to be equally susceptible to drought stress and salt stress (data not shown).
Yet another indication of reduced sensitivity of the antisense lipase transgenic plants to UV is the finding that they were unable to up-regulate PR-1 in response to UV-B treatment. Up-regulation of PR-1 is a well-characterized molecular response to UV radiation (Logemann et al., 1995). Levels of PR-1 transcript in the rosette leaves of UV-B-treated wild-type plants increased relative to untreated control plants within 24 h of initiation of the treatment and remained high throughout the 4-d treatment period (Fig. 10A). However, for transgenic plants (line 4-2A-5), up-regulation of PR-1 transcript was not observed in the leaves of UV-B-treated plants, suggesting that lipase AAD21737.1 is required for this response to UV-B (Fig. 10B). Of note is the fact that up-regulation of PR-1 in UV-B-treated wild-type plants was detectable within 24 h of initiating the treatment (Fig. 10A), whereas UV-B-induced up-regulation of lipase AAD21737.1 was not apparent until 32 h after initiation of the treatment (Fig. 6). This likely reflects the fact that the lipase, although UV-B inducible, is also expressed constitutively in leaves (Fig. 5).
Figure 10.
Effects of UV-B treatment on PR-1 expression in wild-type and transgenic lipase-suppressed Arabidopsis plants. At 3.5 weeks of age, plants were either treated with UV-B for 4 d or maintained under normal growth conditions for 4 d. A, Northern blot of total RNA isolated from the rosette leaves of control and UV-B-treated wild-type plants. B, Northern blot of total RNA isolated from the rosette leaves of control and UV-B-treated transgenic (line 2-3A-5) plants. Corresponding ethidium bromide-stained agarose gels of fractionated RNA are shown. The intensities of the northern-blot bands for the UV-B lanes expressed as a percentage of the intensities of the corresponding control bands are also indicated. The blots were probed with full-length PR-1 cDNA. Each lane contained 10 μg RNA.
Southern analysis indicated that the genome of wild-type Arabidopsis contains only one copy of GenBank sequence AAD21737.1. Specifically, when genomic DNA was cut with EcoRI, a restriction enzyme that does not cut within the genomic sequence corresponding to GenBank sequence AAD21737, only a single restriction fragment was evident in Southern blots probed with the cDNA (data not shown). This finding is consistent with GenBank data indicating that the Arabidopsis genome only contains one copy of this gene. When genomic DNA from the transgenic lines 4-2A-5 and 1-4C-8 was digested with EcoRI, five restriction fragments in addition to the fragment corresponding to the endogenous gene were discernible in Southern blots (data not shown). This can be interpreted as indicating that these lines contain five copies of the antisense transgene. However, Southern blots for transgenic line 3-2C-5 featured only two copies of the transgene (data not shown).
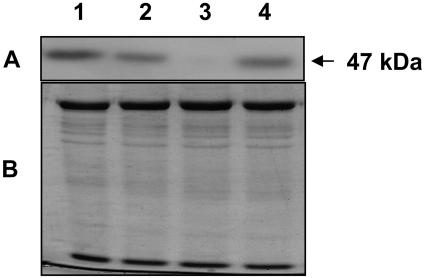
To confirm that expression of the lipase had been suppressed in the transgenic plants, levels of cognate lipase protein were compared in wild-type and transgenic plants by western blotting. Protein was isolated from 3.5-week-old Arabidopsis rosettes, fractionated by SDS-PAGE, transferred onto a membrane, and probed with α-LIP. The antibody recognized a 47-kD polypeptide on the blots, which is the expected size of the lipase (Fig. 11). Suppression of expression relative to corresponding wild-type tissue proved to be strongest in transgenic line 1-4C-8, next strongest in transgenic line 4-2A-5, and least strong in transgenic line 3-2C-5 (Fig. 11). Moreover, the level of suppression in the transgenic lines correlated with the degree to which the phenotype in response to UV-B stress was expressed. For example, increased tolerance to sublethal UV-B stress as demonstrated by reduced propensity to form leaf anthocyanin and decrease leaf chlorophyll was strongest in line 1-4C-8, which exhibited the highest suppression of AAD21737.1 lipase expression, and weakest in line 3-2C-5, the one showing least suppression of AAD21737.1 lipase expression (Fig. 9, B and D).
Figure 11.
Levels of lipase protein in the rosette leaves of 3.5-week-old wild-type and transgenic lipase-suppressed Arabidopsis plants. A, Western blot of rosette leaf homogenate. Lane 1, wild type; lane 2, transgenic line 4-2A-5; lane 3, transgenic line 1-4C-8; lane 4, transgenic line 3-2C-5. The blot was probed with lipase-specific antibody (B), corresponding Coomassie Blue-stained gel of fractionated protein.
DISCUSSION
Two lines of evidence indicate that the lipase AAD21737.1 encoded by GenBank sequence At2g42690 is capable of deesterifying fatty acids from complex lipids. First, the cognate protein contains the lipase consensus sequence [LIV]-X-[LIVAFY]-[LIAMVST]-G-[HYWV]-S-X-G-[GSTAC], which encompasses the active site for lipase-mediated fatty acid deesterification and is a characteristic feature of all known lipases that deesterify fatty acids. Second, the corresponding recombinant protein proved capable of deesterifying fatty acids from phospholipids, galactolipid, and triacylglycerol. However, the recombinant lipase exhibited a 2- to 4-fold higher preference for phospholipid substrate relative to galactolipid and triacylglycerol, indicating that it likely functions as a phospholipase in situ. Moreover, the putative phospholipase may possess molecular species specificity. This was evident from the finding that, although it readily deesterified fatty acids from purified soybean phosphatidylcholine, which contains a mixture of phosphatidylcholine molecular species, the lipase proved incapable of hydrolyzing fatty acids from selected individual molecular species of phosphatidylcholine, including 16:0/20:4 phosphatidylcholine, 20:4/20:4 phosphatidylcholine, 16:0/18:3 phosphatidylcholine, 16:0/18:2 phosphatidylcholine, 18:2/18:2 phosphatidylcholine, and 18:3/18:3 phosphatidylcholine. Although it is possible that this reflects the presence of some unknown factor required for lipase activity that is present in the soybean phosphatidylcholine substrate but not in the individual molecular species substrates, this seems unlikely inasmuch as all of the substrates were purchased as purified products. Moreover, the recombinant lipase proved to be active, although to a lesser degree, on trilinolein and MGDG, which, like soybean phosphatidylcholine, contain a mixture of molecular species. As well, 16:0/18:2 phosphatidylcholine substrate, one of the molecular species tested with lipase AAD21737.1, has been shown to be readily hydrolyzed by recombinant protein corresponding to the Arabidopsis lipase DEFECTIVE IN ANTHER DEHISCENCE1 after emulsification by the same protocol used in this study (Ishiguro et al., 2001). This further strengthens the contention that, as for soybean phosphatidylcholine, emulsifying individual molecular species by sonication in gum Arabic engenders the formation of liposomes. These observations, taken together, suggest, although do not prove, that the phospholipase only deesterifies specific molecular species of phosphatidylcholine.
Although the protein corresponding to Arabidopsis sequence AAD21737.1 appears to be a phospholipase capable of hydrolyzing membrane lipids, western-blot analysis and confocal microscopy indicated that it is localized in the cytosol. This is consistent with the fact that it is composed of mainly hydrophilic amino acids and does not contain any putative transmembrane domains. Nor does it exhibit a plastid- or mitochondrial-targeting sequence. Moreover, the lipase protein proved to be detectable in all organs of the Arabidopsis plant, although it is more abundant in leaves, stems, flowers, and siliques than in seeds. There are several previous reports of lipases that act on membrane lipids being localized in the cytosol, including lipolytic acyl hydrolases (Galliard, 1971), lipases expressed in senescing carnation flowers (Hong et al., 2000) and senescing Arabidopsis leaves (He and Gan, 2002), and PLD (Wang, 2001). It has also been reported that animal phospholipase exists in the cytosol in a nonphosphorylated form, which, upon phosphorylation, is able to associate with membranes and deesterify phospholipids (Shiina and Tazawa, 1986). Accordingly, the finding in this study that sequence AAD21737.1 contains putative phosphorylation sites raises the possibility that, as in animals, the protein may convert from an inactive nonphosphorylated form to an active phosphorylated form capable of associating with membranes.
Transgenic plants with suppressed lipase expression were indistinguishable from wild-type plants, indicating that this lipase is not essential for normal growth and development. As well, transgenic and wild-type plants responded in a similar manner to salt and drought stress. However, the transgenic plants exhibited increased tolerance to sublethal UV-B stress in comparison with wild-type plants. Typical symptoms of UV-B stress include inhibition of growth (Day et al., 2001), decreased biomass (Tosserams et al., 2001), and loss of photosynthetic capability (Bassman et al., 2001). For wild-type plants, the conditions of UV-B stress used in this study resulted in decreased growth, a reduction in biomass and seed yield, reduced levels of leaf chlorophyll, and formation of leaf anthocyanin. For transgenic plants, however, all of these symptoms were reduced in comparison to wild-type plants. Moreover, the degree of increased tolerance to UV-B stress exhibited by the transgenic lines correlated with the degree to which expression of the lipase had been reduced. For example, anthocyanin formation and reduced levels of chlorophyll following exposure to UV-B were least pronounced in line 1-4C-8 exhibiting the highest suppression of lipase expression and more pronounced in line 3-2C-8 showing less suppression of the lipase. That UV-B-induced anthocyanin formation was clearly evident in wild-type plants and suppressed in the transgenic antisense lipase plants is consistent with reports of up-regulated transcript levels for chalcone synthase, a component of the anthocyanin biosynthesis pathway, in response to UV-B (Jenkins et al., 2001; Jordan, 2002). Further evidence linking sequence AAD21737.1 to UV-B comes from the finding that it is strongly up-regulated in wild-type plants subjected to UV-B stress. Low constitutive levels of lipase transcript were detectable by northern blotting in total RNA preparations from all organs of wild-type Arabidopsis plants. However, within 3 to 4 d of the initiation of sublethal UV-B stress, strong up-regulation of lipase transcript was evident in the rosette leaves of wild-type plants coincident with the appearance of in planta symptoms of the stress. This, together with the fact that suppression of AAD21737.1 lipase in transgenic plants delays the development of UV-B stress symptoms, suggests that this phospholipase plays a role in the response of plants to UV-B irradiation.
There is increasing evidence that UV-B-induced changes in gene expression are mediated by a number of distinct signal transduction pathways (Jordan, 2002). One element of this signal transduction is an enhanced titer of reactive oxygen species (ROS; Allan and Fluhr, 1997; AH-Mackerness et al., 2001). These ROS are thought to be formed through the actions of NADPH oxidase and peroxidase(s) and to be involved in UV-B-induced changes in gene expression, including down-regulation of transcripts for photosynthetic proteins (Jordan et al., 1998; Jordan, 2002). ROS do not, however, appear to be required for up-regulation of chalcone synthase transcripts in response to UV-B (Jenkins et al., 2001), which is in keeping with the contention that the effects of UV-B on plants are mediated by a number of signal transduction pathways (Jordan, 2002). That UV-B induces lipid peroxidation in membranes is also well documented (AH-Mackerness et al., 1998; Dawar et al., 1998; Allan and Fluhr, 1997). Peroxidation of membrane fatty acids engenders bilayer instability because the modified fatty acids perturb the packing of lipid molecules (Thompson et al., 1987). Peroxidized phospholipids, like diunsaturated molecular species of phospholipid, are also more prone to lipase-mediated deesterification because their perturbed structure facilitates access of the lipase to the ester bond linking the fatty acid to the glycerol backbone (Thompson et al., 1987). The finding that lipase AAD21737.1 is strongly up-regulated in response to UV-B raises the possibility that it deesterifies peroxidized fatty acids from phospholipids. As shown in this study, it readily deesterifies fatty acids from nonperoxidized phospholipids, and peroxidation of fatty acids by UV-B would render such phospholipids even more prone to deesterification. Free peroxidized fatty acids, like their unperoxidized counterparts, act as detergents in membranes, which, if allowed to accumulate, engender destabilization of bilayer structure and even dissolution of membranes. Indeed, loss of membrane function attributable to lipid peroxidation is known to be a manifestation of UV-B damage (Dawar et al., 1998; Bassman et al., 2001). The amelioration of UV-B damage in transgenic plants with suppressed lipase AAD21737.1 noted in this study may, therefore, reflect reduced deesterification of peroxidized as well as nonperoxidized fatty acids and a corresponding reduction in bilayer destabilization leading to loss of membrane function.
Peroxidized linoleic acid and linolenic acid are also formed in membranes through the octadecanoid pathway. However, levels of these fatty acid hydroperoxides formed through the octadecanoid pathway are normally low, as it is a signal transduction pathway. Moreover, they are further metabolized by additional enzymes in the pathway and ultimately converted to volatile signaling compounds that do not accumulate in membranes (Gardner, 1995). Thus, the operation of this pathway under normal conditions would not be expected to result in perturbation of bilayer structure. However, the high levels of fatty acid hydroperoxides produced in response to UV-B irradiation are likely to be in excess of the throughput capacity of the octadecanoid pathway, resulting in their accumulation in membrane bilayers and ensuing loss of membrane function. Moreover, UV-B-induced formation of fatty acid hydroperoxides is likely to occur in all membranes, not just those that contain a functional octadecanoid pathway.
Yet another indication of reduced sensitivity of the antisense lipase transgenic plants to UV is the finding that they were unable to up-regulate PR-1 in response to UV-B treatment. Up-regulation of PR-1 was clearly evident in the rosette leaves of UV-B-treated wild-type plants but was not discernible in UV-B-treated transgenic plants. UV irradiation is known to simulate some of the effects of pathogen ingression and wounding on gene expression (AH-Mackerness et al., 1999; Roldan-Arjona et al., 2000; Desikan et al., 2001). One such effect is up-regulation of PR-1, attributable to jasmonic acid produced by the octadecanoid pathway (Logemann et al., 1995; Thalmair et al., 1996; AH-Mackerness et al., 1999). Thus, the finding in this study that suppression of the AAD21737.1 lipase inhibits UV-B-induced up-regulation of PR-1 may suggest that this lipase participates in the octadecanoid pathway by deesterifying fatty acid substrate for lipoxygenase. It is to be noted, however, that AAD21737.1 is quite distinct from the Arabidopsis gene denoted DEFECTIVE IN ANTHER DEHISCENCE1, which has been reported to encode a phospholipase A1 catalyzing the initial step in the octadecanoid pathway (Ishiguro et al., 2001).
MATERIALS AND METHODS
Plant Material
Seeds of Arabidopsis, ecotype Columbia, were sown in soil (Premier Pro-Mix BX; Premier Brands, Brampton, ON, Canada), cold treated at 4°C for 2 d, and germinated in a growth chamber at 23°C under 150 μmol m−2 s−1 photosynthetically active radiation in 16-h-light/8-h-dark photoperiods. Seedlings were grown to maturity using the same chamber conditions.
Gene Isolation
Full-length cDNA corresponding to the Arabidopsis GenBank protein sequence, accession number AAD21737.1, was obtained by RT-PCR. Template RNA was isolated from the rosette leaves of 4-week-old Arabidopsis plants according to Davis et al. (1986). The forward (5′CTGGAATTCTATGGATGACGGCGGAAGATATTC3′) and reverse (5′GACTGCAGTCGACATCGATTTTTTTTTTTT3′) primers contained EcoRI and PstI restriction enzyme sites, respectively, to enable in-frame ligation of the PCR product into the protein expression vector pTrc 99a. The reverse transcription reaction mixture contained diethyl pyrocarbonate water, 5× reaction buffer, 10 mm dNTP, 50 mm RNase inhibitor, 5 units Reverse Transcriptase AMV, and 50 mm Random Primer Mix (Roche Diagnostics, Laval, Canada) in a total volume of 100 μL. The reaction was carried out in a programmable thermal cycler (GeneAmp PCR System 2400; Perkin-Elmer, Foster City, CA) at 55°C for 1 h, followed by a 10-min incubation at 65°C to inactivate the reverse transcriptase. The cDNA product was amplified for 35 cycles in 50 μL of reaction mixture containing 10× Tsg buffer, 10 mm dNTPs, 50 pmol each of forward and reverse primer, 15 mm MgCl2, and 5 units Tsg DNA polymerase (BioBasic, Scarborough, Canada). Each cycle consisted of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, extension at 72°C for 2 min, and a final extension at 72°C for 10 min. The PCR product was purified by agarose-gel electrophoresis, digested with EcoRI and PstI, and subcloned into pTrc 99a for sequencing.
Preparation of Recombinant Protein
Recombinant protein was obtained by expressing pTrc 99a containing the cDNA insert corresponding to Arabidopsis GenBank protein sequence, accession number AAD21737.1, in Escherichia coli DH5α. Empty vector was used as a control. Expression was induced by treatment of 50-mL bacterial cultures during log-phase growth with 3 mm isopropylthio-β-galactoside for 6 h at 30°C. The bacteria were then pelleted, resuspended in 1 mL of prechilled TBST lysis buffer (100 mm Tris [pH 8.0], 150 mm NaCl, 1% Triton X-100, 1 mm phenylmethylsulfonyl fluoride), and lysed by sonication (10 8-s pulses in a Branson sonifier cell disruptor 200 [Danbury, CT] set at level 6). The resultant suspension was centrifuged at 14,000g for 10 min at 4°C, and the pellet was washed five times by resuspension in TBST buffer and centrifugation at 14,000g for 10 min. The washed pellet was resuspended in 500 μL of TBST buffer and used directly for assays of lipase activity.
Lipase Assay
Lipase activity was assayed as described by Ishiguro et al. (2001). The reaction mixture contained 50 mm K-phosphate buffer (pH 6.0), 0.2% Triton X-100, 50 μL of crude bacterial extract (corresponding to 5 mg of total protein), and 0.8 mg/mL of specific lipid substrate in a total volume of 500 μL. The lipid substrates were emulsified by sonication in 5% gum Arabic before they were added to the reaction mixture. That the emulsification resulted in formation of discrete lipid particles was confirmed by electron microscopy. A droplet of the emulsified mixture was placed on a formvar-coated copper grid. After 1 min, the excess moisture was absorbed using filter paper, and the grid was positively stained for 1 min with uranyl acetate-saturated 70% ethanol and air-dried. The samples were examined using a Philips 300 transmission electron microscope (Eindhoven, The Netherlands) operating at 60 kV.
The reaction mixture was incubated for 2 h at room temperature on a rotator. The lipids were extracted in 1 mL of chloroform and resuspended in 50 μL of water. Levels of nonesterified fatty acids were measured using the NEFA colorimetric kit (Wako Chemicals, Neuss, Germany). In some experiments, nonesterified fatty acids released during the lipase reaction were identified and quantified by gas chromatography. For this purpose, the chloroform extract of the reaction mixture was dried under nitrogen gas, resuspended in approximately 100 μL of 6:1 (v/v) chloroform:methanol containing 15 μg of heptadecanoic acid internal standard and fractionated by TLC (SIL G-25, 0.25-mm silica gel layer; Macherey-Nagel, Duren, Germany) in petroleum ether:diethylether:acetic acid (7:3:1). The fractionated lipids were identified using authentic standards (dioleoyl l-α-phosphatidylethanolamine, dilinolenin, linolenic acid, trilinolein, cholesteryl arachidonate). The separated free fatty acids, which comigrated with the linolenic acid standard during TLC, were scraped from the plate, eluted from the silica gel in methanol:chloroform:water (2:1:0.8, v/v/v), transmethylated with boron trifluoride-methanol to form fatty acid methyl esters, and analyzed using a Hewlett-Packard HP 5890 series II gas chromatograph (Palo Alto, CA) equipped with a Supelco 2330 fused silica capillary column (15 m × 0.25 mm, i.d., 0.25 μm film thickness), a split inlet sleeve packed with 10% OV-1 on Chromosorb-W (78.5 × 6.3 mm), and a flame ionization detector.
Subcellular Fractionation
Cytosolic and microsomal membrane fractions were isolated from the rosette leaves of 3.5-week-old Arabidopsis plants. The leaves (20 g) were homogenized at 4°C in 60 mL of homogenization buffer [50 mm 4-(2-hydroxyethyl)piperazine-1-propanesulfonic acid (EPPS; pH 7.4), 0.25 m sorbitol, 10 mm EDTA, 2 mm EGTA, 1 mm dithiothreitol (DTT), 10 mm amino-n-caproic acid, 50 μg chymostatin, 4% polyvinylpolypyrrolidone (w/v)] containing 1 mm benzamidine, 1 mm phenylmethylsulfonyl fluoride, and a few drops of antifoam, using an Omni-mixer (eight 3-s bursts at a setting of 10), followed by treatment for 60 s with a Polytron homogenizer (setting of 6; Brinkmann Instruments, Westbury, NY). The homogenate was filtered through four layers of cheesecloth and centrifuged for 20 min at 12,000g. The pellet was discarded, and the supernatant was further centrifuged for 1 h at 305,000g. The resultant pellet of microsomal membranes was resuspended in 2 mL of resuspension buffer (50 mm EPPS [pH 7.0], 0.25 m DTT, 10 mm amino-n-caproic acid, 50 μg chymostatin), and the supernatant (cytosolic fraction) was centrifuged again at 305,000g for 12 h to sediment any residual membrane.
Intact chloroplasts were isolated essentially as described by Kunst (1998). Briefly, 20 g of rosette leaves from 3.5-week-old Arabidopsis plants were floated on ice-cold water for 30 min, blotted dry, and homogenized in 200 mL of homogenization buffer (0.45 m sorbitol, 20 mm Tricine-KOH, 10 mm EDTA, 10 mm NaHCO3, 0.05% NaN3) using an Omni-mixer (three 3-s bursts at maximum speed) and a Polytron homogenizer (three 3-s bursts at maximum speed). The homogenate was filtered through Miracloth (Calbiochem, San Diego) and centrifuged at 5,500 rpm in an SS-34 Sorvall rotor (DuPont Instruments, Newtown, CT) for 30 s. The pellets were resuspended in 1 mL of ice-cold 1× resuspension buffer (RB; 2× RB: 0.6 m sorbitol, 40 mm Tricine-KOH, 10 mm MgCl2, 5 mm EDTA, 0.05% NaN3), overlaid on a Percoll gradient (30 mL of Percoll and 30 mL of 2× RB), and centrifuged at 13,300g for 6 min using an SS-34 Sorvall rotor. The Percoll gradient was prepared beforehand by centrifuging the solution in two Oakridge tubes using an SS-34 Sorvall rotor for 30 min at 43,500g without brakes and stored on ice until use. Intact and broken chloroplasts corresponding to upper and lower diffuse bands, respectively, were removed with a Pasteur pipette. Intact chloroplasts were further fractionated into stroma and chloroplastic membranes (thylakoids and chloroplast envelope) by resuspension in lysis buffer (LB; 10 mm Tricine, 5 mm MgCl2), incubation on ice for 30 min in the dark, and centrifugation at 12,000g for 10 min yielding stroma (the supernatant) and a pellet of chloroplastic membranes.
Preparation of Transgenic Plants
Suppression of the endogenous Arabidopsis sequence (GenBank protein accession no. AAD21737.1) was achieved by expressing the corresponding cDNA in the antisense orientation under the regulation of a constitutive promoter in transgenic plants. For this purpose, an expressed sequence tag (EST; 221D24) corresponding to the lipase protein was obtained from the Arabidopsis Biological Resource Center. The EST lacks 213 nucleotides of the 5′ end of the coding region but is otherwise complete. It was subcloned into the binary vector pKYLX71 (Schardl et al., 1987) in the antisense orientation under the regulation of two copies of the constitutive cauliflower mosaic virus promoter (CaMV-35S). The pKYLX71 vector contains a tetracycline-resistance gene in the bacterial replication region as well a kanamycin-resistance gene (NPT) within the T-DNA region (between the right and left border). Binary vector bearing the antisense construct and binary vector alone, which served as a control, were introduced into Agrobacterium tumefaciens LBA 4404 by electroporation, and 4-week-old Arabidopsis plants were infected with transformed A. tumefaciens by vacuum infiltration (Bechtold et al., 1993). The vacuum-infiltrated plants were grown to maturity and the seed harvested.
Transgenic plants were selected by germinating seeds from the vacuum-infiltrated plants on media containing kanamycin. The seeds were surface-sterilized in a solution of 1% sodium hypochlorite and 0.1% Tween 80, rinsed three times in sterile water, and plated on 0.7% agar containing one-half Murashige and Skoog salt (Sigma-Aldrich, Oakville, Canada) and 50 μg/mL of kanamycin. The plates were maintained at 4°C for 2 d and then transferred to a tissue culture chamber operating at 22°C ± 3°C with 16-h-light (150 μmol m−2 s−1 photosynthetically active radiation)/8-h-dark cycles. After 12 d, surviving seedlings were transferred to soil (Pro-mix BX) and grown to maturity in a growth chamber under conditions specified above. Seed was harvested, and the selection process was repeated until homozygous lines exhibiting 100% germination on kanamycin-containing media were obtained.
The number of antisense gene insertions in homozygous transgenic lines was assessed by Southern-blot analysis. Genomic DNA was isolated from the full complement of rosette leaves from 4-week-old wild-type and transgenic plants and digested with the restriction endonuclease EcoRI. The digested products (10 μg DNA) were fractionated on an agarose gel, immobilized on a nylon membrane, and hybridized with 32P-labeled lipase EST 221D24 according to the method described by Wang et al. (2001).
Treatment with UV-B
Wild-type and transgenic plants at 3.5 weeks of age were transferred to a UV-B treatment chamber (187.94 μmol m−2·s−1 photosynthetically active radiation, 2.46 μmol m−2·s−1 UV-B, and 15.04 μmol m−2 s−1 UV-A) operating on a 24-h cycle of 16 h of irradiation followed by 8 h of darkness. Any incidental UV-C radiation from the UV lamps was removed by screening with cellulose acetate. Plants were maintained in the treatment chamber for various periods of time and harvested for analysis at specified time points during the treatment.
Northern- and Western-Blot Analysis
For northern-blot analysis, total RNA (10 μg) was fractionated on 1.0% denaturing formaldehyde-agarose gels and immobilized on Hybond-N+ nylon membrane (Amersham Pharmacia Biotech, Uppsala). The membrane was probed with 32P-labeled cDNA corresponding to the Arabidopsis EST 221D24 using hybridization conditions described by Wang et al. (2001). For western analysis, protein (20 μg) was fractionated on 12% SDS-polyacrylamide gels, and the separated proteins were transferred to polyvinylidene difluoride membrane (Bio-Rad, Hercules, CA). Immunoblotting was carried out according to Wang et al. (2001) using rabbit antiserum against a synthetic peptide corresponding to amino acids 386 to 407 of the lipase (GenBank protein accession no. AAD21737.1) as the primary antibody and alkaline phosphatase-conjugated secondary antibody (Roche Molecular Biochemicals, Basel).
Confocal Microscopy
Sections (2 mm2) of rosette leaves were vacuum-infiltrated with 2% paraformaldehyde in 25 mm K-phosphate buffer (pH 7.2) for 1 h at 4°C, washed three times for 1 h in deionized water, and treated with 1% Tween 20 in phosphate-buffered saline (PBS) for 30 min. The fixed tissue was then washed three times for 20 min in a mixture of 0.2% Gly and 0.2% Tween 20 in PBS, blocked in 2% ovalbumin, 0.2% Gly, and 0.2% Tween 20 in PBS for 30 min, incubated in lipase antiserum (primary antibody; dilution of 1:200 in the blocking solution) for 1 h, and then washed three times for 30 min in blocking solution. The tissue was then incubated for 2 h in blocking solution containing Alexa 400 goat anti-rabbit fluorescein (dilution of 1:400), washed three times for 30 min in blocking solution and for 30 min in PBS. The labeled tissue was then viewed under a laser scanning spectral confocal microscope (Leica TCS SP2; Leica Microsystems Canada, Richmond Hill, Canada).
Chemical Assays
Chlorophyll and anthocyanin were measured as described by Porra et al. (1989) and Lange et al. (1970), respectively. For protein extraction, 300 mg of tissue was ground to powder in liquid nitrogen with a mortar and pestle and mixed with 500 μL of extraction buffer (50 mm EPPS, 0.25 m sorbitol [pH 7.4], 10 mm EDTA, 2 mm EGTA, 1 mm DTT, 10 mm amino-n-caproic acid) in an Eppendorf tube. Protein was assayed according to Bradford (1976).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers At2g42690 and AAD21737.1.
This work was supported by the Natural Sciences and Engineering Research Council of Canada.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.036376.
References
- AH-Mackerness S, John CF, Jordan B, Thomas B (2001) Early signaling components in ultraviolet-B responses: distinct roles for different reactive oxygen species and nitric oxide. FEBS Lett 489: 237–242 [DOI] [PubMed] [Google Scholar]
- AH-Mackerness S, Surplus SL, Blake P, John CF, Buchanan-Wollaston V, Jordan BR, Thomas B (1999) Ultraviolet-B-induced stress and changes in gene expression in Arabidopsis thaliana: role of signaling pathways controlled by jasmonic acid, ethylene and reactive oxygen species. Plant Cell Environ 22: 1413–1423 [Google Scholar]
- AH-Mackerness S, Surplus SL, Jordan BR, Thomas B (1998) Effects of supplementary ultraviolet-B radiation on photosynthetic transcripts at different stages of leaf development and light levels in pea (Pisum sativum L.): role of active oxygen species and antioxidant enzymes. Photochem Photobiol 68: 88–96 [Google Scholar]
- Allan AC, Fluhr R (1997) Two distinct sources of elicited reactive oxygen species in tobacco epidermal cells. Plant Cell 9: 1552–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MM, McCarty RE, Zimmer EA (1974) The role of galactolipids in spinach chloroplast lamellar membranes. I. Partial purification of bean leaf galactolipid lipase and its action on sub-chloroplast particles. Plant Physiol 53: 699–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassman JH, Robberecht R, Edwards GE (2001) Effects of enhanced UV-B radiation on growth and gas exchange in Populus deltoides Bartr. X Marsh. Int J Plant Sci 162: 103–110 [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. CR Acad Sci Paris Life Sciences 316: 1194–1199 [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method of the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Davis LG, Dibner MD, Battey JF (1986) Basic Methods in Molecular Biology. Elsevier Publishing, New York, NY, pp 130–135
- Dawar S, Vani T, Singhal GS (1998) Stimulation of antioxidant enzymes and lipid peroxidation by UV-B irradiation in thylakoid membrane of wheat. Biol Plant 41: 65–73 [Google Scholar]
- Day TA, Ruhland CT, Xiong FS (2001) Influence of solar ultraviolet-B radiation on Antarctic terrestrial plants: results from a 4-year study. J Photochem Photobiol B 62: 78–87 [DOI] [PubMed] [Google Scholar]
- Derewenda ZS, Derewenda U (1991) Relationships among serine hydrolases: evidence for a common structural motif in triacylglyceride lipases and esterases. Biochem Cell Biol 69: 842–851 [DOI] [PubMed] [Google Scholar]
- Desikan R, Mackerness SA-H, Hancock JT, Neill SJ (2001) Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol 127: 159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman-Silvestre I, Bureau J, Trémolières A, Paulin A (1989) Changes in membrane phospholipids and galactolipids during the senescence of cut carnations: connection with ethylenic rise. Plant Physiol Biochem 27: 931–937 [Google Scholar]
- Fan L, Sheng S, Wang X (1997) Antisense suppression of phospholipase D retards abscisic acid- and ethylene-promoted senescence of postharvest Arabidopsis leaves. Plant Cell 9: 2183–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galliard T (1971) The enzymatic deacylation of phospholipids and galactolipids in plants. Purification and properties of a lipolytic acyl hydrolase from potato tubers. Biochem J 121: 379–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner HW (1995) Biological roles and biochemistry of the lipoxygenase pathway. Hortic Sci 30: 197–205 [Google Scholar]
- He Y, Gan SA (2002) Gene encoding an acyl hydrolase is involved in leaf senescence in Arabidopsis. Plant Cell 14: 805–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Wang TW, Hudak KA, Schade F, Froese CD, Thompson JE (2000) An ethylene-induced cDNA encoding a lipase expressed at the onset of senescence. Proc Natl Acad Sci USA 97: 8717–8722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro S, Kawai-Oda A, Ueda J, Nishida I, Okada K (2001) The DEFECTIVE IN ANTHER DEHISCENCE1 gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13: 2191–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaniuga Z, Gemel J (1984) Galactolipase activity and free fatty acid levels in chloroplasts – novel approach to characteristics of chilling sensitivity of plants. FEBS Lett 171: 55–58 [Google Scholar]
- Jenkins GI, Long JC, Wade HK, Shenton MR, Bibikova TN (2001) UV and blue light signalling: pathways regulating chalcone synthase gene expression in Arabidopsis. New Phytol 151: 121–131 [DOI] [PubMed] [Google Scholar]
- Jordan BR (2002) Molecular response of plant cells to UV-B stress. Funct Plant Biol 29: 909–916 [DOI] [PubMed] [Google Scholar]
- Jordan BR, James P, A-H-Mackerness S (1998) Factors affecting UV-B induced changes in Arabidopsis thaliana gene expression: role of development, protective pigments and the chloroplast signal. Plant Cell Physiol 39: 769–778 [DOI] [PubMed] [Google Scholar]
- Kunst L (1998) Preparation of physiologically active chloroplasts from Arabidopsis. In JM Martin-Zapater, J Salinas, ed, Methods in Molecular Biology, Arabidopsis Protocols, Vol 82. Humana Press, Totowa, NJ, pp 43–48 [DOI] [PubMed]
- Lange H, Shropshire W Jr, Mohr H (1970) An analysis of phytochrome-mediated anthocyanin synthesis. Plant Physiol 47: 649–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann E, Wu S-C, Schroeder J, Schmelzer E, Somssich IE, Hahlbrock K (1995) Gene activation by UV light, fungal elicitor or fungal infection in Petroselinum crispum is correlated with repression of cell cycle-related genes. Plant J 6: 865–876 [DOI] [PubMed] [Google Scholar]
- Matos AR, d'Arcy-Lamet A, Franca M, Zuily-Fodil Y, Pham-Thi AT (2000) A patatin-like protein with galactolipase activity is induced by drought stress in Vigna unguiculata leaves. Biochem Soc Trans 28: 779–781 [PubMed] [Google Scholar]
- Munnik T, Irvine RF, Musgrave A (1998) Phospholipid signaling in plants. Biochim Biophys Acta 1389: 222–272 [DOI] [PubMed] [Google Scholar]
- O'Sullivan JN, Warwick NWM, Dalling MJ (1987) A galactolipase activity associated with the thylakoids of wheat leaves (Triticum aestivum L.). J Plant Physiol 131: 393–404 [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Roldan-Arjona T, Garcia-Ortiz M-V, Ruiz-Rubio M, Ariza RR (2000) cDNA cloning expression and functional characterization of an Arabidopsis thaliana homologue of the Escherichia coli DNA repair enzyme endonuclease III. Plant Mol Biol 44: 43–52 [DOI] [PubMed] [Google Scholar]
- Sahsah Y, Campos P, Gareil M, Zuily-Fodil Y, Pham-Thi AT (1998) Enzymatic degradation of polar lipids in Vigna unguiculata leaves and influence of drought stress. Physiol Plant 104: 577–586 [Google Scholar]
- Sastry PS, Kates M (1964) Hydrolysis of monogalactosyl and digalactosyl diglycerides by specific enzymes in runner-bean leaves and influence of drought stress. Biochemistry 14: 1280–1287 [DOI] [PubMed] [Google Scholar]
- Schardl CL, Byrd AD, Benzio G, Altschuler MA, Hildebrand DF, Hunt AG (1987) Design and construction of a versatile system for the expression of foreign genes in plants. Gene 61: 1–11 [DOI] [PubMed] [Google Scholar]
- Scherer GF (1995) The functional relationship of plant lipid-derived second messengers and plant lipid-activated protein kinase. Biochem Soc Trans 23: 871–875 [DOI] [PubMed] [Google Scholar]
- Senda K, Yoshioka H, Doke N, Kawakita K (1996) A cytosolic phospholipase A2 from potato tissues appears to be patatin. Plant Cell Physiol 37: 347–353 [DOI] [PubMed] [Google Scholar]
- Shiina T, Tazawa M (1986) Regulation of membrane excitation by protein phosphorylation in Nitellopsis obtusa. Protoplasma 134: 60–61 [Google Scholar]
- Thalmair M, Bauw G, Thiel S, Doehring T, Langebartels C, Sandermann H Jr (1996) Ozone and ultraviolet B effects on the defense-related proteins beta-1,3-glucanase and chitinase in tobacco. J Plant Physiol 148: 222–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JE, Froese CD, Madey E, Smith MD, Hong Y (1998) Lipid metabolism during plant senescence. Prog Lipid Res 372: 119–141 [DOI] [PubMed] [Google Scholar]
- Thompson JE, Legge RL, Barber RF (1987) The role of free radicals in senescence and wounding. New Phytol 105: 317–344 [DOI] [PubMed] [Google Scholar]
- Tosserams M, Visser A, Groen M, Kalis G, Magendans E, Rozema J (2001) Combined effects of CO2 concentration and enhanced UV-B radiation on faba bean. Plant Ecol 154: 195–210 [DOI] [PubMed] [Google Scholar]
- Wang TW, Lu L, Wang D, Thompson JE (2001) Isolation and characterization of senescence-induced cDNAs encoding deoxyhypusine synthase and eukaryotic translation initiation factor 5A from tomato. J Biol Chem 276: 17541–17549 [DOI] [PubMed] [Google Scholar]
- Wang X (2001) Plant phospholipases. Annu Rev Plant Physiol Plant Mol Biol 52: 211–231 [DOI] [PubMed] [Google Scholar]
- Xie Z, Ho WT, Exton JH (2000) Association of the N- and C-terminal domains of phospholipase D: contribution of the conserved HKD motifs to the interaction and the requirement of the association for Ser/Thr phosphorylation of the enzyme. J Biol Chem 275: 24962–24969 [DOI] [PubMed] [Google Scholar]