Abstract
In resurrection plants and yeast, trehalose has a function in stress protection, but the absence of measurable amounts of trehalose in other plants precludes such a function. The identification of a trehalose biosynthetic pathway in angiosperms raises questions on the function of trehalose metabolism in nonresurrection plants. We previously identified a mutant in the Arabidopsis trehalose biosynthesis gene AtTPS1. Plants homozygous for the tps1 mutation do not develop mature seeds (Eastmond et al., 2002). AtTPS1 expression analysis and the spatial and temporal activity of its promoter suggest that this gene is active outside the seed-filling stage of development as well. A generally low expression is observed in all organs analyzed, peaking in metabolic sinks such as flower buds, ripening siliques, and young rosette leaves. The arrested tps1/tps1 embryonic state could be rescued using a dexamethasone-inducible AtTPS1 expression system enabling generation of homozygous mutant plants. When depleted in AtTPS1 expression, such mutant plants show reduced root growth, which is correlated with a reduced root meristematic region. Moreover, tps1/tps1 plants are retarded in growth and remain generative during their lifetime. Absence of Trehalose-6-Phosphate Synthase 1 in Arabidopsis plants precludes transition to flowering.
Trehalose (α-d-glucopyranosyl-[1,1] -α-d-glucopyranoside) accumulation has been observed in a variety of species, most typically in anhydrobionts, which are able to survive complete dehydration. However, trehalose was not detected in plant species, with the exception of stress-induced trehalose accumulation in resurrection plants such as Selaginella lepidophylla (Crowe et al., 1992). Recent isolation of trehalose biosynthetic genes from Arabidopsis (Blazquez et al., 1998; Vogel et al., 1998; Müller et al., 2001) has led to the identification of a trehalose biosynthesis pathway in many, if not all, angiosperms. The first trehalose biosynthetic step is catalyzed by trehalose-6-P synthase (TPS), which converts Glc-6-P and UDP-Glc into trehalose-6-P (T-6-P). The second step is catalyzed by T-6-P phosphatase (TPP), which hydrolyzes T-6-P and releases trehalose. This biosynthesis pathway is similar in plants and yeast (Saccharomyces cerevisiae), where this metabolism was first described (Cabib and Leloir, 1958). Trehalose is readily hydrolyzed into two Glc units by trehalase (TRE), which is present in all organs of Arabidopsis and Glycine max (Müller et al., 2001).
The nonreducing disaccharide trehalose is highly resistant to nonenzymatic hydrolysis (Paiva and Panek, 1996) and is known to stabilize proteins during dehydration (Crowe et al., 1992). However, trehalose levels in nonresurrecting plants are barely detectable and insufficient for this function. It was proposed that trehalose metabolism may play a regulatory role in these plant species (Goddijn and Smeekens, 1998). The ability to utilize available sugars depends on trehalose biosynthesis and is most likely linked to the T-6-P intermediate (Eastmond et al., 2002; Schluepmann et al., 2003). Different TPS and TPP overexpressing transgenic plants displaying increased resistance to abiotic stress have been described (Holmström et al., 1996; Romero et al., 1997; Pilon-Smits et al., 1998; Goddijn and Van Dun, 1999; Paul et al., 2001; Garg et al., 2002; Jang et al., 2003; Schluepmann et al., 2003), but resistance phenotype in these plants did not correlate with trehalose accumulation (Jang et al., 2003). Observed effects on growth phenotypes in these transgenic lines were not attributed to increased trehalose levels. Tobacco (Nicotiana tabacum) overexpressing TPS from yeast (ScTPS1) or Escherichia coli (OTSA) display retarded growth and lancet-shaped dark green leaves (Goddijn and Van Dun, 1999). Furthermore, photosynthesis per unit leaf area is increased in OTSA-overexpressing tobacco plants, whereas it is reduced in OTSB (E. coli TPP) overexpressors (Paul and Pellny, 2003). Similarly, transgenic Arabidopsis plants expressing OTSA display opposing phenotypes to plants expressing OTSB and E. coli T-6-P hydrolase (TREC) (Schluepmann et al., 2003). TREC directly cleaves T-6-P into Glc and Glc-6-P without a trehalose intermediate. In OTSA-overexpressing plants, T-6-P levels are increased, whereas in OTSB- and TREC-expressing plants, T-6-P is reduced. Hence, studies with transgenic plants overexpressing microbial enzymes suggest that T-6-P may be a key regulator of metabolism.
The function of plant trehalose biosynthetic enzymes was investigated by identification of mutations in the Arabidopsis trehalose biosynthetic TPS1 gene, which was reported to complement yeast mutants defective in the homologous gene functions (Blazquez et al., 1998; Vogel et al., 1998; Müller et al., 2001). Reverse genetics identified transposon insertions in the AtTPS1 gene. The tps1-1 allele is located in the second exon and tps1-2 in the first exon, as described previously (Eastmond et al., 2002); a third allele obtained from the SAIL (Syngenta Arabidopsis Insertion Library) collection has a T-DNA insertion in the last exon of AtTPS1. All insertions display an embryonic growth arrest in the homozygous state. These embryo lethal phenotypes of homozygous plants carrying the tps1 mutation demonstrated that the trehalose biosynthetic pathway is essential for Arabidopsis embryo maturation (Eastmond et al., 2002).
We investigated the function of AtTPS1 at other stages of plant development. We analyzed AtTPS1 expression during development of wild-type plants and examined the effect of TPS1 deletion throughout Arabidopsis development. AtTPS1 gene expression is increasing during wild-type embryo development in concordance with its essential function during Arabidopsis embryo maturation. The presence of AtTPS1 mRNA is not restricted to the seed-filling stage but is observed in seedlings, roots, leaves, stems, flowers, and siliques. Expression is peaking in metabolic sink organs, including flower buds, ripening siliques, and young rosette leaves. TPS1 may therefore be important beyond the seed-filling process as well.
We used a dexamethasone (DEX)-inducible AtTPS1 system to study the tps1/tps1 phenotype throughout the plant life cycle. Such a system is essential to generate viable mutant seeds. Homozygous tps1 seedlings display delayed growth and short-root phenotypes compared to wild-type and heterozygous seedlings. tps1 mutant plants transgenic for the inducible AtTPS1 construct remain in the vegetative growth phase in the absence of transgene induction. DEX application induces the switch to flowering; however, the mutant plants flower several weeks later than wild type and develop many flowering stems. Comparable phenotypes were observed in mutant plants complemented with a bacterial TPS1 enzyme (OTSA). Hence, besides its necessity for seed maturation, AtTPS1 function is also important for vegetative growth and transition to flowering.
RESULTS
Cloning and Functional Analysis of the AtTPS1 Promoter
The function of plant trehalose metabolism in angiosperms was studied using mutants in the first step of Arabidopsis trehalose biosynthesis, identified using a reverse genetic approach (Eastmond et al., 2002). Homozygous tps1 plants arrest during the seed-filling stage of embryonic development, whereas heterozygous plants appear like wild type. Application of trehalose, T-6-P, or Suc does not allow rescue of tps1/tps1 embryos, which precludes study of the vegetative mutant phenotype. A 3.1-kb AtTPS1 promoter fragment and a 5.9-kb AtTPS1 gene fragment were isolated from an Arabidopsis genomic library. These fragments were combined in a wild-type complementation construct (AtTPS1::AtTPS1), which was then transformed to tps1-2 heterozygous mutant plants. Siliques of several T1 plants showed a mutant embryo frequency of one-fifteenth, in accordance with complementation by a single transgene locus. Complemented offspring seeds and plants were indistinguishable from wild type. Thus, expression of the wild-type AtTPS1 sequence fully rescues the tps1/tps1 embryonic defects, indicating that the 3.1-kb promoter is able to drive functional AtTPS1 gene expression during the essential stages of Arabidopsis embryo maturation.
AtTPS1 Is Expressed throughout the Plant
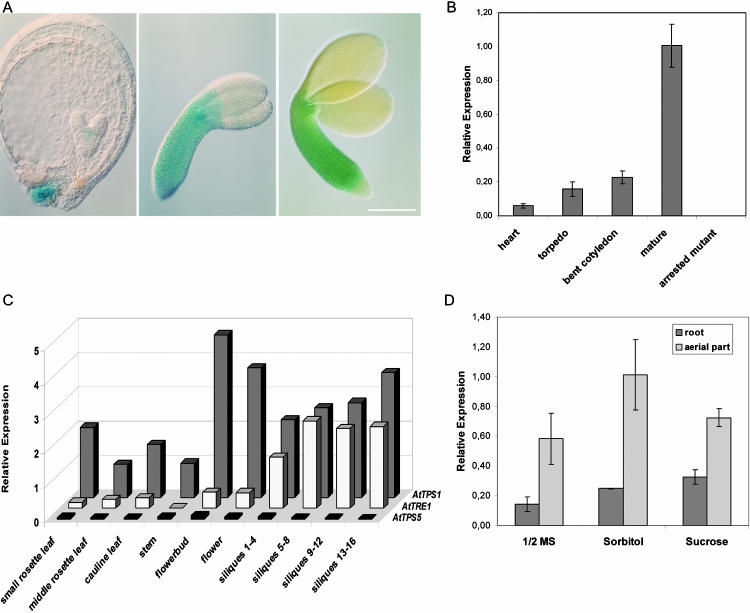
The 3.1-kb AtTPS1 promoter region was fused to the β-glucuronidase reporter gene (GUS) and 27 independent transgenic lines were generated, of which 11 were studied in more detail. In the AtTPS1::GUS-expressing transgenic lines, GUS activity was detected in developing seeds, where it appeared already in the shoot apical meristem of heart-stage embryos (Fig. 1A). Staining is also observed in the funiculus attachment region. The early embryonic expression might explain the growth retardation in tps1 mutant embryos prior to the developmental arrest (Eastmond et al., 2002). From torpedo stage onward, expression expands throughout the hypocotyl; in mature embryos, faint expression is also observed in cotyledon tips (Fig. 1A). In other plant organs, AtTPS1-specific GUS expression could not be detected. Sensitive quantitative PCR (Q-PCR) analysis was used with an AtTPS1-specific Taqman probe. All gene-specific expression levels were normalized for AtACTIN2 expression. Q-PCR confirms the strong increase of AtTPS1 expression during embryo maturation and its absence in tps1/tps1 embryos (Fig. 1B). In wild-type Arabidopsis seedlings and plant organs isolated from plants at principal growth stage 8, flowering plants with siliques ripening (Boyes et al., 2001), AtTPS1 expression is observed in all tissues analyzed. It peaks in metabolic sink organs, including flower buds, ripening siliques, and young rosette leaves (Fig. 1C). AtTPS1 mRNA level in roots is about one-third the level in the arial part of seedlings grown on half-strength Murashige and Skoog medium (MS; Fig. 1D). Presence of 1% Suc (29 mm) in the medium enhances seedling growth and increases the expression level slightly, as does sorbitol.
Figure 1.
Histochemical localization of GUS activity in AtTPS1::GUS transgenic plants during Arabidopsis seed development. From left to right in A, heart, torpedo, and mature stage. Bar = 1 mm. B, Relative AtTPS1 expression analyzed by Q-PCR in developing embryos; C, in distinct organs during plant growth of Arabidopsis; and D, in root and arial tissues of 8-d-old seedlings grown on half-strength MS, sorbitol, or Suc. Relative expression of AtTPS5 and AtTRE1 is displayed in C as a fraction of the AtTPS1 level in stem. Silique numbering was based upon appearance of the first silique below the lowest flower, with further numbering downward (n = 3).
In Arabidopsis, 11 TPS gene homologs are found, divided into classes I and II (Leyman et al., 2001). AtTPS1 belongs to class I. As an example of a class II gene, we studied the expression of TPS5. The expression level of TPS5 is low in all organs analyzed, when compared to the AtTPS1 expression level (Fig. 1C). These data further support the specificity of the primer-probe sets used. Similar to AtTPS1, TRE (AtTRE1) is expressed ubiquitously in Arabidopsis, and the TRE1 mRNA levels increase in ripening siliques. However, in young leaves and flowers, it does not correlate with increased TPS1 levels (Fig. 1C).
The requirement of a functional AtTPS1 during Arabidopsis embryo maturation is reflected in AtTPS1 gene expression, which increases during wild-type seed development. AtTPS1 is also expressed throughout the plant life cycle, peaking in metabolic sink organs. TPS1 might therefore play a role beyond the seed-filling process as well.
Conditional Rescue of the tps1 Mutation
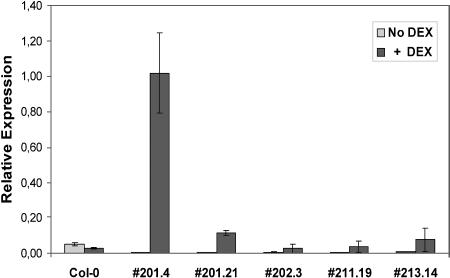
Inducible AtTPS1 expression would enable rescue of the tps1 embryonic defect and thus allow study of the AtTPS1 role throughout plant development. For this, the DEX-inducible system (Aoyama and Chua, 1997) was modified by replacing the 35S cauliflower mosaic virus promoter with the UBIQUITIN10 promoter of Arabidopsis (Sun and Callis, 1997) and inserting the AtTPS1 gene behind the upstream activation sequence to facilitate AtTPS1 expression during all stages of embryo development upon DEX application. Heterozygous mutant tps1-2 plants were transformed with this inducible TPS1 (ind-TPS1) construct. In total, 37 independent inducible lines were obtained, which were selected on phosphinotricin for the transposon insertion in tps1-2, on hygromycin for the inducible AtTPS1 transgene construct, and on DEX for generation of viable seeds. Fourteen T1 lines showed a mutant seed frequency of one-fifteenth instead of one-third, indicative of complementation by a single transgene locus. Viable seeds were harvested from these lines and all flower stems were removed, after which the T1 plants were repotted in fresh soil and grown without DEX induction. The newly formed siliques contained mutant embryos arrested in the torpedo stage of development corresponding to the tps1/tps1 embryonic arrested state, suggesting that the system is inducible, as expected. Four independent inducible mutant lines containing a single transgene insertion were selected to study in more detail. Selection was based on antibiotic resistance segregation and Southern-blot analysis (data not shown). Q-PCR analysis confirmed the absence of AtTPS1 expression in these lines under noninducing conditions and the presence of AtTPS1 mRNA following DEX treatment (Fig. 2). Control plants expressing inducible GUS and heterozygous TPS1/tps1 plants expressing the ind-TPS1 construct are indistinguishable from wild type both in the absence and presence of DEX, showing that DEX treatment is not affecting the plant phenotype nor causing an overexpression phenotype (data not shown). Thus, the DEX-inducible AtTPS1 system can rescue the arrested embryonic phenotype and is switchable, allowing study of the mutant phenotype beyond the seed maturation stage.
Figure 2.
Relative AtTPS1 expression analyzed by Q-PCR in wild-type (Col-0) and ind-TPS1 transgenic tps1/tps1 plants. Plants were grown for 1 month on soil, where DEX was added during watering of the plants (n = 3).
Deleting AtTPS1 Perturbs Root and Shoot Growth
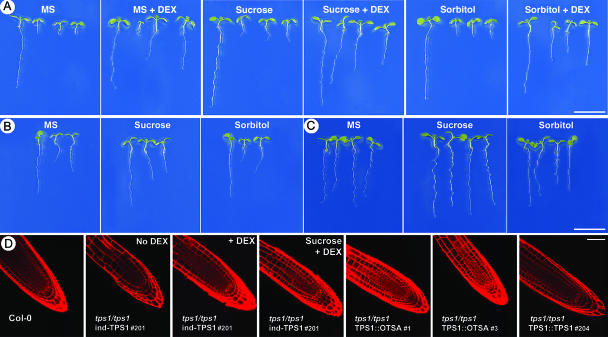
Rescued homozygous mutant seeds germinate in the absence of further DEX induction of the AtTPS1 gene. Mutant tps1 seedlings display a short root (Fig. 3A, MS) compared to wild type. DEX induction of AtTPS1 partially restores root length, whereas it does not affect wild type (Fig. 3A, MS + DEX). Exogenous sugar application of 1% (29 mm) Suc or sorbitol (osmotic control) does not stimulate root growth (Fig. 3, Suc or Sorbitol + DEX). Confocal microscopy shows that the cell patterning in tps1/tps1 roots is not affected (Fig. 3D). By contrast, the length of the meristematic region is much reduced in tps1/tps1 roots versus wild type. This is indicated by the presence of enlarged cells, which are visible immediately above the short meristematic region in the mutant picture. Conversely, the photograph of wild-type roots at this stage display only the meristematic region consisting of small cells (Fig. 3D, Col-0). Partial complementation is obtained in tps1/tps1 plants expressing E. coli TPS (OTSA, driven by the 3.1-kb AtTPS1 promoter), resulting in an intermediate root-length phenotype (TPS1::OTSA) (Fig. 3, B and D). Mutant plants expressing the wild-type TPS1 gene (AtTPS1::AtTPS1) display normal roots (Fig. 3, C and D).
Figure 3.
Root phenotype of 8-d-old seedlings grown on half-strength MS or half-strength MS containing 29 mm (1%) Suc or 29 mm sorbitol. A, Transgenic tps1/tps1 inducible AtTPS1 seedlings grown in the presence or absence of 10 μm DEX as indicated. In each section from left to right: Col-0 and ind-TPS1 lines 201, 211, and 213. B, Col-0 and transgenic tps1/tps1 AtTPS1::OTSA lines 1 and 3 in each section from left to right, respectively. C, Col-0 versus tps1/tps1 seedlings complemented with AtTPS1::AtTPS1. From left to right: Col-0 and TPS1::TPS1 lines 201, 205, and 204. Bar = 1 cm. D, Detail of root tips by confocal microscopy following propidium iodine staining. Root meristematic region in wild type is represented by files with small cells. In the tps1 mutant, this region is very short; cells immediately elongate and become large mature root cells. Bar = 0.1 mm.
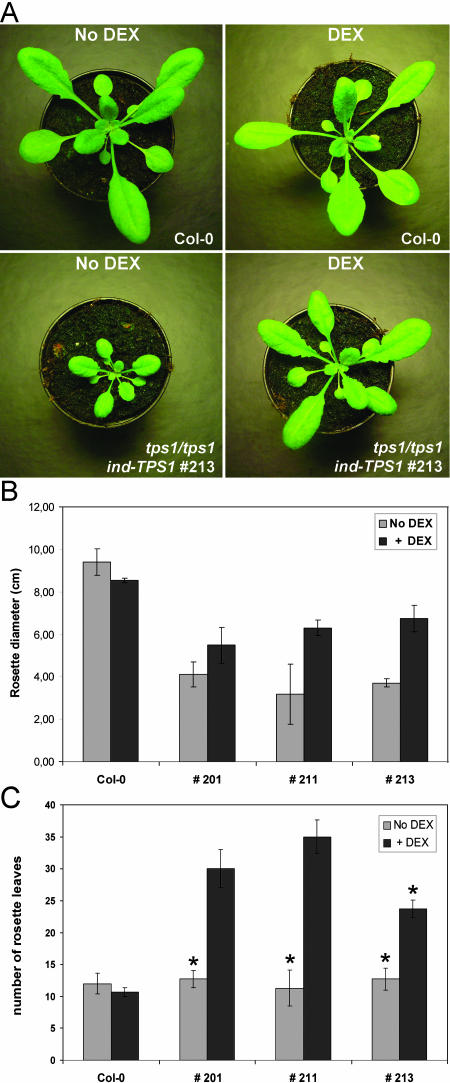
Following seedling establishment, the mutant plants are retarded in growth in the absence of DEX induction. Plants develop small, rounded rosette leaves, often with brownish leaf margins (Fig. 4A, left). The rosette diameter is largely reduced compared to wild type. Under inductive conditions, the rosette diameter is much increased; however, leaf and petiole elongation is still somewhat behind wild type grown under similar conditions (Fig. 4, A, right, and B). Without induction, the leaf initiation rate in tps1 mutant plants is comparable to wild type. This is reflected by a similar number of rosette leaves compared to wild type at the stage where wild type has developed its first floral bud (Fig. 4C, No DEX). Mutant plants grown in the presence of DEX application have a significantly increased leaf number at the time of the first floral bud appearance (Fig. 4C, +DEX).
Figure 4.
A, Wild type (top) and tps1/tps1 ind-TPS1 transgenic plants (bottom) grown for 1 month on soil in long-day light regime. Left, No DEX induction; right, plants grown with DEX treatment. B, Rosette diameters (cm) of 1-month-old soil-grown plants (as depicted in A) with and without DEX application. Wild type (Col-0) versus tps1/tps1 transgenic for ind-TPS1 lines 201, 211, and 213 (n = 3). C, Number of rosette leaves of soil-grown plants at the time when in wild type the first floral bud appeared. The same plant lines were used as described in B. No DEX, Growth without DEX. +DEX, Number of rosette leaves of plants grown in presence of DEX, at appearance of the first floral bud. Asterisk indicates plants without floral bud development.
AtTPS1 Is Essential for Floral Transition
Floral transition in tps1 mutant plants is dependent on DEX application (Fig. 5A). In the absence of DEX, the vegetative state is maintained until the plants die (>6 months after germination). All four AtTPS1-inducible mutant lines tested flower exclusively upon induction. In the presence of DEX, transition from vegetative to generative phase is delayed (Fig. 4C). Continuously induced mutant plants flower several weeks later than similarly treated wild-type plants. Such induced mutant plants showed a reduced apical dominance, resulting in the production of many flowering stems with small siliques and, regularly, aerial rosette formation (Fig. 5A, +DEX). Therefore, it is difficult to generate sufficient quantities of viable tps1/tps1 offspring seed. Growth under continuous light, short-day, or long-day conditions with or without application of 1% Suc does not affect flowering time; neither does vernalization of the seeds for 6 weeks prior to germination. This late-flowering phenotype of tps1 mutant plants might be a result of altered expression of inflorescence initiator genes. However, the expression of flowering-time genes CO, SOC1, FLC, FT, and LFY is not altered in the induced or noninduced mutant plants compared to wild type (Q-PCR analysis on 12-d-old seedlings grown in long-day conditions on half-strength MS or half-strength MS with 10 μm DEX; data not shown).
Figure 5.
Flowering phenotypes. A, tps1/tps1 plants expressing inducible AtTPS1, grown without (left) and with (right) DEX induction. B, Col-0 versus tps1/tps1 carrying the AtTPS1::OTSA transgene. C, Col-0 versus tps1/tps1 complemented with AtTPS1::AtTPS1. Bar = 5 cm.
The tps1/tps1 lines carrying the AtTPS1::OTSA transgene showed a range of phenotypes from wild type to much delayed in flowering time (Fig. 5B). Mutant plants expressing AtTPS1::AtTPS1 are fully complemented for seed maturation, root length, growth, and flowering time (Figs. 3C and 5C). Hence, besides its essential role in seed maturation, AtTPS1 activity is also required for normal growth and transition from the vegetative to the generative state in Arabidopsis.
DISCUSSION
TPS Gene Expression in Arabidopsis
The function of trehalose metabolism in nonresurrection plants is enigmatic. The work of Eastmond et al. (2002) and Schluepmann et al. (2003) afforded a glance at a possible signaling function. AtTPS1 is essential for Arabidopsis seed maturation (Eastmond et al., 2002), since tps1/tps1 embryos are developmentally arrested at the torpedo stage. Viable tps1/tps1 seeds can be obtained by introduction of E. coli OTSA under control of the AtTPS1 promoter, indicating that T-6-P is necessary for complementation (Schluepmann et al., 2003). The OTSA protein is a biosynthetically active enzyme that consists of a catalytic domain only (Goddijn and Van Dun, 1999). AtTPS1 contains an additional domain next to the catalytic domain. This C-terminal domain of AtTPS1 may harbor as-of-yet unknown functions. Absence of this domain in OTSA precludes such additional functions, which may explain partial complementation of the tps1 mutant phenotype by OTSA. Here, we further investigated the role of AtTPS1 throughout the Arabidopsis life cycle.
In the Arabidopsis genome, a total of 11 TPS homologs are present, which can be divided into two classes based upon their sequence homology (Leyman et al., 2001). Within each class, the DNA sequences are highly conserved, which requires use of gene-specific probes for quantitative measurements of gene expression levels. We developed gene-specific Taqman probes to determine the expression of AtTPS1 (class I) and AtTPS5 (class II) representatives by Q-PCR (see “Materials and Methods”).
The requirement for a functional TPS1 during Arabidopsis embryo maturation is reflected by AtTPS1 gene expression. Furthermore, AtTPS1 is expressed at generally low levels in all Arabidopsis organs tested, in spite of the presence of 10 additional TPS homologs. The question then arises whether these homologs are all biosynthetically active in distinct expression domains or whether they are part of a multimeric trehalose-synthesizing protein complex. Only for TPS7 and TPS8 was biosynthetic activity tested by complementation in the yeast tps1Δ mutant. Whereas AtTPS1 complements such a yeast mutant, complementation was not detected for AtTPS7 or AtTPS8 (Vogel et al., 2001). However, this does not exclude in planta TPS biosynthetic activity mediated by these proteins.
AtTPS1 expression in Arabidopsis is highest in metabolically active organs such as young rosette leaves, flower buds, ripening siliques, and maturing embryos (Fig. 1, B and C). In wild-type embryos, Suc levels are highest at the seed-filling stage when growth is arrested in the tps1 mutant (Eastmond et al., 2002), suggesting an involvement in metabolism for AtTPS1 or T-6-P. Carbon status itself only slightly affects AtTPS1 expression levels in seedling tissues (Fig. 1D). Feeding experiments with 8-d-old seedlings supplied with Suc for 0.5 h up to 24 h similarly did not significantly affect TPS1 expression (data not shown). The large induction of AtTRE1 expression after fertilization in ripening siliques (Fig. 1C) coincides with TPS1 induction in these organs. Conversely, AtTRE1 is not increased in floral organs and young rosette leaves, as is AtTPS1. Thus, increasing AtTPS1 expression does not seem to be linked to AtTRE1 induction.
AtTPS5 mRNA levels remain relatively low in the plant organs analyzed (Fig. 1C). The public Arabidopsis microarray database (available at http://www.arabidopsis.org/links/microarrays.jsp) shows that TPS1 expression is generally low and stable. TPS5, 7, 8, 10, and, especially, TPS11 are in general significantly expressed and respond to diverse treatments or conditions. These data suggest that the diverse TPS homologs from classes I and II might have different spatial and temporal regulations. The ubiquitous presence of AtTPS1 mRNA in Arabidopsis organs and the pronounced phenotypes observed in tps1 mutant plants suggests an important role for the TPS1 protein.
AtTPS1 Enzyme Activity Is Required for Growth
The effect of the tps1 mutation beyond seed development was studied using a DEX-inducible AtTPS1 system to generate viable tps1/tps1 seeds. Rescued tps1/tps1 seeds are able to germinate and grow in the absence of AtTPS1 induction. Therefore, TPS1 does not seem to be required for seed germination. However, we cannot exclude residual TPS1 protein in the germinating mutant seed, which might remain from the rescued maturing seeds. Established tps1/tps1 seedlings display a short root containing a reduced meristematic region (Fig. 3). In wild-type seedlings, AtTPS1 is expressed at low levels in roots (Fig. 1D), consistent with microarray data from root cell files, where AtTPS1 is expressed exclusively in root stele tissues (Birnbaum et al., 2003). The tps1/tps1 root has a normal structure (Fig. 3D), thus AtTPS1 is probably not involved in developmental patterning.
Arabidopsis Floral Transition Requires AtTPS1 Function
Vegetative tps1 mutant plants are retarded in growth, with small rosette leaves. In the absence of functional TPS1, no transition from vegetative to generative development occurs. AtTPS1 induction in ind-TPS1 mutant lines can overcome this developmental arrest, but induced plants have a reduced apical dominance, and flowering is delayed compared to wild type. Some tps1 mutant lines carrying the AtTPS1::OTSA transgene show delayed flowering also, whereas others are similar to wild type. Apparently, E. coli OTSA expression under the control of the AtTPS1 promoter is capable of complementing seed maturation and floral transition phenotypes, whereas root growth is only partially complemented. Possibly, normal root growth requires a higher level of TPS1 activity than seed maturation and floral transition. Alternatively, OTSA may lack additional functions present in AtTPS1, such as phosphorylation by protein kinases, 14-3-3 protein binding (Moorhead et al., 1999), or allosteric control sites. In yeast, E. coli OTSA is able to restore T-6-P levels in tps1Δ S. cerevisiae, but this does not lead to a full restoration of growth on Glc, suggesting a role for the TPS1 protein in the control of Glc influx into glycolysis (Bonini et al., 2000, 2003). The UBIQUITIN10 promoter from Arabidopsis drives AtTPS1 expression in the TPS1-inducible mutant lines. Possibly, this promoter is insufficiently active in important cell types, which might explain the observed phenotypes under inductive conditions. It is unlikely that ectopic AtTPS1 expression causes the observed phenotypes since strong ectopic TPS enzyme activity in 35S::OTSA transgenic plants does not result in short-root and late-flowering phenotypes (Schluepmann et al., 2003).
Factors known to be involved in floral transition are light quality, photoperiod or GA, and signaling pathways requiring vernalization (Simpson and Dean, 2002). Several of these stimuli were applied in an attempt to overcome the late-flowering phenotype. Growth on 1% Suc, which is the optimal concentration to stimulate flowering (Ohto et al., 2001), or elevated light conditions to enhance carbon availability in these plants do not shorten the late flowering. Also, seed vernalization for 6 weeks was ineffective in enhancing flowering time. Furthermore, no significant change in expression of flowering-time genes (CO, SOC1, FLC, FT, and LFY) was detected in noninduced versus induced mutant lines or versus wild-type plants. This suggests that the tps1 mutation acts downstream or parallel to these floral integrators.
Taken together, the AtTPS1 expression data and the phenotypes observed for the tps1 mutant suggest an essential role for AtTPS1 throughout the Arabidopsis life cycle. In tps1 mutants, the root meristematic region is dramatically decreased, leaf growth is reduced, and floral transition is absent. Clearly, other TPS homologs are unable to complement AtTPS1 function. Biosynthesis of T-6-P by the AtTPS1::OTSA construct can overcome many, but not all, of the tps1 mutant phenotypes. Previously it was shown that T-6-P, the product of TPS activity, is required for plant growth (Schluepmann et al., 2003). T-6-P levels control utilization of available carbon in a way that is not yet understood. TPS1 likely regulates the metabolic status of the plant through T-6-P biosynthesis. Therefore, the observed phenotypes in the tps1 mutant might have a metabolic basis. Suc addition does not rescue tps1 mutant phenotypes because T-6-P is required for carbon to be utilized for normal growth. Identification of targets of T-6-P and methodology for determining cellular T-6-P levels would be most helpful in further understanding the role of trehalose metabolism in plants.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis Columbia (Col-0) and tps1-2 were obtained from En/Spm transposon insertion collection (Tissier et al., 1999) as described by Eastmond et al. (2002). After seed stratification for 3 d at 4°C, plant material was transferred to 16-h-light/8-h-dark (long-day) growth conditions at 22°C and 200 μmol m−2 s−1 irradiance. In vitro growth and selection were performed on half-strength MS (Murashige and Skoog, 1962) solidified with 0.8% agarose. For resistance marker selection, 20 mg L−1 hygromycin, 10 mg L−1 phosphinotricin (PPT), or 50 mg L−1 kanamycin were added to the medium. For Basta (PPT) selection on soil, plants were sprayed twice with 75 mg L−1 PPT, 0.01% Silwet L77 solution at about a 3-d interval. For DEX treatment, 10 μm were supplied in the in vitro growth medium or in water for watering soil grown plants.
Constructs and Plant Transformation
AtTPS1 cDNA (accession no. Y08568) was cloned by PCR (Titan One Tube RT-PCR System; Roche, Basel), which was used to screen a LambdaGem-11 phage library (Promega, Madison, WI) of Arabidopsis genomic DNA (Col-0). A 3,110-bp (5′ untranslated region ATG; total fragment was 5,007 bp) promoter containing XhoI-fragment and a 5,886-bp (ATG-3′ untranslated region; total fragment was 6,716 bp) TPS1 gene-containing fragment were cloned into pBluescript II KS+ (Stratagene, La Jolla, CA).
The AtTPS1::AtTPS1 complementation construct was obtained by fusing the complete TPS1 promoter fragment from pBlue into pBin19 (KpnI/BstEII). In this binary vector (SpeI/blunted XbaI), the TPS1 gene was introduced (SpeI/SmaI).
For the AtTPS1::OTSA construct, a 3,067-bp AtTPS1 promoter fragment (EcoRI/blunted BstEII) was cloned into EcoRI/blunted Sse8387I from pMog 1423 (provided by Zeneca, Leiden, The Netherlands) to drive Escherichia coli OTSA gene expression.
The ind-TPS1 construct was created in pTA7002 vector (Aoyama and Chua, 1997), where the 35S cauliflower mosaic virus promoter was replaced by 1,020-bp AtUBQ10 promoter (accession no. L05361) from p3325 (Sun and Callis, 1997). Into PmeI (blunted and T-overhang created by Taq-polymerase)/Sse8387I of pTA7002 vector, the UBQ10 promoter fragment (BamHI, blunted with added A-overhang/Sse8387I) was ligated, thereby creating pTA-UBQ10::GVG. Next, a 5,934-bp genomic AtTPS1 (BstEII, blunted/XbaI) fragment was inserted into vector sites XhoI (blunted)/SpeI. As an inducible control vector (ind-GUS), the GUSA reporter gene (1,812 bp, SalI/NheI fragment from pCambia1381) was inserted into XhoI/SpeI of pTA-UBQ10::GVG.
In pCambia1303, the NcoI/BstEII GUSA fragment was replaced by GUSA-GFP (green fluorescent protein) fragment from pCambia1381, creating pCambia1383. The TPS1 promoter was PCR amplified (using reverse primer and tpsNcoI5′u primer 5′-ttccagccatgggctcacacca-3′), from which a 3.1-kb EcoRI/NcoI fragment was inserted into the corresponding vector sites giving the AtTPS1::GUS reporter construct.
The floral dip method (Clough and Bent, 1998) was used for transformation of plant material with Agrobacterium tumefaciens strain GVG2260.
DNA Extraction, RNA Extraction, and PCR Analysis
Genomic DNA was isolated using the cetyl-trimethyl-ammonium bromide method (Doyle and Doyle, 1990) or the quick-prep method (Cheung et al., 1993). For genotyping the OTSA complementing lines, PCR detection was performed using primers (tps4d, 5′-tgtgagcgtatgcctggaaataag-3′; tps471u, 5′-agcccatcctatccatctg-3′; and EnRB544, 5′-ggcttgtgtggaacttactatg-3′, located in the transposon right border) discriminating between wild-type TPS1 (557-bp product) and transposon-inserted tps1-2 (1,060-bp product).
Qiaex RNeasy plant columns (Qiagen USA, Valencia, CA) were used to isolate RNA for Q-PCR analysis. Small-scale RNA isolation from embryos was performed with the PureScript plant RNA extraction kit (Gentra, Minneapolis) as described previously (Eastmond et al., 2002). For Q-PCR analysis, total RNA was DNAseI (DNA-Free; Ambion, Austin, TX) treated to remove genomic DNA. Absence of DNA was analyzed by performing a PCR reaction (40 cycles, similar to the real-time PCR program) on the DNAseI-treated RNA using Taq-DNA polymerase. Sixty units of M-MLV Reverse Transcriptase (Promega, Madison, WI) were used according to the manufacturer's protocol on 1 μg of total RNA to synthesize cDNA with 0.5 μg of odT16V (custom oligo from Invitrogen, Carlsbad, CA) and 0.5 μg random hexamer (Invitrogen). After cDNA synthesis, the sample volume was increased to 200 μL with TE buffer (10 mm Tris, pH 8.0, 1 mm EDTA), from which 5 μL were used as template per Q-PCR reaction. The ABI-prism 7700 Sequence Detection System (PE-Applied Biosystems, Foster City, CA) was used for the real-time PCR measurements. Per reaction, 12.5 μL of Taqman Universal PCR Master Mix (PE-Applied Biosystems), 250 nm gene-specific Taqman probe, and 900 nm of each gene-specific primer (see below) were supplied in a 25-μL reaction volume. Q-PCR was performed according to the standard Taqman Universal PCR Master Mix protocol (PE-Applied Biosystems). Relative quantitation of gene expression is based on the comparative CT method (User Bulletin No. 2: ABI PRISM 7700 Sequence Detection System, 1997) using AtACTIN2 as a calibrator reference. Relative expressions are represented as a fraction from the highest level of each gene, unless otherwise mentioned.
Gene-Specific Primer-Probe Pairs and Their Efficiencies
AtTPS1 (At1G78580) specific: AtTPS1-probe (5′-FAM-atctccttggctcacctgacgacgtc-TAMRA-3′); AtTPS1-F (5′-tgggtcgtactcgcaccaa-3′) and AtTPS1-R (5′-tttgcttccttgagaagctcg-3′); Efficiency: 1.90. AtACT2 (At3g18780) specific: AtACT2-probe (5′-FAM-aagtcttgttccagccctcgtttgtgc-TAMRA-3′); AtACT2-F (5′-gctgagagattcagatgccca-3′) and AtACT2-R (5′-atggaagctgctggaatccac-3′); Efficiency: 1.97. AtTPS5 (At4g17770) specific: AtTPS5-probe (5′-FAM-tcccaagaatatcgtgtacctcgtcagtgg-TAMRA-3′); AtTPS5-F (5′-ccgcgaaacaatcgaaatct-3′) and AtTPS5-R (5′-ttcccagtctgttccatcatttg-3′); Efficiency: 1.93. AtTRE1 (At4G24040) specific: AtTRE1-probe (5′-FAM-ttcgtctcagatccctccggcttcc-TAMRA-3′); AtTRE1-F (5′-gctgcaccacgaaccagtaga-3′) and AtTRE1-R (5′-ttcttcgttctccacgttgga-3′); Efficiency: 1.98. Flowering-time gene pairs are described by El-Din El-Assal et al. (2003). Fluorogenic probes were obtained from Sigma-genosys (Cambridge, UK), and all custom primers were from Invitrogen (Paisley, UK).
Microscopy and β-Glucuronidase Assay
Siliques were opened and embryos excised using precision forceps. For Nomarski images, embryos were taken in chloralhydrate [chloralhydrate:water:glycerol = 8:3:1 (v/v)], placed between an object and a covering slide, after which photos were directly made using Nomarski optics on a Zeiss photomicroscope (Jena, Germany). In case of GUS localization, the excised seeds and embryos were incubated in X-Gluc buffer {5× GUS-buffer, 100 mL: 250 mg X-Gluc, 0.1 g Triton X-100, 82.3 mg K-ferricyanide [K3Fe(CN)6, M = 329.9 g mol−1], 105.6 mg K-ferrocyanide [K4Fe(CN)6-3H2O, M = 422.4 g mol−1] in 50 mm phosphate buffer} at 37°C for 10 h before clearing in chloralhydrate and Nomarski imaging. For confocal laser scanning microscopy (Leica, Wetzlar, Germany), seedlings were stained with propidium iodine (1 μg mL−1).
Acknowledgments
We thank the Photography and Microscopy department from Utrecht University for their expertise.
This work was supported by the Dutch Science Foundation, CW-STW (grant no. 349–4657).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.039743.
References
- Aoyama T, Chua NH (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Blazquez MA, Santos E, Flores CL, Martinez-Zapater JM, Salinas J, Gancedo C (1998) Isolation and molecular characterization of the Arabidopsis TPS1 gene, encoding trehalose-6-phosphate synthase. Plant J 13: 685–689 [DOI] [PubMed] [Google Scholar]
- Bonini B, Van Dijck P, Thevelein JM (2003) Uncoupling of the glucose growth defect and the deregulation of glycolysis in Saccharomyces cerevisiae tps1 mutants expressing trehalose-6-phosphate-insensitive hexokinase from Schizosaccharomyces pombe. Biochim Biophys Acta 1606: 83–93 [DOI] [PubMed] [Google Scholar]
- Bonini B, Van Vaeck C, Larsson C, Gustafsson L, Ma P, Winderickx J, Van Dijck P, Thevelein JM (2000) Expression of Escherichia coli OTSA in a Saccharomyces cerevisiae tps1 mutant restores trehalose 6-phosphate levels and partly restores growth and fermentation with glucose and control of glucose influx into glycolysis. Biochem J 350: 261–268 [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Gorlach J (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabib E, Leloir LF (1958) The biosynthesis of trehalose phosphate. J Biol Chem 231: 259–275 [PubMed] [Google Scholar]
- Cheung WY, Hubert N, Landry BS (1993) A simple and rapid DNA microextraction method for plant, animal, and insect suitable for RAPD and other PCR analyses. PCR Methods Appl 3: 69–70 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Crowe J, Hoekstra F, Crowe LM (1992) Anhydrobiosis. Annu Rev Physiol 54: 579–599 [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12: 13–15 [Google Scholar]
- Eastmond PJ, van Dijken AJH, Spielman M, Kerr A, Tissier AF, Dickinson HG, Jones JDG, Smeekens SC, Graham IA (2002) Trehalose-6-phosphate synthase 1, which catalyses the first step in trehalose synthesis, is essential for Arabidopsis embryo maturation. Plant J 29: 225–235 [DOI] [PubMed] [Google Scholar]
- El-Din El-Assal S, Alonso-Blanco C, Peeters AJM, Wagemaker C, Weller JL, Koornneef M (2003) The role of cryptochrome 2 in flowering in Arabidopsis. Plant Physiol 133: 1504–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg AK, Kim JK, Owens TG, Ranwala AP, Choi YD, Kochian LV, Wu RJ (2002) Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc Natl Acad Sci USA 99: 15898–15903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddijn O, Smeekens S (1998) Sensing trehalose biosynthesis in plants. Plant J 14: 143–146 [DOI] [PubMed] [Google Scholar]
- Goddijn OJ, Van Dun K (1999) Trehalose metabolism in plants. Trends Plant Sci 4: 315–319 [DOI] [PubMed] [Google Scholar]
- Holmström K-O, Mäntylä E, Welin B, Mandal A, Palva ET, Tunnela OE, Londesborough J (1996) Drought tolerance in tobacco. Nature 379: 683–684 [Google Scholar]
- Jang IC, Oh SJ, Seo JS, Choi WB, Song SI, Kim CH, Kim YS, Seo HS, Choi YD, Nahm BH, et al (2003) Expression of a bifunctional fusion of the Escherichia coli genes for trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase in transgenic rice plants increases trehalose accumulation and abiotic stress tolerance without stunting growth. Plant Physiol 131: 516–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyman B, Van Dijck P, Thevelein JM (2001) An unexpected plethora of trehalose biosynthesis genes in Arabidopsis thaliana. Trends Plant Sci 6: 510–513 [DOI] [PubMed] [Google Scholar]
- Moorhead G, Douglas P, Cotelle V, Harthill J, Morrice N, Meek S, Deiting U, Stitt M, Scarabel M, Aitken A, et al (1999) Phosphorylation-dependent interactions between enzymes of plant metabolism and 14-3-3 proteins. Plant J 18: 1–12 [DOI] [PubMed] [Google Scholar]
- Müller J, Aeschbacher RA, Wingler A, Boller T, Wiemken A (2001) Trehalose and trehalase in Arabidopsis. Plant Physiol 125: 1086–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 473–497 [Google Scholar]
- Ohto M, Onai K, Furukawa Y, Aoki E, Araki T, Nakamura K (2001) Effects of sugar on vegetative development and floral transition in Arabidopsis. Plant Physiol 127: 252–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva C, Panek AD (1996) Biotechnological applications of the disaccharide trehalose. Biotechnol Annu Rev 2: 293–314 [DOI] [PubMed] [Google Scholar]
- Paul M, Pellny T, Goddijn O (2001) Enhancing photosynthesis with sugar signals. Trends Plant Sci 6: 197–200 [DOI] [PubMed] [Google Scholar]
- Paul MJ, Pellny TK (2003) Carbon metabolite feedback regulation of leaf photosynthesis and development. J Exp Bot 54: 539–547 [DOI] [PubMed] [Google Scholar]
- Pilon-Smits EAH, Terry N, Sears T, Kim H, Zayed A, Hwang S, Dun K, Voogd E, Verwoerd TC, Krutwagen RWHH, et al (1998) Trehalose-producing transgenic tobacco plants show improved growth performance under drought stress. J Plant Physiol 152: 525–532 [Google Scholar]
- Romero C, Bellés JM, Vayaz JL, Serrano R, Culianez-Macià FA (1997) Expression of the yeast trehalose-6-phosphate synthase gene in transgenic tobacco plants: pleiotropic phenotypes include drought tolerance. Planta 201: 293–297 [DOI] [PubMed] [Google Scholar]
- Schluepmann H, Pellny T, van Dijken A, Smeekens S, Paul M (2003) Trehalose 6-phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana. Proc Natl Acad Sci USA 100: 6849–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG, Dean C (2002) Arabidopsis, the Rosetta Stone of flowering time? Science 296: 285–289 [DOI] [PubMed] [Google Scholar]
- Sun CW, Callis J (1997) Independent modulation of Arabidopsis thaliana polyubiquitin mRNAs in different organs and in response to environmental changes. Plant J 11: 1017–1027 [DOI] [PubMed] [Google Scholar]
- Tissier AF, Marillonnet S, Klimyuk V, Patel K, Torres MA, Murphy G, Jones JDG (1999) Multiple independent defective suppressor-mutator transposon insertions in Arabidopsis: a tool for functional genomics. Plant Cell 11: 1841–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel G, Aeschbacher RA, Müller J, Boller T, Wiemken A (1998) Trehalose-6-phosphate phosphatases from Arabidopsis thaliana: identification by functional complementation of the yeast tps2 mutant. Plant J 13: 673–683 [DOI] [PubMed] [Google Scholar]
- Vogel G, Fiehn O, Jean-Richard-dit-Bressel L, Boller T, Wiemken A, Aeschbacher RA, Wingler A (2001) Trehalose metabolism in Arabidopsis: occurrence of trehalose and molecular cloning and characterization of trehalose-6-phosphate synthase homologues. J Exp Bot 52: 1817–1826 [DOI] [PubMed] [Google Scholar]