Abstract
Auxins are hormones important for numerous processes throughout plant growth and development. Plants use several mechanisms to regulate levels of the auxin indole-3-acetic acid (IAA), including the formation and hydrolysis of amide-linked conjugates that act as storage or inactivation forms of the hormone. Certain members of an Arabidopsis amidohydrolase family hydrolyze these conjugates to free IAA in vitro. We examined amidohydrolase gene expression using northern and promoter-β-glucuronidase analyses and found overlapping but distinct patterns of expression. To examine the in vivo importance of auxin-conjugate hydrolysis, we generated a triple hydrolase mutant, ilr1 iar3 ill2, which is deficient in three of these hydrolases. We compared root and hypocotyl growth of the single, double, and triple hydrolase mutants on IAA-Ala, IAA-Leu, and IAA-Phe. The hydrolase mutant phenotypic profiles on different conjugates reveal the in vivo activities and relative importance of ILR1, IAR3, and ILL2 in IAA-conjugate hydrolysis. In addition to defective responses to exogenous conjugates, ilr1 iar3 ill2 roots are slightly less responsive to exogenous IAA. The triple mutant also has a shorter hypocotyl and fewer lateral roots than wild type on unsupplemented medium. As suggested by the mutant phenotypes, ilr1 iar3 ill2 imbibed seeds and seedlings have lower IAA levels than wild type and accumulate IAA-Ala and IAA-Leu, conjugates that are substrates of the absent hydrolases. These results indicate that amidohydrolases contribute free IAA to the auxin pool during germination in Arabidopsis.
Although auxins are essential regulators of many aspects of plant growth and development, our understanding of how levels of this hormone are controlled remains incomplete. One component of auxin homeostasis is conjugation of the auxin indole-3-acetic acid (IAA) to different moieties, including esterification to sugars and amide linkage to amino acids and peptides. IAA-Leu, IAA-Ala, IAA-Asp, IAA-Glu, and IAA-Glc have been identified in Arabidopsis seedlings (Tam et al., 2000; Kowalczyk and Sandberg, 2001). Several IAA-peptide conjugates have been identified in bean seeds (Bialek and Cohen, 1986; Walz et al., 2002) and Arabidopsis (Walz et al., 2002); in fact, IAA-peptide conjugates are the major IAA conjugates in Arabidopsis seeds (Ljung et al., 2002; Park and Cohen, 2003). Different IAA conjugates apparently have specific functions in plants, such as storage, transport, or inactivation of IAA (Cohen and Bandurski, 1982; Bartel et al., 2001). In general, endogenous IAA conjugates that are biologically active and hydrolyzed in plants may function as auxin storage forms, whereas conjugates inactive in bioassays may have roles in IAA degradation (Bartel et al., 2001; Ljung et al., 2002).
Several Arabidopsis screens have uncovered IAA-amino acid conjugate-resistant mutants that help to delineate conjugate functions (Bartel and Fink, 1995; Campanella et al., 1996; Davies et al., 1999; Lasswell et al., 2000; Magidin et al., 2003; LeClere et al., 2004). Two of these mutants, ilr1 and iar3, are defective in IAA-amino acid conjugate hydrolases (Bartel and Fink, 1995; Davies et al., 1999). The ILR1 and IAR3 enzymes hydrolyze IAA conjugates in vitro (Bartel and Fink, 1995; Davies et al., 1999; LeClere et al., 2002) and are similar to members of the M40 class of bacterial carboxypeptidases (Barrett et al., 2003) that cleave a variety of small molecules, including IAA-Asp (Chou et al., 1996, 1998), benzoylglycine (Hani and Chan, 1995), acetylated amino acids (Sakanyan et al., 1993), and secreted human odorant precursors (Natsch et al., 2003). Degenerate PCR and sequence similarity searches revealed five additional ILR1-like genes (ILL1, ILL2, ILL3, ILL5, and ILL6) in the Arabidopsis amidohydrolase family (Davies et al., 1999; LeClere et al., 2002). Of the seven genes, ILR1, ILL1, ILL2, and IAR3 encode enzymes that can cleave IAA-amino acids at least to some extent, ILL3 and ILL6/GR1 encode enzymes that show no activity on IAA-l-amino acids in vitro, and ILL5 is an apparent pseudogene in the Columbia ecotype (Col-0) accession (Davies et al., 1999; LeClere et al., 2002) for which no cDNAs have been reported. ILR1, ILL2, and IAR3 all have Km values and catalytic efficiencies suggesting that they could function at the expected physiological concentrations of the conjugates (LeClere et al., 2002). Members of the amidohydrolase family are found in both monocots and dicots (LeClere et al., 2002), and an ILR1-like Arabidopsis suecica amidohydrolase that is active on IAA-Ala and IAA-Gly has been described (Campanella et al., 2003a, 2003b).
Although the ilr1 and iar3 mutants are less sensitive than wild type to inhibition of root elongation by exogenous IAA-amino acid conjugates (Bartel and Fink, 1995; Davies et al., 1999), no morphological abnormalities in the absence of conjugates have been reported in either single hydrolase mutant. This observation may reflect functional redundancy among family members. It may be necessary to disrupt multiple genes to determine the roles of the amidohydrolases, and consequently those of IAA conjugates, in plant growth and development.
Here we show that Arabidopsis plants defective in three of the amidohydrolases active on IAA-amino acids have phenotypes suggestive of low endogenous auxin levels, including shorter hypocotyls and fewer lateral roots than wild type on unsupplemented medium. Moreover, ilr1 iar3 ill2 seedlings have lower IAA levels and higher levels of the IAA conjugates that are substrates for these enzymes. Comparison of the single, double, and triple mutant responses to IAA conjugates and amidohydrolase gene expression patterns suggests that each hydrolase may play both unique and overlapping roles in auxin homeostasis.
RESULTS
Hydrolase Genes Have Distinct Expression Patterns
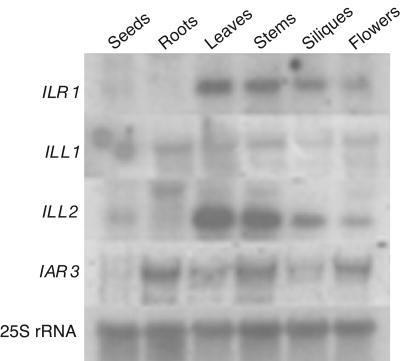
Four of the seven Arabidopsis amidohydrolase genes, ILR1, IAR3, ILL1, and ILL2, encode proteins that are active on IAA-amino acid conjugates in vitro (LeClere et al., 2002). To determine the tissues in which the hydrolases might be active in vivo, we examined expression using northern analyses of ILR1, ILL1, and ILL2 genes with gene-specific probes and compared expression to that of IAR3 (Davies et al., 1999). All four IAA-conjugate hydrolase genes are expressed in leaves, stems, siliques, and flowers. However, only ILL2 and possibly ILR1 messages were detected in seeds, and only IAR3 and ILL1 were detected in roots (Fig. 1).
Figure 1.
Tissue specificity of amidohydrolase gene expression. Total RNA prepared from the indicated Arabidopsis tissues was analyzed by gel-blot hybridization with antisense RNA probes prepared from ILR1, ILL1, and ILL2 or with a DNA probe that detects the 25S rRNA. Previously determined IAR3 hybridization (Davies et al., 1999) is shown for comparison.
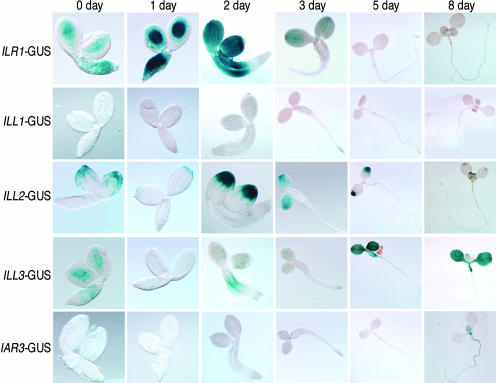
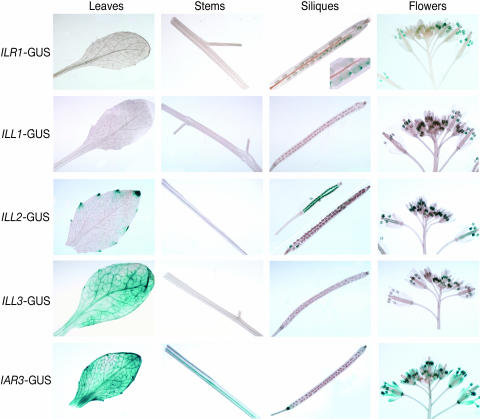
To further explore the expression profiles of the amidohydrolase genes, we constructed transgenic lines containing hydrolase promoter elements driving expression of the β-glucuronidase (GUS) reporter gene (see “Materials and Methods”). We examined staining in mature embryos (0 d) and 1-, 2-, 3-, 5-, and 8-d-old seedlings (Fig. 2). Leaves, stems, siliques, and flowers from adult plants were also stained and examined (Fig. 3). ILR1-GUS was detected in central regions of cotyledons and in the hypocotyl, the radicle of mature embryos, and 1-d-old seedlings (Fig. 2). Staining was not visible in leaves or stems but was detected in the micropyle of siliques and in pollen (Fig. 3). ILL2-GUS was detected in the distal tips of cotyledons and seedling leaves (Fig. 2) and appeared concentrated in the hydathodes of leaves from mature plants (Fig. 3). ILL2-GUS staining was not visible in stems but was seen in pollen, ovules, and developing seeds (Fig. 3). ILL3-GUS staining was visible in cotyledons, leaves, and hypocotyls of seedlings (Fig. 2) and in leaves and pollen of mature plants (Fig. 3). IAR3-GUS stained primarily in vascular tissue, as seen in 8-d-old cotyledons and roots (Fig. 2) and in adult leaves, stems, siliques, and petals (Fig. 3). IAR3-GUS also was detected in hydathodes and in silique abscission zones and funicles (Fig. 3). ILL1-GUS expression was detected only in pollen (Fig. 3).
Figure 2.
GUS staining of amidohydrolase promoter-GUS fusions in seedlings. Seedlings grown under continuous white light were harvested on the indicated day and placed in staining buffer (see “Materials and Methods”). Columns indicate days after imbibition, with 0 d representing embryos from dry seed.
Figure 3.
GUS staining of amidohydrolase promoter-GUS fusions in mature plants. Parts from 47-d-old plants grown in soil under continuous light were harvested and immediately placed in staining buffer (see “Materials and Methods”).
Hydrolase Mutants Have Varied Responses to Exogenous IAA Conjugates
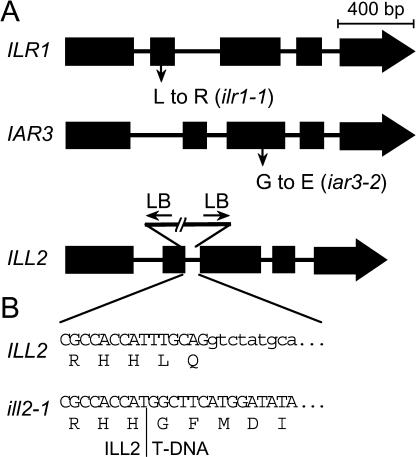
Although forward genetic approaches uncovered ilr1 and iar3 mutants (Bartel and Fink, 1995; Davies et al., 1999), the roles of the other Arabidopsis IAA-conjugate hydrolases have been inferred only from biochemical studies (LeClere et al., 2002). These studies revealed that ILL2 is the most active of the amidohydrolases in vitro. Northern (Fig. 1) and promoter-reporter analyses (Fig. 2) of ILL2 expression suggest that ILL2 and ILR1 are present in partially overlapping tissues, consistent with the possibility that these two hydrolases might have overlapping functions. To explore this possibility, we isolated an ill2 mutant from the Arabidopsis Knockout Facility (Madison, WI). The ill2-1 mutant has a T-DNA insertion near the 3′ end of the second exon (+644 bp from the start codon; Fig. 4). ill2-1 is likely to be a null allele; the T-DNA insertion was accompanied by a 42-bp deletion that includes an exon-intron junction and is in a region of the protein that is highly conserved among the seven Arabidopsis amidohydrolase proteins. We crossed ill2 plants to both ilr1-1 and iar3-2 mutants (Bartel and Fink, 1995; Davies et al., 1999) and used PCR and restriction digestion to identify homozygous lines of ilr1 ill2, iar3 ill2, and ilr1 iar3 ill2.
Figure 4.
Nature of the triple hydrolase mutant alleles. A, Gene models for ILR1, IAR3, and ILL2; exons are represented by black boxes and introns by thin lines. The locations of the missense mutations in ilr1-1 and iar3-2 are designated by arrows. The triangle represents the site of the T-DNA insertion in ill2-1. Left border (LB) T-DNA sequence is located at both 5′ and 3′ ends of the insertion. B, ILL2 and ill2-1 sequence at the 5′ junction of the T-DNA insertion in exon 2.
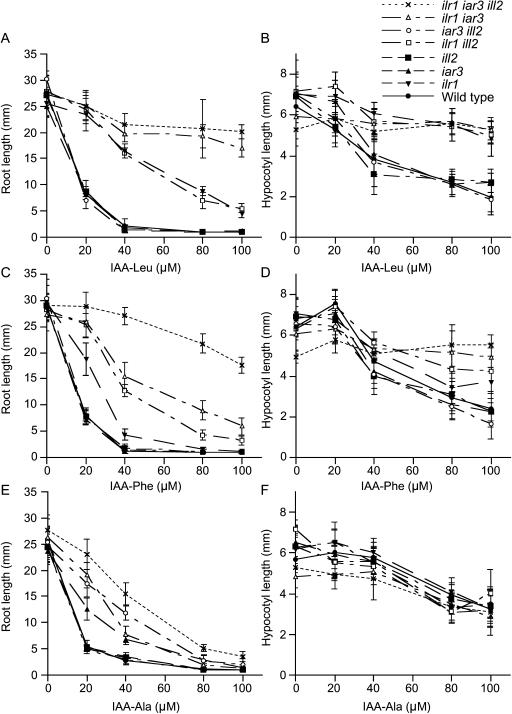
To determine the relative in vivo importance of ILR1, IAR3, and ILL2 hydrolysis on various IAA-amino acid conjugates, we assayed root and hypocotyl elongation of the single, double, and triple hydrolase mutants on IAA-Leu, IAA-Phe, and IAA-Ala (Fig. 5). These conjugates cause a dose-dependent inhibition of wild-type root and hypocotyl elongation, providing a quantitative in vivo assay for hydrolase function. In this assay, iar3, ill2, and iar3 ill2 mutant roots retained wild-type sensitivity to the inhibition of root elongation caused by IAA-Leu; ilr1 and ilr1 ill2 mutant roots were less sensitive than wild type to IAA-Leu; and the ilr1 iar3 and ilr1 iar3 ill2 mutant roots were essentially IAA-Leu insensitive (Fig. 5A). These results indicate that ILR1 and IAR3, but not ILL2, are important for IAA-Leu hydrolysis in roots.
Figure 5.
Hydrolase mutant responses to growth on IAA-Leu, IAA-Phe, or IAA-Ala. Root (A, C, and E) or hypocotyl (B, D, and F) lengths of mutant and wild-type Ws seedlings were measured after 8-d growth under yellow-filtered light on media containing the indicated concentration of IAA-Leu (A and B), IAA-Phe (C and D), or IAA-Ala (E and F). Error bars represent sds from the means (n ≥ 12).
When we similarly assayed IAA-Phe responsiveness, we found that iar3, ill2, and iar3 ill2 mutant roots had wild-type sensitivity to the inhibition of root elongation caused by IAA-Phe, whereas the ilr1, ilr1 ill2, ilr1 iar3, and ilr1 iar3 ill2 mutant roots displayed progressively decreasing IAA-Phe sensitivity (Fig. 5C). These results demonstrate that ILR1 is most important for IAA-Phe hydrolysis in roots, followed by IAR3, and then ILL2.
We also examined the inhibition of hypocotyl elongation caused by IAA-Leu and IAA-Phe (Fig. 5, B and D). Every hydrolase mutant defective in ILR1 was less sensitive than wild type to elongation inhibition by IAA-Leu, whereas mutants with functional ILR1 had wild-type hypocotyl responses. Thus, only ILR1 cleaves exogenous IAA-Leu in the hypocotyl. The hydrolase mutant hypocotyl lengths on IAA-Phe resembled the pattern seen with root lengths on IAA-Phe, showing that all hydrolases can hydrolyze IAA-Phe in hypocotyls, at the relative levels described above for roots.
We found that ilr1, ill2, and ilr1 ill2 mutant roots had wild-type sensitivity to the inhibition of root elongation caused by IAA-Ala (Fig. 5E). As previously shown (Davies et al., 1999), iar3 mutant roots are less IAA-Ala sensitive than wild type. We found that the double hydrolase mutants ilr1 iar3 and iar3 ill2 were less sensitive than the single iar3 mutant and that the triple ilr1 iar3 ill2 mutant roots were the least IAA-Ala sensitive of all the genotypes (Fig. 5E). IAR3 is thus more important than ILR1 and ILL2 for IAA-Ala hydrolysis in roots. Interestingly, roots of all of the hydrolase mutants remained sensitive to IAA-Ala at high concentrations (Fig. 5E). In contrast to the results seen with IAA-Leu and IAA-Phe, hydrolase mutant hypocotyls all remained sensitive to the inhibition of elongation caused by IAA-Ala (Fig. 5F). These results are consistent with the possibility that Arabidopsis seedlings may sense IAA-Ala even in the absence of its hydrolysis.
ilr1 iar3 ill2 Seedlings Display Low Auxin Phenotypes
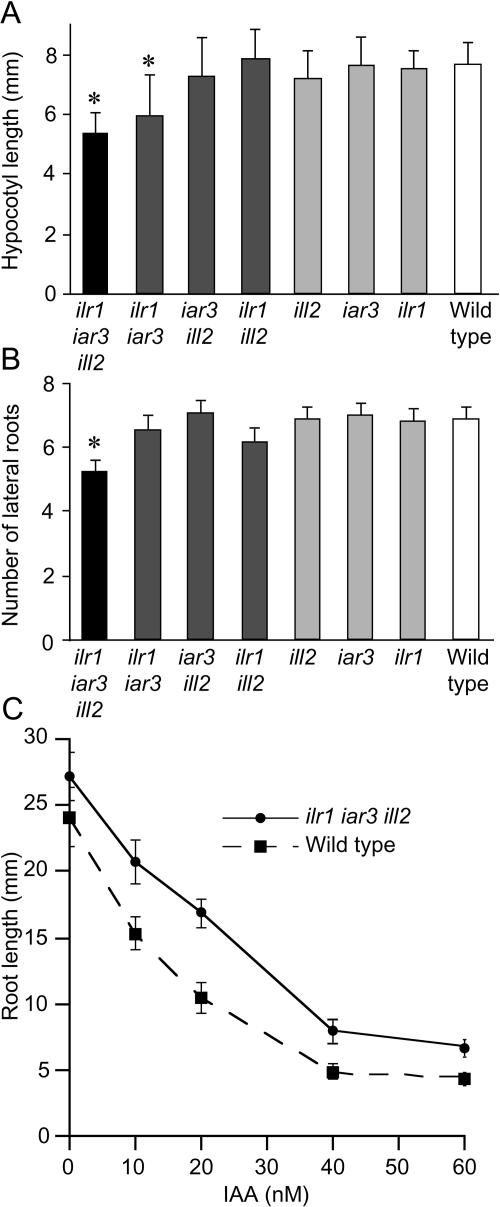
Analysis of ilr1 iar3 ill2 seedlings grown in the absence of exogenous hormones revealed several phenotypes suggestive of low auxin levels. ilr1 iar3 ill2 and ilr1 iar3 mutants have shorter hypocotyls than wild-type plants when grown in the light (Fig. 6A) but not when grown in the dark (data not shown). ilr1 iar3 ill2 seedlings also have slightly fewer lateral roots than wild type (Fig. 6B). These phenotypes are also found in plants with reduced auxin responsiveness, including the auxin-response mutants axr1 (Estelle and Somerville, 1987), axr4 (Hobbie and Estelle, 1995), and ibr5 (Monroe-Augustus et al., 2003). Arabidopsis plants expressing the Pseudomonas savastanoi indoleacetic acid-Lys synthetase gene (iaaL) have shorter hypocotyls than wild type in the light (Jensen et al., 1998), likely because of decreased IAA levels due to increased IAA conjugation. Tobacco plants expressing iaaL have 5-fold less free IAA than wild type, and these plants also display low-auxin phenotypes including reduced apical dominance and reduced rooting (Romano et al., 1991).
Figure 6.
ilr1 iar3 ill2 seedlings have shorter hypocotyls and fewer lateral roots than wild type and are less sensitive to IAA. A, Hypocotyl elongation on unsupplemented medium. Eight-day-old mutant and wild-type Ws seedlings grown on PNS in yellow-filtered light were removed from the agar, and the length of the hypocotyl was measured. Error bars indicate sds of the means (n ≥ 12); asterisks indicate values significantly different from wild type (two-tailed t test; P < 0.001). B, Number of lateral roots on unsupplemented medium. Lateral roots were counted on 10-d-old seedlings grown in yellow-filtered light. Error bars indicate ses of the means (n ≥ 12); asterisks indicate values significantly different from wild type (two-tailed t test; P < 0.001). C, Root elongation on IAA. Root length of ilr1 iar3 ill2 and wild-type seedlings on IAA after 8 d in yellow-filtered light. Error bars indicate sds from the means (n ≥ 10). Root lengths of ilr1 iar3 ill2 were significantly different from wild-type lengths at all IAA concentrations shown (two-tailed t test; P < 0.001).
Triple mutant roots are less sensitive than wild-type roots to elongation inhibition caused by exogenous IAA (Fig. 6C). ilr1 iar3 ill2 plants also are less sensitive than wild type to other natural and synthetic auxins, including indole-3-butyric acid (IBA), 1-naphthaleneacetic acid, and 2,4-dichlorophenoxyacetic acid (data not shown). This reduced sensitivity may reflect a lower level of IAA present in ilr1 iar3 ill2 plants, or may be due to an ability to conjugate exogenous auxin without subsequent hydrolysis, resulting in more efficient inactivation of applied auxins. We therefore decided to directly measure IAA and IAA-amino acid conjugate levels in the triple mutant.
ilr1 iar3 ill2 Mutants Have Reduced IAA Levels
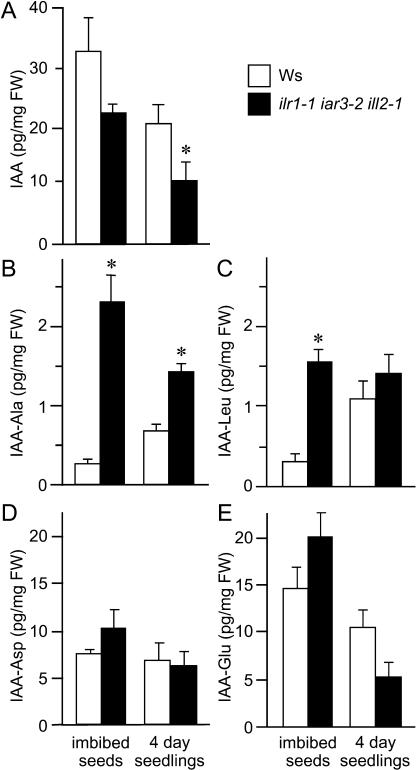
Conjugates of IAA with the amino acids Ala, Leu, Asp, and Glu have been detected in young Arabidopsis seedlings (Tam et al., 2000; Kowalczyk and Sandberg, 2001). We compared IAA and IAA-amino acid levels in wild type and ilr1 iar3 ill2 triple mutant to determine whether the hydrolase mutant has reduced endogenous auxin levels. We found that IAA levels were reduced in the triple mutant by 33% and 47% in imbibed seeds and seedlings, respectively (Fig. 7A). Also consistent with a conjugate hydrolysis defect, ilr1 iar3 ill2 imbibed seeds had 8- and 5-fold higher levels of IAA-Ala (Fig. 7B) and IAA-Leu (Fig. 7C), respectively, than wild type. The elevation in IAA-Ala levels was still significant in 4-d-old triple mutant seedlings (Fig. 7B). In contrast, the ilr1 iar3 ill2 mutant does not accumulate significantly more IAA-Asp or IAA-Glu than wild type (Fig. 7, D and E). The in vivo accumulation of conjugates in the mutant mirrors the in vitro specificity of the enzymes. IAA-Ala is an efficient in vitro substrate of IAR3 and ILL2, and IAA-Leu is efficiently hydrolyzed by ILR1, but IAA-Asp and IAA-Glu are not efficiently hydrolyzed in vitro by any of the amidohydrolases (LeClere et al., 2002).
Figure 7.
ilr1 iar3 ill2 mutants have lower IAA levels and higher IAA-Ala and IAA-Leu levels than wild type. Levels for IAA (A), IAA-Ala (B), IAA-Leu (C), IAA-Asp (D), and IAA-Glu (E) were measured in ilr1 iar3 ill2 and wild-type Ws dry seeds imbibed for 4 d and 4-d-old seedlings grown in light at 22°C after the imbibition period. IAA or conjugate levels are shown as picogram per milligram fresh weight (FW) based on three to five biological replicates. Error bars display sds from the means and asterisks indicate values significantly different from wild type (one-tailed t test; P < 0.05).
DISCUSSION
Four of seven related amidohydrolases encoded in the Arabidopsis genome cleave IAA-amino acid conjugates in vitro (Bartel and Fink, 1995; Davies et al., 1999; LeClere et al., 2002). Northern and promoter-GUS expression analyses show that these amidohydrolases are present at distinct times throughout plant growth and development, perhaps allowing plants to utilize stored IAA when needed. Our observation that the ilr1 iar3 ill2 triple mutant accumulates IAA-Ala and IAA-Leu while displaying reduced levels of IAA (Fig. 7) provides the first direct evidence to our knowledge that these enzymes hydrolyze endogenous IAA-amino acid conjugates in plants.
The high ILR1-GUS accumulation in germinating seeds (Fig. 2) is consistent with previous reverse transcription (RT)-PCR detection of ILR1 mRNA in 1-d-old seedlings (Campanella et al., 2003b) and suggests a role for ILR1 in liberating IAA from conjugates during germination. The low level of ILR1-GUS expression in roots is interesting in light of the finding that IAA-Leu, the preferred substrate of ILR1 (LeClere et al., 2002), accumulates preferentially in Arabidopsis roots (Kowalczyk and Sandberg, 2001). Responses of the hydrolase mutants to IAA conjugates show that ILR1 is most important for IAA-Leu and IAA-Phe responsiveness in roots, and that ILR1 is the only hydrolase needed for IAA-Leu responsiveness in hypocotyls (Fig. 5). The in vitro hydrolysis profile of ILR1 is quite different from the other enzymes, having a different pH optimum, metal cofactor requirement, and substrate preference (LeClere et al., 2002), suggesting that ILR1 may act under different circumstances than the other hydrolases.
The bacterially expressed ILL2 protein hydrolyzes IAA-Ala very efficiently (LeClere et al., 2002). The single ill2 mutant has wild-type sensitivity to IAA conjugates (Fig. 5), suggesting that ILR1 and IAR3 can compensate for loss of ILL2 activity and explaining why ill2 mutants have not been isolated in forward genetic screens for altered conjugate responsiveness. However, double mutant analyses indicate that ILL2 contributes to IAA-Ala and IAA-Phe responsiveness in the absence of ILR1 or IAR3 (Fig. 5). ILL2-GUS is expressed in germinating seedlings, particularly in the distal tips of the cotyledons and developing leaves (Fig. 2), suggesting that ILL2 may act with ILR1 to provide free IAA to germinating seedlings. ILL2-GUS expression is concentrated in the hydathodes of mature leaves (Fig. 3). This hydathode staining is reminiscent of that seen with the auxin-responsive DR5-GUS reporter (Guo et al., 2002), suggesting that ILL2 might be responsible for releasing some of the IAA that DR5-GUS senses during development. Interestingly, the DR5 reporter is derived from the soybean GH3 promoter (Ulmasov et al., 1997), and GH3 is a member of a family that includes IAA-adenylating enzymes (Staswick et al., 2002) likely to function in IAA-amino acid formation. In pea stem segments, applied IAA-Ala is rapidly hydrolyzed followed by conjugation of the released IAA to Asp (Hangarter and Good, 1981), suggesting coordination between conjugate hydrolysis and formation in some tissues. Such coordination could also account for the possibly reduced levels of IAA-Glu that we detect in triple mutant seedlings (Fig. 7E).
IAR3-GUS accumulates in vascular tissues (Figs. 2 and 3) consistent with a role for IAR3 in the hydrolysis of conjugates from the vascular tissue or in release of IAA important for vascular formation or patterning (Davies, 1995). Double and triple hydrolase mutant IAA-conjugate responses show that IAR3 is important for IAA-Ala hydrolysis in roots, but only contributes measurably to IAA-Leu and IAA-Phe responsiveness in the absence of ILR1. IAR3 hydrolyzes IAA conjugates with small side chains, such as IAA-Ala and some jasmonic acid conjugates in vitro (LeClere et al., 2002), suggesting that it may be important in both auxin and wound responses.
The insensitivity of ilr1 iar3 ill2 roots and hypocotyls to high levels (100 μm) of IAA-Leu and IAA-Phe (Fig. 5) suggests that ILR1, IAR3, and ILL2 are the only enzymes cleaving endogenous IAA-Leu during seedling development. This insensitivity also supports the view that these conjugates have auxin activity solely by virtue of their ability to be cleaved to free IAA. In contrast, ilr1 iar3 ill2 roots and hypocotyls remain sensitive to high concentrations of IAA-Ala (Fig. 5), suggesting that some IAA-Ala is still hydrolyzed in the triple mutant or that IAA-Ala has auxin effects independent of its hydrolysis. There are several candidates for enzymes that might hydrolyze IAA-Ala in the ilr1 iar3 ill2 mutant. Although ILL1 can hydrolyze IAA-Ala in vitro, kinetic parameters suggest that this hydrolase would not contribute to IAA-Ala hydrolysis in vivo (LeClere et al., 2002), and ILL1 is very weakly expressed (Figs. 1–3). As ILL1 is less than 1 kb upstream of ILL2, a conventional ill1 ill2 double mutant obtained by crossing would be difficult to obtain. Similarly, ILL5 is only approximately 3 kb downstream of IAR3. The putative hydrolase encoded by ILL5 has not been tested in vitro because a cDNA has not been isolated; the Col-0 version of ILL5 contains a polymorphism in a splice acceptor site that would be predicted to abolish splicing (Davies et al., 1999). Alternatively, it is possible that the responsiveness of the triple hydrolase mutant to IAA-Ala results from proteins in different enzyme families that can hydrolyze IAA-Ala in Arabidopsis, or that the iar3-2 mutation retains some activity. Most intriguing are reports that IAA-Ala can act as an auxin without being hydrolyzed to release free IAA in tomato cell culture (Hangarter et al., 1980); it is possible that hydrolysis-independent IAA-Ala activity has been uncovered in the triple hydrolase mutant.
Aside from ILR1, IAR3, and ILL2, the functions of the remaining Arabidopsis amidohydrolase genes remain unknown. ILL3-GUS stains in shoots of seedlings and leaves and pollen of adult plants. As ILL3 does not hydrolyze IAA-amino acid conjugates in the in vitro conditions that have been tested (LeClere et al., 2002), the function of ILL3 is not known.
In addition to reduced responses to exogenous conjugates, the ilr1 iar3 ill2 mutant has shorter hypocotyls and fewer lateral roots than wild type on unsupplemented medium (Fig. 6). These phenotypes are also seen in some mutants with reduced auxin responsiveness (Estelle and Somerville, 1987; Hobbie and Estelle, 1995; Monroe-Augustus et al., 2003) and suggest that the hydrolysis of endogenous IAA conjugates provides seedlings with some of the IAA necessary for normal hypocotyl growth and lateral root development. In addition, ilr1 iar3 ill2 seedlings are less sensitive than wild type to the inhibition of primary root elongation by exogenous IAA (Fig. 6C), perhaps because the reduced conjugate hydrolysis activity in the mutant results in a greater capacity to inactivate applied auxin or because the mutant seedlings have lower IAA levels than wild type (Fig. 7A). These lower IAA levels are likely due to a deficiency in IAA-conjugate hydrolysis, as evidenced by the IAA-Ala and IAA-Leu that accumulate in ilr1 iar3 ill2 imbibed seeds and seedlings (Fig. 7).
Mechanisms that plants use to regulate the hydrolases remain largely unknown. The ILR1, IAR3, and ILL2 enzymes contain N-terminal signal sequences and C-terminal endoplasmic reticulum (ER)-retention signals (Bartel and Fink, 1995; Davies et al., 1999), suggesting that they may function in the ER and that conjugates would enter the ER prior to their hydrolysis. Proteins that transport conjugates into the ER or IAA out of the ER remain to be identified. Analysis of in vitro hydrolase activity has shown that Mn2+ and Co2+ can enhance hydrolysis (Bartel and Fink, 1995; Davies et al., 1999; LeClere et al., 2002). Moreover, the genes defective in several IAA-conjugate response mutants implicate metal homeostasis as important for in vivo conjugate responsiveness. The ilr2 mutant, which is defective in an apparently cytoplasmic protein, has enhanced ATP-dependent Mn2+ transport, suggesting that ILR2 may function as an inhibitor of a metal transporter (Magidin et al., 2003). The iar1 mutant is defective in a membrane protein (Lasswell et al., 2000) similar to the ZRT, IRT-like protein family of metal transporters (Guerinot, 2000), further implicating metal homeostasis in hydrolase activity. As screens for Arabidopsis mutants with reduced IAA conjugate sensitivity are not saturated, it is possible that additional regulatory mechanisms could be uncovered through this approach.
The ilr1 iar3 ill2 seedling morphological phenotypes provide evidence for the importance of IAA-conjugate hydrolysis during early seedling growth. However, ilr1 iar3 ill2 seedlings grow normally except for the shorter hypocotyl and slightly fewer lateral roots, indicating that other free IAA sources are available to germinating seedlings. Indeed, the total amount of IAA-Ala and IAA-Leu in wild-type seedlings is relatively low (5%–10%) compared to the level of free IAA (Fig. 7); thus these conjugates can only be one of several potential IAA sources during initial seedling growth. Auxin from the shoot promotes lateral root formation in germinating seedlings (Casimiro et al., 2001; Bhalerao et al., 2002). Arabidopsis seedlings already have significant IAA biosynthetic capacity 3 d after germination (Bhalerao et al., 2002), so de novo biosynthesis is likely to supply some of this IAA. β-Oxidation of IBA in peroxisomes (Zolman et al., 2000; Bartel et al., 2001) may provide another source. Indeed, IBA-response mutants that are likely to have reduced IBA to IAA conversion also show defects in lateral root production following germination (Zolman et al., 2001; Zolman and Bartel, 2004). It is possible that IBA to IAA conversion or induced de novo synthesis compensates for decreased IAA-conjugate hydrolysis in the triple hydrolase mutant and that conjugate hydrolysis or induced synthesis can compensate for decreased IBA β-oxidation in certain IBA-response mutants. Hydrolysis of IAA-containing proteins or other types of high molecular mass IAA conjugates, which are abundant in Arabidopsis seeds (Ljung et al., 2002; Walz et al., 2002), might also contribute to the free IAA pool during germination. It will be interesting to identify the enzymes controlling this hydrolysis. This system of biochemical redundancy is likely to be a safeguard that ensures the supply of critically important auxin to the plant. Future efforts to disrupt multiple inputs to the free IAA pool may allow determination of the relative importance of the various pathways that regulate auxin homeostasis.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Plants from the Wassilewskija (Ws) and Col-0 accessions were used. For phenotypic assays, seeds were surface sterilized (Last and Fink, 1988) and grown aseptically on plant nutrient medium containing 0.5% (w/v) Suc (PNS; Haughn and Somerville, 1986) solidified with 0.6% agar. Plates were sealed with gas-permeable Leukopor surgical tape (LecTec, Minnetonka, MN). For hormone-response assays, IAA and IAA-l-amino acid conjugates (Aldrich, St. Louis) were diluted into medium from 100 mm stocks in ethanol. Plates were incubated under yellow long-pass filters to slow the breakdown of indolic compounds (Stasinopoulos and Hangarter, 1990) with constant illumination (25–45 μE m−2 s−2) at 22°C. After 8 d, plants were removed from the agar and hypocotyl and primary root lengths were measured, or after 10 d the number of lateral roots was counted under a dissecting microscope. Primordia emerged from the body of the main root were counted as a lateral root. Plants transferred to soil (Metromix 200, Scotts, Marysville, OH) were grown at 22°C to 25°C under cool-white fluorescent bulbs (Sylvania, Danvers, MA) with continuous illumination.
Mutant Isolation
The ill2-1 allele (in the Ws accession) was isolated from T-DNA lines from the Arabidopsis Knockout Facility (Madison, WI). Pooled genomic DNA from the 140,000 kanamycin-resistant T-DNA lines was amplified with the T-DNA left border (LB) oligonucleotide JL-202 (Krysan et al., 1999) and either an oligonucleotide 3′ of ILL2, 42E9-22 (5′-GTCGTGAAACTTCTCAAGGTCTTTGTAA-3′) or an oligonucleotide 5′ of ILL2, 42E9-23 (5′-AGTTTCTCCCCCATTTATGCTATTTACAG-3′). The resulting PCR products were separated on a 1% agarose gel and transferred to Nytran membrane (Midwest Scientific, Valley Park, MO). A 32P-labeled probe of the 42E9-22 + 42E9-23 PCR product was used to detect ILL2-specific PCR products by Southern hybridization (Ausubel et al., 1999). Individual plants were isolated after additional amplifications of the T-DNA line subpools (Krysan et al., 1999). Homozygous ill2-1 plants were identified by PCR amplification with the oligonucleotides JL-202 and 42E9-22. Heterozygous plants were identified by PCR amplification using the oligonucleotides 42E9-24 (5′-GATATGGATGCTTTGCCTATTCAG-3′) and 42E9-25 (5′-TCTTTGCTCCACTCAAACCTTCCTCAGCTG-3′). The precise site of the insertion was identified by sequencing the JL-202 + 42E9-22 PCR product following sequential ethanol, polyethylene glycol, and ethanol precipitations (Ausubel et al., 1999). A PCR product was also obtained with JL-202 and 42E9-9 (5′-CTCAGTTTGACTTTCCAACTACTCCTTT-3′), an oligonucleotide 5′ of the T-DNA insertion, and this product was also sequenced. No amplification resulted when oligonucleotides from the T-DNA right border were paired with oligonucleotides 5′ of the T-DNA insertion site, indicating that a T-DNA rearrangement occurred during insertion, leaving left-border sequence at both 5′ and 3′ ends. Sequencing results showed that the last 6 bp of the second ILL2 exon and the first 36 bp of the second intron are either missing in ill2-1 or incorporated into the T-DNA insertion. Homozygous lines of ill2-1 crossed to ilr1-1 (Bartel and Fink, 1995), iar3-2 (Davies et al., 1999), and ilr1-1 iar3-2, all in the Ws accession, were identified by PCR analysis in the progeny of the F2 plants. PCR-based identification of the ilr1-1 and iar3-2 alleles was described previously (Bartel and Fink, 1995; Davies et al., 1999).
Northern Analysis
Total RNA was prepared as described (Nagy et al., 1988) from the following tissues of the Col-0 accession: mature dry seeds, roots of 14-d-old plants grown on 3MW gel blot paper (Midwest Scientific) on PNS, above ground parts of 14-d-old plants grown in soil, and stems, siliques, and flowers of 29-d-old plants. RNA was analyzed using a NorthernMax kit according to the recommendations of the manufacturer (Ambion, Austin, TX). Four micrograms of total RNA per lane were separated on a 1% agarose gel containing 0.37 m formaldehyde and transferred to Bright-Star Plus nylon membranes (Ambion).
Antisense RNA probes (Riboprobe in vitro Transcription Systems; Promega, Madison, WI) were used to detect hydrolase mRNAs. For each probe, the corresponding cDNA (Bartel and Fink, 1995; Davies et al., 1999) subcloned into pBluescript KS (+) was linearized by digestion with PstI (ILL1), SacII (ILR1 and ILL2), or EcoRI (IAR3). Linearized plasmids were used as templates to synthesize 32P-labeled antisense RNA probes with T3 (ILR1, ILL2) or T7 (ILL1, IAR3) RNA polymerase. A 25S ribosomal DNA probe 32P-labeled using random 12-mer oligonucleotide primers (Ausubel et al., 1999) was used to verify that equal amounts of RNA were loaded in each lane. After prehybridizing in ULTRAhyb Hybridization buffer (Ambion), the probe was hybridized overnight at 65°C and washed according to the manufacturer's instructions.
Promoter-GUS Vector Construction
A genomic clone containing the adjacent ILL1 and ILL2 genes was isolated by colony hybridization from an Arabidopsis cosmid library (Olszewski et al., 1988) using an ILL1 cDNA probe. This clone was digested with various restriction enzymes, and fragments containing ILL1 and ILL2 were identified via Southern analysis and subcloned into pBluescript. The ends of a 2.1-kb NcoI fragment containing the ILL1 promoter and a 1-kb NcoI-HindIII fragment containing the ILL2 promoter were filled in with T4 DNA polymerase and ligated into SmaI-cut pBI101.3 (Jefferson et al., 1987) to form ILL1-promoter GUS (ILL1-GUS) or ILL2-promoter GUS (ILL2-GUS) fusions. An 8.7-kb SacI fragment containing the ILR1 promoter was mutagenized to contain a BglII site with the oligonucleotide 5′-GATGACAAAGAAGCTCCCAGATCTGAAATCCATTCTGATTTC-3′ (altered residues underlined) 6 bp after the ILR1 start codon (bold). The 3.9-kb BglII-SalI promoter fragment was inserted into BamHI-SalI-cut pBI101 (Jefferson et al., 1987), forming an ILR1-promoter GUS fusion (ILR1-GUS). To create the IAR3 promoter GUS fusion, a HindIII fragment (1.9 kb) containing the IAR3 promoter was mutagenized with the oligonucleotide 5′-CAATCAATCGAATCCGAGATCTAGAATGAGTTTCTTCAAATGGG-3′(altered residues underlined) to form an XbaI site immediately preceding the start codon (bold). The resulting 1.7-kb HindIII-XbaI fragment was ligated into HindIII-XbaI-cut pBI101 to make IAR3-GUS. A 2.6-kb HindIII fragment containing the ILL3 promoter was mutagenized with the oligonucleotide 5′-GAAGAGCTACGATTGAGGAAGGATCCATGTCAAATTTGAAGTC-3′ (altered residues underlined) to contain a BamHI site after the start codon (bold). The resulting 2.5-kb HindIII-BamHI fragment was ligated into BamHI-HindIII-cut pBI101.2 (Jefferson et al., 1987) to create the ILL3-promoter GUS fusion (ILL3-GUS). Constructs were sequenced with a primer from the GUS gene to confirm the correct junctional sequences and electroporated into Agrobacterium tumefaciens strain GV3101 (Koncz and Schell, 1986), which was used to transform Col-0 using the floral dip method (Clough and Bent, 1998). For each construct, at least 12 independent transformants were selected for the ability to develop on medium containing 12 μg/mL kanamycin. Progeny of these plants were tested for GUS expression (see below). For IAR3-GUS, 6 of 6 lines tested showed similar staining patterns with variable intensity. For ILR1-GUS, 4 of 8 lines tested showed detectable staining; these had similar patterns with variable intensity. For ILL3-GUS, 3 of 6 lines tested showed detectable staining; these had similar patterns with variable intensity. Expression analyses were performed using homozygous progeny of kanamycin-resistant transformants showing representative staining patterns.
Examination of Promoter-GUS Expression
For analysis of GUS expression, tissues were harvested into 5-bromo-4-chloro-3-indolyl β-d-glucuronide cyclohexylamine (X-gluc) buffer containing 100 mm NaPO4 pH 7.0, 0.5 mm K3[Fe(CN)6], 0.5 mm K4[Fe(CN)6]3H2O, 10 mm EDTA, 0.01% Triton X-100, and 0.5 mg/mL X-gluc. For dry seeds, seed coats were removed with a razor blade under a dissecting microscope before transfer to X-gluc buffer. For 1-d-old seedlings, seeds were germinated in water overnight before transfer to X-gluc buffer and seed coats were removed. For 2- to 9-d-old seedlings, seedlings were grown on PNS and transferred to X-gluc buffer. For mature plants, seedlings were grown on PNS for 10 d, then transferred to soil and grown at 22°C under constant illumination. When plants were 47 d old, parts were harvested into X-gluc buffer. Samples were incubated in X-gluc buffer at 37°C in the dark. Staining was observed in some tissues as early as 6 h after immersion in X-gluc buffer, but staining was allowed to continue for 2 d to allow comparison of samples. Chlorophyll was removed by sequential incubations in 50% ethanol, 100% ethanol, and 50% ethanol for several hours at each step. After rehydration, samples were transferred to 50% glycerol for mounting on glass slides. Samples were photographed using a Leica FLUO III stereoscope (Wetzlar, Germany) and Polaroid DMC le Low Light v. 1.5 software (Polaroid, Cambridge, MA). No GUS staining was observed in similarly treated untransformed plants.
Analysis of IAA and IAA Conjugate Levels
For hormone measurements, five batches of surface sterilized seeds were imbibed in water in the dark for 48 h at 4°C and for 48 h at 22°C, and then separately germinated under white light at 22°C on vertically oriented Murashige and Skoog plates solidified with 0.7% agar and supplemented with 1% Suc for 4 additional days. Three to five individual samples were harvested at the seed or seedling stage, and the tissue was homogenized in liquid nitrogen. For each sample, 10 to 50 mg tissue was processed as previously described for free IAA (Ljung et al., 2001) or for IAA conjugates (Kowalczyk and Sandberg, 2001).
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Acknowledgments
We thank Rosie Tellez for northern analysis; Rosie Tellez, David Goetz, and Susan Lee for GUS vector construction; and the Arabidopsis Biological Research Center at Ohio State University for the cosmid library. We are grateful to Mary Ellen Lane for microscope use and to Raquel Adham, Diana Dugas, Melanie Monroe-Augustus, Dereth Phillips, Jeanne Rasbery, Andrew Woodward, Bethany Zolman, and an anonymous reviewer for critical comments on the manuscript. R.A.R. and S.L. were recipients of Houston Livestock Show and Rodeo scholarships.
This work was supported by the National Institutes of Health (grant no. R29–GM54749 to B.B.), by the Robert A. Welch Foundation (grant no. C–1309 to B.B.), and by the Swedish Research Council and the Swedish Foundation for Strategic Research (grant to G.S.). Partial support was provided by an NIH Training Grant (grant no. T32–GM08362 to S.L.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.039677.
References
- Ausubel F, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1999) Current Protocols in Molecular Biology. Greene Publishing Associates and Wiley-Interscience, New York
- Barrett AJ, Tolle DP, Rawlings ND (2003) Managing peptidases in the genomic era. Biol Chem 384: 873–882 [DOI] [PubMed] [Google Scholar]
- Bartel B, Fink GR (1995) ILR1, an amidohydrolase that releases active indole-3-acetic acid from conjugates. Science 268: 1745–1748 [DOI] [PubMed] [Google Scholar]
- Bartel B, LeClere S, Magidin M, Zolman BK (2001) Inputs to the active indole-3-acetic acid pool: de novo synthesis, conjugate hydrolysis, and indole-3-butyric acid β-oxidation. J Plant Growth Regul 20: 198–216 [Google Scholar]
- Bhalerao RP, Eklöf S, Ljung K, Marchant A, Bennett M, Sandberg G (2002) Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant J 29: 325–332 [DOI] [PubMed] [Google Scholar]
- Bialek K, Cohen JD (1986) Isolation and partial characterization of the major amide-linked conjugate of indole-3-acetic acid from Phaseolus vulgaris L. Plant Physiol 80: 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanella JJ, Bakllamaja V, Restieri T, Vomacka M, Herron J, Patterson M, Shahtaheri S (2003. a) Isolation of an ILR1 auxin conjugate hydrolases homolog from Arabidopsis suecica. Plant Growth Regul 39: 175–181 [Google Scholar]
- Campanella JJ, Ludwig-Mueller J, Bakllamaja V, Sharma V, Cartier A (2003. b) ILR1 and sILR1 amidohydrolase homologs differ in expression pattern and substrate specificity. Plant Growth Regul 41: 215–223 [Google Scholar]
- Campanella JJ, Ludwig-Mueller J, Town CD (1996) Isolation and characterization of mutants of Arabidopsis thaliana with increased resistance to growth inhibition by indoleacetic acid-amino acid conjugates. Plant Physiol 112: 735–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro I, Marchant A, Bhalerao RP, Beeckman T, Dhooge S, Swarup R, Graham N, Inzé D, Sandberg G, Casero PJ, et al (2001) Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13: 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J-C, Kuleck GA, Cohen JD, Mulbry WW (1996) Partial purification and characterization of an inducible indole-3-acetyl-L-aspartic acid hydrolase from Enterobacter agglomerans. Plant Physiol 112: 1281–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J-C, Mulbry WW, Cohen JD (1998) The gene for indole-3-acetyl-L-aspartic acid hydrolase from Enterobacter agglomerans: molecular cloning, nucleotide sequence, and expression in Escherichia coli. Mol Gen Genet 259: 172–178 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cohen JD, Bandurski RS (1982) Chemistry and physiology of the bound auxins. Annu Rev Plant Physiol 33: 403–430 [Google Scholar]
- Davies PJ (1995) Plant Hormones. Kluwer Academic Publishers, Dordrecht, The Netherlands, p 833
- Davies RT, Goetz DH, Lasswell J, Anderson MN, Bartel B (1999) IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. Plant Cell 11: 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estelle MA, Somerville C (1987) Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Mol Gen Genet 206: 200–206 [Google Scholar]
- Guerinot ML (2000) The ZIP family of metal transporters. Biochim Biophys Acta 1465: 190–198 [DOI] [PubMed] [Google Scholar]
- Guo FQ, Wang R, Crawford NM (2002) The Arabidopsis dual-affinity nitrate transporter gene AtNRT1.1 (CHL1) is regulated by auxin in both shoots and roots. J Exp Bot 53: 835–844 [DOI] [PubMed] [Google Scholar]
- Hangarter RP, Good NE (1981) Evidence that IAA conjugates are slow-release sources of free IAA in plant tissues. Plant Physiol 68: 1424–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hangarter RP, Peterson MD, Good NE (1980) Biological activities of indoleacetylamino acids and their use as auxins in tissue culture. Plant Physiol 65: 761–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hani EK, Chan VL (1995) Expression and characterization of Campylobacter jejuni benzoylglycine amidohydrolase gene (hippuricase) in Escherichia coli. J Bacteriol 177: 2396–2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughn GW, Somerville C (1986) Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol Gen Genet 204: 430–434 [Google Scholar]
- Hobbie L, Estelle M (1995) The axr4 auxin-resistant mutants of Arabidopsis thaliana define a gene important for root gravitropism and lateral root initiation. Plant J 7: 211–220 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PJ, Hangarter RP, Estelle M (1998) Auxin transport is required for hypocotyl elongation in light-grown but not dark-grown Arabidopsis. Plant Physiol 116: 455–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J (1986) The promoter of the TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204: 383–396 [Google Scholar]
- Kowalczyk M, Sandberg G (2001) Quantitative analysis of indole-3-acetic acid metabolites in Arabidopsis thaliana. Plant Physiol 127: 1845–1853 [PMC free article] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Sussman MR (1999) T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11: 2283–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasswell J, Rogg LE, Nelson DC, Rongey C, Bartel B (2000) Cloning and characterization of IAR1, a gene required for auxin conjugate sensitivity in Arabidopsis. Plant Cell 12: 2395–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Last RL, Fink GR (1988) Tryptophan-requiring mutants of the plant Arabidopsis thaliana. Science 240: 305–310 [DOI] [PubMed] [Google Scholar]
- LeClere S, Rampey RA, Bartel B (2004) IAR4, a gene required for auxin conjugate sensitivity in Arabidopsis, encodes a pyruvate dehydrogenase E1α homolog. Plant Physiol 135: 989–999 [DOI] [PMC free article] [PubMed]
- LeClere S, Tellez R, Rampey RA, Matsuda SPT, Bartel B (2002) Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J Biol Chem 277: 20446–20452 [DOI] [PubMed] [Google Scholar]
- Ljung K, Bhalerao RP, Sandberg G (2001) Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J 28: 465–474 [DOI] [PubMed] [Google Scholar]
- Ljung K, Hull AK, Kowalczyk M, Marchant A, Celenza J, Cohen JD, Sandberg G (2002) Biosynthesis, conjugation, catabolism and homeostasis of indole-3-acetic acid in Arabidopsis thaliana. Plant Mol Biol 50: 309–332 [DOI] [PubMed] [Google Scholar]
- Magidin M, Pittman JK, Hirschi K, Bartel B (2003) ILR2, a novel gene regulating IAA conjugate sensitivity and metal transport in Arabidopsis thaliana. Plant J 35: 523–534 [DOI] [PubMed] [Google Scholar]
- Monroe-Augustus M, Zolman BK, Bartel B (2003) IBR5, a dual-specificity phosphatase-like protein modulating auxin and abscisic acid responsiveness in Arabidopsis. Plant Cell 15: 2979–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy F, Kay SA, Chua N-H (1988) Analysis of gene expression in transgenic plants. in SB Gelvin, RA Schilperoort, eds, Plant Molecular Biology Manual. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp B4: 1–29
- Natsch A, Gfeller H, Gygax P, Schmid J, Acuna G (2003) A specific bacterial aminoacylase cleaves odorant precursors secreted in the human axilla. J Biol Chem 278: 5718–5727 [DOI] [PubMed] [Google Scholar]
- Olszewski NE, Martin FB, Ausubel FM (1988) Specialized binary vector for plant transformation: expression of the Arabidopsis thaliana AHAS gene in Nicotiana tabacum. Nucleic Acids Res 16: 10765–10782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Cohen JD (2003) Partial characterization of major IAA conjugates in Arabidopsis (abstract). In Plant Biology 2003, American Society of Plant Biologists, Rockville, MD, p 669
- Romano CP, Hein MB, Klee HJ (1991) Inactivation of auxin in tobacco transformed with the indoleacetic acid-lysine synthetase gene of Pseudomonas savastanoi. Genes Dev 5: 438–446 [DOI] [PubMed] [Google Scholar]
- Sakanyan V, Desmarez L, Legrain C, Charlier D, Mett I, Kochikyan A, Savchenko A, Boyen A, Falmagne P, Pierard A, et al (1993) Gene cloning, sequence analysis, purification, and characterization of a thermostable aminoacylase from Bacillus stearothermophilus. Appl Environ Microbiol 59: 3878–3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasinopoulos TC, Hangarter RP (1990) Preventing photochemistry in culture media by long-pass light filters alters growth of cultured tissues. Plant Physiol 93: 1365–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I, Rowe ML (2002) Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14: 1405–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam YY, Epstein E, Normanly J (2000) Characterization of auxin conjugates in Arabidopsis. Low steady-state levels of indole-3-acetyl-aspartate, indole-3-acetyl-glutamate, and indole-3-acetyl-glucose. Plant Physiol 123: 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walz A, Park S, Slovin JP, Ludwig-Müller J, Momonoki YS, Cohen JD (2002) A gene encoding a protein modified by the phytohormone indoleacetic acid. Proc Natl Acad Sci USA 99: 1718–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Bartel B (2004) An Arabidopsis indole-3-butyric acid-response mutant defective in PEROXIN6, an apparent ATPase implicated in peroxisomal function. Proc Natl Acad Sci USA 101: 1786–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Silva ID, Bartel B (2001) The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid β-oxidation. Plant Physiol 127: 1266–1278 [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Yoder A, Bartel B (2000) Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156: 1323–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]