Abstract
RGA (repressor of ga1-3) and GAI (gibberellin insensitive) are negative regulators of plant hormone gibberellin (GA) signaling in Arabidopsis. The GA-deficient mutant ga1-3 is a nongerminating, extreme dwarf that flowers late and produces male-sterile flowers. The rga and gai null alleles interact synergistically to rescue vegetative growth and floral initiation in ga1-3, indicating that RGA and GAI are major repressors for these processes. However, rga and gai in combination cannot rescue seed germination or floral development in ga1-3. RGA and GAI belong to the DELLA subfamily within the GRAS family of plant regulatory proteins. Three additional DELLA proteins RGL1, RGL2, and RGL3 are present in Arabidopsis. Previous studies provided evidence that RGL2 and possibly RGL1 control seed germination. To investigate further the function of the RGL genes, we examined the expression profiles of all 5 DELLA protein genes by real-time PCR. RGA and, to a lesser extent, GAI mRNAs were expressed ubiquitously in all tissues, whereas RGL1, 2, and 3 transcripts were present at high levels only in germinating seeds and/or flowers and siliques. Using the newly isolated rgl1, rgl2, and rgl3 T-DNA insertion mutants, we demonstrated that RGL2 is the major repressor in seed germination. We further provided evidence that RGA, RGL1, and RGL2 are all involved in modulating floral development. Interestingly, RGL2 expression is regulated not only at the transcript level. We showed that RGL2 protein in imbibed seeds is rapidly degraded by GA treatment and that the F-box protein SLY1 is required for RGL2 degradation to occur.
Bioactive gibberellins (GAs) are phytohormones that are essential for many processes throughout the life of a plant. Seed germination, vegetative growth, and floral and seed development, for example, all require GAs (Davies, 1995). The importance of GAs is clearly illustrated by the Arabidopsis ga1-3 mutant. This mutant contains a large deletion in the GA1 gene, which encodes ent-copalyl diphosphate synthase, the enzyme catalyzing the first committed step in GA biosynthesis (Sun and Kamiya, 1994). The large reduction in bioactive GAs in ga1-3 leads to a GA-deficient phenotype characterized by dark green leaves and severe dwarfism (Koornneef and van der Veen, 1980; Silverstone et al., 2001). The ga1-3 plant also exhibits defects in all the developmental processes regulated by GAs: This mutant is impaired in root growth and trichome initiation, has reduced apical dominance, and fails to flower under short-day conditions (Wilson et al., 1992; Chien and Sussex, 1996; Silverstone et al., 1997; Fu and Harberd, 2003). Under long-day conditions, floral initiation in ga1-3 is delayed, and the mutant flowers are male-sterile (Koornneef et al., 1983; Wilson et al., 1992). Also, ga1-3 seeds cannot germinate without the application of GA (Koornneef et al., 1983).
Loss-of-function mutations in RGA (repressor of ga1-3) and GAI (GA insensitive) can suppress some of the effects of GA deficiency, suggesting that RGA and GAI negatively regulate a subset of GA responses in Arabidopsis (Peng et al., 1997; Silverstone et al., 1997). Fusions of either RGA or GAI to the green fluorescent protein (GFP) are localized to the nuclei of cells in transgenic Arabidopsis (Silverstone et al., 2001; Fleck and Harberd, 2002). Thus, RGA and GAI may function as transcriptional regulators that directly or indirectly repress the expression of GA-induced genes. Through studies of the rga-24 and gai-t6 null alleles in a ga1-3 background, RGA and GAI proteins were demonstrated to have overlapping functions in repressing many vegetative growth processes (e.g. leaf expansion, apical dominance, abaxial trichome development, and stem elongation) as well as floral initiation (Dill and Sun, 2001; King et al., 2001). Although both RGA and GAI regulate these processes, the contribution of RGA is greater than that of GAI (Dill and Sun, 2001). Interestingly, seed germination and floral development are not restored by removing both RGA and GAI functions in the ga1-3 background, suggesting that neither RGA nor GAI plays a major role in controlling these two GA-dependent processes (Dill and Sun, 2001).
GAI and RGA belong to the DELLA subfamily within the GRAS family of plant regulatory proteins (Pysh et al., 1999). GAI and RGA are distinguished from other GRAS family members by an N-terminal DELLA domain (Peng et al., 1997; Silverstone et al., 1998). This domain is involved in modulating the activity of the RGA and GAI proteins in response to GA (Peng et al., 1997; Dill et al., 2001). Deleting 17 amino acids (named the DELLA motif) in the N-terminal region of either GAI or RGA confers a GA-unresponsive dwarf phenotype in Arabidopsis (Koornneef et al., 1985; Peng et al., 1997; Dill et al., 2001). It has also been demonstrated that GA de-represses its signaling pathway by inducing the degradation of RGA and GAI (Silverstone et al., 2001; Dill et al., 2004) and the DELLA motif in RGA is essential for its GA-dependent proteolysis (Dill et al., 2001). Deletion of the DELLA motif in RGA stabilizes the mutant rga protein (rga-Δ17), converting it into a GA-unresponsive, constitutively active repressor of GA signaling (Dill et al., 2001).
Recent studies have indicated that GA-induced degradation of RGA and GAI requires the SLEEPY1 (SLY1) protein, a positive regulator of GA signaling in Arabidopsis (McGinnis et al., 2003; Dill et al., 2004). The loss-of-function sly1-10 mutant is a GA-insensitive dwarf (Steber et al., 1998) that accumulates high levels of RGA and GAI proteins (McGinnis et al., 2003; Dill et al., 2004). Furthermore, a combination of rga and gai null alleles completely suppresses the sly1-10 dwarf phenotype. Cloning of SLY1 revealed that this gene encodes an F-box containing protein, which is likely a component of the ubiquitin E3 ligase SCF complex (McGinnis et al., 2003). In response to the GA signal, SCFSLY1 may target RGA for ubiquitination and subsequent degradation by the 26S proteasome.
With the sequencing of the Arabidopsis genome (The Arabidopsis Genome Initiative, 2000), three other members of the DELLA subfamily were identified: RGA-LIKE 1, 2, and 3 (RGL1, RGL2, and RGL3; Dill and Sun, 2001; Lee et al., 2002; Wen and Chang, 2002). While RGA and GAI exhibit approximately 82% identity at the amino acid level, RGL1 (At1g66350), RGL2 (At3g03450), and RGL3 (At5g17490) are 53% to 62% identical to RGA and GAI (Dill and Sun, 2001; Lee et al., 2002; Wen and Chang, 2002). Because of the presence of the DELLA domain, the RGL genes have been hypothesized, like GAI and RGA, to encode negative regulators of the GA signaling pathway. It has been further proposed that one or more of the RGL genes may be responsible for regulating GA responses such as seed germination and flower development, which are not dramatically affected by RGA or GAI (Dill and Sun, 2001).
To date, two groups studying RGL1 and RGL2 have provided evidence for the roles of these genes in GA signaling, although some of their conclusions are not the same. Wen and Chang (2002) showed that transgenic Arabidopsis lines expressing a dominant 35S:rgl1 transgene (containing the DELLA motif deletion) exhibit a GA-unresponsive dwarf phenotype, similar to that of plants carrying the dominant gai-1 or rga-Δ17 allele with the DELLA motif deletion. This finding suggested that endogenous RGL1 may modulate vegetative growth in wild-type plants, although ectopic expression of the dominant rgl1 gene by the constitutive 35S promoter may contribute to some of the transgenic plant phenotype. This study also suggested that RGL1 regulates seed germination because seeds of a transgenic line in which endogenous RGL1 expression is silenced germinate in the presence of the GA biosynthesis inhibitor paclobutrazol (PAC; Wen and Chang, 2002). In contrast, Lee et al. (2002) studied Ds insertion alleles of RGL1 and RGL2 and determined that RGL2, but not RGL1, is important in regulating seed germination. The discrepancy between these two studies could be explained by the following possibilities. The PAC-resistant phenotype in germinating seeds of the silenced rgl1 line could be due to cosuppression of the RGL2 and RGL3 genes because expression of these genes was not examined in the mutant seed (Wen and Chang, 2002). Alternatively, the rgl1 allele with a Ds insertion might be a leaky allele because the Ds insertion is located 68 bp upstream of the ATG translational start codon (Lee et al., 2002). However, the unpublished RNA blot data suggest that this Ds insertion eliminates the RGL1 gene expression (Lee et al., 2002). Therefore, the role of RGL1 in seed germination needs to be re-examined. Also, the role of RGL3 in GA signaling and the DELLA protein(s) that are responsible for modulating flower development have not been determined.
In this study, we employed quantitative PCR (qPCR) to examine the developmental expression profiles of all 5 DELLA protein genes. We also isolated new knockout alleles of RGL1, RGL2, and RGL3 from T-DNA insertion mutant pools to investigate further the function of the RGL genes. Our data indicated that RGL2 is the major DELLA protein controlling seed germination. In addition, by generating multiple rga/rgl mutant combinations in the ga1-3 background, we provided strong evidence that RGL1 and RGL2, in combination with RGA, contribute to the regulation of male fertility in developing flowers.
RESULTS
Developmental Expression Profiles of RGA, GAI, RGL1, RGL2, and RGL3
RGA and GAI are the major repressors controlling GA-stimulated vegetative growth and the transition to the reproductive phase (Dill and Sun, 2001; King et al., 2001). However, removal of RGA and GAI function does not appear to affect seed germination or flower development significantly. Recent studies provided evidence that two other DELLA proteins, RGL2 and perhaps RGL1, play a role in repressing GA signaling during seed germination (Lee et al., 2002; Wen and Chang, 2002). Although RGA and GAI mRNAs are detected in all tissues throughout plant development (Silverstone et al., 1998; Lee et al., 2002), expression of RGL1, RGL2, and RGL3 appears to show tissue specificity (Lee et al., 2002; Wen and Chang, 2002). However, most of the expression data for RGA, GAI, and RGL genes came from RNA-blot analyses or RT-PCR, and only limited expression patterns for RGL1 and RGL2 were obtained using in situ hybridization or a promoter-GUS transgene. Furthermore, some of the results from previous studies are not consistent with each other. Therefore, a more detailed analysis of the expression pattern of these genes is necessary to identify their roles in modulating GA response in a given tissue during development.
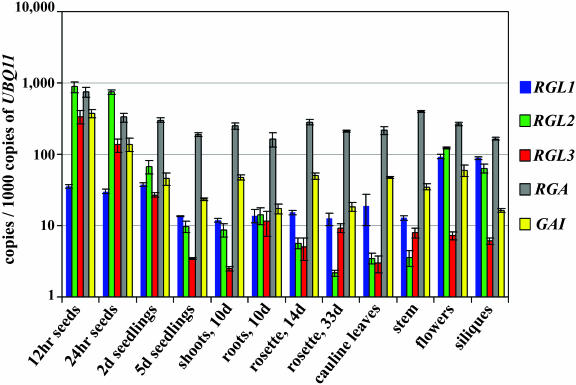
We employed the sensitive, real-time PCR technique using gene-specific primers to quantify transcript levels of RGA, GAI, and the RGLs in different tissues and organs at various developmental stages. Among several housekeeping genes examined, only the expression of the UBIQUITIN11 gene (UBQ11) remained fairly constant in most of the tissues across a wide range of developmental stages, when compared to 18S rRNA (Supplemental Table I, available at www.plantphysiol.org). A few tissues did show small variations in UBQ11 transcript levels: they are approximately 3- or 4-fold higher in 33-d-old rosette, stems, flowers, and cauline leaves. We decided to use UBQ11 instead of 18S rRNA as a control to normalize the samples because we found that the real-time qPCR using cDNA as a template gave more reproducible results than the qRT-PCR using the total RNA as a template. Figure 1 and Supplemental Table II show the cDNA copy number of each gene per 103 copies of UBQ11 cDNA in different tissue samples. Consistent with previous RNA-blot data (Silverstone et al., 1998), RGA was ubiquitously expressed and was present at more than 160 copies/103 copies of UBQ11 in all tissues examined (Fig. 1). GAI was moderately expressed (approximately 50 copies) in most tissues, with highest mRNA levels in 12-h-imbibed seeds (>300 copies) and low levels in 5-d-old seedlings, roots, 33-d-old rosette leaves, and siliques (16–24 copies). RGL1, 2, and 3 genes were expressed at higher levels in germinating seeds, young seedlings, and/or flowers and siliques but only produced low amounts of transcripts in most vegetative tissues (Fig. 1). RGL1 transcripts were present at approximately 30 copies in 12-h- and 24-h-imbibed seeds and 2-d-old seedlings and approximately 90 copies in flowers and siliques. RGL2 mRNA accumulated at approximately 800 copies in germinating seeds, an extremely high level compared to other tissues, and was also high in 2-d-old seedlings (68 copies), flower clusters (123 copies), and siliques (64 copies). RGL3 was only expressed highly in 12-h- and 24-h-imbibed seeds (337 and 136 copies). The expression profiles of these genes are consistent with the previous finding that RGA and GAI are the major repressors in vegetative growth. The fact that RGA is expressed more highly than the other DELLA protein genes in vegetative tissues also supports the previous conclusion that RGA plays a predominant role in repressing GA signaling. All of the DELLA protein genes, except RGL1, were expressed at high levels (>100 copies) in imbibed seeds, suggesting that these four genes may be involved in regulating germination. In flower clusters and siliques, RGL1, RGL2, and RGA transcripts were present at high amounts (most are >90 copies). GAI transcript also accumulated to a moderate 59 copies in flowers. Therefore, RGL1, RGL2, RGA, and perhaps GAI may be important for flower and silique development.
Figure 1.
Transcript levels of RGL1, RGL2, RGL3, RGA, and GAI throughout development. The level of gene expression in each tissue sample, as determined by qPCR, is shown as the number of cDNA copies per 103 copies of UBQ11 cDNA. The tissues and developmental stages tested were, from left to right in the graph, 12-h- and 24-h-imbibed seeds, 2-d-old and 5-d-old seedlings, 10-d-old shoots, roots of 10-d-old seedlings, 14-d-old rosettes, and the following tissues from 33-d-old plants: rosette leaves, cauline leaves, stems, flower clusters, and siliques. The means of three experiments ±se are shown. Although UBQ11 mRNA levels are constant in most tissues, they are approximately 3- to 4-fold higher in 33-d-old rosettes, flowers, stems, and cauline leaves (Supplemental Table I). Therefore, the expression levels of the DELLA protein genes in this figure are slightly underestimated in these tissues.
Isolation of New T-DNA Insertion rga, rgl1, rgl2, and rgl3 Alleles
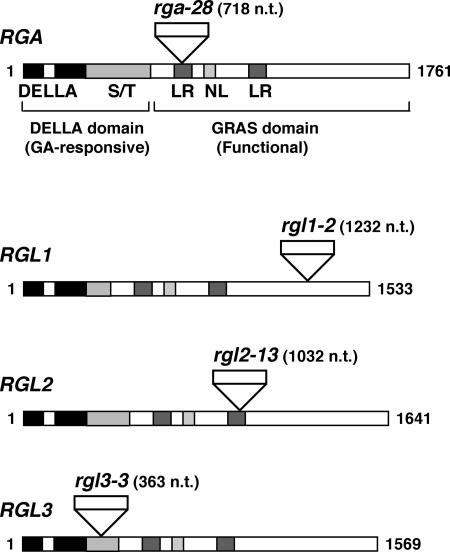
To help define the role of each RGL gene in regulating GA signaling and plant development, we isolated rgl1-2, rgl2-13, rgl3-3, and a new rga-28 allele by screening the T-DNA insertion mutant collection (Columbia [Col-0] background) from the Salk Institute (La Jolla, CA) using a PCR-based approach (Alonso et al., 2003). DNA sequence analysis indicated that in rgl1-2, the T-DNA insertion includes 116 extra unknown nucleotides and is located 1,232 nucleotides after the translational start site in RGL1 (Fig. 2). The T-DNA insertions in rgl2-13, rgl3-3, and rga-28 are 1032, 363, and 718 nucleotides, respectively, after the translational start site in each gene and include no extra sequence. RGL1, RGL2, RGL3, and RGA do not contain any introns, and their ORFs are 1533, 1641, 1569, and 1761 nucleotides, respectively (Fig. 2). Analysis of the previously isolated rga alleles indicated that the C-terminal GRAS domain is essential for the function of RGA as a GA signaling repressor (Silverstone et al., 1998; A.L. Silverstone and T.-p. Sun, unpublished data). The GRAS domain of the DELLA proteins is also highly conserved (Wen and Chang, 2002; Lee et al., 2002), suggesting that this region is important for the function of all DELLA proteins. The newly isolated rga and rgl alleles should not encode any functional proteins because the T-DNA is inserted in the middle of the coding sequence (Fig. 2). No wild-type transcript was detected in these mutants by RT-PCR using primers spanning the T-DNA insertions, although some transcripts were detected when using primers that amplified the 5′ sequences upstream of the insertion (data not shown). We will refer to the three new knockout rgl alleles as rgl1, rgl2, and rgl3 in the rest of the paper. The homozygous single mutants, rga-28, rgl1, rgl2, and rgl3, have a wild-type-like phenotype (data not shown). This result is consistent with the previous finding that rga-24 or gai-t6 single null mutants have nearly wild-type morphology due to functional redundancy of the DELLA protein family (Peng et al., 1997; Dill and Sun, 2001).
Figure 2.
Locations of the T-DNA insertion sites in the new alleles of rga and rgls. The schematic diagram of each gene shows the encoded functional domains and motifs as labeled: DELLA, DELLA motif; S/T, polymeric Ser and/or Thr; LR, Leu heptad repeat; NL, nuclear localization signal. The numbers refer to the nucleotide length of the coding region of each gene and the insertion site of each T-DNA. n.t., nucleotide.
rgl2, But Not rgl1 or rgl3, Confers PAC Resistance during Germination
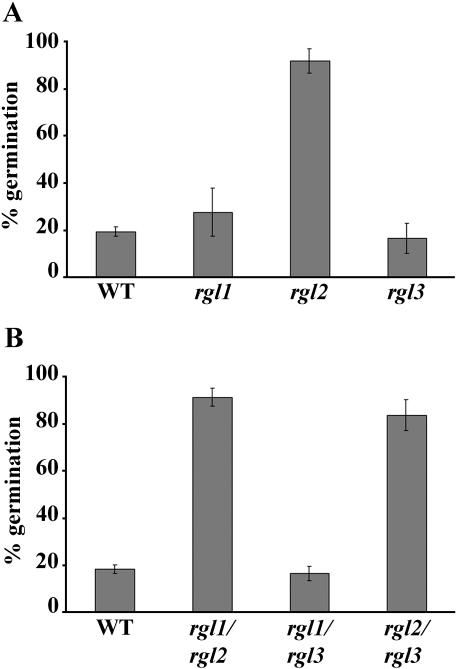
To study the role of the RGLs in seed germination, we tested the germination percentage of rgl1, rgl2, and rgl3 in the presence of 120 μm PAC. PAC inhibits GA-dependent germination by blocking GA biosynthesis (Takahashi et al., 1991). Figure 3A shows that only rgl2, but not rgl1 or rgl3, conferred PAC resistance during seed germination. We have generated a set of homozygous double mutants by genetic crossing among rgl1, rgl2, and rgl3 and examined whether there is a synergistic interaction among these alleles in seed germination as observed between rga and gai null alleles in vegetative growth (Dill and Sun, 2001). However, the rgl1/rgl2 and rgl2/rgl3 double mutants had a similar germination percentage as that of rgl2 (Fig. 3B), indicating that RGL2 is the primary GA signaling repressor during seed germination.
Figure 3.
Germination of rgl2 seeds is resistant to PAC. Seeds of wild-type plants (WT) and of homozygous single (A) and double (B) rgl mutants were treated with 120 μm PAC. The germination percentages in A and B are the means of three separate experiments. Error bars indicate the se of the mean. For each experiment, approximately 100 to 160 seeds were scored per genotype.
A loss-of-function mutation in RGL2 was shown to rescue the germination defect of the ga1-3 mutant seed (Lee et al., 2002). We therefore introduced the ga1-3 allele into single rgl1, rgl2, rgl3, and rga-28 mutants. Because the original ga1-3 mutation is in the Landsberg erecta (Ler) ecotype, we first made a ga1-3 line that had been crossed for six generations into Col-0. We found that only rgl2 rescued the germination defect of ga1-3, whereas rgl1, rgl3, or rga individually did not promote ga1-3 germination (data not shown). These results are consistent with the finding of Lee et al. (2002), showing that RGL2 plays a major role in repressing seed germination.
Regulation of Floral Development by RGA, RGL1, and RGL2
The ga1-3 mutant is male-sterile with poorly developed stamens and petals (Koornneef et al., 1983), indicating that GA is essential in controlling flower development in Arabidopsis. However, the appropriate level of GA signaling is important because wild-type plants overdosed with GA also have reduced fertility (Jacobsen and Olszewski, 1993). The flowers produced by the rga-24/gai-t6 double null mutant have lower fertility than wild type, although a combination of rga and gai null mutations was unable to suppress the male-sterility of ga1-3 (Dill and Sun, 2001). The reduced fertility of the rga/gai double null mutant mimics the GA overdose phenotype, suggesting that RGA and GAI play only a minor role in flower development (Dill and Sun, 2001). The high levels of RGL1 and RGL2 transcripts in flower clusters and siliques (Fig. 1) suggest that these two RGL genes may be important for floral and seed development. If any of the RGLs is the major repressor in GA-regulated floral development, we would expect to observe a reduced fertility phenotype when the given gene is mutated. We found that none of the single rgl mutants showed floral defects (data not shown). Because of functional redundancy, the role of the RGL genes may be better revealed in multiple mutant backgrounds. However, none of the double or triple homozygous rgl mutants showed any visible floral or fertility defect (data not shown), indicating that RGA and/or GAI also must be inactivated to affect flower development.
The phenotypic effects of rga and gai null mutations are more evident in the ga1-3 background, indicating that RGA and GAI are more active in repressing GA signaling in GA-deficient conditions (Dill and Sun, 2001; King et al., 2001). If this is also true for the RGLs, we would expect to reveal the physiological function of the RGLs more readily in the ga1-3 background. We therefore analyzed a number of GA-regulated traits in single and multiple rga-28 and rgl mutants in the GA-deficient ga1-3 (Col-0) background. These included rosette leaf size (rosette radius), flowering time, final stem height, flower morphology, and fertility (average number of seeds per silique). The rgl3 allele was not included in this study because RGL3 mRNA was present at a very low level in reproductive tissues (Fig. 1; Supplemental Table II).
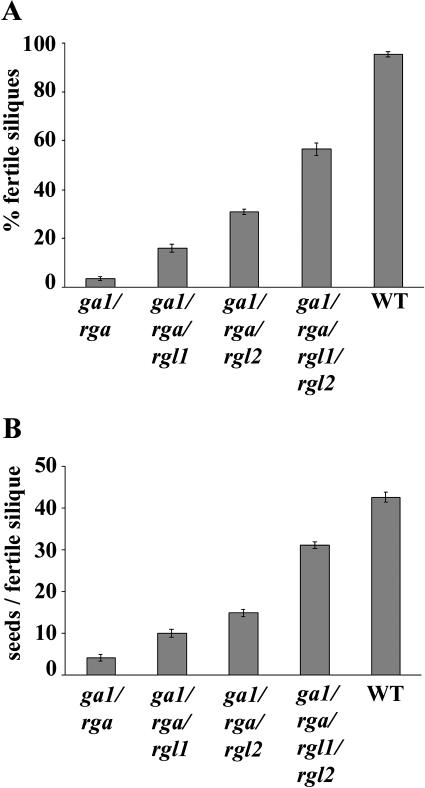
In the Col-0 background, the ga1-3 mutant exhibited a dwarf and sterile phenotype, similar to that seen in the Ler background. The only difference was that this mutant did bolt slightly, with a long delay between flowering and bolting (Table I). The phenotype of ga1-3 (Col-0) is similar to another knockout ga1 allele in the Col-0 background, which was identified from the T-DNA insertion mutant collection at the Salk Institute (M. Goellner-Mitchum and T.-p. Sun, unpublished data). The slightly bolting phenotype of ga1-3 in the Col-0 background is consistent with the previous observation that the erecta (er) mutation in Ler enhances a GA-related dwarf phenotype (Fridborg et al., 2001). Figure 4A and Table I show that rgl1and rgl2—singly or in combination—did not rescue the extreme dwarfism and sterility of ga1-3. This is consistent with our observation that the RGL1 and RGL2 transcripts are present at very low levels in leaves and stems (Fig. 1) and also with previous findings that RGA and GAI are the major repressors in vegetative growth (Dill and Sun, 2001; King et al., 2001). In the Ler ecotype background, loss-of-function rga mutations partially rescue vegetative growth and floral induction, but not fertility of ga1-3 (Silverstone et al., 1997). We found that in the Col-0 background, the knockout rga-28 allele has a similar effect in restoring vegetative growth and floral initiation of ga1-3 (Fig. 4B; Table I). Unexpectedly, rga-28 also partially rescued the floral defect. In contrast to the flowers of ga1-3, the ga1-3/rga-28 mutant flowers contained well-developed petals and more elongated stamen filaments and produced some viable pollen (Fig. 4D). A few (3%) of the flowers on the primary inflorescence stem of ga1-3/rga-28 did produce fertile siliques with very few seeds (an average of four seeds/fertile silique) (Fig. 5, A and B). The homozygous triple mutants ga1/rga/rgl1 and ga1/rga/rgl2 produced more fertile siliques (16% and 31%, respectively) in comparison with the double mutant ga1/rga (Figs. 4C and 5). The homozygous quadruple mutant ga1/rga/rgl1/rgl2 showed the highest fertility among all the mutant combinations (57% fertile siliques with an average of 31 seeds/fertile silique; Fig. 5, A and B). When comparing the stamen filaments among these mutant flowers, the quadruple mutant had the longest stamen filaments that reached the stigma surface, whereas the filaments of the double (ga1/rga) and triple (ga1/rga/rgl1 and ga1/rga/rgl2) mutants were of intermediate length, but longer than in ga1-3 (Fig. 4D). These results indicated that there is an additive interaction among rga, rgl1, and rgl2 in restoring stamen development and fertility. As predicted by the expression patterns of RGL1 and RGL2 (Fig. 1), rgl1 and rgl2 mutations have little effect on the vegetative growth processes examined. The rosette radius, flowering time, and final stem height of the triple and quadruple mutants are similar to those of the double mutant ga1/rga (Fig. 4B; Table I).
Table I.
Flowering time, rosette size, and height of homozygous mutants and wild type
| Genotype | Days to Flower | Maximum Rosette Radius | Final Height |
|---|---|---|---|
| mm | cm | ||
| ga1-3 | 42.4 ± 1.5 | 16.1 ± 0.4 | 1.50 ± 0.16a |
| ga1-3/rgl1 | 38.6 ± 1.1 | 16.4 ± 0.5 | 2.31 ± 0.23a |
| ga1-3/rgl2 | 43.8 ± 1.9 | 16.0 ± 0.2 | 1.92 ± 0.25a |
| ga1-3/rgl1/rgl2 | 38.8 ± 1.3 | 15.5 ± 0.5 | 2.32 ± 0.26a |
| ga1-3/rga-28 | 27.0 ± 0.5 | 21.9 ± 0.5 | 18.21 ± 0.31 |
| ga1-3/rga-28/rgl1 | 29.1 ± 0.4 | 24.6 ± 0.9 | 20.18 ± 0.30 |
| ga1-3/rga-28/rgl2 | 29.8 ± 0.4 | 19.7 ± 0.6 | 17.08 ± 0.32 |
| ga1-3/rga-28/rgl1/rgl2 | 29.5 ± 0.5 | 23.8 ± 1.4 | 17.13 ± 0.36 |
| Wild type | 20.2 ± 0.5 | 27.8 ± 0.8 | 32.32 ± 0.59 |
The values shown are means ± se for 11–18 plants per genotype.
Height 105 d after planting, rather than the final height. Unlike the mutants homozygous for rga-28, which all began bolting immediately after flowering, these plants did not begin bolting until day 65 or later. Only a fraction of the plants (35%, 75%, 38%, and 76% for ga1-3, ga1-3/rgl1, ga1-3/rgl2, and ga1-3/rgl1/rgl2, respectively) had bolted by the end of the experiment.
Figure 4.
Phenotypic effects of loss-of-function rga and rgl mutations in a GA-deficient background. A, ga1-3 and rgl mutant combinations without rga-28. B, ga1-3, rgl, and rga-28 mutant combinations and a wild-type control. C, Primary inflorescence stems of mutants and wild type, as labeled. D, Flowers of mutant plants and wild type, as labeled. The front-most floral organs (sepal, petal, and in some cases stamen) were removed to expose the interior of the flower. Wild-type and homozygous mutant plants were grown on soil under long-day conditions. The plants shown in panels (A) and (B) are 73 d old, except for wild type, which is 44 d old.
Figure 5.
Effect of the rga, rgl1, and rgl2 mutations on fertility in a ga1-3 background. A, Percent of fertile siliques on the primary inflorescence. B, Number of seeds per fertile silique on the primary inflorescence. Seeds were counted for a minimum of 17 fertile siliques for each wild-type plant and for all possible fertile siliques for each mutant plant. Both A and B show the means ± se for 14 to 16 plants per genotype.
GA-Induced RGL2 Degradation
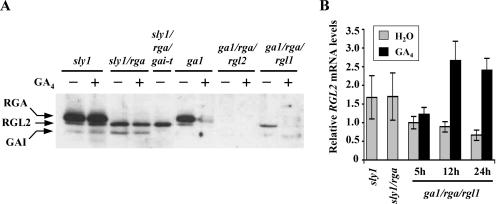
Our qPCR data revealed that expression of the DELLA protein genes, especially for RGL1, 2, and 3, is regulated developmentally at the transcript level (Fig. 1). In the case of RGA and GAI activity, another regulatory mechanism appears to be essential. The endogenous RGA and GAI proteins are degraded rapidly in response to GA treatment, and the proteolysis of these proteins requires a functional SLY1, which is likely a component of the ubiquitin E3 ligase complex SCFSLY1 (Silverstone et al., 2001; McGinnis et al., 2003; Dill et al., 2004). To examine whether RGL2 protein stability is affected by GA and the loss-of-function sly1-10 allele, we first analyzed RGL2 protein levels in imbibed seeds by immunoblot analysis. Imbibed seeds were tested because the RGL2 mRNA level is high in this tissue and RGL2 is the major regulator during germination. Furthermore, seeds with the ga1-3 mutant background were chosen for this analysis, because RGA protein levels are elevated under GA-deficient conditions (Silverstone et al., 2001; McGinnis et al., 2003). If GA affects RGL2 protein stability, RGL2 would accumulate to a higher level in this GA-deficient background. Using polyclonal anti-RGA antibodies, we detected a protein of approximately 61 kD in addition to RGA (64 kD) in ga1-3 (Fig. 6A). A protein with the same mobility was also present in the ga1-3/rga-28/rgl1 mutant (–GA sample) but was missing in ga1-3/rga-28/rgl2 (Fig. 6A), suggesting that this protein is RGL2. Interestingly, this protein (presumably RGL2) disappeared after 5 h of GA treatment in both ga1 and ga1/rga/rgl1 seeds. In addition, in the sly1-10 mutant background (sly1-10, sly1-10/rga, and sly1-10/rga/gai-t6), RGL2 accumulated at higher levels and was unaffected after GA treatment. These results suggest that RGL2, like RGA and GAI, is targeted for GA-induced degradation by SLY1. The effects of GA and sly1-10 on the RGL2 protein levels are not due to changes in the amounts of the RGL2 transcript (Fig. 6B).
Figure 6.
SLY1 regulates GA-induced RGL2 degradation. A and B, protein or RNA was isolated from imbibed seeds of various homozygous mutants as labeled. Before harvesting the tissues, the seeds were imbibed in water for 2 d, and then treated for 5, 12, or 24 h with 10 μm GA4 (+) or water (–). A, The protein blot contains 60 μg total proteins from imbibed seeds of homozygous mutants (±GA for 5 h) and was probed with polyclonal anti-RGA antibodies from rat. B, Relative RGL2 mRNA levels determined by qRT-PCR. At least three reactions were performed for each sample, and the RGL2 mRNA level was normalized using 18S rRNA. The means ± se are shown. The value of RGL2 mRNA in 5-h water-treated ga1-3/rga/rgl1 is arbitrarily set to 1.0.
Figure 6A also shows that the RGL2 signals detected by the polyclonal anti-RGA antibodies were significantly lower than those of RGA in imbibed seeds. Using recombinant His-tagged RGA and RGL2 proteins in immunoblot analysis, we found that the affinity of the polyclonal anti-RGA antibodies to RGA is approximately 3-fold of the affinity to RGL2 (data not shown). Therefore, RGL2 is present at a lower level than RGA in imbibed seeds, even though RGL2 plays a major role in repressing seed germination. In addition, the amounts of RGL2 were elevated when RGA protein was absent (sly1-10 versus sly1-10/rga, ga1-3 versus ga1-3/rga/rgl1; Fig. 6A), suggesting that the cell has a regulatory mechanism to sense and modulate different DELLA protein levels to achieve proper levels of GA signaling. Alternatively, the changes in RGL2 protein levels may be due to differences in developmental stages caused by the absence of RGA.
Interaction of RGLs and SLY1 in Yeast Two-Hybrid Assays
Previous studies demonstrated that RGA and GAI directly interact with SLY1 in yeast two-hybrid assays and that a dominant mutation (sly1-d) affecting the C-terminal region of SLY1 enhances the interaction with RGA and GAI (Dill et al., 2004). We tested whether RGLs also interact with SLY1 and sly1-d in this yeast two-hybrid system, where SLY1 and sly1-d were expressed as fusion proteins with the LexA DNA binding domain (DB), and RGL1, RGL2, RGL3, and RGA were fused to the Gal4 transcription activation domain (AD). Expression of two reporter genes (His-3 and LacZ) was assayed as previously described (Dill et al., 2004). We found that RGL1 and RGL3 showed a weak interaction with SLY1, similar to RGA (Table II). However, we were unable to detect an interaction between RGL2 and SLY1. Interestingly, sly1-d interacted much more strongly with RGL1, RGL2, and RGL3 than SLY1 did (Table II), consistent with the earlier finding that the sly1-d mutation allows for a greater interaction between sly1 and RGA (Dill et al., 2004). These results support the idea that SLY1 recruits RGL1, RGL2, and RGL3 for GA-induced degradation.
Table II.
Interaction of RGLs and RGA with SLY1 and sly1-d in yeast two-hybrid assays
| LexA DB Fusion | Gal4 AD Fusion | His− Media + 3-ATa | β-gal Unitsb |
|---|---|---|---|
| mm | |||
| SLY1 | RGL1 | 2 | 1.5 ± 0.2 |
| SLY1 | RGL2 | 0 | 0.1 ± 0.1 |
| SLY1 | RGL3 | 5 | 1.4 ± 0.2 |
| SLY1 | RGA | 2 | 0.7 ± 0.1 |
| sly1-d | RGL1 | 5 | 7.1 ± 1.3 |
| sly1-d | RGL2 | 30 | 43.0 ± 5.9 |
| sly1-d | RGL3 | 30 | 34.0 ± 1.5 |
| sly1-d | RGA | 60 | 118.7 ± 16.1 |
| LexA | RGL1 | 1 | 1.0 ± 0.0 |
| LexA | RGL2 | 0 | 0.1 ± 0.1 |
| LexA | RGL3 | 0 | 0.1 ± 0.0 |
| LexA | RGA | 0 | 0.1 ± 0.1 |
| SLY1 | Gal4 | −c | Ndd |
| sly1-d | Gal4 | − | Nd |
The relative growth on His− plates containing 3-aminotriazole (3-AT) at 0, 1, 2, 5, 10, 30, and 60 mm. 3-AT is a competitive inhibitor of the His3 enzyme. The experiment was repeated twice.
β-gal activity (units). For each sample, at least three independent enzyme assays were performed and the means ± se are shown.
No growth on His− plates at 0 mm 3-AT.
Nd, Not determined.
DISCUSSION
To study the potential function of the RGL genes and to identify the specific DELLA protein genes that control floral development in Arabidopsis, we surveyed the developmental expression profiles of all 5 DELLA protein genes and isolated and characterized new knockout rga, rgl1, rgl2, and rgl3 mutants. Our results demonstrate that RGA, RGL1, and RGL2 together are important for modulating GA-regulated floral development, while RGL2 plays a major role during seed germination in Arabidopsis.
In the case of RGA and GAI, transcript levels do not necessarily reflect protein levels or activity, and ubiquitin-mediated proteolysis serves as a crucial mechanism for the regulation of RGA and GAI activity. However, our qPCR-generated expression data provided a starting point for the investigation of the potential function of each RGL gene and supported the idea of overlapping functions for DELLA subfamily members. Imbibed seeds contained high levels of RGL2, RGL3, RGA, and GAI transcripts, suggesting that they all might regulate germination. However, rga and gai null alleles—singly or in combination—cannot rescue the germination defect of ga1-3 (Silverstone et al., 1997). Using the newly isolated knockout rgl mutants, we demonstrated that RGL2 plays the most important role (among the DELLA proteins) for regulating seed germination. Our results support the previous data of Lee et al. (2002) showing that only rgl2, but not rgl1, affects germination of ga1-3 seed. However, GA or PAC dose response curves for seed germination need to be performed to determine whether other DELLA proteins may also contribute to this process. It is possible that additional DELLA proteins do play some roles in modulating germination, especially under different environmental conditions that may affect seed dormancy. It is interesting that RGA, GAI, and RGL3 transcripts—like RGL2 transcripts—accumulate to high levels in imbibed seeds (Fig. 1), and RGA protein is also relatively abundant in comparison to RGL2 (Fig. 6A). These results suggest that RGL2 may have a higher specific activity than RGA in repressing the germination of imbibed seeds. Alternatively, RGL2 and RGA may express in different tissue- or cell-types, and therefore may regulate different developmental aspects of germinating seed.
In this study, we found that the rga-28 mutation in the Col-0 background restored petal development and also slightly rescued the stamen defect and male infertility of ga1-3. In contrast, rga alleles in the Ler background have no effect on the floral defect or male sterility of ga1-3 (Silverstone et al., 1997; Dill and Sun, 2001). The er mutation in Ler enhances dwarf phenotypes of GA-deficient or GA-insensitive mutants (Fridborg et al., 2001), although the molecular mechanism involved has not been elucidated. A loss-of-function rga mutation may more readily suppress the floral defect of ga1-3 in the Col-0 ecotype (ER) than in the Ler background, because Col-0 does not contain the er mutation. Alternatively, additional modifier gene(s) may be present in different ecotype backgrounds. Our mutant analysis also revealed that rgl1 and rgl2, in combination with rga, significantly increased the stamen filament growth, anther development, and fertility of ga1-3 flowers. However, the quadruple mutant ga1/rga/rgl1/rgl2 still did not reach wild-type levels of fertility (Fig. 5). Thus, there is a high degree of functional redundancy in controlling flower development. It remains to be determined whether GAI and/or RGL3 also contribute to this regulation. Our qPCR data would predict that GAI, but not RGL3, is more likely to be involved because RGL3 mRNA is present only at a very low level in flowers and siliques. Recently, GA has also been shown to affect pollen tube growth (Singh et al., 2002). Future studies will be needed to elucidate the potential role of all the DELLA protein genes in this process. Therefore, in contrast to germination and vegetative growth, which are mainly controlled by 1 or 2 DELLA protein genes, floral development appears to have more complex regulation.
Studies of RGA and GAI orthologs in several crops indicate that the function of these DELLA proteins in repressing GA signaling is highly conserved between dicots and monocots (Richards et al., 2001; Olszewski et al., 2002; Gomi and Matsuoka, 2003). Interestingly, unlike Arabidopsis, only one functional ortholog of DELLA protein is present in rice (SLR1; Ikeda et al., 2001) and one in barley (SLN1; Chandler et al., 2002). Consequently, GA-independent stem growth is achieved by removing only SLR1 or SLN1, respectively, in these species. Similar to RGA, SLN1 and SLR1 are responsive to GA-induced proteolysis (Gubler et al., 2002; Itoh et al., 2002). The transcript levels of RGA and SLR1 are slightly increased by GA, probably due to a feedback mechanism (Silverstone et al., 1998; Itoh et al., 2002).
Recent studies showed that RGL2 mRNA levels in imbibed ga1-3 seeds are reduced after GA treatment for 48 h at 23°C (Lee et al., 2002). This result led to the conclusion that GA promotes germination by reducing the amount of RGL2 transcript. However, we found that in imbibed ga1-3 seeds, RGL2 protein disappeared after only 5 h of GA treatment, and that the loss-of-function sly1 mutation resulted in a high RGL2 protein level (Fig. 6A). In contrast, 5- to 24-h GA treatment or the sly1 mutation did not significantly alter the RGL2 transcript levels. Moreover, SLY1 and RGL2 interacted in a yeast two-hybrid assay. These data supported the idea that GA induces germination by causing proteolysis of RGL2 and that SLY1 targets RGL2 for degradation. The reduction of RGL2 mRNA levels following a prolonged GA treatment seen in the previous study (Lee et al., 2002) may be a secondary effect of GA-induced germination. Our interpretation is also based on the observation that during wild-type seed germination, RGL2 mRNA levels are elevated following imbibition but reduced when germination occurs (Lee et al., 2002; Fig. 1). After treating the imbibed ga1-3 seeds with GA for 48 h at 23°C, germination would have occurred. In summary, RGL2 gene expression is regulated at both transcript and protein levels during seed germination. Prior to the germination of wild-type imbibed seeds, elevation of endogenous GA biosynthesis (Ogawa et al., 2003) followed by GA-dependent rapid proteolysis of RGL2 (and other DELLA proteins) may be required to promote seed germination.
It is not clear why Arabidopsis contains five DELLA proteins whereas only one DELLA protein is present in rice and in barley. Nevertheless, results from our study and previous studies are beginning to reveal the complex regulation of these functionally overlapping genes for fine-tuning GA-regulated development in Arabidopsis. Isolation and characterization of DELLA proteins in additional species will allow us to determine whether complex developmental control of dicots requires multiple DELLA proteins. Alternatively, the differences in the DELLA-protein copy numbers may simply be the consequence of gene duplication events that occurred after the divergence of eudicots and monocots.
MATERIALS AND METHODS
Plant Lines
For all of the mutant characterization experiments, the Columbia-0 (Col-0) genetic background was used as the wild type. The rgl1-2, rgl2-13, rgl3-3, and rga-28 lines were isolated in this study by PCR screening of a collection of T-DNA insertion lines generated in the Col-0 background at the Salk Institute (Alonso et al., 2003; see below). Crossing ga1-3, originally in Landsberg erecta (Ler), to Col-0 six successive times yielded the ga1-3 line (ER homozygous) used in this study. This ga1-3 line was then used to generate double, triple, and quadruple mutants with the rga-28 and rgl alleles. Homozygous mutants were identified by PCR using allele-specific primers (Supplemental Table III). For protein-blot analysis, several homozygous single, double, and triple mutant lines (sly1-10, sly1-10/rga-24 and sly1-10/rga-24/gai-t6; McGinnis et al., 2003; Dill et al., 2004) that are in the Ler background were also included.
Identification of T-DNA Insertion Mutant Lines
The rga-28, rgl1-2, rgl2-13, and rgl3-3 mutants were isolated by screening the multidimensional DNA pools of T-DNA insertion mutant populations using a PCR-based method, as described previously (Alonso et al., 2003). The T-DNA left and right border primers (Alonso et al., 2003) and degenerate primers (primer 355 for rgl mutants and primer 244 for rga mutants) were used for PCR (Supplemental Table III). Mutant lines were identified by analyzing PCR products by DNA-blot analyses using gene-specific cDNA probes for each gene of interest, and the location of each T-DNA insertion in each mutant was determined by DNA sequence analysis of the PCR products. Homozygous mutant plants were identified by PCR using allele-specific primers (Supplemental Table III). To detect any remaining transcripts produced in each mutant, RT-PCR was performed using primers that span the T-DNA insertion (Supplemental Table III) or are specific for sequences upstream of the insertion.
Plant Growth Conditions
Plants were grown on MetroMix 200 soil (Scotts-Sierra Horticultural Products, Marysville, OH) at 22°C under long-day conditions (16 h light/8 h darkness). To promote the germination of GA-deficient seeds, all seeds for the characterization study (except wild type) were treated with 50 μm GA4 at 4°C for 7 d and rinsed thoroughly with water before planting. Wild-type seeds were incubated in water at 4°C for 3 d prior to planting. The first clear appearance of a flower bud that was visible to the naked eye was taken to mark the flowering time. The length of the longest rosette leaf was considered to be the maximum rosette radius.
Quantitative, Real-Time PCR
Total RNA was extracted from approximately 150 to 200 mg samples of the following wild-type tissues: seeds imbibed in water for 12 or 24 h under continuous light; 2- and 5-d-old seedlings; shoots and roots of 10-d-old seedlings; entire aerial portions (rosettes) of 14-d-old plants; and rosette leaves, cauline leaves, stem tissue, flower clusters, and siliques from 33-d-old plants. Seeds (for 12 and 24 h imbibed samples) were washed in 0.02% Triton X-100 and rinsed with sterile water before plating on filter paper for imbibition under continuous light. At the 24 h time-point, most of the seeds had begun to germinate, as evidenced by a cracked seed coat or protruding radicle. Seedlings for the 2-, 5-, and 10-d time points were grown on plates of 1× Murashige and Skoog medium containing 2% sucrose and 0.8% agar. Seedlings for the 10-d time point were grown vertically to facilitate the recovery of root tissue. The stem tissue included only internodes, without the nodes. Flower clusters and siliques were at various stages of development.
RNA was extracted from seeds using the RNAqueous kit plus Plant RNA Isolation Aid (Ambion, Austin, TX). An RNeasy Plant Mini kit (QIAgen, Valencia, CA) was used for the remaining samples, with buffer RLT for all samples except siliques, for which buffer RLC was employed. DNA was removed via either an on-column DNase treatment for the QIAgen kit or a separate treatment with RQ1 DNase (Promega, Madison, WI) for the samples extracted with the Ambion kit. RT-PCR was first performed using primers that span an intron in AtGA3ox1 (Supplemental Table III) to confirm that each RNA sample was free of genomic DNA contamination. For real-time PCR, the First Strand cDNA Synthesis kit (Roche, Diagnostics, Mannheim, Germany) was used to make cDNA from 1 μg of RNA in a 20 μL reaction volume. Each cDNA sample was diluted 1:20 in water, and 2 μL of this dilution was used as template for qPCR. Half-reactions (10 μL each) were performed with the Lightcycler FastStart DNA Master SYBR Green I kit (Roche) on a Roche LightCycler real-time PCR machine, according to the manufacturer's instructions. UBIQUITIN11 (UBQ11, At4g05050) was used as a control in qPCR, because its transcript levels remained similar across the tissues and developmental stages tested when compared with the 18S rRNA by quantitative RT-PCR (qRT-PCR) using the LightCycler RNA amplification kit SYBR Green I (Roche; Supplemental Table I). Gene-specific primers for detecting transcripts of UBQ11, 18S, RGL1, RGL2, RGL3, RGA, and GAI are listed in Supplemental Table III. Primers were used at a final concentration of 0.5 μm each, and the annealing temperature was 55°C in all cases. The qRT-PCR for 18S and UBQ11 contained 5 pg and 50 ng of RNA template, respectively, as well as a final MgCl2 concentration of 6 mm. The qPCR reactions for RGL1 included a final MgCl2 concentration of 3 mm. Reactions for RGL3 were performed with 5 mm MgCl2, and the remaining qPCR reactions (for RGL2, RGA, GAI, and UBQ11) included 4 mm MgCl2. PCR products for RGL1, RGL2, RGL3, RGA, and GAI and a linearized plasmid for UBQ11 were used to generate standard curves. The number of gene-specific cDNA copies was determined for each sample, normalized using the UBQ11 cDNA level, and averaged over three replicates.
Germination Assays
For germination inhibition assays, a stock of 50 mm PAC (PhytoTechnology Laboratories, Shawnee Mission, KS) in 95% ethanol was diluted to 120 μm PAC in 0.01% Tween 20. Seeds were washed with 0.02% Triton X-100, rinsed with sterile water, and spread onto filter paper saturated with the 120 μm PAC solution, in petri dishes. As a negative control (–PAC), the seeds were incubated with 0.01% Tween 20 and 0.23% ethanol. After 7 d of incubation at 22°C under 16 h light (100 μE)/8 h darkness, radicle protrusion was scored as germination. Without PAC treatment, seeds of all genotypes germinated. Only seeds harvested at the same time were used for each experiment, because the percent germination could vary significantly depending on the age of the seeds.
To test the effect of rga or rgls in restoring ga1-3 germination, the seeds were rinsed once with sterile water and spread onto filter paper saturated with sterile water. Germination was scored after 4 d of incubation under the same conditions as in the PAC assays.
Analysis of RGL2 Protein and Transcript Levels by Immunoblot Analysis and Real-Time RT-PCR
Seeds of homozygous mutants ga1-3, ga1-3/rga-28/rgl1-2, ga1-3/rga-28/rgl2-13, sly1-10, sly1-10/rga-24, and sly1-10/rga-24/gai-t6 were sprinkled onto filter paper that was saturated with sterile water in petri dishes and imbibed for 2 d under continuous light of 100 μE at 22°C. Imbibed seeds were then treated with 10 μm GA4 (+) or water (–) for 5 h (all genotypes) and 12 h or 24 h (only for ga1-3/rga-28/rgl1) before harvesting. Under these conditions, none of the seeds (±GA for 5 or 12 h) in the ga1-3 background germinated, except ga1-3/rga/rgl2. After 24 h GA treatment, approximately 20% of ga1-3/rga/rgl1 seeds germinated. For seeds in the sly1 background, only a small fraction of the seeds germinated, and we only harvested ungerminated seeds for protein and transcript analysis. Total proteins were extracted from the 5 h ± GA samples and analyzed by immunoblot analysis using anti-RGA antibodies from rat (DUR18) as described previously (McGinnis et al., 2003). Ponceau staining was used to confirm equal loading.
RNA was extracted from the imbibed ga1-3/rga-28/rgl1-2 seeds (±GA for 5 h, 12 h, and 24 h), sly1-10 and sly1-10/rga-24 seeds (–GA, 5 h) as described in the “Quantitative Real-Time PCR” section. The RGL2 transcript levels were analyzed by qRT-PCR using a Roche LightCycler and the LightCycler RNA Amplification kit SYBR Green I (Roche) according to the manufacturer's instructions. Gene-specific primers for RGL2 and 18S rRNA (Supplemental Table III) were used in the qRT-PCR with the annealing temperature at 55°C in 7 mm MgCl2 for RGL2, and 6 mm MgCl2 for 18S. 5 pg or 25 ng of total RNA was used as a template in each qRT-PCR reaction (10 μL volume) for 18S rRNA and RGL2 RNA, respectively. A no-template control was routinely included to confirm the absence of DNA or RNA contamination. Relative transcript levels of RGL2 in all samples were normalized using 18S rRNA because UBQ11 expression is affected by the GA treatment. The reactions were performed three times using freshly diluted RNA samples in each set of reactions.
Yeast Two-Hybrid Assays
Yeast two-hybrid assays were performed using Saccharomyces cerevisiae strain L40 as described previously (Dill et al., 2004). The bait and prey protein fusions were expressed as LexA DNA-binding domain (DB) and Gal4 activation domain (AD) fusions using the yeast plasmid expression vectors pLexA-NLS (Vojtek et al., 1993) and pACTII (Li et al., 1994), respectively. pLexA-SLY1 and pLexA-sly1-d were made previously (Dill et al., 2004), and pGal4-RGL1 (in pACT) was a gift from Dr. Caren Chang (Wen and Chang, 2002). The Gal4-RGL2 and Gal4-RGL3 constructs were made by amplifying the coding regions of RGL2 and RGL3 from Col-0 genomic DNA with PCR primers that incorporate NcoI and BamHI sites (for RGL2) or BglII site (for RGL3) in the correct reading frame (Supplemental Table III). The PCR DNA fragments were digested with NcoI and BamHI (for RGL2) or BglII (for RGL3) and subcloned into the NcoI/BamHI sites or BamHI site of pACTII.
Distribution of Materials
Upon requests, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes. The only exception is that we will not be able to distribute the anti-RGA antibodies from rat (DUR18) because we only have a very limited amount.
Supplementary Material
Acknowledgments
We thank Aron Silverstone and Dong Wang for helpful comments on the manuscript. We also thank Caren Chang for providing the Gal4 AD-RGL1 plasmid. We are grateful to Melissa Goellner-Mitchum for help with real-time PCR and for providing the UBQ11 clone used to generate a standard curve.
This work was supported by the National Science Foundation (grant nos. IBN–0078003 and IBN–0235656).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.039578.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-Wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Chandler PM, Marion-Poll A, Ellis M, Gubler F (2002) Mutants at the Slender1 locus of barley cv Himalaya: molecular and physiological characterization. Plant Physiol 129: 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien JC, Sussex IM (1996) Differential regulation of trichome formation on the adaxial and abaxial leaf surfaces by gibberellins and photoperiod in Arabidopsis thaliana (L.) Heynh. Plant Physiol 111: 1321–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies PJ, ed (1985) Plant Hormones: Physiology, Biochemistry, and Molecular BIology. Kluwer Academic Publishers, Dordrecht, The Netherlands
- Dill A, Jung H-S, Sun T-p (2001) The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc Natl Acad Sci USA 98: 14162–14167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Sun T-p (2001) Synergistic de-repression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159: 777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Thomas SG, Hu J, Steber CM Sun T-p (2004) The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16: 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleck B, Harberd NP (2002) Evidence that the Arabidopsis nuclear gibberellin signalling protein GAI is not destabilized by gibberellin. Plant J 32: 935–947 [DOI] [PubMed] [Google Scholar]
- Fridborg I, Kuusk S, Moritz T, Sundberg E (2001) The Arabidopsis protein SHI represses gibberellin responses in Arabidopsis and barley. Plant Physiol 127: 937–948 [PMC free article] [PubMed] [Google Scholar]
- Fu X, Harberd NP (2003) Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421: 740–743 [DOI] [PubMed] [Google Scholar]
- Gomi K, Matsuoka M (2003) Gibberellin signalling pathway. Curr Opin Plant Biol 6: 489–493 [DOI] [PubMed] [Google Scholar]
- Gubler F, Chandler P, White R, Llewellyn D, Jacobsen J (2002) GA signaling in barley aleurone cells: control of SLN1 and GAMYB expression. Plant Physiol 129: 191–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J (2001) Slender rice, a constitutive gibberellin response mutant is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13: 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H, Ueguchi-Tanaka M, Sato Y, Ashikari M, Matsuoka M (2002) The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14: 57–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE (1993) Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 5: 887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King K, Moritz T, Harberd N (2001) Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 159: 767–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Elgersma A, Hanhart CJ, van Loenen MEP, van Rijn L, Zeevaart JAD (1985) A gibberellin insensitive mutant of Arabidopsis thaliana. Physiol Plant 65: 33–39 [Google Scholar]
- Koornneef M, van der Veen JH (1980) Induction and analysis of gibberellin-sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theor Appl Genet 58: 257–263 [DOI] [PubMed] [Google Scholar]
- Koornneef M, van Eden J, Hanhart CJ, de Jongh AMM (1983) Genetic fine-structure of the GA-1 locus in the higher plant Arabidopsis thaliana (L.) Heynh. Genet Res Camb 41: 57–68 [Google Scholar]
- Lee S, Cheng H, King KE, Wang W, He Y, Hussain A, Lo J, Harberd NP, Peng J (2002) Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev 16: 646–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Elledge SJ, Peterson CA, Bales ES, Legerski RJ (1994) Specific Association Between the Human DNA Repair Proteins XPA and ERCC1. Proc Natl Acad Sci USA 91: 5012–5016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis KM, Thomas SG, Soule JD, Strader LC, Zale JM, Sun T-p, Steber CM (2003) The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15: 1120–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, Yamaguchi S (2003) Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15: 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski N, Sun T-p, Gubler F (2002) Gibberellin signaling:biosynthesis, catabolism, and response pathways. Plant Cell Supplement: S61–S80 [DOI] [PMC free article] [PubMed]
- Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP (1997) The Arabidopsis GAI gene defines a signalling pathway that negatively regulates gibberellin responses. Genes Dev 11: 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN (1999) The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J 18: 111–119 [DOI] [PubMed] [Google Scholar]
- Richards DE, King KE, Ait-ali T, Harberd NP (2001) How gibberellin regulates plant growth and development: a molecular genetic analysis of gibberellin signaling. Annu Rev Plant Physiol Plant Mol Biol 52: 67–88 [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Ciampaglio CN, Sun T-p (1998) The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10: 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Jung H-S, Dill A, Kawaide H, Kamiya Y, Sun T-p (2001) Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13: 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Mak PYA, Casamitjana Martínez E, Sun T-p (1997) The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics 146: 1087–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh DP, Jermakow AM, Swain SM (2002) Gibberellins are required for seed development and pollen tube growth in Arabidopsis. Plant Cell 14: 3133–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber CM, Cooney S, McCourt P (1998) Isolation of the GA-response mutant sly1 as a suppressor of ABI1-1 in Arabidopsis thaliana. Genetics 149: 509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T-p, Kamiya Y (1994) The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6: 1509–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Phinney BO, MacMillan J, eds (1991) Gibberellins. Springer-Verlag, New York Inc., New York
- The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Vojtek AB, Hollenberg SM, Cooper JA (1993) Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell 74: 205–214 [DOI] [PubMed] [Google Scholar]
- Wen C-K, Chang C (2002) Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 14: 87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RN, Heckman JW, Somerville CR (1992) Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol 100: 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.