Abstract
Acute myeloid leukemia (AML) is associated with poor clinical outcome and the development of more effective therapies is urgently needed. G protein-coupled receptors (GPCRs) represent attractive therapeutic targets, accounting for approximately 30% of all targets of marketed drugs. Using next-generation sequencing, we studied the expression of 772 GPCRs in 148 genetically diverse AML specimens, normal blood and bone marrow cell populations as well as cord blood-derived CD34-positive cells. Among these receptors, 30 are overexpressed and 19 are downregulated in AML samples compared with normal CD34-positive cells. Upregulated GPCRs are enriched in chemokine (CCR1, CXCR4, CCR2, CX3CR1, CCR7 and CCRL2), adhesion (CD97, EMR1, EMR2 and GPR114) and purine (including P2RY2 and P2RY13) receptor subfamilies. The downregulated receptors include adhesion GPCRs, such as LPHN1, GPR125, GPR56, CELSR3 and GPR126, protease-activated receptors (F2R and F2RL1) and the Frizzled family receptors SMO and FZD6. Interestingly, specific deregulation was observed in genetically distinct subgroups of AML, thereby identifying different potential therapeutic targets in these frequent AML subgroups.
Introduction
G protein-coupled receptors (GPCRs) represent the largest family of membrane receptors with an estimated number of 800 members in human. They are key transducers that bind a vast diversity of ligands allowing the cells to adapt to their environment by regulating a wide variety of physiological processes including the control of blood pressure, heart rate, digestive processes, hormone secretion, cell growth and migration as well as vision and olfaction. Binding to their ligands leads to conformational rearrangements promoting the engagement and modulation of many distinct downstream signaling effectors that are both G protein-dependent and independent.1, 2
Several GPCRs are critical for cell proliferation and survival, and can be aberrantly expressed in cancer cells.3, 4 For example, PAR1 is overexpressed in invasive breast carcinomas5 or advanced-stage prostate cancer.6, 7 Likewise, the chemokine receptor CXCR4 has an important role in tumor metastasis and angiogenesis.3, 4 Moreover, the Wnt target gene, Lgr5, identified as a marker of intestinal stem cells, is implicated in mouse intestinal tumorigenesis8, 9 and its expression is also associated with poor clinical outcome in colorectal cancer.10
Although GPCRs are targets for approximately 30% of all marketed drugs,11 only a very limited number of agonists or antagonists acting through these receptors are currently used for cancer therapy.12 Notable exceptions include peptide antagonists of gonadotropin releasing-hormone receptor in prostate cancer,12 a somatostatin receptor agonist (Octreotide) and a growth hormone receptor antagonist (Pegvisomant) for neuroendocrine tumors,12 as well as Vismodegib, an antagonist of smoothened (SMO), approved by the US Food and Drug Administration (FDA) to treat advanced basal cell carcinoma.13
In acute myeloid leukemia (AML), CXCR4 overexpression has been associated with poor outcome.14, 15, 16 Moreover, in vivo studies have shown that the use of AMD3465, a small molecule antagonist of CXCR4, increases the mobilization of AML cells into the peripheral blood and improves the efficacy of chemotherapy.17 This activity has been explored in a phase 1/2 clinical study showing that the addition of CXCR4 antagonists to chemotherapy is possible in AML and might improve the remission rate.18 The role of GPCRs in mouse leukemic cells was also suggested in a transcriptome analysis of two related leukemia clones which differ in their stem cell frequency.19 This study revealed that genes encoding GPCRs were the most differentially expressed between the two clones compared with other classes of genes (~22% versus ~5%).
Despite these punctual observations, an exhaustive assessment of GPCR expression in human AML is lacking. To address this issue, we sequenced the transcriptome of a large cohort of AML samples and herein report the expression pattern of GPCRs in 148 AML samples.
Materials and methods
Human primary leukemic and cord blood cells
The 148 AML samples of the Leucegene cohort used for this study (described in Supplementary Table 1) were collected by the Banque de cellules leucémiques du Québec (BCLQ) with an informed consent and approval of the project by the Research Ethics Board of the Maisonneuve-Rosemont Hospital and Université de Montréal. The genetic subgroups of the AML samples included in this study are listed in Supplementary Table 2. Cord blood samples (n=12) collected with an informed consent were provided by Héma-Québec and pre-enriched for CD34+ cells before being sorted for the CD34 APC+/CD45RA PE− cell population as previously described.20 The Cancer Genome Atlas (TCGA) RNA-Seq AML dataset has been downloaded from the TCGA website (https://tcga-data.nci.nih.gov/tcga/tcgaDownload.jsp) in November 2013 and the associated clinical informations obtained from Cancer Genome Atlas Research Network.21
Sorting of normal bone marrow and peripheral blood cell populations
Three unsorted fresh bone marrow samples from healthy donors were purchased from Lonza (Lonza 1M-125; Basel, Switzerland). Red blood cell lysis was performed prior to resuspending the cells in phosphate-buffered saline, 0.1% bovine serum albumin and DNase 10 μg/ml. Subpopulations were sorted on a BD Aria II sorter (BD Biosciences, San Jose, CA, USA) based on published surface marker combinations (Supplementary Methods and Supplementary Table 3). Peripheral blood was collected from healthy donors with informed consent, subjected to red blood cell lysis and subsequently sorted on the basis of the following sorting strategy: Granulocytes (SSChigh, CD33+), B cells (Lymphocyte gate FSClow, SSClow, CD19+), T cells (Lymphocyte gate FSClow, SSClow, CD3+), Monocytes (Monocyte gate FSChigh, SSClow/med, CD14bright, CD33med), total white blood cells (WBC). Sorting purity was checked on aliquots after sorting, and cells were counted, resuspended in TRIzol reagent and stored at −80 °C until RNA isolation was performed according to the manufacturer's instructions. An additional purification step on RNeasy mini columns (Qiagen 74104; Hilden, Germany) was performed to optimize RNA quality, which was subsequently tested on an Agilent bioanalyzer 2100 (Agilent, Santa Clara, CA, USA). A minimum of 50 000 cells was used for RNA Sequencing (RNA-Seq), which was performed as described below.
Sequencing and RNA-Seq data analysis
RNA-Seq was performed using an Illumina HiSeq 2000 instrument (Illumina, San Diego, CA, USA). Libraries were prepared according to the manufacturer's recommendations (Illumina). RefSeq annotations were based on the UCSC January 27th 2011 version. The alignment to reference genome (hg19) was carried out using the CASAVA 1.8.2 package and Eland v2 mapping software and bioinformatic analyses were performed as described earlier.22
Statistical analysis
RNA-Seq data in Reads Per Kilobase per Million mapped reads (RPKM) were transformed to lRPKM (log2(RPKM+1)), where +1 was added to avoid excessive variations due to very small values. Log transformation was performed to avoid overrepresentation of extreme values. Highly expressed GPCRs were selected using a threshold of 3.5 lRPKM (or 10.35 RPKM) as shown in Supplementary Figure 1A. The variability of expression between samples was determined by calculating the coefficient of variation, a ratio between the standard deviation and the mean expression value. As illustrated in Supplementary Figure 1B, genes with a coefficient of variation smaller than the threshold (50%, gray area) are considered as GPCRs with low variability in their expression. Upregulated and downregulated GPCRs were described as those having a difference in median expression between AML and normal CD34+ cells greater than 1 and less than −1, respectively.
GPCR subfamily enrichment analysis
Grouping of GPCR subfamilies was based on the International Union of Basic and Clinical Pharmacology (IUPHAR) database classification downloaded from the website in July 2014 (http://www.guidetopharmacology.org/). To complete the classification and subdivide the class A group into further subgroups, the GRAFS phylogenetic classification of GPCRs was also used.23 Taste receptors have been added along with vomeronasal receptors, opsins and three orphan GPCRs (GPR137B, TAPT1 and XPR1). Overall, GPCRs were classified into 18 subfamilies (Supplementary Table 4).
The GPCR subfamily enrichment analysis in the upregulated or downregulated groups was performed using a Fisher's exact test. Significance (two-tailed P-value) was calculated using the function FET of the add-in Fisherexact downloaded from http://www.obertfamily.com/software/fisherexact.html. The receptors associated with a specific AML genetic subgroup were identified by calculating a difference in mean expression level (lRPKM) between samples with and without the genetic abnormality. An arbitrary difference of 1.5 lRPKM and a significant Student's t test (P-value<0.05) were used as cutoff levels to identify differentially expressed GPCRs in the different genetically defined subgroups of the studied AML cohort.
Results
GPCRs expression in human AML and normal CD34-positive cells
Using RNA-Seq, we have evaluated the expression of GPCRs in 148 AML samples and compared it with that observed in normal cord blood-derived CD34+CD45RA− hematopoietic stem and progenitor cells (hereafter called CD34+ cells) and in normal bone marrow and peripheral blood cell populations. The 772 GPCRs analyzed in this study comprise all GPCRs included in the IUPHAR database, as well as 370 olfactory, 24 taste and 4 vomeronasal receptors. Information about the receptor subfamilies and the 18 subgroups based on their ligands is provided in Supplementary Tables 4, 5 and 6. Overall, 240 GPCRs were expressed at ⩾1 lRPKM (used as an arbitrary threshold) in at least one AML sample (Supplementary Table 5). Expression was above 3.5 lRPKM (highly expressed) for 111 and above 6.7 lRPKM (very highly expressed) for 19 receptors.
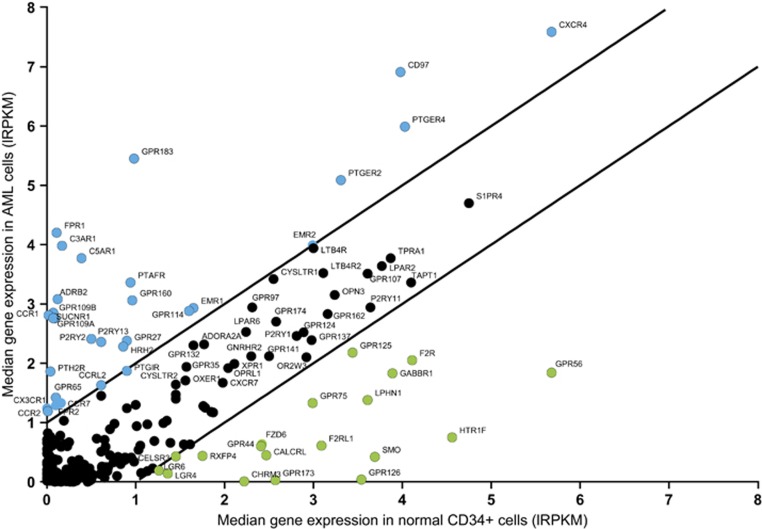
We first ranked the various GPCRs according to their median expression level from highest to lowest in AML (Figure 1) and in CD34+ cells (Supplementary Figure 2). Using the threshold of 3.5 lRPKM, the most highly expressed GPCRs in AML cells are in decreasing order: CXCR4, CD97, PTGER4, GPR183, PTGER2, S1PR4, FPR1, EMR2, C3AR1, LTB4R, TPRA1, C5AR1, LPAR2, LTB4R2 and GPR107. This contrasts with the expression profiles observed in normal CD34+ cells where the rank order of expression is: GPR56, CXCR4, S1PR4, HTR1F, F2R, TAPT1, PTGER4, CD97, GABBR1, TPRA1, LPAR2, SMO, P2RY11, LPHN1, GPR107 and GPR126. In addition to the different order of expression levels observed among highly expressed GPCRs, some receptors that are not found in the most highly expressed ones in normal CD34+ cells are clearly overexpressed in AML (Figure 1).
Figure 1.
Identification of overexpressed GPCRs in AML. RNA-Seq was used to determine expression levels of 772 GPCRs in 148 AML samples of the Leucegene cohort and 12 samples of normal cord blood-derived CD34+ CD45RA− cells. The 50 GPCRs with the highest median expression levels in AML samples are presented in the heatmap. The 15 GPCRs that have a higher expression level as defined in Supplementary Figure 1A are in bold. NK, normal karyotype; Int.abn., Intermediate abnormal karyotype; MLL+, AML with MLL translocations; EVI1+, AML with EVI1 rearrangements; Complex, AML with three or more unrelated clonal chromosomal abnormalities. Boxes in green, blue or gray represent one sample with NUP98-NSD1 fusion, 17p deletion or an insufficient number of metaphases respectively. RNA-Seq data were transformed to lRPKM (log2(RPKM+1)).
Real-time quantitative RT-PCR studies confirmed RNA-Seq results for 10 selected GPCRs and revealed a robust correlation between both methods (Supplementary Figure 3 and Supplementary Table 7). We also confirmed high protein expression levels of selected GPCRs for which validated antibodies were available, using flow cytometry as shown in Supplementary Figures 4 and 5. With the exception of CD97, which is uniformly highly expressed in all samples tested, GPCR expression was distributed unequally within each patient sample highlighting the possibility of defining AML subpopulations with these protein markers (Supplementary Figure 5).
Deregulated GPCRs in AML belong to specific receptor subfamilies
When compared with normal cord blood-derived CD34+ cells, 30 GPCRs are upregulated in AML specimens (blue dots in Figure 2). The most highly expressed GPCRs in AML are CXCR4, CD97, PTGER4, PTGER2, EMR2, GPR183, FPR1, C3AR1 and C5AR1. Except for FPR1 and C5AR1, these GPCRs show little inter-specimen variability (Figure 1 and Supplementary Table 5). Likewise, 19 GPCRs are less expressed in AML cells (green dots in Figure 2). GPR56, HTR1F, SMO and GPR126 are most discriminatory of normal CD34+ cells. Most GPCRs are equally expressed in AML and normal CD34+ cells (black dots in Figure 2).
Figure 2.
Relation between GPCR expression in AML and in normal cord blood-derived CD34+ cells. The median gene expression level of 772 GPCRs in AML cells (y axis) is represented against their expression in normal cord blood-derived CD34+ cells (x axis). The 30 upregulated GPCRs in AML (blue dots) have a difference in median expression level between AML and normal CD34+ cells greater than 1. The 19 downregulated GPCRs in AML (green dots) have a difference in median expression less than −1. GPCRs represented in black dots are not differentially expressed between AML and normal CD34+ cells. RNA-Seq data were transformed to lRPKM (log2(RPKM+1)).
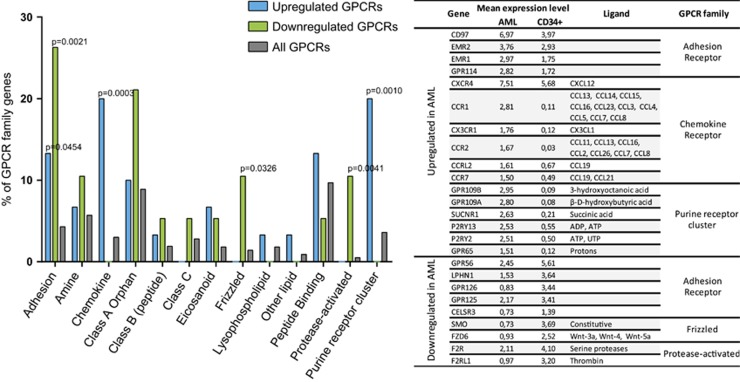
Class enrichment analyses showed that both AML upregulated and downregulated GPCRs are enriched in adhesion GPCRs compared with their representation in the genome indicating that this subfamily of receptors is highly deregulated in AML compared with the overall GPCR family. Chemokine and purine receptors were overrepresented in AML upregulated genes, whereas protease-activated GPCRs and Frizzled family members were overrepresented among the AML downregulated transcripts indicating that these subclasses of receptors are disproportionally affected in the disease state (Figure 3).
Figure 3.
GPCR subfamily distribution of upregulated and downregulated GPCRs in AML. The left panel shows the proportion of genes upregulated or downregulated in AML and all GPCRs into different subfamilies of GPCRs (adhesion, amine, chemokine and so on). The P-values are indicated for significant families by Fisher's exact tests. The right panel shows individual GPCRs of enriched groups. Values indicated in second and third columns correspond to the receptor mean expression level in AML and CD34+ cells. All represented GPCRs have mean expression levels in AML significantly different from their mean in normal CD34+ cells, P-values<0.005. The Fisher's exact test was performed between the upregulated or downregulated group and remaining GPCRs, i.e., all GPCRs excluding differentially expressed genes.
GPCRs are differentially expressed in distinct AML genetic subgroups
We next studied GPCR expression levels in relation to the most frequent AML genetic subgroups represented in this cohort, that are AML with t(8;21)(q22;q22), inv(16)(p13.1q22) or MLL translocations, and normal karyotype AML with NPM1, DNMT3A or FLT3-ITD mutations (Supplementary Table 2).
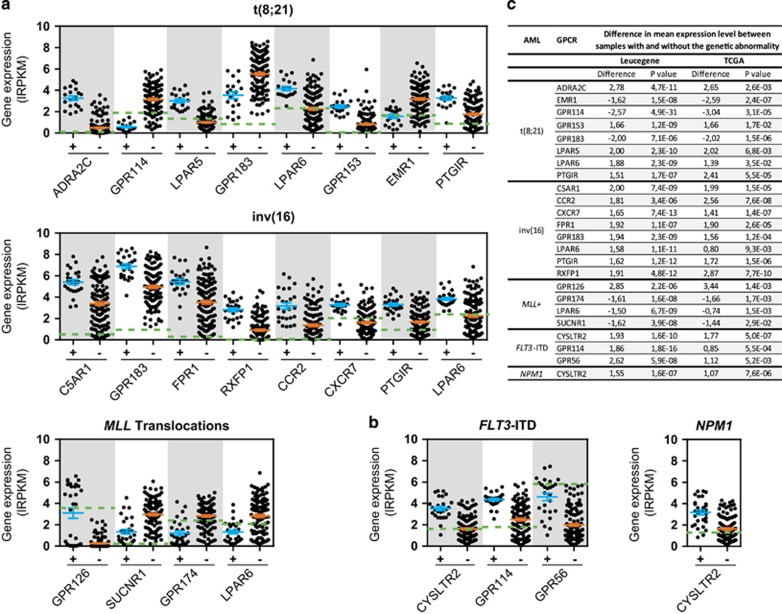
A GPCR expression fingerprint was observed for AML samples with t(8;21), inv(16) and MLL translocations (Figure 4a and Supplementary Figure 6A). For example, eight GPCRs were specifically upregulated or downregulated in the AML subgroup with the t(8;21) translocation. These included the adrenergic receptor ADRA2C, the orphan receptor GPR153 and the lipid receptors LPAR5, LPAR6 and PTGIR (all upregulated) as well as the adhesion GPCRs, EMR1 and GPR114 and the oxysterol-binding receptor, GPR183 (downregulated). Overexpression of eight other GPCRs occurs in the inv(16) AML subgroup. These are C5AR1, CCR2, CXCR7/ACKR3, FPR1, GPR183, RXFP1, PTGIR and LPAR6 (Figure 4a). AML with MLL translocations were associated with an upregulation of GPR126 and a downregulation of GPR174, SUCNR1 and LPAR6 (Figure 4a, bottom panel). In addition, GPR126 expression differed between subtypes of MLL rearranged leukemias according to the translocation partners, being overexpressed at a high level in AML samples with the MLL-MLLT4, MLL-ELL and MLL-SEPT9 fusions and not expressed in the majority of AML samples with the MLL-MLLT3 and MLL-MLLT1 fusions (Supplementary Figure 7). FLT3-ITD mutated samples revealed an upregulation of CYSLTR2, also observed in NPM1-mutated samples, and of the adhesion GPCRs, GPR114 and GPR56 (Figure 4b and Supplementary Figure 6B). These results were validated in an independent AML dataset of 160 samples available from The Cancer Genome Atlas (TCGA) project which comprised 7 samples with t(8;21), 12 with inv(16), 11 with MLL translocations and normal karyotype AML with FLT3-ITD (n=22), NPM1 (n=43) or DNMT3A mutations (n=30) (Figure 4c). DNMT3A-mutated samples did not reveal any significant GPCR expression fingerprint when analyzed in the TCGA cohort.
Figure 4.
GPCR expression level analysis in AML of different genetic subgroups. Expression of deregulated GPCRs in AML samples with (a) t(8;21), inv(16) and MLL translocations and (b) normal karyotype with FLT3-ITD or NPM1 mutations. Differentially expressed GPCRs are defined as having a difference in mean expression higher or equal to 1.5 lRPKM between samples with (+) and without (−) the genetic abnormality and a significant Student's t test (P<0.05). Only GPCRs that were validated in the TCGA dataset are shown. Data are expressed as individual sample expression value and means ±1 s.e.m. for all samples. Mean expression of GPCRs in normal CD34+ cells is illustrated with a green dashed line. (c) List of GPCRs with a significant difference in mean expression level in AML samples of representative genetic subgroups in Leucegene and TCGA cohorts. RNA-Seq data were transformed to lRPKM (log2(RPKM+1)).
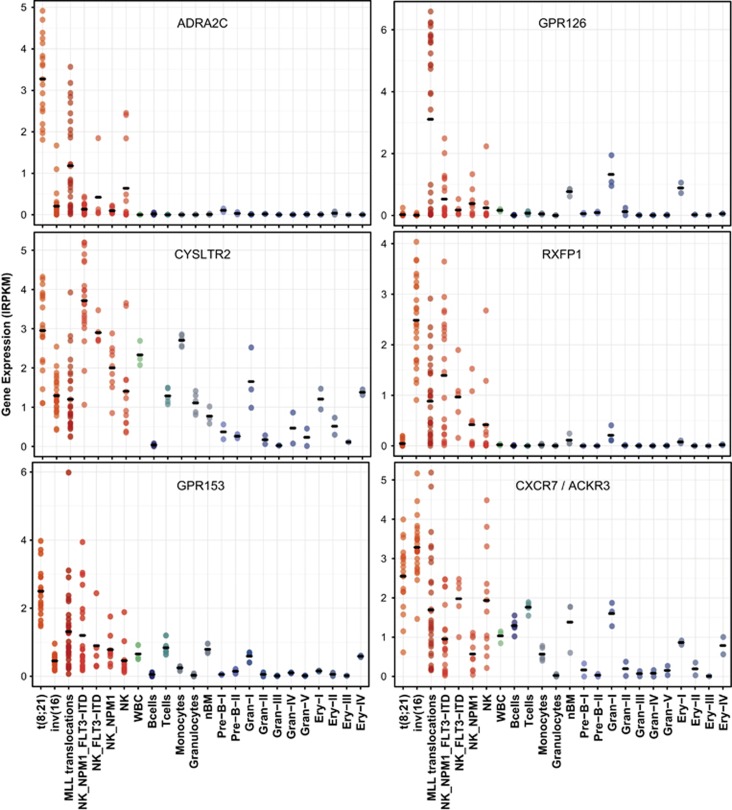
Ideal therapeutic targets should be expressed on leukemic cells but not on normal bone marrow and blood hematopoietic cells. Accordingly, we analyzed the genes upregulated in genetic subgroups by comparing their expression in AML cells with expression levels in normal mature blood cells and bone marrow erythroid, myeloid and B-cell precursors. Interestingly, ADRA2C, GPR126, CYSLTR2, RXFP1, GPR153 and CXCR7/ACKR3 maintained their significant overexpression in specific AML genetic subgroups when compared with normal cell populations (Figure 5 and Supplementary Table 9).
Figure 5.
Expression levels of GPCRs overexpressed in specific AML genetic subgroups in AML, normal blood and bone marrow cells. Expression levels of GPCRs previously identified as overexpressed in specific genetic subgroups of AML are compared with their expression in normal blood and bone marrow cell populations. Represented GPCRs, i.e., ADRA2C and GPR153 in AML with t(8;21), GPR126 in AML with MLL translocations, CYSLTR2 in normal karyotype AML with NPM1 or FLT3-ITD mutations, and RXFP1 and CXCR7/ACKR3 in AML with inv(16), show a significant difference in mean expression between the specific AML genetic subgroup and each normal cell population identified (Student's t test (P<0.05)). AML samples are represented by red dots and normal cell samples by green, blue or gray dots. Data are expressed as individual sample expression value and means for all samples. Normal blood and bone marrow cell populations were prepared as described in the Materials and methods section. RNA-Seq data were transformed to lRPKM (log2(RPKM+1)). NK, normal karyotype; WBC, white blood cells; nBM, normal bone marrow.
Discussion
Our results revealed that 30 GPCRs are overexpressed in AML samples compared with normal CD34+ cells. These receptors are enriched in the chemokine (CCR1, CXCR4, CCR2, CX3CR1, CCR7 and CCRL2), adhesion (CD97, EMR1, EMR2 and GPR114) and purine (including P2RY2 and P2RY13) receptor subfamilies. This list includes GPCRs previously described as important for AML cell biology, such as CXCR4,24 as well as GPCRs that have a role in hematopoietic stem cell engraftment, such as C3AR1 and PTGER2.25, 26 In addition, 19 receptors are downregulated in AML cells including adhesion GPCRs like LPHN1, GPR125, GPR56, CELSR3 and GPR126, protease-activated receptors (F2R and F2RL1) and the Frizzled family members SMO and FZD6.
Among these deregulated GPCRs, prostaglandin receptors are of particular interest. Indeed, both PTGER4 and PTGER2 are among the most highly expressed GPCRs differentiating AML from normal CD34+ cells. Preliminary results have shown that PGE2 can increase cyclic AMP production by human leukemic cells through EP2 (encoded by PTGER2) but not EP4 receptors. Further functional studies are needed to confirm these observations.27 Interestingly, highly selective EP2 antagonists (TG4-155 and TG6-10-1) have recently been developed and might therefore be available for pre-clinical studies.28 It is worth noting that different studies have highlighted the importance of PGE2, acting through four GPCRs, PTGER1–4, in the progression of many cancers including colorectal, gastric, lung or breast cancers.29
Chemokine receptors found to be overexpressed in AML specimens such as CXCR4, CCR7, CCR1, CCR2, CX3CR1 and CCRL2 are also of potential interest. Except for CXCR4,24 their role in AML has never been invoked. In other hematological cancers, CX3CR1 and its ligand, CX3CL1, have been proposed to be involved in the interaction between chronic lymphocytic leukemia cells and their microenvironment,30 and CCR1 has a crucial role in the pathogenesis of myeloma-associated bone disease. Interestingly, CCX721, a selective CCR1 inhibitor, improves osteolytic bone lesions in a preclinical mouse model of this disease.31 This compound is analogous to CCX354-C, an oral CCR1 antagonist that has been evaluated in clinical trial for human rheumatoid arthritis.32
The purine receptor family members overexpressed in AML include GPR109A, GPR109B, SUCNR1, P2RY2, P2RY13 and GPR65. To our knowledge, none of these receptors have been involved previously in hematologic malignancies. However, GPR109A is silenced in colon cancer and primary breast tumor tissues where it acts as a tumor suppressor.33 In contrast to these findings, the receptor is overexpressed in squamous cell cancers compared with normal keratinocytes.34 P2RY2 mediates prostate cancer cell migration and metastasis,35 and its activation leads to increased proliferation of melanoma36 and lung tumor cells.37 This observation is context-dependent because P2RY2 activation induces apoptosis in human colorectal carcinoma cell lines.38 The pH-sensing receptor GPR65 has also been reported to be overexpressed in a significant proportion of kidney, ovarian, colon and breast tumors.39 Recent findings have also highlighted that a member of this GPCR family, P2Y14, is critical for stress hematopoiesis.40
Adhesion GPCRs are a family of receptors characterized by a long extracellular domain and an autoproteolytic site leading to an extracellular N-terminal fragment and a transmembrane C-terminal fragment that remain noncovalently associated after cleavage. These receptors have been shown to have a crucial role in development, cell migration and are increasingly being invoked as deregulated in numerous cancers.41 They represent another group of GPCRs for which we observed a strong expression in AML. Notably, CD97 is expressed at high levels in 100% of AML studied to date at a much higher level than found in normal CD34+ cells. CD97 is upregulated in a variety of other malignancies, including glioblastoma,42 digestive and thyroid cancers.43 GPR56 and GPR114 are specifically overexpressed in FLT3-ITD-mutated samples and GPR126 is most specific to AML with MLL translocations. Notably, GPR56 has recently been identified as a leukemia stem cell marker, and its expression correlates with poor prognosis in AML.44, 45
Two protease-activated GPCRs F2R or PAR1 and F2RL1 or PAR2 are downregulated in AML. PAR1 has a well-established role in thrombosis, hemostasis and inflammatory diseases. It is also involved in promoting growth and in the angiogenesis and metastasis processes of several malignancies.4 Unlike AML samples, most solid tumors show an upregulation of PAR1 expression. Our findings are in accordance with the results of a recent study that demonstrated that PAR1 expression is downregulated in primary AML cells.46 Furthermore, Par1 homozygous null mutant mice develop leukemia with a shorter latency than control animals in secondary transplantation experiments.46 PAR2 is involved in migration and/or proliferation of many cancer cells, including breast,47 pancreatic48 or colon49 cancer and seems to have a role in tumor angiogenesis through vascular endothelial growth factor production in cancer cells.50
Two frizzled class receptors SMO and FZD6 are also downregulated in all AML samples except AML with EVI1 rearrangements, which have a FZD6 median expression close to CD34+ cells. SMO and Patched are two receptors mediating Hedgehog signaling. This pathway is aberrantly activated in many solid tumors including basal cell carcinoma.51 This feature has been exploited for the development of Vismodegib, an antagonist of SMO.13 However, in contrast to several other cancers, SMO expression in AML cells is five times lower than in CD34+ cells in our study. Vismodegib is currently being investigated in a phase II clinical trial (www.clinicaltrials.gov, NCT02073838) in AML patients in combination with ribavirin, because it could potentially overcome the resistance of leukemic cells to ribavirin.52
It is tempting to speculate that the GPCR members that are differently expressed in frequent AML genetic subgroups and more specifically expressed in AML cells compared with normal hematopoietic cells could be exploited for the development of novel therapeutic approaches. In this context, ADRA2C, which is upregulated in AML with t(8;21), could be an interesting target to explore, because many agonists and antagonists of this receptor are available, including several FDA-approved drugs such as the antihypertensive drug clonidine and the antidepressant Mirtazapine. CYSLTR2, a receptor for the inflammatory mediators cysteinyl leukotrienes, is also overexpressed in FLT3-ITD- and NPM1-mutated samples. In aggressive breast tumors, this gene has been described as an independent prognostic factor in combination with CYSLTR1.53 Selective agonist (NMLTC4)54 and antagonists, such as HAMI3379(ref. 55) or BayCysLT2,56 have been developed for CYSLTR2. Other interesting targets are the GPCR members that are overexpressed in AML with inv(16) or AML M4 (Supplementary Table 8). For example, selective antagonists of the chemokine receptor CCR2, such as CCX140-B, studied in diabetic mice and tested in clinical trials for patients with diabetic nephropathy (www.clinicaltrials.gov, NCT01447147 and NCT01440257) might be explored for anti-leukemic activity.57
In conclusion, we provide the first comprehensive transcriptome analysis of GPCRs in AML, which reveals that these surface receptors are potential novel therapeutic targets in AML. Using available agonists/antagonists to leukemia-enriched GPCRs, specific drugs can now be tested in preclinical models and, when active, in clinical trials for AML therapeutics.
Acknowledgments
We acknowledge Muriel Draoui for project coordination, Geneviève Boucher and Patrick Gendron from IRIC bioinformatics platform for data processing, the team of the Banque de cellules leucémiques du Québec (BCLQ), who provided and characterized all AML samples included in the Leucegene cohort, Pierre Chagnon and Marianne Arteau at the IRIC genomics platform for RNA sequencing, Christian Le Gouill and Mireille Hogue for providing cDNA for LTB4R and CXCR4 and Monique Lagacé for revision of the manuscript. This work was supported by the Government of Canada through Genome Canada and the Ministère de l'enseignement supérieur, de la recherche, de la science et de la technologie du Québec through Génome Québec to GS, JH and SL, and from the Canadian Institute for Health Research to MB. GS and JH are recipients of research chairs from the Canada Research Chair program and Industrielle-Alliance (Université de Montréal) respectively. MB holds a Canada research chair in Signal Transduction and Molecular Pharmacology. The Banque de cellules leucémiques du Québec (BCLQ) is supported by grants from the Cancer Research Network of the Fonds de recherche du Québec-Santé (FRQS). RNA-Seq read mapping and transcript quantification were performed on the supercomputer Briaree from Université de Montréal, managed by Calcul Québec and Compute Canada. The operation of this supercomputer is funded by the Canada Foundation for Innovation (CFI), NanoQuébec, Réseau de médecine génétique appliquée (RMGA) and the Fonds de recherche du Québec - Nature et technologies (FRQ-NT). AM, CP and VPL were supported by a fellowship from the Cole Foundation. CP was also supported by a post-doctoral fellowship from the German Cancer Aid. Accession codes: GSE49642, GSE52656, GSE62190, GSE66917 and GSE67039.
Author contributions
AM analyzed GPCR expression profile and performed flow cytometry and quantitative RT-PCR experiments, generated figures, tables and supplementary material and co-wrote the paper; SL was responsible for supervision of the bioinformatics team and of statistical analyses; CP performed sorting and sequencing of normal bone marrow and peripheral blood cell populations, contributed to flow cytometry experiments and edited the manuscript; VPL analyzed mutations of all AML samples and revised the manuscript; MB contributed to project conception, provided expertise in the field of GPCRs and edited the manuscript; GS contributed to project conception and edited the manuscript; JH contributed to project conception, analyzed the cytogenetic studies, provided all AML samples and co-wrote the paper.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Blood Cancer Journal website (http://www.nature.com/bcj)
Supplementary Material
References
- Wisler JW, Xiao K, Thomsen AR, Lefkowitz RJ. Recent developments in biased agonism. Curr Opin Cell Biol 2014; 27: 18–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier M. Unraveling the structural basis of GPCR activation and inactivation. Nat Struct Mol Biol 2013; 20: 539–541. [DOI] [PubMed] [Google Scholar]
- O'Hayre M, Degese MS, Gutkind JS. Novel insights into G protein and G protein-coupled receptor signaling in cancer. Curr Opin Cell Biol 2014; 27: 126–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsam RT, Gutkind JS. G-protein-coupled receptors and cancer. Nat Rev Cancer 2007; 7: 79–94. [DOI] [PubMed] [Google Scholar]
- Even-Ram S, Uziely B, Cohen P, Grisaru-Granovsky S, Maoz M, Ginzburg Y et al. Thrombin receptor overexpression in malignant and physiological invasion processes. Nat Med 1998; 4: 909–914. [DOI] [PubMed] [Google Scholar]
- Kaushal V, Kohli M, Dennis RA, Siegel ER, Chiles WW, Mukunyadzi P. Thrombin receptor expression is upregulated in prostate cancer. Prostate 2006; 66: 273–282. [DOI] [PubMed] [Google Scholar]
- Daaka Y. G proteins in cancer: the prostate cancer paradigm. Sci STKE 2004; 2004: re2. [DOI] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007; 449: 1003–1007. [DOI] [PubMed] [Google Scholar]
- Schepers AG, Snippert HJ, Stange DE, van den Born M, van Es JH, van de Wetering M et al. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science 2012; 337: 730–735. [DOI] [PubMed] [Google Scholar]
- Merlos-Suarez A, Barriga FM, Jung P, Iglesias M, Cespedes MV, Rossell D et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 2011; 8: 511–524. [DOI] [PubMed] [Google Scholar]
- Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov 2006; 5: 993–996. [DOI] [PubMed] [Google Scholar]
- Innamorati G. Molecular approaches to target GPCRs in cancer therapy. Pharmaceuticals 2011; 4: 567–589. [Google Scholar]
- Axelson M, Liu K, Jiang X, He K, Wang J, Zhao H et al. U.S. Food and Drug Administration approval: vismodegib for recurrent, locally advanced, or metastatic basal cell carcinoma. Clin Cancer Res 2013; 19: 2289–2293. [DOI] [PubMed] [Google Scholar]
- Rombouts EJ, Pavic B, Lowenberg B, Ploemacher RE. Relation between CXCR-4 expression, Flt3 mutations, and unfavorable prognosis of adult acute myeloid leukemia. Blood 2004; 104: 550–557. [DOI] [PubMed] [Google Scholar]
- Spoo AC, Lubbert M, Wierda WG, Burger JA. CXCR4 is a prognostic marker in acute myelogenous leukemia. Blood 2007; 109: 786–791. [DOI] [PubMed] [Google Scholar]
- Konoplev S, Rassidakis GZ, Estey E, Kantarjian H, Liakou CI, Huang X et al. Overexpression of CXCR4 predicts adverse overall and event-free survival in patients with unmutated FLT3 acute myeloid leukemia with normal karyotype. Cancer 2007; 109: 1152–1156. [DOI] [PubMed] [Google Scholar]
- Zeng Z, Shi YX, Samudio IJ, Wang RY, Ling X, Frolova O et al. Targeting the leukemia microenvironment by CXCR4 inhibition overcomes resistance to kinase inhibitors and chemotherapy in AML. Blood 2009; 113: 6215–6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uy GL, Rettig MP, Motabi IH, McFarland K, Trinkaus KM, Hladnik LM et al. A phase 1/2 study of chemosensitization with the CXCR4 antagonist plerixafor in relapsed or refractory acute myeloid leukemia. Blood 2012; 119: 3917–3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm BT, Briau M, Austin P, Faubert A, Boucher G, Chagnon P et al. RNA-seq analysis of 2 closely related leukemia clones that differ in their self-renewal capacity. Blood 2011; 117: e27–e38. [DOI] [PubMed] [Google Scholar]
- Macrae T, Sargeant T, Lemieux S, Hebert J, Deneault E, Sauvageau G. RNA-Seq reveals spliceosome and proteasome genes as most consistent transcripts in human cancer cells. PloS One 2013; 8: e72884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013; 368: 2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavallee VP, Gendron P, Lemieux S, D'Angelo G, Hebert J, Sauvageau G. EVI1-rearranged acute myeloid leukemias are characterized by distinct molecular alterations. Blood 2015; 125: 140–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol 2003; 63: 1256–1272. [DOI] [PubMed] [Google Scholar]
- Peled A, Tavor S. Role of CXCR4 in the pathogenesis of acute myeloid leukemia. Theranostics 2013; 3: 34–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysoczynski M, Reca R, Lee H, Wu W, Ratajczak J, Ratajczak MZ. Defective engraftment of C3aR-/- hematopoietic stem progenitor cells shows a novel role of the C3a-C3aR axis in bone marrow homing. Leukemia 2009; 23: 1455–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggatt J, Singh P, Sampath J, Pelus LM. Prostaglandin E2 enhances hematopoietic stem cell homing, survival, and proliferation. Blood 2009; 113: 5444–5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denizot Y, Donnard M, Truffinet V, Malissein E, Faucher JL, Turlure P et al. Functional EP2 receptors on blast cells of patients with acute leukemia. Int J Cancer J 2005; 115: 499–501. [DOI] [PubMed] [Google Scholar]
- Jiang J, Quan Y, Ganesh T, Pouliot WA, Dudek FE, Dingledine R. Inhibition of the prostaglandin receptor EP2 following status epilepticus reduces delayed mortality and brain inflammation. Proc Natl Acad Sci USA 2013; 110: 3591–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Dubois RN. Eicosanoids and cancer. Nat Rev Cancer 2010; 10: 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti E, Bertolotto M, Deaglio S, Tripodo C, Ribatti D, Audrito V et al. A novel role of the CX3CR1/CX3CL1 system in the cross-talk between chronic lymphocytic leukemia cells and tumor microenvironment. Leukemia 2011; 25: 1268–1277. [DOI] [PubMed] [Google Scholar]
- Dairaghi DJ, Oyajobi BO, Gupta A, McCluskey B, Miao S, Powers JP et al. CCR1 blockade reduces tumor burden and osteolysis in vivo in a mouse model of myeloma bone disease. Blood 2012; 120: 1449–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tak PP, Balanescu A, Tseluyko V, Bojin S, Drescher E, Dairaghi D et al. Chemokine receptor CCR1 antagonist CCX354-C treatment for rheumatoid arthritis: CARAT-2, a randomised, placebo controlled clinical trial. Ann Rheum Dis 2013; 72: 337–344. [DOI] [PubMed] [Google Scholar]
- Elangovan S, Pathania R, Ramachandran S, Ananth S, Padia RN, Lan L et al. The niacin/butyrate receptor GPR109A suppresses mammary tumorigenesis by inhibiting cell survival. Cancer Res 2014; 74: 1166–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez Y, Benavente CA, Meyer RG, Coyle WR, Jacobson MK, Jacobson EL. Nicotinic acid receptor abnormalities in human skin cancer: implications for a role in epidermal differentiation. PloS One 2011; 6: e20487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WH, Qiu Y, Zhang HQ, Liu Y, You JF, Tian XX et al. P2Y2 receptor promotes cell invasion and metastasis in prostate cancer cells. Br J Cancer 2013; 109: 1666–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White N, Ryten M, Clayton E, Butler P, Burnstock G. P2Y purinergic receptors regulate the growth of human melanomas. Cancer Lett 2005; 224: 81–91. [DOI] [PubMed] [Google Scholar]
- Schafer R, Sedehizade F, Welte T, Reiser G. ATP- and UTP-activated P2Y receptors differently regulate proliferation of human lung epithelial tumor cells. Am J Physiol Lung Cell Mol Physiol 2003; 285: L376–L385. [DOI] [PubMed] [Google Scholar]
- Hopfner M, Maaser K, Barthel B, von Lampe B, Hanski C, Riecken EO et al. Growth inhibition and apoptosis induced by P2Y2 receptors in human colorectal carcinoma cells: involvement of intracellular calcium and cyclic adenosine monophosphate. Int J Colorectal Dis 2001; 16: 154–166. [DOI] [PubMed] [Google Scholar]
- Sin WC, Zhang Y, Zhong W, Adhikarakunnathu S, Powers S, Hoey T et al. G protein-coupled receptors GPR4 and TDAG8 are oncogenic and overexpressed in human cancers. Oncogene 2004; 23: 6299–6303. [DOI] [PubMed] [Google Scholar]
- Cho J, Yusuf R, Kook S, Attar E, Lee D, Park B et al. Purinergic P2Y(1)(4) receptor modulates stress-induced hematopoietic stem/progenitor cell senescence. J Clin Invest 2014; 124: 3159–3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann J, Aust G, Arac D, Engel FB, Formstone C, Fredriksson R et al. International Union of Basic and Clinical Pharmacology. XCIV. Adhesion G protein-coupled receptors. Pharmacol Rev 2015; 67: 338–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feve M, Saliou JM, Zeniou M, Lennon S, Carapito C, Dong J et al. Comparative expression study of the endo-G protein coupled receptor (GPCR) repertoire in human glioblastoma cancer stem-like cells, U87-MG cells and non malignant cells of neural origin unveils new potential therapeutic targets. PloS One 2014; 9: e91519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward Y, Lake R, Martin PL, Killian K, Salerno P, Wang T et al. CD97 amplifies LPA receptor signaling and promotes thyroid cancer progression in a mouse model. Oncogene 2013; 32: 2726–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppert K, Takenaka K, Lechman ER, Waldron L, Nilsson B, van Galen P et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med 2011; 17: 1086–1093. [DOI] [PubMed] [Google Scholar]
- Pabst C, Bergeron A, Lavallee VP, Yeh J, Gendron P, Norddahl GL et al. GPR56 identifies primary human acute myeloid leukemia cells with high repopulating potential in vivo. Blood 2016; 127: 2018–2027. [DOI] [PubMed] [Google Scholar]
- Baumer N, Krause A, Kohler G, Lettermann S, Evers G, Hascher A et al. Proteinase-Activated Receptor 1 (PAR1) Regulates Leukemic Stem Cell Functions. PloS One 2014; 9: e94993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, Shenoy SK, Lefkowitz RJ, DeFea K. Constitutive protease-activated receptor-2-mediated migration of MDA MB-231 breast cancer cells requires both beta-arrestin-1 and -2. J Biol Chem 2004; 279: 55419–55424. [DOI] [PubMed] [Google Scholar]
- Shi K, Queiroz KC, Stap J, Richel DJ, Spek CA. Protease-activated receptor-2 induces migration of pancreatic cancer cells in an extracellular ATP-dependent manner. J Thromb Haemost 2013; 11: 1892–1902. [DOI] [PubMed] [Google Scholar]
- Zhou B, Zhou H, Ling S, Guo D, Yan Y, Zhou F et al. Activation of PAR2 or/and TLR4 promotes SW620 cell proliferation and migration via phosphorylation of ERK1/2. Oncol Rep 2011; 25: 503–511. [DOI] [PubMed] [Google Scholar]
- Dutra-Oliveira A, Monteiro RQ, Mariano-Oliveira A. Protease-activated receptor-2 (PAR2) mediates VEGF production through the ERK1/2 pathway in human glioblastoma cell lines. Biochem Biophys Res Commun 2012; 421: 221–227. [DOI] [PubMed] [Google Scholar]
- Ruch JM, Kim EJ. Hedgehog signaling pathway and cancer therapeutics: progress to date. Drugs 2013; 73: 613–623. [DOI] [PubMed] [Google Scholar]
- Zahreddine HA, Culjkovic-Kraljacic B, Assouline S, Gendron P, Romeo AA, Morris SJ et al. The sonic hedgehog factor GLI1 imparts drug resistance through inducible glucuronidation. Nature 2014; 511: 90–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson C, Liu J, Ehrnstrom R, Manjer J, Jirstrom K, Andersson T et al. Cysteinyl leukotriene receptor expression pattern affects migration of breast cancer cells and survival of breast cancer patients. Int J Cancer 2011; 129: 9–22. [DOI] [PubMed] [Google Scholar]
- Yan D, Stocco R, Sawyer N, Nesheim ME, Abramovitz M, Funk CD. Differential signaling of cysteinyl leukotrienes and a novel cysteinyl leukotriene receptor 2 (CysLT(2)) agonist, N-methyl-leukotriene C(4), in calcium reporter and beta arrestin assays. Mol Pharmacol 2011; 79: 270–278. [DOI] [PubMed] [Google Scholar]
- Wunder F, Tinel H, Kast R, Geerts A, Becker EM, Kolkhof P et al. Pharmacological characterization of the first potent and selective antagonist at the cysteinyl leukotriene 2 (CysLT(2)) receptor. Br J Pharmacol 2010; 160: 399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnini C, Accomazzo MR, Borroni E, Vitellaro-Zuccarello L, Durand T, Folco G et al. Synthesis of cysteinyl leukotrienes in human endothelial cells: subcellular localization and autocrine signaling through the CysLT2 receptor. FASEB J 2011; 25: 3519–3528. [DOI] [PubMed] [Google Scholar]
- Sullivan T, Miao Z, Dairaghi DJ, Krasinski A, Wang Y, Zhao BN et al. CCR2 antagonist CCX140-B provides renal and glycemic benefits in diabetic transgenic human CCR2 knockin mice. Am J Physiol Renal Physiol 2013; 305: F1288–F1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.