Abstract
Major intrinsic proteins (MIPs) are a family of membrane channels that facilitate the bidirectional transport of water and small uncharged solutes such as glycerol. The 35 full-length members of the MIP family in Arabidopsis are segregated into four structurally homologous subfamilies: plasma membrane intrinsic proteins (PIPs), tonoplast intrinsic proteins (TIPs), nodulin 26-like intrinsic membrane proteins (NIPs), and small basic intrinsic proteins (SIPs). Computational methods were used to construct structural models of the putative pore regions of various plant MIPs based on homology modeling with the atomic resolution crystal structures of mammalian aquaporin 1 and the bacterial glycerol permease GlpF. Based on comparisons of the narrow selectivity filter regions (the aromatic/Arg [ar/R] filter), the members of the four phylogenetic subfamilies of Arabidopsis MIPs can be classified into eight groups. PIPs possess a uniform ar/R signature characteristic of high water transport aquaporins, whereas TIPs are highly diverse with three separate conserved ar/R regions. NIPs possess two separate conserved ar/R regions, one that is similar to the archetype, soybean (Glycine max) nodulin 26, and another that is characteristic of Arabidopsis NIP6;1. The SIP subfamily possesses two ar/R subgroups, characteristic of either SIP1 or SIP2. Both SIP ar/R residues are divergent from all other MIPs in plants and other kingdoms. Overall, these findings suggest that higher plant MIPs have a common fold but show distinct differences in proposed pore apertures, potential to form hydrogen bonds with transported molecules, and amphiphilicity that likely results in divergent transport selectivities.
Members of the major intrinsic protein (MIP) family form a large and diverse group of membrane proteins that facilitate the bidirectional transport of water and some small solutes across cellular membranes (Agre et al., 2002; Thomas et al., 2002). MIPs are widely distributed in organisms from bacteria to higher eukaryotes, and these proteins are especially abundant in plants, with 35 full-length genes present in Arabidopsis (Johanson et al., 2001; Quigley et al., 2001). Members of the plant MIP family have been implicated in cell elongation and development, changes in hydraulic conductivity in response to environmental cues, and numerous other processes that require rapid transmembrane movements of water (for review, see Johansson et al., 2000; Maurel et al., 2002; Tyerman et al., 2002).
MIP family members in Arabidopsis are subdivided into four subfamilies: the plasma membrane intrinsic proteins (PIPs; 13 genes), the tonoplast intrinsic membrane proteins (TIPs; 10 genes), the nodulin 26-like intrinsic membrane proteins (NIPs; 9 genes), and the small basic intrinsic proteins (SIPs; 3 genes) (Weig et al., 1997; Johanson et al., 2001; Quigley et al., 2001). By contrast, mammals have only two functional subfamilies of MIPs: water-specific aquaporins and solute-transporting glyceroporins and aquaglyceroporins (Agre et al., 2002). This disparity leads to the question of whether the greater structural diversity of plant MIPs results in distinct transport properties that reflect their biological roles in solute transport and water relations during development and stress adaptation.
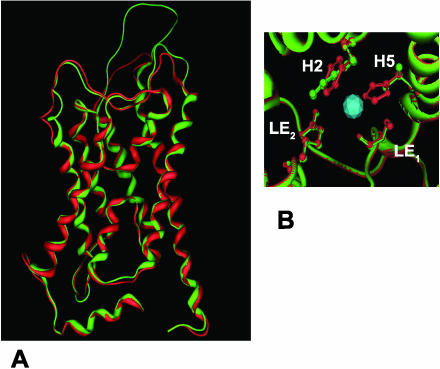
The structures of aquaporin 1 (AQP1), a mammalian water-specific aquaporin, and the Escherichia coli glyceroporin GlpF have been solved at atomic resolution by x-ray crystallography (Fu et al., 2000; Sui et al., 2001). The structures show that the overall topologies of these two archetypical MIPs are remarkably similar, which supports the hypothesis that MIP family members share a common ancestor (Reizer et al., 1993) and adopt a common hourglass fold (Jung et al., 1994). The hourglass MIP fold is characterized by six membrane-spanning α-helices that are arranged in a right-handed bundle and five interhelical loop regions (A–E) that form the extracellular and cytoplasmic vestibules (Fig. 1A). Loops B and E contain the highly conserved NPA (Asn-Pro-Ala) motifs and form two half-helices that meet at the center of the pore to create one of the two major constrictions in the channel, the NPA region (Fig. 1, B and C). A second narrower constriction, the aromatic/Arg (ar/R) region, is formed toward the extracellular vestibule, 8 Å above the NPA region. The ar/R region consists of a tetrad formed by two residues from helices 2 (H2) and 5 (H5; Fig. 1A) and two residues from loop E (Fig. 1A, LE1 and LE2). The ar/R region and the NPA constrictions form two hydrophilic nodes within AQP1 and GlpF, with each forming intimate contacts with conductant (either water or glycerol). The ar/R region constitutes the principal selectivity filter of the AQP1 and GlpF channels (Fu et al., 2000; Sui et al., 2001; Thomas et al., 2002).
Figure 1.
Topology of MIPs and architecture of the ar/R selectivity region. A, The general topology conserved in the MIP family is shown. Each NPA half-repeat is color coordinated with transmembrane α-helices 1 to 3 shown and the first NPA half-helix (loop B) shown in blue and yellow, respectively, and transmembrane α-helices 4 to 6 and the second NPA half helix (loop E) shown in red and white, respectively. The relative positions of the four residues on H2, H5, and loop E (LE1 and LE2) that comprise the ar/R tetrad are indicated. B, The assembly of these structural elements into the MIP hourglass fold is shown using the crystal structure of bovine AQP1 (Sui et al., 2001). Three of the four waters traversing the AQP1 pore are shown as space-filling aqua spheres. The extracellular (Ex) and cytosolic (Cyto) sides are indicated, and the position of the lipid bilayer is shown as a solid bar. C, Representation of the ar/R and NPA constrictions in the AQP1 pore showing the disposition of the ar/R tetrad (magenta) and the NPA Asn side chains (white and yellow) relative to waters traversing the pore. D, Comparison of the ar/R selectivity filters of AQP1 and the glyceroporin GlpF (Fu et al., 2000) with conductant (water or glycerol) bound. Residue side chains are colored blue for basic hydrophilic, yellow for hydrophobic, and white for neutral hydrophilic. For glycerol, the hydrocarbon backbone is magenta, and the hydroxyl groups are aqua.
Structural information on plant MIPs is limited, but low-resolution analysis by cryoelectron microscopy supports the contention that they adopt a topology similar to other MIPs (Daniels et al., 1999). In this study, plant MIPs were analyzed by homology modeling utilizing the AQP1 and GlpF templates and were grouped according to similarities within the pore selectivity regions. The results show that plant MIPs can be separated into eight groups based on differences within the ar/R selectivity region of the pore of the model structures. Several structures show considerable divergence from the aquaporin versus glyceroporin paradigm, suggesting a greater functional diversity of the plant MIPs compared to their mammalian and microbial counterparts.
RESULTS
AQP1 and GlpF: Paradigms for MIP Function
In AQP1, the ar/R region is formed by Phe-58 (H2), His-182 (H5), Cys-191 (LE1), and Arg-197 (LE2; Fig. 1). The presence of Phe at H2 and the conserved His at H5 constrains the AQP1 pore to 2.8 Å, permitting the flux of water (diameter 2.4 Å) and the exclusion of bulkier solutes. Single-file passage through the ar/R constriction requires the removal of the solvation shell of the water molecule, a process that is thermodynamically unfavorable. The ar/R region of AQP1 is proposed to overcome this energy barrier by forming three hydrogen bonds to each transported water molecule contributed by His-182 (ε2 nitrogen), Arg-197 (guanido hydrogens), and Cys-191 (backbone carbonyl; Sui et al., 2001; Fig. 1D). Thus, the AQP1 ar/R filter provides as many hydrogen bonds to water as are formed in solution, contributing to a high intrinsic water transport rate (Zeidel et al., 1992).
By contrast, the ar/R tetrad of GlpF is composed of two hydrophobic (Trp-48 [H2] and Phe-200 [LE1]) and two hydrophilic (Gly-191 [H5] and Arg-206 [LE2]) residues (Fu et al., 2000). The substitution of Gly for His at H5 provides a wider pore aperture (3.6 Å) to accommodate the larger glycerol molecule and allows the close approach of the Phe-200 at position LE1 (Fig. 1D). This allows the Phe and Trp residues at LE1 and H2 to form a hydrophobic wedge that interacts with the hydrocarbon backbone of glycerol. The conserved Arg at LE2 simultaneously forms hydrogen bonds with the hydrophilic hydroxyl groups of glycerol (Fig. 1). In addition to providing the amphipathic properties necessary to interact with glycerol, the increase in the hydrophobicity in the GlpF ar/R tetrad forms a barrier to water transport because of a decreased capacity to form compensatory hydrogen bonds (Fu et al., 2000).
A remarkable feature of the AQP1 and GlpF structures is the high similarity of the α-carbon backbone conformation (root mean square [rms] deviation = 2.12 Å; Table I), despite a low overall degree of sequence similarity (22% identity). The conservation of the hourglass MIP fold among proteins with divergent sequences lends the family to structure prediction methods such as threading and homology modeling, with the latter technique able to make predictions within 2 Å of experimental structures (Schoonman et al., 1998). To test the validity of the application of this approach to MIP family proteins, structural alignment and homology modeling of the GlpF primary sequence utilizing the AQP1 structural coordinates (Sui et al., 2001) as a template and the modeling of the AQP1 primary sequence utilizing the GlpF structural coordinates (Fu et al., 2000) as a template were performed.
Table I.
Comparison of GlpF and AQP1 sequences by homology modeling
| rms Deviation
|
|||
|---|---|---|---|
| Structural Elementa | AQP1 versus GlpF Structuresb | AQP1 Structure versus AQP1 Modelc | GlpF1 Structure versus GlpF Modeld |
| Å | |||
| Full α-carbon backbone | 2.12 | 6.27 | 5.30 |
| H2, H5, loop E | 1.29 | 1.28 | 1.59 |
| Pore-forming residues | 1.04 | 1.07 | 2.28 |
Comparisons were made of the full-length α-carbon backbone, as well as H2, H5, and NPA loop E and the residues facing the pore of AQP1 and GlpF based on Fu et al. (2000) and Sui et al. (2001).
rms deviation of superimposed AQP1 (Sui et al., 2001) and GlpF (Fu et al., 2000) atomic structures.
rms deviation of superimposition of AQP1 experimental structure and AQP homology model utilizing the GlpF structure as a modeling template.
rms deviation of superimposition of GlpF experimental structure and GlpF homology model utilizing the AQP1 structure as a modeling template.
The results of these reciprocal modeling experiments are shown in Table I. The α-carbon backbones of the models show a reasonable overall similarity with the experimental structures, with the major differences in structure residing in the loops and other regions that are not found within the pore. These are presumably a result of the fact that GlpF has an extended C-loop sequence characteristic of bacterial glyceroporins that is not found in AQP1 and most MIPs. However, comparison of the pore regions and the structural elements (H2, H5, and loop E) comprising the ar/R region show excellent agreement between model and experimental structures (rms deviation of 1–2 Å; Table I). The rms deviation of the actual ar/R residues of experimental and model structures was less than 1 Å for both GlpF and AQP1.
Homology Modeling Plant MIPs: Nodulin 26 as a Test Case
Nodulin 26 is an aquaglyceroporin with low intrinsic water conductance (Rivers et al., 1997; Dean et al., 1999; Wallace et al., 2002) that is positively regulated by phosphorylation on Ser-262 (Guenther et al., 2003). The nodulin 26 pore is also permeated by some uncharged solutes, such as glycerol, whereas other test solutes (urea and acetamide) are excluded (Rivers et al., 1997). A homology model was produced for nodulin 26 by alignment with the AQP1 and GlpF structures followed by three-dimensional model building and energy minimization. A database of 10 distinct protein models was generated that showed good overall alignment with the α-carbon backbone of the AQP1 and GlpF templates (rms deviation from 0.41–2.5 Å). The calculated phi/psi backbone angles and side-chain rotamers were evaluated to discard structures with high-energy, disallowed bonds, and the best model was selected for analysis.
The α-carbon backbone of the nodulin 26 homology model shows excellent superposition with the AQP1 (0.92 Å) and GlpF (1.69 Å) experimental structures, particularly within the membrane spanning helices and pore-forming loops B and E (Fig. 2A). The ar/R regions are superimposible, but the ar/R tetrad of nodulin 26 reveals significant differences from the AQP1 and GlpF structures (Fig. 2B). The nodulin 26 ar/R region is composed of Trp-77 (H2), Val-197 (H5), Ala-206 (LE1), and Arg-212 (LE2). The Trp at H2 and Arg at LE2 are similar to the glycerol interacting residues of the GlpF ar/R; however, the Val residue at H5 is an unusual substitution compared to the AQP1 and GlpF templates, and an aliphatic side chain is commonly found in this position in NIPs and TIPs. Comparison of the AQP1 and nodulin 26 ar/R regions by modeling (Fig. 3) suggests that the replacement of His (AQP1) with Val at H5 also results in a wider ar/R region that could accommodate the larger glycerol molecule. This is supported by analysis of the proposed pore architecture of the nodulin 26 homology model using the HOLE program (Smart et al., 1993), which shows that the ar/R of both proteins represents the narrowest constriction but that the nodulin 26 pore is wider (Fig. 4).
Figure 2.
Structural analysis of the nodulin 26 homology model. A, Superposition of the experimental AQP1 structure (red) and the nodulin 26 homology model (green) is shown viewed perpendicular to the pore of the protein. B, The superposed positions of the ar/R tetrad for AQP1 (red) and nodulin 26 (green) are shown viewed perpendicular to the plane of the membrane. The positions of the ar/R residues from H2, H5, and loop E (LE1 and LE2) are indicated.
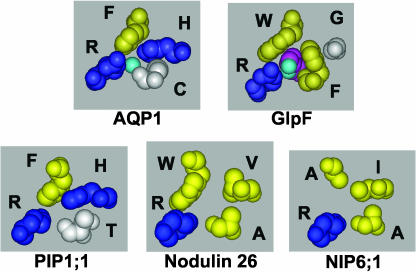
Figure 3.
Comparison of the ar/R selectivity regions of the NIP and PIP subfamilies. Space-filling side-chain residues of the ar/R selectivity region of AtPIP1;1, soybean nodulin 26, and AtNIP6;1 shown compared to AQP1 and GlpF with conductant bound. The single-letter amino acid code appears along side each residue, and residue side chains are colored blue for basic hydrophilic, yellow for hydrophobic, and white for neutral hydrophilic. The ar/R filter in each projection is viewed perpendicular to the plane of the bilayer from the extracellular vestibule.
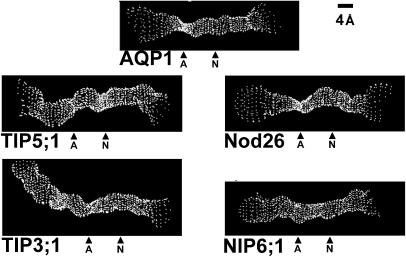
Figure 4.
Analysis of the putative pore dimensions of representative plant MIPs compared with AQP1. The dimensions of the pore regions of AQP1 and the molecular models of nodulin 26, NIP6;1, TIP3:1, and TIP5;1 were analyzed by using the HOLE program (Smart et al., 1993) and were visualized on InsightII. The pores are viewed perpendicular to the pore axis and are labeled with respect to the location of the ar/R (A) and NPA (N) regions.
Modeling suggests that the Val side chain at H5, along with the Ala at LE1, pack against the roof of the pore, such that it presents a hydrophobic surface to complement the Trp at H2 (Fig. 3). The resulting hydrophobic surface could provide a site of interaction with the glycerol hydrocarbon backbone, while Arg LE2 would form hydrogen bonds with the hydroxyl groups, similar to GlpF. Further, the nodulin 26 ar/R would have a reduced ability to form hydrogen bonds with transported water, possibly accounting for a low intrinsic water flux rate observed for the protein (Dean et al., 1999).
ar/R Subgroups of Plant MIPs
Analyses of the 35 MIP genes based on structural alignment show that the ar/R regions of plant MIPs adhere to the higher PIP, TIP, NIP, and SIP divisions (Supplemental Figs. 1–4) but can be further subdivided into eight ar/R groups (Table II). Homology models of representative plant MIPs were generated using both GlpF and AQP1 as structural templates. Similar alignments of putative pore-forming residues were obtained with either template (Table III). However, since AQP and Arabidopsis MIP proteins apparently lack the extended loop C structure of GlpF, AQP1 was chosen as the primary modeling template for comparison.
Table II.
Distribution of specific Arabidopsis MIP gene products among the eight ar/R subcategories
| ar/R Subgroupa | Members in Arabidopsisb |
|---|---|
| PIP | PIP1;1 1;2 1;3 1;4 1;5 PIP2;1 2;2 2;3 2;4 2;5 2;6 2;7 2;8 |
| TIP Group I | TIP1;1, TIP1;2, TIP1;3 |
| TIP Group II | TIP2;1 TIP2;2, TIP2;3 TIP3;1 TIP3;2 TIP4;1 |
| TIP Group III | TIP5;1 |
| NIP Group I (Nodulin 26-like) | NIP1;1, NIP1;2, NIP2;1, NIP3;1, NIP4;1, NIP4;2 |
| NIP Group II | NIP5;1 NIP6;1 NIP7;1 |
| SIP Group I | SIP1;1, SIP1;2 |
| SIP Group II | SIP2;1 |
Each of the eight ar/R subgroups based on sequence alignment and homology modeling are listed.
Members of each ar/R subgroup in Arabidopsis based on the nomenclature of Quigley et al. (2001) and Johanson et al. (2001).
Table III.
Conserved ar/R signatures of Arabidopsis MIPs
| ar/Ra Subgroup | H2b | H5 | LE1 | LE2 | rms Deviationc |
|---|---|---|---|---|---|
| Å | |||||
| PIP | F | H | T | R | 0.91 |
| TIP | |||||
| Group I | H | I | A | V | 0.67 |
| Group IIA | H | I | G | R | 1.03 |
| Group IIB | H | I | A | R | 0.72 |
| Group III | N | V | G | C | 1.56d |
| NIP | |||||
| Group I | W | I, V | A | R | 0.92 |
| Group II | A | I, V | A or G | R | 0.97 |
| SIP | |||||
| Group I | T | F, V, Ie | P | I | 1.39 |
| Group II | S | H | G | A | 1.86 |
The bold designations represent each of the subfamilies of plant MIPs as described by Quigley et al. (2001) and Johanson et al. (2001). Under each heading, ar/R subcategories are listed based on the representative member of each subfamily.
The conserved consensus residue at each position of the ar/R tetrad based on sequence alignment and homology modeling is listed.
The rms deviation of the α-carbon backbone of the homology model of the indicated MIP from the backbone of the AQP1 template is listed. Similar homology modeling results were obtained with GlpF (rms deviation 1.5–2 Å; data not shown).
The TIP5;1 homology model shows a shorter α-helix 1(21 residues; Supplemental Fig. 3) compared to the AQP1 template.
While SIP Group I ar/R regions generally possess an aliphatic hydrophobic residue at the H5 position, AtSIP1;2 possesses a Phe substitution.
NIP Subfamily
Nodulin 26 is the most well-studied NIP with respect to functional properties. However, other proteins that segregate into this MIP subfamily and show similar functional properties (i.e. low water transport, aquaglyceroporins) have been identified in legume (Guenther and Roberts, 2000) as well as nonlegume tissues (Weig et al., 1997; Weig and Jakob, 2000). Structural alignment of the nine full-length Arabidopsis NIPs reveals that there are two different ar/R signatures that differ principally at the H2 position (Table III; Supplemental Fig. 1). Six Arabidopsis NIPs possess a conserved ar/R characteristic of the nodulin 26 ar/R tetrad discussed above (designated NIP Group I), whereas three (NIP5;1, NIP6;1, and NIP7;1) have a divergent ar/R tetrad (Table III) with the substitution of an Ala for Trp at position H2 (designated NIP Group II). None of the proteins in NIP Group II have been functionally characterized. Analysis of the proposed NIP6;1 pore by homology modeling (Fig. 3) and with HOLE (Fig. 4) shows an ar/R region with a much wider aperture (7 Å) than AQP1 and nodulin 26 (Fig. 4), suggesting that NIP6;1 might accommodate larger solutes. Besides differences in the ar/R region, NIP6;1 and NIP5;1 also possess a substitution within the NPA motif in loop E, with the conserved Ala being replaced by the bulkier Val residue (i.e. NPV). This would result in an NPA constriction (Fig. 4) of slightly smaller aperture and increased hydrophobicity compared to other classical aquaporins and glyceroporins.
PIP Subfamily
The PIP subfamily consists of 13 members in Arabidopsis that are broken into two subgroups: PIP1 and PIP2 (Quigley et al., 2001). The PIP family is homogeneous with respect to the residues in the ar/R region (Tables II and III; Supplemental Fig. 2). An examination of the ar/R region from the PIP1;1 homology model (Fig. 3) shows striking similarity to AQP1, with the conservation of Phe (H2) and His (H5) residues as well as the Arg residue at LE2. The LE1 residue is Thr rather than Cys (Fig. 3B). However, since the LE1 residue binds water through a carbonyl from the peptide backbone (Sui et al., 2001), this substitution likely would not affect transport significantly. Thus, all ar/R determinants required for high conductance water transport are conserved in PIPs.
TIP Subfamily
The TIP family is the second largest MIP subfamily in Arabidopsis, consisting of 10 full-length genes (Quigley et al., 2001). TIPs show the most diversity within putative pore regions with three different ar/R subgroups: TIP Group I (TIP1;1–3 gene products), TIP Groups IIa (TIP2;1–3 gene products) and IIb (TIP3;1, TIP3;2, and TIP4;1 gene products), and TIP Group III (a single member TIP5;1) (Tables II and III; Supplemental Fig. 3).
Comparison of the homology models (Fig. 5) of representative TIPs from Groups I and II show that the ar/R regions have a conserved His residue at the H2 position and a conserved Ile residue in the H5 position. Group III (TIP5;1) also possesses an aliphatic side chain (Val) at the H5 position, but His at the H2 position is replaced by an Asn residue. These observations suggest that the polarity of the H2 and H5 positions of the ar/R tetrad of TIPs is reversed from PIPs and most mammalian water-selective aquaporins such as AQP1 (Fig. 5).
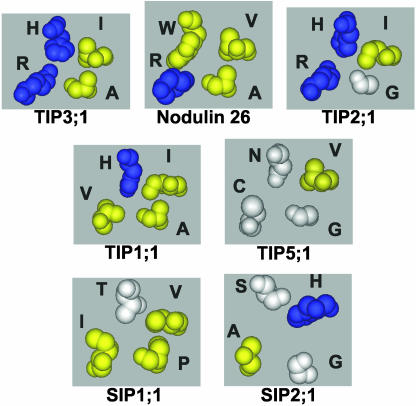
Figure 5.
Comparison of the ar/R selectivity regions of the TIP and SIP subfamilies. Space-filling side-chain residues of the ar/R selectivity region are shown from homology models of representative subgroups of the TIP and the SIP subfamilies. For comparison, the ar/R region of nodulin 26 is included. Residues are color coordinated according to charge and hydrophilicity as described in the legend for Figure 1.
The loop E residues of the ar/R regions of Group II TIPs are similar to other aquaporins and contain the highly conserved Arg residue at LE2 and either an Ala (Group IIB) or a Gly (Group IIA) at the LE1 position. By contrast, Group I and III TIPs possess unusual substitutions within the loop E positions (Table III; Fig. 5). Unlike virtually all other MIPs, Group I TIPs have a Val substitution for the highly conserved Arg at LE2, a substitution that is likely to affect the hydrogen bonding ability of this critical side chain.
TIP Group III is novel, with the conserved Arg at LE2 replaced by a smaller, uncharged Cys residue and a small flexible Gly residue at LE1. Together with Asn at the H2 position and Val at the H5 position, this would result in an unusual ar/R region with a larger apparent pore aperture (Fig. 4) and a reduced ability to form hydrogen bonds with transported solutes. These data suggest that TIP5;1 may have transport properties that are unlike conventional MIPs.
SIP Subfamily
Unlike other plant MIP subfamilies, the generation of homology models for the SIP family members is difficult because the sequence identity between these proteins and the AQP1 template is only about 15%. Since the elements of the ar/R and NPA regions showed a greater degree of identity to the AQP1 model, these were used as constraints in the alignment to model the SIP structures. The resulting model showed a reasonable alignment with AQP1 (Supplemental Fig. 4) with a calculated rms deviation of <2 Å (Table III).
An analysis of the putative ar/R regions of SIPs suggests that two different combinations of residues are formed, characteristic of either SIP Group I (SIP1;1 and SIP1;2) or SIP Group II (SIP2;1) (Table III). Group I shows a hydrophobic residue at H5 (Val in SIP1;1 and Phe in SIP1;2) similar to the aliphatic side chain exhibited by NIP and TIP subgroups (Fig. 5). The remaining SIP Group I ar/R residues are unique, with a Thr at position H2 and conserved Pro and Ile residues at LE1 and LE2, respectively. Overall, this creates a putative ar/R region (Fig. 5) with greater hydrophobic character compared to other MIPs. By contrast, the SIP Group II ar/R region is distinct from the other plant MIP subgroups with Ser and His residues at H2 and H5, respectively, as well as Gly and Ala at positions LE1 and LE2, generating a more open and hydrophilic ar/R region (Fig. 5). The SIP family also contains substitutions at the first NPA sequence (loop B), with the SIP1 family possessing the NPT sequence and the SIP2 family having the NPL sequence in place of the characteristic NPA motif.
DISCUSSION
The ar/R region is a critical site that influences MIP transport selectivity. MIP superfamily members from animal and microbial species generally possess one of two types of ar/R regions, with the conserved features of either aquaporins or glyceroporins (Thomas et al., 2002). By contrast, sequence alignment and molecular homology modeling of the MIPs of Arabidopsis suggests greater structural diversity in pore-determinant sequences, and the various MIPs can be separated into eight distinct ar/R groups that likely aid in determining the pore architecture and functional properties of each protein.
TIP and NIP Subgroups
A conserved feature of all NIPs and TIPs characterized, and a trait that distinguishes them from PIPs and most mammalian and microbial aquaporins, is the presence of a conserved aliphatic residue (either Val or Ile) at position H5. TIPs can further be distinguished from NIPs by the residues found at the H2 position. In almost all MIPs, H2 is a hydrophobic aromatic residue (either Trp or Phe), but in TIPs a hydrophilic residue occupies this position. In the case of Groups I and II, the H2 residue is a highly conserved His. The conserved Ile/His pair at the H5 and H2 positions of these TIPs contrasts with most water-specific aquaporins, such as mammalian AQP1, in that the H2 and H5 positions appear to be reversed (compare AQP1 [Fig. 3] and the TIPs [Fig. 5]). Nevertheless, analysis of the TIP molecular models suggests that the ar/R region is the zone of narrowest constriction (Fig. 4) and likely forms a filter of selectivity. In this regard, it is interesting to consider that MIPs are proposed to have arisen from an ancient gene duplication event (Reizer et al., 1993) and that H2 is homologous to the symmetrically related H5 (Fig. 1A). In the case of plant TIPs, the inversion of these residues relative to most animal and microbial aquaporins would conserve the properties of the ar/R selectivity filter, such that residues form similar contacts with transported water in three-dimensional space despite having a distinct location in the primary sequence.
A comparison of the NIP ar/R Group I (nodulin 26-like) with those of TIP ar/R Group II (Table III) shows that the key difference is the substitution of a Trp for His at the H2 position. A comparison of the ar/R regions of these subgroups (Fig. 5) shows that the presence of the Trp at H2 combined with the Val at H5 and the Ala at LE1 results in a wider and more hydrophobic ar/R region that provides a nonpolar surface (H2 and H5) for interaction with the hydrocarbon backbone of glycerol as well as a hydrophilic surface (LE1 carbonyl and LE2 Arg side chain) to facilitate hydrogen bonding of glycerol as well as transported water. This proposed mechanism sharply contrasts that of mammalian and microbial aquaglyceroporins that have an ar/R signature that is similar to E. coli GlpF with a Gly at H5 and an aromatic Phe or Tyr at LE1 and could represent a divergent solution to the formation of an aquaglyceroporin channel in plants. Since all characterized members of the NIP family are aquaglyceroporins (Rivers et al., 1997; Dean et al., 1999; Guenther and Roberts, 2000; Weig and Jakob, 2000), it is argued that the H2 position may be a critical determinant of selectivity. This is supported by the observation that the substitution of the conserved Trp at H2 with a TIP-like His results in the conversion of NIPs to water-selective aquaporins (Wallace et al., 2002).
The ar/R region of Group II NIPs (NIP6;1-like; Table III) differs from nodulin 26 principally by the substitution of an Ala for the highly conserved Trp at H2. This substitution results in the loss of the ar/R constriction, which becomes wide enough (7 Å) to transport larger solutes. Another, unusual conserved feature of the NIP6;1 is the presence of an Ala to Val residue at the NPA constriction region, increasing the hydrophobicity of this region of the pore. Consistent with these observations, preliminary analysis of the transport properties of NIP6;1 shows that it is a glyceroporin with an extremely low water flux rate (I.S. Wallace and D.M. Roberts, unpublished data). Whether larger solutes are transported remains to be determined.
An interesting observation of the TIP Group I ar/R is the presence of the highly unusual Val for Arg at the LE2 position of the ar/R. This Arg residue is conserved in the vast majority of the members of the MIP superfamily (Park and Saier, 1996; Froger et al., 1998; Engel et al., 2000) and plays a multifunctional role in the selectivity of the pore, providing hydrogen bonds to transported water in water-selective aquaporins (Sui et al., 2001) and to glycerol in glyceroporins (Fu et al., 2000), as well as serving as an electrostatic barrier to the transport of positive ions (de Groot and Grubmuller, 2001). In comparison to other plant aquaporins, the modeled ar/R region of Group I TIPs shows a reduced capacity to form hydrogen bonds with transported water due to the loss of Arg at LE2. Based on the proposed mechanism of the ar/R in facilitating high water flux (Sui et al., 2001), it could be argued that proteins of this group would exhibit a reduced unitary water conductance compared to aquaporins with a conserved Arg LE2. Certainly, the substitution of a Val for Arg at this residue in other aquaporins abrogates water transport (Borgnia et al., 1999). However, it is clear that the archetype of this subgroup, AtTIP1;1, forms a robust water channel upon expression in Xenopus ooctyes, and it is impermeable to ions and other solutes (Maurel et al., 1993). Careful analysis of the unitary water transport rate and elucidation of the structure of a member of TIP Group I will help resolve how this unusual proposed pore structure results in high water transport.
The most divergent member of the TIP family is the TIP5;1 protein, which has a unique ar/R tetrad not found in any other MIP. While functional data for TIP5;1 is presently not available, the putative pore and ar/R selectivity region of TIP5;1 appears to be large enough to accommodate substrates larger than water, and it may represent a multiselective transport MIP.
PIP Subgroup
Based on homology modeling, the PIP family stands alone as the plant MIP family that most resembles mammalian and microbial aquaporins with respect to conservation of the ar/R selectivity filter. They are the only plant MIPs to possess the characteristic conserved His at H5, which has been implicated as providing both steric and hydrogen bonding character to the ar/R region leading to water selectivity (Sui et al., 2001). Consistent with this observation, functional analysis of most PIP proteins shows that they form aquaporins upon expression in Xenopus oocytes (Kammerloher et al., 1994; Johansson et al., 1998; Chaumont et al., 2000; Li et al., 2000; Katsuhara et al., 2002). However, a disparity in the transport rate associated with the two major subfamilies has been noted, with the PIP1 subclass forming channels with low water channel activity and those of the PIP2 subclass forming high conductance water channels (Chaumont et al., 2000). The distinction is intriguing because PIP1 and PIP2 not only have identical ar/R regions but also share complete sequence identity with respect to other residues lining the putative pore. These residues are highly similar to those in AQP1. These observations would suggest that the two classes of PIPs should have identical transport selectivities and rates. In this regard it is important to note that the ar/R region may be one of several determinants that control water or solute flow through the MIP pore. For example, water permeability through plasma membrane water channels appears to be subject to regulation by other factors, including protein phosphorylation (Johansson et al., 1996, 1998), as well as by pH and divalent cations (Gerbeau et al., 2002). How these factors affect PIP structure and function and regulate the flux through the pore remains to be determined.
Additionally, the question of the water selectivity of PIPs remains an interesting one. In the case of a PIP1 derivative from tobacco (Nicotiana tabacum; NtAQP1), a significant glycerol flux has been documented upon expression in Xenopus (Biela et al., 1999). By contrast, such an activity was found lacking in the PIP1 protein ZmPIP1b from Zea mays (Chaumont et al., 2000), even though these two proteins show 85% overall amino acid identity, including complete identity of putative pore-forming residues, and high similarity to AQP1 within these regions. Further structural and functional analysis of this protein is necessary to determine the basis for this discrepancy and whether regions outside of the pore region exhibit a conformational influence of the pore/selectivity of PIPs.
SIP Subgroup
The SIPs are a unique subset of MIPs characterized by an unusually high pI due to the large amount of basic residues at their carboxyl termini (Johanson and Gustavsson, 2002). Based on sequence similarity, the SIP family is divided into two subclasses, SIP1 (with two members) and SIP2 (with a single member), which are conserved in Arabidopsis, Z. mays, and other plant species (Chaumont et al., 2001; Johanson et al., 2001; Quigley et al., 2001). Neither the transport function of the SIPs nor their cellular and subcellular expression patterns have been reported. From expressed sequence tag information, SIP1 appears to be expressed throughout the plant (Johanson and Gustavsson, 2002), whereas SIP2 is principally expressed in roots.
As originally pointed out by Johanson and Gustavsson (2002) in their analysis of SIP1, this subfamily is distantly related in sequence to the MIP family with many sequence substitutions in putative functional residues, suggesting that they perform functions that are divergent from the rest of the MIP family. This hypothesis is supported by homology modeling of both SIP1 and SIP2, which shows that while they may adopt an overall MIP fold, the ar/R selectivity regions are unique with none of the conserved features of known aquaporins or glyceroporins. Further, the first NPA repeat of each SIP is also distinct, with a conserved Thr or Cys (NPT in SIP1;1 and NPC 1;2) or Leu (NPL in SIP2) replacing the Ala. Given the high degree of divergence of this plant MIP subfamily from NIPs, PIPs, and TIPs, it is unclear whether the paradigm regarding MIP structure and function applies, and the transport function and structural organization of this unusual class of MIPs await biochemical characterization.
CONCLUSION
The ar/R region has been shown to account for many of the functional properties of MIPs, and in this study, we have classified the large number of plant MIPs according to their ar/R regions in an attempt to gain an understanding of the structural basis for plant MIP diversity. We propose that plant MIPs can be grouped into eight different ar/R groups, some of which adhere to the classical aquaporin/MIP structures and some of which are completely divergent and likely have functions distinct from classical aquaporins and glyceroporins. It is important to note that these models are static structures based on two crystal structure templates (AQP1 and GlpF) and do not take into account the dynamics and flexibility of the pore residues. For example, molecular dynamic simulations and comparisons of AQP1 and GlpF show that the latter possesses greater flexibility with the pore residues shifting during glycerol transport (de Groot and Grubmuller, 2001; Tajkhorshid et al., 2002). As a final consideration, transport through several MIPS is subject to regulation by stimuli such as phosphorylation and pH, which add another dimension to the complexity of water and solute transport in plants. The affects of these factors on MIP pore structure and function remain to be addressed.
MATERIALS AND METHODS
All homology models were constructed using the Molecular Operating Environment (MOE 2002.03; Chemical Computing Group, Montreal). Target sequences were aligned with AQP1 and GlpF using MOE's multiple sequence and structural alignment algorithm (Kelly, 1996; Kelly and Labute, 1996) with the structural alignment tool enabled and the blosum62 substitution matrix. This alignment of MIP sequences was based on sequence as well as structural homology to the AQP1 (Sui et al., 2001) and GlpF (Fu et al., 2000) experimental structures. Accession numbers for Arabidopsis target sequences used in this study were: AtPIP1;1, CAB71073; AtPIP1;2, AAC28529; AtPIP1;3, AAF81320; AtPIP1;4, AAF02782; AtPIP1;5, T05378; AtPIP2;1, CAB67649; AtPIP2;2, AAD18142; AtPIP2;3, AAD18141; AtPIP2;4, BAB09839; AtPIP2;5, T06738; AtPIP2;6, AAC79629; AtPIP2;7, CAA17774; AtPIP2;8, AAC64216; AtTIP1;1, AAD31569; AtTIP1;2, BAB01832; AtTIP1;3, T01947; AtTIP2;1, BAB01264; AtTIP2;2, F71442; AtTIP2;3, BAB09071; AtTIP3;1, AAF18716; AtTIP3;2, AAF97261; AtTIP4;1, AAC42249; AtTIP5;1, T12999; AtSIP1;1, AAF26804; AtSIP1;2, BAB09487; AtSIP2;1, CAB72165; AtNIP1;1, AAM51272; AtNIP1;2, T05028; AtNIP2;1, T02327; AtNIP3;1, NP_174472; AtNIP4;1, BAB10360; AtNIP4;2, BAB10361; AtNIP5;1, T04053; AtNIP6;1, AAF14664; and AtNIP7;1, AAF30303. The accession number for soybean (Glycine max) nodulin 26 is CAA28471. The protein databank (pdb) accession numbers for the AQP1 and GlpF structural coordinates from x-ray crystallography are 1J4N and 1FX8, respectively.
Three-dimensional model building was performed using the MOE homology program (Kelly, 1996) based on a segment matching procedure (Levitt, 1992) and a best intermediate algorithm with the option to refine each individual structure enabled. A database of 10 structures that were each individually refined to an rms gradient of 1 Å was generated. Comparative analysis of this database of structures showed that differences between them reside almost exclusively within the nonpore-forming loop regions and that the membrane spanning and NPA loops were superimposible.
The stereochemical quality of the models was assessed by using Ramachandran plot analysis and structural analysis using the Protein Report function of the MOE Protein Structure Evaluation, which searches for disallowed bond angles, bond lengths, and side-chain rotamers. In all cases, the models had one or fewer residues in the disallowed region of the Ramachandran plot, and these residues were present in putative loop regions in extramembrane regions that do not contribute to the formation of MIP pores. The models were further analyzed by superposition of each model from the database with experimental structures (AQP1 or GlpF) to determine which models had the smallest rms deviation. The model that suited these criteria was selected for use in all further structural analysis.
The pore regions of various MIPs were analyzed with the HOLE program (Smart et al., 1993), which uses a simulated annealing algorithm to find the best trajectory of a sphere with variable radius through the pore of a protein. The AMBER van der Waals radius file was used during the run. Both a vector and initial point in the channel must be specified to start the program. An initial vector of <0,0,1> was used since all of the structures were oriented along the z axis. To determine an initial point in the channel, two points were taken from the NPA and ar/R regions and averaged. The output file was imaged on a Silicon Graphics workstation (Mountain View, CA) with the InsightII software (Biosym, San Diego).
Supplementary Material
Acknowledgments
We thank Dr. A. Raelene Lawrence of the Chemical Computing Group, Montreal, for assistance with the MOE software package. We would like to thank Drs. Hong Guo and Elias Fernandez of the Department of Biochemistry, Cellular, and Molecular Biology for helpful comments and suggestions during the course of this study.
This work was supported by the National Science Foundation (grant no. MCB–0237219), the National Institutes of Health (grant no. RR018470–01), and the American Society of Plant Biologists (Summer Undergraduate Fellowship to I.S.W.).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.033415.
References
- Agre P, King LS, Yasui M, Guggino WB, Ottersen OP, Fujiyoshi Y, Engel A, Nielsen S (2002) Aquaporin water channels—from atomic structure to clinical medicine. J Physiol 542: 3–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biela A, Grote K, Otto B, Hoth S, Hedrich R, Kaldenhoff R (1999) The Nicotiana tabacum plasma membrane aquaporin NtAQP1 is mercury-insensitive and permeable for glycerol. Plant J 18: 565–570 [DOI] [PubMed] [Google Scholar]
- Borgnia MJ, Kozono D, Calamita G, Maloney PC, Agre P (1999) Functional reconstitution and characterization of AqpZ, the E. coli water channel protein. J Mol Biol 291: 1169–1179 [DOI] [PubMed] [Google Scholar]
- Chaumont F, Barrieu F, Jung R, Chrispeels MJ (2000) Plasma membrane intrinsic proteins from maize cluster in two sequence subgroups with differential aquaporin activity. Plant Physiol 122: 1025–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumont F, Barrieu F, Wojcik E, Chrispeels MJ, Jung R (2001) Aquaporins constitute a large and highly divergent protein family in maize. Plant Physiol 125: 1206–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels MJ, Chrispeels MJ, Yeager M (1999) Projection structure of a plant vacuole membrane aquaporin by electron cryo-crystallography. J Mol Biol 294: 1337–1349 [DOI] [PubMed] [Google Scholar]
- Dean RM, Rivers RL, Zeidel ML, Roberts DM (1999) Purification and functional reconstitution of soybean nodulin 26: an aquaporin with water and glycerol transport properties. Biochemistry 38: 347–353 [DOI] [PubMed] [Google Scholar]
- de Groot BL, Grubmuller H (2001) Water permeation across biological membranes: mechanism and dynamics of aquaporin-1 and GlpF. Science 294: 2353–2357 [DOI] [PubMed] [Google Scholar]
- Engel A, Fujiyoshi Y, Agre P (2000) The importance of aquaporin water channel protein structures. EMBO J 19: 800–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froger A, Tallur B, Thomas D, Delamarche C (1998) Prediction of functional residues in water channels and related proteins. Protein Sci 7: 1458–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D, Libson A, Miercke LJW, Weitzman C, Nollert P, Krucinski J, Stroud RM (2000) Structure of a glycerol conducting channel and the basis for its selectivity. Science 290: 481–486 [DOI] [PubMed] [Google Scholar]
- Gerbeau P, Amodeo G, Henzler T, Santoni V, Ripoche P, Maurel C (2002) The water permeability of Arabidopsis plasma membrane is regulated by divalent cations and pH. Plant J 30: 71–81 [DOI] [PubMed] [Google Scholar]
- Guenther JF, Chanmanivone N, Galetovic MP, Wallace IS, Cobb JA, Roberts DM (2003) Phosphorylation of nodulin 26 on serine 262 enhances water permeability and is regulated developmentally and by osmotic signals. Plant Cell 15: 981–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther JF, Roberts DM (2000) Water-selective and multifunctional aquaporins from Lotus japonicus nodules. Planta 210: 741–748 [DOI] [PubMed] [Google Scholar]
- Johanson U, Gustavsson S (2002) A new subfamily of major intrinsic proteins in plants. Mol Biol Evol 19: 456–461 [DOI] [PubMed] [Google Scholar]
- Johanson U, Karlsson M, Johansson I, Gustavsson S, Sjovall S, Fraysse L, Weig AR, Kjellbom P (2001) The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiol 126: 1358–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson I, Karlsson M, Johanson U, Larsson C, Kjellbom P (2000) The role of aquaporins in cellular and whole plant water balance. Biochim Biophys Acta 1465: 324–342 [DOI] [PubMed] [Google Scholar]
- Johansson I, Karlsson M, Shukla VK, Chrispeels MJ, Larsson C, Kjellbom P (1998) Water transport activity of the plasma membrane aquaporin PM28A is regulated by phosphorylation. Plant Cell 10: 451–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson I, Larsson C, Ek B, Kjellbom P (1996) The major integral proteins of spinach leaf plasma membranes are putative aquaporins and are phosphorylated in response to Ca2+ and apoplastic water potential. Plant Cell 8: 1181–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JS, Preston GM, Smith BL, Guggino WB, Agre P (1994) Molecular structure of the water channel through aquaporin CHIP. The hourglass model. J Biol Chem 269: 14648–14654 [PubMed] [Google Scholar]
- Kammerloher W, Fischer U, Piechottka GP, Schaffner AR (1994) Water channels in the plant plasma membrane cloned by immunoselection from a mammalian expression system. Plant J 6: 187–199 [DOI] [PubMed] [Google Scholar]
- Katsuhara M, Akiyama Y, Koshio K, Shibasaka M, Kasamo K (2002) Functional analysis of water channels in barley roots. Plant Cell Physiol 43: 885–893 [DOI] [PubMed] [Google Scholar]
- Kelly K (1996) Multiple sequence and structural alignment in MOE. Chemical Computing Group. http://www.chemcomp.com/Journal_of_CCG/Features/align.htm
- Kelly K, Labute P (1996) The A* search and applications to sequence alignment. Chemical Computing Group. http://www.chemcomp.com/Journal_of_CCG/Articles/astar.htm
- Levitt M (1992) Accurate modeling of protein conformation by automatic segment matching. J Mol Biol 226: 507–533 [DOI] [PubMed] [Google Scholar]
- Li L, Li S, Tao Y, Kitagawa Y (2000) Molecular cloning of a novel water channel from rice: its products expression in Xenopus oocytes and involvement in chilling tolerance. Plant Sci 154: 43–51 [DOI] [PubMed] [Google Scholar]
- Maurel C, Javot H, Lauvergeat V, Gerbeau P, Tournaire C, Santoni V, Heyes J (2002) Molecular physiology of aquaporins in plants. Int Rev Cytol 215: 105–148 [DOI] [PubMed] [Google Scholar]
- Maurel C, Reizer J, Schroeder JI, Chrispeels MJ (1993) The vacuolar membrane protein γ-TIP creates water specific channels in Xenopus oocytes. EMBO J 12: 2241–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Saier MH, Jr. (1996) Phylogenetic characterization of the MIP family of transmembrane channel proteins. J Membr Biol 153: 171–180 [DOI] [PubMed] [Google Scholar]
- Quigley F, Rosenberg JM, Shachar-Hill Y, Bohnert HJ (2001) From genome to function: the Arabidopsis aquaporins. Genome Biol 3: 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reizer J, Reizer A, Saier MH Jr (1993) The MIP family of integral membrane channel proteins: sequence comparisons, evolutionary relationships, reconstructed pathway of evolution, and proposed functional differentiation of the two repeated halves of the proteins. Crit Rev Biochem Mol Biol 28: 235–257 [DOI] [PubMed] [Google Scholar]
- Rivers RL, Dean RM, Chandy G, Hall JE, Roberts DM, Zeidel ML (1997) Functional analysis of nodulin 26, an aquaporin in soybean root nodule symbiosomes. J Biol Chem 272: 16256–16261 [DOI] [PubMed] [Google Scholar]
- Schoonman MJL, Knegtel RMA, Grootenhuis PDJ (1998) Practical evaluation of comparative modeling and threading methods. Comput Chem 22: 369–375 [DOI] [PubMed] [Google Scholar]
- Smart OS, Goodfellow JM, Wallace BA (1993) The pore dimensions of gramicidin A. Biophys J 65: 2455–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui H, Han BG, Lee JK, Walian P, Jap BK (2001) Structural basis of water-specific transport through the AQP1 water channel. Nature 414: 872–878 [DOI] [PubMed] [Google Scholar]
- Tajkhorshid E, Nollert P, Jensen MO, Miercke LJW, O'Connell J, Stroud RM, Schulten K (2002) Control of selectivity of the aquaporin water channel family by global orientational tuning. Science 296: 525–530 [DOI] [PubMed] [Google Scholar]
- Thomas D, Bron P, Ranchy G, Duchesne L, Cavalier A, Rolland JP, Raguenes-Nicol C, Hubert JF, Haase W, Delamarche C (2002) Aquaglyceroporins, one channel for two molecules. Biochim Biophys Acta 1555: 181–186 [DOI] [PubMed] [Google Scholar]
- Tyerman SD, Niemietz CM, Bramley H (2002) Plant aquaporins: multifunctional water and solute channels with expanding roles. Plant Cell Environ 25: 173–194 [DOI] [PubMed] [Google Scholar]
- Wallace IS, Wills DM, Guenther JF, Roberts DM (2002) Functional selectivity for glycerol of the nodulin 26 subfamily of plant membrane intrinsic proteins. FEBS Lett 523: 109–112 [DOI] [PubMed] [Google Scholar]
- Weig A, Deswarte C, Chrispeels MJ (1997) The major intrinsic protein family of Arabidopsis has 23 members that form three distinct groups with functional aquaporins in each group. Plant Physiol 114: 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weig AR, Jakob C (2000) Functional identification of the glycerol permease activity of Arabidopsis thaliana NLM1 and NLM2 proteins by heterologous expression in Saccharomyces cerevisiae. FEBS Lett 481: 293–298 [DOI] [PubMed] [Google Scholar]
- Zeidel ML, Ambudkar SV, Smith BL, Agre P (1992) Reconstitution of functional water channels in liposomes containing purified red cell CHIP28 protein. Biochemistry 31: 7436–7440 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.