Abstract
Prokaryotic systematics provides the fundamental framework for microbiological research but remains a discipline that relies on a labour- and time-intensive polyphasic taxonomic approach, including DNA-DNA hybridization, variation in 16S rRNA gene sequence and phenotypic characteristics. These techniques suffer from poor resolution in distinguishing between closely related species and often result in misclassification and misidentification of strains. Moreover, guidelines are unclear for the delineation of bacterial genera. Here, we have applied an innovative phylogenetic and taxogenomic approach to a heterogeneous actinobacterial taxon, Rhodococcus, to identify boundaries for intrageneric and supraspecific classification. Seven species-groups were identified within the genus Rhodococcus that are as distantly related to one another as they are to representatives of other mycolic acid containing actinobacteria and can thus be equated with the rank of genus. It was also evident that strains assigned to rhodococcal species-groups are underspeciated with many misclassified using conventional taxonomic criteria. The phylogenetic and taxogenomic methods used in this study provide data of theoretical value for the circumscription of generic and species boundaries and are also of practical significance as they provide a robust basis for the classification and identification of rhodococci of agricultural, industrial and medical/veterinary significance.
It is common knowledge that prokaryotes are widely distributed in nature though the lack of understanding about their abundance and the scale of their diversity feature among the major challenges facing microbiologists1,2,3. Prokaryotic systematics is a fundamental scientific discipline which, inter alia, provides the framework for determining the extent of diversity and underpins research into the ecological, industrial and medical importance of prokaryotes. Fundamental to the current practice of polyphasic taxonomy is the definition of taxa at different ranks in the taxonomic hierarchy. The term species, for instance, is generally defined as a group of closely related strains evolved from a common ancestor and which have a degree of phenotypic consistency, ≥70% pairwise DNA-DNA hybridization (DDH) values, ca. >98.7% identity between their 16S rRNA gene sequences and a high mutual phenetic similarity4,5,6. However, the value of phenotyping and DDH is limited, not least by a lack of reproducibility and compatibility of results between different laboratories7,8 while 16S rRNA gene sequences tend to provide insufficient resolution to distinguish between closely related species9,10. Moreover, it is not possible to apply the ‘polyphasic’ approach to unculturable bacteria3,11, the so called ‘microbial dark matter’.
The limitations of current approaches to prokaryotic systematics have been addressed by several workers who have pressed the need to embrace the genome10,12,13,14,15,16,17. Indeed, the advent of inexpensive whole genome sequencing technologies and associated bioinformatic tools are promoting a step change in taxonomic practice, notably the availability of new metrics for delineating species10,12,18. In contrast, prokaryotic genera remain loosely defined, typically based on monophyly of strains with an average sequence divergence <6% in16S rRNA gene phylogenies3. Only limited attempts have been made to define generic boundaries between prokaryotes19.
Here, we have applied a range of genomic approaches to clarify the taxonomy of the genus Rhodococcus; the long and chequered taxonomic history of this genus has been addressed in several authoritative reviews20,21,22. The genus is classified in the family Nocardiaceae23 of the order Corynebacteriales24. The former encompasses other mycolic acid containing taxa such as the genera Gordonia, Nocardia, Smaragdicoccus and Williamsia and the latter more distantly related genera including Corynebacterium and Segniliparus. The genus Rhodococcus currently contains nearly 50 species with validly published names which fall into several 16S rRNA gene lineages, notably ones corresponding to the Rhodococcus corynebacterioides, Rhodococcus equi, Rhodococcus erythropolis and Rhodococcus rhodochrous clades25,26. 16S rRNA phylogeny indicates the presence of up to nine distinct groups within this genus and highlights widespread taxonomic ambiguities within this taxon27. Furthermore, a number of gene clusters have been found to vary between major rhodococcal clades, emphasizing extensive variation at the genomic level27. Similarly, phylogenetic groups of rhodococcal species have been detected based on other genes, such as alkB28 and from the analysis of a limited number of rhodococcal genomes29. Thus, there is a clear need to further unravel taxonomic relationships within the genus Rhodococcus, particularly given the importance of R. equi, a facultative intracellular pathogen of animals, especially foals30, Rhodococcus fascians, a phytopathogen of numerous dicotyledonous plants31 and R. erythropolis which is capable of numerous industrially significant bioconversions and biodegradations32. To embed rhodococcal taxonomy within a genomic framework, we present here an analysis of 100 rhodococcal strains and 15 representatives from related genera. These analyses revealed the existence of seven species-groups within the genus Rhodococcus that are as distantly related to one another as they are to other Corynebacteriales genera, thereby confirming the need for a significant revision of rhodococcal systematics. These analyses also highlight widespread misclassification and misidentification of rhodococci within the genus. However, most importantly, the results of this study show that the taxogenomic approach has the potential to resolve complex taxonomic questions both at the intrageneric and supra-species (intra-family) level.
Results
Rhodococcus, a highly polyphyletic taxon
To investigate the genomic heterogeneity within Rhodococcus, we sequenced the genomes of 15 strains representing different taxa previously classified within the genus, including the type strains of “Rhodococcus hoagii” (priority type strain for R. equi26), Rhodococcus corynebacterioides, Rhodococcus gordoniae, Rhodococcus kunmingensis, Rhodococcus kroppenstedtii, Rhodococcus opacus, Rhodococcus pyridinivorans, Rhodococcus phenolicus, Rhodococcus qingshengii, Rhodococcus ruber and Rhodococcus rhodochrous (the type species of the genus), representatives from two previously identified R. equi subgroups25 and the unclassified strain Rhodococcus sp. AJR001. The genome sequences of 85 strains belonging to the genus Rhodococcus were retrieved from GenBank (July 2015), including two strains previously sequenced by us33,34 (Supplementary Table 1). We also included 15 publicly available genomes of representatives of related genera classified within the order Corynebacteriales both for comparative analyses and as outgroups (Supplementary Table 1). The resultant 115 genomes were re-annotated by the RAST pipeline35 to have an equivalence of annotation and were compared using EDGAR36 to calculate the core genome. Information on the size of assemblies, GC content and number of coding sequences, RNA genes and GenBank accession numbers is provided in Supplementary Table 1.
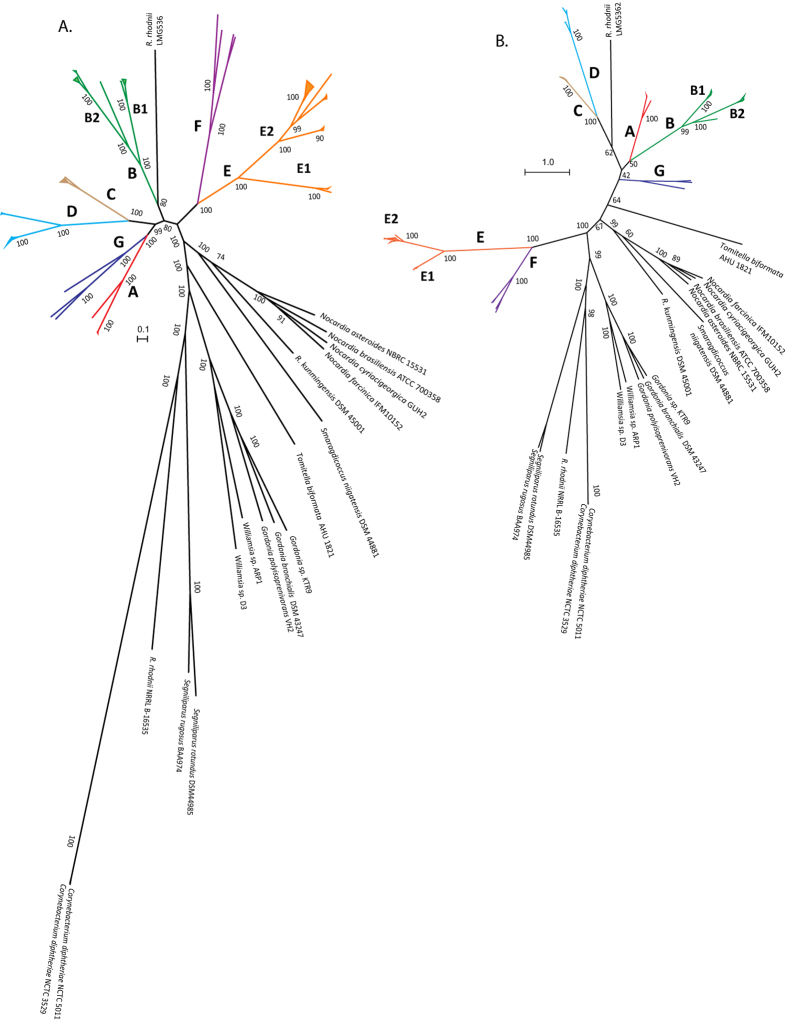
A maximum-likelihood (ML) tree was constructed from a concatenated sequence alignment of codons from the core genes (255 genes) after stripping the start codons, stop codons as well as any codon with missing data using the best-fit codon substitution model (SCHN05 + F + I + G4). The rhodococci were clearly separated into seven distinct clusters and three singletons in the phylogenetic tree (Fig. 1A; Supplementary Fig. 1). R. equi formed a distinct group (group A) together with R. defluvii, a result consistent with those of previous analyses26,33. Despite its frequent association with R. equi in 16S rRNA gene trees25,26,37, the type strain of R. kunmingensis was recovered as a singleton that was loosely associated with group A in the phylogenetic tree.
Figure 1.
Un-rooted radial maximum-likelihood phylogenetic trees derived from (A) concatenated codon alignment of the core genome (scale bar represents nucleotide substitutions per codon site) and (B) a subset of amino acids from 400 broadly conserved prokaryotic proteins. The scale bar shows normalized fraction of total branch lengths as described by Segata et al.38.
The species assigned to the R. rhodochrous group (B, Rhodococcus sensu stricto) were subdivided into two major subgroups with the exception of the type strain of R. phenolicus which formed a phyletic line separate from each of the subgroups. In addition, Rhodococcus triatomae formed a distinct group (group G) together with two unclassified rhodococci (Fig. 1A; Supplementary Fig. 1). The two Rhodococcus rhodnii strains, symbionts in the gut of Rhodnius prolixus, a vector of Chagas disease, were also recovered as singletons; R. rhodnii NRRL B-16535T was more closely related to Corynebacterium diphtheriae and Segniliparus strains than to other rhodococci. Rhodococcus species that formed sub-groups within the R. erythropolis clade in the 16S rRNA phylogeny of Jones et al.25 were separated into three distinct groups, C, D and a relatively distant group E. All of the R. erythropolis strains formed a single taxon, group D (Fig. 1A). The type strains of R. corynebacterioides and R. kroppenstedtii formed group F together with two unclassified rhodococci.
ML phylogenies were also reconstructed from a computationally selected subset of amino-acid sequences from 400 broadly conserved prokaryotic proteins38 (Fig. 1B; Supplementary Fig. 2) and the protein sequence alignment of the core genomes (Supplementary Fig. 3). Significantly, these trees confirm that the genus Rhodococcus, as presently defined, is polyphyletic and includes at least seven distinct species-groups. These results also show that 16S rRNA gene sequences have insufficient resolution to deduce precise inter-species relatedness within the genus Rhodococcus. A phylogenetic tree from 16S rRNA sequences extracted from the genomes confirms this conclusion (Supplementary Fig. 4).
Taxogenomic separation of rhodococci into seven robust species-groups and identification of intrageneric and supraspecific boundaries
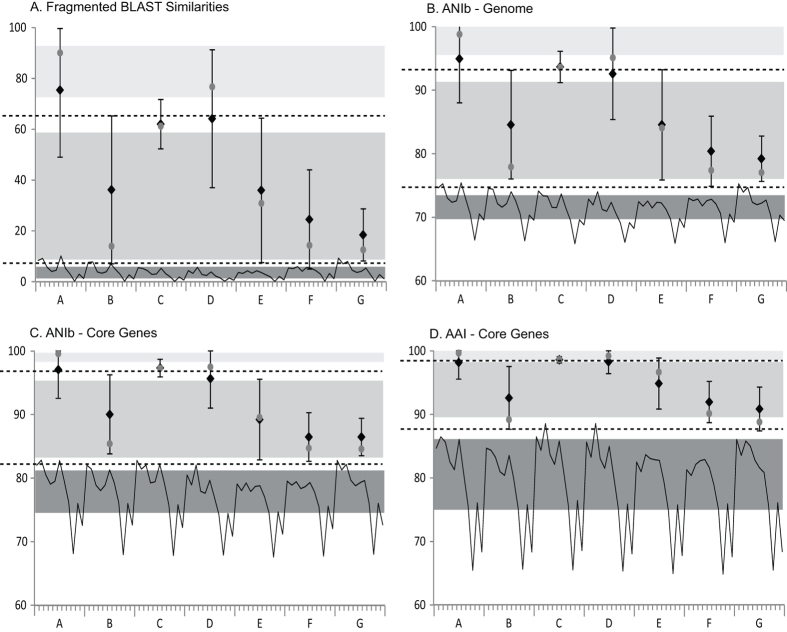
The similarity matrix derived from the pairwise BLAST-based fragmented genome analysis supported the phylogenetic group structure (Figs 2A and 3A; Supplementary Table 2a). The mean similarity score (fragmented BLAST similarity, FBS) varied between 18.86 ± 10.26 and 75.82 ± 26.37 based on the diversity within each rhodococcal group. The pairwise similarity scores between the rhodococcal species-groups are between 2.49 ± 0.23 and 10.16 ± 0.50. However, two strains of group G showed slightly higher similarities (a score up to 11.31) with some strains in group A and vice versa.
Figure 2.
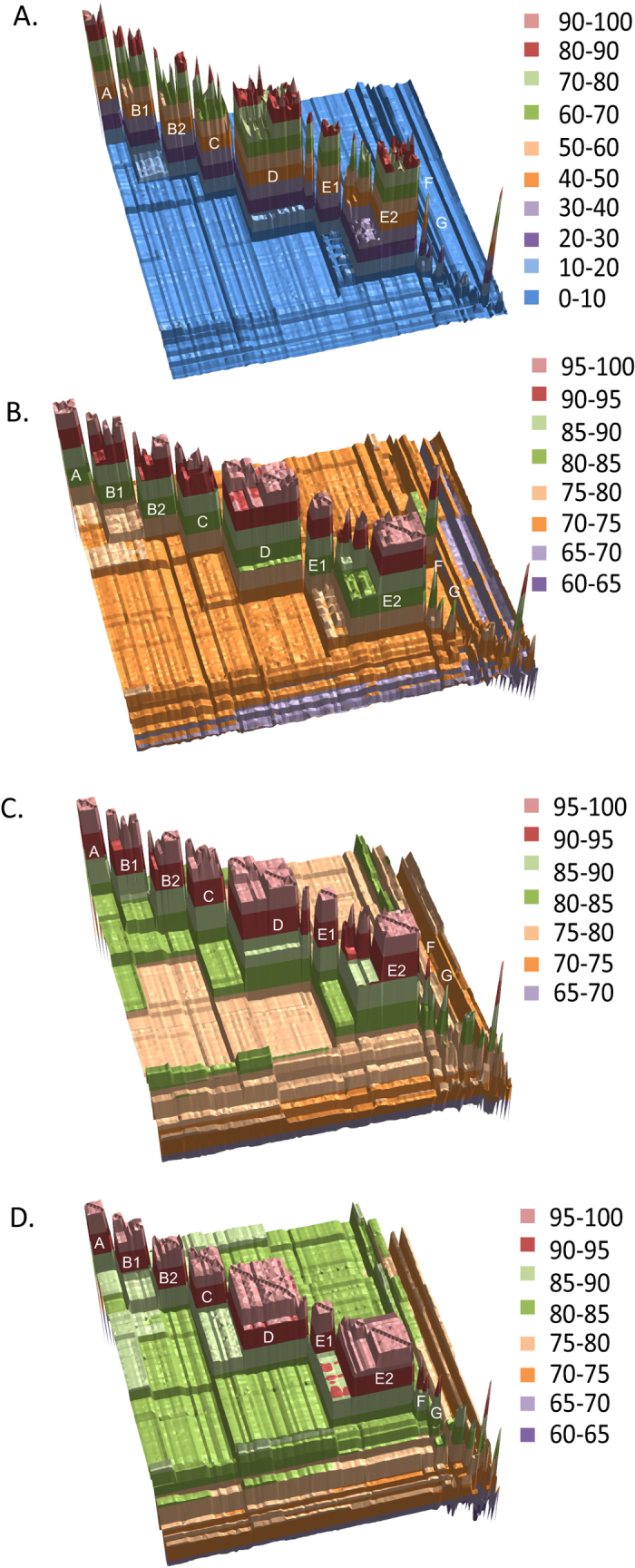
3D graphical representation of pairwise similarity matrices obtained by (A) fragmented BLAST searches (FBS values), (B) genomic average nucleotide identities (ANIb-G values), (C) average nucleotide identities among core genes (ANIb-C values) and (D) average amino-acid identities from the core genes (AAI values). Rhodococcus species-groups A-G are labelled whilst the reference genera are plotted at the lower right hand corner.
Figure 3.
Average taxogenomic values (filled diamonds), (A) fragmented BLAST similarities (FBS), (B) genomic average nucleotide identities (ANIb-G), (C) average nucleotide identities among core genes (ANIb-C) and (D) average amino-acid identities from the core genes (AAI) with standard deviations. The median values are shown with filled circles. Average pairwise similarities with standard deviations within species, within groups of species (excluding similarity among members assigned to the same species), and between different groups are marked in light, intermediate and dark grey colour, respectively. The average diversity between different individual groups is plotted at the bottom for each group against species-groups (A–G), Nocardia, Gordonia, Corynebacterium diphtheriae, Williamsia and Segniliparus, respectively (excluding self-values).
These results are consistent with the BLAST-based pairwise average nucleotide identities from the whole genome sequences (ANIb-G). An ANIb-G value of ≥75% (79.20 ± 3.56–94.92 ± 6.92) was observed between strains within each of the species-groups, apart from group E where the values were marginally lower (down to 74.71%) between some strains (Figs 2B and 3B; Supplementary Table 2b). Similarly, the strains of species-group G share slightly higher ANIb-G values with the members of group A and vice versa (75.47 ± 0.33–75.33 ± 0.27). Multiple strains between rhodococcal species-groups A and B and groups A and C also showed >75% ANIb-G values. ANIb values calculated from the nucleotide sequences of the 255 core genes (ANIb-C) underlined the taxonomic integrity of these phylogenetic groups though similarity values were relatively higher than their corresponding ANIb-G values (Figs 2C and 3C; Supplementary Table 2c). The ANIb-C values within each rhodococcal species-group are >84% (86.39 ± 2.95–97.24 ± 1.39) though some strains from the two subgroups within group E showed slightly lower ANIb-C values (down to 81.1%).
An average amino acid identity (AAI) of 87.75–100% (mean, 90.85 ± 3.46–98.58 ± 0.50) was observed between two individuals of the same species-group from the core 255 protein sequences (Figs 2D and 3D; Supplementary Table 2d). The AAI values between different species-groups are <87% (80.42 ± 0.36–86.53 ± 0.17) except for groups C and D where they were slightly higher (88.60 ± 0.28).
The mean FBS score within a species was 83.45 ± 6.91 and between other members within a species-group 32.67 ± 24.29, resulting in a potential species threshold of 66.75 (Fig. 3A). A mean ANI-G value of 98.02 ± 0.84 was observed between strains within a species and corresponding 83.47 ± 7.30 values within each predicted genus suggesting a boundary of approximately 94% between species within the same genus (Fig. 3B). Although ANI-C and AAI values (99.11 ± 0.42 and 99.44 ± 0.76 within species and 89.28 ± 5.79 and 93.92 ± 4.22 within species-groups, respectively) clearly separated strains within and between species-groups (Fig. 3C,D), the species thresholds were much higher (96.88 and 98.41, respectively) with narrower buffer zones; hence, FBS and ANIb-G appear to be the most useful tools for delineating Rhodococcus species.
The mean FBS and ANIb-G values between different species-groups were found to be 3.64 ± 1.86 and 71.63 ± 1.86 with a suggested generic boundary of approximately 6.9 and 74.8, respectively (Fig. 3A,B). The ANI-C and AAI thresholds for delineating genera from the core genome were relatively higher that were around 82.3 and 87.8, respectively.
The taxogenomic similarities between members of the different Rhodococcus species-groups are comparable to corresponding similarities between these taxa and the representatives of the related genera (Figs 2A–D and 3A–D). Similar FBS scores were observed between the Rhodococcus species-groups (2.49 ± 0.23–10.16 ± 0.50) as between them and the genera Gordonia, Nocardia and Williamsia (1.51 ± 0.13–5.36 ± 0.84) while the C. diphtheriae and Segniliparus strains have relatively distant values (0.13 ± 0.05–1.23 ± 0.25). The ANIb-G, ANIb-C and AAI values between the rhodococcal species-groups are also comparable to those against the other genera though C. diphtheriae remains quite distant from all of these taxa (Figs 2A–D and 3A–D). These results clearly show that each of the rhodococcal species-groups can be considered to represent a distinct taxon (Fig. 2A–D and Fig. 3A–D; Supplementary Table 2a–d).
Species-group A – R. equi cluster
The R. equi strains formed a distinct compact cluster together with the type strain of R. defluvii, a result consistent with our previous study33. These strains are primarily associated with foal disease and opportunistic human pathogenicity, with the exception of R. defluvii. The strains within this species-group shared 3,457 genes (63.6–73.5% of the total coding sequences). The genome size and GC content of these strains fell within the narrow range of 4.97–5.65 Mb and 68.5–68.8 mol%, respectively (Fig. 4). As expected, the R. hoagii/R. equi genomes are very closely related with a FBS similarity score of >82.6, ANIb-G values of >98.5% and ANIb-C and AAI values of >99.3% (Fig. 2A–D). Digital DNA-DNA hybridisation (dDDH, species cut-off ≥70%) values between these strains were >90.8% (Supplementary Table 3), these results are in line with the assignment of these strains to the same species25. ANIb-G values between the R. equi and R. defluvii strains are approximately 83%, and the corresponding ANIb-C and AAI values are ~89% and 93%, respectively. The dDDH value between the R. hoagii/R. equi strains and the R. defluvii strain is 27 ± 3%, a result consistent with their classification as separate species.
Figure 4. A 3D distribution of genome size, GC content and fraction of shared genes within each species-group (Supplementary Table 1).
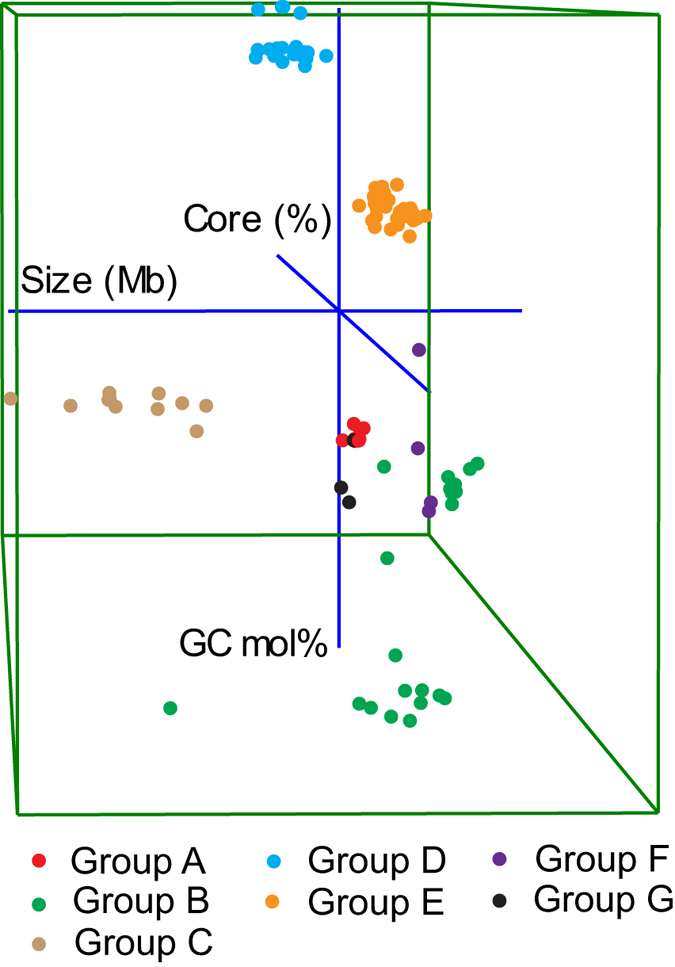
The three axes are shown in blue in the centre of the plot and are labelled. The individuals belonging to seven species-groups are shown in different colours.
Jones et al.25 assigned R. equi strains to two subgroups based on the amplification of repetitive elements (rep-PCR), amplified 16S ribosomal DNA restriction analysis (ARDRA) and numerical taxonomic data25. However, phylogenetic and taxogenomic analyses (Figs 1 and 2A–D) of representatives of these subgroups (strains C7T and N1288 from subgroup 1 and N1295 and N1301 from subgroup 2) did not show any evidence of subgroup structure (Figs 1A–B and 2A–D and Supplementary Figs 1–3), indicating that the division of strains into subgroups in the earlier analyses was probably more apparent than real.
Species-group B – Rhodococcus sensu stricto cluster
This taxon can be considered to represent Rhodococcus sensu stricto as it includes the type strain of the type species of the genus, R. rhodochrous DSM 43241T. This species-group was found to encompass a diverse set of mainly environmental isolates which were divided into two subgroups, B1 and B2 (Fig. 1A,B and 2A–D). The mean FBS score within this species-group was 36.59 ± 29.08 and varied from 8.09–93.77 between individual pairs of strains. The average ANIb-G, ANIb-C and AAI values were 84.55 ± 8.54, 89.95 ± 6.21and 92.59 ± 4.93, respectively (Fig. 3A–D). The size of the genomes, average GC mol% and the shared fraction of genes within the group also show a clear subdivision of strains into the two subgroups (Fig. 4). However, two strains from subgroup B2, namely R. rhodochrous BKS6-46 and Rhodococcus sp. R4, had slightly larger genome sizes and varied in the fraction of the shared genes within the group (Supplementary Table 1).
Group B1 includes a Rhodococcus aetherivorans strain, five R. ruber strains, including the type strain, three unclassified rhodococci and a strain identified as R. rhodochrous (Fig. 1A,B, Supplementary Figs 1 and 2,Supplementary Table 1). The dDDH values (Supplementary Table 3) between representative strains of this taxon indicated the presence of three predicted species within this subgroup, a conclusion that is supported by pairwise ANIb-G and ANIb-C values (cut-off value of >94%; Supplementary Tables 1 and 2). The AAI from the core genes are >98% between individuals within these predicted species. One of the predicted species included four strains identified previously as Rhodococcus sp. BCP1, Rhodococcus sp. EsD8, R. aetherivorans IcdP1 and R. rhodochrous ATCC 21198 and a second one R. ruber strains DSM 43338T, IEGM 231, P25, Chol-4 and Rhodococcus sp. P14. The strain ‘R. ruber’ BKS 20–38 is clearly misidentified and may represent a third putative species given marginal taxogenomic similarities (ANIb genome, 94.76%; ANIb core genes, 97.21%; AAI core genes, 98.08%; dDDH, 60.3 ± 2.82) when compared with R. ruber DSM 43338T. The genome of strain BKS 20–38 is relatively large (6.13 Mb) and has a slightly lower GC content (69.7 mol%) compared with the other R. ruber strains (5.30–5.99 Mb; 70.2–70.7 mol%).
Group B2 includes the type strains of R. rhodochrous, R. gordoniae and R. pyridinivorans (Fig. 1A,B, Supplementary Figs 1 and 2, Supplementary Table 1), the taxogenomic analyses support the recognition of three species within this subgroup Supplementary Figs 1 and 2, (Supplementary Tables 1–3). The four unclassified strains in this subgroup can be assigned to known species. Rhodococcus sp. R1101 showed 85.7 ± 2.5 dDDH, >84 FBS score and >98% ANIb-G, ANIb-C and AAI similarities with R. gordoniae DSM 44689T hence it can be assigned to this species. Similarly, taxogenomic values above the accepted species delineation thresholds were found between strains Rhodococcus sp. Chr-9, Rhodococcus sp. R4, Rhodococcus sp. P52 and R. pyridinivorans DSM 44555T. R. phenolicus DSM 44812T does not belong to either of these subgroups (Fig. 1A,B), consistent with comparative taxogenomic values with individuals from each of the subgroups (ANIb genome, ANIb core genes and AAI core genes <90%, and dDDH between 21.9–23.0 ± 2.35; Supplementary Tables 1–3). ‘R. rhodochrous’ ATCC 21198 is taxogenomically distant from R. rhodochrous DSM 43241T and is clearly misidentified.
Species-group C – Rhodococcus opacus cluster
This group of environmental isolates encompasses eleven strains, five Rhodococcus opacus, two Rhodococcus wratislaviensis, one Rhodococcus jostii, one Rhodococcus imtechensis and two unclassified rhodococci (Supplementary Table 1). These strains were found to have genome sizes that varied between 7.8–10.4 Mb and GC contents between 66.8–67.9 mol% but nevertheless formed a distinct but diffused cluster based on both of these properties and on the fraction of shared genes (Fig. 4). dDDH values support the circumscription of four species within this group (Supplementary Table 3). The first of these taxa includes three strains, R. jostii RHA1, Rhodococcus sp. JVH1 and Rhodococcus sp. DK17, the second five strains, including R. opacus DSM 43205T, M213 and PD630, R. wratislaviensis IFP 2016 and R. imtechensis RKJ300T. The dDDH value between the type strains of R. opacus and R. imtechensis is 81.2 ± 2.7, the corresponding ANIb-G value >96% and the ANIb-C and AAI values ~98.9%, results which indicate that R. imtechensis RKJ300T represents a later heterotypic synonym of R. opacus DSM 43205T. ‘R. wratislaviensis’ IFP 2016 is clearly misidentified as it is well separated from R. wratislaviensis NBRC 100605T based on a dDDH value of 57.5 ± 2.8, a FBS score of <58 and an ANIb-G value of <93%; the ANIb-C and AAI values were ~97.0% and ~98.5%, respectively. ‘R. opacus’ strain R7 is also misclassified as the matrices show that it is a bona fide R. wratislaviensis strain. In turn, ‘R. opacus’ B4 probably represents a distinct species according to the taxogenomic analyses (Supplementary Tables 1–3). These results highlight the extent of misclassification and misidentification of rhodococcal strains thereby underlining the difficulty of classifying such strains reliably on the basis of traditional taxonomic criteria.
Species-group D – Rhodococcus erythropolis cluster
This group of environmental isolates is compact and clearly defined based on genome size, GC content and the fraction of shared genes (Fig. 4). It encompasses 22 strains which fall into three species based on dDDH values (Supplementary Table 3), one of which includes four R. erythropolis, three R. qingshengii and two unclassified strains (Supplementary Tables 1 and 2). This taxon includes the type strain of R. qingshengii which shares a dDDH value of >80% with other strains (Supplementary Table 3). We obtained the partial sequences of 16S rRNA, catA and gyrB genes of this strain from GenBank (accession numbers DQ090961.1, KF500432.1 and KF374699.1, respectively) and confirmed the authenticity of the strain in a BLAST search that showed 100% coverage and identity with the sequenced R. qingshengii strain. The mean FBS score and ANIb-G values between the strains within this taxon were 83.04 and 98.01%, indicating that they may be reclassified as R. qingshengii.
Six strains classified as R. erythropolis, including the well-studied strain PR4, formed a second species within this species-group together with two strains of unclassified Rhodococcus spp., one of which was previously identified as R. opacus and the other as R. rhodochrous (Supplementary Tables 1–3). The 16S rRNA gene sequence of strain PR4 is identical to that of R. erythropolis DSM 43066T (accession number: KJ476725.1). Hence the strains within this taxon belong to the species R. erythropolis. The third species within this group encompasses three strains, two of which have not previously been assigned species names while the remaining strain was described as R. rhodochrous (Supplementary Tables 1–3).
Species-group E – Rhodococcus fascians cluster
The strains in this cluster can be divided into two subgroups based on phylogenetic and taxogenomic data (Figs 1, 2, 3 and Supplementary Tables 1–3), a result in line with an earlier report29. The strains within this taxon, which include plant pathogens and some environmental isolates, have a genome size ranging between 5.17–6.24 Mb and a fairly narrow GC content (64.1–64.7 mol%; Fig. 4). Seven strains, including two unclassified strains, formed a subgroup which corresponds to clade II as defined by Creason et al.29; this taxon encompasses two species (Supplementary Tables 1–3). The remaining sixteen R. fascians strains, including the type strain (LMG3623T), formed the second subgroup together with five unclassified strains (Supplementary Tables 1–3) that matches clade I in the above study29. These strains can be assigned to six predicted species based on dDDH, FBS score of >70 and ANIb-G values of >94%; five of these taxa correspond to taxa delineated by Creason et al.29.
Minor Rhodococcus taxa
The remaining rhodococcal strains were assigned to two small groups, F and G and three singletons. Group F includes four strains, namely R. corynebacterioides DSM 20151T, R. kroppenstedtii DSM 44908T and two unclassified strains, each representing a distinct species according to the taxogenomic data (Supplementary Tables 1–3). Group G encompasses three strains, R. triatomae BKS 15–14 and two unclassified rhodococci which belong to three predicted species (Supplementary Tables 1–3).
Discussion
There is increasing evidence that current approaches to prokaryotic systematics will be enriched by the inclusion of whole genome sequencing data10,12,13,14,15,16,17. In particular, new metrics have been suggested for species delineation, as exemplified by an ANI cut-off of >94% and dDDH values of >70% to identify strains within a species4,39,40,41,42. A multi-gene phylogenetic approach applied to members of the class Clostridia indicated that they could be reclassified into multiple species that belonged to novel genera43. Here, we have built upon such studies by applying a comprehensive genomic approach to delineate species within the genus Rhodococcus that include strains of agricultural, industrial and medical/veterinary significance. The genetic heterogeneity within this genus has become increasingly clear, particularly in the light of a succession of 16S rRNA gene sequence analyses21,22,25,27,44. However, the number and composition of distinct lineages varied between these studies thereby indicating the need to re-examine relationships within this genus using genomic methods. Our genomic analyses of 100 Rhodococcus strains highlighted the presence of at least seven species-groups and three singletons (Fig. 1A,B; Supplementary Figs 1–3). It is particularly significant that these taxa are as distant from one another as they are from other genera classified in the family Nocardiaceae (Figs 2A–D and 3A–D) and should thus be recognised as putatively novel genera. These seven lineages were also identified in the 16S rRNA phylogeny from 641 most reliable sites (Supplementary Fig. 4); however, the resolution was very limited at the species level.
This integrated genomic approach identified clear intrageneric and supraspecific boundaries for a reliable delineation of species and genera (Fig. 3). FBS scores are average pairwise normalized BLAST similarity scores calculated using a non-overlapping 500 bp fragment size45. This approach is faster when a large number of genomes are compared. However, a more accurate matrix can be obtained using a computationally extensive approach with smaller fragment size and an overlapping sliding window. Rhodococcus species-groups, as well as different species within species-groups, are well separated using the FBS cut-off values of 6.9 and 66.75, respectively.
ANI was first calculated from the conserved genes for a robust resolution of prokaryotic species with minimum effect of horizontal gene transfer42. An ANI value of ~94% was suggested to correspond to an experimental DDH value of 70% for species separation. In this study, the ANIb-C threshold, calculated from 255 core genes, was relatively high (~96.88%) for species delineation. This value may be affected by the size of the core genome analysed, which is dependent on the number of genomes in the dataset as well as the criterion for defining orthologous genes. However, the approach of splicing the genome into 1020 bp fragments followed by BLAST-search against other genomes46 appears to be more pragmatic. The ANI is calculated from pairwise BLASTN matches with >30% sequence identity and ≥70% alignable length and an ANI value of 95% corresponds to the 70% DDH for species delineation46. The ANIb-G cut-off value to define rhodococcal species is ~94% (Fig. 3B), which is consistent with previous reports of defining an ANI cut-off of >94% to identify strains within a species5,36,37. The ANIb-G threshold for separating potential genera is ~74.8%.
It has been proposed that AAI derived from the conserved genes should be incorporated into prokaryotic taxonomy as AAI provides more robust resolution than ANI between divergent strains47,48. The AAI thresholds from the 255 core genes are 87.8% and 98.41% for separating potential genera and species, respectively. Again, these values may be affected by the number of genes in the core genome, as described for ANIb-C. The species designations with cut-off values from different matrices are also supported by dDDH values which are based on the genome to genome distance calculation that mimics the experiment based DDH values39,40. However, it will be important to use these taxogenomic indices and suggested thresholds in conjunction with robust genome based phylogenies.
Qin et al.19 suggested that ANI values are not suitable for separating genera19 and that a genus should be defined by a shared percentage of conserved proteins of at least 50%. Here, we have applied a more robust approach that uses fragmented BLAST similarity scores, ANI values and phylogenies assembled from universal proteins and the core genome, and found that ANIb-G values can reliably distinguish between rhodococci assigned to different species-groups. In contrast, the fraction of shared genes could be below 50% for diverse species-groups (Supplementary Table 1).
In this study, the genome sequences of 75 strains that represented 20 rhodococcal species, including 18 type strains, were analysed together with 25 strains that were unclassified at the species level (Supplementary Table 1). The taxogenomic analyses indicate that these strains should be classified into 31 species. Species-group E (R. fascians), the most underspeciated taxon, includes eight presumptive species thereby reinforcing previous work where strains classified as R. fascians were separated into different, albeit closely related species29. Similarly, some strains classified as R. erythropolis, R. opacus, R. rhodochrous, R. ruber and R. wratislaviensis were found to be sufficiently taxogenomically distinct to be separated into different species (Supplementary Tables 1–3). ‘R. opacus’ NRRL B-24011, ‘R. rhodnii’ LMG 5362, ‘R. rhodochrous’ ATCC 17895 and ‘R. rhodochrous’ NRRL B-1306 were also shown to be misclassified as they are more closely related to strains in taxonomically distinct species-groups than the corresponding type strains (Fig. 1A,B, Supplementary Table 1). The genomic analyses challenge the retention of R. imtechensis as a distinct species since the type strain of this taxon clearly belongs to the established species R. opacus. It is also significant that the taxogenomic approach allowed many of the unclassified strains to be assigned to validly published Rhodococcus species, as exemplified by the assignment of Rhodococcus strains BCP1 and EsD8 to R. aetherivorans, strain R1101 to R. gordoniae, strains Chr-9, R4 and P52 to R. pyridinivorans, strains JVH1 and DK17 to R. jostii, strain 311R to R. erythropolis, and strains PML 026 and JG-3 to R. fascians. Therefore, this study provides a proof of concept for the integration of genomics in prokaryotic systematics for a reliable, robust and stable classification of prokaryotic species.
Despite multiple calls to revisit complex rhodococcal taxonomy20,21,22,25,27,29, a recent study based on the analyses of fewer rhodococcal genome sequences presented an alternative phylogenomic view even though similar species-groups were recovered49. In contrast, the present study is based on more extensive and comprehensive phylogenomic and taxogenomic analyses of a larger genomic dataset, including more type strains. The results of this study clearly support the separation of rhodococci into multiple presumptive genera. The taxogenomic analyses (Figs 1 and 2) unequivocally support the proposal that R. equi be classified in the genus Prescottella as Prescottella equi25,50, and the subsequent conclusion that R. defluvii belongs to this taxon and should be classified as Prescottella defluvii33. Complex nomenclatural problems have delayed the formal validation of the names of these taxa51. Five genes that encode hypothetical proteins are specific to this presumptive novel genus according to BLAST searches in the NCBI nucleotide and protein sequence databases (Supplementary Table 4). In this context, it is interesting to note that Myoviridae phage E3 infects R. equi strains but not other rhodococci or mycolic acid containing actinobacteria52.
An extensive literature search of phenotypic data acquired on type strains representing each of the species-groups did not reveal any characteristics that could be unambiguously weighted to distinguish between them, a problem compounded by the fact that most validly published rhodococcal species are based on the descriptions of single strains22. Previously, we have noted that few standard chemotaxonomic characteristics are available to distinguish Rhodococcus strains from other genera classified in the family Nocardiaceae, such as Nocardia and Smaragdicoccus25. It can, therefore, be concluded that the taxogenomic approaches employed here reveal stable clustering of representative rhodococci that could not be gleamed using traditional taxonomic criteria. Even so, it is interesting to note that none of the group A Prescottella strains, including additional isolates previously investigated53,54, use L-arabinose, cellobiose, maltose, mannitol, sorbitol and trehalose as sole carbon sources, features shared only with the type strain of R. triatomae (a representative of Group G).
In essence, the phylogenetic and taxogenomic data show that strains assigned to rhodococcal species-groups are under-speciated and that many have been misclassified, results that highlight problems associated with the use of current polyphasic approaches to resolve relationships between closely related taxa. These findings are of theoretical value as they provide an insight into matrices that can be used to define generic and species boundaries. The outcomes of this study are also of practical value as they provide a sound basis for improving the classification and identification of rhodococci of agricultural, industrial and medical/veterinary significance, as exemplified by strains assigned to the R. equi, R. erythropolis and R. fascians species-groups. Importantly, this case study provides tangible evidence that step changes can be made in prokaryotic systematics by “embracing the genome”. Further, it can be anticipated that phylogenetic and taxogenomic procedures will revolutionise the classification and identification of other taxonomically complex actinobacterial taxa, notably the genus Streptomyces. Indeed, genome based classification of prokaryotes are likely to become the norm as increasing numbers of whole genomes become available, especially through co-ordinated projects, notably the Genome Encyclopaedia of Bacteria and Archea (GEBA; http://jgi.doe.gov/our-science/science-programs/microbial-genomics/phylogenetic-diversity/).
Methods
Bacterial strains and genome sequencing
Fifteen strains: “Corynebacterium hoagii”/R. hoagii DSM 20295T, R. corynebacterioides DSM 20151T, R. equi N1288, N1295 and N1301, R. gordoniae DSM 44689T, R. kroppenstedtii DSM 44908T, R. kunmingensis DSM 45001T, R. opacus DSM 43205T, R. phenolicus DSM 44812T, R. pyridinivorans DSM 44555T, R. qingshengii JCM 15477T, R. rhodocorous DSM 43241T, R. ruber DSM 43338T and Rhodococcus strain AJR001were cultured in 5 ml Brain-Heart Infusion broth (Oxoid) at 28 °C for 48 hours. Genomic DNA was extracted from 1.5 ml culture of each of the strain using an UltraClean® Microbial DNA Isolation Kit (MoBio).
The genome sequencing of R. kunmingensis DSM 45001T, R. equi strains N1288, N1295 and N1301were performed on a Roche GS Junior instrument and reads were assembled into contigs using the GS de novo assembler (Roche) and previously defined criteria34. The remaining genomes were sequenced on an Illumina MiSeq instrument and the reads were assembled using the CLC Genomic Workbench (Qiagen), as previously defined33. The whole genome shotgun sequences of all the strains have been deposited at DDBJ/EMBL/GenBank, the accession numbers are provided in the Supplementary Table 1.
The genome sequences of R. equi C7T and R. defluvii Ca11T that we have previously sequenced were also included in the analyses33,34. We also obtained 59 genome sequences of 14 rhodococcal species and 25 genomes of unclassified rhodococci from GenBank (Supplementary Table 1). Representative strains of the genera Gordonia, Nocardia, Segniliparus, Smardigococcus, Tomitella and Williamsia were also included together with two C. diphtheriae genomes55,56 as an outgroup (Supplementary Table 1).
Computational analyses
A BLAST-based pairwise average nucleotide identity (ANIb) was calculated from the nucleotide sequences using Jspecies57. A matrix of whole genome BLAST-based similarity scores was generated using GEGENEES45 using the fast algorithm with a BLAST fragment size of 500 bp. All 115 genome sequences were annotated using the RAST pipeline35 to give an equivalence of annotation for the comparative genomic analyses. A subset of amino acids from 400 broadly conserved proteins in prokaryotes was extracted for phylogenetic reconstruction using PhyloPhlAn38 with modified MUSCLE58 section to compute 16 iterations for refinement of multiple sequence alignment. The best fit substitution model was selected for the final alignment of 3,797 amino acids (VT + F + G4) and a maximum likelihood (ML) tree was generated with 1,000 SH-aLRT (SH-like approximate likelihood ratio test) and ultrafast bootstrap iterations using IQ-Tree59,60.
The annotated genome sequences were compared using EDGAR36 to calculate the core genome and the number of genes shared within each phylogenetic group. For a more comprehensive phylogenetic reconstruction, the nucleotide sequences of 255 core genes were concatenated after removing start and stop codons. A codon based alignment was performed on the concatenated sequence using MUSCLE58 in MEGA61 with 2 iterations due to computational constraints. The codons with the missing data were striped and a ML tree was generated using the best fit codon substitution model (SCHN05 + F + I + G4) with 1,000 SH-aLRT and ultrafast bootstrap replicates using IQ-Tree59,60. Another ML tree was constructed using the LG + F + I + G4 amino acid substitution model and 10,000 SH-aLRT and ultrafast bootstrap iterations59,60 from a concatenated protein sequence alignment of the core genes after removing the sites with missing data and poorly aligned regions using GBLOCKS62.
16S rRNA sequences were extracted from 107 of the 115 genomes where the size of the annotated gene was ≥1000 bp. The sequences were aligned using MUSCLE58 and the gaps were removed using GBLOCKS62, resulting in 641 most reliable sites in the final alignment. A ML tree was constructed using the GTR + I + G4 model with 10,000 SH-aLRT and ultrafast bootstrap iterations using IQ-Tree59,60. All phylogenetic trees were visualized using the web based program, Interactive Tree Of Life (iTOL)63.
The digital DNA-DNA hybridization values were calculated using GGDC 2.139 between representatives of each of the groups that were identified in the phylogenetic and other genomic analyses. A 3D plot from the GC content, genome sizes and the fraction of shared genes within each rhodococcal group (Supplementary Table 1) was generated using PAST64.
Additional Information
How to cite this article: Sangal, V. et al. Next-generation systematics: An innovative approach to resolve the structure of complex prokaryotic taxa. Sci. Rep. 6, 38392; doi: 10.1038/srep38392 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
The authors would like to thank the NU-OMICS facility for assistance in genome sequencing and Jimmy Gibson for IT assistance.
Footnotes
Author Contributions V.S., M.G., P.A.H. and I.C.S. conceived and designed the study. A.L.J. contributed selected strains for sequencing and phenotypic data on rhodococcal species-groups. V.S., E.C.S. and J.B. analysed the genomic data. All authors were involved in drafting and finalizing the manuscript.
References
- Kallmeyer J., Pockalny R., Adhikari R. R., Smith D. C. & D’Hondt S. Global distribution of microbial abundance and biomass in subseafloor sediment. Proc Natl Acad Sci USA 109, 16213–16216, doi: 10.1073/pnas.1203849109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman W. B., Coleman D. C. & Wiebe W. J. Prokaryotes: the unseen majority. Proc Natl Acad Sci USA 95, 6578–6583 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarza P. et al. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol 12, 635–645, doi: 10.1038/nrmicro3330 (2014). [DOI] [PubMed] [Google Scholar]
- Kim M., Oh H. S., Park S. C. & Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64, 346–351, doi: 10.1099/ijs.0.059774-0 (2014). [DOI] [PubMed] [Google Scholar]
- Oren A. & Garrity G. M. Then and now: a systematic review of the systematics of prokaryotes in the last 80 years. Antonie van Leeuwenhoek 106, 43–56, doi: 10.1007/s10482-013-0084-1 (2014). [DOI] [PubMed] [Google Scholar]
- Sneath P. H. A. Numerical taxonomy. In: Bergey’s Manual of Systematic Bacteriology Vol. 1 (eds Boone D. R., Castenholz R. W. & Garrity G. M. ) 39–42 (Springer-Verlag, 2001). [Google Scholar]
- Achtman M. & Wagner M. Microbial diversity and the genetic nature of microbial species. Nat Rev Microbiol 6, 431–440, doi: 10.1038/nrmicro1872 (2008). [DOI] [PubMed] [Google Scholar]
- Moore E. R., Mihaylova S. A., Vandamme P., Krichevsky M. I. & Dijkshoorn L. Microbial systematics and taxonomy: relevance for a microbial commons. Res Microbiol 161, 430–438, doi: 10.1016/j.resmic.2010.05.007 (2010). [DOI] [PubMed] [Google Scholar]
- Park G. et al. Evaluation of four methods of assigning species and genus to medically important bacteria using 16S rRNA gene sequence analysis. Microbiol Immunol 59, 285–298, doi: 10.1111/1348-0421.12254 (2015). [DOI] [PubMed] [Google Scholar]
- Sangal V., Nieminen L., Tucker N. P. & Hoskisson P. A. Revolutionising systematics through next-generation sequencing. In: Methods in Microbiology: Bacterial taxonomy Vol. 41 (eds Goodfellow M., Sutcliffe I. C. & Chun J. ) Ch. 5, 75–101 (Elsevier, 2014). [Google Scholar]
- Hedlund B. P., Dodsworth J. A. & Staley J. T. The changing landscape of microbial biodiversity exploration and its implications for systematics. Syst Appl Microbiol 38, 231–236, doi: 10.1016/j.syapm.2015.03.003 (2015). [DOI] [PubMed] [Google Scholar]
- Chun J. & Rainey F. A. Integrating genomics into the taxonomy and systematics of the Bacteria and Archaea. Int J Syst Evol Microbiol 64, 316–324, doi: 10.1099/ijs.0.054171-0 (2014). [DOI] [PubMed] [Google Scholar]
- Rosselló-Móra R. & Amann R. Past and future species definitions for Bacteria and Archaea. Syst Appl Microbiol 38, 209–216, doi: 10.1016/j.syapm.2015.02.001 (2015). [DOI] [PubMed] [Google Scholar]
- Sutcliffe I. C. Challenging the anthropocentric emphasis on phenotypic testing in prokaryotic species descriptions: rip it up and start again. Front Genet 6, 218, doi: 10.3389/fgene.2015.00218 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe I. C., Trujillo M. E. & Goodfellow M. A call to arms for systematists: revitalising the purpose and practices underpinning the description of novel microbial taxa. Antonie van Leeuwenhoek 101, 13–20, doi: 10.1007/s10482-011-9664-0 (2012). [DOI] [PubMed] [Google Scholar]
- Thompson C. C. et al. Microbial taxonomy in the post-genomic era: rebuilding from scratch? Arch Microbiol 197, 359–370, doi: 10.1007/s00203-014-1071-2 (2015). [DOI] [PubMed] [Google Scholar]
- Whitman W. B. The need for change: embracing the genome. In: Methods in Microbiology: Bacterial taxonomy Vol. 41 (eds Goodfellow M., Sutcliffe I. C. & Chun J. ) Ch. 1, 1–12 (Elsevier, 2014). [Google Scholar]
- Klenk H. P. & Göker M. En route to a genome-based classification of Archaea and Bacteria? Syst Appl Microbiol 33, 175–182, doi: 10.1016/j.syapm.2010.03.003 (2010). [DOI] [PubMed] [Google Scholar]
- Qin Q. L. et al. A proposed genus boundary for the prokaryotes based on genomic insights. J Bacteriol 196, 2210–2215, doi: 10.1128/JB.01688-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousfield I. J. & Goodfellow M. The ‘rhodochrous’ complex and its relationship with allied taxa. In: The Biology of the Nocardiae (eds Goodfellow M., Brownell G. H. & Serrano J. A. ) 39–65 (Academic Press, 1976). [Google Scholar]
- Goodfellow M., Alderson G. & Chun J. Rhodococcal systematics: problems and developments. Antonie van Leeuwenhoek 74, 3–20 (1998). [DOI] [PubMed] [Google Scholar]
- Jones A. L. & Goodfellow M. Genus IV Rhodococcus (Zopf 1891) emended.Goodfellow, Alderson and Chun 1998a. In: Bergey’s Manual of Systematic Bacteriology Vol. 5: The Actinobacteria, Part A (eds Goodfellow M. et al. ) 437–464 (Springer, 2012). [Google Scholar]
- Goodfellow M. Nocardiaceae (Castellani and Chalmers 1919) emend. Zhi, Li and Stackebrandt 2009. In: Bergey’s Manual of Systematic Bacteriology Vol. 5: The Actinobacteria, Part A (eds Goodfellow M. et al. ) 376–496 (Springer, 2012). [Google Scholar]
- Goodfellow M. & Jones A. L. Order V. Corynebacteriales ord. nov. In: Bergey’s Manual of Systematic Bacteriology Vol. 5 The Actinobacteria, Part A (eds Goodfellow, M. et al. ) 235–243 (Springer, 2012). [Google Scholar]
- Jones A. L., Sutcliffe I. C. & Goodfellow M. Prescottia equi gen. nov., comb. nov.: a new home for an old pathogen. Antonie van Leeuwenhoek 103, 655–671, doi: 10.1007/s10482-012-9850-8 (2013). [DOI] [PubMed] [Google Scholar]
- Kämpfer P., Dott W., Martin K. & Glaeser S. P. Rhodococcus defluvii sp. nov., isolated from wastewater of a bioreactor and formal proposal to reclassify [Corynebacterium hoagii] and Rhodococcus equi as Rhodococcus hoagii comb. nov. Int J Syst Evol Microbiol 64, 755–761, doi: 10.1099/ijs.0.053322-0 (2014). [DOI] [PubMed] [Google Scholar]
- Gürtler V., Mayall B. C. & Seviour R. Can whole genome analysis refine the taxonomy of the genus Rhodococcus? FEMS Microbiol Rev 28, 377–403 (2004). [DOI] [PubMed] [Google Scholar]
- Táncsics A. et al. The detection and phylogenetic analysis of the alkane 1-monooxygenase gene of members of the genus Rhodococcus. Syst Appl Microbiol 38, 1–7, doi: 10.1016/j.syapm.2014.10.010 (2015). [DOI] [PubMed] [Google Scholar]
- Creason A. L., Davis E. W., Putnam M. L., Vandeputte O. M. & Chang J. H. Use of whole genome sequences to develop a molecular phylogenetic framework for Rhodococcus fascians and the Rhodococcus genus. Front Plant Sci 5, 406, doi: 10.3389/fpls.2014.00406 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott J. F. Rhodococcus equi: an animal and human pathogen. Clin Microbiol Rev 4, 20–34 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goethals K., Vereecke D., Jaziri M., Van Montagu M. & Holsters M. Leafy gall formation by Rhodococcus fascians. Annu Rev Phytopathol 39, 27–52, doi: 10.1146/annurev.phyto.39.1.27 (2001). [DOI] [PubMed] [Google Scholar]
- de Carvalho C. C. & da Fonseca M. M. Degradation of hydrocarbons and alcohols at different temperatures and salinities by Rhodococcus erythropolis DCL14. FEMS Microbiol Ecol 51, 389–399, doi: 10.1016/j.femsec.2004.09.010 (2005). [DOI] [PubMed] [Google Scholar]
- Sangal V. et al. Genomic analyses confirm close relatedness between Rhodococcus defluvii and Rhodococcus equi (Rhodococcus hoagii). Arch Microbiol 197, 113–116, doi: 10.1007/s00203-014-1060-5 (2015). [DOI] [PubMed] [Google Scholar]
- Sangal V., Jones A. L., Goodfellow M., Sutcliffe I. C. & Hoskisson P. A. Comparative genomic analyses reveal a lack of a substantial signature of host adaptation in Rhodococcus equi (“Prescottella equi”). Pathog Dis 71, 352–356, doi: 10.1111/2049-632X.12126 (2014). [DOI] [PubMed] [Google Scholar]
- Aziz R. K. et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9, 75, doi: 10.1186/1471-2164-9-75 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom J. et al. EDGAR: a software framework for the comparative analysis of prokaryotic genomes. BMC Bioinformatics 10, 154, doi: 10.1186/1471-2105-10-154 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. X. et al. Rhodococcus kunmingensis sp. nov., an actinobacterium isolated from a rhizosphere soil. Int J Syst Evol Microbiol 58, 1467–1471, doi: 10.1099/ijs.0.65673-0 (2008). [DOI] [PubMed] [Google Scholar]
- Segata N., Bornigen D., Morgan X. C. & Huttenhower C. PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat Commun 4, 2304, doi: 10.1038/ncomms3304 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auch A. F., Klenk H. P. & Göker M. Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand Genomic Sci 2, 142–148, doi: 10.4056/sigs.541628 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auch A. F., von Jan M., Klenk H. P. & Göker M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci 2, 117–134, doi: 10.4056/sigs.531120 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis K. T., Ramette A. & Tiedje J. M. The bacterial species definition in the genomic era. Philos Trans R Soc Lond B 361, 1929–1940 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis K. T. & Tiedje J. M. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA 102, 2567–2572, doi: 10.1073/pnas.0409727102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N. & Galperin M. Y. A genomic update on clostridial phylogeny: Gram-negative spore formers and other misplaced clostridia. Environ Microbiol 15, 2631–2641, doi: 10.1111/1462-2920.12173 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey F. A., Burghardt J., Kroppenstedt R. M., Klatte S. & Stackebrandt E. Phylogenetic analysis of the genera Rhodococcus and Nocardia and evidence for the evolutionary origin of the genus Nocardia from within the radiation of Rhodococcus species. Microbiology 141, 523–528 (1995). [Google Scholar]
- Agren J., Sundstrom A., Hafstrom T. & Segerman B. Gegenees: fragmented alignment of multiple genomes for determining phylogenomic distances and genetic signatures unique for specified target groups. PLoS One 7, e39107, doi: 10.1371/journal.pone.0039107 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goris J. et al. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57, 81–91, doi: 10.1099/ijs.0.64483-0 (2007). [DOI] [PubMed] [Google Scholar]
- Konstantinidis K. T. & Tiedje J. M. Towards a genome-based taxonomy for prokaryotes. J Bacteriol 187, 6258–6264, doi: 10.1128/JB.187.18.6258-6264.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Rivera L. D. & Konstantinidis K. T. Bypassing cultivation to identify bacterial species. ASM Microbe Magazine 9, 111–118 (2014). [Google Scholar]
- Anastasi E. et al. Pangenome and phylogenomic analysis of the pathogenic actinobacterium Rhodococcus equi. Genome Biol Evol, doi: 10.1093/gbe/evw222 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A. L., Sutcliffe I. C. & Goodfellow M. Proposal to replace the illegitimate genus name Prescottia Jones et al. 2013 with the genus name Prescottella gen. nov. and to replace the illegitimate combination Prescottia equi Jones et al. 2013 with Prescottella equi comb. nov. Antonie van Leeuwenhoek 103, 1405–1407, doi: 10.1007/s10482-013-9924-2 (2013). [DOI] [Google Scholar]
- Goodfellow M., Sangal V., Jones A. L. & Sutcliffe I. C. Charting stormy waters: A commentary on the nomenclature of the equine pathogen variously named Prescottella equi, Rhodococcus equi and Rhodococcus hoagii. Equine Vet J, doi: 10.1111/evj.12399 (2015). [DOI] [PubMed] [Google Scholar]
- Salifu S. P. et al. Genome and proteome analysis of phage E3 infecting the soil-borne actinomycete Rhodococcus equi. Environ Microbiol Rep 5, 170–178, doi: 10.1111/1758-2229.12028 (2013). [DOI] [PubMed] [Google Scholar]
- de La Pena-Moctezuma A., Prescott J. F. & Goodfellow M. Attempts to find phenotypic markers of the virulence plasmid of Rhodococcus equi. Can J Vet Res 60, 29–33 (1996). [PMC free article] [PubMed] [Google Scholar]
- Goodfellow M., Beckham A. R. & Barton M. D. Numerical classification of Rhodococcus equi and related actinomycetes. J Appl Bacteriol 53, 199–207 (1982). [DOI] [PubMed] [Google Scholar]
- Sangal V., Tucker N. P., Burkovski A. & Hoskisson P. A. The draft genome sequence of Corynebacterium diphtheriae bv. mitis NCTC 3529 reveals significant diversity between the primary disease-causing biovars. J Bacteriol 194, 3269, doi: 10.1128/JB.00503-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangal V., Tucker N. P., Burkovski A. & Hoskisson P. A. Draft genome sequence of Corynebacterium diphtheriae biovar intermedius NCTC 5011. J Bacteriol 194, 4738, doi: 10.1128/JB.00939-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M. & Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106, 19126–19131, doi: 10.1073/pnas.0906412106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32, 1792–1797, doi: 10.1093/nar/gkh340 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh B. Q., Nguyen M. A. & von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol 30, 1188–1195, doi: 10.1093/molbev/mst024 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L. T., Schmidt H. A., von Haeseler A. & Minh B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32, 268–274, doi: 10.1093/molbev/msu300 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G. & Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol 33, 1870–1874, doi: 10.1093/molbev/msw054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G. & Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56, 564–577, doi: 10.1080/10635150701472164 (2007). [DOI] [PubMed] [Google Scholar]
- Letunic I. & Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44, W242–W245. doi: 10.1093/nar/gkw290 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer Ø., Harper D. A. T. & Ryan P. D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol Electron 4, 9 pp (2001). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.