Abstract
Cyclins are primary regulators of the activity of cyclin-dependent kinases, which are known to play critical roles in controlling eukaryotic cell cycle progression. While there has been extensive research on cell cycle mechanisms and cyclin function in animals and yeasts, only a small number of plant cyclins have been characterized functionally. In this paper, we describe an exhaustive search for cyclin genes in the Arabidopsis genome and among available sequences from other vascular plants. Based on phylogenetic analysis, we define 10 classes of plant cyclins, four of which are plant-specific, and a fifth is shared between plants and protists but not animals. Microarray and reverse transcriptase-polymerase chain reaction analyses further provide expression profiles of cyclin genes in different tissues of wild-type Arabidopsis plants. Comparative phylogenetic studies of 174 plant cyclins were also performed. The phylogenetic results imply that the cyclin gene family in plants has experienced more gene duplication events than in animals. Expression patterns and phylogenetic analyses of Arabidopsis cyclin genes suggest potential gene redundancy among members belonging to the same group. We discuss possible divergence and conservation of some plant cyclins. Our study provides an opportunity to rapidly assess the position of plant cyclin genes in terms of evolution and classification, serving as a guide for further functional study of plant cyclins.
Progression of the eukarytotic cell cycle is primarily controlled by a family of Ser/Thr protein kinases known as cyclin-dependent kinases (CDKs). The catalytic activity of CDKs is dependent on cyclin binding and activation, and can be further regulated by several additional mechanisms. These include protein phosphorylation/dephosphorylation, direct binding of CDK inhibitor protein (CKI) and CDK subunit (CKS), proteolysis, and intracellular trafficking (Morgan, 1995; Bourne et al., 1996; King et al., 1996; Nakayama, 1998; Peters, 1998; Rossi and Varotto, 2002). The first cyclins were identified in marine invertebrates as proteins with an oscillating abundance during the cell cycle (Evans et al., 1983). Subsequent studies indicated that cyclins, as essential regulators of CDKs, not only activate CDKs by changing the conformation at their catalytic sites, but also contribute to the selection of CDK substrates, subcellular localization, and regulation of protein stability (Booher et al., 1989; Peeper et al., 1993; Mironov et al., 1997; Potuschak and Doerner, 2001; Criqui and Genschik, 2002).
The basic cell cycle machinery appears to be conserved in all eukaryotes (Nasmyth, 1996; Novak et al., 1998). A variety of cyclins, CDKs, CKIs, and homologs of the retinoblastoma protein and the E2F transcription factors have been identified in both animals and plants (Mironov et al., 1999). In multicellular organisms such as animals and plants, development requires spatial and temporal control of cell division, so the cell cycle must be integrated into a complex system of histogenesis and organogenesis. In particular, the fact that plant cells are not mobile means that local cell division is critical for morphogenesis (Meijer and Murray, 2001). In addition, plants have several characteristics that are different from animals, including postembryonic organogenesis, indeterminate growth, sessile life style, and dramatic alteration of growth and development in response to environmental changes. Therefore, it is reasonable to postulate that plant-specific regulatory pathway(s) of cell division may exist. The current knowledge of the molecular regulatory mechanisms of cell cycle progression is primarily based on results from yeast and Drosophila genetics, as well as animal biology and biochemistry (Okayama et al., 1996). Compared to the extensive studies of cell division mechanisms in yeast and animals (Nigg, 1993; Chen et al., 2000; Fung et al., 2002), the mechanisms underlying the plant cell cycle are just beginning to be understood (De Veylder et al., 2003).
A large number of cyclin genes have been cloned from various organisms. On the basis of sequence similarity, expression pattern, and protein activity during the cell cycle, cyclins have been grouped into several classes. In animals, at least 13 classes (A to L and T) of cyclins have been described (Nakamura et al., 1995; Pines, 1995). Since the first discovery of plant cyclin genes in 1991 (Hata et al., 1991), more than 60 cyclin genes have been isolated from various plant species (Renaudin et al., 1996; Ito, 2000). According to their sequence similarity to animal cyclins, these cyclins have been classified as A-, B-, C-, D-, H-, and L-type cyclins (Renaudin et al., 1996; Yamaguchi et al., 2000; Barroco et al., 2003). Some cyclin genes, such as SOLO DANCERS (SDS) from Arabidopsis, are distantly related to all other cyclins (Azumi et al., 2002), suggesting that they may belong to distinct lineages. However, detailed phylogenetic analysis and classification of plant cyclins are still lacking.
To provide clues to the relationship between the cell cycle and the regulatory mechanism underlying plant development, it is important to investigate the spatial expression patterns of key cell cycle regulators, including cyclins. There have been some studies describing the expression of some Arabidopsis cyclin genes during the cell cycle (Mironov et al., 1999; Richard et al., 2001; Menges et al., 2002). The spatial-temporal expression patterns of a few Arabidopsis cyclin genes have been analyzed, including the studies of developmental expression pattern of CycA2;1 and CycB1;1 (Ferreira et al., 1994; Colon-Carmona et al., 1999; Burssens et al., 2000). However, no systematic study of expression patterns of the majority of Arabidopsis cyclin genes has been reported.
The available information on animal and plant cyclins raises several questions. Do plant genomes encode the same classes of cyclins as in animals? How are plant and animal cyclins related to each other? Do the genes with similar sequences and close phylogenetic relationships also have similar expression patterns? What fraction of cyclin genes might be functionally redundant? Recently, whole genome sequences of Arabidopsis and rice have been published, providing an excellent opportunity to study plant cyclins extensively (Arabidopsis Genome Initiative, 2000; Goff et al., 2002; Yu et al., 2002). In this paper, we describe an extensive search for Arabidopsis cyclins and phylogenetic studies of these proteins. Furthermore, we report results from expression analyses of these genes using both microarray and reverse transcription (RT)-PCR methods. We present phylogenetic analyses of Arabidopsis cyclins and their putative ortholog(s) from other plants. Our results indicate that flowering plants possess 10 classes of cyclins, including five classes that have not been found in animals. In addition, some phylogenetically related genes in Arabidopsis exhibit very similar expression patterns, suggesting potential functional redundancy among them. Moreover, we discuss evolutionary implications of our analyses.
RESULTS
The Arabidopsis Genome Codes for at Least 50 Cyclin-Like Proteins
To identify cyclin genes in the Arabidopsis genome, BLAST searches were performed against the Arabidopsis AGI protein database (see “Materials and Methods”). Since different types of cyclins have very low levels of sequence similarity, we used representatives of all previously published cyclins from plants and animals as query sequences, with a cutoff of the E-value at 1e-005. Our BLAST searches identified several distinctive classes of cyclins in Arabidopsis. Members within the same class are usually very similar and could be detected easily during the BLAST search, while members of other classes were often not detected. For example, when we used any of the A- or B-type cyclins as a query, we could not find cyclins other than these two types. Conversely, when we used a cyclin of another type as a query, such as SDS or a D- or H-type cyclin, rarely could we find an A- or B-type cyclin with an E-value lower than 1e-005. These findings suggest that the Arabidopsis genome encodes divergent types of cyclins. For this reason, we used all detected Arabidopsis cyclins, as well as sequences that had E-values greater than 1e-005, as queries for further searches until we no longer recovered any new cyclin-like sequences. Sequences that lack a cyclin domain according to Pfam domain analysis were eliminated before further analysis.
After BLAST search and Pfam domain analysis, a total of 50 putative cyclin proteins (referred to as cyclins hereafter for convenience) were obtained from the Arabidopsis protein database. Thirty-six of these have been described previously (Renaudin et al., 1996; Yamaguchi et al., 2000; Barroco et al., 2003) and 14 are new (see Supplemental Table I). Previous studies indicated that cyclins contain a conserved 250-amino acid region called cyclin core (Nugent et al., 1991), which has two domains: cyclin_N and cyclin_C. The cyclin_N domain is about 100-amino acid long and contains the CDK-binding site; it is also called the cyclin box and is found in all known cyclins. The cyclin_C domain is less conserved. Some known cyclins, such as the human cyclins G1 and G2, have the cyclin_N domain but not the cyclin_C domain (Horne et al., 1996), suggesting that the cyclin_C domain may not be critical for function. All 50 Arabidopsis cyclin proteins contain one or both of the cyclin_N and cyclin_C domains. Forty-nine of the 50 proteins contain the cyclin_N domain; 31 of these also have the cyclin_C domain, whereas the remaining 18 have only the cyclin_N domain (Fig. 2). The CycD5;1 protein sequence in The Arabidopsis Information Resource database contains only the cyclin_N domain. However, a careful inspection of its coding sequence suggests that a frameshift mutation may have occurred downstream of the cyclin_N domain, resulting in the loss of the cyclin_C domain due to truncation (this was confirmed by PCR and sequencing; data not shown). One of the 50 proteins, encoded by the At2g41830 locus, lacks the cyclin_N domain, although a divergent cyclin_C domain (E-value = 0.00019) is present in this protein. Because the cyclin_N domain is present in all animal cyclins and most Arabidopsis ones and is more highly conserved than the cyclin_C domain, we decided to use cyclin_N for phylogenetic analysis. Therefore, At2g41830 was not included in the dataset for phylogenetic analysis.
Figure 2.
Phylogenetic relationships and domain structure of the 49 Arabidopsis cyclins. With 0.01 as the E-value cutoff, almost all of the A-, B-, D-, and SDS-type of cyclins contain both cyclin_N and cyclin_C domains, whereas all other types of cyclins lack cyclin_C domain. D-box was found in nine A- and nine B-cyclins.
The Arabidopsis Cyclins Can Be Grouped into 10 Types
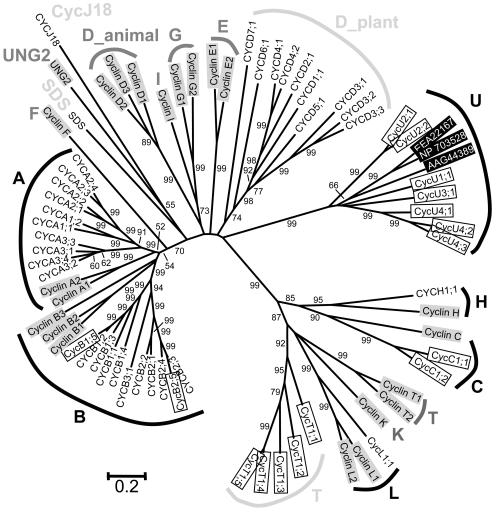
It is known that the human genome codes for at least 22 cyclins, which have been divided into 13 types on the basis of function and sequence analysis (Nakamura et al., 1995; Pines, 1995; Fig. 1). To gain an understanding of the evolutionary relationship between Arabidopsis cyclins and nonplant cyclins and to identify a basis for classifying newly uncovered members, phylogenetic analysis was performed for cyclins from the Arabidopsis and human genomes. Preliminary phylogenetic results (not shown) suggest that Arabidopsis may have four types of cyclins that do not have corresponding human cyclins. These putative novel types were used as queries to search the National Center for Biotechnology Information nonredundant database and several protist cyclins were recovered. Two representatives of the protist cyclins were included in the final phylogenetic analysis to provide additional nonplant comparisons for Arabidopsis cyclins. Our results confirm the previous designation of the A-, B-, C-, H-, and L-type cyclins in Arabidopsis and support their orthologous relationships with the respective human cyclins (Fig. 1). In addition, several newly identified genes are well-supported members of these known types: CycB1;5 (At1g34460), CycB2;5 (At1g20590), CycC1;1 (At5g48640), and CycC1;2 (At5g48630).
Figure 1.
Unrooted NJ tree of the Arabidopsis and human cyclins, with bootstrap values higher than 50% shown for each clade. Ten and 13 families of cyclins are recognizable in Arabidopsis and human, respectively, five of which (A, B, C, H, and L) are shared by both species. The remaining five types of Arabidopsis cyclins are named as CycJ18-, D_plant-, T-, SDS-, and L-type, respectively. Twenty-two proteins from human are highlighted by gray boxes. Three proteins from protists are in inverted boxes: NP_703528, EAA22167, and AAG44389 from Plasmodium falciparum, P. yoelii yoelii, and Trypanosoma cruzi, respectively. Eighteen newly named Arabidopsis cyclins are in open boxes.
However, although the Arabidopsis D-type cyclins probably form a single clade (see below for further analysis of the D-type cyclins), an orthologous relationship between Arabidopsis and human D-type cyclins is not supported by the phylogenetic analysis. Therefore, we regard the plant D-type cyclins as plant specific and designate them members of the D_plant type. Three other Arabidopsis cyclin genes were previously designated as CycT or CycT-like because they were found to be similar in sequence and possibly in function to animal T-type cyclins (Barroco et al., 2003). However, our analysis indicates that they and two newly identified genes form a separate clade related to the human K/L/T-types, although not orthologous to them. Nevertheless, because of the possible functional similarity and to avoid nomenclature confusion, we have kept the T-type designation. In addition, the phylogenetic analysis also supports the designation of another new type of Arabidopsis cyclins, called U-type, which is related to cyclins from protists, but lack close homologs in animals (Fig. 1 and data not shown). Therefore, this may represent an ancient type that has been lost in the animal lineage. Pfam analysis indicated that cyclin_C domain is absent from all of the T- and U-type cyclins.
Phylogenetically, A- and B-type cyclins are more closely related to each other than to other types. Also, the human K/L/T- and Arabidopsis T-types of cyclins are distantly related to the C- and H-type cyclins, whereas the U-type cyclins might be even more distantly related. In addition to these and the D-type cyclins, there are two more cyclins, SDS and CycJ18 (Abrahams et al., 2001; Azumi et al., 2002), which are quite isolated from others and thus need to be treated as two separate classes. In this paper, we call them SDS- and CycJ18-type cyclins, respectively (Fig. 1). Our analysis also indicates that Arabidopsis lacks clear orthologs of the human E-, F-, and G-type cyclins, and the UNG2 (Uracil DNA Glycosylase 2) protein. Our maximum parsimony (MP) trees based on the same data set are not in conflict with the neighbor joining (NJ) tree shown in Fig. 1, but have lower bootstrap values for a number of nodes (data not shown).
In addition to the cyclin core, some cyclins also contain a Destruction box (D-box), which is involved in cyclin proteolysis by the ubiquitin-dependent proteasome pathway (Glotzer et al., 1991; Renaudin et al., 1996; Peters, 1998; Vandepoele et al., 2002). Some cyclins may have another motif called PEST region, which is rich in Pro (P), Glu (E), Ser (S), and Thr (T) residues, and is a marker for unstable proteins (Rogers et al., 1986; Rechsteiner and Rogers, 1996). The presence of these motifs is consistent with cyclin function, which requires rapid degradation to terminate CDK activity at a specific point during the cell cycle. In this study, putative D-boxes were found in all A-type cyclins except for CycA3;3 and all B-type cyclins except for CycB2;5 and CycB3;1 (Fig. 2). Putative PEST regions were also found in 45 out of the 49 Arabidopsis cyclins (Supplemental Table I).
Expression Profiles of Arabidopsis Cyclin Genes
Since gene expression patterns can provide important clues for gene function, we employed both microarray and RT-PCR experiments to comprehensively characterize gene expression profiles of the Arabidopsis cyclin genes. Out of the 49 Arabidopsis cyclin genes, the probes of 43 are available in the GeneChip Arabidopsis ATH1 Genome Array (Affymetrix, Santa Clara, CA). The average normalized signal strengths of the genes are presented in Table I. As described by X. Zhang et al. (unpublished data), if the average of Log Data of duplicate ([Log2 X1 + Log2X2]/2) of a gene is smaller than Log2 50, the expression of that gene is regarded as not reliably detected (designated as “A”). Twenty-five of the 43 genes show expression in all the tissues examined; 10 other genes are expressed in several tissues, but are detected at very low levels or not at all in others. Three genes have very specific expression patterns, with CycU2;2 and CycU4;3 reliably detected only in roots, and SDS in anthers. The expression of five remaining genes, namely CycA2;1, CycA3;3, CycD4;2, CycD6;1, and CycU4;2, was not detected with confidence in any of the tissues examined.
Table I.
Microarray data for 43 Arabidopsis cyclin genes
| Gene Name | Lf | Rt | St | At | If | S12 | Sl |
|---|---|---|---|---|---|---|---|
| CYCA1;1 | 61.16 | 111.21 | 237.78 | 585.71 | 1015.40 | 278.99 | 132.36 |
| CYCA1;2 | 93.26 | 50.76 | A | 231.20 | 120.75 | 73.73 | 64.73 |
| CYCA2;1 | A | A | A | A | A | A | A |
| CYCA2;2 | 50.40 | 70.92 | 105.46 | 204.06 | 135.70 | 58.95 | 77.46 |
| CYCA2;3 | 84.78 | 85.36 | 94.67 | 233.34 | 347.50 | 121.73 | 141.53 |
| CYCA2;4 | 80.67 | 129.26 | 83.37 | 174.35 | 435.55 | 197.10 | 151.59 |
| CYCA3;1 | 70.51 | 75.63 | 161.80 | 213.04 | 308.10 | 143.95 | 109.20 |
| CYCA3;2 | 173.21 | 232.03 | 443.66 | 314.33 | 376.12 | 259.35 | 238.54 |
| CYCA3;3 | A | A | A | A | A | A | A |
| CYCA3;4 | 85.40 | 81.61 | 92.97 | 147.36 | 211.75 | 98.49 | 89.51 |
| CYCB1;1 | A | 60.48 | 65.68 | 373.84 | 606.44 | 159.36 | 75.95 |
| CYCB1;3 | 79.61 | 186.89 | 199.46 | 361.86 | 462.58 | 114.82 | 171.46 |
| CYCB1;4 | 63.66 | 204.00 | 192.55 | 409.66 | 548.98 | 162.97 | 159.09 |
| CYCB1;5 | A | 65.46 | 57.79 | 340.22 | 384.61 | 79.85 | 50.15 |
| CYCB2;1 | A | 69.37 | 60.06 | 158.43 | 217.62 | 64.89 | 60.94 |
| CYCB2;2 | A | 50.36 | 85.49 | 289.34 | 278.10 | 62.61 | A |
| CYCB2;4 | A | 59.24 | 80.70 | 236.77 | 391.24 | 85.29 | 56.92 |
| CYCB2;5 | A | 67.53 | 65.90 | 363.97 | 307.71 | 183.49 | 80.86 |
| CYCB3;1 | 87.06 | 92.35 | 94.89 | 326.01 | 277.78 | 86.05 | 86.81 |
| CYCC1;1 | 116.20 | 115.10 | 115.07 | 252.60 | 278.43 | 220.50 | 194.30 |
| CYCC1;2 | 277.69 | 182.99 | 298.36 | 306.71 | 310.78 | 261.33 | 220.91 |
| CYCD1;1 | A | 64.34 | 388.12 | 313.84 | 320.91 | 261.45 | 308.58 |
| CYCD2;1 | 244.04 | 263.86 | 183.39 | 187.88 | 117.56 | 243.34 | 166.11 |
| CYCD3;1 | 80.84 | 229.41 | 184.20 | 471.59 | 984.59 | 281.69 | 223.30 |
| CYCD3;2 | 430.59 | 218.61 | 484.88 | 1207.75 | 1904.40 | 1066.72 | 731.27 |
| CYCD3;3 | 157.38 | 214.08 | 145.72 | 644.98 | 1825.93 | 760.65 | 426.65 |
| CYCD4;1 | A | A | A | 94.74 | 77.94 | 61.53 | A |
| CYCD4;2 | A | A | A | A | A | A | A |
| CYCD5;1 | 130.33 | 64.07 | 100.85 | 97.49 | 137.50 | 82.77 | 74.31 |
| CYCD6;1 | A | A | A | A | A | A | A |
| CYCH;1 | 60.01 | 63.64 | 65.82 | 100.10 | 153.77 | 74.52 | 73.47 |
| CYCT1;3 | 223.51 | 315.81 | 240.95 | 329.95 | 340.42 | 367.21 | 234.20 |
| CYCT1;4 | 133.58 | 200.93 | 138.63 | 236.31 | 229.62 | 190.28 | 144.74 |
| CYCT1;5 | 201.61 | 302.84 | 194.10 | 403.17 | 349.49 | 260.29 | 213.83 |
| CYCL1;1 | 323.24 | 303.93 | 217.44 | 487.00 | 369.71 | 282.94 | 235.32 |
| CYCU1;1 | 1392.74 | 51.72 | 339.69 | 201.77 | 133.75 | 162.92 | 263.68 |
| CYCU2;1 | 51.35 | 367.69 | 217.01 | A | 82.08 | 93.27 | 111.55 |
| CYCU2;2 | A | 89.49 | A | A | A | A | A |
| CYCU3;1 | 335.85 | 408.43 | 665.14 | 187.32 | 350.08 | 275.63 | 447.49 |
| CYCU4;1 | 150.79 | 157.15 | 228.05 | 76.17 | 165.93 | 292.97 | 377.70 |
| CYCU4;2 | A | A | A | A | A | A | A |
| CYCU4;3 | A | 152.21 | A | A | A | A | A |
| SDS | A | A | A | 101.60 | A | A | A |
The numbers present the average of two biological replicates. A, absence, as defined for expression below the value of 50; Lf, leaf; Rt, root; St, stem; At, anther; If, inflorescence; S12, stage 12 flower; Sl, silique.
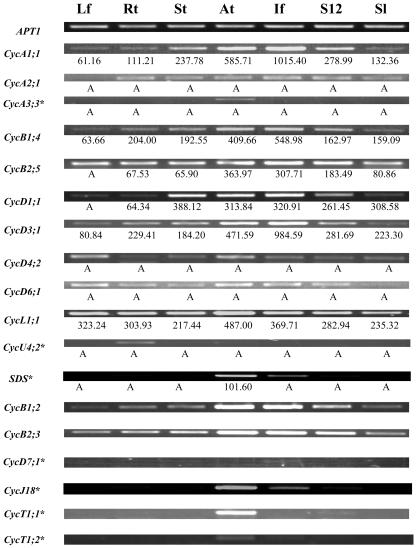
RT-PCR experiments were performed to verify the microarray data and to obtain the expression patterns of several other cyclin genes, for which probes are not available on the Affymetrix GeneChip. BLAST searches were performed to verify the specificity of all primer sequences (Table II), and the sizes of all RT-PCR bands were as expected. As shown in Figure 3, the RT-PCR results are in good agreement with the microarray data. It is worth noting that RT-PCR is very sensitive and able to detect gene expression even when a gene cannot be reliably detected by the microarray study. For example, an RNA in situ hybridization experiment has shown that the SDS gene is expressed in both male and female meiotic cells (Azumi et al., 2002), and RT-PCR indicated expression in anthers, young inflorescences, and stage-12 flowers. The young inflorescence sample contained a small fraction of stage-9 flowers, which have male meiotic cells, and the very faint band in stage-12 flowers was possibly caused by some contamination of stage-11 flowers, which contains female meiotic cells, during RNA sample preparation. RT-PCR was also performed for another five genes, which had very low (“A”) expression values (less than 50) in all the tissues tested from our microarray experiment. For CycA3;3 and CycU4;2 (At5g07450), 29 cycles of PCR yielded no band, but more PCR cycles (34 cycles) detected specific expression of CycA3;3 in anthers and CycU4;2 in roots. For the other three genes, CycD6;1 and CycD4;2 showed weak ubiquitous expression, and CycA2;1 showed broad expression except in leaf. Taken together, our results and similar studies from members in our lab (X. Zhang et al., unpublished data; W. Zhang et al., unpublished data) illustrated that the microarray data are quite reliable.
Table II.
Primers for RT-PCRs
| Gene Name
|
Primer
|
Sequence
|
Predicted Size
|
|---|---|---|---|
| bp | |||
| APT1a | OMC571 | F 5′-TCCCAGAATCGCTAAGATTGCC-3′ | 479 |
| OMC572 | R 5′-CCTTTCCCTTAAGCTCTG-3′ | ||
| CYCA1;1b | OMC1384 | F 5′-TCCTCACAAAGTTGCTTCTTCACC-3′ | 354 |
| OMC1385 | R 5′-TCAGGTCGCTTCTTAGCCTCAG-3′ | ||
| CYCA2;1 | OMC1394 | F 5′-AAACGAAGCGTGTTGCTAGACCG-3′ | 447 |
| OMC1395 | R 5′-GCACTTGCACCATATAGCTAGTTGAAGG-3′ | ||
| CYCA3;3b | OMC1165 | F 5′-GGCGGAAATTCGACATAAGCTAC-3′ | 543 |
| OMC1166 | R 5′-TGTGCAACCCTTATGAACCGTC-3′ | ||
| CYCB1;2b | OMC1171 | F 5′-GCTCCTCTCGTTGATGGTTTGAAG-3′ | 501 |
| OMC1172 | R 5′-CACCGCAGCCAAATGGTTATC-3′ | ||
| CYCB1;4 | OMC1386 | F 5′-TTGCGAAGAAGGCGAAACAAC-3′ | 467 |
| OMC1387 | R 5′-TCCCAACAGCTGAAGCTCTCTTC-3′ | ||
| CYCB2;3b | OMC1167 | F 5′-AGGACCGACAAGAAGAGCACTAAG-3′ | 430 |
| OMC1168 | R 5′-AACCGCAGCCAAAGGATTATTC-3′ | ||
| CYCB2;5b | OMC1169 | F 5′-ATGTCATTCCTTGCGGTTCATC-3′ | 450 |
| OMC1170-2 | R 5′-CATGCCAGTAGCTGTTCTTCGTTG-3′ | ||
| CYCD1;1b | OMC1370 | F 5′-CGGGTACCTTTCTCGGGTTCTTTATC-3′ | 694 |
| OMC1371 | R 5′-AATGATGAGCTTCGTCTCTCCCAAC-3′ | ||
| CYCD3;1 | OMC1390 | F 5′-TGGGACTTAAGAACAATGCTCACTGG-3′ | 516 |
| OMC1391 | R 5′-CTACGATTGCCCATGGCAGATG-3′ | ||
| CYCD4;2 | OMC1380 | F 5′-TGACGCTCTATAATGGCTGAATTTATGG-3′ | 385 |
| OMC1381 | R 5′-CCAATGATAAACAAGCAACAGCCAAC-3′ | ||
| CYCD6;1b | OMC1583 | F 5′-TACTTCCATAGCCTCAAGTCCTCTGC-3′ | 528 |
| OMC1584 | R 5′-CTCGAAAGAAGCAAAGAGAAGTGCC-3′ | ||
| CYCD7;1 | OMC1173 | F 5′-ATCTACTCTGCGAAGAATCTTGGC-3′ | 543 |
| OMC1174 | R 5′-TATAACTCGTAACCGCGTTCACAC-3′ | ||
| CYCT1;1 | OMC1175 | F 5′-GGTGACGTCGTCTTTGTCTCGTAC-3′ | 449 |
| OMC1176 | R 5′-AAGCCATGTCTCGGAACAACG-3′ | ||
| CYCT1;2 | OMC1382 | F 5′-AGCTCTCTGTTTCTTCCCGTTTGG-3′ | 694 |
| OMC1383 | R 5′-CGATGAGCCTCCTTCAATATCAGAAC-3′ | ||
| CYCL1;1 | OMC1344 | F 5′-GAAACCTACGGGCAGCAGCATACACTAC-3′ | 449 |
| OMC1345 | R 5′-GCAGGTTCTGTTTCAGCGATTCTATTGC-3′ | ||
| CYCU4;2b | OMC1376 | F 5′-CGATCAAGAACCAATGGCTGAG-3′ | 517 |
| OMC1377 | R 5′-TGGAGAGTACATCGTCCTCATAACC-3′ | ||
| SDSc | OMC1164 | F 5′-TCTAGTTTCAAGCTTTCGTACGGAG-3′ | 1,144 |
| OMC700-2 | R 5′-TGGATCCCTTAGCACAGTCCCGCATATA-3′ | ||
| CYCJ18 | OMC1378 | F 5′-GCAGAACATTGGCTATTGCAACC-3′ | 442 |
| OMC1379 | R 5′-GGAATTCGTACTGCTGTTTAGGCAC-3′ |
PCR annealing temperature is 56°C for this gene.
PCR annealing temperature is 58°C for these genes, and it is 60°C for all the other genes.
PCR extension time is 1 h 20 min for these genes, and it is 50 min for all the other genes.
Figure 3.
Analysis of expression of some cyclin genes using RT-PCR. The APT1 gene was used as an internal control. Thirty-four cycles PCR reactions were performed for those genes with asterisks (*), while 29 cycles were done for all the other genes. The microarray data, if available, are listed below the corresponding RT-PCR bands to show the general consistency of results from these two methods and the high sensitivity of RT-PCR. The abbreviation for the organs is the same as in Table I.
The microarray and RT-PCR results together present global expression profiles of all the Arabidopsis cyclins in several major tissues. Sometimes, the RT-PCR experiments detected low-level expression as faint bands when the gene was regarded as not reliably detected from the microarray analysis. In these cases, we adopted RT-PCR results due to the high sensitivity of RT-PCR. Arabidopsis cyclins can be classified into four groups according to their expression profiles. The largest group is composed of 31 genes that are expressed in all tissues examined; the second group contains eight genes whose expression were detected in the majority, but not all, of the tissues. The third group includes seven genes with very specific expression: CycA3;3, CycD7;1, CycT1;1 (At1g35440), and CycT1;2 (At4g19560) are exclusively expressed in anthers; while CycU2;2 (At3g60550), CycU4;2, and CycU4;3 (At5g61650) are exclusively expressed in roots. The last group contains SDS, CycJ18, and CycD4;1, the three genes that are expressed only in reproductive organs, i.e. anthers, inflorescences, and stage-12 flowers of this study.
Cyclins from Higher Plants
BLAST searches against the GenBank protein databases yielded more than 200 cyclin-like proteins from various plant species. A more detailed analysis (see “Materials and Methods”) revealed that approximately a quarter of them are duplicates, alleles, or partial sequences of other proteins. Some others do not contain a detectable cyclin_N domain and thus were excluded from further analysis. Among the remaining 127 proteins, the majority contains both cyclin_N and cyclin_C domains and shares high similarity to one of the A-, B-, or D-type cyclins in Arabidopsis. The rice (Oryza sativa) genome codes for at least 44 cyclins, most of which share significant similarity to at least one Arabidopsis cyclin. Because the cyclin-like proteins from other plants fall into one of the ten types revealed in Arabidopsis, we believe that our classification of the Arabidopsis cyclins into 10 classes is also valid for other plants. However, since the regions that could be used for phylogenetic analysis are very short if all members are considered, we analyzed phylogenetic relationships using sequences in five subsets, each of which contains phylogenetically related cyclins. As many as possible reliable residues, most of which are from the cyclin_N and cylin_C domains, were included into the final estimations for each subset.
A-Type Cyclins
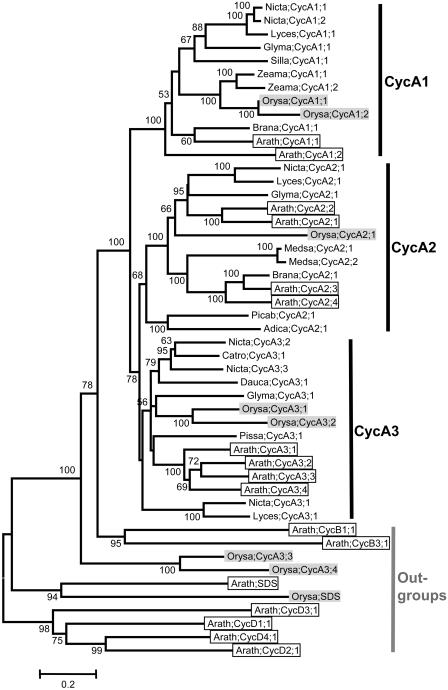
A total of 49 plant cyclins were included in our CycA_plant data set, of which 41 are A-type, two are B-type, two are SDS-type, and four are D-type cyclins. Structurally, most of the A-type cyclins contain the LVEVxEEY motif, a conserved region that has been proposed as the signature of all A-type cyclins (Renaudin et al., 1996; Chaubet-Gigot, 2000; Supplemental Table I). In some sequences, however, one or several of these residues are replaced by amino acids with very similar chemical properties. For example, the Val (V) at the second position has been replaced by an Ile (I) in Arath;CycA1;1, and the two Glus (EE) at the sixth and seventh positions by Asps (DD) in Arath;CycA2;2. Since these substitutions are not very frequent, we still regard the LVEVxEEY motif as a universal signature for A-type cyclins.
In the phylogenetic tree of the A-type cyclins, three previously identified groups, i.e. CycA1, CycA2, and CycA3, are recognizable, and the relationships between these three groups are (CycA1,(CycA2, CycA3)) (Fig. 4). Two rice cyclins, Orysa;CycA3;3 and Orysa;CycA3;4, which contain MEELVYGF and MADVAYVF, respectively, instead of the conserved LVEVxEEY motif, were not resolved as members of CycA1, CycA2, or CycA3 group; they formed a separate clade outside of the A- and B-type cyclins, suggesting that they may not be true A-type cyclins. Among the remaining 39 A-type cyclins, 11 of the 13 CycA1 and 13 of the 16 CycA3 group members possess an Ala (A) at the fifth position (the “x”), while all of the 13 CycA2 group members have a Ser (S) at this position (Supplemental Table I).
Figure 4.
NJ tree of 49 A-, two B-, two SDS-, and four D-type cyclins from plants. Forty-seven core members of A-type were clustered into three groups, i.e. CycA1, CycA2, and CycA3, while the remaining two CycA-like proteins from rice, Orasa;CycA3;3 and Orasa;CycA3;3, were resolved as sisters to all other A- and B-type cyclins. Sequences from Arabidopsis are in open boxes, while those from rice are in light shading. Cyclin nomenclature is according to Renaudin et al. (1996). Adica, Adiantum capillus; Arath, Arabidopsis; Brana, Brassica napus; Catro, Catharantus roseus; Dauca, Daucus carota; Glyma, Glycine max; Lyces, Lycopersicon esculentum; Medsa, Medicago sativa; Nicta, Nicotiana tabacum; Orysa, Oryza sativa; Picba, Picea abies; Pissa, Pisum sativum; Silla, Silene latifolia; Zeama, Zea mays. For more information, such as synonym(s), accession number, locus name, and signature motif, please see the Supplemental Table I.
As shown in Figure 4, it is also obvious that each of the CycA1, CycA2, and CycA3 groups is composed of sequences from well-studied species such as eudicotyledenous Arabidopsis, Glycine max, Nicotiana tabacum, Lycopersicon esculentum, and the monocotyledonous rice. This result strongly supports the idea that the common ancestor of each of these three groups predates the separation of monocots and eudicots. In addition, two cyclin-like proteins from the pteridophyte Adiantum capillus and the gymnospermous Picea abies, Adica;CycA2;1 and Picab;CycA2;1, were resolved as outgroups of all other angiosperm A2-type cyclins, suggesting that the split of A-type cyclins into three groups may have occurred even earlier, possibly before the emergence of vascular plants.
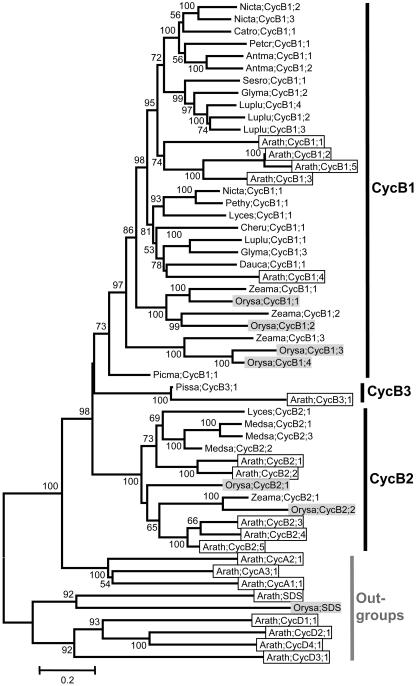
B-Type Cyclins
Our CycB-plant data set includes 45 B-type cyclins and nine outgroups. Phylogenetic analysis indicated that the plant B-type cyclins fall into three distinct groups: CycB1, CycB2, and CycB3 (Fig. 5), although the relationships among them are not well resolved. The CycB1 group, which contains more than two-thirds of the B-type cyclins, is by far the largest of the three. The typical cyclin B signature, the (H/Q)x(K/R/Q)(F/L) motif, is present in all B-type cyclins investigated. However, to make the signature motif of B-type cyclins comparable to that of A-type cyclins, we expanded the four residues mentioned above to the same eight positions found in the A-type cyclins. Interestingly, we found that the majority of plant B-type cyclins contains a L(I/V)(E/D)VHx(K/R)(F/L) motif, although significant variation does exist at some positions (Supplemental Table I). For CycB2 and CycB3 group members, the consensus sequence of this region are LIEVHxK(F/L) and LIEVHFK(F/L), respectively, while, for CycB1 group members, there are amino acid substitution at all the eight positions.
Figure 5.
NJ tree of 45 B-type cyclins from plants, with two A-, two SDS-, and four D-type cyclins as outgroups. Three major groups, CycB1, CycB2, and CycB3, are recognizable, although the relationships among them are not resolved. Sequences from Arabidopsis are in open boxes, while those from rice in light shading. Cyclin nomenclature is according to Renaudin et al. (1996). Antma, Antirrhinum majus; Arath, Arabidopsis; Catro, Catharantus roseus; Cheru, Chenopodium rubrum; Dauca, Daucus carota; Glyma, Glycine max; Luplu, Lupinus luteus; Lyces, Lycopersicon esculentum; Medsa, Medicago sativa; Nicta, Nicotiana tabacum; Orysa, Oryza sativa; Petcr, Petroselinum crispum; Pethy, Petunia x hybrida; Picma, Picea mariana; Pissa, Pisum sativum; Sesro, Sesbania rostrata; Zeama, Zea mays. For more information, such as synonym(s), accession number, locus name, and signature motif, please see the Supplemental Table I.
Like the A-type cyclins, the CycB1 and CycB2 groups also contain representatives from both monocotyledonous and eudicotyledonous species, suggesting that these two lineages were present before the split of monocots and eudicots. The placement of Picma;CycB1;1 at the base of all other B1-type cyclins further suggests that the CycB1 and CycB2 lineages predate the origin of seed plants. However, in our BLAST searches, we could not find a CycB3 group member from a monocotyledonous species, thus the evolutionary history of this group remains unclear.
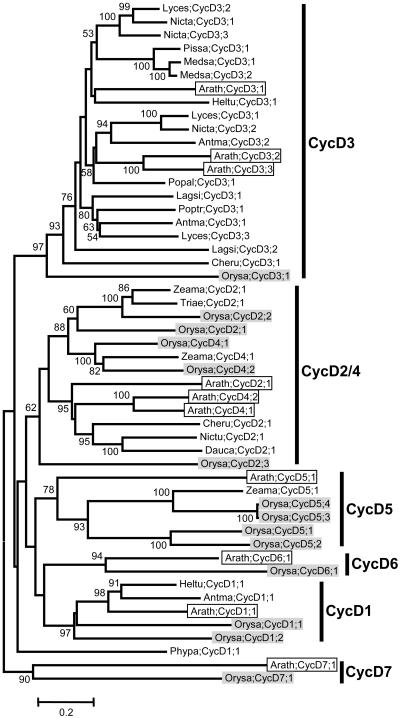
D-Type Cyclins
As indicated in Figures 1 and 2, the monophyly of the Arabidopsis D-type cyclins and the internal relationships within them were not well resolved. In particular, the Arabidopsis CycD6;1 and CycD7;1 proteins seem to be quite divergent from all other D-type cyclins. Investigation of the protein sequences also failed to find a conserved motif corresponding to the LVEVxEEY motif of A-type cyclins. Despite this, the previously reported LxCx(D/E) motif at the N terminus are present in almost all the D-type cyclins (Supplemental Table I). In the phylogenetic tree of the CycD_plant data set, six groups are recognizable, although the detailed relationships among them are obscure (Fig. 6). Within each of the CycD1, CycD3, CycD5, CycD6, and CycD7 groups, members from Arabidopsis are all clustered with their putative orthologs from other plants. Putative ortholog(s) were found from both monocotyledonous and eudicotyledonous species, suggesting that the common ancestors of these groups predates the separation of monocots and eudicots. However, distinctive orthologs could not be identified for the CycD2 and CycD4 subtypes, suggesting that the Arabidopsis CycD2 and CycD4 genes may be the result of recent duplications since the divergence of eudicots. Nevertheless, the existence of a number of cyclins related to both CycD2 and CycD4 from other eudicots and monocots support an origin of the combined CycD2/CycD4 group before the split of monocots and eudicots.
Figure 6.
Unrooted NJ tree of 51 D-type cyclins from plants. Six groups of cyclins, CycD1-, CycD2-/CycD4-, CycD3-, CycD5-, CycD6-, and CycD7-type cyclins, are recognizable, although the relationships among then are not resolved. Sequences from Arabidopsis are in open boxes, while those from rice are in light shading. Cyclin nomenclature is according to Renaudin et al. (1996). Antma, Antirrhinum majus; Arath, Arabidopsis; Cheru, Chenopodium rubrum; Dauca, Daucus carota; Hiltu, Helianthus tuberosus; Lagsi, Lagenaria siceraria; Lyces, Lycopersicon esculentum; Medsa, Medicago sativa; Nicta, Nicotiana tabacum; Orysa, Oryza sativa; Pissa, Pisum sativum; Phypa, Physcomitrella patens; Popal, Populus alba; Poptr, Populus tremula x Populus tremuloides; Triea, Triticum aestivum; Zeama, Zea mays. For more information, such as synonym(s), accession number, locus name, and signature motif, please see the Supplemental Table I.
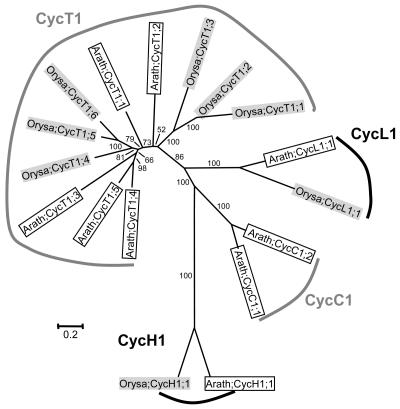
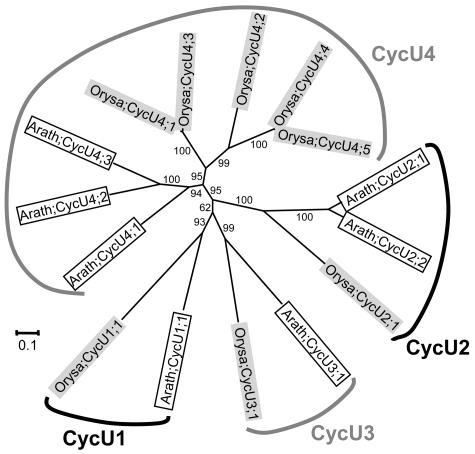
H-, L-, T-, U-, and SDS-Type Cyclins
Except for C- and CycJ18-type, cyclins of all types were also found in at least one more plant species other than Arabidopsis. However, as shown in Figures 4, 5, 7, and 8, the plant H-, L-, T-, U-, and SDS-type cyclins are detected only from Arabidopsis and rice, the two model species whose whole genomes have been sequenced. This suggests that cyclins of these types are possibly expressed either at very low levels or in specific organs, thereby reducing their chances of being represented in expressed sequence tag collections of less-well-studied species. The Arabidopsis SDS gene, for example, is specifically expressed in meiotic cells and is critical for meiosis (Azumi et al., 2002). Furthermore, since putative orthologous relationships could be detected within the SDS, CycH1, CycL1, CycT1, CycU1, CycU2, CycU3, and CycU4 groups, it is possible that the ancestor of each group predates the separation of monocots and eudicots.
Figure 7.
Unrooted NJ tree of two C-, two H-, two L-, and 11 T-type cyclins from plants, with four monophyletic groups formed for each type. Sequences from Arabidopsis are in open boxes, while those from rice are in light shading. For more information, such as synonym(s), accession number, locus name, and signature motif, please see Supplemental Table I.
Figure 8.
Unrooted NJ tree of 15 U-type cyclins from plants. Four groups, CycU1-, CycU2-, CycU3-, and CycU4-type cyclins, were formed. Sequences from Arabidopsis are in open boxes, while those from rice are in light shading. For more information, such as synonym(s), accession number, locus name, and signature motif, please see Supplemental Table I.
Compared with the A-, B-, D-, and SDS-type, cyclins of other types are generally short in length and do not contain an easily detectable cyclin_C domain. Despite this, some of them possess conserved motifs that could be regarded as a signature of each type of cyclins. Most of the newly identified U-type cyclins, for example, contain a conserved Y(L/A)(E/A)RI(F/A)(R/K)(Y/F) motif at the positions of the A-type LVEVxEEY motif (Supplemental Table I). Similarly, a relatively conserved motif, (L/I)(Q/R)D(L/V)G(M/I)RL, seems to exist in most T-type cyclins.
DISCUSSION
Plants Possess a Large and Complex Family of Cyclins
In this study, we have identified and analyzed a large number of cyclins from Arabidopsis, rice, and many other species. Our results show that the cyclin family in plants has a large number of members that can be divided into 10 groups. Nieduszynski et al. (2002) analyzed the genomic sequences of Caenorhabditis elegans, Drosophila melanogaster, and human and found one A-type cyclin in invertebrates and two in vertebrates. The number of B-type cyclins varies from two in D. melanogaster to three in humans and four in C. elegans. The total number of cyclins in the C. elegans genome is 34 (Plowman et al., 1999), and the number in human is at least 22. Plants can adopt dramatically different alternative developmental pathways and must integrate cell cycle progression, growth, and development in response to environmental cues (Boniotti and Griffith, 2002), suggesting that they may have acquired distinctive cyclin genes. The plant cell cycle also has unique features compared to the animal cell cycle, including the need to form unique cytoskeletal structures called preprophase band and phragmoplast, the formation of diffuse mitotic spindles, and distinctive cytokinesis (Cyr and Palevitz, 1995; Smith, 1999; Breyne et al., 2002). So, it is not surprising if plant genomes code for more cyclins than animal genomes. Reports of the persistence of some cyclins to late cell division (Mews et al., 1997) and the existence of a plant-unique CDK type, CDKB (Joubes et al., 2000), already provide evidence for the existence of plant-specific cyclins. However, as suggested previously (John et al., 2001), only after we collect all the information on cyclin function can we really understand the full meaning of plant cyclin diversity.
As shown in Figure 1, there are 10 and 13 classes of cyclins in Arabidopsis and human genomes, respectively. Five types (A-, B-, C-, H-, and L-types) are shared by both species, with K- and T-types being closely related to L-type. The human E-, F-, G-, I-, and UNG2-type cyclins lack clear orthologs in Arabidopsis, whereas plants have the CycJ18-, T-, SDS-, and U-type cyclins that are not found in human. Both animals and plants have D-type cyclins, but the affinity between them was not supported in our phylogenetic analysis (Fig. 1). In addition, the greater numbers of A- and B-type cyclins than those in animals and the lack of certain animal types suggest that some of the A- and B-types may assume the functions that are carried out by the animal-specific cyclins. For example, though plants lack E-type cyclins which are involved in G1/S checkpoint control in animals, it was reported recently that Nicta;CycA3;2 can control cell division and differentiation (Yu et al., 2003), functions that are analogous to those of cyclin E in animals.
Although only five types of cyclins are shared between Arabidopsis and human, all 10 types identified in Arabidopsis were supported by the existence of related cyclins in other plants. As indicated in BLAST searches and phylogenetic analyses, members within each class share significant similarity, while affinities between members of different classes are usually undetectable, except for that between A- and B-type cyclins and among K-, L-, and T-type cyclins. BLAST searches seldom yielded members from other classes, and the only region that was found in all cyclin proteins and can be confidently aligned is the cyclin_N domain. These results suggest that the collection of the plant cyclin proteins can be regarded as a superfamily that is comprised of several small families, such as A/B-, C-, D-, H-, L-, T-, U-, SDS-, and CycJ18-type cyclins, rather than a single huge protein family.
Diverse Expression Patterns of Arabidopsis Cyclins
Overall, the Arabidopsis cyclin family shows diverse expression patterns. A majority of the cyclins are expressed in all tissues tested, with various expression levels. Because cyclins are thought to regulate the cell cycle, our results suggest that all of the tissues tested have some actively dividing cells, although we cannot rule out the possibility that some cyclins might be expressed in nondividing cells and play different roles, as suggested for some CDKs (Barroco et al., 2003). In addition, for most of the identified types of cyclins, there is no specific expression pattern associated with that type, suggesting that most types of cyclins function in a variety of tissues. This notion is in agreement with studies of some A-, B-, and D-type cyclins, which are important for the mitotic cell cycle and/or for mitotic growth (Chaubet-Gigot, 2000; Ito, 2000; Meijer and Murray, 2000; Meszaros et al., 2000). Analogously, because several members of the T- and newly defined U-type cyclons exhibit broad expression patterns, they are likely to have generally functions in most or all tissues. Because most of the tissues we analyzed contain multiple cell types, it is possible that genes that appear to have similar expression patterns from microarray and RT-PCR experiments may have different patterns at the cellular level. In situ hybridization and/or reporter gene studies will need to be performed to obtain such detailed spatial expression patterns.
In several types with multiple members, some members exhibit highly specific expression patterns. Three genes, CycA3;3, CycT1;1, and CycT1;2, showed anther-specific expression, suggesting a specific function in the anther. Similarly, the fact that three U-type genes are specifically expressed in the root suggests that they have a specific function in the root. In addition, two very divergent genes, SDS and CYCJ18, exhibited unique expression patterns, suggesting that they also have specialized functions. The SDS gene has a highly restricted expression only in meiotic cells and has been shown to be required for chromosome pairing and synapsis during meiotic prophase I but not for mitotic growth. The expression pattern of CYCJ18 resembles that of SDS, suggesting that it is also involved in male reproduction, perhaps even meiosis. Further experiments are required to test this hypothesis.
In many cases, the most closely related cyclins also exhibit very similar expression patterns, suggesting possible functional redundancy between the highly similar genes. These genes include CycA1;1 and CycA1;2; CycA2;3 and CycA2;4; CycB1;2 and CycB1;5; CycB2;1 and CycB2;2; CycB2;3 and CycB2;5; CycC1;1 and CycC1;2; CycD3;3 and CycD3;2; CycT1;4 (At4g19600) and CycT1;5 (At5g45190); and CycU4;2 and CycU4;3. The pair CycU4;2 and CycU4;3 exhibit specific expression in roots. In addition, microarray results from another group (Birnbaum et al., 2003) showed that these two genes also fall into a group of genes that were differentially regulated across root subzones, suggesting that they have specific roles in root growth or function. For another pair, CycB1;2 and CycB1;5, since CycB1;5 is the only member in B-type cyclins which contains only cyclin_N domain, it is possible that CycB1;5 is a pseudogene. Also, the expression of CycB1;5 detected by the microarray experiments could be for CycB1;2 due to cross-hybridization. Currently, the sds mutant is the only reported single-gene mutant that exhibits phenotypic changes. The scarcity of cyclin mutants supports the hypothesized functional redundancy between closely related cyclins. Functional redundancy among cyclin genes has been well characterized in the budding yeast, which has six B-type cyclins that have overlapping and specific functions (Miller and Cross, 2001). In several previous functional studies of Arabidopsis cyclins, transgenic plants with ectopic expression and/or overexpression of a cyclin gene were usually generated to study the in vivo function of those cyclins. For example, CycD2;1 can increase root growth rate when it is overexpressed in tobacco (Cockcroft et al., 2000). Ectopic expression of CycD3;1 in Arabidopsis trichomes can induce both DNA replication and cell division (Schnittger et al., 2002a). It is worth noting that functions of some closely related members, even those with similar expression pattern, may still be different. For example, the ectopic expression of CycB1;2, but not CycB1;1, in Arabidopsis trichomes induced mitotic division (Schnittger et al., 2002b). Furthermore, members of three pairs of closely related genes, CycA2;1 and CycA2;2; CycD4;1 and CycD4;2; and CycU2;1 (At2g45080) and CycU2;2, exhibit different expression patterns. It is possible that both functional redundancy and functional specificity exist among plant cyclin groups.
Expression profiling within different tissues is only the first step to understanding the function of cyclin genes. Our results suggest that many, or even most, cyclin genes in Arabidopsis have similar sequences and expression patterns and may be functionally redundant. Alternatively, some of these genes may have different expression patterns within the tissues that we analyzed, but such differences could not be detected using microarray and RT-PCR experiments. Further detailed studies of developmental expression, cell-type expression, and cell cycle-phase specific expression patterns will provide more clues for functional prediction of cyclin genes. Expression profiles of all the Arabidopsis cyclin genes and the detailed phylogenetic analysis of plant cyclins provide useful information for future research. Those cyclins with specific expression patterns can be the focus of functional studies for their possible roles in specific tissues. Also, closely related genes with similar expression patterns can be tested for functional redundancy using double/triple mutants. Moreover, detailed analysis of newly identified plant-specific types of cyclins may potentially uncover plant-specific functions of cyclins, perhaps in the regulation of unique aspects of the plant cell cycle. Finally, an orthologous relationship between genes from Arabidopsis and other plants can form a basis for functional comparison using multiple approaches.
MATERIALS AND METHODS
Data Retrieval, Domain Identification, and Phylogenetic Analysis
A search of the Arabidopsis cyclin proteins was performed by using the BLASTP program against the AGI proteins database on The Arabidopsis Information Resource Web site (http://www.arabidopsis.org/Blast/), with various published plant and animal cyclins as query sequences and with the E-value cutoff set as 1e-005. Programs utilized on the Floral Genome Project Website (FGP-MINE; http://fgp.bio.psu.edu/cgi-bin/fgpmine/fgp_family_list.cgi; Wall et al., unpublished data) were applied to obtain cyclin-like proteins from rice and other plants. All the other plant cyclin-like proteins were obtained from PsiBLAST searches at the National Center for Biotechnology Information Web site (http://www.ncbi.nlm.nih.gov/BLAST/) against the nonredundant database, with one to a few representatives from each of the major classes as query sequences. Preliminary phylogenetic analysis was conducted to choose closely related sequences from the same species, and those sequences were further compared at both protein and DNA levels to identify duplicates, alleles, and partial sequences. Sequences that share higher than 95% identity at the DNA level were regarded as likely alleles (Zhang et al., 2001), unless they were previously reported to be different genes. Duplicates, alleles, and partial sequences were excluded from further analyses. Following preliminary phylogenetic analysis with Arabidopsis and human cyclins, members of novel Arabidopsis clades were used as queries to detect other possible nonplant homologs, and three protist sequences were added to the data set.
Protein sequences were analyzed in the Pfam HMM database to find cyclin-specific domains (http://pfam.wustl.edu/hmmsearch.shtml), with E-value = 0.01 as the cutoff. Proteins containing detectable cyclin_N or cyclin_C domains were regarded as cyclins; otherwise, they were excluded from the data set. Since all but one plant sequence had the cyclin_N domain while only approximate half of them contain the cyclin_C domain, we used only the cyclin_N domain for the genome-wide analysis. For analysis of individual families, however, longer regions that could be aligned with confidence were used. Sequence alignments were generated with CLUSTALX 1.81, with BLOSUM 30 as the protein weight matrix (Henikoff and Henikoff, 1992). Several values for gap opening penalty and gap extension penalty were tried to identify the commonly resolved domain. A combination of gap opening penalty = 6.0 and gap extension penalty = 0.1 was finally adopted that enabled reasonable alignment among conserved domains with few gaps. Phylogenetic analyses of cyclin proteins were carried out using the NJ, MP, and maximum likelihood (ML) methods in MEGA 2.1 (Kumar et al., 2001), PAUP* 4.10b (Swofford, 2001), and PHYLIP 3.6a3 (Felsenstein, 2002), respectively. NJ analyses were done with the pairwise deletion option selected and with Poisson correction set for distance model. For parsimony analysis, 100 replicates of random stepwise addition with tree-bisection-reconnection branch swamping were performed by using Heuristic Search. Support for each node was tested with bootstrap analysis, 1,000 replicates for NJ and 100 for MP and ML, using random input order for each replicate. Since the MP and ML trees do not disagree with the NJ trees in topology, we present only the NJ trees in this paper.
Six data sets were generated to estimate the relationships of cyclin genes, i.e. Cyc_N_all, CycA_plant, CycB_plant, CycD_plant, CycCHLT_plant, and CycU_plant. Cyc_N_all contained the cyclin_N domains of all the cyclins that we have identified and was used to estimate cyclin relationships at the genomic level. Three sets of analyses were performed for this data set: (1) comparison of Arabidopsis and human cyclins (Fig. 1); (2) phylogeny of Arabidopsis cyclins (Fig. 2); and (3) preliminary classification of plant cyclins (data not shown). The other data sets contain the information of full-length sequences and were used to determine phylogenetic relationships of plant A-/SDS-, B-/SDS-, D-, C-/H-/L-/T-, and U-type cyclins, respectively (alignments will be provided when requested). For each data set, only regions with reliable homology were used in the final analysis.
In addition to the cyclin_N and cyclin_C domains, some cyclins also contain a D-box region and one or several PEST motifs. To identify the D-box in Arabidopsis cyclins, a program on the website (http://bioinfo.weizmann.ac.il/danag/d-box/form.html) was used, and the output D-box information was also further verified by doing alignment with the consensus of published D-box sequence (Renaudin et al., 1996; Vandepoele et al., 2002). In addition, PESTfind software (http://emb1.bcc.univie.ac.at/embnet/tools/bio/) was used to detect the existence of PEST motifs for all the Arabidopsis cyclins.
Microarray Experiments
All seven tissues, including roots, stems, leaves, young inflorescences (stage1-8), anthers (stage 4–6), stage-12 flowers, and siliques were prepared from Arabidopsis Landsberg erecta. RNA was extracted using RNeasy Plant Kit (Qiagen USA, Valencia, CA), and the subsequent cRNA preparation and microarray hybridization were performed according to Affymetrix GeneChip Expression Analysis Overview (Affymetrix). Hybridization was performed twice using RNA samples that were extracted from two independently grown populations (two biological replicates) for each of the seven tissues. After the conversion of scan data to probewise expression levels, the data were further normalized using quantile normalization (Bolstad et al., 2003) and then converted to probeset expression levels using the R-Affy package in Bioconductor (version 1.3.25, Irizarry, Gautier, and Bolstad). Log250 was chosen as the cutoff for reliable detection of gene expression. Genes not reliably detected are designated by “A.” Pearson's correlation coefficient for the two replicates was 95.5% for anthers and ranged from 98.0% to 99.4% for the other six tissues. Expression of cyclins was analyzed for various tissues. The final signal intensity reflects the relative expression level of a gene.
RT-PCR
Plant materials of the same stages used for microarrays were harvested and ground in liquid nitrogen, and total RNA was isolated according to manufacturer's protocol using RNeasy mini kit (Qiagen USA). The amount of total RNA was determined by UV spectrophotometry. Total RNA (1 μg) was treated with 1 unit of DNase I (Invitrogen, Carlsbad, CA) prior to RT-PCR to remove residual DNA contamination. The first strand cDNA was synthesized using SuperScriptII reverse transcriptase (Invitrogen) and about one tenth was used as a template for RT-PCR. Thirty-four cycles of PCR amplification were used for seven genes including SDS, CycJ18, CycA3;3, CycD7;1, CycT1;1, CycT1;2, and CycU4;2. Twenty-nine cycles were done for all the other PCR reactions. Control PCRs without reverse transcriptase did not produce any PCR bands. ADENINE PHOSPHORIBOSYL TRANSFERASE 1 (APT1) (Moffatt et al., 1994) gene was used as an internal control. The primers used for RT-PCR are listed in Table I. PCR products were fractionated on 1% agarose gels containing ethidium bromide and photographed under UV light.
Note Added in Proof
The U-type cyclins described here are the same as the P-type cyclins that were reported in a recent paper describing the molecular and phylogenetic analysis of a novel type of Arabidopsis cylcins (Torres Acosta J, de Almeida Engler J, Raes J, Magyar Z, DeGroodt R, Inze D, De Veylder L [2004] Cell Mol Life Sci 61: 1485–1497). For the Arabidopsis proteins the correspondence of names is: CycU1;1=CycP2;1; CycU2;1=CycP3;1; CycU2;2=CycP3;2; CycU3;1=CycP1;1; CycU4;1=CycP4;1; CycU4;2=CycP4;3; CycU4;3=CycP4;2. The designation of P-type cyclins is supported by molecular results and should be used for future work. Additional sequences from plants and protists are also included in the phylogenetic analysis reported by Torres Acosta et al. (2004).
Supplementary Material
Acknowledgments
We thank A. Omeis and J. Wang for plant care and K. Wall for help with BLAST search against the rice genome. We are grateful for helpful comments from D. Zhao, W. Ni, and L. Quan.
This work was supported by the National Institutes of Health (grant no. RO1 GM63871), the National Science Foundation (grant nos. MCB-0092075, IBN-0077832, and DBI-0115684), the National Natural Science Foundation of China (grant no. 30130030 to Anmin Lu), the Intercollege Graduate Program in Plant Physiology at the Pennsylvania State University (G.W.), and by funds from the Department of Biology and the Huck Institutes of Life Sciences at the Pennsylvania State University. This is the Floral Genome Project's publication number 20.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.040436.
References
- Abrahams S, Cavet G, Oakenfull EA, Carmichael JP, Shah ZH, Soni R, Murray JA (2001) A novel and highly divergent Arabidopsis cyclin isolated by complementation in budding yeast. Biochim Biophys Acta 1539: 1–6 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Azumi Y, Liu D, Zhao D, Li W, Wang G, Hu Y, Ma H (2002) Homolog interaction during meiotic prophase I in Arabidopsis requires the SOLO DANCERS gene encoding a novel cyclin-like protein. EMBO J 21: 3081–3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroco RM, De Veylder L, Magyar Z, Engler G, Inze D, Mironov V (2003) Novel complexes of cyclin-dependent kinases and a cyclin-like protein from Arabidopsis thaliana with a function unrelated to cell division. Cell Mol Life Sci 60: 401–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193 [DOI] [PubMed] [Google Scholar]
- Boniotti MB, Griffith ME (2002) “Cross-talk” between cell division cycle and development in plants. Plant Cell 14: 11–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booher RN, Alfa CE, Hyams JS, Beach DH (1989) The fission yeast cdc2/cdc13/suc1 protein kinase: regulation of catalytic activity and nuclear localization. Cell 58: 485–497 [DOI] [PubMed] [Google Scholar]
- Bourne Y, Watson MH, Hickey MJ, Holmes W, Rocque W, Reed SI, Tainer JA (1996) Crystal structure and mutational analysis of the human CDK2 kinase complex with cell cycle-regulatory protein CksHs1. Cell 84: 863–874 [DOI] [PubMed] [Google Scholar]
- Breyne P, Dreesen R, Vandepoele K, De Veylder L, Van Breusegem F, Callewaert L, Rombauts S, Raes J, Cannoot B, Engler G, et al (2002) Transcriptome analysis during cell division in plants. Proc Natl Acad Sci USA 99: 14825–14830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burssens S, de Almeida Engler J, Beeckman T, Richard C, Shaul O, Ferreira P, Van Montagu M, Inze D (2000) Developmental expression of the Arabidopsis thaliana CycA2;1 gene. Planta 211: 623–631 [DOI] [PubMed] [Google Scholar]
- Chaubet-Gigot N (2000) Plant A-type cyclins. Plant Mol Biol 43: 659–675 [DOI] [PubMed] [Google Scholar]
- Chen KC, Csikasz-Nagy A, Gyorffy B, Val J, Novak B, Tyson JJ (2000) Kinetic analysis of a molecular model of the budding yeast cell cycle. Mol Biol Cell 11: 369–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockcroft CE, den Boer BG, Healy JM, Murray JA (2000) Cyclin D control of growth rate in plants. Nature 405: 575–579 [DOI] [PubMed] [Google Scholar]
- Colon-Carmona A, You R, Haimovitch-Gal T, Doerner P (1999) Technical advance: spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J 20: 503–508 [DOI] [PubMed] [Google Scholar]
- Criqui MC, Genschik P (2002) Mitosis in plants: how far we have come at the molecular level? Curr Opin Plant Biol 5: 487–493 [DOI] [PubMed] [Google Scholar]
- Cyr RJ, Palevitz BA (1995) Organization of cortical microtubules in plant cells. Curr Opin Cell Biol 7: 65–71 [DOI] [PubMed] [Google Scholar]
- De Veylder L, Joubes J, Inze D (2003) Plant cell cycle transitions. Curr Opin Plant Biol 6: 536–543 [DOI] [PubMed] [Google Scholar]
- Evans T, Rosenthal ET, Youngblom J, Distel D, Hunt T (1983) Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell 33: 389–396 [DOI] [PubMed] [Google Scholar]
- Felsenstein J (2002) PHYLIP (Phylogeny Inference Package) Version 3.6a3. Distributed by the author. Department of Genetics, University of Washington, Seattle
- Ferreira PC, Hemerly AS, Engler JD, van Montagu M, Engler G, Inze D (1994) Developmental expression of the arabidopsis cyclin gene cyc1At. Plant Cell 6: 1763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung TK, Siu WY, Yam CH, Lau A, Poon RY (2002) Cyclin F is degraded during G2-M by mechanisms fundamentally different from other cyclins. J Biol Chem 277: 35140–35149 [DOI] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW (1991) Cyclin is degraded by the ubiquitin pathway. Nature 349: 132–138 [DOI] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100 [DOI] [PubMed] [Google Scholar]
- Hata S, Kouchi H, Suzuka I, Ishii T (1991) Isolation and characterization of cDNA clones for plant cyclins. EMBO J 10: 2681–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG (1992) Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA 89: 10915–10919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne MC, Goolsby GL, Donaldson KL, Tran D, Neubauer M, Wahl AF (1996) Cyclin G1 and cyclin G2 comprise a new family of cyclins with contrasting tissue-specific and cell cycle-regulated expression. J Biol Chem 271: 6050–6061 [DOI] [PubMed] [Google Scholar]
- Ito M (2000) Factors controlling cyclin B expression. Plant Mol Biol 43: 677–690 [DOI] [PubMed] [Google Scholar]
- John PCL, Mews M, Moore R (2001) Cyclin/CDK complexes: their involvement in cell cyclin pregression and mitotic division. Protoplasma 216: 119–142 [DOI] [PubMed] [Google Scholar]
- Joubes J, Chevalier C, Dudits D, Heberle-Bors E, Inze D, Umeda M, Renaudi JP (2000) CDK-related protein kinases in plants. Plant Mol Biol 43: 607–620 [DOI] [PubMed] [Google Scholar]
- King RW, Deshaies RJ, Peters JM, Kirschner MW (1996) How proteolysis drives the cell cycle. Science 274: 1652–1659 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M (2001) MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17: 1244–1245 [DOI] [PubMed] [Google Scholar]
- Meijer M, Murray JA (2001) Cell cycle controls and the development of plant form. Curr Opin Plant Biol 4: 44–49 [DOI] [PubMed] [Google Scholar]
- Meijer M, Murray JAH (2000) The role and regulation of D-type cyclins in the plant cell cycle. Plant Mol Biol 43: 621–633 [DOI] [PubMed] [Google Scholar]
- Menges M, Hennig L, Gruissem W, Murray JA (2002) Cell cycle-regulated gene expression in Arabidopsis. J Biol Chem 277: 41987–42002 [DOI] [PubMed] [Google Scholar]
- Meszaros T, Miskolczi P, Ayaydin F, Pettko-Szandtner A, Peres A, Magyar Z, Horvath GV, Bako L, Feher A, Dudits D (2000) Multiple cyclin-dependent kinase complexes and phosphatases control G2/M progression in alfalfa cells. Plant Mol Biol 43: 595–605 [DOI] [PubMed] [Google Scholar]
- Mews M, Sek FJ, Moore R, Volkmann D, Gunning BES, John PCL (1997) Mitotic cyclin distribution during maize cell division: implications for the sequence diversity and function of cyclins in plants. Protoplasma 200: 128–145 [Google Scholar]
- Miller ME, Cross FR (2001) Cyclin specificity: how many wheels do you need on a unicycle? J Cell Sci 114: 1811–1820 [DOI] [PubMed] [Google Scholar]
- Mironov V, Van Montagu M, Inze D (1997) Regulation of cell division in plants: an Arabidopsis perspective. Prog Cell Cycle Res 3: 29–41 [DOI] [PubMed] [Google Scholar]
- Mironov VV, De Veylder L, Van Montagu M, Inze D (1999) Cyclin-dependent kinases and cell division in plants- the nexus. Plant Cell 11: 509–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt BA, McWhinnie EA, Agarwal SK, Schaff DA (1994) The adenine phosphoribosyltransferase-encoding gene of Arabidopsis thaliana. Gene 143: 211–216 [DOI] [PubMed] [Google Scholar]
- Morgan DO (1995) Principles of CDK regulation. Nature 374: 131–134 [DOI] [PubMed] [Google Scholar]
- Nakamura T, Sanokawa R, Sasaki YF, Ayusawa D, Oishi M, Mori N (1995) Cyclin I: a new cyclin encoded by a gene isolated from human brain. Exp Cell Res 221: 534–542 [DOI] [PubMed] [Google Scholar]
- Nakayama K (1998) Cip/Kip cyclin-dependent kinase inhibitors: brakes of the cell cycle engine during development. Bioessays 20: 1020–1029 [DOI] [PubMed] [Google Scholar]
- Nasmyth K (1996) At the heart of the budding yeast cell cycle. Trends Genet 12: 405–412 [DOI] [PubMed] [Google Scholar]
- Nieduszynski CA, Murray J, Carrington M (2002) Whole-Genome Analysis of Animal A- and B-Type Cyclins. Genome Biol 3: RESEARCH0070.1-0070.8 [DOI] [PMC free article] [PubMed]
- Nigg EA (1993) Targets of cyclin-dependent protein kinases. Curr Opin Cell Biol 5: 187–193 [DOI] [PubMed] [Google Scholar]
- Novak B, Csikasz-Nagy A, Gyorffy B, Nasmyth K, Tyson JJ (1998) Model scenarios for evolution of the eukaryotic cell cycle. Philos Trans R Soc Lond B Biol Sci 353: 2063–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent JH, Alfa CE, Young T, Hyams JS (1991) Conserved structural motifs in cyclins identified by sequence analysis. J Cell Sci 99: 669–674 [DOI] [PubMed] [Google Scholar]
- Okayama H, Nagata A, Jinno S, Murakami H, Tanaka K, Nakashima N (1996) Cell cycle control in fission yeast and mammals: identification of new regulatory mechanisms. Adv Cancer Res 69: 17–62 [DOI] [PubMed] [Google Scholar]
- Peeper DS, Parker LL, Ewen ME, Toebes M, Hall FL, Xu M, Zantema A, van der Eb AJ, Piwnica-Worms H (1993) A- and B-type cyclins differentially modulate substrate specificity of cyclin-cdk complexes. EMBO J 12: 1947–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM (1998) SCF and APC: the Yin and Yang of cell cycle regulated proteolysis. Curr Opin Cell Biol 10: 759–768 [DOI] [PubMed] [Google Scholar]
- Pines J (1995) Cyclins and cyclin-dependent kinases: a biochemical view. Biochem J 308: 697–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowman GD, Sudarsanam S, Bingham J, Whyte D, Hunter T (1999) The protein kinases of Caenorhabditis elegans: a model for signal transduction in multicellular organisms. Proc Natl Acad Sci USA 96: 13603–13610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak T, Doerner P (2001) Cell cycle controls: genome-wide analysis in Arabidopsis. Curr Opin Plant Biol 4: 501–506 [DOI] [PubMed] [Google Scholar]
- Rechsteiner M, Rogers SW (1996) PEST sequences and regulation by proteolysis. Trends Biochem Sci 21: 267–271 [PubMed] [Google Scholar]
- Renaudin JP, Doonan JH, Freeman D, Hashimoto J, Hirt H, Inze D, Jacobs T, Kouchi H, Rouze P, Sauter M, et al (1996) Plant cyclins: a unified nomenclature for plant A-, B- and D-type cyclins based on sequence organization. Plant Mol Biol 32: 1003–1018 [DOI] [PubMed] [Google Scholar]
- Richard C, Granier C, Inze D, De Veylder L (2001) Analysis of cell division parameters and cell cycle gene expression during the cultivation of Arabidopsis thaliana cell suspensions. J Exp Bot 52: 1625–1633 [PubMed] [Google Scholar]
- Rogers S, Wells R, Rechsteiner M (1986) Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science 234: 364–368 [DOI] [PubMed] [Google Scholar]
- Rossi V, Varotto S (2002) Insights into the G1/S transition in plants. Planta 215: 345–356 [DOI] [PubMed] [Google Scholar]
- Schnittger A, Schobinger U, Bouyer D, Weinl C, Stierhof YD, Hulskamp M (2002. a) Ectopic D-type cyclin expression induces not only DNA replication but also cell division in Arabidopsis trichomes. Proc Natl Acad Sci USA 99: 6410–6415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger A, Schobinger U, Stierhof YD, Hulskamp M (2002. b) Ectopic B-type cyclin expression induces mitotic cycles in endoreduplicating Arabidopsis trichomes. Curr Biol 12: 415–420 [DOI] [PubMed] [Google Scholar]
- Smith LG (1999) Divide and conquer: cytokinesis in plant cells. Curr Opin Plant Biol 2: 447–453 [DOI] [PubMed] [Google Scholar]
- Swofford DL (2001) PAUP: Phylogenetic Analysis Using Parsimony* and Other Methods. Version 4.0 beta., Sinauer, Sunderland, Massachusetts
- Vandepoele K, Raes J, De Veylder L, Rouze P, Rombauts S, Inze D (2002) Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14: 903–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Fabian T, Sauter M, Bhalerao RP, Schrader J, Sandberg G, Umeda M, Uchimiya H (2000) Activation of CDK-activating kinase is dependent on interaction with H-type cyclins in plants. Plant J 24: 11–20 [DOI] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92 [DOI] [PubMed] [Google Scholar]
- Yu Y, Steinmetz A, Meyer D, Brown S, Shen WH (2003) The tobacco A-type cyclin, Nicta;CYCA3;2, at the nexus of cell division and differentiation. Plant Cell 15: 2763–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Pond SK, Gaut BS (2001) A survey of the molecular evolutionary dynamics of twenty-five multigene families from four grass taxa. J Mol Evol 52: 144–156 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.