Abstract
There is a long-going debate on the genetic origin of Chinese Muslim populations, such as Uygur, Dongxiang, and Hui. However, genetic information for those Muslim populations except Uygur is extremely limited. In this study, we investigated the genetic structure and ancestry of Chinese Muslims by analyzing 15 autosomal short tandem repeats in 652 individuals from Dongxiang, Hui, and Han Chinese populations in Gansu province. Both genetic distance and Bayesian-clustering methods showed significant genetic homogeneity between the two Muslim populations and East Asian populations, suggesting a common genetic ancestry. Our analysis found no evidence of substantial gene flow from Middle East or Europe into Dongxiang and Hui people during their Islamization. The dataset generated in present study are also valuable for forensic identification and paternity tests in China.
Chinese Muslim populations refer to ten officially recognized Muslim ethnic groups, which are Uygur, Dongxiang, Hui, Bo’an, Kazakh, Kirghiz, Salar, Tatar, Tajik, and Uzbek. The origin of those populations via demic diffusion involves mass movement of people or simple cultural diffusion is a long-going debate.
According to historical materials, Islam was first introduced to China some 1,400 years ago in Tang Dynasty (618–907 AD) by a large number of soldiers, merchants and political emissaries from Arabia and Persia (nowadays Middle East)1. Chinese Muslim populations are believed to be decedents of those immigrants. Uygur has already been proven to be a typical admixture of East Asian and European by genome-wide scan2. However, Dongxinag, Bo’an, and Hui in Gansu and Ningxia have the common physical features of East Asians (Mongoloid type)3,4,5. Xie and Shan6 also detected the genetic similarity between Hui and Han Chinese using two autosomal short tandem repeats (STRs) TH01 and D13S317. The paternal Y chromosomal STR clustering has put Hui of Liaoning and Ningxia into the group of Han Chinese and Tibeto-Burman populations7. About 24–30% Y chromosomes of Salar, Bo’an, and Dongxiang belong to East Asian specific haplogroup O3-M122. The Central Asian, South Asian, and European prevalent Y chromosomal lineage R-M17 also comprises 17%, 26%, and 28% of Salar, Bo’an, and Dongxiang, respectively8.
The origin of Chinese Muslim populations likely involved massive assimilation of indigenous ethnic groups9. But previous studies with limited genetic markers and small sample size have not been able to give a clear answer to the genetic ancestry of those Muslim populations. Therefore, we analyzed 15 autosomal STRs in 652 individuals of Dongxiang, Hui, and the co-resident Han Chinese populations in Linxia, Gansu province to explore the genetic diversity of Chinese Muslims and to test population affinities and the level of admixture. Dongxiang and Hui are typical of contemporary Chinese Muslim communities. The comprehensive comparison of those two populations with worldwide Muslims and non-Muslims will shed more light on the origin of Chinese Muslims.
Methods
We collected blood samples of 163 and 219 unrelated individuals from two Muslim populations Dongxiang and Hui in Linxia, Gansu province. We also collected blood samples of 270 unrelated individuals from Han Chinese in Linxia for comparison purpose. Our study was approved by the Ethical Committee of Gansu Institute of Political Science and Law. The study was conducted in accordance with the human and ethical research principles of Gansu Institute of Political Science and Law. All individuals were adequately informed and signed their informed content before their participation. For each sample, genomic DNA was extracted according to the Chelex-100 method and proteinase K protocol10. 15 most widely used forensic loci were amplified simultaneously using AmpFlSTR Sinofiler PCR Amplification Kit (Applied Biosystems, Foster City, CA, USA) at the D8S1179, D21S11, D7S820, CSF1PO, D3S1358, D13S317, D16S539, D2S1338, D19S433, vWA, D18S51, D5S818, FGA, D6S1043 and D12S391 STR loci. The PCR products were analyzed with the 3500XL DNA Genetic Analyzer and Genemapper ID-X software (Applied Biosystems, Foster City, CA, USA).
Allele frequency, heterozygosity, polymorphism information content (PIC), power of discrimination (PD), probability of paternity exclusion (PPE), and other forensic parameters were calculated using PowerStatesV12 (http://www.promega.com/) and Cervus 3.011. Tests for Hardy–Weinberg equilibrium (HWE) were performed in Arlequin v3.5.1.312 using a likelihood ratio test and an exact test to prevent miscalling STR genotypes or biased sampling. Since the statistical analyses in this study were on the basis of Bayesian-clustering algorithm, raw genotypic data of 13 STRs (excluding D6S1043 and D12S391) from 45 populations (13793 individuals) all around the world were extracted to determine population affinity13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39. Average number of pairwise differences, pairwise Fst, Slatkins linearized Fst, and coancestry coefficients were all calculated in Arlequin v3.5.1.3 using genotype data12. The detailed population genetic structure was performed using model-based clustering method implemented in Structure 2.3.440,41 under assumptions of admixture and correlated allele frequencies. Each run used 100,000 estimation iterations for K = 2 to 8 after a 20,000 burn-in length with several replicates. Posterior probabilities for each K were computed for each set of runs. Graphical display for Matrix plot of genetic distance and population structure were carried out in R statistical software v3.0.242 and Distruct v1.143.
Results
Forensic parameter analysis
The genotype data for the three populations Dongxiang, Hui, and Han Chinese was given in Supplementary Table 1. The allele frequency distributions and forensic parameters are listed in Table 1 and Supplementary Table 2. The Ho ranged from 0.688 at CSF1PO locus in Hui to 0.914 at D6S1043 locus in Dongxiang while the He ranged from 0.704 at D3S1358 locus in Han to 0.883 at D6S1043 locus in Dongxiang. In the test of HWE, the genotype frequency distributions showed no significant deviations from expectations except p-value of 0.030 at D19S433 locus in Hui. PIC of all selected loci ranged from 0.652 at D3S1358 in Han to 0.868 at D6S1043 in Dongxiang. The values of DP were in the range of 0.861 at D3S1358 in Han to 0.969 in Dongxiang and Hui. The highest PPE was found at D6S1043 locus in Dongxiang (0.824), with the lowest found at CSF1PO locus in Hui (0.410). The most polymorphic loci in all three populations were highly discriminating, which demonstrates that this set of loci will be useful for forensic identification.
Table 1. Forensic statistical parameters of the 15 autosomal short tandem repeats from Dongxiang, Hui, and Han Chinese populations in Linxia.
| CSF1PO | D12S391 | D13S317 | D16S539 | D18S51 | D19S433 | D21S11 | D2S1338 | D3S1358 | D5S818 | D6S1043 | D7S820 | D8S1179 | FGA | vWA | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dongxiang | Ho | 0.693 | 0.853 | 0.755 | 0.785 | 0.810 | 0.834 | 0.871 | 0.822 | 0.810 | 0.767 | 0.914 | 0.791 | 0.847 | 0.847 | 0.847 |
| He | 0.733 | 0.844 | 0.800 | 0.787 | 0.868 | 0.817 | 0.839 | 0.868 | 0.729 | 0.778 | 0.883 | 0.793 | 0.836 | 0.860 | 0.802 | |
| PIC | 0.688 | 0.824 | 0.767 | 0.751 | 0.852 | 0.791 | 0.817 | 0.851 | 0.680 | 0.742 | 0.868 | 0.758 | 0.812 | 0.843 | 0.769 | |
| DP | 0.878 | 0.952 | 0.930 | 0.920 | 0.965 | 0.940 | 0.950 | 0.963 | 0.862 | 0.908 | 0.969 | 0.920 | 0.948 | 0.962 | 0.920 | |
| PPE | 0.418 | 0.700 | 0.518 | 0.572 | 0.617 | 0.664 | 0.737 | 0.641 | 0.617 | 0.539 | 0.824 | 0.583 | 0.688 | 0.688 | 0.688 | |
| NE-1P | 0.679 | 0.478 | 0.578 | 0.601 | 0.424 | 0.534 | 0.489 | 0.428 | 0.688 | 0.611 | 0.392 | 0.592 | 0.505 | 0.441 | 0.576 | |
| NE-2P | 0.502 | 0.311 | 0.400 | 0.423 | 0.268 | 0.360 | 0.321 | 0.270 | 0.514 | 0.432 | 0.243 | 0.414 | 0.333 | 0.281 | 0.398 | |
| NE-PP | 0.318 | 0.139 | 0.219 | 0.241 | 0.107 | 0.178 | 0.146 | 0.110 | 0.334 | 0.247 | 0.091 | 0.233 | 0.160 | 0.115 | 0.219 | |
| NE-I | 0.115 | 0.043 | 0.071 | 0.080 | 0.032 | 0.058 | 0.046 | 0.033 | 0.121 | 0.084 | 0.026 | 0.076 | 0.049 | 0.035 | 0.071 | |
| NE-SI | 0.414 | 0.340 | 0.369 | 0.378 | 0.325 | 0.357 | 0.343 | 0.325 | 0.417 | 0.383 | 0.316 | 0.374 | 0.346 | 0.330 | 0.368 | |
| P-Value | 0.094 | 0.738 | 0.595 | 0.970 | 0.313 | 0.384 | 0.894 | 0.172 | 0.375 | 0.198 | 0.755 | 0.504 | 0.896 | 0.623 | 0.178 | |
| Hui_Linxia | Ho | 0.688 | 0.839 | 0.808 | 0.776 | 0.858 | 0.849 | 0.831 | 0.826 | 0.708 | 0.749 | 0.831 | 0.807 | 0.817 | 0.895 | 0.775 |
| He | 0.728 | 0.853 | 0.813 | 0.785 | 0.860 | 0.819 | 0.835 | 0.853 | 0.720 | 0.777 | 0.877 | 0.791 | 0.817 | 0.871 | 0.803 | |
| PIC | 0.678 | 0.835 | 0.784 | 0.750 | 0.843 | 0.794 | 0.813 | 0.833 | 0.668 | 0.743 | 0.862 | 0.757 | 0.791 | 0.855 | 0.771 | |
| DP | 0.877 | 0.961 | 0.937 | 0.914 | 0.960 | 0.937 | 0.951 | 0.959 | 0.873 | 0.916 | 0.969 | 0.918 | 0.938 | 0.965 | 0.931 | |
| PPE | 0.410 | 0.674 | 0.614 | 0.556 | 0.710 | 0.694 | 0.658 | 0.649 | 0.440 | 0.508 | 0.655 | 0.613 | 0.632 | 0.785 | 0.554 | |
| NE-1P | 0.690 | 0.457 | 0.552 | 0.601 | 0.440 | 0.528 | 0.496 | 0.462 | 0.701 | 0.608 | 0.406 | 0.593 | 0.538 | 0.417 | 0.573 | |
| NE-2P | 0.518 | 0.294 | 0.375 | 0.423 | 0.280 | 0.355 | 0.326 | 0.298 | 0.530 | 0.428 | 0.253 | 0.415 | 0.363 | 0.262 | 0.395 | |
| NE-PP | 0.339 | 0.126 | 0.197 | 0.240 | 0.115 | 0.174 | 0.150 | 0.130 | 0.352 | 0.241 | 0.098 | 0.233 | 0.184 | 0.103 | 0.215 | |
| NE-I | 0.124 | 0.039 | 0.063 | 0.080 | 0.035 | 0.056 | 0.048 | 0.040 | 0.130 | 0.083 | 0.029 | 0.077 | 0.059 | 0.031 | 0.069 | |
| NE-SI | 0.418 | 0.334 | 0.360 | 0.378 | 0.330 | 0.355 | 0.345 | 0.335 | 0.423 | 0.383 | 0.320 | 0.375 | 0.357 | 0.323 | 0.367 | |
| P-Value | 0.344 | 0.968 | 0.394 | 0.100 | 0.492 | 0.030 | 0.287 | 0.225 | 0.714 | 0.564 | 0.268 | 0.255 | 0.969 | 0.639 | 0.258 | |
| Han_Linxia | Ho | 0.741 | 0.847 | 0.800 | 0.748 | 0.844 | 0.870 | 0.802 | 0.844 | 0.703 | 0.785 | 0.900 | 0.789 | 0.830 | 0.856 | 0.814 |
| He | 0.735 | 0.849 | 0.807 | 0.775 | 0.857 | 0.830 | 0.824 | 0.860 | 0.704 | 0.766 | 0.874 | 0.769 | 0.832 | 0.857 | 0.810 | |
| PIC | 0.689 | 0.830 | 0.777 | 0.738 | 0.839 | 0.808 | 0.799 | 0.843 | 0.652 | 0.728 | 0.859 | 0.734 | 0.809 | 0.839 | 0.780 | |
| DP | 0.883 | 0.956 | 0.932 | 0.915 | 0.961 | 0.942 | 0.945 | 0.964 | 0.861 | 0.904 | 0.965 | 0.910 | 0.947 | 0.960 | 0.934 | |
| PPE | 0.494 | 0.689 | 0.599 | 0.507 | 0.684 | 0.735 | 0.606 | 0.684 | 0.432 | 0.572 | 0.795 | 0.579 | 0.655 | 0.706 | 0.626 | |
| NE-1P | 0.676 | 0.468 | 0.563 | 0.619 | 0.449 | 0.506 | 0.512 | 0.444 | 0.714 | 0.632 | 0.411 | 0.622 | 0.507 | 0.448 | 0.558 | |
| NE-2P | 0.501 | 0.303 | 0.386 | 0.440 | 0.288 | 0.335 | 0.340 | 0.283 | 0.544 | 0.452 | 0.257 | 0.442 | 0.335 | 0.287 | 0.381 | |
| NE-PP | 0.318 | 0.134 | 0.207 | 0.258 | 0.122 | 0.158 | 0.159 | 0.119 | 0.363 | 0.268 | 0.101 | 0.255 | 0.160 | 0.120 | 0.202 | |
| NE-I | 0.115 | 0.041 | 0.066 | 0.087 | 0.037 | 0.050 | 0.052 | 0.036 | 0.139 | 0.092 | 0.030 | 0.088 | 0.050 | 0.037 | 0.065 | |
| NE-SI | 0.412 | 0.336 | 0.364 | 0.385 | 0.332 | 0.348 | 0.352 | 0.330 | 0.433 | 0.391 | 0.321 | 0.388 | 0.347 | 0.332 | 0.362 | |
| P-Value | 0.954 | 0.331 | 0.265 | 0.543 | 0.639 | 0.560 | 0.523 | 0.928 | 0.122 | 0.618 | 0.172 | 0.396 | 0.191 | 0.398 | 0.828 |
Interpopulation genetic distances
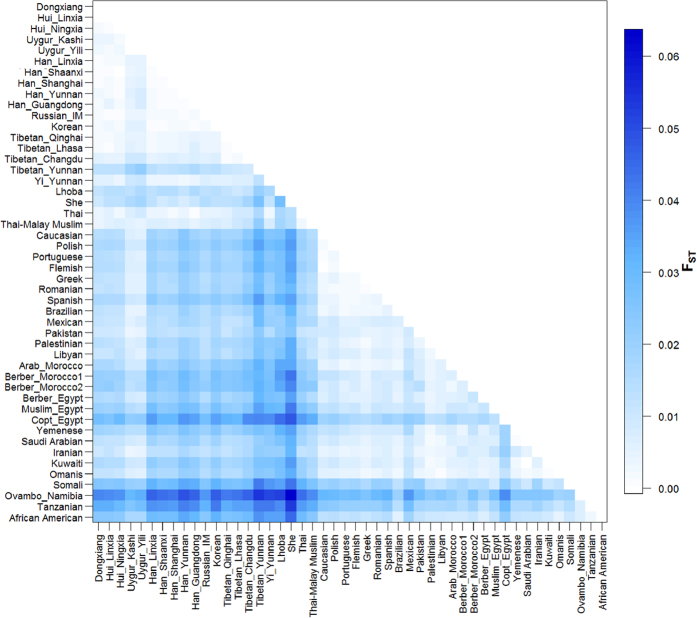
We performed various parameters of genetic distances to infer population structure (Fig. 1 and Supplementary Table 3). Chinese Muslim populations Dongxiang and Hui showed the largest pairwise genetic distances with populations from Africa and Middle East. The smallest genetic distances were noted for Chinese Muslim populations with East Asian populations, especially Han Chinese. Dongxiang showed nonsignificant pairwise Fst difference from Hui in Linxia and Ningxia, Han Chinese in Linxia, Shaanxi, Shanghai, and Guangdong, and Tibetan in Lhasa (p > 0.005). The genetic divergence of Dongxiang and those populations are relatively small (pairwise Fst < 0.002 and Slatkin linearized Fst < 0.003). Hui in Linxia also showed nonsignificant genetic difference with Hui in Ningxia, Uygur in Yili, Han Chinese in Shaanxi and Yunnan, Russian in Inner Mongolia, and Tibetan in Lhasa. Hui in Ningxia also did not differ from all five Han Chinese populations in this study. However, almost all the pairwise Fst differences between Dongxiang and Hui with European, Middle Eastern, and African populations are all above 0.01. The average pairwise differences exhibit the very similar pattern. The two Uygur populations statistically differed from all other populations. The genetic distances of Uygur with East Asian, European, and most Middle Eastern populations are almost the same, indicating Uygur is an admixed population.
Figure 1. Plots of pairwise Fst of Dongxinag, Hui, and Han Chinese in Linxia and other 45 worldwide populations.
Clustering by structure analysis
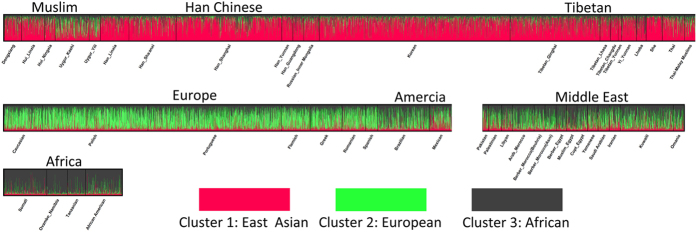
Analysis of genetic distance failed to support the genetic affinity between Chinese Muslim populations Dongxiang and Hui with Middle Eastern or European populations. We then employed a cluster based algorithm to further clarify population genetic structure at individual level. According to the highest posterior probabilities, the most suitable K was observed at K = 3 (Supplementary Table 4). The clustering showed a very clear geographic pattern (Fig. 2). East Asian, European, and African populations belong to cluster 1, 2, and 3, respectively. Middle Eastern populations seem like to be admixture of African and European populations. Uygur populations shared a similar degree of membership with East Asian and European (35% to 40%), which is consistent with genetic distance analysis and previous reports2. The case of Uygur shows clearly that the 15 STRs and Structure analysis have enough power to delineate the ancestry of populations. The proportion of membership of Dongxiang and Hui in cluster 1 reaches 58.3% to 63.9%. Although this proportion is about 10% lower than Han Chinese (66.8% to 74.4%), it still fall into the general pattern of East Asian populations ranging from 57.8% (Lhoba) to 80.3% (She) (Supplementary Table 4). It’s possible that some individuals of Dongxiang and Hui might have excess affinity with West Eurasians as they are living closely together with Uygur and Central Asians (Supplementary 6). However, the majority of Dongxiang and Hui samples share very similar membership with other East Asian populations, revealing a common genetic makeup.
Figure 2. Estimated population genetic structure of Dongxinag, Hui, and Han Chinese in Linxia and other 45 worldwide populations.
Discussion
The origin of Chinese Muslim populations via demic diffusion or simple cultural diffusion has long been a hot debate. Previous genetic studies with limited markers and small sample size often came to contradictory conclusions. In this study, we focused on the genetic makeup and ancestry clustering of Dongxiang and Hui using 15 autosomal STRs genotyped from more than 600 individuals. The two Chinese Muslim populations Dongxiang and Hui showed significant genetic homogeneity with co-resident Han Chinese in Linxia and other East Asian populations rather than with European or Middle Eastern populations, which support a simple cultural diffusion for the origin of Dongxiang and Hui in China. This cultural transformation phenomenon has also been observed in other Muslim populations. Although the Utsat people in Hainan Island are thought to be descendants of the Champa Kingdom and have been officially recognized as Hui nationality, they are genetically much closer to the Hainan indigenous ethnic groups than to the Cham and other mainland Southeast Asian populations9. The spread of Islam in the Indian subcontinent was also proven to be predominantly cultural diffusion associated with minor gene flow from West Asia and Arabia by analyzing autosomal STRs44,45, mitochondrial DNA46,47, and Y chromosome47. Autosomal STRs also reveal common genetic ancestry of the Thai-Malay Muslims and Thai Buddhists36. In this context, cultural transformation has shaped worldwide Muslim populations.
Additional Information
How to cite this article: Yao, H.-B. et al. Genetic evidence for an East Asian origin of Chinese Muslim populations Dongxiang and Hui. Sci. Rep. 6, 38656; doi: 10.1038/srep38656 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We would like to thank Dr. Masood S.H. Abu Halima, Dr. Andrea Berti, and Dr. Phuvadol Thanakiatkrai for sharing STR genotype data with us. This work was supported by the Natural Science Foundation of Gansu province (1308RJZA190), Scientific Research Project for Colleges of Gansu province (2014A-085), National Excellent Youth Science Foundation of China (31222030), National Natural Science Foundation of China (91131002, 31260252, 31671297, 31071098, and 30760097), Shanghai Rising-Star Program (12QA1400300), MOE University Doctoral Research Supervisor’s Funds (20120071110021), and MOE Scientific Research Project (113022A). C.C.W is supported by Max Planck Society. The research leading to these results has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No 646612) granted to Martine Robbeets.
Footnotes
Author Contributions H.B.Y., H.L. and C.C.W. supervised the study. H.B.Y. and X.T. collected the samples and did the experiments. C.C.W. and S.L. analyzed the data. C.C.W. wrote the manuscript. L.S., S.Q.W., B.Z., L.K., L.J. and H.L. were involved in discussions and manuscript revisions. All authors reviewed the manuscript.
References
- Gladney D. C. Ethnic identity in China: the making of a Muslim minority nationality. (Harcourt Brace College, 1998). [Google Scholar]
- Xu S., Huang W., Qian J. & Jin L. Analysis of genomic admixture in Uyghur and its implication in mapping strategy. Am J Hum Genet 82, 883–94 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D. Y. & Dai Y. J. Physical characteristics of Bonan Nationality from Gansu province, northwest China. Acta Anthropologica Sinica 9, 56–63 (1990). [Google Scholar]
- Dai Y. J. & Yang D. Y. Research on the physical characteristics of Dongxiang Nationality in Gansu province, northwest China. Acta Anthropologica Sinica 10, 127–134 (1991). [Google Scholar]
- Zheng L. B. et al. Physical characters of Hui Nationality in Ningxia. Acta Anthropologica Sinica 16, 11–21 (1997). [Google Scholar]
- Xie X. D. & Shan X. M. The DNA evidence of the origin of Hui. Researches on the Hui 3, 75–78 (2002). [Google Scholar]
- Zhang F., Li H., Huang L., Lu Y. & Hu S. Similarities in Patrilineal Genetics between the Han Chinese of Central China and Chaoshanese in Southern China. Commun Contemp Anthropol 4, e2 (2010). [Google Scholar]
- Wang W., Wise C., Baric T., Black M. L. & Bittles A. H. The origins and genetic structure of three co-resident Chinese Muslim populations: the Salar, Bo’an and Dongxiang. Hum Genet 113, 244–52 (2003). [DOI] [PubMed] [Google Scholar]
- Li D. N. et al. Substitution of Hainan indigenous genetic lineage in the Utsat people, exiles of the Champa kingdom. J Syst Evol 51, 287–294 (2013). [Google Scholar]
- Zhou Y. Q., Zhu W., Liu Z. P. & Wu W. Q. A quick method of extraction of DNA by Chelex-100 from trace bloodstains, Fudan Univ J Med Sci 30, 379–380 (2003). [Google Scholar]
- Marshall T. C., Slate J., Kruuk L. E. & Pemberton J. M. Statistical confidence for likelihood-based paternity inference in natural populations. Mol Ecol 7, 639–655 (1998). [DOI] [PubMed] [Google Scholar]
- Excoffier L., Laval G. & Schneider S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinform Online 1, 47–50 (2005). [PMC free article] [PubMed] [Google Scholar]
- Yan J. et al. Genetic analysis of 15 STR loci on Chinese Tibetan in Qinghai Province. Forensic Sci Int 169, e3–6 (2007). [DOI] [PubMed] [Google Scholar]
- Zhu B. F. et al. Population genetic analysis of 15 autosomal STR loci in the Russian population of northeastern Inner-Mongolia, China. Mol Biol Rep 37, 3889–3895 (2010). [DOI] [PubMed] [Google Scholar]
- Wu Y. M., Zhang X. N., Zhou Y., Chen Z. Y. & Wang X. B. Genetic polymorphisms of 15 STR loci in Chinese Han population living in Xi’an city of Shaanxi Province. Forensic Sci Int Genet 2, e15–18 (2008). [DOI] [PubMed] [Google Scholar]
- Chen J. G. et al. Population genetic data of 15 autosomal STR loci in Uygur ethnic group of China. Forensic Sci Int Genet 6, e178–9 (2012). [DOI] [PubMed] [Google Scholar]
- Zhu B. F., Shen C. M., Wu Q. J. & Deng Y. J. Population data of 15 STR loci of Chinese Yi ethnic minority group. Leg Med (Tokyo) 10, 220–224 (2008). [DOI] [PubMed] [Google Scholar]
- Zhu J. et al. Population data of 15 STR in Chinese Han population from north of Guangdong. J Forensic Sci 50, 1510–1511 (2005). [PubMed] [Google Scholar]
- Wang Z. et al. Genetic polymorphisms of 15 STR loci in Chinese Hui population. J Forensic Sci 50, 1508–1509 (2005). [PubMed] [Google Scholar]
- Zhu B. et al. Genetic analysis of 15 STR loci of Chinese Uigur ethnic population. J Forensic Sci 50, 1235–1236 (2005). [PubMed] [Google Scholar]
- Li C. et al. Genetic polymorphism of 17 STR loci for forensic use in Chinese population from Shanghai in East China. Forensic Sci Int Genet 3, e117–118 (2009). [DOI] [PubMed] [Google Scholar]
- Rerkamnuaychoke B. et al. Thai population data on 15 tetrameric STR loci-D8S1179, D21S11, D7S820, CSF1PO, D3S1358, TH01, D13S317, D16S539, D2S1338, D19S433, vWA, TPOX, D18S51, D5S818 and FGA. Forensic Sci Int 158, 234–237 (2006). [DOI] [PubMed] [Google Scholar]
- Alenizi M., Goodwin W., Ismael S. & Hadi S. STR data for the AmpFlSTR Identifiler loci in Kuwaiti population. Leg Med (Tokyo) 10, 321–325 (2008). [DOI] [PubMed] [Google Scholar]
- Coudray C. et al. Population genetic data of 15 tetrameric short tandem repeats (STRs) in Berbers from Morocco. Forensic Sci Int 167, 81–86 (2007). [DOI] [PubMed] [Google Scholar]
- Forward B. W., Eastman M. W., Nyambo T. B. & Ballard, R. E. AMPFlSTR Identifiler STR allele frequencies in Tanzania, Africa. J Forensic Sci 53, 245 (2008). [DOI] [PubMed] [Google Scholar]
- Muro T. et al. Allele frequencies for 15 STR loci in Ovambo population using AmpFlSTR Identifiler Kit. Leg Med (Tokyo) 10, 157–159 (2008). [DOI] [PubMed] [Google Scholar]
- Immel U. D., Erhuma M., Mustafa T., Kleiber M. & Klintschar M. Population genetic analysis in a Libyan population using the PowerPlex 16 system. Int Congr Ser 1288, 421–423 (2006). [Google Scholar]
- Coudray C., Guitard E., EL-Chennawi F., Larrouy G. & Dugoujon J. M. Allele frequencies of 15 short tandem repeats (STRs) in three Egyptian populations of different ethnic groups. Forensic Sci Int 169, 260–265 (2007). [DOI] [PubMed] [Google Scholar]
- Coudray C. et al. Allele frequencies of 15 tetrameric short tandem repeats (STRs) in Andalusians from Huelva (Spain). Forensic Sci Int 68, e21–24 (2007). [DOI] [PubMed] [Google Scholar]
- Marian C. et al. STR data for the 15 AmpFlSTR identifiler loci in the Western Romanian population. Forensic Sci Int 170, 73–75 (2007). [DOI] [PubMed] [Google Scholar]
- Sánchez-Diz P., Menounos P. G., Carracedo A. & Skitsa I. 16 STR data of a Greek population. Forensic Sci Int Genet 2, e71–72 (2008). [DOI] [PubMed] [Google Scholar]
- Mertens G. et al. Flemish population genetic analysis using 15 STRs of the Identifiler® kit. Int Congr Ser 1288, 328–330 (2006). [Google Scholar]
- Hernández-Gutiérrez S., Hernández-Franco P., Martínez-Tripp S., Ramos-Kuri M. & Rangel-Villalobos H. STR data for 15 loci in a population sample from the central region of Mexico. Forensic Sci Int 151, 97–100 (2005). [DOI] [PubMed] [Google Scholar]
- Ossmani H. E., Talbi J., Bouchrif B. & Chafik A. Allele frequencies of 15 autosomal STR loci in the southern Morocco population with phylogenetic structure among worldwide populations. Leg Med (Tokyo) 11, 155–158 (2009). [DOI] [PubMed] [Google Scholar]
- Halima M. S. A., Bernal L. P. & Sharif F. A. Genetic variation of 15 autosomal short tandem repeat (STR) loci in the Palestinian population of Gaza Strip. Leg Med (Tokyo) 11, 203–204 (2009). [DOI] [PubMed] [Google Scholar]
- Kutanan W., Kitpipit T., Phetpeng S.& Thanakiatkrai, Forensic STR loci reveal common genetic ancestry of the Thai-Malay Muslims and Thai Buddhists in the deep Southern region of Thailand. J Hum Genet 59, 675–681 (2014). [DOI] [PubMed] [Google Scholar]
- Butler J. M., Schoske R., Vallone P. M., Redman J. W. & Kline M. C. Allele frequencies for 15 autosomal STR loci on U.S. Caucasian, African American, and Hispanic populations. J Forensic Sci 48, 908–911 (2003). [PubMed] [Google Scholar]
- Pereira L. et al. PopAffiliator: online calculator for individual affiliation to a major population group based on 17 autosomal short tandem repeat genotype profile. Int J Legal Med 125, 629–636 (2011). [DOI] [PubMed] [Google Scholar]
- Kang L. et al. Genetic structures of the Tibetans and the Deng people in the Himalayas viewed from autosomal STRs. J Hum Genet 55, 270–277 (2010). [DOI] [PubMed] [Google Scholar]
- Falush D., Stephens M. & Pritchard J. K. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubisz M. J., Falush D., Stephens M. & Pritchard J. K. Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9, 1322–1332 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team: R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, 2013. [Google Scholar]
- Rosenberg N. A. Distruct: A program for the graphical display of population structure. Mol Ecol Notes 4, 137–138 (2004). [Google Scholar]
- Eaaswarkhanth M. et al. Diverse genetic origin of Indian Muslims: evidence from autosomal STR loci. J Hum Genet 54, 340–348 (2009). [DOI] [PubMed] [Google Scholar]
- Eaaswarkhanth M. et al. Microsatellite diversity delineates genetic relationships of Shia and Sunni Muslim populations of Uttar Pradesh, India. Hum Biol 81, 427–445 (2009). [DOI] [PubMed] [Google Scholar]
- Terreros M. C. et al. North Indian Muslims: enclaves of foreign DNA or Hindu converts? Am J Phys Anthropol 133, 1004–1012 (2007). [DOI] [PubMed] [Google Scholar]
- Eaaswarkhanth M. et al. Traces of sub-Saharan and Middle Eastern lineages in Indian Muslim populations. Eur J Hum Genet 18, 354–363 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.