If we could extract, purify, and visualize the intact DNA molecules from chloroplasts, what would those molecules look like? Most would expect to find circular DNA molecules the size of the chloroplast genome. By contrast, however, only a small fraction of the DNA obtained from chloroplasts is found as genome-sized circles. The reasons for this profound discrepancy are the subject of this article. I will trace the history of research on chloroplast DNA (cpDNA) to elucidate why it has taken more than 30 years to realize that the circle is not the principal form of DNA in chloroplasts and to examine the relationship between the chloroplast genome and the segregating genetic unit or chromosome in chloroplasts. The critical finding is that the chloroplast chromosome can contain many genome equivalents. I will also discuss the coupling between chromosomal replication and segregation in chloroplasts.
To avoid confusion, I define some terms. The genome is the entire complement of genetic material in a virus, prokaryote, mitochondrion, or chloroplast or the haploid nuclear genetic complement of a eukaryotic species. A genome equivalent is the amount of DNA in a single copy of a genome. A chromosome is the segregating genetic unit that carries either a subset or all of the genome into each daughter cell or organelle during division. For example, the ant Myrmecia pilosula has a single pair of homologous chromosomes in the nucleus (Crosland and Crozier, 1986). The amount of DNA in a single chromosome equals the amount of DNA in the genome. In most ants and most eukaryotes, however, the DNA content of a single nuclear chromosome is less than that of the genome, and the DNA content of a segregating genetic unit never exceeds that of the genome. The genome in most bacteria, such as Escherichia coli, is contained on a single DNA-containing body known as the nucleoid. As a nucleoid segregates to daughter cells upon division, that nucleoid represents a bacterial chromosome. In contrast with what is found in the eukaryotic nucleus, however, the amount of DNA per bacterial chromosome in rapidly dividing cells is typically several to many times larger than one genome equivalent. As I will show, the situation in chloroplasts is similar to that in bacteria, and the size of the chloroplast chromosome is rarely as small as one genome equivalent.
CHROMOSOMAL DNA BEFORE THE IDENTIFICATION OF CHLOROPLAST DNA MOLECULES
There is a strong belief that a chromosome from any source should contain no more DNA than one genome equivalent. The first chromosomes to be studied were those in the nucleus of eukaryotes, where cytological and genetic evidence indicated that a gamete, typically being haploid, contains a single genome. Later for bacteria, the small size of chromosomes limited cytological investigation, so only genetic evidence was informative. The monoploid nature of genetics in model prokaryotic organisms combined with the circular autoradiographic images of large E. coli DNA molecules in the early 1960s led to the conclusion that the chromosome in bacteria was contained on a single circular DNA molecule (Cairns, 1963; Bendich, 2001). It was only recently that we learned that bacterial chromosomes can consist of many copies of their genome. Rapidly dividing cells of E. coli can contain an average of ∼11 genome equivalents per cell (the range is ∼5 to 18) (Akerlund et al., 1995). Because approximately equal amounts of DNA are partitioned to daughter cells during cytokinesis (Woldringh, 1976), none of the cells in the population inherit a chromosome containing a single genome equivalent. The DNA content per cell decreases as the culture continues to grow exponentially, illustrating variability in the number of genome equivalents per chromosome. When deproteinized DNA is examined by fluorescence microscopy, most individual cells of E. coli and other bacteria contain a bunched-up form of DNA several times larger than one genome equivalent (Hinnebusch and Bendich, 1997). I conclude that because none of this information was available in 1972, when isolated cpDNA was first visualized, there was no reason then to challenge the concept of a strictly genome-sized chromosome in bacteria or to embrace the possibility of a complex and branched form of chromosomal DNA in their endosymbiotic descendant, the chloroplast.
THE BROKEN CIRCLES THEORY
Kolodner and Tewari (1972) were the first to utilize DNase treatment of chloroplasts before DNA extraction, so that contamination of cpDNA with nuclear DNA was avoided. Their article is probably the most influential paper in cpDNA research. They found that the size of the pea chloroplast genome determined by DNA reassociation kinetics measurements matched the length of the single size class of circular cpDNA molecules observed by electron microscopy (EM). Circles represented a substantial fraction (37%) of the cpDNA molecules that they measured, and because all the linear molecules were shorter than the genome size, linear molecules were attributed to breakage of the circles during extraction. This report established the Broken Circles theory for cpDNA that has dominated the field until the present day. Using similar procedures for cpDNA isolation and EM with several plant species, Herrmann et al. (1975) found as much as 80% circular cpDNA molecules in some preparations, and they too adopted the Broken Circles theory. EM was again used to provide additional examples of plants with circular cpDNA (Kolodner and Tewari, 1975a). The results of these studies were entirely consistent with those for bacteria and seemed to definitively establish the chloroplast chromosome as a genome-sized circle.
Subsequent restriction fragment mapping and genome sequencing data yielded circular genomic maps and served to solidify and generalize the conclusions in these early investigations. How then, could the early investigators have missed the bulk of the cpDNA that we now find in complex branched forms of multigenomic size (Oldenburg and Bendich, 2004)? We suspect that the very strong expectation that the chromosome in vivo should be a simple genome-sized circle, as in bacteria, may have led to the physical or conceptual exclusion from analysis of complex forms. Before examination by EM, a high-speed centrifugation step was performed, which may have removed the largest forms of cpDNA from the sample to be analyzed (Kolodner and Tewari, 1972; Herrmann et al., 1975). Another possibility is that complex forms of DNA much larger than the size of genomic circles might have been taken as nuclear DNA contamination (or uninterpretable structures) and excluded from analysis.
The use of in-gel procedures for the extraction and purification of DNA coupled with pulsed-field gel electrophoresis (PFGE) permits analysis of megabase-sized chromosomal DNA molecules. The first to adapt these procedures for cpDNA were Deng et al. (1989), who found gel bands representing an oligomeric series of genomic sizes for several plants. They subscribed to the Broken Circles theory and concluded that “the chloroplast DNA exists as a mixture of circular forms, whether or not the chloroplast DNA in the pulsed-field gels is present as circular or linear molecules.” Our work then showed that the PFGE bands contained linear cpDNA molecules, and we suggested that they and the analogous bands of Deng et al. (1989) contained cpDNA that was artifactually and randomly linearized during extraction (Bendich and Smith, 1990).
The Broken Circles theory was first invoked to explain the extreme difficulty in obtaining genome-sized circular forms of mitochondrial DNA (mtDNA) from fungi (Bendich, 1993). Williamson (2002) describes how dogged adherence to the Broken Circles theory impeded progress toward the realization that the genome in yeast mitochondria is actually borne on linear DNA. He bemoans his refusal to even measure the linear mtDNA molecules that filled his electron microscopic images. I suggested that the strong expectation in the late 1960s that fungal mtDNA should be circular derived chiefly from the discovery in the mid 1960s that circular molecules were the predominant form of mtDNA from many metazoan animals (Bendich, 1993).
When I reported complex cpDNA forms several to many times the size of the genome, I interpreted them as representing either catenated or otherwise clustered circular molecules of genome size (Bendich, 1991), exemplifying the pernicious power of the Broken Circles theory. It was only when we produced high-resolution digital images, rather than the low-resolution images produced by image intensification, that the ethidium-stained cpDNA appeared as branched linear forms including molecules longer than the genome size (Oldenburg and Bendich, 2004). Much or most of the cpDNA subjected to PFGE remains immobile, and in 1995 the consensus was that this DNA was “trapped in the agarose as large circles which cannot move into the gel” (Backert et al., 1995). It is now clear, however, that the immobility of most of this DNA is due to its complex structure, with only a minor fraction due to relaxed circular molecules trapped by impediments (Oldenburg and Bendich, 2004).
To summarize, the Broken Circles theory was formulated to provide an excuse for why we failed to find most of the DNA in the form we expected to find, both for chloroplasts and mitochondria. Furthermore, although it was long known that mapping alone cannot be used to distinguish linear from circular chromosomes (Stahl and Steinberg, 1964; Stahl, 1967; Bendich, 2001), these techniques (sequence assembly is a mapping technique) commonly provided the only data used to assign the circular form to these organellar chromosomes.
THE END OF THE CIRCLE
The ends of linear, genome-sized cpDNA molecules could be located either at specific sites within the genome or at random sites, as expected from circles broken during extraction. Deng et al. (1989) concluded that the ends were random because they did not detect any restriction fragment length polymorphism between unfractionated spinach cpDNA and the cpDNA from PFGE bands containing one to four lengths of the genome. The absence of differences between total cpDNA and the PFGE bands, however, is also expected if all forms of cpDNA (except a minor circular form) had the same, specific ends. Specific ends were found for unfractionated maize cpDNA, as well as the linear monomeric and oligomeric forms, thus providing the evidence that linear cpDNA is the most common form of DNA within chloroplasts (Oldenburg and Bendich, 2004). Because the ends were near putative origins of replication, analysis of molecular structure can also lead to insights concerning the mechanism of cpDNA replication.
THE REPLICATION OF CHLOROPLAST DNA: A REVISED VIEW
It is remarkable how little is known of even the most basic aspects of cpDNA replication. The current standard model for cpDNA replication is based largely on inferences drawn from electron micrographs of cpDNA produced 30 years ago (Kolodner and Tewari, 1975b). Furthermore, that cpDNA was obtained from nonmeristematic tissues, and it is now known that cpDNA replication is largely or entirely restricted to meristematic cells (Kuroiwa, 1991; Fujie et al., 1994). It should therefore come as no surprise that the scheme presented below differs completely from the standard model.
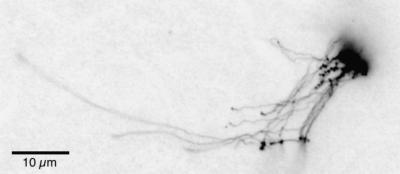
The standard model involves initiation at two sites forming D-loops, merging of the D-loops to form a θ or Cairns replication intermediate, and conversion to a rolling circle. According to this model, the start site for the rolling circle would be 180° from the D-loop origin of replication, at the terminus for θ replication where the bidirectional replication forks converge (Kolodner and Tewari, 1975b; Heinhorst and Cannon, 1993). Thus, the end sequence of the rolling circle tail would not be near the origin. Because, however, the ends of linear molecules are near putative origins, and very little maize cpDNA is found in circular form (Oldenburg and Bendich, 2004), the standard model is no longer tenable for maize. The recombination-dependent replication process for cpDNA depicted in Figure 1A accounts for the branched, multigenomic structures (Figure 1B) that can represent most of the DNA in plastids and involves no circular forms of cpDNA. Figure 2 shows an enormous structure typical of the cpDNA found in the meristematic cells at the base of a maize leaf. These structures contain long fibers emanating from a dense core of DNA. After digestion with a restriction endonuclease that cleaves once per genome, the DNA mass of such structures decreases by 50 to 60%, the average length of the peripheral fibers decreases by 40 to 50%, but the core persists (Oldenburg and Bendich, 2004). Thus, the core is not comprised of catenated circles, and its DNA is (presumably) very tightly branched. The challenge now is to determine the structure of the core to elucidate the DNA replication mechanism.
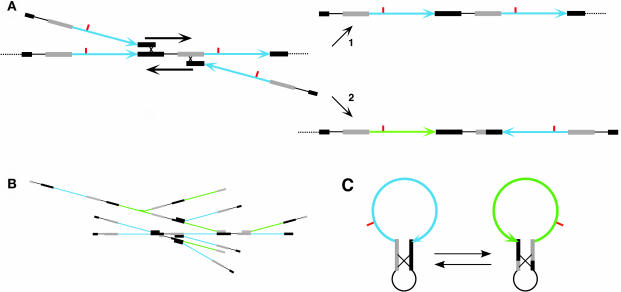
Figure 1.
Chloroplast DNA Structure and Recombination-Dependent DNA Replication.
(A) The end of a monomeric genome recombines with another molecule and initiates replication. Replication procedes to the right to generate product 1, an all-blue head-to-tail concatemer. Digestion with a restriction endonuclease that cleaves the genome once, at the site marked in red, yields a genome-size fragment. An alternative recombination initiates leftward replication to generate product 2, a head-to-tail concatemer containing an inverted (green) segment, that yields a larger-than-genome-sized fragment. Bold arrows indicate the direction of replication. The IRs are indicated by thick gray and black lines.
(B) A multigenomic structure produced by recombination-dependent replication.
(C) Circular forms of the genome produced by intramolecular recombination (flipping). Note that the larger-than-genome-sized fragment predicted by the map of product 2, and detected by Oldenburg and Bendich (2004), cannot be generated from genome-sized circles.
Figure 2.
A Multigenomic Chloroplast DNA Structure.
This ethidium-stained structure was obtained from meristematic tissue of maize. Similarly complex cpDNA forms were found for every plant species examined, including watermelon and pea (Bendich, 1991), Arabidopsis (Rowan et al., 2004), wheat, and Medicago truncatula. No such complex forms were reported for pea and maize (Kolodner and Tewari, 1972, 1975a).
It must be emphasized that the scheme in Figure 1A is based entirely on structural analysis without benefit from genetics. It is intended merely to focus attention on linear genomes, rather than circular molecules, to guide future research. Figure 1A does, however, show how the two isomeric forms of the chloroplast genome in most plants can be produced without invoking flipping: intramolecular recombination between the large inverted repeat sequences (IRs) in a circular cpDNA molecule (Figure 1C). Flipping was proposed as the mechanism responsible for the equimolar alternative fragment maps for species with IRs (Palmer, 1983, 1985). The mapping data were obtained before the advent of PFGE, so that the sizes of the largest fragments could not be determined accurately. Recent mapping employing PFGE and rare-cutting enzymes revealed a larger-than-genome-size fragment for maize cpDNA that cannot be derived from the circular forms in Figure 1C (Oldenburg and Bendich, 2004). This large fragment appeared in approximately half-stoichiometric amount, relative to other fragments. Half-stoichiometric fragments were an essential observation in the older mapping experiments that led to the proposed flipping. Figure 1A shows the linear maps, including the long half-stoichiometric fragments that render flipping untenable. The only form of the cpDNA that can lead to the observed data is the head-to-tail linear concatemer (Oldenburg and Bendich, 2004). Using EM, results with the denaturation mapping technique were interpreted to indicate a head-to-head linkage between neighboring genomic units in most (extremely rare) circular dimers of cpDNA from lettuce and spinach (Kolodner and Tewari, 1979). The same dumbbell forms, however, would result from head-to-tail dimers containing an inversion of the large single copy region in one of the genomic units (Oldenburg and Bendich, 2004). I conclude that all concatemeric forms of cpDNA are likely composed of head-to-tail units.
To summarize, it is replication driven by recombination that leads to the cpDNA branching, head-to-tail concatemers, and inversion isomers. The isomers cannot result from flipping. The small amount of cpDNA found in circular form may actually serve no function that depends on circularity, but may represent an incidental byproduct of the recombination events required for cpDNA replication and repair, as we suggested for mtDNA circles (Oldenburg and Bendich, 1998).
CHROMOSOMES IN CHLOROPLASTS ARE COMPLEX IN STRUCTURE AND VARIABLE IN SIZE
In a eukaryotic cell, the G2 phase of the cell cycle separates the replication of nuclear DNA in the S phase from segregation of the duplicated chromosomes in the M phase. Activities in the G2 phase ensure that only once-replicated and precisely replicated chromosomes are inherited by daughter cells. Such activities are absent in E. coli cells that divide every 25 min at 37° and require 40 min to replicate their genome (Helmstetter, 1996), so that chromosomal segregation occurs before DNA replication is complete and the DNA content among cells is quite variable (Akerlund et al., 1995). I suggest that it is the absence of such activities in the chloroplast that allows both the segregation to daughter organelles of chromosomes containing multigenomic amounts of DNA, including incompletely replicated genomes, and the large variability in DNA content among chloroplasts in the population. In the terms of Murray (2004), the coupling between chromosomal replication and segregation is flexible for the chloroplast and tight for the cell that harbors the chloroplast.
Flexible coupling for chloroplasts is best illustrated in Scenedesmus quadricauda, a chlorococcal alga with a single chloroplast. In one daughter cell, the chloroplast contains nine nucleoids each carrying from 1 to 20 genome equivalents (assuming a 200-kb genome size), and the other daughter contains five nucleoids with 4 to 19 genomes (Zachleder et al., 1995). Only one of the 30 nucleoids among the two pairs of daughter cells analyzed contained as little as one genome equivalent.
I propose the following rules for the chromosomes in chloroplasts. (1) The replication of cpDNA begins at one of several potential replication origins on linear cpDNA molecules, but most of the cpDNA is produced via recombination-dependent replication (RDR) that does not require firing of an origin. This RDR process is similar to those for herpes simplex virus (HSV) and bacteriophage T4, both of which contain a genome borne on linear DNA molecules (Kreuzer and Morrical, 1994; Jackson and DeLuca, 2003). A T4-like RDR process was also proposed for the mtDNA in plants (Oldenburg and Bendich, 1998). (2) Replicating cpDNA structures consist of branched, multigenomic forms, and it is these complex cpDNA forms that comprise the chromosomes in chloroplasts. For both HSV and T4, replication intermediates are multigenomic structures that are processed down to genome-sized units before packaging into virus particles because of the need to maximize the number of infectious particles per genome. There is no such need for chloroplasts that divide into only two daughter organelles, and so chloroplast chromosomes are inherited without processing. (3) There is usually more than one genome equivalent in a chloroplast chromosome, just as in rapidly dividing cells of E. coli. After cpDNA replication ceases, and the number of chloroplasts per cell increases to the final level in nonmeristematic cells, the number of genome equivalents per chromosome decreases. This plasticity of DNA content per chromosome is also observed for E. coli as the culture ages, but contrasts with the constancy found in the nucleus of plant cells.
THE CHLOROPLAST NUCLEOID: A REVISED VIEW
I now apply these rules to reinterpret cytological observations of the chloroplast nucleoid that are puzzling when a circular genome-sized molecule is assumed to represent the chloroplast chromosome. Chlamydomonas is a single-celled alga containing one chloroplast and has served as the principal organism for studying chloroplast inheritance. Vegetative cells of C. reinhardtii contain ∼75 copies of the chloroplast genome distributed unequally among 8 to 10 nucleoids (Armbrust, 1998). If the nucleoid is the segregating genetic unit, as proposed by VanWinkle-Swift (1980), we have a conundrum. How do we reconcile the small number of nucleoids (chromosomes) with the large number of genomes if each genome exists as a discrete circle (also a possible chromosome)? The problem of defining a chromosome intensifies when the 8 to 10 nucleoids coalesce to one large nucleoid in the zygote. The conundrum evaporates, however, when we consider the nucleoid as one complex containing many genomes linked by recombination junctions. During the resting stage that follows zygote formation, origin-independent replication would produce the cpDNA to be partitioned among the four haploid zygospores issued from the zygote.
A long-standing cytological puzzle is the nature of the cpDNA in nucleoids of fibrous form, including a two-dimensional beaded ring that lies just under the plastid envelope, and a three-dimensional skein distributed throughout the chloroplast (Coleman, 1978; Kuroiwa et al., 1981). How can such structures be composed of discrete circles of cpDNA? I suggest that all such forms contain single, complex cpDNA molecules of the type shown in Figure 2, rather than a series of circles tethered to a sinuous membrane (Kuroiwa, 1991).
During the development of chloroplasts from proplastids in wheat (Miyamura et al., 1986) and Arabidopsis thaliana (Fujie et al., 1994), nucleoids first increase in size and then fragment and disperse. What changes at the level of cpDNA molecules accompany these cytological changes? In one scheme, there is an initial increase in the number of genome-sized circles at clustered membrane attachment sites. Then the circles are scattered as the thylakoid membranes enlarge and ramify throughout the expanding and greening chloroplast (Kuroiwa, 1991). I now consider an alternative scheme based on RDR.
As the first foliage leaf of Arabidopsis develops from cells in the shoot apical meristem, the number of genomes per plastid increases from ∼40 (3 d after seeds are sown) to 600 at day 7, when the leaf is <0.5 mm in length (Fujie et al., 1994). The nucleoids increase in size during this interval, and then decrease in size and disperse within the chloroplast by day 10. In the expanded leaf at day 38, the 4′,6-diamidino-2-phenylindole fluorescence of nucleoids is greatly reduced, the cpDNA has changed from structures such as shown in Figure 2 to much smaller forms, and many chloroplasts contain no detectable DNA (Rowan et al., 2004). Similarly, most chloroplasts from mature maize leaves contain no DNA at all (D.J. Oldenburg and A.J. Bendich, unpublished data). I propose that the size and branching of complex cpDNA forms reflects their activity in RDR. As replication winds down, and forks reach the ends of template strands, DNA is released from the complex and borne away on membranes. Although the demise of cpDNA occurs long before the onset of leaf senescence and appears to be developmentally programmed in both Arabidopsis and maize, the generality of these observations for other species remains to be assessed.
To summarize, the variability in chromosomal DNA molecules among and within plastids is only perplexing if one assumes a genome-sized chromosome and a tight coupling between chromosomal replication and segregation, as in the nucleus. The variability is not at all surprising if the coupling is flexible, as in bacteria. The disappearance of cpDNA from mature, nonmeristematic leaf cells may be surprising, but such cells no longer need chloroplast chromosomes as vehicles of inheritance or apparently to encode gene products.
CONCLUDING REMARKS
There are lessons to be learned about how to interpret data that initially seem at odds with popular thinking. Chief among them is that complex DNA structures should not be dismissed simply because they do not meet our expectations for a chromosomal DNA molecule: simple in form and genomic in size. The complex cpDNA should not have been removed either physically or conceptually before analysis. The ploidy paradox (few segregating genetic units but many genomes per chloroplast) should have directed us toward chromosomes of multigenomic size. Instead, the rare circular forms were a distraction. Finally, although mapping cannot establish chromosomal topology (linear head-to-tail concatemers and linear T4 phage DNA molecules have circular maps), a circular map continues to be the principal or only evidence used to conclude that a chromosome is circular both for chloroplasts and bacteria.
Acknowledgments
I thank George Miklos and Delene Oldenburg for critically reading the manuscript and Delene Oldenburg for producing the figures. This work was supported by a grant from the USDA (2002-35301-12021).
References
- Akerlund, T., Nordstrom, K., and Bernander, R. (1995). Analysis of cell size and DNA content in exponentially growing and stationary-phase batch cultures of Escherichia coli. J. Bacteriol. 177, 6791–6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbrust, E.V. (1998). Uniparental inheritance of chloroplast genomes. In The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas, S. Merchant, ed (Amsterdam: Kluwer), pp. 93–113.
- Backert, S., Dörfel, P., and Börner, T. (1995). Investigation of plant organellar DNAs by pulsed-field gel electrophoresis. Curr. Genet. 28, 390–399. [DOI] [PubMed] [Google Scholar]
- Bendich, A.J. (1991). Moving pictures of DNA released upon lysis from bacteria, chloroplasts, and mitochondria. Protoplasma 160, 121–130. [Google Scholar]
- Bendich, A.J. (1993). Reaching for the ring: The study of mitochondrial genome structure. Curr. Genet. 24, 279–290. [DOI] [PubMed] [Google Scholar]
- Bendich, A.J. (2001). The form of chromosomal DNA molecules in bacterial cells. Biochimie 83, 177–186. [DOI] [PubMed] [Google Scholar]
- Bendich, A.J., and Smith, S.B. (1990). Moving pictures and pulsed-field gel electrophoresis show linear DNA molecules from chloroplasts and mitochondria. Curr. Genet. 17, 421–425. [DOI] [PubMed] [Google Scholar]
- Cairns, J. (1963). The chromosome of Escherichia coli. Cold Spring Harb. Symp. Quant. Biol. 28, 43–46. [DOI] [PubMed] [Google Scholar]
- Coleman, A.W. (1978). Visualization of chloroplast DNA with two fluorochromes. Exp. Cell Res. 114, 95–100. [DOI] [PubMed] [Google Scholar]
- Crosland, M.W.J., and Crozier, R.H. (1986). Myrmecia pilosula, an ant with only one pair of chromosomes. Science 231, 1278–1281. [DOI] [PubMed] [Google Scholar]
- Deng, X.-W., Wing, R.A., and Gruissem, W. (1989). The chloroplast genome exists in multimeric forms. Proc. Natl. Acad. Sci. USA 86, 4156–4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujie, M., Kuroiwa, H., Kawano, S., Mutoh, S., and Kuroiwa, T. (1994). Behavior of organelles and their nucleoids in the shoot apical meristem during leaf development in Arabidopsis thaliana L. Planta 194, 395–405. [DOI] [PubMed] [Google Scholar]
- Heinhorst, S., and Cannon, G.C. (1993). DNA replication in chloroplasts. J. Cell Sci. 104, 1–9. [Google Scholar]
- Helmstetter, C.E. (1996). Timing of synthetic activities in the cell cycle. In Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology, H.E. Umbarger, ed (Washington, DC: American Society for Microbiology), pp. 1627–1629.
- Herrmann, R.G., Bohnert, H.-J., Kowallik, K.V., and Schmitt, J.M. (1975). Size, conformation and purity of chloroplast DNA of some higher plants. Biochim. Biophys. Acta 378, 305–317. [DOI] [PubMed] [Google Scholar]
- Hinnebusch, B.J., and Bendich, A.J. (1997). The bacterial nucleoid visualized by fluorescence microscopy of cells lysed within agarose: Comparison of Escherichia coli and spirochetes of the genus Borrelia. J. Bacteriol. 179, 2228–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, S.A., and DeLuca, N.A. (2003). Relationship of herpes simplex virus genome configuration to productive and persistent infections. Proc. Natl. Acad. Sci. USA 100, 7871–7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner, R., and Tewari, K.K. (1972). Molecular size and conformation of chloroplast deoxyribonucleic acid from pea leaves. J. Biol. Chem. 247, 6355–6364. [PubMed] [Google Scholar]
- Kolodner, R., and Tewari, K.K. (1975. a). The molecular size and conformation of the chloroplast DNA from higher plants. Biochim. Biophys. Acta 402, 372–390. [DOI] [PubMed] [Google Scholar]
- Kolodner, R., and Tewari, K.K. (1975. b). Chloroplast DNA from higher plants replicates by both the Cairns and the rolling circle mechanism. Nature 256, 708–711. [DOI] [PubMed] [Google Scholar]
- Kolodner, R., and Tewari, K.K. (1979). Inverted repeats in chloroplast DNA from higher plants. Proc. Natl. Acad. Sci. USA 76, 41–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzer, K.N., and Morrical, S.W. (1994). Initiation of DNA Replication. In Molecular Biology of Bacteriophage T4, J.D. Karam, ed (Washington DC: American Society for Microbiology Press), pp. 28–42.
- Kuroiwa, T. (1991). The replication, differentiation, and inheritance of plastids with emphasis on the concept of organelle nuclei. Int. Rev. Cytol. 128, 1–61. [Google Scholar]
- Kuroiwa, T., Suzuki, T., Ogawa, K., and Kawano, S. (1981). The chloroplast nucleus: Distribution, number, size, and shape, and a model for the multiplication of the chloroplast genome during chloroplast development. Plant Cell Physiol. 22, 381–396. [Google Scholar]
- Miyamura, S., Nagata, T., and Kuroiwa, T. (1986). Quantitative fluorescence microscopy on dynamic changes of plastid nucleoids during wheat development. Protoplasma 133, 66–72. [Google Scholar]
- Murray, A.W. (2004). Recycling the cell cycle: Cyclins revisited. Cell 116, 221–234. [DOI] [PubMed] [Google Scholar]
- Oldenburg, D.J., and Bendich, A.J. (1998). The structure of mitochondrial DNA from the liverwort, Marchantia polymorpha. J. Mol. Biol. 276, 745–758. [DOI] [PubMed] [Google Scholar]
- Oldenburg, D.J., and Bendich, A.J. (2004). Most chloroplast DNA of maize seedlings in linear molecules with defined ends and branched forms. J. Mol. Biol. 335, 953–970. [DOI] [PubMed] [Google Scholar]
- Rowan, B.A., Oldenburg, D.J., and Bendich, A.J. (2004). The demise of chloroplast DNA in Arabidopsis. Curr. Genet., in press. [DOI] [PubMed]
- Palmer, J.D. (1983). Chloroplast DNA exists into two orientations. Nature 301, 92–93. [Google Scholar]
- Palmer, J.D. (1985). Comparative organization of chloroplast genomes. Annu. Rev. Genet. 19, 325–354. [DOI] [PubMed] [Google Scholar]
- Stahl, F.W. (1967). Circular genetic maps. J. Cell. Physiol. 70 (suppl.), 1–12. [DOI] [PubMed] [Google Scholar]
- Stahl, F.W., and Steinberg, C.M. (1964). The theory of formal phage genetics for circular maps. Genetics 50, 531–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanWinkle-Swift, K.P. (1980). A model for the rapid vegetative segregation of multiple chloroplast genomes in Chlamydomonas: Assumptions and predictions of the model. Curr. Genet. 1, 113–125. [DOI] [PubMed] [Google Scholar]
- Williamson, D. (2002). The curious history of yeast mitochondrial DNA. Nat. Rev. Genet. 3, 475–481. [DOI] [PubMed] [Google Scholar]
- Woldringh, C.L. (1976). Morphological analysis of nuclear separation and cell division during the life cycle of Escherichia coli. J. Bacteriol. 125, 248–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachleder, V., Kawano, S., and Kuroiwa, T. (1995). The course of chloroplast DNA replication and its relationship to other reproductive processes in the chloroplast and nucleocytoplasmic compartment during the cell cycle of the alga Scenedesmus quadricauda. Protoplasma 188, 245–251. [Google Scholar]