Abstract
Allelic expression variation of nonimprinted autosomal genes has recently been uncovered in mouse hybrids and humans. The allelic expression variation is attributed to differences in noncoding DNA sequences and does not involve epigenetic regulation or gene imprinting. This expression variation is suggested to play important roles in determining phenotypic diversity. Virtually nothing is known about such allele-specific expression variation in a hybrid plant where two alleles are compared in the same genetic context. We examined parental transcript accumulation in maize (Zea mays) hybrids using allele-specific RT-PCR analysis. Among 15 genes analyzed, 11 showed differences at the RNA level, ranging from unequal expression of the two alleles (biallelic) to expression of a single allele (monoallelic). Maternal or paternal transmission had little effect on the allele-specific transcript ratio of nearly all genes analyzed, suggesting that parent-of-origin effect was minimal. We analyzed the allelic difference in genetically contrasting hybrids and hybrids under high planting density and drought stress. Whereas a genetically improved modern hybrid expressed both alleles, a less improved old hybrid frequently showed mono-allelic expression. Furthermore, the two alleles in the hybrid responded differentially to abiotic stresses. The results of allele-specific regulation in different tissues in responding to environment and stress suggest an unequivalent function of the parental alleles in the hybrid, which may have an impact on heterosis.
INTRODUCTION
Nucleotide sequence variation can potentially alter protein function if it occurs in the coding region and causes a qualitative difference. Alternatively, regulatory allelic variants can affect the level of gene expression and result in quantitative variants. Both types of allelic variation may produce functional changes and affect the phenotype of an organism. Naturally occurring allelic diversity in plants has been suggested to be an important genetic component for phenotypic variation (Doebley and Lukens, 1998; Buckler and Thornsberry, 2002). In contrast with induced mutations that eliminate or cause a large reduction of a functional gene product, naturally occurring allelic variation modulates gene products and may be an underlying mechanism for quantitative trait variation (Tanksley, 1993; Yano and Sasaki, 1997; Mackay, 2001). A large amount of nucleotide sequence data has been generated as facilitated by recent advances in DNA sequencing technology. The sequence data reveals that nucleotide sequence variation widely exists within a species. The maize (Zea mays) genome has an especially high level of DNA sequence polymorphism, approximately an order of magnitude higher than that in humans (Sunyaev et al., 2000; Bhattramakki et al., 2002; Buckler and Thornsberry, 2002; Ching et al., 2002). The types of allelic variants range from single nucleotide polymorphisms (SNPs), insertion/deletions (InDels), to large variants in structure involving several kilobases of DNA fragments (Fu and Dooner, 2002). These allelic variants may play a role in gene regulation and affect the expression level. However, the relationship of allelic expression differences resulting from changes in a regulatory region and the resultant phenotype is poorly understood because of its complexity and the lack of efficient methodology (Cowles et al., 2002; Glazier et al., 2002).
Recent studies have shown that allelic variation of expression level occurs frequently in mammals. It has been reported in mouse hybrids (Cowles et al., 2002) and humans (Yan et al., 2002) that alleles of nonimprinted autosomal genes are not expressed equally at the transcript level, and such allelic expression variation has been suggested as a regulatory mechanism involved in genetic diseases. Therefore, the difference in the regulatory regions of a gene could affect the pattern of gene expression and cause phenotypic variation. The importance of this allelic expression variation in determining phenotypic diversity has been reviewed recently by Knight (2004). Virtually nothing is known regarding such allelic variation of gene expression at the transcript level in maize. The high level of allelic variants in maize could lead to a high level of allelic expression variation. This hypothesis can be tested by comparing the two alleles in the maize hybrid where the alleles are exposed to a common genetic and environmental context.
Furthermore, for nearly a century, geneticists and breeders have known that the progeny of crosses between unrelated individuals often exhibit hybrid vigor or heterosis, a phenomenon of the superior performance of hybrid progeny in comparison with their inbred parents (Shull, 1908). The genetic difference between the inbred parents presumably contributes to the genetic basis of heterosis. The allelic sequence differences between inbreds have been characterized by analysis of SNPs or InDels (Helentjaris et al., 1985; Smith and Smith, 1992). Very little is known regarding the allelic differences at the transcript level in the hybrid.
In this study, we used RT-PCR to measure the proportion of transcript contributed by each parental allele expressed in the maize hybrid. We further analyzed the allele-specific transcript variation of hybrids of recent versus past development. Because yield gains of modern hybrids are primarily attributable to the genetic improvement in tolerance to both biotic and abiotic stresses (Janick, 1999; Duvick, 2001), we sought to compare allelic expression in a modern and an old hybrid and hybrids under high planting density and drought stress. Results from this study showed the predominance of allelic expression variation at the accumulated transcript level in maize hybrids, different allelic expression patterns between the two hybrids analyzed, and allelic variation in responding to environmental stresses.
RESULTS
Allele-Specific Transcript Accumulation in Hybrids
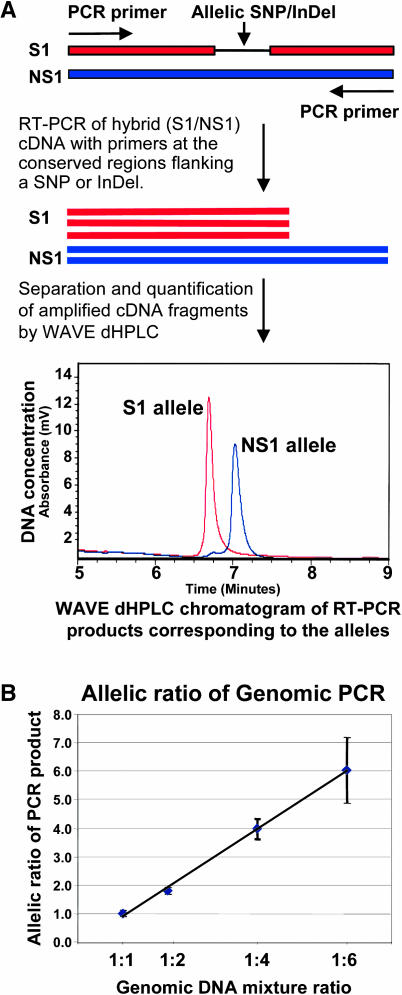
To compare the expression level of the two parental alleles in maize hybrids, we developed a method to measure the allele-specific transcript. This method involved RT-PCR of hybrid cDNA, separation, and then quantification of allele-specific cDNA fragments using the WAVE denaturing HPLC (dHPLC) system. The WAVE dHPLC system separates DNA fragments by fragment size (InDels) or denaturing temperature (SNPs) (Guo et al., 2003; Figure 1A). It is important to note that transcript levels measured by this technology reflect only the steady state accumulated mRNA amount and not the rate of transcription or posttranscriptional effects. The accuracy of this method in measuring the allele-specific transcript was tested by PCR analysis of a series of genomic DNA mixtures from two inbred parents with known ratios (1:1, 1:2, 1:4, and 1:6). The allelic ratio after amplification showed a linear relationship with the synthetic fold and was not significantly different from the original genomic DNA mixture ratio (Figure 1B).
Figure 1.
Schematic Illustration of Allele Expression Analysis Using the WAVE dHPLC System and Genomic PCR as Control.
(A) RT-PCR and allele-specific cDNA quantification. The parental alleles of a gene are cloned and sequenced to find an allelic polymorphism, either SNPs or InDels, shown as the thin line. RT-PCR is then performed with hybrid cDNA using primers designed at the conserved region between the alleles. The RT-PCR products were separated and quantified by the WAVE dHPLC system. The longer DNA fragments corresponding to one parental allele have higher affinity than the shorter DNA fragments and, therefore, take a longer time to be eluted from the WAVE column. (In the case of SNPs, the allele-specific fragments were separated by differential melting temperature.) Chromatogram traces for each PCR were generated by UV detection. When both alleles are expressed, the two types of cDNA sequences could form two types of heteroduplex (on the left of the chromatogram in some hybrids if any) in addition to the two types of homoduplexes (corresponding to two alleles). The two types of heteroduplexes are eluted earlier than the homoduplexes because of the low affinity to the column and shown as one or two peaks depending on the separation condition. Peak areas corresponding to the homoduplexes and heteroduplexes were calculated by WAVEMAKER software. The x axis is the time in minutes when the DNA fragments are eluted out. The y axis is the UV absorbency unit measuring the DNA concentration or expression level. This analysis quantifies the allele-specific transcript in a relative ratio and does not measure the absolute transcript level expressed in the hybrid.
(B) Allelic ratio of PCR product from genomic DNA mixture series. The genomic PCR was used to test whether PCR amplification maintained the allelic ratio with different proportions of the allelic genomic DNA. Genomic DNA from parental inbreds S1 and NS1 were mixed according to the ratio of S1:NS1 as 1:1, 1:2, 1:4, and 1:6. We used gene-specific primers (LTP was used because of the available genomic DNA sequences and its dynamic allelic expression differences; see Results) for the PCR analysis. The amplified genomic DNA fragments, a mixture of the two parental alleles, were separated and quantified by the WAVE dHPLC system. Three replicates were done for each mixture, and the means of the genomic allelic ratio (NS1:S1) are shown with standard deviation (error bar). The allelic ratios of the genomic PCR product are not significantly different (P > 0.05) from the corresponding DNA mixture ratio.
Although we designed the PCR primers at the conserved regions between alleles, we conducted genomic DNA PCR analysis with hybrid S1/NS1 for four genes to test for any PCR amplification bias for either allele, which may confound the allele expression data. The greatest deviation from the expected allelic ratio of 1.0 in the hybrid genomic DNA PCR analysis was 0.85 and was not significantly different from 1.0 (Table 1). Therefore, the allelic transcript ratios generated from the cDNA PCR are not likely because of any amplification bias. Subsequently, we used an allelic ratio cutoff of 0.85 (or 1.18, the inverse of 0.85) as a threshold for allelic transcript ratios deviating significantly from 1.0. We also tested whether the allelic transcript ratio was confounded by amplification of gene family members during RT-PCR. We used the PCR primer sequences in BLAST (Altschul et al., 1990) searches against the Pioneer/DuPont EST database to confirm that the primer sets did not have any significant matches to other genes. Amplicon sequencing and WAVE chromatograms confirmed that only one PCR product was amplified with each primer set.
Table 1.
Allele-Specific Transcript Ratio in Hybrid S1/NS1 Grown in Different Environments
| Mean Allelic Ratio NS:S (sd)
|
||||
|---|---|---|---|---|
| Gene Annotation | April Planting | May Planting | June Planting | Genomic PCR |
| LTP (Lipid transfer protein) | Monoallelic | 0.92 (0.05) | Monoallelic | 0.89 (0.04) |
| GGT (Geranyl geranyl transferase) | 3.67 (0.28)**,a | 4.15 (0.30)**,a | 4.58 (0.31)**,a | 0.85 (0.07) |
| GAB (Glu/Asp binding peptide) | 0.63 (0.13)**,a | 0.62 (0.13)**,a | 0.71 (0.10)*,a | – |
| AP2 (AP2 domain containing protein) | 1.55 (0.12)**,a | 1.74 (0.14)**,a,b | 1.92 (0.17)**,b | – |
| Histone H2B | 1.45 (0.10)**,a | 1.42 (0.09)**,a | 1.22 (0.06)**,b | 1.09 (0.03) |
| ARDA (Auxin repressed dormancy associated protein) | 0.87 (0.04)a | 2.30 (0.20)**,b | 1.36 (0.08)**,a | – |
| PRP (Pro-rich protein) | 1.28 (0.07)**,a | 1.34 (0.08)**,b | 1.41 (0.09)**,b | – |
| Histone H4 | 0.83 (0.05)**,a | 0.82 (0.06)*,a | 0.86 (0.04)a | 1.04 (0.03) |
| ZmPIP1-3 (Plasma membrane integral protein) | 0.92 (0.03)a | 0.80 (0.07)**,a | 0.93 (0.02)a | – |
| Ribo-S29 (Ribosomal S29) | 1.00 (0.00)a | 0.94 (0.02)a | 0.95 (0.01)a | – |
| UCR (Ubiquinol-cytochrome C) | 1.06 (0.02)a | 1.05 (0.01)a | 1.01 (0.00)a | – |
| PGS (Putative glutathione) | ∼1 | ∼1 | ∼1 | – |
| Ribo-S4 (Ribsomal S4 protein) | <1 | <1 | <1 | – |
| ZmPIP2-1 (Plasma membrane integral protein) | ∼1 | ∼1 | ∼1 | – |
| MSP (Met synthase reductase iron-sulfur subunit) | >1 | >1 | >1 | – |
The allelic transcript ratio (NS1:S1) was compared with the expected ratio of equal allelic expression 1.0. The mean, standard error (in parenthesis), and P values in the t test were based on three RT-PCR replicates. *, P < 0.05; **, P < 0.01 (italicized). The allele expression of four genes could not be quantified on the WAVE because of the limited separation of the alleles (e.g., PGS shown in Figure 2C). We estimated the allelic ratio by the peak height of the two alleles as NS:S ∼1, <1, or >1. Planting dates were April 1, May 8, and June 3. Planting date treatments with different letters (a and b) are significantly different (shown in boldface); treatments with the same letter are not significantly different. The dash indicates that data were not available. In the case of monoallelic expression of LTP, only the NS1 allele was expressed.
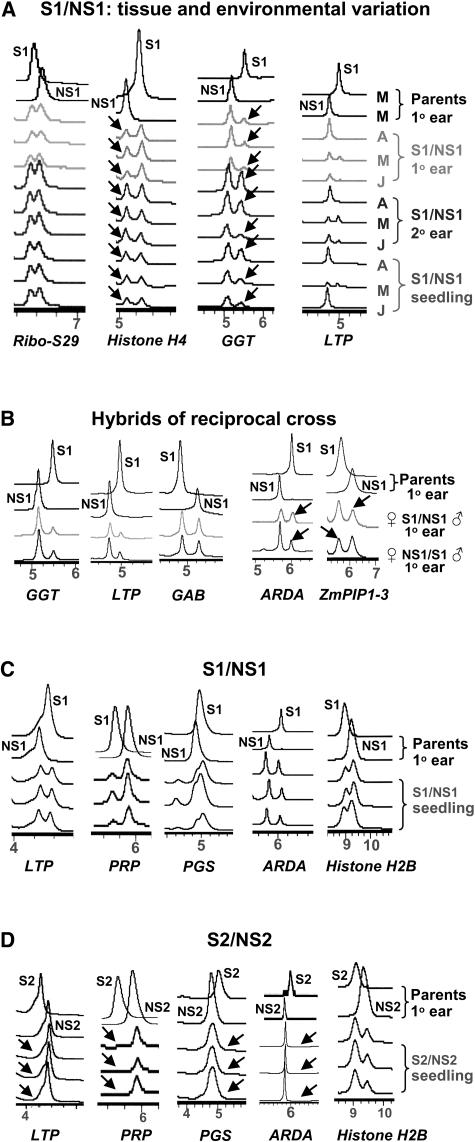
We analyzed gene expression using immature ears, prepollination of hybrid 3394 (S1/NS1), which were grown in three environments (see Methods). We first selected a random set of genes for this study from the Pioneer/DuPont EST databases. We then obtained partial sequences of these genes for S1 and NS1 alleles. Fifteen genes were selected for allelic expression analysis based on the criteria of presence of allele sequence polymorphisms resolvable by the WAVE dHPLC system and expression in immature ear tissue. The selection of these genes was therefore random in a sense that it involved little biological input from the perspective of this study. It is important to note that only partial regions of these genes (not the entire gene) were analyzed to find allele-specific polymorphism as a marker (see supplemental data online). Very often, the allelic sequence polymorphisms were located at the 3′ untranslated region. The relative allele expression in the hybrid is measured as a ratio of the transcript amount from the NS1 parental allele relative to that from the S1 parental allele. An equal allele expression in the hybrid would give a transcript ratio of NS1:S1 = 1.0. Allele-specific transcript ratios for nine genes expressed in this hybrid deviated from 1.0 (P < 0.05) in at least one environment, indicating unequal transcript accumulation between the two alleles (Table 1). The allelic variation ranged from near equal expression (biallelic) to only one allele (monoallelic) being expressed at a detectable level.
When comparing hybrids that were grown in different environments, we found five genes exhibited significant environmental variation in their allelic ratio (Table 1). The most dramatically affected genes were LTP (Lipid transfer protein) and ARDA (Auxin repressed dormancy associated protein). The LTP gene showed monoallelic expression in one environment but biallelic expression in others (Figure 2A). These genes are known to respond to stress (Treviño and O'Connell, 1998; Park and Han, 2003; Yubero-Serrano et al., 2003). Based on our microarray expression profiling experiments (our unpublished data), we know that these two genes also showed significantly reduced expression under abiotic stress in maize immature ear tissue. The allelic transcript data indicates that different alleles of LTP and ARDA responded differentially in different environments. Because only one biological sample was analyzed in each environment, some of the environmental variation could be confounded with variation between plants, although each sample consisted of three pooled individual plants.
Figure 2.
WAVE Chromatograms Showing Allelic Variation in Transcript Accumulation in Hybrids.
The x axis is time in minutes when the cDNA fragments are eluted from the column. The y axis is the DNA concentration measured by UV absorbency. (The y axis plots are not shown to allow for better juxtaposition of the allelic traces for comparison.) The parent samples were used as allele references.
(A) Allele expression in different tissues and growing environments. Each sample consisted of tissue pooled from three individual plants. A, April 1 planting; M, May 8 planting; J, June 3 planting. 1°, primary ear; 2°, secondary ear. The parent samples were from the May planting and are shown as allele references. Arrows indicate the relative allele expression changes.
(B) Expression of five genes is shown as examples in hybrids of reciprocal cross, in which the female parent is written on the left and the male on the right. The first three genes (left) are examples of genes in which the allele expression pattern is not significantly different between the reciprocal hybrids. The two genes on the right exhibited significant difference in allele-specific transcript ratio (P < 0.05) when alleles transmitted maternally versus paternally (Table 3).
(C) Allele expression patterns in the S1/NS1 hybrid.
(D) Allele expression patterns in the S2/NS2 hybrid. Contrasting expression patterns (biallelic versus monoallelic expression) between the two hybrids are shown in the first four genes (see arrows). The last gene, Histone H2B, which exhibited biallelic expression, is shown as a control to illustrate that the monoallelic expression of other genes in hybrid S2/NS2 was not an artifact of RNA or cDNA quality because the same RNA and cDNA samples were used in all genes analyzed.
Allele expression variation in different tissue types and developmental stages was examined with four genes, Ribo-S29 (Ribosomal S29 protein), Histone H4, GGT (Geranyl geranyl transferase), and LTP and in the following three tissues: primary ear, secondary immature ears (prepollination), and seedlings of the hybrid. We sampled these four genes based on their detectable expression in all three tissues. The Ribo-S29 gene demonstrated equal allelic expression. Ribo-S29 and LTP did not show significant difference in different tissues. GGT and Histone H4 exhibited developmental variation in allele expression (i.e., the allelic difference was greater in the primary immature ear compared with the other two tissues) (Figure 2A, Table 2). The primary ear is developmentally more advanced compared with the secondary immature ear and seedling. Although with a limited number of genes and tissues, the results may suggest a differential regulation of the two parental alleles in these tissues and a trend toward increased allele expression difference as development progresses.
Table 2.
Allelic Variation in Different Tissues of Hybrid S1/NS1
| Gene Annotation | Tissue Types | Mean Allelic Ratio NS:S (sd) | Developmental Stage Comparison |
|---|---|---|---|
| GGT | 1° Ear | 4.13** (0.30) | a |
| 2° Ear | 2.40* (0.20) | b* | |
| Seedling | 2.27** (0.20) | b* | |
| Histone H4 | 1° Ear | 0.84** (0.05) | a |
| 2° Ear | 1.19* (0.05) | b** | |
| Seedling | 1.17 (0.09) | b** |
The mean, standard error, and P values in the t test were based on three environmental replicates. 1°, primary ear; 2°, secondary ear; seedling, V4 leaf stage. *, P < 0.05; **, P < 0.01. Tissue type treatments with different letters (a and b) are significantly different; treatments with the same letter are not significantly different.
Allele-Specific Transcript Accumulation in Hybrids of Reciprocal Crosses
To test whether a parent-of-origin effect was responsible for the expression differences of the alleles, we analyzed hybrids of reciprocal crosses, S1/NS1 and NS1/S1, where the first inbred denotes the female parent. We analyzed one biological replicate of each genotype that consisted of three individual plants. Thirteen of the 15 genes showed the same expression pattern in primary ears of reciprocal hybrids (Table 3, Figure 2B), suggesting that there is no parent-of-origin effect for the majority of genes. The allele-specific transcript ratio of ARDA and ZmPIP1-3 (Plasma membrane integral protein), however, showed statistically significant difference between reciprocal hybrids, exhibiting maternally preferred expression.
Table 3.
Allele-Specific Transcript Ratio in Hybrids of Reciprocal Cross
| Gene Annotation | Mean Allelic Ratio NS:S (sd)
|
|
|---|---|---|
| S1/NS1 | NS1/S1 | |
| LTP | 1.72 (0.14)**,a | 1.47 (0.11)*,a |
| GGT | 4.28 (0.30)**,a | 3.30 (0.26)**,a |
| GAB | 0.55 (0.15)**,a | 0.59 (0.14)**,a |
| AP2 | 1.83 (0.16)**,a | 1.86 (0.16)**,a |
| Histone H2B | 1.56 (0.12)**,a | 1.61 (0.12)**,a |
| ARDA | 1.29 (0.07)*,a | 2.51 (0.22)**,b |
| PRP | 1.26 (0.07)**,a | 1.26 (0.07)**,a |
| Histone H4 | 0.95 (0.02)a | 0.93 (0.02)a |
| ZmPIP1-3 | 0.75 (0.08)**,a | 1.02 (0.02)b |
| Ribo-S29 | 0.97 (0.01)a | 0.93 (0.02)a |
| UCR | 0.92 (0.03)a | 0.76 (0.10)a |
| PGS | >1 | >1 |
| Ribo-S4 | <1 | <1 |
| ZmPIP2-1 | ∼1 | ∼1 |
| MSP | >1 | >1 |
See notes in Table 1. The allelic transcript ratio (NS1:S1) was compared with the expected ratio of equal allelic expression 1.0. The mean, standard error (in parenthesis), and P values in the t test were based on three RT-PCR replicates. *, P < 0.05, **, P < 0.01 (italicized). Treatments (reciprocal hybrids) with different letters (a and b) are significantly different (P < 0.05; in boldface); treatments with the same letter are not significantly different.
Allele Expression in a Modern and an Old Hybrid
To examine the historical prevalence of allelic difference in transcript accumulation, we analyzed two contrasting hybrids, a modern hybrid 3394 (S1/NS1) and a less improved hybrid 3306 (S2/NS2), which was one of the earliest (1960s) commercial single cross hybrids released by Pioneer Hi-Bred. The hybrid S2/NS2 is typical of the germplasm that farmers used in the late 1960s, whereas hybrid S1/NS1 was developed in late 1980s to early 1990s, during which time the management practices had changed dramatically, which resulted in an average of 50% yield increase. Therefore, these two hybrids were developed under very different management practices, such as increased fertilizer usage and planting density in hybrid S1/NS1 development.
Seven of the 15 genes analyzed in hybrid S1/NS1 had resolvable allele sequence polymorphisms in hybrid S2/NS2. Allele-specific expression of these seven genes was analyzed in seedlings of both hybrids grown in the greenhouse (see Methods). Three biological replications were sampled; each consisted of three individual plants. Distinct gene expression patterns were found between the two hybrids (Figures 2C and 2D). Whereas the modern, improved hybrid S1/NS1 expressed both alleles of all 15 genes, the less improved hybrid S2/NS2 expressed only a single allele in four of the seven genes tested. The monoallelic expression in the S2/NS2 hybrid was not because of the absence of expression in one parent because the gene was expressed in both inbred parents.
Allele-Specific Response to Density and Drought Stress
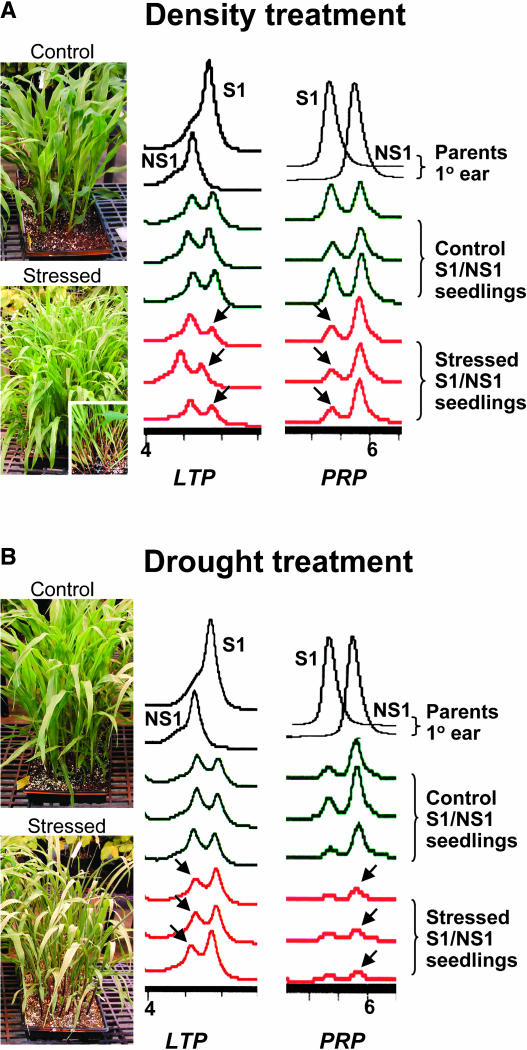
Stress-related genes might respond to stress by either increasing or decreasing their transcript levels. We conducted experiments to investigate whether the two parental alleles showed any allelic differences in response to stress. We treated seedlings of both hybrids S1/N1 and S2/NS2 with drought and high-density stresses (Figure 3; see Methods) and assayed allele-specific transcript accumulation for the 15 genes assayed previously. Among all the genes examined in hybrid S1/NS1, four genes in the drought treatment and two genes in the density treatment were significantly different between stressed and control samples. The allele-specific transcript ratio of two genes, LTP and PRP (Pro-rich protein), showed significant difference between stressed and controls in both density and drought treatments (Table 4, Figure 3). LTP and PRP are both known to be stress responsive (Treviño and O'Connell, 1998; He et al., 2002; Yubero-Serrano et al., 2003); indeed, among all 15 genes, these two genes showed the most dynamic allelic differences in response to stress. First, the two alleles of the hybrid responded differentially to the same stress. For instance, under density stress, the transcript level of the S1 LTP and PRP alleles decreased more than the NS1 alleles. As shown in Table 4, the allele-specific transcript ratio of NS1:S1 changed from 0.84 to 1.54 (LTP) and 1.44 to 3.42 (PRP), respectively, from control to density stress. Second, the same allele responded differentially to different types of stresses. Transcripts of the S1 LTP and PRP alleles decreased more than the NS1 alleles under density stress, resulting in a larger transcript ratio of NS1:S1 as compared with the control. Conversely, under drought stress, the NS1 transcripts decreased more, as shown by the smaller transcript ratio of NS1:S1 as compared with the control (Table 4, Figure 3). Studies of human genes show that environmental cues are often mediated through transcriptional regulation, with cis-regulatory polymorphisms among alleles likely contributing to the genetic basis for a diversity of genotype–environment interactions (Rockman and Wray, 2002). Consistent with our results, genes that are known to be stress or environment responsive, such as LTP and PRP, also showed the most dynamic allele-specific stress responses. Because of the monoallelic expression in the S2/NS2 hybrid, it was not applicable to assay the change of allele-specific transcript ratio for four of the seven genes. However, in some cases, genes that had monoallelic expression in the control showed no expression of either allele under stress (data not shown). The other three genes expressed in S2/NS2 did not show any significant difference in the allele-specific transcript accumulation between stressed and control tissues (Table 4).
Figure 3.
Allele Expression Variation in Response to Density and Drought Stresses.
(A) Density treatment. Plants showed symptoms of thin and tall stature and leaf senesces (inset) in the density stressed treatment.
(B) Drought treatment. Leaf rolling can be seen in the drought-stressed seedlings as compared with the control. For expression analysis, three plants were pooled as one sample, and three biological replicates are shown. Expression of the two alleles in the hybrid was compared relative to each other. Arrows indicate the relative allele expression changes in Discussion. The seedlings were at the V4 leaf stage. The control plants in the drought treatment experiment appeared to experience some degree of density stress as shown by the lower expression of the inbred S1 PRP allele compared with the control in the density experiment. This could be because the drought experiment was planted at 50 plants per flat, and the control in the density experiment was planted at 20 plants per flat. However, the differential response of the two alleles to the drought stress treatment can still be observed. Parents were used as allele references and not treated with any stress.
Table 4.
Allele-Specific Transcript Ratio in Hybrids under Density and Drought Stresses
| S1/NS1 Hybrid | S2/NS2 Hybrid | ||
|---|---|---|---|
| Gene Annotation | Stress Treatment | Allelic Ratio NS:S (cDNA RT-PCR) | Allelic Ratio NS:S (cDNA RT-PCR) |
| LTP | Controla,* | 0.84 (0.05)* | Monoallelic (NS2) |
| Densityb | 1.54 (0.11) | Monoallelic (NS2) | |
| Controla,* | 1.16 (0.05) | Monoallelic (NS2) | |
| Droughtb | 0.69 (0.11)* | Monoallelic (NS2) | |
| GGT | Controla | 4.37 (0.30)** | – |
| Densitya | 3.90 (0.29)** | – | |
| Controla | 3.26 (0.26)** | – | |
| Droughta | 3.16 (0.26)** | – | |
| GAB | Controla | 0.57 (0.15)** | – |
| Densitya | 0.55 (0.16)** | – | |
| Controla,* | 0.57 (0.15)** | – | |
| Droughtb | 0.70 (0.10)** | – | |
| AP2 | Controla | 1.22 (0.06)* | 0.88 (0.04) |
| Densitya | 1.51 (0.11)* | 0.88 (0.09) | |
| Controla | 1.46 (0.11)* | 0.90 (0.04) | |
| Droughta | 1.53 (0.12)** | 1.12 (0.04)* | |
| Histone H2B | Controla | 1.15 (0.18) | 0.50 (0.17)** |
| Densitya | 1.45 (0.10)* | 0.52 (0.17)** | |
| Controla | 1.37 (0.09)** | 0.52 (0.17)** | |
| Droughta | 1.42 (0.10)** | 0.51 (0.17)** | |
| ARDA | Controla | 1.10 (0.19) | Monoallelic (NS2) |
| Densitya | 1.33 (0.08)** | Monoallelic (NS2) | |
| Controla | 1.54 (0.11)** | Monoallelic (NS2) | |
| Droughta | 1.49 (0.11)** | Monoallelic (NS2) | |
| PRP | Controla,* | 1.44 (0.11) | 9.59 (0.39)** |
| Densityb | 3.42 (0.26)** | Monoallelic (NS2) | |
| Controla,* | 2.98 (0.25)** | Monoallelic (NS2) | |
| Droughtb | 1.88 (0.16)** | Monoallelic (NS2) | |
| Histone H4 | Controla | 0.87 (0.05) | – |
| Densitya | 0.89 (0.04) | – | |
| Controla | 0.87 (0.04) | – | |
| Droughta | 1.12 (0.03)* | – | |
| ZmPIP1-3 | Controla | 0.61 (0.14)* | – |
| Densitya | 0.96 (0.00) | – | |
| Controla | 0.80 (0.06)** | – | |
| Droughta | 0.73 (0.09)** | – | |
| Ribo-S29 | Controla | 0.80 (0.11) | – |
| Densitya | 0.99 (0.00) | – | |
| Controla,* | 1.02 (0.09) | – | |
| Droughtb | 1.48 (0.11)* | – | |
| UCR | Controla | 0.94 (0.09) | – |
| Densitya | 1.00 (0.07) | – | |
| Controla | 1.06 (0.07) | – | |
| Droughta | 0.93 (0.07) | – | |
| PGS | Control | <1 | Monoallelic (NS2) |
| Density | <1 | Monoallelic (NS2) | |
| Control | <1 | Monoallelic (NS2) | |
| Drought | <1 | Monoallelic (NS2) | |
| Ribo-S4 | Control | >1 | ∼1 |
| Density | >1 | ∼1 | |
| Control | ∼1 | ∼1 | |
| Drought | >1 | ∼1 | |
| ZmPIP2-1 | Control | >1 | – |
| Density | >1 | – | |
| Control | >1 | – | |
| Drought | >1 | – | |
| MSP | Control | >1 | – |
| Density | >1 | – | |
| Control | ∼1 | – | |
| Drought | >1 | – |
See notes in Table 1. The allelic transcript ratio (NS1:S1) was compared with the expected ratio of equal allelic expression 1.0. The mean, standard error (in parenthesis), and P values in the t test were based on three biological replicates. *, P < 0.05; **, P < 0.01 (italicized). Stress treatments with different letters (a and b) are significantly different (in boldface); treatments with the same letter are not significantly different. Significant level between stress treatments (denoted by a and b) was for hybrid S1/NS1 only. Hybrid S2/NS2 showed no significant differences between treatments (partly because of the monoallelic expression of the genes). The dash indicates that data were not available. In the case of monoallelic expression, the allele in parenthesis is the expressed allele.
DISCUSSION
The Predominance of Genes Exhibiting Allelic Difference in Transcript Accumulation in Maize Hybrids
We analyzed allele-specific transcript accumulation in maize hybrids by using RT-PCR and dHPLC. Because a comparison between alleles is made within the same genotype (the hybrid), rather than comparing alleles from different genotypes, the relative expression of the alleles is less complicated by factors such as environment and genetic background, which affect both alleles equally. Because both parental alleles are simultaneously amplified using identical primers designed in conserved regions, PCR produces equal amplification of both alleles. Using a threshold cutoff of allele-specific transcript ratio below 0.85 or above 1.18, based on hybrid genomic DNA PCR, we found that 11 of the 15 (73%) genes showed significant differences between the parental alleles. Although the number of genes examined is small, they were selected by the presence of allelic sequence polymorphisms and otherwise represent a random set of genes. We have previously found allelic expression level and timing differences in the maize endosperm (a triploid tissue) of several hybrids, including B73/Mo17 (Guo et al., 2003). These results demonstrated the predominance of allelic expression variation at the accumulated RNA level in maize hybrids of different genetic backgrounds and tissue types. A study in mouse hybrids (Cowles et al., 2002) has showed that ∼10% of genes analyzed (7 of 69 genes) showed significant allelic expression difference ranging from 1.5-fold to fourfold. Yan et al. (2002) have shown in human genes that 6 out of 13 genes have allele-specific expression of 1.3- to 4.3-fold difference. Similar results were reported in a large-scale allele-specific expression analysis of human genes using Affymetrix oligo array (Lo et al., 2003). The higher percentage of genes that showed allelic expression variation in maize than in animals could be because of the highly polymorphic nature of the maize genome. However, different methodologies used in these studies or sample size variation may also contribute to the differences.
Allelic difference in the accumulated transcript level could result from various regulatory mechanisms, including transcriptional and posttranscriptional regulations. Differential mRNA degradation, for instance, is a posttranscriptional mechanism that controls transcript abundance. Gene regulation by RNA degradation appears to be limited to only a small fraction of genes and involves specific functional categories in Arabidopsis thaliana (Gutiérrez et al., 2002). Although RNA degradation is a possible mechanism, such regulation would involve regions with allelic sequence variation responsible for the allele specificity of the RNA degradation. Although other possibilities cannot be excluded, presence of allelic regulatory variants, such as cis-acting sequence polymorphisms, might cause allelic difference in transcriptional regulation. Variants of these regulatory regions may respond differently to developmental and environmental cues and result in allele-specific expression in different tissue types, different environments, or stress response.
The significance of gene expression differences to phenotypic variation has been reported in plants. Two genes controlling quantitative traits have previously been isolated, teosinte branched1 in maize (Wang et al., 1999) and the tomato (Lycopersicon esculentum) fruit weight fw2.2 gene (Cong et al., 2002). The phenotypic variation controlled by both genes is regulated by variation in expression, either in the level or timing of expression rather than from protein coding differences between alleles. Recent studies in mouse hybrids (Cowles et al., 2002) and humans (Yan et al., 2002; Lo et al., 2003) have proposed that such allelic expression differences may be a regulatory mechanism responsible for genetic diseases and human variability. If allelic variation in the regulatory region were responsible for the allele-specific transcript differences observed in this study, such regulatory allelic variants would exist to a great extent in natural populations. By this assumption, our results would be consistent with the finding of numerous trans-acting factors that modulate gene expression, which are hypothesized to be an important mechanism for quantitative variation (Guo and Birchler, 1994). Because in the hybrid the two alleles are exposed to the common trans-acting regulatory factors, the allelic sequence variation of the cis-regulatory elements may cause a target gene to interact or bind differentially with the transcriptional factors, thus resulting in differential transcription between alleles. However, more direct evidence is needed to determine the regulatory mechanism of the allelic expression differences. Whether such allelic variation indeed causes any phenotypic variation remains to be tested.
We examined the possibility that a parent-of-origin effect is responsible for the allele-specific expression variation observed in this study. The allele-specific transcript ratio for a majority of genes tested was not affected whether the allele was inherited maternally or paternally. Therefore, parental effect did not play a significant role in allele-specific transcript variation. However, two genes, ARDA and ZmPIP1-3, showed significant differences in their allele-specific transcript accumulation upon maternal versus paternal transmission, exhibiting a maternally preferred expression. The maternal effect could be because of cytoplasmic effects or genomic imprinting. Both possibilities need to be tested. Genomic imprinting in sporophytic tissue of plants has not been reported to date. However, using the RT-PCR/dHPLC method, we recently discovered an imprinted gene in the maize endosperm (Guo et al., 2003).
Allelic Expression between Hybrids S1/NS1 and S2/NS2
Commercial maize breeding involves selection of hybrid genotypes that exhibit high yield in both stressful and high yield environments (Janick, 1999). The yield gains of improved hybrids are primarily because of the genetic improvement in tolerance to both biotic and abiotic stresses (Duvick, 2001). In contrast with the biallelic expression in the new hybrid S1/NS1, the old hybrid S2/NS2 tended to show monoallelic expression. The contrasting allele-specific expression patterns between the two hybrids make it tempting to speculate that some of the genetic improvement through breeding may involve selection for regulatory allelic variants that respond to stress. Because NS2 is in the parentage of NS1 and S2 is in the parentage of S1, the NS1 and S1 alleles could be either a modified NS2 and S2 allele, respectively, or entirely different alleles. Whether alleles of modern hybrids are more responsive to stress still remains to be determined. It is worth noting that depending on the nature of the allelic difference in protein function, biallelic expression could be advantageous to monoallelic expression in some cases. Further studies involving more hybrids and a larger number of genes may be necessary to confirm whether such allelic difference can be generalized among old versus modern hybrids and to determine the significance of this phenomenon.
Differential Expression of the Parental Alleles in Maize Hybrids and Implication to Heterosis
The allele-specific expression variation in different tissue types, environments, and stress conditions may suggest a differential role for the alleles in hybrid development and interaction with the environment. It is possible that the functional diversity of the two parental alleles in the hybrid may have an impact on hybrid performance.
Recent studies in maize (Fu and Dooner, 2002; Song and Messing, 2003) show that some genes can be present in one inbred but missing in the other and suggest that the complementary or synergetic effect from such allelic diversity may be the underlying mechanism of heterosis. Although consistent with this concept of allelic complementation in hybrids, our data also suggest that a gene present in one inbred allele may not always be expressed or expressed in the correct spatial or temporal pattern to fulfill the complementation role to the other inbred allele in the hybrid. It is important to note that complementation at the level of gene regulation and the level of protein function is not mutually exclusive.
The differential expression between the alleles could potentially result in hybrids surpassing the inbred parents in expression in different dimensions, such as (1) expression level, (2) expression timing/duration, and (3) response to developmental and environmental cues. It has been reported in human gene expression that complementation of alleles produces expression patterns beyond the range of the parents in spatial and temporal patterns (Rockman and Wray, 2002). It remains to be tested whether such expression indeed occurs in maize hybrids.
In summary, this study shows that the allelic expression variation occurred frequently in maize hybrids. The data suggest that the two parental alleles in maize hybrids may be regulated differentially during plant development and in response to environmental signals. Although only a small number of genes were analyzed using one each of the hybrid, distinct allelic expression patterns were found between a modern and an old hybrid. This work demonstrates that the maize hybrid is an excellent system to study allele expression variation because alleles are compared within the same genotype of a hybrid and equally affected by genetic background or environmental factors.
METHODS
Plant Materials
Two commercial maize (Zea mays) hybrids, S1/NS1 (Pioneer hybrid 3394; Johnston, IA) and S2/NS2 (Pioneer hybrid 3306) were developed in the 1990s and 1960s, respectively. The inbred parents are proprietary lines of Pioneer Hi-Bred. The parental inbreds S1 and S2 are Iowa Synthetic Stiff Stalk lines; the NS1 and NS2 inbreds are Non-Stiff Stalk lines (Labate et al., 1997). S2 is in the parentage of S1, and both are derived from the same public line plus other Stiff Stalk public and proprietary lines. The NS2 inbred is derived from a cross of two first-cycle lines out of Midwestern dent open pollinated populations. The NS1 inbred is a Non-Stiff Stalk line of complex parentage involving NS2. These inbred parents of both hybrids are adapted to the central U.S. cornbelt and have similar maturity.
Experimental Design
Hybrid 3394 and parental inbreds were planted in the field on April 1, May 8, and June 3, 1997. The different planting dates were used as three environmental treatments. We collected three seedlings at the V4 leaf stage, three primary and three secondary prepollinated immature ears at the V19 stage (before silks emerged from the husks), and pooled as one sample from each environmental treatment. The secondary ear was harvested at the same time the primary ear was harvested. In 1999, hybrids of reciprocal crosses were grown together, and primary immature ears were harvested using the same sampling protocol as above.
The high planting density and drought stress experiments were conducted in the greenhouse for hybrids 3394 and 3306. For the density experiment, plants were grown in three replicated flats at a density of 20 plants per flat (11 × 21 inches) for the control and 250 plants per flat for the high-density stress treatment. Tissue was collected for RNA expression analysis at the V4 leaf stage. At this stage, the high-density plants displayed stress symptoms, including senescence of the first leaf and yellowing of the second leaf. Control plants appeared normal. For the drought experiment, we grew hybrids at a density of 50 plants per flat in three replicated flats. At the V4 leaf stage, irrigation was withheld for drought stress, whereas the control was irrigated regularly. The stressed plants started leaf rolling 4 d after the last irrigation. Tissue was sampled on the fifth day, 4 h after all plants showed leaf rolling and reached permanent wilting. From each replicate, three whole seedlings were pooled to make one RNA sample for gene expression analysis, and three biological replicates were sampled. All tissue samples were frozen immediately in liquid N2 and stored at −80°C.
RT-PCR and Genomic PCR Analysis Using the WAVE dHPLC System
Total RNA was extracted using TriPure reagent (Roche Molecular Biochemicals, Indianapolis, IN) according to the manufacturer's protocol. Protocols for RNA preparation and RT-PCR analysis were described previously (Guo et al., 2003). We used gene-specific primers to obtain the cDNA from each inbred parent by RT-PCR with Pwo DNA polymerase (Roche). The PCR products were then sequenced to identify allele-specific sequence polymorphisms between the inbred lines that would allow separation of the two parental alleles on the WAVE dHPLC system (Transgenomic, Omaha, NE). Gene-specific primers were designed in conserved regions between alleles that flank a sequence polymorphism, either an SNP or an InDel, to minimize amplification preference of either allele and to optimize the amplicon for analysis on the WAVE. (Sequences for amplicons are provided in the supplemental data online.) Thirty-cycle PCR was performed with cDNA from hybrids. Three PCR replicates were performed for each RNA sample. The RT-PCR products were then separated and quantified by the WAVE dHPLC system (Figure 1A). Detailed WAVE dHPLC analysis has been described previously (Guo et al., 2003).
Genomic DNA PCR Analysis
Genomic DNA was isolated from immature ear tissue of hybrids and inbreds using the protocol described by Saghai-Maroof et al. (1984). We performed 30 cycles of PCR with 400 ng of genomic DNA using PCR primers designed to the conserved regions between the alleles. The PCR conditions and the analysis of the allele-specific genomic PCR products using the WAVE dHPLC system were the same as in the cDNA analysis. Although we designed RT-PCR primers at the allelic conserved regions to reduce amplification bias, we performed PCR with hybrid genomic DNA as a control for amplification variation of the two alleles. An allelic ratio of 1.0 is expected from the PCR product of hybrid genomic DNA if the two alleles are equally amplified and one dose of each allele is present. Genomic DNA from the hybrid was tested for four genes, and a significant deviation from the 1:1 ratio was not observed. These data were used to determine a ratio threshold.
Accession Numbers and PCR Primer Sequences
LTP (AY103916.1), 5′-TGTGTCATGCATCAGACTGGCACATAATAA-3′ and 5′-CAAACCCAAGGACATTGAGGTGCTG-3′; GGT (AY104360), 5′-GGCCACAGCCAATG-3′ and 5′-ATTCGACGGAGAAATAGC-3′; GAB (AY104589), 5′-GCCACACCTACGACGAGT-3′ and 5′-TAACAACCAGAGCACAGGACC-3′; AP2 (BU09871), 5′-TACAACTCCTGGACACAGCTA-3′ and 5′-TTGTAAAGAGCAGCACG-3′; Histone H2B (X57313), 5′-CGCGAGATCCAGACCT-3′ and 5′-CGACGAGAACATCAACGATTT-3′; ARDA (AY103814), 5′-GACGGCGGCAGCTACAAG-3′ and 5′-GAAGACGCTGCGCCACA-3′; PRP (AY106116), 5′-GGGCCAACGTCCTG-3′ and 5′-GATCGATGGGCGTG-3′; Histone H4 (M13370), 5′-CCGTCACCTACACCGAGCAC-3′ and 5′-ATACCGCACTGCAGTTCTACA-3′; ZmPIP1-3 (AF326487), 5′-CCGTTCAAGAGCAGGTC-3′ and 5′-TGAAGGGAAGAGAGATGGAAG-3′; Ribo-S29 (AF457936), 5′-GCAACGCCAAGGACATT-3′ and 5′-GCGGACTTAAATAGCAGAGTT-3′; UCR (M77224), 5′-CCTGCTGTGCTGTTATA-3′ and 5′-CGCACTTTGGCAT-3′; PGS (AJ302784), 5′-GTTGAAAATGTAGGGAA-3′ and 5′-GCCATCTCCTAGCTATTC-3′; Ribo-S4 (AF015522), 5′-GCTGCTGCCAAGGCATAAGTT-3′ and 5′-GCTGCAAACGCTGTTCAAGAA-3′; ZmPIP2-1 (AY243801), 5′-AGGCGACCCGAACCAACC-3′ and 5′-ATGGCGGGCGACCTAC-3′; MSP (CF023931), 5′-CGGTACCTGTTTTACTAC-3′ and 5′-GCAGATGAAACGATG-3′.
Supplementary Material
Acknowledgments
The authors thank Xiaofeng Yang, Suling Zhao, Jo Dieter, Erin McLaughlin, Guo-Hua Miao, Dave Ritland, Lynn Heetland, Shifu Zhen, and Susan Nilles for support with this work. The authors specially thank Olga Danilevskaya for valuable discussions and Michael Muszynski for critical reading of the manuscript.

Online version contains Web-only data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Mei Guo (mei.guo@pioneer.com).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.022087.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Bhattramakki, D., Dolan, M., Hanafey, M., Wineland, R., Vaske, D., Register, J.C., III, Tingey, S.V., and Rafalski, A. (2002). Insertion-deletion polymorphisms in 3′ regions of maize genes occur frequently and can be used as highly informative genetic markers. Plant Mol. Biol. 48, 539–547. [DOI] [PubMed] [Google Scholar]
- Buckler, E.S., and Thornsberry, J.M. (2002). Plant molecular diversity and applications to genomics. Curr. Opin. Plant Biol. 5, 107–111. [DOI] [PubMed] [Google Scholar]
- Ching, A., Caldwell, K.S., Jung, M., Dolan, M., Smith, O.S., Tingey, S., Morgante, M., and Rafalski, A.J. (2002). SNP frequency, haplotype structure and linkage disequilibrium in elite maize inbred lines. BMC Genet. 3, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong, B., Liu, J., and Tanksley, S.D. (2002). Natural alleles at a tomato fruit size quantitative trait locus differ by heterochronic regulatory mutations. Proc. Natl. Acad. Sci. USA 99, 13606–13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles, C.R., Hirschhorn, J.N., Altshuler, D., and Lander, E.S. (2002). Detection of regulatory variation in mouse genes. Nat. Genet. 32, 432–437. [DOI] [PubMed] [Google Scholar]
- Doebley, J., and Lukens, L. (1998). Transcriptional regulators and the evolution of plant form. Plant Cell 10, 1075–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvick, D.N. (2001). Biotechnology in the 1930s: The development of hybrid maize. Nat. Genet. Rev. 2, 69–74. [DOI] [PubMed] [Google Scholar]
- Fu, H., and Dooner, H.K. (2002). Intraspecific violation of genetic colinearity and its implications in maize. Proc. Natl. Acad. Sci. USA 99, 9573–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazier, A.M., Nadeau, J.H., and Aitman, T.J. (2002). Finding genes that underlie complex traits. Science 298, 2345–2349. [DOI] [PubMed] [Google Scholar]
- Guo, M., and Birchler, J.A. (1994). Trans-acting dosage effects on the expression of model gene systems in maize aneuploids. Science 266, 1999–2002. [DOI] [PubMed] [Google Scholar]
- Guo, M., Rupe, M.A., Danilevskaya, O.N., Yang, X., and Hu, Z. (2003). Genome-wide mRNA profiling reveals heterochronic allelic variation and a new imprinted gene in hybrid maize endosperm. Plant J. 36, 30–44. [DOI] [PubMed] [Google Scholar]
- Gutiérrez, R.A., Ewing, R.M., Cherry, J.M., and Green, P.J. (2002). Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: Rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc. Natl. Acad. Sci. USA 99, 11513–11518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, C.Y., Zhang, J.S., and Chen, S.Y. (2002). A soybean gene encoding a proline-rich protein is regulated by salicylic acid, an endogenous circadian rhythm and by various stresses. Theor. Appl. Genet. 104, 1125–1131. [DOI] [PubMed] [Google Scholar]
- Helentjaris, T., King, G., Slocum, M., Siedenstrang, C., and Wegman, S. (1985). Restriction fragment length polymorphisms as probes for plant diversity and their development as tools for applied plant breeding. Plant Mol. Biol. 5, 109–118. [DOI] [PubMed] [Google Scholar]
- Janick, J. (1999). Exploitation of heterosis: Uniformity and stability. In The Genetics and Exploitation of Heterosis in Crops, J.G. Coors and S. Pandey, eds (Madison, WI: American Society of Agronomy, Crop Science Society of America), pp. 319–333.
- Knight, J.C. (2004). Allele-specific gene expression uncovered. Trends Genet. 20, 113–116. [DOI] [PubMed] [Google Scholar]
- Labate, J.A., Lamkey, K.R., Lee, M., and Woodman, W.W. (1997). Molecular genetic diversity after reciprocal recurrent selection in BSSS and BCCB1 maize populations. Crop Sci. 37, 416–423. [Google Scholar]
- Lo, H.S., Wang, Z., Hu, Y., Yang, H.H., Gere, S., Buetow, K.H., and Lee, M.P. (2003). Allelic variation in gene expression is common in the human genome. Genome Res. 13, 1855–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay, T.F.C. (2001). Quantitative trait loci in Drosophila. Nat. Rev. Genet. 2, 11–20. [DOI] [PubMed] [Google Scholar]
- Park, S., and Han, K.H. (2003). An auxin-repressed gene (RpARP) from black locust (Robinia pseudoacacia) is posttranscriptionally regulated and negatively associated with shoot elongation. Tree Physiol. 12, 815–823. [DOI] [PubMed] [Google Scholar]
- Rockman, M.V., and Wray, G.A. (2002). Abundant raw material for cis-regulatory evolution in humans. Mol. Biol. Evol. 19, 1991–2004. [DOI] [PubMed] [Google Scholar]
- Saghai-Maroof, M.A., Soliman, K.M., Jorgensen, R.A., and Allard, R.W. (1984). Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. USA 81, 8014–8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shull, G.H. (1908). The composition of a field of maize. Am. Breed. Assoc. Rep. 4, 296–301. [Google Scholar]
- Smith, O.S., and Smith, J.S.C. (1992). Measurement of genetic diversity among hybrids: A comparison of isozymic, RFLP, pedigree, and heterosis data. Medica 37, 53–60. [Google Scholar]
- Song, R.T., and Messing, J. (2003). Gene expression of a gene family in maize based on noncollinear haplotypes. Proc. Natl. Acad. Sci. USA 100, 9055–9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunyaev, S.R., Lathe, W.C., III, Ramensky, V.E., and Bork, P. (2000). SNP frequencies in human genes: An excess of rare alleles and differing modes of selection. Trends Genet. 16, 335–337. [DOI] [PubMed] [Google Scholar]
- Tanksley, S. (1993). Mapping polygenes. Annu. Rev. Genet. 27, 205–233. [DOI] [PubMed] [Google Scholar]
- Treviño, M.B., and O'Connell, M.A. (1998). Three drought-responsive members of the nonspecific lipid-transfer protein gene family in Lycopersicon pennellii show different developmental patterns of expression. Plant Physiol. 116, 1461–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R., Stec, A., Hey, J., Lukens, L., and Doebley, J. (1999). The limits of selection during maize domestication. Nature 398, 236–239. [DOI] [PubMed] [Google Scholar]
- Yan, H., Yuan, W., Velculescu, V.E., Vogelstein, B., and Kinzler, K.W. (2002). Allelic variation in human gene expression. Science 297, 1143. [DOI] [PubMed] [Google Scholar]
- Yano, M., and Sasaki, T. (1997). Genetic and molecular dissection of quantitative traits in rice. Plant Mol. Biol. 35, 145–153. [PubMed] [Google Scholar]
- Yubero-Serrano, E.M., Moyano, E., Medina-Escobar, N., Munoz-Blanco, J., and Caballero, J.L. (2003). Identification of a strawberry gene encoding a non-specific lipid transfer protein that responds to ABA, wounding and cold stress. J. Exp. Bot. 54, 1865–1877. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.