Abstract
Sugars have been shown to regulate transcription of numerous genes in plants. Sucrose controls translation of the group S basic region leucine zipper (bZIP)-type transcription factor ATB2/AtbZIP11 (Rook et al., 1998a). This control requires the unusually long 5′ untranslated region (UTR) of the gene. Point mutations and deletions of the 5′UTR have uncovered the sequences involved. A highly conserved upstream open reading frame (uORF) coding for 42 amino acids is essential for the repression mechanism. It is conserved in 5′UTRs of bZIP transcription factors from other Arabidopsis thaliana genes and many other plants. ATB2/AtbZIP11 is normally expressed in association with vascular tissues. Ectopic expression of a 5′UTR construct shows that the sucrose repression system is functional in all tissues. AtbZIP2 is another Arabidopsis bZIP transcription factor gene harboring the conserved uORF, which is regulated similarly via sucrose-induced repression of translation. This suggests a general function of the conserved uORF in sucrose-controlled regulation of expression. Our findings imply the operation of a sucrose-sensing pathway that controls translation of several plant bZIP transcription factor genes harboring the conserved uORF in their 5′UTRs. Target genes of such transcription factors will then be regulated in sucrose-dependent way.
INTRODUCTION
Plants are autotrophic organisms and synthesize sugars for growth and storage de novo. Sugars can also function as hormone-like signaling molecules that adjust metabolism, growth, and development of plants. Sugar signaling operates essentially at all phases of the plant life cycle and dominates plant metabolism. Most sugar signaling effects appear to be mediated through transcriptional control; changes in the sugar concentration cause induction or repression of gene transcription (Koch, 1996). However, sugar sensing also affects gene expression posttranscriptionally by changing mRNA stability, translation, or protein stability (Chan and Yu, 1998; Rook et al., 1998a; Cheng et al., 1999; Yanagisawa et al., 2003).
The dominant sugars, sucrose, and hexoses activate different cellular processes in plants. Investigations of developing seeds suggest that hexoses control metabolism and growth, whereas sucrose regulates differentiation and storage (Wobus and Weber, 1999). Sucrose and hexoses are sensed via different not yet fully understood signaling pathways (Smeekens, 2000; Rolland et al., 2002). Hexoses are thought to be sensed either in a hexokinase-dependent or -independent manner, the latter possibly involving hexose transporters (Smeekens, 2000; Rolland et al., 2002). Recently, the catalytic function of hexokinase was separated from the sensory function of the enzyme, showing that downstream metabolism is not involved in hexokinase signaling (Moore et al., 2003).
It is now well established that molecular sucrose is sensed via independent pathways. Such pathways have not yet been unraveled, but sucrose transporters with unusual long cytosolic loops may act as receptors (Lalonde et al., 1999). Downstream signaling possibly involves sucrose nonfermenting-like kinases (Purcell et al., 1998; Tiessen et al., 2003).
Molecular sucrose specifically regulates transcription of genes. Such genes are not activated to the same extent by the sucrose breakdown products glucose and fructose. They encode patatin, rolC, and UDP-glucose pyrophosphorylase (Wenzler et al., 1989; Jefferson et al., 1990; Yokoyama et al., 1994; Ciereszko et al., 2001). Sucrose represses a proton-sucrose symporter in sugar beet (Beta vulgaris) (Chiou and Bush, 1998). Moreover, the phenotypic deviations caused by constitutive expression of the homeodomain leucine zipper transcription factor gene ATHB13 are induced specifically by sucrose (Hanson et al., 2001). Disaccharide sensing that is independent from metabolism was observed in experiments with the nonmetabolizable sucrose analogs palatinose and turanose. These analogs affect the expression of the vine hexose transporter VvHT1, α-amylase in barley (Hordeum vulgare) embryos, and ADP-glucose pyrophosphorylase in potato (Solanum tuberosum) (Loreti et al., 2000; Atanassova et al., 2003; Tiessen et al., 2003). However, such sucrose analogs were shown to activate signaling pathways different from sucrose as well (Sinha et al., 2002; Roitsch et al., 2003).
A posttranscriptional regulation of expression mediated by sucrose was observed for the group S basic region leucine zipper (bZIP) transcription factor ATB2/AtbZIP11. Sucrose regulation of ATB2/AtbZIP11 expression takes place at the level of translation; elevated sucrose concentrations repress translation of the transcription factor (Rook et al., 1998a). Thus, ATB2/AtbZIP11 is the first known gene being translationally repressed by sucrose. Interestingly, ATB2/AtbZIP11 transcription is stimulated by sugars and light. Transcription is repressed in darkness in a COP1 and DET1-dependent manner (Rook et al., 1998b). ATB2/AtbZIP11 is prominently expressed in the vascular tissues of seedlings and young vegetative tissues. It is highly induced in funiculi of fertilized ovules (Rook et al., 1998a). This expression pattern and the translational regulation by sucrose suggest that ATB2/AtbZIP11 might coordinate metabolism-associated processes in newly established sinks (Rook et al., 1998a).
Translational control has been observed for plant genes stimulated by light, hormones, and programmed cell death (Bailey-Serres, 1999). Next to ATB2/AtbZIP11, only one other metabolite-induced translational control system in plants has been described. Polyamines trigger translational repression of an S-adenosyl-l-Met decarboxylase gene (Hanfrey et al., 2002).
Elements involved in translational control are mostly found in untranslated regions (UTRs). The 5′caps and the 3′poly(A) determine translational efficiency in a more general way. The efficiency of translation is further controlled by various features of the 5′UTR, such as length, secondary structures, upstream start codons (uAUGs) or open reading frames (uORFs), internal ribosome entry sites, and binding sites for regulatory proteins (Wilkie et al., 2003). The 5′UTR of the ATB2/AtbZIP11 mRNA proved to be necessary for sucrose-induced repression of translation (SIRT) (Rook et al., 1998a). The unusually long ATB2/AtbZIP11-5′UTR (547 bp; Rook et al., 1998b) contains four uORFs (uORF1 to uORF4). The upstream ORF2 encodes a peptide of 42 amino acids. Interestingly, this peptide is highly conserved in uORFs in some other bZIP 5′UTRs in Arabidopsis thaliana as well as in other dicotyledonous and monocotyledonous plants. Moreover, the overlapping uORF1 of ATB2/AtbZIP11 is conserved in some 5′UTRs encoding the conserved uORF2. Deletions and point mutations in the 5′UTR allowed us to investigate the involvement of the uORFs in sucrose-induced translational repression. Ectopic expression of a marker gene construct shows that the sucrose-induced repression system functions in all tissues of the seedling beyond the vascular expression pattern of ATB2/AtbZIP11. We propose bZIP transcription factors harboring the conserved uORF in their 5′UTR to be translationally controlled by sucrose and confirm this suggestion for other Arabidopsis bZIP transcription factors encoding the conserved uORF in a different 5′UTR context.
RESULTS
The Sucrose-Induced Repression System Controls Translation in All Tissues
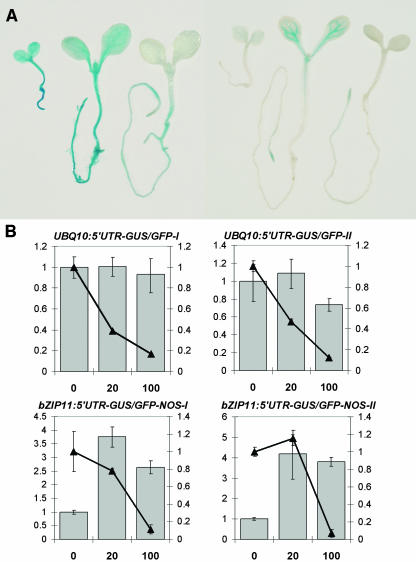
SIRT of ATB2/AtbZIP11 was observed only in association with veins because of the tissue specificity of the ATB2/AtbZIP11 promoter (Rook et al., 1998a). To test the tissue specificity of the repression system, we expressed an ATB2/AtbZIP11-5′UTR–controlled β-glucuronidase (GUS)/green fluorescent protein (GFP) chimera terminated by the ATB2/AtbZIP11-3′-UTR ectopically by the Arabidopsis POLYUBIQUITIN10 promoter (UBQ10) (Sun and Callis, 1997). The resulting construct, UBQ10:5′UTR-GUS/GFP, was transformed to Arabidopsis (ecotype Columbia-0 [Col-0]). Histochemical staining of 5-d-old seedlings of the selected lines UBQ10:5′UTR-GUS-I and -II show that the UBQ10-promoter is ubiquitously active. Seedlings grown without sucrose or with 20 mM show GUS expression in root and shoot (Figure 1A). A weak repression of GUS activity in the cotyledons is observed in seedlings grown in 20 mM sucrose, whereas seedlings grown in 100 mM sucrose do not show GUS activity in the shoot. Some residual GUS expression is still observed in roots of seedlings grown in 100 mM sucrose (Figure 1A), which is repressed at higher sucrose concentrations (200 mM sucrose, see Supplemental Figure S1 online).
Figure 1.
SIRT in Transgenic Seedlings Expressing the ATB2/AtbZIP11-5′UTR Controlled Marker Gene.
Seedlings were grown in half-strength MS medium containing 0, 20, or 100 mM sucrose in liquid culture for 5 d in constant light.
(A) Histochemical staining of seedlings. Left three seedlings, UBQ10:5`UTR-GUS/GFP-I; right three seedlings, bZIP11:5′UTR-GUS. Leftmost seedling in row grown in half-strength MS medium without sucrose; middle, 20 mM sucrose; right, 100 mM sucrose.
(B) Quantification of GUS activity and mRNA accumulation in lines UBQ10:5′UTR-GUS/GFP-I and bZIP11-5′UTR-GUS/GFP-NOS. Measured GUS activities of seedling extracts was adjusted to total protein content. GUS activities (line and filled triangles) are shown relative to GUS activity of seedlings grown without sucrose. Expression of the marker gene mRNA was quantified by quantitative PCR and normalized to the expression of the ACTIN2 gene. Transcript accumulation of the marker gene mRNA (gray bars) is shown as relative expression compared with the marker gene mRNA level of seedlings grown without sucrose. Left axis, relative expression; right axis, relative GUS activity. Error bars represent standard deviation of three measurements.
For comparison, histological staining of Arabidopsis Col-0 seedlings harboring bZIP11:5′UTR-GUS (Rook et al., 1998a) is presented, which shows the normal expression of the ATB2/AtbZIP11 gene (Figure 1A). Intense GUS activity in the veins is observed in seedlings grown in 20 mM sucrose, whereas repression of GUS activity occurs in seedlings grown in 100 mM sucrose (Figure 1A) as presented previously for the C24 ecotype (Rook et al., 1998a). Sucrose-induced repression of the GUS marker gene was obtained for all transgenic lines within 24 h of incubation in 100 mM sucrose (see Supplemental Figure S1 online; A. Wiese, unpublished data). Quantification of GUS activity in UBQ10:5′UTR-GUS seedlings and comparison to the level of GUS-mRNA show that the repression is not because of changes in the mRNA levels because only GUS activity decreases with increasing sucrose (Figure 1B). This shows that the GUS activity is controlled by SIRT. The ectopic expression with UBQ10:5′UTR-GUS shows a higher sensitivity against the sucrose repression than observed with the ATB2/AtbZIP11 promoter. Here, repression takes place already at lower sucrose concentrations (Figure 1B). Such differences in sensitivity might be because of differences in the local sucrose concentration in different tissues. Quantification of GUS activity relative to the protein content showed a threefold to fivefold higher activity in the roots compared with the shoots. Such high GUS levels in the roots can be repressed efficiently by sucrose as well (see Supplemental Figure S1 online).
SIRT occurs in all tissues of the seedling and might be a general sucrose control mechanism. Similar 5′UTR constructs to those tested for UBQ10-promoter were also constructed with the 35S promoter of Cauliflower mosaic virus. Such 35SCaMV:5′UTR-GUS/GFP constructs allow SIRT in transgenic Arabidopsis seedlings. Thus, relatively high mRNA levels as they are normally obtained with the 35SCaMV-promoter also show SIRT.
The ATB2/AtbZIP11-5′ Leader Is Necessary and Sufficient for SIRT
In previous work, it was shown that deletion of the 5′UTR results in a loss of SIRT (Rook et al., 1998a). The 5′UTR was necessary for sucrose repression, but a possible function of the ATB2/AtbZIP11-3′UTR dependent on the 5′UTR was not excluded. Thus, it was tested whether the 5′UTR by itself is sufficient for SIRT. For this, a GFP/GUS chimera-coding region terminated by the nopaline synthase (NOS) 3′UTR extracted from pCAMBIA1304 was fused to the ATB2/AtbZIP11-promoter and 5′UTR. The construct bZIP11:5′UTR-GUS/GFP-NOS was transformed to Arabidopsis, ecotype Col-0, and independent homozygous lines were selected for analysis. GUS activity of the transgenic lines is repressed in seedlings grown in 100 mM sucrose, whereas RNA levels are comparable to the seedlings grown on lower sucrose concentration, indicating a repression of translation (Figure 1B). This shows that the exchange of the ATB2/AtbZIP11-3′UTR does not affect the SIRT mechanism. Hence, the 5′UTR is necessary and sufficient for repression.
Translation of the Conserved uORF2 Is Required for SIRT
The ATB2/AtbZIP11 5′UTR harbors four uORFs. Starting from the 5′-end of the mRNA, these overlapping uORFs are designated uORF1 (18 amino acids), uORF2 (42 amino acids), uORF3 (5 amino acids), and uORF4 (19 amino acids) (Figure 2A). An additional internal start codon (AUG) is present in uORF2. This uORF can be translated as the long uORF2a, starting at the first AUG codon, or as uORF2b, starting at the internal AUG (28 amino acids).
Figure 2.
uORF Involvement in SIRT.
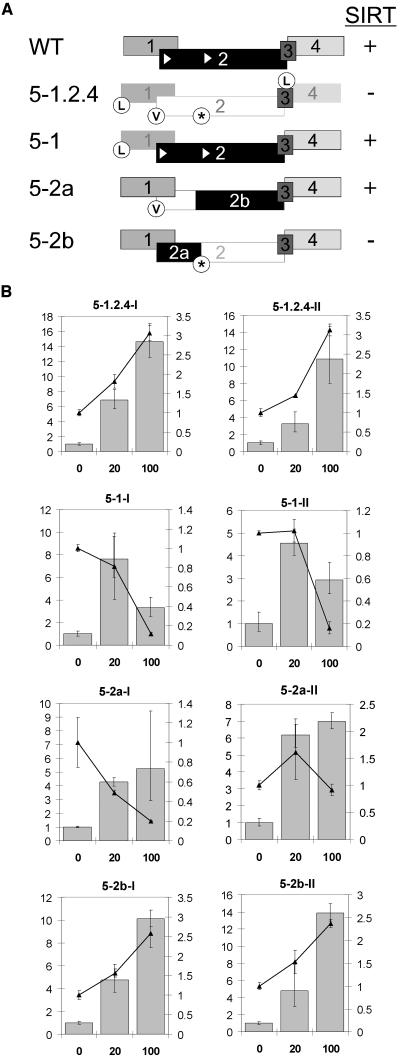
(A) Schematic illustration of the ATB2/AtbZIP11-5′-UTR. Shown are uORF arrangement and effects of the introduced point mutations. asterisk, stop codon; L, Lys; V, Val. White triangles represent start codons in uORF2.
(B) Analysis of SIRT mediated by 5′UTRs with different point mutations. Seedlings were grown for 5 d in constant light in 0, 20, or 100 mM sucrose. Results presented as described in legend of Figure 1.
uORFs are known to affect translation (Morris and Geballe, 2000); therefore, their involvement in SIRT was investigated. Point mutations disrupting the start codons of the uORFs in the 5′ leader of ATB2/AtbZIP11 were introduced to investigate the importance of translation of the uORFs for the SIRT mechanism. Start codons were exchanged to either stop codons or alternative amino acid codons (Table 1). These mutated 5′UTRs were inserted into the bZIP11:5′UTR-GUS construct (Rook et al., 1998a) and transformed into Arabidopsis. Independent homozygous lines for each construct were selected, and 5-d-old seedlings grown in liquid culture were analyzed for SIRT. Results for two representative lines are shown in Figure 2B.
Table 1.
Start Codon Point Mutations Introduced into the ATB2/AtbZIP11-5′UTR
| Name | Mutated uORF(s) | AUG Exchange |
|---|---|---|
| 5-1 | 1 | AAG (L) |
| 5-2ab | 2a/b | 5-2a: GTG (V)/5-2b: TAG (*) |
| 5-2a | 2a | GTG (V) |
| 5-2b | 2b | TAG (*) |
| 5-1.2.4 | 1,2,4 | 5-1, 5-2ab, and uORF4: AAG (L) |
The effect of the exchange on the codon is shown in parentheses. Asterisk, stop codon; L, Lys; V, Val.
First, a construct was created, in which translation of uORF1, 2, and 4 was disrupted (bZIP11:5-1.2.4-GUS I and II) by mutation of the respective start codons (Table 1, Figure 2A). In transgenic seedlings harboring this construct, GUS activities remain high when grown in 100 mM sucrose, whereas the marker gene mRNA accumulates as expected (Figure 2B). In these lines, SIRT is absent, suggesting that one or more of these uORFs is involved in the sucrose repression of translation. Moreover, the small uORF3 alone does not mediate SIRT.
Next, we introduced point mutations in single uORFs to identify the ones involved in SIRT. The ATB2/AtbZIP11 uORF1 and uORF2 show conservation in their sequence with uORFs in other plant bZIP-5′UTRs (see below), and our investigations were concentrated on those. The exchange of the start codon (AUG) of uORF1 to AAG (Lys) in construct bZIP11:5-1-GUS (Table 1, Figure 2A) still allows repression of GUS activity in seedlings grown in 100 mM sucrose. This is because of SIRT because the mRNA of the marker gene does not reflect the repression of GUS activity (line bZIP11:5-1-GUS, Figure 2B).
The first AUG of the highly conserved uORF2 (AUG2a) was converted to a GUG (Val) in 5′UTR 5-2a (Table 1, Figure 2A). Transgenic seedling of lines bZIP11:5-2a-GUS I and II showed repression of GUS activity when grown in 100 mM sucrose (Figure 2B). Marker gene mRNA levels did not reflect this repressive effect (Figure 2B); thus, SIRT occurs normally. Remarkably, the exchange of the internal putative start codon AUG2b (5′UTR 5-2b) to a stop codon (Table 1, Figure 2A) destroys SIRT. Seedlings harboring bZIP11:5-2b-GUS and grown in 100 mM sucrose show an unrepressed GUS activity (Figure 2B). Thus, SIRT activity is dependent on translation of the conserved uORF2. Only translation of the highly conserved C-terminal part is essential for SIRT because usage of the first start codon is not required. In the following, the uORF2 of ATB2/AtbZIP11 and any orthologous uORF from other plants or other Arabidopsis genes will be named the Sucrose Control uORF (SC-uORF).
The SC-uORF Is Translated in Vitro
After the identification of the SC-uORF, we tested whether this uORF is translated to the SC-peptide in vitro. For this purpose, the ATB2/AtbZIP11-5′UTR was subcloned EcoRV/NcoI into pBluescript II KS+, resulting in construct pb5 (Figure 3A). The first 43 bp of the 5′UTR are absent in this construct, which does not affect the uORFs. The 5′UTR sequence was transcribed with T3 polymerase and translated with 35S-labeled Met in wheat germ lysate. Translation of pb5 results in a peptide with a molecular weight of ∼4 kD consistent with the calculated molecular weight of the translated uORF2 (Figure 3B, lane 2). Translation of the pBluescript II KS+ vector alone showed that the additional larger peptides are not resulting from translation of the 5′UTR (Figure 3B, lane 4). To verify the identity of the translated peptide as derived from uORF2, mutations were introduced in the 5′UTR. Deletion of the first 172 bp of the 5′UTR, including uORF1, and modification of the uAUG2a into a better context (ACACATGTCT) (Lutcke et al., 1987) in construct pb5-AUGc (Figure 3A) results in high-level translation of a peptide of the same size as with the wild-type 5′UTR (Figure 3B, lane 1). This indicates that the in vitro translation starts at the first start codon of the conserved uORF. The point mutation of the 5′UTR 5-2a exchanging AUG2a to a Val codon (Table 1, Figure 2A) in construct pb5-2a results in a loss of the 4-kD SC-peptide (Figure 3B, lane 3).
Figure 3.
In Vitro Translation of the ATB2/AtbZIP11-5′UTR.
(A) In vitro translation constructs of the ATB2/AtbZIP11-5′UTR in pBluescript II KS+. The overlapping uORFs are indicated. pb5, wild-type ATB2/AtbZIP11 5′UTR; pb5-AUGc, 5′UTR with exchange of AUG2a to an improved context; pb5-2a, point mutation in AUG2a, removing the first start codon of uORF2.
(B) Gel electrophoresis of coupled in vitro transcription/translation products. Extracts were separated in a 16.5% Tris-tricine SDS gel, and 35S-Met radiolabeled products were analyzed by a PhosphorImager after 2 weeks of exposure. Molecular weights of a protein standard are indicated at the left in kD. Lane 1, pb5-AUGc; lane 2, pb5; lane 3, pb5-2a; lane 4, pBluescript II KS+ vector.
The SC-uORF Is Conserved among Plants
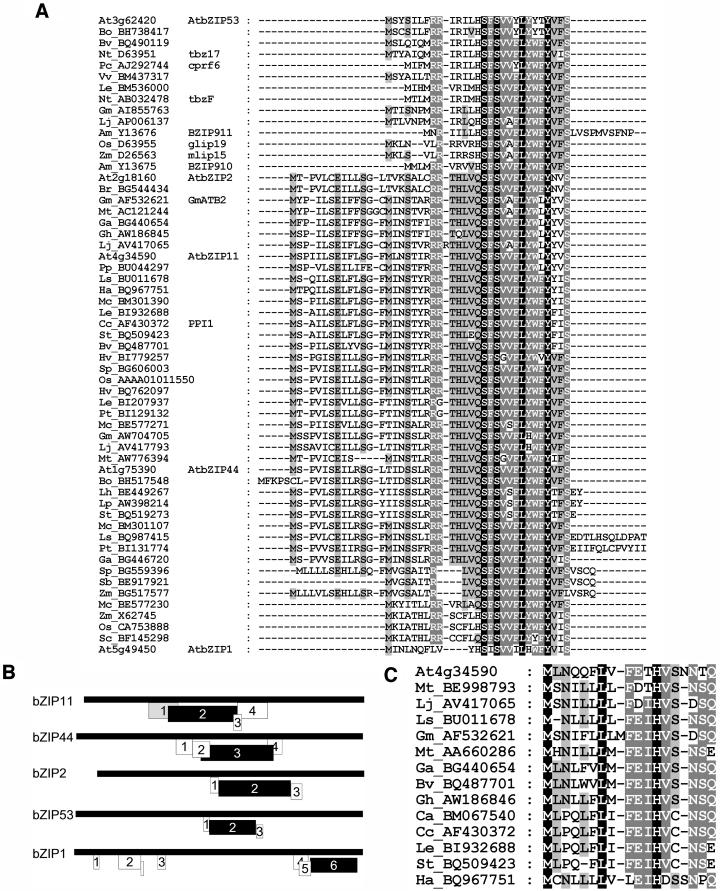
Protein BLAST searches with the uORF2 amino acid sequence against translated databases (Altschul et al., 1990) revealed that uORF2 of ATB2/AtbZIP11 (SC-uORF) shows high amino acid sequence conservation to other plant uORFs (Figure 4A). SC-uORFs were found upstream of four other bZIP transcription factor genes in Arabidopsis and also in bZIP genes of other dicotyledonous and monocotyledonous plant species (Figures 4A and 4B). Remarkably, these bZIP transcription factors all belong to the group S of bZIP transcription factors (Jakoby et al., 2002). Table 2 shows 21 identified ESTs, cDNAs, or genomic sequences coding for an SC-uORF followed by the complete group S bZIP transcription factor coding sequence. SC-uORFs were not observed in any other genes in the databases. In Arabidopsis, only 5 of the 17 identified S-group bZIP transcription factors (Jakoby et al., 2002) harbor the SC-uORF.
Figure 4.
Conservation of uORFs in Group S bZIP-5′UTRs.
(A) Alignment of the conserved SC-uORF amino acid sequences identified in BLAST searches. Black box with white letter, 100% identity; dark gray box with white letter, 80% identity; gray box with black letter, 60% identity. ESTs carry their accession number and a shortcut of the species. Am, Antirrhinum majus (snapdragon); At, Arabidopsis (thale cress); Bo, Brassica oleracea (cabbage); Br, B. rapa (birdsrape mustard); Bv, Beta vulgaris (sugar beet); Cc, Capsicum chinense (pepper); Ga, Gossypium arboreum (cotton tree); Gh, G. hirsutum (cotton); Gm, Glycine max (soybean); Ha, Helianthus annuus (sunflower); Hv, Hordeum vulgare (barley); Le, Lycopersicon esculentum (tomato); Lh, L. hirsutum (wild tomato); Lj, Lotus japonicus; Lp, L. pennellii (wild tomato); Ls, Latuca sativa (lettuce serriola); Mc, Mesembryanthemum crystallinum (ice plant); Mt, Medicago trunculata (barrel medic); Nt, Nicotiana tabacum (tobacco); Os, Oryza sativa (rice); Pc, Petroselinum crispum (parsley); Pp, Prunus persica (peach); Pt, Populus tremula × P. tremuloides (poplar); Sb, Sorghum bicolor (broomcorn); Sc, Secale cereale (rye); Sp, S. propinquum (sorghum); St, Solanum tuberosum (potato); Vv, Vitis vinifera (vine); Zm, Zea mays (maize).
(B) Schematic illustration of uORF organization in 5′UTRs of the five Arabidopsis group S bZIP-genes, encoding the SC-uORF (indicated as black box). The conserved uORF1 of ATB2/AtbZIP11 is presented in gray. uORFs are numbered starting from the 5′end of UTR. An upstream AUG in AtbZIP1 is indicated as a small unnumbered box.
(C) Amino acid sequence alignment of conserved uORFs overlapping the SC-uORF. ESTs carry their accession number and a shortcut of the species. Black box with white letter, 100% identity; dark gray box with white letter, 80% identity; gray box with black letter, 60% identity.
Table 2.
Data of 5′UTRs Harboring the SC-uORF
| Plant | Gene Name | Accession Number | Intercistronic Spacer Length (nt) | SC-uORF Length (aa) | SC-uORF pI |
|---|---|---|---|---|---|
| Arabidopsis thaliana (thale cress) | ATB2/AtbZIP11 | At4g34590 | 255 | 42 | 9.69 |
| Arabidopsis | AtbZIP44 | At1g75390 | 164 | 41 | 9.97 |
| Arabidopsis | AtbZIP2 | At2g18160 | 116 | 41 | 8.84 |
| Arabidopsis | AtbZIP53 | At3g62420 | 182 | 28 | 9.82 |
| Arabidopsis | AtbZIP1 | At5g49450 | 10 | 25 | 6.69 |
| Nicotiana tabaccum (tobacco) | tbz17 | D63951 | 240 | 28 | 10.27 |
| N. tabaccum | tbzF | AB032478 | 162 | 25 | 10.90 |
| Zea mays (maize) | mLIP15 | D26563 | 170 | 26 | 11.00 |
| Z. mays | OCSBF-1 | X6245.1 | 188 | 28 | 9.78 |
| Antirrhinum majus (snapdragon) | BZIP910 | Y13675.1 | 166 | 25 | 10.90 |
| A. majus | BZIP911 | Y13676.1 | 130 | 32 | 8.36 |
| Oryza sativa (rice) | glip19 | D63955 | 169 | 26 | 11.57 |
| O. sativa | – | AAAA01011550 | 254 | 41 | 9.98 |
| Glycine max (soybean) | GmATB2 | AF532621 | 228 | 41 | 9.10 |
| Petroselinum crispum (parsley) | cprf6 | AJ292744 | 138 | 25 | 10.27 |
| Lycopersicon esculentum (tomato) | – | BI207937 | 114 | 41 | 8.36 |
| Medicago trunculata (barrel medic) | – | AC121244 | 198 | 42 | 9.10 |
| Gossypium arboreum (tree cotton) | – | BG446720 | 89 | 41 | 8.27 |
| Lotus japonicus | – | AP006137 | 115 | 28 | 10.90 |
| Capsicum chinense (pepper) | PPI1 | AF430372 | 188 | 41 | 9.69 |
Only EST/DNA sequences were chosen that code for the complete group S bZIP coding sequence. SC-uORF distance, nucleotides (nt) following SC-uORF stop codon until main bZIP AUG start codon; SC-uORF length: number of amino acids (aa) encoded by the uORF; SC-uORF pI, pI calculated with the Compute pI/Mw tool, http://www.expasy.org/tools/pi_tool.html.
SC-uORFs are present in long and short versions; they vary in length between 41 to 53 and 22 to 32 amino acids (Figure 4A, Table 2), respectively. SC-uORFs show amino acid identities between 28 and 97%. The coding sequence shows variability in codon usage in all third wobble positions of the codons. An effect of a conserved RNA sequence structure is therefore unlikely. All SC-uORFs are highly conserved in their C terminus; the shorter uORFs lack the N terminus present in the longer uORFs. Both types of uORFs are found within and between plant species. The distances of the SC-uORF stop codon to the main bZIP reading frame start codon are in the range from 71 (BQ987415, Figure 4A) to 255 nucleotides (ATB2/AtbZIP11, Table 2).
The peptide sequences, encoded by SC-uORFs have a high, calculated isoelectric point (Table 2). These basic peptides mainly contain two to three consecutive Arg (Figure 4A), a feature also found in the translation inhibiting uORF β2-adrenergic receptor upstream peptide (BUP), which inhibits the translation of the β2-adrenergic receptor (Parola and Kobilka, 1994). Interestingly, the coding sequence of the SC-uORF includes one rare Arg codon, cgc or cgg, which occurs with a frequency of 3.8 or 4.8 per 1000 (Table 3; Nakamura et al., 2000). Such rare codons might be involved in the repression mechanism as was shown for a rare Leu codon in frog (Xenopus laevis) Connexin41 mRNA (Meijer and Thomas, 2003).
Table 3.
Frequency of Arg Codons in Arabidopsis (Nakamura et al., 2000)
| Codon | Frequency/1000 |
|---|---|
| CGU | 9.0 |
| CGC | 3.8 |
| CGA | 6.3 |
| CGG | 4.3 |
| AGA | 18.3 |
| AGG | 10.9 |
So far, we identified a maximum of three long uORFs and one or two short uORFs in rice (Oryza sativa), Lotus japonicus, and ice plant (Mesembryanthemum crystallinum). Arabidopsis encodes five S-group bZIP genes harboring the SC-uORF, ATB2/AtbZIP11, AtbZIP2, AtbZIP44, AtbZIP53, and AtbZIP1. The AtbZIP1 5′UTR shows features not observed in any of the other genes. Its short SC-uORF sequence lacks consecutive Arg, is not basic (pI 6.69, Table 2), and its distance to the start codon of the bZIP coding sequence is only 10 nucleotides long (Table 2).
Figure 4B shows the uORF organization of all five AtbZIP 5′UTRs coding for the conserved uORF. The number of encoded uORFs varies between three and six. Other uORFs overlap the highly conserved SC-uORF in all plant species. In some SC-uORF encoding 5′UTRs, further inspection revealed conservation of an overlapping uORF upstream of the SC-uORF. In ATB2/AtbZIP11, it is represented by the uORF1 coding sequence (Figures 4B and 4C). These 16– to 18–amino acid long uORFs show 33 to 100% amino acid identity to each other. They overlap the highly conserved SC-uORF sequence in a different reading frame over ∼20 nucleotides (Figure 4B).
The Intercistronic Region Is Important for SIRT
The distance between the stop codon of the SC-uORF and the bZIP start generally ranges between 71 and 255 bp (Table 1). Such sequences may be important for the SIRT mechanism as well. This was investigated by partial deletions of the ATB2/AtbZIP11 5′UTRs (Figure 5A). The four overlapping uORFs are located within the middle part of the ATB2/AtbZIP11 5′UTR (Figure 5A). The 5′UTR starting from NheI (−401 from bZIP11-AUG) sequence was divided into two parts: the uORF containing the 5′-end part and the downstream 3′-end (Figure 5A).
Figure 5.
Effects of Deletions in the ATB2/AtbZIP11 5′UTR on SIRT.
(A) Schematic illustration of ATB2/AtbZIP11-5′-UTR, the uORF arrangement, and deletions in lines bZIP11:4-11-5′UTR-GUS and bZIP11:4-23-5′UTR-GUS.
(B) Analysis of SIRT mediated by 5′UTRs with deletions. Seedlings were grown for 5 d in constant light in 0, 20, or 100 mM sucrose. Results presented are as described in legend of Figure 1.
In deletion 4-11, 166 bp of the 3′-end part of the leader were removed, starting immediately after the stop codon of uORF4 (Figure 5A). Thus, all uORFs are present in this construct. Deletion 4-23 represents the 5′-end deletion of the uORF-containing part. Only the uORF-free 3′ terminal 169 bp are present, starting with the uORF4 stop codon (Figure 5A).
The truncated 5′UTRs 4-11 and 4-23 were inserted into the bZIP11:5′UTR-GUS (Rook et al., 1998a), resulting in bZIP11:4-23UTR-GUS and bZIP11:4-11UTR-GUS. These were transformed into Arabidopsis (ecotype Col-0). The construct lacking the spacer (bZIP11:4-11-GUS) results in high GUS activity in seedlings grown in 100 mM sucrose, and this is mirrored by the GUS-mRNA level. Here, SIRT is completely abolished. However, the intercistronic region by itself does not mediate SIRT. Transgenic seedlings harboring bZIP11:4-23-GUS do not show SIRT when grown in 100 mM sucrose. Under the experimental conditions, control line bZIP11:5′UTR-GUS showed SIRT as expected (data not shown).
SC-uORF Containing 5′UTRs Mediate SIRT
SIRT in ATB2/AtbZIP11 requires translation of its conserved SC-uORF. We tested whether other bZIP genes, coding for such a SC-uORF in their 5′UTR show SIRT. The Arabidopsis group S bZIP transcription factor AtbZIP2 (GBF5) encodes uORFs in its 5′UTR similar to ATB2/AtbZIP11, including the conserved SC-uORF (Figure 4B). Arabidopsis (ecotype Col-0) was transformed with the AtbZIP2 gene in which 81 bp of the AtbZIP2-coding sequence were translationally fused to the GUS-marker gene, replacing the major part of the AtbZIP2-coding sequence (bZIP2:5′UTR-GUS; Figure 6A). Furthermore, a similar construct with a deletion of the 5′UTR was constructed and tested. Construct bZIP2:Δ5′UTR-GUS resembles bZIP2:5′UTR-GUS but carries a deletion of the 5′UTR from the 5′end up to position −86 bp relative to the AtbZIP2 start codon.
Figure 6.
SIRT in AtbZIP2.
(A) Schematic illustration of GUS-marker gene constructs of AtbZIP2. bZIP2:5′UTR-GUS, AtbZIP2 5′UTR, and 17 amino acids of the AtbZIP2 coding sequence translationally fused to GUS coding sequence. bZIP2:Δ5′UTR-GUS carries a deletion of the 5′UTR from the 5′end up to position −86 from relative to the main start codon.
(B) Analysis of SIRT on AtbZIP2. Seedlings were grown for 5 d in constant light in 0, 20, or 100 mM sucrose. Results presented are as described in legend of Figure 1.
In transgenic homozygous seedlings harboring bZIP2:5′UTR-GUS, GUS-mRNA increases with the sucrose concentration, whereas the GUS activity is sucrose repressed. SIRT is lost in transgenic seedlings expressing the 5′UTR-marker gene construct with deletion of the major part of the 5′UTR, including all uORFs (line bZIP2:Δ5′UTR-GUS). GUS activity increases with sucrose concentration and the GUS-mRNA level. 5′UTR-dependent SIRT was also observed for the SC-uORF–containing AtbZIP44 gene, which is expressed in floral tissues (B. Wobbes, unpublished data).
DISCUSSION
Sucrose specifically represses translation of AtB2/AtbZIP11. This repressive effect is not mediated by glucose or fructose, used separately or in combination, nor by the sucrose-to-hexose ratio (Rook et al., 1998a; A. Wiese, unpublished data). Here, we uncovered the cis elements involved in sucrose repression.
The Conserved SC-uORF Mediates SIRT
Sucrose-induced repression of ATB2/AtbZIP11 translation was shown to depend on the 5′UTR of the gene (Rook et al., 1998a). This region is necessary and sufficient for SIRT and was investigated in more detail.
Point mutations in uORF2 start codons in the 5′UTR of this gene revealed that SIRT requires the translation of this uORF, named the SC-uORF. Remarkably, BLAST searches revealed a strong conservation of the ATB2/AtbZIP11-SC-uORF encoded peptide in 5′UTRs of group S bZIP transcription factors (Jakoby et al., 2002) of all plant species. In these 5′UTRs, the SC-uORF is arranged together with at least two other uORFs. Usually the SC-uORF overlaps with other uORFs.
Five Arabidopsis bZIP transcription factors harbor the SC-uORF, and SIRT was observed in 5-d-old seedlings for ATB2/AtbZIP11 and AtbZIP2. Also, for AtbZIP44, SIRT occurs in sucrose fed floral organs (data not shown).
These Arabidopsis bZIP transcription factors code for long conserved uORFs (41 to 42 amino acids). Many of the SC-uORF sequences in plants are shorter (22 to 32 amino acids), including those in the 5′UTRs of AtbZIP53 and AtbZIP1 (Figure 4A). These represent mainly the more conserved C-terminal part of the ATB2/AtbZIP11 uORF2. Such a shorter conserved uORF was created by mutation of the first AUG in uORF2 of ATB2/AtbZIP11 (AUG2a, bZIP11:5-2a-GUS). This mutation still allows SIRT, suggesting that the N-terminal 14 amino acids of this uORF are not essential for SIRT. This first part of the SC peptide shows high amino acid sequence conservation, and its importance is currently unclear. In lines bZIP11:5-2b-GUS, a stop codon replaces the second AUG codon of uORF2 (AUG2b), and this mutation destroys SIRT. Thus, initiation at AUG2b is sufficient for SIRT to occur. Most likely, in planta the larger SC peptide is synthesized, as was observed in in vitro translation studies (Figure 3B). Importantly, AtbZIP2 and AtbZIP44 SC-uORF sequences do not contain the internal AUG (Figure 4A), precluding internal initiation of translation in these uORFs. Only long SC peptides can be translated from these SC-uORFs. Most likely, uORF2 translation is initiated at the first AUG2a. In case of abrogation of AUG2a (bZIP11:5-2a-GUS), the shorter SC peptide initiating at AUG2b still mediates SIRT. Therefore, we propose that both long and short conserved translated SC-uORFs mediate SIRT. The existence of the conserved SC-uORFs in bZIP transcription factors was noted before (Martinez-Garcia et al., 1998; Strathmann et al., 2001; Yang et al., 2001). Here, we propose a function of the conserved SC-uORF in sucrose-induced translational control.
The conserved uORF1 in the 5′UTR of ATB2/AtbZIP11 is not involved in SIRT. Repression occurs normally when translation of uORF1 is prevented by mutation of the start codon (line bZIP11-5-1-GUS). Moreover, such an additional conserved uORF is also absent in AtbZIP2 and AtbZIP44, supporting the assumption that it is not required for SIRT. This conserved uORF is only present in some of the identified plant bZIP 5′UTRs (Figure 4C), and its function remains unclear.
The Repression Mechanism
Different mechanisms of uORF control on translation of the downstream main ORF have been described. Usually, the nascent uORF peptide acts in cis on translation (Damiani and Wessler, 1993; Degnin et al., 1993; Lohmer et al., 1993). uORFs can hinder translation of the downstream ORF by forcing the ribosomes to reinitiate after termination of the uORF. Such uORFs can also cause stalling of the translational machinery during peptide elongation or at termination. Ribosome stalling seems to depend on the amino acid sequence of the uORF. Amino acid sequence-dependent uORFs are thought to inhibit translation via ribosome stalling by interaction with RNA or protein components of the translational machinery (Vilela and McCarthy, 2003).
The remarkable high conservation of the SC-uORF amino acid sequence implies a sequence dependence of the encoded SC peptides as shown for uORFs of fungal carbamoyl phosphate synthases and the cytomegalovirus UL4 gene (Degnin et al., 1993; Wang and Sachs, 1997). Comparison of all known sequence-dependent uORFs did not allow the identification of conserved domains that might be involved in interaction with the translational apparatus or RNA (Tenson and Ehrenberg, 2002). Interestingly, the SC-uORF and the conserved BUP-uORFs in mammals both contain two to three consecutive Arg. A cis-like action of the BUP was suggested by binding directly to the mRNA of origin after its translation and hindering further scanning of ribosomes. Mutating the consecutive Arg in the BUP peptide reduced the repressive effect of the peptide, probably because of abrogation of the inhibitory BUP interaction with the mRNA (Parola and Kobilka, 1994).
The length of the intercistronic region between the SC-uORF and the main start codon varies from 71 to 255 bp (Table 1) with the exception of AtbZIP1, which harbors a spacer of only 10 bp. The length of the intercistronic region is important for reinitiation efficiency, which was observed to increase with the length of the intercistronic spacer (Child et al., 1999). Deletion of the intercistronic spacer of ATB2/AtbZIP11 shows a clear requirement of this region for the SIRT. The intercistronic region alone has no repressive effect; thus, interaction of the intercistronic region with the uORF region is required to mediate repression. A similar interdependence of uORF and intercistronic spacer was described for the Arg/Lys transporter gene cat-1, where induced translation of an uORF opens an internal ribosome entry site, which promotes translation of the major ORF (Yaman et al., 2003). Similarly, the translation of the SC-uORF might induce an inhibitory RNA structure or expose a binding site for an inhibitory RNA binding protein.
A sucrose-activated signal transduction process somehow affects the initiation of the SC-uORF translation or the SC peptide activity in such a way that translation is repressed. The molecular details of this process remain to be uncovered.
Physiological Consequences of SIRT
The physiological importance of metabolite-induced translational regulation was recently shown for plant S-adenosylmethionine decarboxylase genes (Hanfrey et al., 2002). These genes encode a highly conserved uORF of 50 to 54 amino acids (small uORF) overlapped by a short uORF of 2 to 4 amino acids (tiny uORF) (Franceschetti et al., 2001). Polyamines repress translation of this enzyme under physiological conditions in the plant. Deletion or disruption of the conserved small uORF in the gene causes a loss of translational control and results in severe growth perturbations in transgenic tobacco (Nicotiana tabacum) because of unrepressed translation of the enzyme (Hanfrey et al., 2002). ATB2/AtbZIP11 is the only other known metabolite-dependent translational control system involving a conserved uORF in plants. Constitutive overexpression of ATB2/AtbZIP11 in Arabidopsis by a CaMV35S-promoter construct lacking the translational control of the 5′UTR causes severe growth phenotypes, including lethality and sterility (our unpublished results). Thus, loss of control of ATB2/AtbZIP11 translation may cause such severe effects as well.
The bZIP transcription factor ATB2/AtbZIP11 is expressed in vascular tissues of seedlings. In these tissues, sucrose-induced translational repression was shown (Rook et al., 1998a). Expression of the ATB2/AtbZIP11 5′UTR-GUS-construct under control of the UBQ10-promoter showed that SIRT is active in all tissues of the seedling. Thus, SIRT could control other bZIP transcription factors with different tissue-specific expression patterns.
Plants undergo 5- to 10-fold changes in sucrose concentration over the diurnal period (Farrar et al., 2000), and SIRT represents a flexible way to respond to such changes. ATB2/AtbZIP11 expression is light dependent. Photosynthesis results in increased sucrose levels (Farrar and Farrar, 1987) and activation of sucrose transport processes. The association of the ATB2/AtbZIP11 expression with vascular tissues suggests a function in assimilate partitioning. At low or intermediate sucrose concentrations, ATB2/AtbZIP11 is translated, and higher sucrose levels switch off ATB2 synthesis and affect target gene expression. The target genes of ATB2/AtbZIP11 and the physiological importance of SIRT on the expression of ATB2/AtbZIP11 are currently under investigation.
Many previously characterized S-group transcription factors harboring the SC-uORF are induced by low temperature, salt stress, abscisic acid, ethylene, indoleacetic acid, jasmonic acid, pathogen attack, or senescence (Aguan et al., 1993; Kusano et al., 1995; Yang et al., 2001; Lee et al., 2002). Such conditions often promote changes of sucrose concentrations, which in turn could affect the translation of bZIP proteins and activation of downstream target genes. SC-uORF–containing genes can be regulated by different stimuli, but gene activity is overridden and/or fine-tuned by SIRT, depending on the prevailing sucrose concentration.
Plant bZIP transcription factors have been shown to homodimerize or heterodimerize. The group S bZIP factor CPRF6 heterodimerizes with the group C bZIP factor CPRF2 (Onate et al., 1999; Rugner et al., 2001; Strathmann et al., 2001). The snapdragon (Antirrhinum majus) bZIP910 and bZIP911 were found to bind to hybrid c-box/g-box ACGT elements as homodimers, whereas heterodimers of these transcription factors show low affinity for these boxes (Martinez-Garcia et al., 1998). Thus, a whole range of mechanisms are involved in controlling activity of bZIP transcription factors, including transcription, translation, posttranslational modifications, and homodimerization or heterodimerization with other transcription factors. Such a system allows a flexible, multiresponsive regulation of bZIP target genes.
In conclusion, we propose SIRT to act on a set of plant bZIP transcription factors encoding the SC-uORF. Interaction of transcriptional activation and translational control allows these regulatory genes to respond in a flexible way to rapidly changing stimuli that affect sucrose levels in cells or tissues.
METHODS
Plant Transformation and Growth Conditions
Wild-type Arabidopsis thaliana (ecotype Col-0) were grown in a growth chamber at 22°C under a 16-h-light/8-h-dark cycle with a relative humidity of 80%. Floral dip transformations (Clough and Bent, 1998) were performed by using Agrobacterium tumefaciens strains GVG 2260 and EHA105. For in vitro cultures, seeds were surface sterilized by liquid or vapor-phase (Clough and Bent, 1998) methods. For surface sterilization, seeds were soaked in 96% ethanol for 5 min and 10 min in 20% commercial bleach and washed four times in sterile bidistilled water. After surface sterilization, seeds were stratified for 2 d at 4°C. Transformed plants were selected by the bar resistance of the T-DNA on solid half-strength MS medium with vitamins (Duchefa, Haarlem, The Netherlands) containing 15 mg/L of phosphinotricine. Homozygous lines were selected and analyzed for GUS- or GFP-mRNA and for GUS activity. Results of two representative independent lines are presented and named line I and line II of each construct. Transgenic seedlings of selected lines were grown for 5 d in liquid half-strength MS medium with vitamins (Duchefa). Filter-sterilized sucrose was added to autoclaved medium at indicated concentrations. After imbibition, seedlings were grown under continuous light at 22°C for 5 d with agitation (150 rpm).
Marker Gene Constructs
Molecular cloning techniques were performed as described (Sambrook et al., 1998). For constructing UBQ10:5′UTR-GUS/GFP, the GUS coding sequence in the ATB2-GUS construct in pBluescript SK− (pbPGA) (Rook et al., 1998a) was exchanged for a GFP-GUS chimera coding region by restriction of pbPGA with BamHI and of pCambia1304 (Cambia, Canberra, Australia) with BstEII. The overlapping ends were filled in with T4-DNA polymerase. Both vectors were digested with NcoI, and the isolated GFP-GUS coding-sequence was inserted into pbPGA, resulting in pbPGGA. The Arabidopsis UBQ10 promoter (PUBQ10, upstream sequence of At4g05320 −389 to −1307 from ATG) was isolated PstI/BamHI from the vector p3325 (Norris et al., 1993) and ligated into PstI/BamHI opened binary vector pGreenII0229 (Hellens et al., 2000), resulting in pGreenII0229-PUBQ10. pbPGGA was digested with XbaI, for isolation of the ATB2/AtbZIP11 marker gene construct, which was introduced into the XbaI-digested vector pGreenII0229-PUBQ10. In this construct, 196 bp of the ATB2/AtbZIP11 promoter are included.
The exchange of the ATB2/AtbZIP11-3′UTR was performed by SpHI restriction of pCambia1304 and after fill in. The GFP-GUS chimera-coding sequence fused to the nos 3′UTR was isolated by a second restriction with NcoI. Construct pbPGA (Rook et al., 1998a) was opened with NotI, and sticky ends were filled in by Klenow treatment. NcoI restriction released the GUS-coding sequence and the ATB2/AtbZIP11 3′UTR from pbPGA, and the GFP-GUS chimera coding sequence with nos-poly(A) was inserted. The whole construct with ATB2/AtbZIP11 promoter and 5′UTR was inserted via XhoI and SacI restriction sites into the binary vector pGreenII0229 (Hellens et al., 2000).
For construction of AtbZIP2-GUS constructs, the 5′ and 3′UTRs were amplified by PCR, subcloned in pGEM-T easy (Promega, Madison, WI), and sequenced. The 3′UTR was amplified with primer bZIP2 3′UTR-F, 5′-GCTAGCTGATTAATAAAATTAATTAAAATAATTAGATG-3′, containing an NheI site (underlined) and 3′UTR-R, 5′-TCAAATCTACCAAGTCAAATTTCACCGCTAGCGGGCCC-3′, with NheI and ApaI sites (underlined). This fragment was isolated with NheI from pGEM-T easy and inserted in pCambia1301. The 5′UTR was amplified with primer bZIP2+5′UTR-F, 5′-AAGCTTGTTAACCTCTTCCTTATCTCCTTAAAA-3′, containing HindIII and HpaI (underlined) and bZIP2 5′UTR-R with NcoI (underlined), 5′-CCATGGTGACGACGGAGTCCGACG-3′, a fragment of 501-bp 5′UTR, and the first 81 bp of the coding sequence. The 5′UTR was isolated with HindIII/NcoI from a pGEM-T easy subclone and inserted in the HindIII/NcoI-digested pCambia1301 carrying the GUS coding sequence, resulting in 5′UTR-bZIP2 CDS-GUS-3′UTR in translational fusion. The AtbZIP2 promoter was isolated with BalI from BAC F8D23 (4.4 kb) and subcloned in the SmaI site of pBluescript II SK+; subsequently, the 2213-bp AtbZIP2 promoter fragment was isolated with Eco72I and EarI. pCambia1301 5′UTR-bZIP2 CDS-GUS-3′UTR was digested with HpaI. After a partial digestion with EarI, this construct was ligated to the 2.2-kb Eco72I/EarI promoter fragment. Finally, the entire promoter-5′UTR-GUS-3′UTR was isolated from pCambia1301 with ApaI/BamHI and ligated in the ApaI/BamHI-digested pGreenII0229 (Hellens et al., 2000), resulting in vector bZIP2:5′UTR-GUS.
For construction of the AtbZIP2-5′UTR deletion construct, bZIP2:Δ5′UTR-GUS primer bZIP2 5′UTR-R (described above) and bZIP2Δ 5′UTR-F, 5′-AAGCTTCAGACAGATCATAAAAAAAAACCAAAC-3′, with a HindIII site (underlined) were used to amplify a fragment consisting of 86 bp 5′UTR and 81 bp of the coding sequence. The 5′UTR-Δ was digested with HindIII/NcoI from a sequenced pGEM-T easy subclone and exchanged for the HindIII/NcoI fragment of pCambia1301. pCambia1301 5′UTR-Δ-bZIP2 CDS-GUS-3′UTR was digested with HindIII, polished with T4, and ligated to the 2.2-kb Eco72I/EarI, T4-polished AtbZIP2 promoter fragment. Finally, the entire AtbZIP2-promoter-5′UTR-Δ-GUS-3′UTR was digested from pCambia1301 with ApaI/BamHI and ligated in the ApaI/BamHI-digested pGreenII0229 (Hellens et al., 2000), resulting in vector bZIP2:Δ5′UTR-GUS.
Deletion Constructs
Deletions of the 5′UTR were generated by PCR. The PCR products were NcoI/NheI cloned into the pNE03. The partially deleted leader segments were NcoI/NheI cloned from pNE03 into the PGA vector. The promoter-leader-GUS-trailer was KpnI/NotI cloned into pGreenII0229 (Hellens et al., 2000). Primers used for the 3′-end deletion 4-11 were NE03 DraIII-SmaI, 5′-GGAACAAGAGTCCACTATTA-3′, and NE4-11, 5′-TATCCATGGCTAGGGTTTTGTTGTAATTATGCG-3′. Primers used for the 5′-deletion were GUSA-leader ATB2, 5′-GTCCACCAGGTGTTCGGCGTGGTG-3′, and 4-23, 5′-TATAGCTAGCTAGTTTCTTTTCAAATTTCTCTTCTTCG-3′.
Point Mutation Constructs
An EcoRV fragment from pbPGA was cloned into pBluescript II KS+ (Stratagene, La Jolla, CA), creating pNE03. Point mutations were created in the pNE03 vector with the QuikChange Multi Site-Directed mutagenesis kit (Stratagene) as described by the manufacturer. After checking the desired mutations by sequencing, the NcoI/NheI restriction fragment was cloned into the pbPGA vector. Subsequently, the KpnI/NotI fragment, containing the ATB2/AtbZIP11-promoter-5′UTR-GUS-3′UTR, was fused into pGreenII0229 (Hellens et al., 2000). Primers used for mutagenesis were NE05mutorf1 5′-AAACATTGAAGCTTAATCAGC-3′, NE05mutorf2 5′-TTGAGACACGTGTCTCCAATA-3′, NE05mutorf2A 5′-TTTCTCTCTGGGTTTTAGTTAAATTCCA-3′, and NE05mutorf4 5′-TATGTCTCAAGATCTCTGAAC-3′.
Constructs for in Vitro Translation
For in vitro translation, the ATB2/AtbZIP11 5′UTR from +4 to −496 (relative to start codon) was isolated from vector PGA (Rook et al., 1998a) in an EcoRV and NcoI restriction. After fill in of the NcoI overlapping ends, the fragment was subcloned into EcoRV-opened pBluescript II KS+ (Stratagene), resulting in pb5. In the same manner for pb5-2a, the 5′UTR of the construct bZIP11:52a-GUS was subcloned into pBluescript II KS+ (Stratagene). The AUG context in the construct pb5-AUGc was modified using PCR on pbPGA (Rook et al., 1998a) with the following primers: primer 1, 5′-CACAATGGCTCCAATAATACTCAGTGAG-3′ (exchanged start codon context underlined); and primer 2, 5′-TCCATGGAGTAACACACAAACAAAAACAG-3′, for amplification up to the included NcoI restriction site (underlined) at the end of the 5′UTR. The PCR product was subcloned in pGEM-T easy (Promega). After sequencing, the fragment was isolated by EcoRI restriction and ligated into pBluescript II KS+ (Stratagene), resulting in pb5-AUGc. Sequencing confirmed the correct orientation of the ATB2/AtbZIP11 5′ UTR and its modifications to the T3 promoter.
GUS Chemistry and Quantification
Plant material was stained using GUS buffer consisting of 0.1 M NaH2PO4, 0.1 M Na2HPO4, 0.5 mM ferricyanide, 0.5 mM ferrocyanide, 10 mM EDTA, 0.1% (v/v) Triton X-100, and 1 mg/mL of 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid. For GUS quantification, seedlings were ground using a drill with a metal pistil in an eppendorf tube or in a mortar with liquid nitrogen. The glucuronidase activity was quantified using the fluorometric 4-methylumbelliferyl-β-d-glucuronide (MUG) assay (Jefferson et al., 1987). Seedlings were ground in extraction buffer (50 mM Na2PO4, pH 7.0, 10 mM dithiothreitol, 1 mM EDTA, pH 8.0, 0.1% sodium lauryl sarcosine, and 0.1% Triton X-100). The suspension was centrifuged 1 min at 14,000 rpm, and 27 μL of prewarmed assay buffer of 37°C (22 mg MUG in 50 mL of extraction buffer) was added to 3 μL of extract. The reaction was incubated at 37°C for 1 h and stopped by adding 270 μL of 0.2M Na2CO3. Fluorescence was measured with a Fluostar Galaxy (BMG, Offenburg, Germany) at λ355/λ460. Protein content was determined using the Bradford assay (Bradford, 1976) to compare activity to protein units.
In Vitro Translation Analysis
Coupled in vitro transcription/translation reactions were performed with the coupled transcription/translation wheat germ lysate system (Promega) according to the manufacturer with T3 polymerase, 1 μg of plasmid, and 10 μCi of [35S]Met (Amersham Pharmacia, Buckinghamshire, UK) per 50 μL of translation mixture. Incubation was performed for 90 min at 30°C. Translation reaction was directly mixed with (2×) loading buffer consisting of 100 mM Tris-HCl, pH 7.5, 4% SDS, 0.2% bromophenol blue, 15% glycerol, and 200 mM β-mercaptoethanol. The mixture was heated at 95°C for 4 min. Sample mixes were then microfuged (15,000g for 1 min). Qualitative analysis of the translated products was performed by separation on a 16.5% Tris/tricine polyacrylamide precast gel (Bio-Rad, Hercules, CA) (Schagger and von Jagow, 1987). Radiolabeled translation products were visualized on a PhosphorImager (Molecular Dynamics, Sunnyvale, CA) after 10 d of exposure. Protein size standard was prestained broad range (Bio-Rad).
RT-PCR Measurements
Relative quantification of the marker gene messenger levels was performed using Taqman relative quantitative real-time PCR technology on the ABI 7700 LightCycler (Applied Biosystems, Foster City, CA) with gene-specific primers and probes labeled 5′ FAM and 3′ TAMRA, using the 2× Taqman Q-PCR mix (Applied Biosystems). Primers were used at concentrations of 900 μM and probes at 250 μM. The 5′UTR-GUS-fusion was amplified with 5′-TTGTGTGTTACTCCATGGTCCG-3′ and 5′-CCCAGGCCGTCGAGTTTT-3′ and detected with the probe 5′-CCTGTAGAAACCCCAACCCGTGAAATCA-3′; alternatively, GUS was detected in PCR with primer GUS forward 5′-AACCCCAACCCGTGAAATC-3′, GUS reverse 5′-CACAGTTTTCGCGATCCAGAC-3′, and the GUS probe 5′-ACTCGACGGCCTGTGGGCATTC-3′. Expression of the GUS/GFP-chimera coding sequence was performed with GFP forward, 5′-ACGGCATCAAAGCCAACTTC-3′, 5′-TCAGCGAGTTGCACGCC-3′, and GFP-probe 5′-AGACCCGCCACAACATCGAAGAC-3′. Actin2 (At3g18780) mRNA expression was detected as reference with Act forward 5′-GCTGAGAGATTCAGACTGCCCA-3′, Act reverse 5′-CACAGTTTTCGCGATCCAGAC-3′, and the Act probe 5′-AAGTCTTGTTCCAGCCCTCGTTTGTGG-3′.
All primers and probes were designed using the Primer Express version 1.0 software of Applied Biosystems. Q-PCR results were analyzed with SDS version 1.7 software from Applied Biosystems. Results for AtbZIP2 were calculated using equation  (control-sample)/
(control-sample)/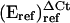 (control-sample) as described by (Pfaffl, 2001). Relative quantification for all other primer-probe combinations was sufficiently similar to Actin2 to use the ΔΔCt method (user bulletin 2, ABI Prism 7700 sequence detection system; Applied Biosystems). Primer-probe efficiencies for all primer-probe sets were determined according to equation E = 10(−1/slope) as described by Rasmussen (2001).
(control-sample) as described by (Pfaffl, 2001). Relative quantification for all other primer-probe combinations was sufficiently similar to Actin2 to use the ΔΔCt method (user bulletin 2, ABI Prism 7700 sequence detection system; Applied Biosystems). Primer-probe efficiencies for all primer-probe sets were determined according to equation E = 10(−1/slope) as described by Rasmussen (2001).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers At4g34590 (AtbZIP11), At1g75390 (AtbZIP44), At2g18160 (AtbZIP2), At3g62420 (AtbZIP53), At5g49450 (AtbZIP1), D63951 (tbZ17), AB032478 (tbzF), D26563 (mLIP15), X6245.1 (OCSBF-1), Y13675.1 (BZIP910), Y13676.1 (BZIP911), D63955 (glip19), AF532621 (GmATB2), AJ292744 (cprf6), BI207937, AC121244, BG446720, AP006137, and AF430372 (PPI1).
Supplementary Material
Acknowledgments
We thank A. Thomas for helpful discussions on uORFs and translational regulation. This work was supported in part by European Union Marie Curie fellowship HPMF-CT-2001-01460 to A.W.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Anika Wiese (a.wiese@bio.uu.nl).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.019349.
References
- Aguan, K., Sugawara, K., Suzuki, N., and Kusano, T. (1993). Low-temperature-dependent expression of a rice gene encoding a protein with a leucine-zipper motif. Mol. Gen. Genet. 240, 1–8. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Atanassova, R., Leterrier, M., Gaillard, C., Agasse, A., Sagot, E., Coutos-Thevenot, P., and Delrot, S. (2003). Sugar-regulated expression of a putative hexose transport gene in grape. Plant Physiol. 131, 326–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Serres, J. (1999). Selective translation of cytoplasmic mRNAs in plants. Trends Plant Sci. 4, 142–148. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Chan, M.T., and Yu, S.M. (1998). The 3′ untranslated region of a rice alpha-amylase gene mediates sugar-dependent abundance of mRNA. Plant J. 15, 685–695. [DOI] [PubMed] [Google Scholar]
- Cheng, W.H., Taliercio, E.W., and Chourey, P.S. (1999). Sugars modulate an unusual mode of control of the cell-wall invertase gene (Incw1) through its 3′ untranslated region in a cell suspension culture of maize. Proc. Natl. Acad. Sci. USA 96, 10512–10517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Child, S.J., Miller, M.K., and Geballe, A.P. (1999). Translational control by an upstream open reading frame in the HER-2/neu transcript. J. Biol. Chem. 274, 24335–24341. [DOI] [PubMed] [Google Scholar]
- Chiou, T.J., and Bush, D.R. (1998). Sucrose is a signal molecule in assimilate partitioning. Proc. Natl. Acad. Sci. USA 95, 4784–4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciereszko, I., Johansson, H., and Kleczkowski, L.A. (2001). Sucrose and light regulation of a cold-inducible UDP-glucose pyrophosphorylase gene via a hexokinase-independent and abscisic acid-insensitive pathway in Arabidopsis. Biochem. J. 354, 67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Damiani, R.D., Jr., and Wessler, S.R. (1993). An upstream open reading frame represses expression of Lc, a member of the R/B family of maize transcriptional activators. Proc. Natl. Acad. Sci. USA 90, 8244–8248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnin, C.R., Schleiss, M.R., Cao, J., and Geballe, A.P. (1993). Translational inhibition mediated by a short upstream open reading frame in the human cytomegalovirus gpUL4 (gp48) transcript. J. Virol. 67, 5514–5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrar, J., Pollock, C., and Gallagher, J. (2000). Sucrose and the integration of metabolism in vascular plants. Plant Sci. 154, 1–11. [DOI] [PubMed] [Google Scholar]
- Farrar, S.C., and Farrar, J. (1987). Effects of photon fluence rate on carbon partitioning in barley source leaves. Plant Physiol. Biochem. 25, 541–548. [Google Scholar]
- Franceschetti, M., Hanfrey, C., Scaramagli, S., Torrigiani, P., Bagni, N., Burtin, D., and Michael, A.J. (2001). Characterization of monocot and dicot plant S-adenosyl-l-methionine decarboxylase gene families including identification in the mRNA of a highly conserved pair of upstream overlapping open reading frames. Biochem. J. 353, 403–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanfrey, C., Franceschetti, M., Mayer, M.J., Illingworth, C., and Michael, A.J. (2002). Abrogation of upstream open reading frame-mediated translational control of a plant S-adenosylmethionine decarboxylase results in polyamine disruption and growth perturbations. J. Biol. Chem. 277, 44131–44139. [DOI] [PubMed] [Google Scholar]
- Hanson, J., Johannesson, H., and Engstrom, P. (2001). Sugar-dependent alterations in cotyledon and leaf development in transgenic plants expressing the HDZhdip gene ATHB13. Plant Mol. Biol. 45, 247–262. [DOI] [PubMed] [Google Scholar]
- Hellens, R.P., Edwards, E.A., Leyland, N.R., Bean, S., and Mullineaux, P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42, 819–832. [DOI] [PubMed] [Google Scholar]
- Jakoby, M., Weisshaar, B., Droge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., and Parcy, F. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7, 106–111. [DOI] [PubMed] [Google Scholar]
- Jefferson, R., Goldsbrough, A., and Bevan, M. (1990). Transcriptional regulation of a patatin-1 gene in potato. Plant Mol. Biol. 14, 995–1006. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, K.E. (1996). Carbohydrate-modulated gene expression in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 509–540. [DOI] [PubMed] [Google Scholar]
- Kusano, T., Berberich, T., Harada, M., Suzuki, N., and Sugawara, K. (1995). A maize DNA-binding factor with a bZIP motif is induced by low temperature. Mol. Gen. Genet. 248, 507–517. [DOI] [PubMed] [Google Scholar]
- Lalonde, S., Boles, E., Hellmann, H., Barker, L., Patrick, J.W., Frommer, W.B., and Ward, J.M. (1999). The dual function of sugar carriers. Transport and sugar sensing. Plant Cell 11, 707–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.J., Lee, M.Y., Yi, S.Y., Oh, S.K., Choi, S.H., Her, N.H., Choi, D., Min, B.W., Yang, S.G., and Harn, C.H. (2002). PPI1: A novel pathogen-induced basic region-leucine zipper (bZIP) transcription factor from pepper. Mol. Plant Microbe Interact. 15, 540–548. [DOI] [PubMed] [Google Scholar]
- Lohmer, S., Maddaloni, M., Motto, M., Salamini, F., and Thompson, R.D. (1993). Translation of the mRNA of the maize transcriptional activator Opaque-2 is inhibited by upstream open reading frames present in the leader sequence. Plant Cell 5, 65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti, E., Alpi, A., and Perata, P. (2000). Glucose and disaccharide-sensing mechanisms modulate the expression of alpha-amylase in barley embryos. Plant Physiol. 123, 939–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutcke, H.A., Chow, K.C., Mickel, F.S., Moss, K.A., Kern, H.F., and Scheele, G.A. (1987). Selection of AUG initiation codons differs in plants and animals. EMBO J. 6, 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia, J.F., Moyano, E., Alcocer, M.J., and Martin, C. (1998). Two bZIP proteins from Antirrhinum flowers preferentially bind a hybrid C-box/G-box motif and help to define a new sub-family of bZIP transcription factors. Plant J. 13, 489–505. [DOI] [PubMed] [Google Scholar]
- Meijer, H.A., and Thomas, A.A. (2003). Ribosomes stalling on uORF1 in the Xenopus Cx41 5′ UTR inhibit downstream translation initiation. Nucleic Acids Res. 31, 3174–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, B., Zhou, L., Rolland, F., Hall, Q., Cheng, W.H., Liu, Y.X., Hwang, I., Jones, T., and Sheen, J. (2003). Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300, 332–336. [DOI] [PubMed] [Google Scholar]
- Morris, D.R., and Geballe, A.P. (2000). Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 20, 8635–8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, Y., Gojobori, T., and Ikemura, T. (2000). Codon usage tabulated from international DNA sequence databases: Status for the year 2000. Nucleic Acids Res. 28, 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris, S.R., Meyer, S.E., and Callis, J. (1993). The intron of Arabidopsis thaliana polyubiquitin genes is conserved in location and is a quantitative determinant of chimeric gene expression. Plant Mol. Biol. 21, 895–906. [DOI] [PubMed] [Google Scholar]
- Onate, L., Vicente-Carbajosa, J., Lara, P., Diaz, I., and Carbonero, P. (1999). Barley BLZ2, a seed-specific bZIP protein that interacts with BLZ1 in vivo and activates transcription from the GCN4-like motif of B-hordein promoters in barley endosperm. J. Biol. Chem. 274, 9175–9182. [DOI] [PubMed] [Google Scholar]
- Parola, A.L., and Kobilka, B.K. (1994). The peptide product of a 5′ leader cistron in the beta 2 adrenergic receptor mRNA inhibits receptor synthesis. J. Biol. Chem. 269, 4497–4505. [PubMed] [Google Scholar]
- Pfaffl, M.W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, 2002–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell, P.C., Smith, A.M., and Halford, N.G. (1998). Antisense expression of a sucrose nonfermenting1 related protein kinase sequence in potato results in decreased expression of sucrose synthase in tubers and loss of sucrose inducibility of sucrose synthase transcripts in leaves. Plant J. 14, 195–202. [Google Scholar]
- Rasmussen, R. (2001). Quantification on the LightCycler. In Rapid Cycle Real-Time PCR: Methods and Applications, S. Meuer, C. Wittwer, and K. Nakagawara, eds (Heidelberg: Springer Press), pp. 21–34.
- Roitsch, T., Balibrea, M.E., Hofmann, M., Proels, R., and Sinha, A.K. (2003). Extracellular invertase: Key metabolic enzyme and PR protein. J. Exp. Bot. 54, 513–524. [DOI] [PubMed] [Google Scholar]
- Rolland, F., Moore, B., and Sheen, J. (2002). Sugar sensing and signaling in plants. Plant Cell 14 (suppl.), S185–S205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook, F., Gerrits, N., Kortstee, A., van Kampen, M., Borrias, M., Weisbeek, P., and Smeekens, S. (1998. a). Sucrose-specific signalling represses translation of the Arabidopsis ATB2 bZIP transcription factor gene. Plant J. 15, 253–263. [DOI] [PubMed] [Google Scholar]
- Rook, F., Weisbeek, P., and Smeekens, S. (1998. b). The light-regulated Arabidopsis bZIP transcription factor gene ATB2 encodes a protein with an unusually long leucine zipper domain. Plant Mol. Biol. 37, 171–178. [DOI] [PubMed] [Google Scholar]
- Rugner, A., Frohnmeyer, H., Nake, C., Wellmer, F., Kircher, S., Schafer, E., and Harter, K. (2001). Isolation and characterization of four novel parsley proteins that interact with the transcriptional regulators CPRF1 and CPRF2. Mol. Genet. Genomics 265, 964–976. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1998). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schagger, H., and von Jagow, G. (1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379. [DOI] [PubMed] [Google Scholar]
- Sinha, A.K., Hofmann, M.G., Romer, U., Kockenberger, W., Elling, L., and Roitsch, T. (2002). Metabolizable and non-metabolizable sugars activate different signal transduction pathways in tomato. Plant Physiol. 128, 1480–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens, S. (2000). Sugar-induced signal transduction in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 49–81. [DOI] [PubMed] [Google Scholar]
- Strathmann, A., Kuhlmann, M., Heinekamp, T., and Droge-Laser, W. (2001). BZI-1 specifically heterodimerises with the tobacco bZIP transcription factors BZI-2, BZI-3/TBZF and BZI-4, and is functionally involved in flower development. Plant J. 28, 397–408. [DOI] [PubMed] [Google Scholar]
- Sun, C.W., and Callis, J. (1997). Independent modulation of Arabidopsis thaliana polyubiquitin mRNAs in different organs and in response to environmental changes. Plant J. 11, 1017–1027. [DOI] [PubMed] [Google Scholar]
- Tenson, T., and Ehrenberg, M. (2002). Regulatory nascent peptides in the ribosomal tunnel. Cell 108, 591–594. [DOI] [PubMed] [Google Scholar]
- Tiessen, A., Prescha, K., Branscheid, A., Palacios, N., McKibbin, R., Halford, N.G., and Geigenberger, P. (2003). Evidence that SNF1-related kinase and hexokinase are involved in separate sugar-signalling pathways modulating post-translational redox activation of ADP-glucose pyrophosphorylase in potato tubers. Plant J. 35, 490–500. [DOI] [PubMed] [Google Scholar]
- Vilela, C., and McCarthy, J.E. (2003). Regulation of fungal gene expression via short open reading frames in the mRNA 5′untranslated region. Mol. Microbiol. 49, 859–867. [DOI] [PubMed] [Google Scholar]
- Wang, Z., and Sachs, M.S. (1997). Ribosome stalling is responsible for arginine-specific translational attenuation in Neurospora crassa. Mol. Cell. Biol. 17, 4904–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzler, H., Mignery, G., Fisher, L., and Park, W. (1989). Sucrose-regulated expression of a chimeric potato tuber gene in leaves of transgenic tobacco plants. Plant Mol. Biol. 13, 347–354. [DOI] [PubMed] [Google Scholar]
- Wilkie, G.S., Dickson, K.S., and Gray, N.K. (2003). Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem. Sci. 28, 182–188. [DOI] [PubMed] [Google Scholar]
- Wobus, U., and Weber, H. (1999). Sugars as signal molecules in plant seed development. Biol. Chem. 380, 937–944. [DOI] [PubMed] [Google Scholar]
- Yaman, I., Fernandez, J., Liu, H., Caprara, M., Komar, A.A., Koromilas, A.E., Zhou, L., Snider, M.D., Scheuner, D., Kaufman, R.J., and Hatzoglou, M. (2003). The zipper model of translational control: A small upstream ORF is the switch that controls structural remodeling of an mRNA leader. Cell 113, 519–531. [DOI] [PubMed] [Google Scholar]
- Yanagisawa, S., Yoo, S.D., and Sheen, J. (2003). Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 425, 521–525. [DOI] [PubMed] [Google Scholar]
- Yang, S.H., Berberich, T., Sano, H., and Kusano, T. (2001). Specific association of transcripts of tbzF and tbz17, tobacco genes encoding basic region leucine zipper-type transcriptional activators, with guard cells of senescing leaves and/or flowers. Plant Physiol. 127, 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama, R., Hirose, T., Fujii, N., Aspuria, E.T., Kato, A., and Uchimiya, H. (1994). The rolC promoter of Agrobacterium rhizogenes Ri plasmid is activated by sucrose in transgenic tobacco plants. Mol. Gen. Genet. 244, 15–22. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.