Abstract
Leaf initiation in the peripheral zone of the shoot apical meristem involves a transition to determinate cell fate, but indeterminacy is maintained in the vascular cambium, a tissue critical to the continuous growth of vascular tissue in leaves and stems. We show that the orientation of cambial growth is regulated by microRNA (miRNA)-directed cleavage of mRNA from the Nicotiana sylvestris ortholog of PHAVOLUTA (NsPHAV). Loss of miRNA regulation in semidominant phv1 mutants misdirects lateral growth of leaf midveins and stem vasculature away from the shoot, disrupting vascular connections in stem nodes. The phv1 mutation also expands the central zone in vegetative and inflorescence meristems, implicating miRNA and NsPHAV in regulation of meristem structure. In flowers, phv1 causes reiteration of carpel initiation, a phenocopy for loss of CARPEL FACTORY/DICER LIKE1, indicating that miRNA is critical to the termination of indeterminacy in floral meristems. Results point to a common role for miRNA in spatial and temporal restriction of HD-ZIPIII mediated indeterminacy in apical and vascular meristems.
INTRODUCTION
Lateral organ primordia are initiated at the periphery of the shoot apical meristem (SAM) as cell clusters making a fundamental transition to determinacy, a process associated with the downregulation of KNOX genes (Jackson et al., 1994). Leaves attain determinacy and developmental autonomy shortly after emergence (Steeves, 1961), but early morphologists argued that the meristem remained in communication with emerging primordia and played an ongoing role in their development (Wardlaw, 1949). Microsurgical experiments provided direct evidence that the meristem is in fact critical to adaxial development and the initiation of leaf blades at the adaxial/abaxial boundary of the primordium (Sussex, 1955). Exactly how the meristem promotes adaxial development is still uncertain, but it is clearly associated with the HD-ZIPIII transcription factors PHABULOSA (PHB), PHAVOLUTA (PHV), and REVOLUTA (REV), which are expressed in the SAM and in bands extending to the adaxial surface of leaf primordia (McConnell and Barton, 1998; McConnell et al., 2001; Emery et al., 2003; Juarez et al., 2004; Kidner and Martienssen, 2004). Semidominant mutations leading to overexpression of PHB, PHV, or REV in Arabidopsis thaliana expand the adaxial domain of the primordium at the expense of the abaxial domain, changing the position of the adaxial/abaxial junction and producing a correlated shift in the position of blade initiation. Because the mutations occur in a putative sterol binding domain (START), it was initially proposed that the mutant proteins were constitutively active in the absence of the sterol ligand. An alternative explanation came with the subsequent discovery that these mutations also fell within a putative microRNA (miRNA) recognition site located at the boundary between exons 4 and 5 in both genes (Rhoades et al., 2002), which was later shown to be a functional site for mRNA cleavage in vitro (Tang et al., 2003). Analysis of similar dominant mutations in REV provided direct evidence that the adaxialized leaf phenotype results primarily if not solely from the disruption in miRNA regulation (Emery et al., 2003).
The actual mechanism of HD-ZIPIII function remains unknown, but important insights on this have come from studies on REV, which regulates formation of apical and lateral meristems (Talbert et al., 1995; Otsuga et al., 2001) and the development of vascular tissue (Zhong and Ye, 1999, 2001; Ratcliffe et al., 2000) in Arabidopsis. Initially, the vascular role appeared specific to the formation of interfascicular fibers in the stem, but dominant mutations abolishing miRNA regulation revealed an additional role in development of leaf and stem vasculature (Emery et al., 2003). Misexpression of PHB and PHV (McConnell and Barton, 1998) as well as REV (Emery et al., 2003) causes leaf midveins to overproduce xylem on the adaxial side, which eventually envelops the abaxial phloem and produces an adaxialized vein with radial symmetry. The phenotype suggests that the HD-ZIPIII pathway regulates the process by which vascular initials are generated from the indeterminate cells of the vascular cambium. A REV function related to cellular indeterminacy has also emerged from studies on its role in SAM development. Analysis of double mutant combinations in Arabidopsis indicates that REV lies in the same pathway and upstream of the meristem identity genes WUSCHEL (WUS), SHOOTMERISTEMLESS (STM), and CLAVATA1,3 (CLV) (Otsuga et al., 2001). It appears that PHB, PHV, and REV act redundantly here because double mutants lacking PHB and PHV are phenotypically normal, whereas triple mutants lacking all three functions are defective in SAM formation (Emery et al., 2003). Though the HD-ZIPIII pathway is essential for meristem initiation, the recurrent failure and reinitiation of the SAM in wus mutants of Arabidopsis (Laux et al., 1996) demonstrates that it cannot sustain a functional SAM independently. Thus, it appears that HD-ZIPIII genes regulate the initial onset of cellular indeterminacy during SAM formation but that sustained SAM function requires downstream expression of WUS and STM.
This indeterminacy role, both in apical and vascular meristems, raises an intriguing possibility that the HD-ZIPIII pathway regulates leaf adaxial identity by this mechanism as well. A distinguishing feature of the leaf adaxial domain is the competence to generate axillary meristems (McConnell and Barton, 1998), which clearly hinges on the expression of cellular indeterminacy. In fact, experimental overexpression of class I KNOX genes in dicots has demonstrated that the entire adaxial surface of the leaf is competent for expression of meristematic indeterminacy (Sinha et al., 1993; Chuck et al., 1996). The first indication that this might play a role in wild-type leaf development came from molecular analysis of compound leaves in tomato (Lycopersicon esculentum) and pea (Pisum sativum), where expression of meristem identity genes prolongs the period of adaxial indeterminacy, potentiating initiation of lateral leaflets at the adaxial/abaxial boundary of the primordium (TKN1, Hareven et al., 1996; LeT6/TKN2, Janssen et al., 1998; UNIFOLIATA, Gourlay et al., 2000; Bharathan and Sinha, 2001). These meristem genes are not expressed in simple leaves, but the formation of leaf blade primordia at the adaxial/abaxial boundary is in certain respects a similar morphogenic event (Hagemann and Gleissberg, 1996). Recent studies showing ectopic blade formation in response to misexpression of class I KNOX genes in blade-on-petiole1 mutants of Arabidopsis (Ha et al., 2003) and in antisense NsPHAN transgenics of tobacco (McHale and Koning, 2004) have provided the first evidence that indeterminacy plays a role in blade initiation. Loss of NsPHAN in juvenile leaves of Nicotiana promotes highly disorganized patterns of proliferation in the adaxial mesophyll, which delays palisade differentiation and potentiates de novo formation of ectopic leaf blades along the adaxial flank of the midrib. The phenotype suggests that blade formation hinges on a transient state of indeterminacy in the adaxial domain, which is normally suppressed by NsPHAN after initiation of the primary blade.
Do HD-ZIPIII genes operate by a common mechanism in all locations? To address this question, we examined the role of PHAVOLUTA (NsPHAV) in leaf development, vascular patterning, and SAM function in Nicotiana sylvestris. Here, we report the developmental consequence of a mutation (phv1) abolishing miRNA regulation of NsPHAV, which produces alterations in leaf polarity, vascular patterning, and meristem function. The most pronounced vascular phenotype for phv1 occurs in the stem, where lateral proliferation of the cambium is misdirected away from the shoot, causing leaf and stem veins to develop as a series of disconnected loops. This cambial disruption also misdirects lateral growth of leaf midveins in a manner implicating vascular development as a determinant of organ polarity and blade formation. Finally, we show that the phv1 mutation disrupts the organization of vegetative, inflorescence, and floral meristems, revealing a direct role for the HD-ZIPIII pathway and miRNA in the spatial restriction of indeterminacy. The results point to a common mechanism for HD-ZIPIII functions in adaxial identity, vascular patterning, and meristem structure.
RESULTS
The phv1 Mutation Disrupts miRNA Regulation
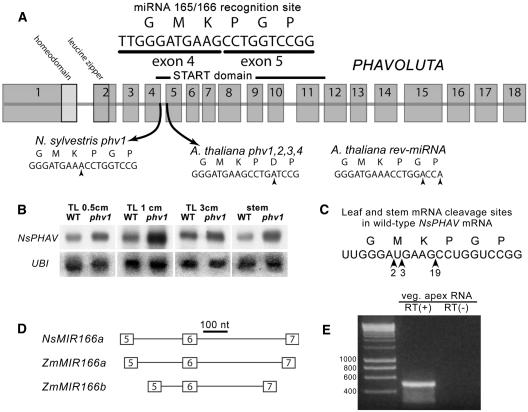
After seed mutagenesis in N. sylvestris with ethyl methanesulfonate, we isolated a semidominant mutant with the leaf phenotype observed in phb, phv, and rev mutants of Arabidopsis (McConnell and Barton, 1998; McConnell et al., 2001; Emery et al., 2003). After four backcrosses to the wild type, genomic DNA sequencing in heterozygous mutant plants revealed a wild-type NsPHAV allele and a mutant NsPHAV allele with a single nucleotide change from G to A at the last position of exon 4. This falls in the center of the miRNA 165/166 recognition site (Rhoades et al., 2002; Tang et al., 2003) but does not change the predicted mRNA translation, suggesting that the leaf phenotype results solely from a disruption in RNA processing. As shown in Figure 1A, the mutant phv alleles in Arabidopsis are also at the miRNA recognition site (five nucleotides downstream) but produce a translational change from Gly to Asp at the second position of exon 5 (McConnell et al., 2001). There are two nucleotide changes in the rev-∂miRNA allele of Arabidopsis (Emery et al., 2003), neither of which changes translation. To determine whether the nucleotide change in N. sylvestris affected regulation of NsPHAV expression, patterns of transcript accumulation were examined by RNA gel blot analysis (Figure 1B). In leaves from 0.5 cm to 3 cm in length and in stem tissue just below the shoot apex, levels of NsPHAV mRNA were elevated in heterozygous mutant plants relative to the wild type, consistent with a disruption in miRNA regulation. Because neither 5′ nor 3′ mRNA cleavage products were visible in RNA gel blots, we employed a modified 5′ RACE analysis (Kasschau et al., 2003) to map in vivo the 5′ ends of cleaved mRNAs in mutant and wild-type plants (Figure 1C). Sequence analysis of 24 wild-type transcripts from leaves and stems revealed that all 24 were cleaved within the 21-nucleotide miRNA recognition site. Nineteen of 24 were cleaved at a central site adjacent to the nucleotide position altered in the phv1 mutants. Transcript mapping in heterozygous mutant leaves showed evidence for altered processing of NsPHAV transcripts. Only 2 of 12 transcripts examined were cleaved within the recognition sequence, which were presumably wild-type molecules, although this could not be confirmed. The remaining 10 were confirmed mutant molecules, all of which terminated at alternative sites 43 and 78 nucleotides upstream, indicating that the phv1 mutation disrupts miRNA directed cleavage of NsPHAV transcripts.
Figure 1.
Gene Structure and Expression Patterns for NsPHAV and NsMIR166a.
(A) PHV gene structure and mutations at the miRNA 165/166 recognition site in Nicotiana and Arabidopsis.
(B) NsPHAV mRNA levels in wild-type and mutant phv1 leaves and stems relative to a ubiquitin (UBI) control.
(C) The ends of 24 wild-type NsPHAV mRNAs were mapped by 5′ RACE analysis. All were cleaved at the miRNA 165/166 site with a strong preference (n = 19) for a central site flanking the G-to-A base substitution in the phv1 mutant. In heterozygous phv1 mutant plants (data not shown), mutant transcripts terminated at alternative sites 43 and 78 nucleotides upstream.
(D) Genomic structure of precursor MIR166 genes in maize and Nicotiana (NsMIR166a), showing the miRNA recognition site (6 box) flanked by two conserved regions. The upstream 5 box represents the predicted foldback region involved in hairpin formation. Downstream conservation in the 7 box suggests that this region is part of the precursor RNA transcript. nt, nucleotides.
(E) RT-PCR amplification with (+) and without (−) reverse transcriptase (RT) using primers specific to nonconserved regions internal to the 5 and 7 boxes confirmed transcription of NsMIR166a in RNA samples from the vegetative shoot apex.
In Arabidopsis (Reinhart et al., 2002; Kidner and Martienssen, 2004) and maize (Zea mays) (Juarez et al., 2004), single stranded miRNAs targeting HD-ZIPIII transcripts are thought to arise from hairpin precursor RNAs transcribed from the MIR165 and MIR166 gene families. Members of the MIR166 family from Arabidopsis, maize, and rice (Oryza sativa) are characterized by three main regions of sequence conservation (5, 6, and 7 boxes), with the 6 box representing the actual recognition sequence and an upstream 5 box representing the predicted foldback region involved in hairpin formation. Downstream conservation in the 7 box suggests that this region is part of the precursor RNA transcript. To determine whether this MIR166 gene structure was conserved in Nicotiana, we attached primer adapters to blunt genomic fragments in N. sylvestris and amplified fragments with homology to the maize 5 box (primer 5F, Juarez et al., 2004). A putative precursor gene with homology to the 5, 6, and 7 boxes of maize was isolated and named NsMIR166a based on its structural similarity to ZmMIR166a (Figure 1D). RT-PCR amplification with primers specific to nonconserved regions internal to the 5 and 7 boxes confirmed transcription of NsMIR166a in RNA samples from the vegetative shoot apex (Figure 1E).
NsPHAV Regulates Adaxial Identity and Midvein Development
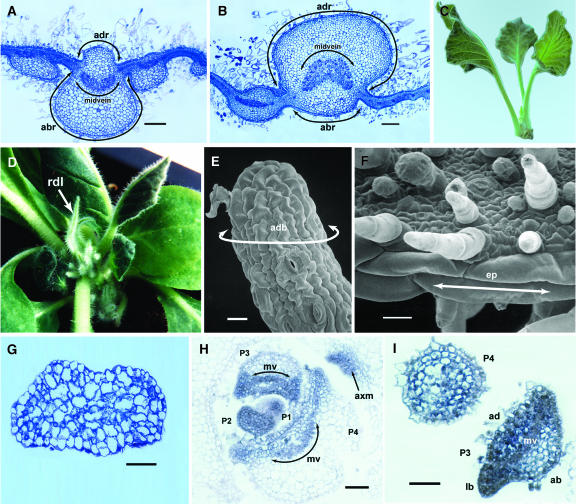
Phenotypic analysis of phv1 heterozygous and homozygous mutant plants is presented in Figure 2. In phv1 heterozygotes and homozygotes, the disruption in leaf polarity is quite mild in juvenile leaves and becomes gradually more severe in the adult phase of growth. Wild-type adult leaves are divided unequally into adaxial (front third) and abaxial (back two-thirds) domains, with an internal adaxial/abaxial boundary marked by a midvein curving in the adaxial direction. Lateral outgrowth of leaf blades occurs at the adaxial/abaxial boundary on the flanks of the primordium (Figure 2A). As in the phb, phv, and rev mutants of Arabidopsis, heterozygous phv1 mutants show an expansion of the adaxial domain at the expense of the abaxial domain, shifting the position of the adaxial/abaxial boundary toward the back of the primordium (Figure 2B). Leaf blades and midveins develop in accordance with this shifted boundary, which results in formation of the cup-shaped leaves shown in Figure 2C. Adult leaves of homozygous phv1 mutants appear to be fully adaxialized rod-like structures, where the absence of an adaxial/abaxial boundary precludes lateral outgrowth of leaf blades (Figure 2D). The presence of blade cell types reveals, however, that these leaves are not actually bladeless. Epidermal cells with crenulated side walls show blade identity wrapping fully around these primordia (Figure 2E), with no evidence of an adaxial/abaxial boundary that is normally marked by elongated files of epidermal cells (Figure 2F). The mesophyll tissue in rod-like leaves also shows no evidence of adaxial/abaxial differentiation, suggesting that they are adaxialized internally (Figure 2G). In addition, they have either diminutive midveins or no midvein at all, consistent with an apparent absence of an internal adaxial/abaxial boundary.
Figure 2.
Leaf Phenotypes and NsPHAV Expression.
(A) and (B) Relative positions of adaxial and abaxial ribs domains (adr and abr, respectively) and midveins in wild-type (A) and heterozygous phv1 mutant (B) leaf primordia. Note the enlarged adaxial domain in the mutant. Bars = 160 μm.
(C) Cup-shaped leaves in heterozygous phv1 mutant.
(D), (E), and (G) Homozygous phv1 mutant rod-like (rdl) leaves (D) with adaxial blade tissue (adb) encircling the primordium (E) and a uniform mesophyll lacking adaxial/abaxial differentiation (G). Bar in (E) = 20 μm; bar in (G) = 80 μm.
(F) Elongated epidermal files (ep) marking the adaxial/abaxial boundary in a wild-type leaf. Note the absence of this boundary in the homozygous mutant (E). Bar in (F) = 20 μm.
(H) and (I) In situ NsPHAV expression in the wild type (H) and heterozygous phv1 mutant (I) shoot apex showing midveins (mv), leaf blades (lb), and leaf primordia at stages P1 to P4. ad, adaxial; ab, abaxial. Bars = 80 μm.
To determine how the loss of miRNA regulation affected patterns of NsPHAV expression, in situ hybridizations were conducted with digoxygenin-labeled probes in wild-type and heterozygous mutant plants (Figures 2H and 2I). In wild-type tissue just above the SAM (Figure 2H), NsPHAV mRNA was detected throughout P1 and P2 primordia and begins to show adaxial localization at the P3 stage along a line coinciding with the developing midvein. At the periphery of the primordium, NsPHAV expression still extends somewhat into the abaxial domain, but with subsequent lateral growth of the midvein, it becomes fully restricted to the adaxial domain. In heterozygous phv1 mutants, NsPHAV mRNA appears to accumulate to higher levels overall (Figure 2I), consistent with results from RNA gel blot analysis (Figure 1B). At the bladeless base of P4 primordia (adult phase), NsPHAV is expressed throughout the primordium. The adjacent P3 section shows a more distal location in the leaf, where leaf blades are initiated at a central position in the primordium, and the midvein is flattened to a straight line (cf. to wild type, Figure 2H). NsPHAV is expressed at high levels in the adaxial domain and is detectable in the abaxial domain, although apparently at lower levels.
NsPHAV Regulates Growth of the Vascular Cambium
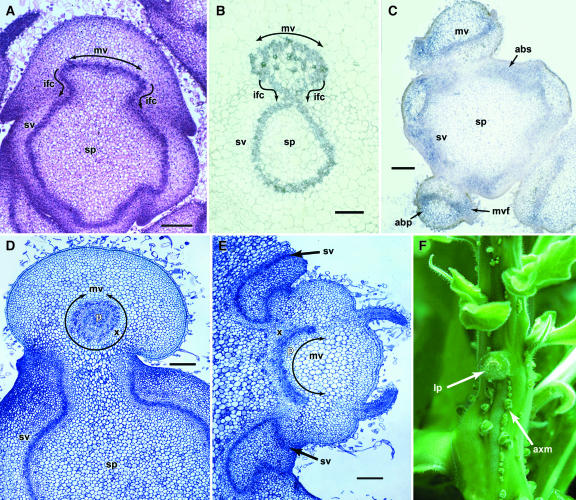
In the region where leaf petioles join the stem (nodes), leaf midveins show characteristic patterns of growth mediated by the vascular cambium, a sheath of meristematic cells positioned between the xylem and phloem (Esau, 1977). The cambium performs two functions, generating new vascular initials within the vein itself (fascicular) but also proliferating outward from the lateral flanks (interfascicular) and toward the shoot, connecting leaf midveins to the vascular cylinder of the stem (Figure 3A). In situ analysis in wild-type nodal regions revealed NsPHAV expression in the cambium of stem vasculature and in leaf midveins (Figure 3B). Mutants also show this pattern of expression, but broad bands of ectopic NsPHAV extend outward from the flanks of leaf midveins and into the abaxial domain of the petiole (Figure 3C). Mutant midveins have a correlated pattern of abnormal growth in this region, failing to curve toward the stem and often turning sharply in the opposite (abaxial) direction (Figures 3C and 3E). In extreme cases, midveins fold backward entirely into a circular structure with xylem surrounding phloem (Figure 3D). Thus, the phv1 mutation appears to alter the lateral course of cambial proliferation, resulting in an abnormal vascular curvature. As in the leaf midveins, ectopic NsPHAV is also observed on the abaxial side of the stem vasculature (Figure 3C) and is likewise associated with misdirected cambial growth. This results in aberrant curvature of stem vasculature away from the shoot, disrupting lateral connections with the leaf midveins (Figure 3E). At the lateral flanks of leaf insertion sites, misdirected stem veins grow downward along the stem surface, inducing lateral outgrowths (Figure 3F), which were confirmed histologically as ectopic axillary meristems (data not shown). There are broad domains of ectopic NsPHAV in this region (Figure 3C), but meristems occur in a discrete linear array directly associated with misdirected vasculature.
Figure 3.
Vascular Phenotypes and NsPHAV Expression.
(A) and (B) Wild-type leaf/stem junction. Paraffin section in (A) shows stem pith (sp) and curvilinear growth of interfascicular vascular cambium (ifc) from the flanks of the leaf midvein (mv) connecting with stem vasculature (sv). Wild-type in situ NsPHAV expression is confined to the cambium (B). Bars = 160 μm.
(C) to (F) Heterozygous phv1 mutant leaf/stem junctions. Ectopic NsPHAV expression in abaxial domains of leaf petioles (abp) and the stem (abs) and on the lateral flanks of leaf midveins (mvf) (C). Circularized midveins with xylem (x) surrounding phloem (p) (D). Aberrant abaxial curvature of leaf midveins (mv) and stem vasculature (sv) (E). Note that leaf midveins fail to connect with stem vasculature ([C], [D], and [E]). Formation of ectopic axillary meristems (axm) beginning at the flanks of the leaf petiole (lp) and extending down the stem (F). Bar in (C) = 160 μm; bars in (D) and (E) = 200 μm.
NsPHAV Regulates the Structure of Vegetative and Reproductive Meristems
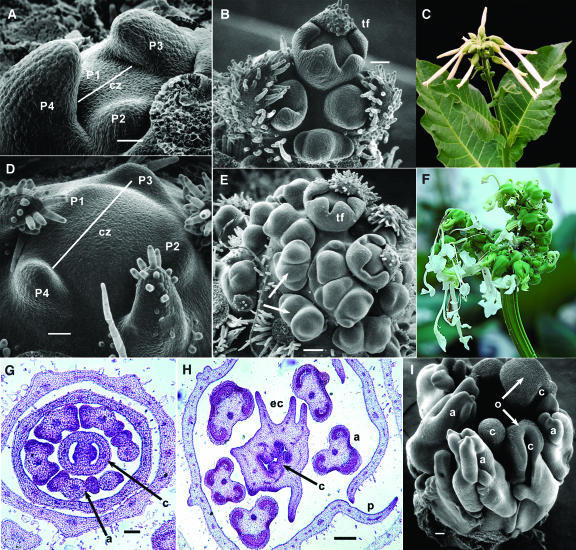
The adult vegetative SAM of wild-type N. sylvestris is shown in Figure 4A, with a central zone measuring around 100 μ between the P3 and P4 leaf primordia. The corresponding SAM in a phv1 mutant (Figure 4D) shows a doubling in the size of the central zone separating P3 and P4 primordia, with no apparent change in overall phyllotaxis. Cell size also remains unchanged in the central zone, indicating that its expansion results from an increase in cell number. In the reproductive phase, the wild-type inflorescence meristem (Figure 4B) is normally subdivided at the apex into a group of floral meristems. The process begins with formation of a terminal flower and then continues in subtending positions to produce a terminal cluster (Figure 4C). In heterozygous phv1 mutants, the inflorescence meristem generates a terminal flower but reiterates the formation of subtending flowers (Figure 4E), producing a highly ramified cluster with 30 to 40 flowers (Figure 4F). In many cases, it appears that floral meristems are initiated in very close proximity and then fuse during development. In addition, it appears that meristems can emerge from existing meristems to produce twin flowers. Floral meristems of phv1 mutants initiate the correct number of floral organs (Figure 4H) but typically over a much broader expanse relative to the wild type (Figure 4G). All floral organs develop abnormally in the mutant. Lateral growth of petals often misdirected abaxially, precluding closure of the corolla. Anthers show fusion of adjoining locules and generate a variety of protrusions. Carpels are affected most severely and in two distinct ways. Whereas in the wild type the carpel primordia curve toward one another and fuse to enclose developing ovules, mutant carpels show an inverted pattern, curving away from one another and leaving the ovules on an exposed surface (Figure 4I). In addition, carpel primordia are reinitiated on existing carpels (Figure 4H), often in successive rounds, and again grow outward in an abaxial direction leaving the ovules exposed.
Figure 4.
Meristem and Flower Phenotypes.
(A) Wild-type adult vegetative meristem showing central (cz; line = 100 μm) and peripheral zones with P1 to P4 leaf primordia.
(B) Wild-type inflorescence meristem showing a terminal flower (tf) and subtending cluster.
(C) Wild-type inflorescence.
(D) phv1 adult vegetative meristem showing an enlarged central zone (cz; line = 200 μm) with P1 to P4 leaf primordia in the peripheral zone.
(E) and (F) phv1 mutant inflorescence meristem showing a terminal flower (tf) (E) and subtending cluster (F) phv1 mutant inflorescence.
(G) Transverse section of a wild-type flower showing anthers (a) and carpels (c).
(H) Transverse section of a phv1 flower showing anthers (a) and formation of ectopic carpel primordia (ec) emerging on the abaxial side of the primary carpel (c). Lateral growth of petals (p) is often misdirected abaxially, precluding corolla fusion.
(I) Scanning micrograph of a phv1 flower showing distorted anthers (a) and inverted curvature in carpels (c), leaving developing ovules (o) on an exposed surface.
Bars in (A) and (D) = 40 μm; bars in (B), (E), (I), (G), and (H) = 100 μm.
NsPHAV Transgenics
Wild-type NsPHAV and mutant phv1 transgenes driven by the 35S promoter were constructed for insertion into wild-type N. sylvestris. Twenty transgenic plants with the 35S:wild-type allele were recovered and self-pollinated to produce T2 progeny. Seven independent transgenic progenies displayed upward curling of the leaf margin (data not shown), a phenotype similar to that observed in semidominant Rolled leaf1 (Rld1) mutants of maize, where it appears to result from adaxialization and a partial reversal of leaf polarity (Nelson et al., 2002). Subsequent work revealed that Rld1 is the maize ortholog of REV and that mutant phenotypes result from a nucleotide change at the 5′ end of the miRNA recognition site (Juarez et al., 2004). In spite of the phenotypic similarity to Rld1 mutants, there was no clear evidence of polarity disruptions in leaves of 35S:NsPHAV transgenics, and the plants displayed no other obvious defects. One of the transgenic progenies segregated a seedling lethal phenotype resulting from loss of the SAM, which may result from cosuppression of PHB, PHV, and REV because triple mutants lacking these functions in Arabidopsis have this phenotype (Emery et al., 2003).
Insertion of the 35S:mutant phv1 transgene into wild-type explants produced hygromycin-resistant calli that remained highly disorganized and failed to produce shoots. Twelve confirmed transgenics and T2 progenies were eventually recovered, but none of these displayed expression of the transgene and had no phenotype. Given the high degree of disorganization in homozygous phv1 mutants and eventual loss of an organized SAM, the results suggest that active 35S:phv1 transgenes are lethal.
DISCUSSION
The existence of noncoding regulatory RNAs in higher plants emerged first from studies of posttranscriptional gene silencing, a sequence specific defense mechanism targeting viral RNAs for destruction (Hamilton and Baulcombe, 1999; Fagard and Vaucheret, 2000). Later it was recognized that miRNAs arising from endogenous genes act as signaling molecules (Llave et al., 2002; Rhoades et al., 2002), and the evidence indicates that they regulate some of the most fundamental aspects of plant morphogenesis (Bartel and Bartel, 2003; Palatnik et al., 2003). Our results show that miRNA regulation of NsPHAV sets the lateral course of growth in the vascular cambium. This promotes vascular connections in nodal regions of the stem and appears to have a direct influence on adaxial/abaxial polarity in leaves. The NsPHAV/miRNA interplay is critical as well in the overall structure and function of vegetative, inflorescence, and floral meristems. The results point to a fundamental role for the HD-ZIPIII pathway in cellular indeterminacy and implicate miRNA as a ubiquitous mechanism for spatial and temporal restriction of this function in higher plants.
Blade Formation and Vascular Development
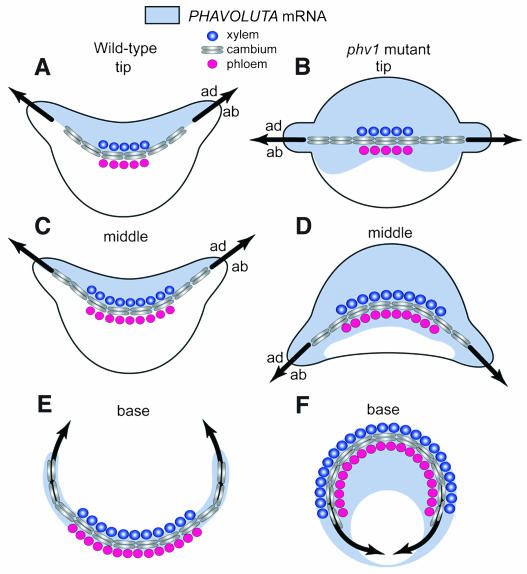
As seen in dominant HD-ZIPIII mutants of Arabidopsis, the phv1 mutation in Nicotiana expands the domain of NsPHAV expression in leaf primordia, enlarging the adaxial domain at the expense of the abaxial domain and shifting the position of the adaxial/abaxial boundary. This proceeds along a gradient of increasing severity from tip to base, shifting blade initiation progressively toward the back of the primordium and eventually precluding blade formation in basal regions, which appear fully adaxialized (Figures 2B, 5B, 5D, and 5F). Midvein alterations follow this same gradient in mutant leaves, first flattening into straight lines across the center of the primordium and then turning fully away from the shoot at the base of the petiole. One thought regarding these correlated changes in vascular development and blade formation was that each was responding directly to changes in leaf polarity. But vascular aberrations persist in the stem of phv1 mutants, well outside the context of developing leaves, a phenotype first observed in the dominant rev-∂miRNA mutants of Arabidopsis (Emery et al., 2003). This indicates that the vascular phenotype arises independently, an interpretation supported by vascular specific expression of NsPHAV in wild-type nodes (Figures 3C and 5E). Thus, it appears that cambial patterning is regulated directly by the NsPHAV/miRNA interplay and that ectopic accumulation of NsPHAV mRNA on the phloem side of vascular tissue is the primary cause of aberrant vascular curvature. Exactly how HD-ZIPIII proteins influence cambial patterning is unknown. The rev-∂miRNA mutation in Arabidopsis apparently causes overproduction of xylem by the fascicular cambium (Emery et al., 2003), whereas here in phv1 mutants, it appears that the lateral course of proliferation for interfascicular growth is the primary aberration.
Figure 5.
Vascular Development and Leaf Polarity.
NsPHAV mRNA accumulates initially throughout wild-type leaf primordia and then localizes to the adaxial domain along a crescent line corresponding to the developing midvein ([A] and [C]). This marks the adaxial/abaxial (ad/ab) boundary, and leaf blades are initiated where this traces to the surface of the primordium. At the base of wild-type petioles (E), NsPHAV expression localizes to the cambium of the midvein, a strip of meristematic tissue proliferating within veins (fascicular) and outward from midveins (interfascicular) along a curvilinear path, which connects to the vascular cylinder of the stem. In heterozygous phv1 mutants ([B], [D], and [F]), loss of miRNA regulation expands the domain of NsPHAV expression, leading to correlated changes in midvein development, leaf polarity, and blade formation, each following a gradient of increasing severity moving from tip to base. Mutant midveins are flattened to straight lines (B), misdirected into abaxial crescents (D), and often circularized at the base (F). Midveins no longer mark the NsPHAV on/off interface ([B] and [D]) but still define the adaxial/abaxial boundary guiding blade formation, suggesting that it is the mutation's effect on the midvein that dictates expansion of the adaxial domain. By positioning negative regulators of HD-ZIPIII expression, such as KANADI and miRNA, the midvein may provide a framework for the onset of adaxial/abaxial polarity in leaves.
An independent cambial function for HD-ZIPIII genes has important implications regarding the connection between vascular development and leaf polarity. If aberrant curvature of stem veins is in fact a cambial disruption, then this is very likely the explanation for the midvein phenotype as well. The idea is supported by the observation that abaxial misdirection of the midvein is most severe at the leaf base, where the cambium is most active (Figures 5B, 5D, and 5F). These changes in midvein architecture could have a significant impact on the overall pattern of HD-ZIPIII expression in leaf primordia and consequently on leaf polarity because negative regulators of HD-ZIPIII expression (KANADI1,2,3, Emery et al., 2003; miRNA166, Juarez et al., 2004) are specifically expressed on the phloem side of leaf vasculature. By positioning these negative regulators, midveins may actually establish the HD-ZIPIII on/off interface in the primordium, thus defining the adaxial/abaxial boundary for blade formation. This is supported by in situ data in wild-type primordia showing adaxial localization for NsPHAV mRNA along a line that coincides with the developing midvein (Figures 2H, 5A, and 5C). Because mutant phv1 mRNA evades miRNA cleavage, it extends well beyond the flattened midvein and into the abaxial domain (Figures 2I and 5B). It is of considerable interest, however, that blade formation remains associated with the displaced midvein, which apparently still defines the adaxial/abaxial boundary. This suggests that it is the mutation's effect on midvein development that actually dictates the extent of adaxial expansion and the altered position for blade initiation, rather than the location of NsPHAV mRNA itself. A possible explanation for this is that the flattened midvein now represents the new position in the primordium where PHB and REV encounter the antagonistic effects of KANADI and miRNA. Expansion of adaxial identity up to this new vascular boundary may result to some extent from concerted misexpression of HD-ZIPIII genes. In this perspective, vascular tissue would emerge as a primary determinant of adaxial/abaxial asymmetry in developing leaves.
Meristematic Indeterminacy
Investigations of STM, WUS, and CLV1,3 functions in Arabidopsis have revealed critical insights on the nature SAM identity. It appears that the WUS and STM functions represent the basal requirement for indeterminacy in the central zone of the SAM (Gallois et al., 2002). Spatial restriction of this program and formation of a peripheral organogenic zone is accomplished through a regulatory loop where WUS induces CLV3 (Fletcher et al., 1999), which in turn limits expression of WUS (Clark et al., 1997; Mayer et al., 1998). In the absence of CLV function, expansion of WUS expression produces an enlarged central zone of meristematic cells. Beginning in the adult phase of growth, we observed a very similar phenotype in the vegetative SAM of the phv1 mutant (Figure 4D), indicating that the NsPHAV/miRNA interaction also participates in the regulation of meristem structure. This is consistent with studies showing that REV is in the same pathway and upstream of the meristem identity genes (WUS, CLV, and STM) (Otsuga et al., 2001) and with the observation that dominant phb mutants partially suppress the stm phenotype in double mutants of Arabidopsis (McConnell and Barton, 1998). Aberrant expansion of the meristematic zone is observed once again during formation of the inflorescence meristem in phv1 mutants (Figure 4E), suggesting that the NsPHAV/miRNA interplay may play its most prominent role during developmental alterations of meristem structure. During this transition, the phv1 mutation allows meristematic activity to spread well beyond the apical dome and down the flanks of the shoot where floral meristems are initiated in a variety of ectopic locations. A similar pattern is observed in floral meristems, where a temporal extension of indeterminacy causes reiteration of carpel formation (Figures 4H and 4I). Carpels are the last organ initiated by the floral meristem, an event normally marking the end of indeterminacy in this structure. Failure to terminate this program in phv1 mutants is a phenotype directly analogous to that resulting from loss of DICER function in caf/dcl1 mutants of Arabidopsis (Jacobsen et al., 1999; Golden et al., 2002; Park et al., 2002), where unregulated proliferation in floral meristems likewise leads to formation of extra carpels. This suggests that the caf/dcl1 mutant phenotype results in part from ectopic HD-ZIPIII expression in the absence of miRNA processing. We conclude that miRNA regulation of the HD-ZIPIII pathway is a fundamental requirement for temporal and spatial regulation of indeterminacy, both in the vegetative and reproductive stages of plant development.
Indeterminacy: A Common Link for HD-ZIPIII Functions
Evidence that HD-ZIPIII genes regulate indeterminacy in meristems raises a central question as to whether they regulate vascular patterning and leaf adaxial identity by this same mechanism. This is certainly a plausible explanation for the vascular function. The vascular cambium in fact represents the only tissue outside the SAM whose function hinges directly on the long term maintenance of an indeterminate cell population. But how would this relate to leaf adaxial identity? One possibility is that adaxial identity begins with a transient projection of meristematic indeterminacy from the SAM to adjoining leaf primordia, mediated partially if not entirely by the HD-ZIPIII pathway. Prolonged adaxial indeterminacy is known to be a fundamental component in compound leaf development (Bharathan and Sinha, 2001), but even in simple leaves, the adaxial domain is initially characterized by meristematic cell divisions and a morphogenic potential to generate lateral outgrowths, features with clear parallels to the SAM (Hagemann and Gleissberg, 1996). The adaxial/abaxial boundary guiding lateral outgrowth of leaf blades could represent an interface where this meristem-like status is truncated by KANADI and miRNA. Support for this hypothesis has emerged from recent studies on ectopic blade formation in Arabidopsis and Nicotiana. Loss of NsPHAN function in Nicotiana and misexpression of the class I KNOX gene NTH20 (KNAT1/BP ortholog) appears to prolong indeterminacy in the adaxial leaf domain, delaying differentiation of palisade and allowing de novo reinitiation of leaf blades along the flanks of the midrib (McHale and Koning, 2004). A very similar phenotype is reported in the blade-on-petiole mutants of Arabidopsis, where ectopic expression of class I KNOX genes also leads to reiteration of blade formation on leaf petioles (Ha et al., 2003). These negative regulators of KNOX expression appear to be essential components in the onset of determinacy in the emerging lamina, marking the termination of its morphogenic phase and the beginning of lamina differentiation.
Perhaps the most striking implication here, however, is that formation of lateral organs at the peripheral boundary of the SAM and initiation of lamina at the adaxial/abaxial boundary in leaf primordia could actually be variations on a common theme. The meristem and leaf phenotypes in phv1 mutants in fact suggest that organ initiation and blade formation are directly related to HD-ZIPIII mediated indeterminacy and that each is taking place at a position where miRNA truncates this program. This would suggest that the SAM does not simply send positional signals to leaf primordia but rather projects an element of its own character, which represents the foundation for leaf adaxial identity and blade formation. Ongoing efforts to define the molecular dynamics of the leaf/meristem interface will undoubtedly provide a wealth of new insights on the mechanisms of plant morphogenesis.
METHODS
Mutagenesis
Seeds of Nicotiana sylvestris were soaked overnight in water at room temperature, exposed to 0.4% ethyl methanesulfonate for 2 h, washed, germinated, and grown to maturity for collection of seeds after self-pollination.
Histology
Plants for histological studies and in situ hybridizations were grown at 27°C under continuous illumination in Metro-Mix 360 (Scotts-Sierra, Marysville, OH) in a Percival growth chamber (Percival Scientific, Perry, IA). Samples were fixed in FAA (50% EtOH, 5% acetic acid, and 3.7% formaldehyde), embedded in Paraplast X-TRA (Oxford Labware, St. Louis, MO), and sectioned at a thickness of 8 μ. For light microscopy, wax sections were stained in 0.1% toluidine blue and photographed with a Zeiss Axiophot microscope (Carl Zeiss, Jena, Germany). Samples for scanning electron microscopy were fixed in FAA, transferred to 100% ethanol, and critical point dried with liquid carbon dioxide in a Polaron pressure chamber (Polaron, Hertfordshire, UK), sputter coated with gold, and photographed with an ISI-SS40 scanning electron microscope (International Scientific Instruments, Prahran, Australia). Samples for in situ hybridization were fixed in FAA, sectioned at 8 μ, affixed to Probe Plus slides (Fisher Scientific, Loughborough, UK), and processed as described in http://www.wisc.edu/genetics/CATG/barton/protocols.html with color substrate incubation overnight. Digoxygenin-labeled probes were synthesized from SP6 or T7 promoters in linearized pCRII TOPO vectors (Invitrogen, Carlsbad, CA) according to the DIG RNA labeling protocol (Roche Molecular Biochemicals, Indianapolis, IN). The NsPHAV probe represented the C-terminal coding region beginning at exon 16 and the 3′ untranslated region.
RNA Gel Blots
Poly(A) RNA was isolated from leaves and stems with the Poly(A) Pure kit (Ambion, Austin, TX), and samples (2 μg) were fractionated in formaldehyde gels, transferred to nylon membranes as described (McHale et al., 1990), and hybridized with 32P-UTP labeled RNA probes synthesized from pCRII TOPO vectors according to the Roche RNA labeling protocol. The NsPHAV probe represented the C-terminal coding region beginning at exon 16 and including the 3′ untranslated region.
5′ RACE Analysis
Poly(A) RNA isolated from leaves and stems was subjected to a modified RACE analysis as described by Kasschau et al. (2003) using the First Choice RLM RACE kit (Ambion). A synthetic RNA adapter was ligated to partial transcripts, reverse transcribed, and amplified by nested PCR with forward adapter primers and reverse NsPHAV primers from exon 6. PCR products were captured in the pCRII TOPO vector and sequenced.
Genomic NsMIR166a Isolation and RT-PCR Analysis
Genomic DNA from N. sylvestris was digested, ligated to a synthetic adapter, and amplified by PCR with an adapter primer in combination with the maize 5 box primer 5F (Juarez et al., 2004) according to manufacturer's recommendations (GenomeWalker kit; BD Biosciences, Palo Alto, CA). Sequence analysis revealed homology to the 5, 6, and 7 boxes of ZmMIR166a and intervening regions of similar length but with no sequence homology. RT-PCR analysis was performed with forward 5′-ATGTTGTCTGGCTCGAGGTC-3′ and reverse 5′-CACCAATCTGCTCTTGTCCA-3′ primers specific to nonconserved regions internal to the 5 and 7 boxes, respectively.
NsPHAV Transgenics
Transgenes representing the full-length coding region of the wild-type and mutant phv1 alleles of NsPHAV were constructed in the binary vector pCAMBIA 1300 (Canberra ACT 2601, Australia) and introduced into wild-type N. sylvestris plants as previously described (McHale and Koning, 2004).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under the following accession numbers: Arabidopsis PHB (Y11122), PHV (Y10922), REV (AF188994), NsPHAV (AY560320), ZmMIR166a (AY501431), ZmMIR166b (AY501432), and NsMIR166a (AY599420).
Acknowledgments
We thank James C. Carrington, Marja C.P. Timmermans, and Neil P. Schultes for technical advice and Regan Huntley for excellent technical assistance. This research was supported by the USDA National Research Initiative Grants 1999-01899 (N.A.M.) and 98-35311-6922 (R.E.K.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Neil A. McHale (neil.mchale@po.state.ct.us).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.021816.
References
- Bartel, B., and Bartel, D. (2003). MicroRNAs: At the root of plant development? Plant Physiol. 132, 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharathan, G., and Sinha, N. (2001). The regulation of compound leaf development. Plant Physiol. 127, 1533–1538. [PMC free article] [PubMed] [Google Scholar]
- Chuck, G., Lincoln, C., and Hake, S. (1996). KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8, 1277–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, S.E., Williams, R.W., and Meyerowitz, E.M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575–585. [DOI] [PubMed] [Google Scholar]
- Emery, J.F., Floyd, S.K., Alvarez, J., Eshed, Y., Hawker, N.P., Izhaki, A., Baum, S.F., and Bowman, J.L. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13, 1768–1774. [DOI] [PubMed] [Google Scholar]
- Esau, K. (1977). Anatomy of Seed Plants. (New York: John Wiley and Sons).
- Fagard, M., and Vaucheret, H. (2000). Systemic silencing signals. Plant Mol. Biol. 43, 285–293. [DOI] [PubMed] [Google Scholar]
- Fletcher, J., Brand, U., Running, M., Simon, R., and Meyerowitz, E. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283, 1911–1914. [DOI] [PubMed] [Google Scholar]
- Gallois, J.L., Woodward, C., Reddy, V.G., and Sablowski, R. (2002). Combined SHOOTMERISTEM-LESS and WUSCHEL trigger ectopic organogenesis in Arabidopsis. Development 129, 3207–3217. [DOI] [PubMed] [Google Scholar]
- Golden, T.A., Schauer, S.E., Land, J.D., Pien, S., Mushegian, A.R., Grossniklaus, U., Meinke, D.W., and Ray, A. (2002). SHORT INTEGUMENTS1/SUSPENSOR1/CARPEL FACTORY, a Dicer homolog, is a maternal effect gene required for embryo development in Arabidopsis. Plant Physiol. 130, 808–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourlay, C.W., Hofer, J.M.I., and Ellis, T.H.N. (2000). Pea compound leaf architecture is regulated by interactions among the genes UNIFOLIATA, COCHLEATA, AFILA, and TENDRIL-LESS. Plant Cell 12, 1279–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha, C.M., Kim, G.T., Kim, B.C., Jun, J.H., Soh, M.S., Ueno, Y., Machida, Y., Tsukaya, H., and Nam, H.G. (2003). The BLADE-ON-PETIOLE1 gene controls leaf pattern formation through the modulation of meristematic activity in Arabidopsis. Development 130, 161–172. [DOI] [PubMed] [Google Scholar]
- Hagemann, W., and Gleissberg, S. (1996). Organogenetic capacity of leaves: The significance of marginal blastozones in angiosperms. Plant Syst. Evol. 199, 121–152. [Google Scholar]
- Hamilton, A.J., and Baulcombe, D.C. (1999). A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286, 950–952. [DOI] [PubMed] [Google Scholar]
- Hareven, K., Gutfinger, T., Parnis, A., Eshed, Y., and Lifschitz, E. (1996). The making of a compound leaf: Genetic manipulation of leaf architecture in tomato. Cell 84, 735–744. [DOI] [PubMed] [Google Scholar]
- Jackson, D., Veit, B., and Hake, S. (1994). Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120, 405–413. [Google Scholar]
- Jacobsen, S.E., Running, M.P., and Meyerowitz, E.M. (1999). Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 126, 5231–5243. [DOI] [PubMed] [Google Scholar]
- Janssen, B.J., Lund, L., and Sinha, N. (1998). Overexpression of a homeobox gene, LeT6, reveals indeterminate features in the tomato compound leaf. Plant Physiol. 117, 771–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez, M.T., Kui, J.S., Thomas, J., Heller, G.A., and Timmermans, M.C.P. (2004). MicroRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428, 84–88. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D., Xie, Z., Allen, E., Llave, C., Chapman, E.J., Krizan, K.A., and Carrington, J.C. (2003). P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 4, 205–217. [DOI] [PubMed] [Google Scholar]
- Kidner, C.A., and Martienssen, R.A. (2004). Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428, 81–84. [DOI] [PubMed] [Google Scholar]
- Laux, T., Mayer, K.F., Berger, J., and Jürgens, G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122, 87–96. [DOI] [PubMed] [Google Scholar]
- Llave, C., Kasschau, K.D., Rector, M.A., and Carrington, J.C. (2002). Endogenous and silencing-associated small RNAs in plants. Plant Cell 14, 1605–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, K., Schoof, H., Haecker, A., Lenhard, M., Jurgens, G., and Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., and Barton, M.K. (1998). Leaf polarity and meristem formation in Arabidopsis. Development 125, 2935–2942. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713. [DOI] [PubMed] [Google Scholar]
- McHale, N.A., Kawata, E.E., and Cheung, A.Y. (1990). Plastid disruption in a thiamine-requiring mutant of Nicotiana sylvestris blocks accumulation of specific nuclear and plastid mRNAs. Mol. Gen. Genet. 221, 203–209. [Google Scholar]
- McHale, N., and Koning, R. (2004). PHANTASTICA regulates development of the adaxial mesophyll in Nicotiana leaves. Plant Cell 16, 1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, J.M., Lane, B., and Freeling, M. (2002). Expression of a mutant maize gene in the ventral leaf epidermis is sufficient to signal a switch of the leaf's dorsoventral axis. Development 129, 4581–4589. [DOI] [PubMed] [Google Scholar]
- Otsuga, D., DeGuzman, B., Prigge, M.J., Drews, G.N., and Clark, S.E. (2001). REVOLUTA regulates meristem initiation at lateral positions. Plant J. 25, 223–236. [DOI] [PubMed] [Google Scholar]
- Palatnik, J.F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carringtion, J.C., and Weigel, D. (2003). Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. [DOI] [PubMed] [Google Scholar]
- Park, W., Li, J., Song, R., Messing, J., and Chen, X. (2002). CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12, 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe, O.J., Riechmann, J.L., and Zhang, J.Z. (2000). INTERFASCICULAR FIBERLESS1 is the same gene as REVOLUTA. Plant Cell 12, 315–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M.W., Reinhart, B.J., Lim, L.P., Burge, C.B., Bartel, B., and Bartel, D.P. (2002). Prediction of plant microRNA targets. Cell 110, 513–520. [DOI] [PubMed] [Google Scholar]
- Reinhart, B.J., Weinstein, E.G., Rhoades, M.W., Bartel, B., and Bartel, D.P. (2002). MicroRNAs in plants. Genes Dev. 16, 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha, N., Williams, R.E., and Hake, S. (1993). Overexpression of the maize homeobox gene KNOTTED1 causes a switch from determinate to indeterminate cell fates. Genes Dev. 7, 787–795. [DOI] [PubMed] [Google Scholar]
- Steeves, T.A. (1961). A study of developmental potentialities of excised leaf primordia in sterile culture. Phytomorphology 1, 346–359. [Google Scholar]
- Sussex, I.M. (1955). Morphogenesis in Solanum tuberosum L.: Apical structure and developmental pattern of the juvenile shoot. Phytomorphology 5, 253–273. [Google Scholar]
- Talbert, P.B., Adler, H.T., Parks, D.W., and Comai, L. (1995). The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development 121, 2723–2735. [DOI] [PubMed] [Google Scholar]
- Tang, G., Reinhart, B.J., Bartel, D.P., and Zamore, P.D. (2003). A biochemical framework for RNA silencing in plants. Genes Dev. 17, 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardlaw, C.W. (1949). Further experimental observations on the shoot apex of Dryopteris aristata Druce. Philos. Trans. R. Soc. Lond. Ser. B 233, 415–451. [Google Scholar]
- Zhong, R., and Ye, Z.H. (1999). IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeodomain-leucine zipper protein. Plant Cell 11, 2139–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., and Ye, Z.H. (2001). Alteration of auxin polar transport in the Arabidopsis ifl1 mutants. Plant Physiol. 126, 549–563. [DOI] [PMC free article] [PubMed] [Google Scholar]