Abstract
Increased demand for BRCA testing is placing pressures on diagnostic laboratories to raise their mutation screening capacity and handle the challenges associated with classifying BRCA sequence variants for clinical significance, for example interpretation of pathogenic mutations or variants of unknown significance, accurate determination of large genomic rearrangements and detection of somatic mutations in DNA extracted from formalin-fixed, paraffin-embedded tumour samples. Many diagnostic laboratories are adopting next-generation sequencing (NGS) technology to increase their screening capacity and reduce processing time and unit costs. However, migration to NGS introduces complexities arising from choice of components of the BRCA testing workflow, such as NGS platform, enrichment method and bioinformatics analysis process. An efficient, cost-effective accurate mutation detection strategy and a standardised, systematic approach to the reporting of BRCA test results is imperative for diagnostic laboratories. This review covers the challenges of BRCA testing from the perspective of a diagnostics laboratory.
Introduction
The demand for BRCA testing has steadily increased since the discovery that women carrying pathogenic BRCA mutations have elevated lifetime ovarian and breast cancer risks.1, 2, 3 In a meta-analysis of pathogenic BRCA mutation penetrance, carriers of BRCA1 and BRCA2 pathogenic mutations were shown to have a cumulative risk of 57 and 49%, respectively, for developing breast cancer and 40 and 18%, respectively, for developing ovarian cancer by 70 years of age.1 In support of this observation, results from a prospective epidemiological study (EMBRACE) showed carriers of BRCA1 and BRCA2 pathogenic mutations have a cumulative risk of 60 and 55%, respectively, for developing breast cancer and 59 and 17%, respectively, for developing ovarian cancer by 70 years of age.2
A contributory factor to the demand for BRCA testing has been heightened public awareness of the consequences, costs and prophylactic options surrounding BRCA testing, an issue highlighted by celebrity publicity.3, 4 For women carrying pathogenic BRCA mutations, routine surveillance for breast cancer is recommended from 25 years of age and prophylactic salpingo-oophorectomy is recommended after 35 years or once childbearing is complete.5, 6 Prophylactic oophorectomies and mastectomies have been shown to reduce cancer incidence compared with chemoprevention or surveillance.7
The increasing demand for BRCA testing is placing a strain on diagnostic laboratories, particularly in those offering rapid genetic testing at the point of diagnosis. For instance, the UK's National Institute for Health and Care Excellence recommends fast-track genetic testing as part of a clinical trial within 4 weeks of a diagnosis of breast cancer.8 Against this backdrop of rising demand, more diagnostic laboratories are adopting next-generation sequencing (NGS) technology for BRCA testing, which offers the potential of fast, scalable, cost-efficient and comprehensive sequencing. In the 2014 BRCA scheme report from the European Molecular Genetics Quality Network (EMQN), 19% of laboratories were using NGS for BRCA testing, an increase from 6% of laboratories from the previous year's scheme (Dr S Patton, EMQN Director, personal communication). The same EQA scheme reports also indicated a reduction in the use of Sanger sequencing alone for BRCA testing: from 83% down to 75% of laboratories.
Adopting NGS in the diagnostic laboratory setting is not straightforward, as the technology is not simple or homogeneous and many potential configurations are possible. Transitioning to NGS also imposes a significant validation overhead for clinical laboratories, as they are compelled to demonstrate that a new assay is sensitive, specific and fit for purpose prior to adoption. This review covers key considerations with respect to NGS and the specific challenges relating to BRCA testing, such as difficulties in interpreting complex BRCA-sequencing data and the issues of testing tumour samples.
BRCA testing: an overview
Genetic testing is undertaken in many countries to detect BRCA1 and BRCA2 sequence variants.6 The selection of candidates appropriate for testing is typically based on national guidelines or by larger international societies.5, 8 A blood sample is typically used for these tests; however, other sample types can be used, for example, buccal scrape.5, 6 Written informed consent should be obtained from all patients prior to storage or analysis of their sample, and genetic counselling is standard practice both prior to the decision to test and at the time results are given to the patient.
Sequence variants in BRCA1 and BRCA2 can be subdivided into three broad classes: single-nucleotide changes, small insertion or deletion events (indels) and large genomic rearrangements (LGRs). Pathogenic BRCA single-nucleotide mutations and small indels are found widely distributed throughout the coding sequence and conserved intronic sequences of both genes. Typically, a very broad spread of pathogenic mutations is present in populations; however, founder pathogenic mutations are present at high frequency in some populations. For example, in the Ashkenazi Jewish population three founder pathogenic mutations (BRCA1 NM_007294.3: c.68_69delAG p.(Glu23Valfs*17), BRCA1 NM_007294.3: c.5266dupC p.(Gln1756Profs*74) and BRCA2 NM_000059.3: c.5946delT p.(Ser1982Argfs*22)) account for the overwhelming majority of clinically relevant pathogenic mutations and are observed at relatively high frequency (~2% in total).9, 10 In addition, in Polish breast and breast-ovarian cancer families, three pathogenic mutations in BRCA1 (NM_007294.3: c.5266dupC, NM_007294.3: c.181T>G p.(Cys61Gly) and NM_007294.3: c.4034delA) were found to account for the majority of pathogenic BRCA mutations.11 More recently, three further pathogenic founder BRCA1 mutations have been observed in a study of 1164 Polish women with unselected breast cancer (NM_007294.3: c.3700_3704del p.(Val1234Glnfs*8), NM_007294.3: c.68_69delAG p.(Glu23Valfs*17) and NM_007294.3: c.5251C>T p.(Arg1751*)).12
BRCA1 contains a number of Alu sequences,13 which are known to mediate the occurrence of LGRs. In BRCA1, LGRs have been detected with varying frequencies among patients with breast or ovarian cancer.14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24 In a study of 805 Dutch families with a known predisposition for breast and/or ovarian cancer, those without identified pathogenic BRCA1 or BRCA2 mutations by conventional mutation screening methods (661) were assessed for BRCA1 germline LGRs.23 A total of 33 families with a deletion or duplication event in BRCA1 were identified, representing 27% of the total 121 pathogenic BRCA1 mutations. In a separate study of unrelated individuals (n=3580) with a family history of breast and ovarian cancer or those with early onset disease (n=934), a 6 kb BRCA1 exon 13 duplication event was identified in 11 families with ancestry links to Northern Britain.22 BRCA2 contains fewer Alu sequences,25 which may explain why fewer rearrangements have been reported for this gene. Examples of BRCA2 LGRs include deletion of exon 2 and an Alu insertion in exon 3, which occurs with relatively high frequency in those patients from North and Central Portugal.15, 26, 27
The interpretation of the results of BRCA1 and BRCA2 screening is made complex by the significant numbers of patients with variants of unknown clinical significance (VUS). VUS are alterations in the DNA sequence of a gene that have an unknown effect on the function of the gene product or on the risk of disease.28 They can include variants in promoter regions, intronic nucleotide changes close to the exon boundary, small in-frame insertions/deletions and missense/synonymous substitutions where there is no firm evidence for a deleterious effect on RNA processing, or protein structure and function. In an analysis of BRCA1 and BRCA2 sequences from 10 000 individuals, 13% were observed to harbour a VUS.29 However, the frequency of VUS should be lower in well-characterised populations, as databases of pathogenic BRCA mutations are updated, allowing reclassification of previous VUS. The finding of a potential VUS during screening requires detailed expert interpretation and as many sources of evidence about a variant should be collated and assessed as possible before coming to a conclusion that is clinically reported.
BRCA testing is commonly performed by direct (Sanger) DNA sequencing. This method is considered the ‘gold standard' of DNA sequencing; technologically reliable, widely available and a relatively simple workflow. The drawbacks of Sanger sequencing are limited throughput and lower cost-effectiveness compared with NGS. In addition, Sanger sequencing cannot detect LGRs, which require alternative polymerase chain reaction (PCR)-based techniques for analysis. Quantitative PCR is a viable technique for detecting LGRs;24 however, this is labour intensive for analysis of all BRCA1 and BRCA2 exons. Instead, the most commonly used method for analysing LGRs is multiplex ligation-dependent probe amplification (MLPA), a technique in which pairs of oligonucleotide probes able to ligate to each other, bind to adjacent positions at the genomic area of interest and amplify only if both probes are bound and ligated in a semi-quantitative manner. Examples of commercially available, research-use-only (RUO) BRCA testing MLPA probes are P002 for BRCA1 and P045 for BRCA2 (MRC-Holland, Amsterdam, The Netherlands). Alternatively, some laboratories have developed custom probes analogous to the MLPA technique,30 or customised PCR assays to detect specific LGRs.27
BRCA testing with NGS technology offers many advantages over Sanger sequencing, including the potential to detect LGRs in a single workflow, although NGS LGR detection has not been fully established in the diagnostic setting. Some of the key advantages and disadvantages associated with NGS are summarised in Table 1.
Table 1. Advantages and disadvantages of next-generation sequencing 31, 32, 33, 34, 35 .
| Advantages | Disadvantages |
|---|---|
| High throughput Able to multiplex Lower cost Automated analysis Uses less DNA Can run in parallel with other genetic tests | Higher start-up cost Mutation-positive test may require Sanger sequencing for confirmation Complex workflow Dedicated data storage and analysis required Reduced sensitivity for large insertions/deletions >20 base pairs |
The following section describes the key considerations with respect to adopting NGS in the diagnostic laboratory.
Next-generation sequencing: choice and complexity
The rapid evolution of massively parallel sequencing technology and NGS platforms is revolutionising the management of inherited diseases, where traditionally molecular diagnostics have been underused due to the issues of cost, time, labour and availability of services. A number of early clinical studies in BRCA testing have shown that NGS offers high sensitivity, specificity and cost-effectiveness compared with current approaches.35, 36, 37, 38, 39, 40, 41
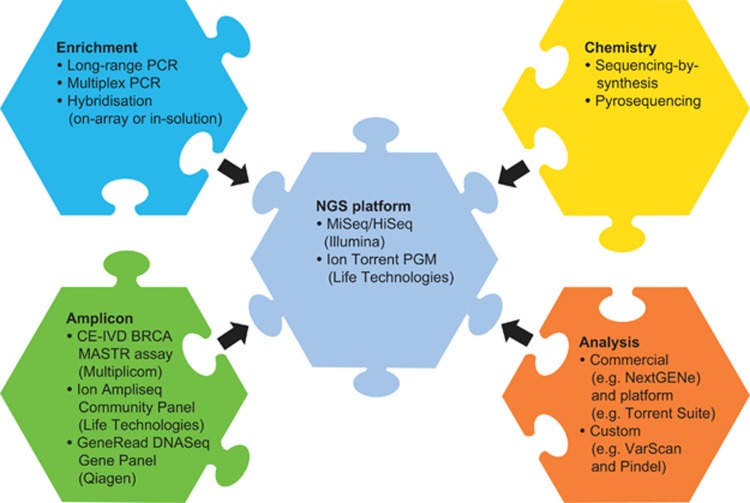
Recently, NGS benchtop platforms have made available gigabase-scale DNA sequencing with relatively short run times (<24 h), for example, MiSeq (Illumina, San Diego, CA, USA) and the Ion Torrent Personal Genome Machine (PGM; Life Technologies, Carlsbad, CA, USA) (refer to Table 2 for examples of NGS platforms). The potential of high-volume analytical throughput makes NGS platforms an increasingly attractive investment for diagnostic laboratories in the clinical setting. However, the choice of NGS platform is only one factor to consider in the total NGS workflow, which is also dependent on other components including enrichment methods, sequencing chemistries and analytical procedures (Figure 1).
Table 2. Examples of next-generation sequencing platforms as of May 2014.
| Platform | Read length (bp) and mode | Run timea (hours) | Number of reads (single or paired; millions) | Data yield (Gb per run) |
|---|---|---|---|---|
| Illumina MiSeq v2 chemistryb | 2 × 150 paired | ~24 | 24–30 (paired) | 4.5–5.1 |
| Illumina MiSeq v3 chemistryb | 2 × 75 paired | ~21 | 44–50 (paired) | 3.3–3.8 |
| Illumina MiSeq v3 chemistryb | 2 × 300 paired | ~56 | 44–50 (paired) | 13.2–15.0 |
| Ion Torrent PGM, 316 Chip v2c | Up to 200 single | 3 | 2–3 (single) | 0.3–0.5 |
| Ion Torrent PGM, 318 Chip v2c | Up to 200 single | 4.4 | 4.0–5.5 (single) | 0.6–1.0 |
| Ion Torrent PGM, 318 Chip v2c | Up to 400 single | 7.3 | 4.0–5.5 (single) | 1.2–2.0 |
| Ion Torrent Proton with PI Chipc | Up to 200 single | 2–4 | 60–80 (single) | Up to 10 |
| Illumina NextSeq 500 (mid output)b | 2 × 150 paired | 26 | Up to 260 (paired) | 32.5–39.0 |
| Illumina HiSeq 2500 (rapid run single flow cell)b | 2 × 150 paired | 40d | Up to 300 (paired) | 75–90 |
Upstream preparatory work not taken into consideration.
Run time based on dual flow cell.
Figure 1.
Components/complexities to consider in the NGS workflow, including the NGS platform, enrichment methods, sequencing chemistries and analytical procedures.
The following section describes some of the key considerations with respect to NGS platforms, chemistries and analysis of data.
Platforms
The MiSeq (Illumina) and Ion Torrent PGM (Life Technologies) are examples of benchtop NGS platforms. Both operate on the principle of sequencing-by-synthesis, in which the addition of nucleotide triphosphates to primed clonal DNA templates are measured. In the case of the MiSeq, these are fluorescently labelled reversible deoxyribonucleoside triphosphates (dNTPs) and sequences are optically read by fluorescence imaging.42 Conversely, the Ion Torrent reads sequences non-optically through the use of semiconductor sequencing technology (Thermo Fisher, Waltham, MA, USA), where pH changes resulting from the addition of dNTPs to the nascent strand are recorded as voltage changes.43 The rapid pace of NGS platform development renders any detailed review out-of-date by the time of publication. Given this, up-to-date online reviews such as the NGS Field Guide from The Molecular Ecologist are important references when choosing an NGS platform.44
Enrichment methods
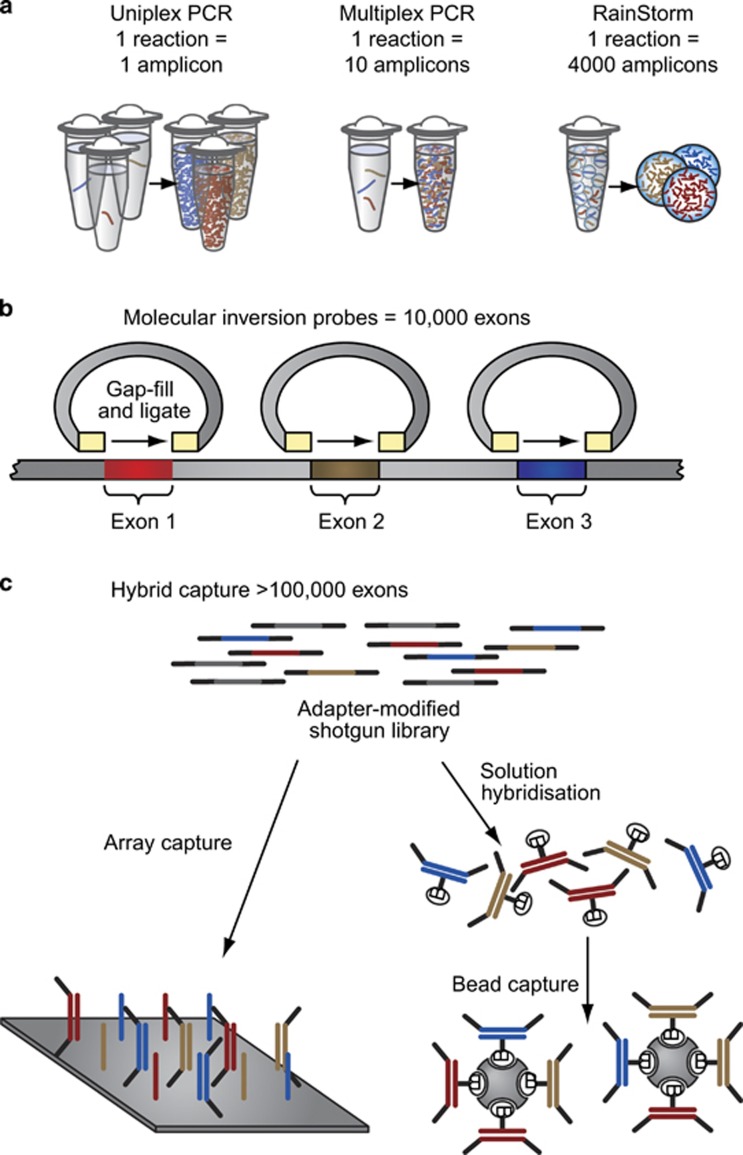
In the clinical setting, NGS is typically used for sequencing specific genes, such as BRCA1 and BRCA2, or panels of genes rather than for sequencing entire genomes, as it is far more cost-effective and time-efficient to target, capture and sequence only the genomic regions of interest. This has led to the development of numerous enrichment methods, most commonly based on PCR or hybridisation approaches (Figure 2).45 It is possible that future decreases in the overall cost for gene sequencing may result in exome sequencing, with a virtual panel being a cost-effective mode of delivery.46
Figure 2.
Enrichment methods. (a) PCR-based approach. Multiplex PCR kits enrich for a specific gene or panel of genes in a small number of PCR amplifications. (b) Molecular inversion probes consist of amplicons containing a universal spacer region flanked by target-specific sequences. Genomic DNA is digested, and the target DNA is PCR-amplified and sequenced. (c) Hybridisation enrichment methods work on the principle of selection using probes complementary to DNA in the genomic area of interest either by surface microarray or in solution with labelled beads.45 Reprinted with permission from Mamanova et al.45
PCR is a well-established pre-sequencing enrichment technique, particularly for use with Sanger sequencing. In the case of NGS, laboratory-developed long-range PCR methods have been used successfully with BRCA testing;47 however, to make full use of high-throughput NGS, a large number of amplicons must be prepared separately and then combined and sequenced together. This has led to the development of commercially available multiplex PCR kits that enrich for a specific gene or panel of genes in a small number of PCR amplifications (Figure 2a). For instance, the CE-IVD BRCA MASTR assay (Multiplicom, Niel, Belgium) amplifies the coding regions of BRCA1 and BRCA2 in 93 amplicons in five multiplex PCR reactions. Other RUO multiplex kits are available for BRCA1 and BRCA2, such as the Ion Ampliseq Community Panel (Thermo Fisher) and GeneRead DNASeq Gene Panel (Qiagen, Hilden, Germany).
Molecular inversion probe (MIP)-based enrichment approaches can provide greater specificity over standard PCR-based approaches.45 MIPs consist of a universal spacer region flanked by sequences specific to either side of the target region (Figure 2b). Once the MIP anneals to the target, the gap between the sequences is filled by a DNA polymerase and ligase. Genomic DNA is digested, and the target DNA is PCR-amplified and sequenced. Although PCR as an NGS enrichment method is highly sensitive, specific and reproducible,45 it is inefficient for NGS of larger genomic targets where other approaches to target enrichment should be considered.
Hybridisation enrichment methods work on the principle of selection using probes complementary to DNA in the genomic area of interest (Figure 2c). On-array capture uses high-density microarrays containing complementary probes, whereas in-solution capture uses complementary probes that are then purified using labelled beads (Figure 2). The in-solution approach has the advantage of being highly scalable and does not require additional equipment associated with processing microarrays. Hybridisation kits are available that target BRCA1 and BRCA2 specifically, such as RUO, in-solution capture kits, HaloPlex and SureSelect (Agilent, Santa Clara, CA, USA). In addition, hybridisation kits are available that include BRCA1 and BRCA2 as part of a larger panel of genes associated with cancer, such as RUO TruSight Cancer Sequencing Panel (Illumina), which targets 94 genes, or NimbleGen Comprehensive Cancer Design (Roche NimbleGen, Madison, WI, USA), which targets 578 genes. It should be noted that hybridisation methods, particularly on-array capture methods, can add additional cost, time and, if the panel of genes is extensive, incidental findings to the overall NGS process.
Bioinformatics analysis and laboratory information management systems
Even benchtop NGS platforms produce large amounts of sequence data that require bioinformatics analysis to align, variant call and filter the data to make them accessible, coherent and comprehensible. Many options are available for a diagnostic laboratory for analysing NGS data; however, choosing appropriate software and thorough validation is crucial in order to obtain accurate and reliable results suitable for clinical application. Commercially available software include NextGENe, CLC Workbench (Qiagen) and those linked to specific platforms, such as the Torrent Suite Software Plugins (Life Technologies). Publicly available and open-source bioinformatics software can also be used to build a custom bioinformatics pipeline. Examples include Pindel, VarScan, GATK-lite and Samtools. It should be noted that input from bioinformaticians into an NGS-based diagnosis is an important consideration for reaching a quality level required for medical analysis.
Tracking and managing clinical samples through a high-throughput NGS workflow requires a laboratory information management system (LIMS) that can support NGS, is configurable and customisable to suit diagnostic laboratories' needs and yet flexible to accommodate changes in testing practice. Numerous commercial systems are available, some of which are tailored to managing NGS workflows in clinical laboratories, such as Exemplar LIMS (Sapio Sciences, Baltimore, MD, USA), Clarity LIMS (GenoLogics, Victoria, BC, Canada) and Sequencing LIMS (Edinburgh Genomics, Edinburgh, UK). Commercial systems tend to involve high setup costs and can require extensive configuration and customisation to address specific laboratory needs. Open-source solutions are also available, such as Galaxy LIMS (Tron, Mainz, Germany).
NGS for detecting large genomic rearrangements
An NGS-based strategy offers the potential to screen for point mutations and LGRs on a single platform and workflow. NGS has been applied to detection of LGRs in DNA from cancers using depth of coverage and a paired-end mapping whole-genome sequencing approach.48, 49
The ability to detect LGRs in NGS data can be limited by the use of an enrichment method. PCR enrichment methods are currently unsuitable for reliable measurement of copy-number variants (CNVs). Hybridisation NGS enrichment methods do offer the potential to detect CNVs affecting targeted regions. However, this requires specialist bioinformatics, and particular problems are encountered in detection of smaller CNVs; <200 bp where sensitivity is low.50, 51 Extensive site-specific validation in the clinical setting is required before NGS can be routinely used for comprehensive CNV detection in a clinical setting for BRCA analysis.
BRCA Testing from tumour samples
Significant frequencies of somatic BRCA1 and BRCA2 pathogenic mutations have been observed in patients with ovarian cancer.52 There is growing evidence that tumours with somatically acquired BRCA1 or BRCA2 pathogenic mutations will respond to drugs that inhibit poly (ADP-ribose) polymerase (PARP).53, 54, 55, 56 Tumour samples, as part of standard pathology practice, are routinely processed and stored as formalin-fixed paraffin-embedded (FFPE) blocks; this presents a number of challenges to the diagnostic laboratory. FFPE samples are typically a variable mix of neoplastic and normal cell tissue (stroma) and the DNA extracted is often limited in quantity, fragmented and of poor quality. In addition, DNA extracted from FFPE samples may contain artefactual sequence alterations arising from formalin crosslinking and deamination of cytosine nucleotides. These problems can be mitigated by the use of shorter amplicons, de-crosslinking steps and treatment with uracil-DNA glycosylase, a DNA repair enzyme, which has been shown to markedly reduce the number of sequence artefacts in damaged FFPE DNA when used prior to PCR amplification.57, 58
The challenges in sequencing FFPE tumour samples are observed in NGS analysis. In a large-scale, prospective, cohort study of the incidence of cancer in a population in Victoria, Australia, an initial pilot phase for the first 488 patients established the feasibility of NGS for profiling mutations in tumours.59 Disproportionate levels of C>T/G>A changes were displayed in the 1–10% allele frequency range, whereas artefacts were less apparent in the 10–25% allele frequency range. Importantly, an example of the dangers of mutational artefacts was shown in one sample, where an activating NRAS p.Gly12Asp mutation was discovered on first screening but not confirmed in the same DNA specimen by subsequent sequencing of uracil-DNA glycosylase-treated FFPE DNA or repeat NGS. This highlights the utility of replicate analysis to confirm the identification of mutations.
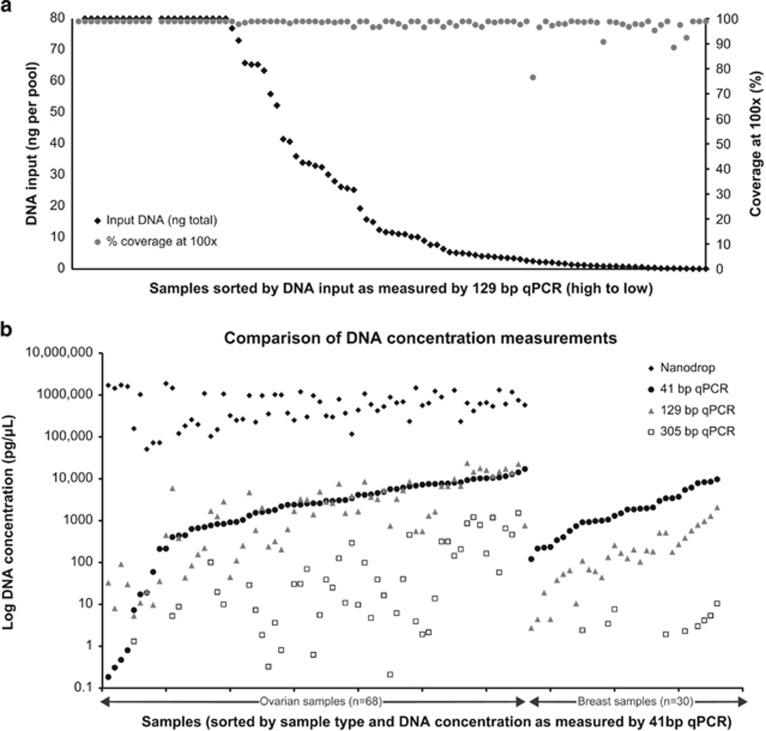
Despite the limitations imposed by DNA extracted from FFPE samples, successful sequencing with NGS has been shown.57, 60, 61 In an NGS study of ovarian (n=68) and breast (n=30) FFPE samples, DNA was amplified using a GeneRead DNAseq Targeted Exon Enrichment Breast Panel (Qiagen) and sequenced using a 2 × 150 bp analysis on a MiSeq platform (Illumina).62 The majority of samples with low DNA yields produced adequate PCR products and sequencing data without any significant deterioration in coverage or read depth until <1 ng of amplifiable DNA was added per primer pool (Figure 3). Among 75 samples with less than optimal DNA input, 32 samples still generated the maximum possible coverage of ~97%, and a further 20 samples generated a coverage of >95% at a minimum read depth of 100 × .62 As the input quantity of DNA diminished, there were still many samples with >90% coverage depth at 100 × , although the frequency of samples with low percentage coverage at 100 × increased. Significant variants were confirmed by Sanger sequencing. A small number of variants were identified as potential artefacts common to FFPE-extracted DNA; however, the majority of these were in the poor-quality low-input DNA samples with lower coverage depth and artefacts could be identified as such by replicate analysis. The conclusion of this study is that routine analysis of BRCA1 or BRCA2 sequences from FFPE breast and ovarian tumours is feasible. However, it is important that tumour BRCA screening should not be substituted for germline BRCA screening in patient groups at high risk of carrying an inherited pathogenic mutation unless proven to be at least as sensitive as germline screening at detecting the full range of inherited pathogenic BRCA1 and BRCA2 mutations.
Figure 3.
(a) NGS percentage 100 × coverage vs DNA input from 98 breast and ovarian cancer FFPE DNA samples. (b) Comparison of DNA concentration measurements using a hgDNA Quantification and QC Kit (KapaBiosystems, Anachem, Luton, UK) and three different amplicon sizes. The ovarian DNA samples were also quantified using a Nanodrop (Thermo Fisher). The 129 bp product was selected to determine the amount of DNA to add into the BRCA panel, as it was the closest measure to the mean amplicon size of all methods being evaluated (GeneRead (Qiagen) V.1: 155 bp (estimated), V.2: 153 bp, Ion AmpliSeq (Life Technologies) ~197 bp). Figure reproduced under the terms of the Creative Commons Attribution License from Ellison et al.62
Interpreting BRCA Test results: dealing with variants of unknown clinical significance
Because of the size of the BRCA1 and BRCA2 genes and the large number of screens carried out by diagnostic laboratories, many BRCA gene VUS have been identified. One study reported a VUS frequency rate for BRCA1 and BRCA2 of 13% for 10 000 consecutive individuals.29 In certain populations, a higher VUS frequency has been reported, such as 21% of alterations reported in patients with African–American ancestry.63 However, initiatives to reclassify BRCA VUS are likely to reduce this number.64, 65, 66 In our experience (St Mary's Hospital, Manchester, UK) the rate of BRCA VUS is ~8%.
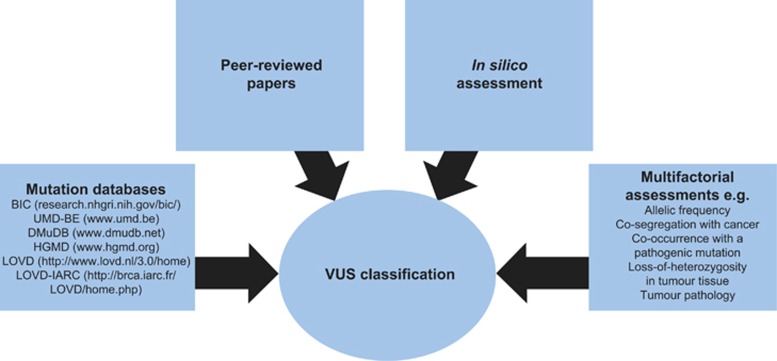
Assessing a BRCA VUS is a complex task (Figure 4), but one that is greatly assisted by pooling of genetic, clinical and histopathological information from a world-wide network of laboratories. There are many data-sharing initiatives aimed at developing sequencing methods and resources to facilitate BRCA1 and BRCA2 variant classification. Examples include the BRCA Challenge (a joint initiative of the Global Alliance for Genomics and Health and the Human Variome Project) and the Evidence-based Network for the Interpretation of Germline Mutant Alleles (ENIGMA).64, 67 There has also been a large-scale collaboration of the International Society for Gastrointestinal Hereditary Tumours (InSiGHT) to develop, test and apply a standardised classification scheme for mismatch repair gene variants in the setting of Lynch Syndrome.68 The guidance from ENIGMA, along with peer-reviewed publications, in silico assessment and information from mutation databases, including BIC (research.nhgri.nih.gov/bic/), UMD-BE (www.umd.be), DMudB (www.dmudb.net) and HGMD (www.hgmd.org), assist a diagnostic laboratory to establish the likely pathogenicity of a BRCA VUS. Caution should be applied when using information from public mutation databases because of the varying levels of curation. After reviewing all of the evidence on variant classification, clinical laboratories are required to provide an opinion for the purpose of clinical decision-making.
Figure 4.
VUS classification for BRCA: a complex task involving many elements.
As part of the assessment of a BRCA VUS, it is essential to clearly and systematically categorise a VUS as to whether it is pathogenic, neutral or of unknown status. This is a critical task, as the risk of miscomprehension is high among VUS uneducated genetic counsellors. In a recent survey exploring genetic counsellors' information preferences on VUS laboratory reports, a minority of respondents expressed concerns about awareness of VUS and appropriate medical recommendations among other health-care professionals.69 The survey also highlighted that the majority of respondents (243/267 (91%)) reported too little information provided on laboratory VUS reports, and that additional information would help contextualise the VUS result for patients. This is supported by a previous survey of breast cancer genetic counselling practices, where only 63% of genetic counsellors felt their patients understood the meaning of a VUS finding.70 It should be noted however, that a minority of genetic counsellors believe interpretation of additional information to guide patient medical management could be problematic, as interpretation was the responsibility of the laboratory conducting the test.69
In 2008, Plon et al71 devised a system of five classes of variants based on the degree of likelihood of pathogenicity, alongside recommendations for clinical management (Table 3). In contrast to an earlier classification system,72 this system subdivided the VUS category by the addition of ‘likely not pathogenic' or ‘likely pathogenic'. The five-class system provides health-care professionals with consistent classification information and clinical recommendations for each variant class. In order to assist with the application of this system, standardised data collection forms for VUS assessment are used by diagnostic laboratories.
Table 3. Five-class system for VUS 71 .
| Class | Description | Likelihood | Clinical management |
|---|---|---|---|
| 5 | Definitely pathogenic | >0.99 | Test at-risk relatives for variant, full high-risk surveillance guidelines |
| 4 | Likely pathogenic | 0.95–0.99 | Test at-risk relatives for varianta, full high-risk surveillance guidelines |
| 3 | Uncertain | 0.05–0.949 | Do not use for predictive testing in at-risk relativesa |
| 2 | Likely not pathogenic or of little clinical significance | 0.001–0.049 | Do not use for predictive testing in at-risk relativesa |
| 1 | Not pathogenic or of no clinical significance | <0.001 | Do not use for predictive testing in at-risk relativesa |
Recommend continuing to test proband for any additional testing modalities available for the disorder in question, for example, rearrangement testing.
Conclusions
As demand for BRCA testing increases, diagnostic laboratories will need to adapt their testing strategies and technologies to deal with the increased sequencing demand. Adoption of NGS can help meet this high demand and reduce the overall unit cost and staff time required for analysis; however, the choice and complexity in adopting NGS requires considerable thought and coordination of multiple-interdependent elements. Interpretation of screening results, particularly those of VUS, requires a thorough, systematic assessment, and consistent, clear reporting is essential for mainstream (non-genetic) disciplines. It is also important for the diagnostic laboratory to be aware of the significant challenges involved with screening tumour DNA, particularly in view of the potential utility of PARP inhibitors in tumours with somatic pathogenic BRCA mutations.
Acknowledgments
The supplement was sponsored by AstraZeneca. Medical writing services were provided by Tom Hudson of iMed Comms, Macclesfield, UK and were funded by AstraZeneca.
Footnotes
AW has received payments for speaker services from AstraZeneca and payments for consultancy from AstraZeneca and Roche Products.
References
- Chen S, Parmigiani G: Meta-analysis of BRCA1 and BRCA2 penetrance. J Clin Oncol 2007; 25: 1329–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavaddat N, Peock S, Frost D et al: Cancer risks for BRCA1 and BRCA2 mutation carriers: results from prospective analysis of EMBRACE. J Natl Cancer Inst 2013; 105: 812–822. [DOI] [PubMed] [Google Scholar]
- Evans DG, Barwell J, Eccles DM et al: The Angelina Jolie effect: how high celebrity profile can have a major impact on provision of cancer related services. Breast Cancer Res 2014; 16: 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluger J, Park A: The Angelina Effect. TIME Magazine 2013; 181: 28–33. [PubMed] [Google Scholar]
- NCCN Clinical Practice Guidelines: Genetic/familial high-risk assessment: breast and ovarian, version 1.2014. J Natl Compr Canc Netw 2014; 12: 1326–1338.25190698 [Google Scholar]
- Balmana J, Diez O, Rubio IT, Cardoso F.: BRCA in breast cancer: ESMO Clinical Practice Guidelines. Ann Oncol 2011; 22 (Suppl 6): vi31–vi34. [DOI] [PubMed] [Google Scholar]
- Bermejo-Perez MJ, Marquez-Calderon S, Llanos-Mendez A: Effectiveness of preventive interventions in BRCA1/2 gene mutation carriers: a systematic review. Int J Cancer 2007; 121: 225–231. [DOI] [PubMed] [Google Scholar]
- NICE Clinical Guidelines: NICE Clinical Guidance 164: Familial Breast Cancer. NICE Clinical Guidelines 2013, Available at www.nice.org.uk/Guidance/CG164.
- Struewing JP, Hartge P, Wacholder S et al: The risk of cancer associated with specific mutations of BRCA1 and BRCA2 among Ashkenazi Jews. N Engl J Med 1997; 336: 1401–1408. [DOI] [PubMed] [Google Scholar]
- Tonin P, Weber B, Offit K et al: Frequency of recurrent BRCA1 and BRCA2 mutations in Ashkenazi Jewish breast cancer families. Nat Med 1996; 2: 1179–1183. [DOI] [PubMed] [Google Scholar]
- Gorski B, Jakubowska A, Huzarski T et al: A high proportion of founder BRCA1 mutations in Polish breast cancer families. Int J Cancer 2004; 110: 683–686. [DOI] [PubMed] [Google Scholar]
- Szwiec M, Jakubowska A, Górski B et al: Recurrent mutations of BRCA1 and BRCA2 in Poland: an update. Clin Genet 2015; 87: 288–292. [DOI] [PubMed] [Google Scholar]
- Smith TM, Lee MK, Szabo CI et al: Complete genomic sequence and analysis of 117 kb of human DNA containing the gene BRCA1. Genome Res 1996; 6: 1029–1049. [DOI] [PubMed] [Google Scholar]
- Rouleau E, Jesson B, Briaux A et al: Rare germline large rearrangements in the BRCA1/2 genes and eight candidate genes in 472 patients with breast cancer predisposition. Breast Cancer Res Treat 2012; 133: 1179–1190. [DOI] [PubMed] [Google Scholar]
- Fachal L, Blanco A, Santamarina M, Carracedo A, Vega A: Large genomic rearrangements of BRCA1 and BRCA2 among patients referred for genetic analysis in Galicia (NW Spain): delimitation and mechanism of three novel BRCA1 rearrangements. PLoS One 2014; 9: e93306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim YK, Lau PT, Ali AB et al: Identification of novel BRCA large genomic rearrangements in Singapore Asian breast and ovarian patients with cancer. Clin Genet 2007; 71: 331–342. [DOI] [PubMed] [Google Scholar]
- Sharifah NA, Nurismah MI, Lee HC et al: Identification of novel large genomic rearrangements at the BRCA1 locus in Malaysian women with breast cancer. Cancer Epidemiol 2010; 34: 442–447. [DOI] [PubMed] [Google Scholar]
- Kang P, Mariapun S, Phuah SY et al: Large BRCA1 and BRCA2 genomic rearrangements in Malaysian high risk breast-ovarian cancer families. Breast Cancer Res Treat 2010; 124: 579–584. [DOI] [PubMed] [Google Scholar]
- Rudnicka H, Debniak T, Cybulski C et al: Large BRCA1 and BRCA2 genomic rearrangements in Polish high-risk breast and ovarian cancer families. Mol Biol Rep 2013; 40: 6619–6623. [DOI] [PubMed] [Google Scholar]
- Pietschmann A, Mehdipour P, Mehdipour P et al: Mutation analysis of BRCA1 and BRCA2 genes in Iranian high risk breast cancer families. J Cancer Res Clin Oncol 2005; 131: 552–558. [DOI] [PubMed] [Google Scholar]
- Moisan AM, Fortin J, Dumont M et al: No evidence of BRCA1/2 genomic rearrangements in high-risk French-Canadian breast/ovarian cancer families. Genet Test 2006; 10: 104–115. [DOI] [PubMed] [Google Scholar]
- The BRCA1 Exon 13 Duplication Screening Group: The exon 13 duplication in the BRCA1 gene is a founder mutation present in geographically diverse populations. Am J Hum Genet 2000; 67: 207–212. [PMC free article] [PubMed] [Google Scholar]
- Hogervorst FB, Nederlof PM, Gille JJ et al: Large genomic deletions and duplications in the BRCA1 gene identified by a novel quantitative method. Cancer Res 2003; 63: 1449–1453. [PubMed] [Google Scholar]
- Montagna M, Dalla PM, Menin C et al: Genomic rearrangements account for more than one-third of the BRCA1 mutations in northern Italian breast/ovarian cancer families. Hum Mol Genet 2003; 12: 1055–1061. [DOI] [PubMed] [Google Scholar]
- Welcsh PL, King MC: BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Hum Mol Genet 2001; 10: 705–713. [DOI] [PubMed] [Google Scholar]
- Ruiz de Garibay G, Gutierrez-Enriquez S, Garre P et al: Characterization of four novel BRCA2 large genomic rearrangements in Spanish breast/ovarian cancer families: review of the literature, and reevaluation of the genetic mechanisms involved in their origin. Breast Cancer Res Treat 2012; 133: 273–283. [DOI] [PubMed] [Google Scholar]
- Peixoto A, Santos C, Rocha P et al: The c.156_157insAlu BRCA2 rearrangement accounts for more than one-fourth of deleterious BRCA mutations in northern/central Portugal. Breast Cancer Res Treat 2009; 114: 31–38. [DOI] [PubMed] [Google Scholar]
- Lindor NM, Goldgar DE, Tavtigian SV et al: BRCA1/2 sequence variants of uncertain significance: a primer for providers to assist in discussions and in medical management. Oncologist 2013; 18: 518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank TS, Deffenbaugh AM, Reid JE et al: Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: analysis of 10,000 individuals. J Clin Oncol 2002; 20: 1480–1490. [DOI] [PubMed] [Google Scholar]
- Casilli F, Tournier I, Sinilnikova OM et al: The contribution of germline rearrangements to the spectrum of BRCA2 mutations. J Med Genet 2006; 43: e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grada A, Weinbrecht K: Next-generation sequencing: methodology and application. J Invest Dermatol 2013; 133: e11. [DOI] [PubMed] [Google Scholar]
- Idris SF, Ahmad SS, Scott MA, Vassiliou GS, Hadfield J: The role of high-throughput technologies in clinical cancer genomics. Expert Rev Mol Diagn 2013; 13: 167–181. [DOI] [PubMed] [Google Scholar]
- Loman NJ, Misra RV, Dallman TJ et al: Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol 2012; 30: 434–439. [DOI] [PubMed] [Google Scholar]
- Mardis ER: Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet 2008; 9: 387–402. [DOI] [PubMed] [Google Scholar]
- Walsh T, Lee MK, Casadei S et al: Detection of inherited mutations for breast and ovarian cancer using genomic capture and massively parallel sequencing. Proc Natl Acad Sci USA 2010; 107: 12629–12633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JE, Carr IM, Sheridan E et al: Genetic diagnosis of familial breast cancer using clonal sequencing. Hum Mutat 2010; 31: 484–491. [DOI] [PubMed] [Google Scholar]
- Chan M, Ji SM, Yeo ZX et al: Development of a next-generation sequencing method for BRCA mutation screening: a comparison between a high-throughput and a benchtop platform. J Mol Diagn 2012; 14: 602–612. [DOI] [PubMed] [Google Scholar]
- De Leeneer K, Hellemans J, De Schrijver J et al: Massive parallel amplicon sequencing of the breast cancer genes BRCA1 and BRCA2: opportunities, challenges, and limitations. Hum Mutat 2011; 32: 335–344. [DOI] [PubMed] [Google Scholar]
- Feliubadalo L, Lopez-Doriga A, Castellsague E et al: Next-generation sequencing meets genetic diagnostics: development of a comprehensive workflow for the analysis of BRCA1 and BRCA2 genes. Eur J Hum Genet 2013; 21: 864–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarabeux J, Zeitouni B, Moncoutier V et al: Streamlined ion torrent PGM-based diagnostics: BRCA1 and BRCA2 genes as a model. Eur J Hum Genet 2014; 22: 535–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castéra L, Krieger S, Rousselin A et al: Next-generation sequencing for the diagnosis of hereditary breast and ovarian cancer using genomic capture targeting multiple candidate genes. Eur J Hum Genet 2014; 22: 1305–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzker ML: Sequencing technologies - the next generation. Nat Rev Genet 2010; 11: 31–46. [DOI] [PubMed] [Google Scholar]
- Rothberg JM, Hinz W, Rearick TM et al: An integrated semiconductor device enabling non-optical genome sequencing. Nature 2011; 475: 348–352. [DOI] [PubMed] [Google Scholar]
- Glenn T 2014 NGS field guide: overview. The Molecular Ecologist 2014. Available at: http://www.molecularecologist.com/next-gen-fieldguide-2014/.
- Mamanova L, Coffey AJ, Scott CE et al: Target-enrichment strategies for next-generation sequencing. Nat Methods 2010; 7: 111–118. [DOI] [PubMed] [Google Scholar]
- Cheon JY, Mozersky J, Cook-Deegan R: Variants of uncertain significance in BRCA: a harbinger of ethical and policy issues to come? Genome Med 2014; 6: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcelik H, Shi X, Chang MC et al: Long-range PCR and next-generation sequencing of BRCA1 and BRCA2 in breast cancer. J Mol Diagn 2012; 14: 467–475. [DOI] [PubMed] [Google Scholar]
- Campbell PJ, Stephens PJ, Pleasance ED et al: Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet 2008; 40: 722–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedev P, Stanciu M, Brudno M: Computational methods for discovering structural variation with next-generation sequencing. Nat Methods 2009; 6: S13–S20. [DOI] [PubMed] [Google Scholar]
- Li J, Lupat R, Amarasinghe KC et al: CONTRA: copy number analysis for targeted resequencing. Bioinformatics 2012; 28: 1307–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord AS, Lee M, King MC, Walsh T: Accurate and exact CNV identification from targeted high-throughput sequence data. BMC Genomics 2011; 12: 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy BT, Timms KM, Carey MS et al: Somatic mutations in BRCA1 and BRCA2 could expand the number of patients that benefit from poly (ADP ribose) polymerase inhibitors in ovarian cancer. J Clin Oncol 2010; 28: 3570–3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu SK, Schelman WR, Wilding G et al: The poly(ADP-ribose) polymerase inhibitor niraparib (MK4827) in BRCA mutation carriers and patients with sporadic cancer: a phase 1 dose-escalation trial. Lancet Oncol 2013; 14: 882–892. [DOI] [PubMed] [Google Scholar]
- Ledermann J, Harter P, Gourley C et al: Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol 2014; 15: 852–861. [DOI] [PubMed] [Google Scholar]
- Lee JM, Hays JL, Annunziata CM et al: Phase I/Ib study of olaparib and carboplatin in BRCA1 or BRCA2 mutation-associated breast or ovarian cancer with biomarker analyses. J Natl Cancer Inst 2014; 106: dju089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmana J, Tung NM, Isakoff SJ et al: Phase I trial of olaparib in combination with cisplatin for the treatment of patients with advanced breast, ovarian and other solid tumors. Ann Oncol 2014; 25: 1656–1663. [DOI] [PubMed] [Google Scholar]
- Do H, Wong SQ, Li J, Dobrovic A: Reducing sequence artifacts in amplicon-based massively parallel sequencing of formalin-fixed paraffin-embedded DNA by enzymatic depletion of uracil-containing templates. Clin Chem 2013; 59: 1376–1383. [DOI] [PubMed] [Google Scholar]
- Do H, Dobrovic A: Dramatic reduction of sequence artefacts from DNA isolated from formalin-fixed cancer biopsies by treatment with uracil- DNA glycosylase. Oncotarget 2012; 3: 546–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SQ, Li J, Tan AY et al: Sequence artefacts in a prospective series of formalin-fixed tumours tested for mutations in hotspot regions by massively parallel sequencing. BMC Med Genomics 2014; 7: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiger MR, Kerick M, Timmermann B et al: Genome-wide massively parallel sequencing of formaldehyde fixed-paraffin embedded (FFPE) tumor tissues for copy-number- and mutation-analysis. PLoS One 2009; 4: e5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerick M, Isau M, Timmermann B et al: Targeted high throughput sequencing in clinical cancer settings: formaldehyde fixed-paraffin embedded (FFPE) tumor tissues, input amount and tumor heterogeneity. BMC Med Genomics 2011; 4: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison G, Huang S, Carr H et al: A reliable method for the detection of BRCA1 and BRCA2 mutations in fixed tumour tissue utilising multiplex PCR-based targeted next generation sequencing. BMC Clin Pathol 2015; 15: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ready K, Gutierrez-Barrera AM, Amos C et al: Cancer risk management decisions of women with BRCA1 or BRCA2 variants of uncertain significance. Breast J 2011; 17: 210–212. [DOI] [PubMed] [Google Scholar]
- Spurdle AB, Healey S, Devereau A et al: ENIGMA—evidence-based network for the interpretation of germline mutant alleles: an international initiative to evaluate risk and clinical significance associated with sequence variation in BRCA1 and BRCA2 genes. Hum Mutat 2012; 33: 2–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo C, Masiello A, Ryan JF, Brody LC: The breast cancer information core: database design, structure, and scope. Hum Mutat 2000; 16: 123–131. [DOI] [PubMed] [Google Scholar]
- Vallee MP, Francy TC, Judkins MK et al: Classification of missense substitutions in the BRCA genes: a database dedicated to Ex-UVs. Hum Mutat 2012; 33: 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRCA Challenge. The Human Variome Project. Available at: http://www.humanvariomeproject.org/brca-challenge.html. (accessed on 06 November 2014).
- Thompson BA, Spurdle AB, Plazzer JP et al: Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat Genet 2014; 46: 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherr CL, Lindor NM, Malo TL, Couch FJ, Vadaparampil ST: A preliminary investigation of genetic counselors' information needs when receiving a variant of uncertain significance result: a mixed methods study. Genet Med 2015; 17: 739–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrucelli N, Lazebnik N, Huelsman KM, Lazebnik RS: Clinical interpretation and recommendations for patients with a variant of uncertain significance in BRCA1 or BRCA2: a survey of genetic counseling practice. Genet Test 2002; 6: 107–113. [DOI] [PubMed] [Google Scholar]
- Plon SE, Eccles DM, Easton D et al: Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat 2008; 29: 1282–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards CS, Bale S, Bellissimo DB et al: ACMG recommendations for standards for interpretation and reporting of sequence variations: revisions 2007. Genet Med 2008; 10: 294–300. [DOI] [PubMed] [Google Scholar]