Abstract
The plasmodesmata and phloem form a symplasmic network that mediates direct cell–cell communication and transport throughout a plant. Selected endogenous RNAs, viral RNAs, and viroids traffic between specific cells or organs via this network. Whether an RNA itself has structural motifs to potentiate trafficking is not well understood. We have used mutational analysis to identify a motif that the noncoding Potato spindle tuber viroid RNA evolved to potentiate its efficient trafficking from the bundle sheath into mesophyll that is vital to establishing systemic infection in tobacco (Nicotiana tabacum). Surprisingly, this motif is not necessary for trafficking in the reverse direction (i.e., from the mesophyll to bundle sheath). It is not required for trafficking between other cell types either. We also found that the requirement for this motif to mediate bundle sheath-to-mesophyll trafficking is dependent on leaf developmental stages. Our results provide genetic evidence that (1) RNA structural motifs can play a direct role in mediating trafficking across a cellular boundary in a defined direction, (2) the bundle sheath–mesophyll boundary serves as a novel regulatory point for RNA trafficking between the phloem and nonvascular tissues, and (3) the symplasmic network remodels its capacity to traffic RNAs during plant development. These findings may help further studies to elucidate the interactions between RNA motifs and cellular factors that potentiate directional trafficking across specific cellular boundaries.
INTRODUCTION
Cell–cell communication is essential for a multicellular organism to coordinate differential gene expression that underlies cell division, cell differentiation, and morphogenesis. Plants have evolved a unique cytoplasmic (symplasmic) network consisting of plasmodesmata and the phloem to enable direct cell–cell communication and transport (Lucas et al., 1993). Studies on the intercellular movement of viruses provided initial experimental evidence that plasmodesmata have the capacity to mediate cell–cell trafficking of viral RNAs in the form of ribonucleoprotein complex (Fujiwara et al., 1993; Ding et al., 1995; Nguyen et al., 1996). Subsequent studies demonstrated that many, but selected, endogenous RNAs can also traffic intercellularly. These include the mRNA of the maize (Zea mays) transcription factor KN1 (Lucas et al., 1995) and the mRNA encoding the sucrose transporter SUT1 (Kühn et al., 1997). In addition, >100 mRNAs appear to be transported in the phloem of pumpkin (Cucurbita maxima). Some of these mRNAs encode proteins with functions ranging from control of meristem development and hormone response to defense response. Importantly, grafting experiments showed that some mRNAs are selectively transported into the shoot apex (Ruiz-Medrano et al., 1999). The potential significance of systemic trafficking of RNAs to regulate development was underscored by the finding that a mutant fusion transcript between LeT6, a tomato (Lycopersicon esculentum) homeodomain protein gene, and PYROPHOSPHATE-DEPENDENT PHOSPHOFRUCTOKINASE gene can traffic long distance from a tomato rootstock into the shoot apex of the scion to regulate leaf developmental patterns (Kim et al., 2001). RNAs yet to be identified traffic through the symplasmic network to induce systemic gene silencing in a plant (Mlotshwa et al., 2002), suggesting that trafficking of specific RNAs can modulate gene expression between distant organs. Although the full spectra of biological functions of systemic RNA trafficking remain to be elucidated, current data support the hypothesis that an RNA-based signaling network controls many fundamental plant processes ranging from development to defense responses (Jorgensen et al., 1998; Ding et al., 1999; Citovsky and Zambryski, 2000; Lucas et al., 2001; Ueki and Citovsky, 2001; Haywood et al., 2002; Jorgensen, 2002; Wu et al., 2002).
Because systemic trafficking of an RNA involves passage through various cellular boundaries, a most important question is how an RNA produced in a cell is recognized and then transported via the symplasmic network to specific cellular sites without being misdelivered or lost. It has been suggested that an RNA may have sequence/structural motifs that interact with cellular factors to confer selectivity (Citovsky and Zambryski, 2000; Lucas et al., 2001; Haywood et al., 2002). Multiple checkpoints have also been proposed at which molecular interactions between an RNA and cellular factors control the targeting of an RNA to specific cells (Ding et al., 2003). To date, however, there is no genetic evidence that an RNA indeed has a motif to mediate trafficking, and there is little information about how the trafficking might be controlled across specific cellular boundaries. Lack of this basic information not only hinders progress on mechanistic studies on the regulation of RNA trafficking but also creates a bottleneck in experimental studies on the functions of such trafficking.
To learn about the molecular basis of selectivity and cell-specific targeting of RNAs, we tested whether an RNA possesses structural motifs to mediate its movement across specific cellular boundaries and investigated the cellular and developmental parameters that control the trafficking. We used trafficking of a pathogenic RNA, the Potato spindle tuber viroid (PSTVd), as an experimental system. Viroids are small, single-stranded, and covalently closed circular RNAs that infect plants. A distinguishing feature of these small RNAs is that they do not encode any proteins for function (Flores et al., 1997, 2000; Diener, 2001). Yet, a viroid RNA can replicate and move systemically throughout an infected plant. Therefore, viroid systemic trafficking offers a simple model in which to test the direct role of RNA structures in trafficking.
PSTVd is 359 nucleotides in length and assumes a rod-shaped secondary structure in its native state (Gross et al., 1978). Depending on the host and the PSTVd strain, PSTVd infection can alter expression of many plant genes important for growth and development (Itaya et al., 2002a; Qi and Ding, 2003a). PSTVd replicates in the nucleus via a rolling-circle mechanism, which starts with transcription of the incoming monomeric, circular (+)-strand RNA into multimeric, linear (−)-strands. The latter then serve as the replication intermediates for the massive production of multimeric, linear (+)-strands that are finally processed into unit-length, mature circular RNAs (Branch and Robertson, 1984). During replication, the (−)-strand RNA is anchored in the nucleoplasm, whereas the (+)-strand RNA can be selectively transported into the nucleolus, presumably for processing (Qi and Ding, 2003b). The (+)-strand PSTVd RNA moves from cell to cell via plasmodesmata (Ding et al., 1996). Systemic movement from organ to organ occurs through the phloem (Palukaitis, 1987; Zhu et al., 2001). This viroid RNA therefore provides an extraordinary system to investigate how an RNA can accomplish intranuclear, cell–cell and organ–organ trafficking altogether.
In this study, we analyzed PSTVd systemic trafficking in tobacco (Nicotiana tabacum) using a combination of cellular, genetic, and molecular approaches. Our results provide genetic evidence that an RNA has motifs to mediate trafficking across a specific cellular boundary in a defined direction and is governed by developmental status of an organ. The conceptual framework and experimental approaches developed in this study should be valuable in investigating the mechanisms and functions of the systemic trafficking of plant endogenous and infectious RNAs and the putative systemic RNA silencing signals.
RESULTS
Two PSTVd Strains Differ in Intercellular Trafficking Capacities
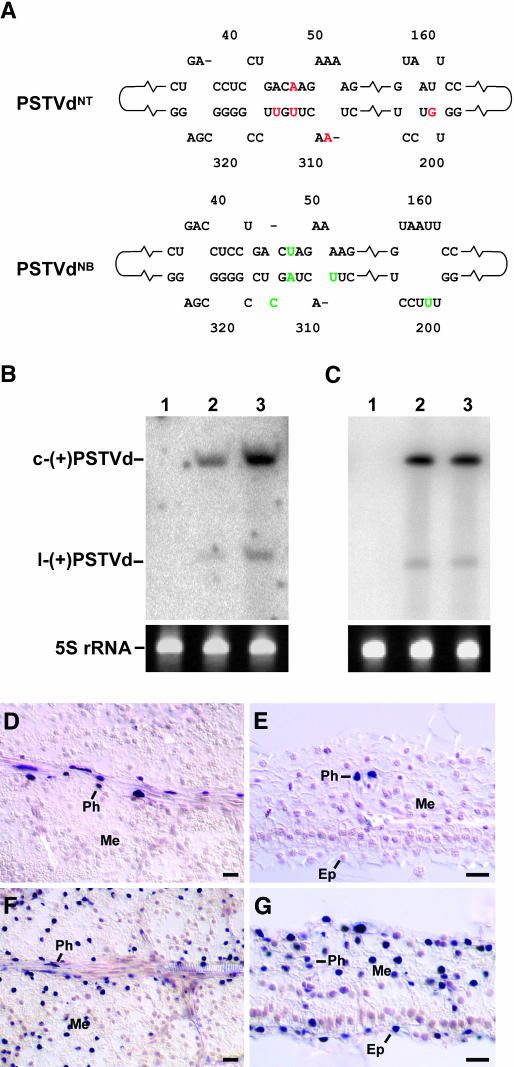
We used two PSTVd strains, PSTVdNT and PSTVdNB, that are infectious in tobacco for analyses. PSTVdNT was derived from the tomato isolate PSTVdKF440-2 by a single nucleotide C259U substitution (Wassenegger et al., 1996). This substitution converted the PSTVdKF440-2 from a noninfectious to an infectious RNA for tobacco. However, in primary PSTVdNT-infected tobacco plants, the accumulation of mature viroid RNA was less efficient than in plants that were maintained for several years by propagating cuttings of the primary infected plants. Moreover, using total RNA isolated from these cuttings as inoculum to infect tobacco plants revealed similar high accumulation levels in RNA gel blots (data not shown) as were detected in the cuttings. These observations suggested the emergence and positive selection of a new tobacco-specific PSTVd variant. Indeed, sequence analysis of the mature PSTVd accumulating in the cuttings uncovered a new PSTVd strain, designated PSTVdNB, that was derived from PSTVdNT by five nucleotide substitutions: G201U, A309U, A47U/U313A, and U315C (Figure 1A; see Supplemental Figure 1 online). To test directly the infection capacity of PSTVdNB in comparison with that of PSTVdNT, we mechanically inoculated their in vitro transcripts into leaves of young tobacco plants. RNA gel blot analysis of systemic leaves at 5 weeks postinoculation demonstrated that PSTVdNB indeed accumulated to a much higher level (approximately fivefold) than did PSTVdNT (Figure 1B). The difference in the accumulation levels of PSTVdNB and PSTVdNT in the leaves could be attributed to differences in their replication efficiency or cellular distribution. To distinguish between these possibilities, we first inoculated protoplasts of tobacco BY2 cells with the in vitro transcripts of these strains and then analyzed their accumulation levels at 3 d postinoculation. Both strains accumulated to similar levels in the infected protoplasts (Figure 1C). Thus, the higher accumulation level of PSTVdNB as compared with that of PSTVdNT in the infected plants likely resulted from differences in their cellular distribution. To test this hypothesis, we performed in situ hybridization, using digoxigenin-labeled riboprobes specific for the (+)-strand RNA of PSTVd, on tissue sections from systemic leaves of mechanically inoculated plants. Such experiments showed that PSTVdNT was present predominantly in the phloem (Figures 1D and 1E). By contrast, PSTVdNB was present in the phloem, mesophyll, and epidermis (Figures 1F and 1G). Sequence analyses confirmed maintenance of the original genomic sequences of these strains in the infected plants (data not shown).
Figure 1.
Phloem Limitation of PSTVdNT in an Infected Tobacco Plant.
(A) Partial nucleotide sequences to illustrate the differences between PSTVdNT and PSTVdNB in the classical pathogenicity (left portion) and right-terminal (right portion) domains. The red nucleotides in PSTVdNT are substituted by the green nucleotides in PSTVdNB through natural mutations during successive passages of infection. See Supplemental Figure 1 online for the complete nucleotide sequences of the two PSTVd strains.
(B) and (C) RNA gel blots showing the accumulation levels of PSTVdNT (lane 2) and PSTVdNB (lane 3) in systemically infected leaves (B) and BY-2 protoplasts (C) of tobacco. There is no viroid accumulation in mock-inoculated plants and protoplasts (lane 1). Bands of 5S rRNA show equal loading of RNA in all lanes. c-(+)PSTVd and l-(+)PSTVd denote circular and linear (+)-PSTVd, respectively. Note the higher accumulation of PSTVdNB than PSTVdNT in systemic leaves (B) and similar accumulation levels of the two in protoplasts (C).
(D) and (E) Paradermal and transverse sections, respectively, of systemically infected young leaves showing the presence of PSTVdNT in the nuclei (purple dots) of the phloem cells (Ph) and its absence from the mesophyll (Me) and epidermis (Ep).
(F) and (G) Paradermal and transverse sections, respectively, of systemically infected young leaves showing the presence of PSTVdNB in the nuclei (purple dots) of all leaf cells. Ep, epidermis; Me, mesophyll; Ph, phloem. Bars = 20 μm.
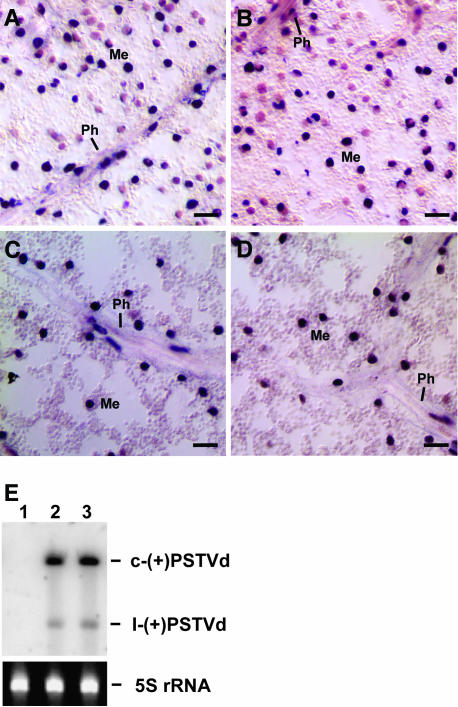
The absence of PSTVdNT in the mesophyll could be attributed to its intrinsic inability (1) to traffic from the phloem into mesophyll or (2) to replicate/accumulate in the mesophyll. As described above, both PSTVdNT and PSTVdNB accumulated equally well in the infected tobacco protoplasts, suggesting that absence of PSTVdNT from the mesophyll tissue was because of a defect in trafficking into the mesophyll and not because of a defect in replication/accumulation in the mesophyll. To further test this hypothesis, we investigated the replication capacity of PSTVdNT as compared with that of PSTVdNB in all tissues of transgenic tobacco expressing their (+)-strand cDNAs under the control of the constitutive 35S promoter of Cauliflower mosaic virus (CaMV). In situ hybridization revealed accumulation of the (−)-strand, replication intermediate form of PSTVdNB and PSTVdNT in all tissues of young and mature leaves using digoxigenin-labeled (+)-strand PSTVd RNA as the probe (Figures 2A to 2D). Dot blot analysis confirmed the specific hybridization of the (+)-PSTVd probe to the (−)-strand PSTVd but not the (+)-strand PSTVd (see Supplemental Figure 2 online). RNA gel blots showed accumulation of the (+)-circular, mature viroid RNAs of both strains (Figure 2E). These data provide conclusive evidence for the replication of PSTVdNT as well as PSTVdNB in all leaf tissues. Sequence analysis confirmed maintenance of the viroid sequences in the transgenic plants. Four transgenic lines were examined, and the results were consistent among these lines. These data establish unequivocally that the absence of PSTVdNT from mesophyll of systemic leaves is attributed to its inability to move from the phloem into the mesophyll.
Figure 2.
Replication of PSTVdNT as Well as PSTVdNB in Leaf Mesophyll of Transgenic Tobacco Plants Expressing Their (+)-Strand cDNAs under the Control of the CaMV 35S Promoter.
(A) and (B) In situ hybridization showing the presence of the (−)-strand RNAs of PSTVdNB and PSTVdNT, respectively, in the mesophyll (Me) and phloem (Ph) cells of a young leaf. Bars = 20 μm.
(C) and (D) In situ hybridization showing the presence of the (−)-strand RNAs of PSTVdNB and PSTVdNT, respectively, in the mesophyll (Me) and phloem (Ph) cells of a mature leaf. Bars = 20 μm
(E) RNA gel blots showing the accumulation levels of PSTVdNT (lane 2) and PSTVdNB (lane 3) in 5th leaves of such transgenic plants. There is no viroid accumulation in similar leaves of a nontransgenic plant (lane 1). Bands of 5S rRNA show equal loading of RNA in all lanes. c-(+)PSTVd and l-(+)PSTVd denote circular and linear (+)-PSTVd, respectively.
Bundle Sheath–Mesophyll Boundary Functions as a Novel Checkpoint for RNA Trafficking during Leaf Development
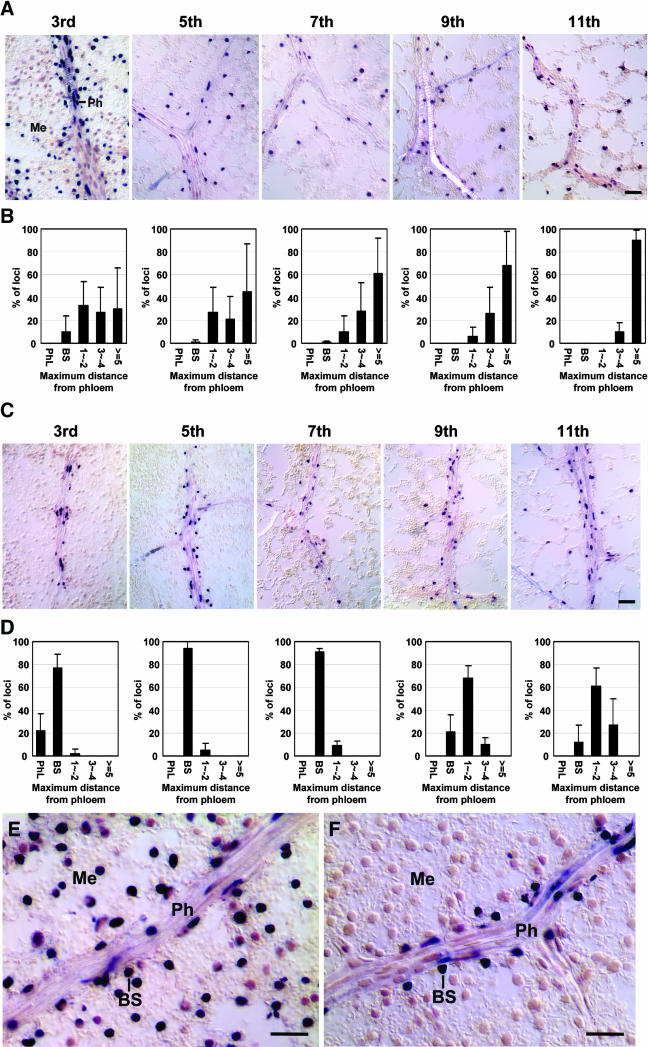
To test the developmental effect of an organ on RNA trafficking, we produced PSTVd RNAs in phloem cells and then determined their cellular localization in leaves of different developmental stages. We generated transgenic tobacco expressing the (+)-strand cDNAs of PSTVdNB and PSTVdNT, respectively, under the control of the Commelina yellow mottle virus (CoYMV) promoter that is active specifically in the phloem companion cells of all organs (Matsuda et al., 2002). Samples were collected from leaves of successive developmental stages of the transgenic plants that had 18 to 20 leaves. Counting from the top of a plant, the leaves sampled were 3rd, 5th, 7th, 9th, and 11th. The 5th leaf under our growth conditions was undergoing photoassimilate sink–source transition (see Methods). Thus, the 3rd leaf was a sink and the 7th and older leaves were source leaves. RNA gel blots showed higher accumulation of PSTVdNB than PSTVdNT in all the leaves tested (see Supplemental Figure 3 online), a pattern consistent with that shown in mechanically inoculated tobacco plants (Figure 1B). In situ hybridization revealed PSTVdNB in all cell types of all leaves, with the viroid signal detected in more cells in older leaves (Figures 3A, 3B, and 3E). By contrast, PSTVdNT was detected predominantly in the phloem and the associated bundle sheath of the 3rd and 5th leaves (Figures 3C, 3D, and 3F). In the 7th to 11th leaves, PSTVdNT became increasingly present in a few layers of mesophyll cells around the vein (Figures 3C and 3D). Its distribution in the nonvascular tissues of such mature leaves, however, was not as extensive as PSTVdNB. Sequence analyses confirmed maintenance of the original genomic sequences of these strains in the transgenic plants. Results obtained from four transgenic lines showed similar patterns. These findings revealed that (1) movement across the bundle sheath–mesophyll boundary is a novel limiting step for an RNA that has exited the phloem to reach the mesophyll, and (2) this limiting role diminishes at later stages, beyond the stage of sink–source transition, of leaf development.
Figure 3.
Cellular Distribution of PSTVdNB and PSTVdNT in Leaves of Successive Developmental Stages of Transgenic Tobacco Expressing the Viroid cDNA under the Control of the Companion Cell-Specific CoYMV Promoter.
(A) In situ hybridization shows presence of the (+)-strand RNA of PSTVdNB in leaf cells from the 3rd through the 11th leaves. Me, mesophyll; Ph, phloem.
(B) Quantitative analysis of the extent of PSTVdNB movement out of the phloem in the corresponding leaves. The data show the percentages of veins and the associated neighboring tissues (y axis) in which the viroid signal is detected in the maximal distance (layers of cells) away from the phloem. Along the x axis, PhL indicates viroid presence in the phloem but not in nonvascular tissues, BS denotes bundle sheath, and the numbers indicate layers of mesophyll cells in paradermal sections. The data are averages of six independent experiments plus standard deviations.
(C) In situ hybridization shows presence of (+)-strand RNA of PSTVdNT predominantly in the phloem and the associated bundle sheath in 3rd to 7th leaves and increasing appearance in a few layers of mesophyll cells in older leaves.
(D) Quantitative analysis of the extent of PSTVdNT movement out of the phloem in the corresponding leaves. See legend for (B) for data presentations.
(E) High-magnification view of a paradermal section from a 3rd leaf showing the presence of the (+)-strand RNA of PSTVdNB in the phloem (Ph), bundle sheath (BS), and mesophyll (Me).
(F) High-magnification view of a paradermal section from a 3rd leaf showing the presence of the (+)-strand RNA of PSTVdNT in the phloem (Ph) and bundle sheath (BS) and its absence from the mesophyll (Me).
Bars = 40 μm in (A) and (C) and 20 μm in (E) and (F).
A Bipartite Motif Is Required and Sufficient to Mediate Bundle Sheath-to-Mesophyll Trafficking in Young Leaves
The differing capacity of PSTVdNT and PSTVdNB to traffic from the bundle sheath to mesophyll in young leaves suggests existence of distinct molecular interactions that control RNA trafficking across specific cellular boundaries. Specifically, a unique RNA motif(s) must be present in PSTVdNB, but is absent from PSTVdNT, to mediate bundle sheath-to-mesophyll trafficking. As illustrated in Figure 1 (also see Supplemental Figure 1 online), PSTVdNB derives from PSTVdNT by five nucleotide substitutions. Notably, the G201U substitution creates a new loop structure in the right-terminal domain as predicted by computing with mfold version 3.0 at 25°C (Water et al., 1994). The other nucleotide changes are clustered in the classical pathogenicity domain, near the interface with the left-terminal domain. Among these substitutions, A309U and U315C alter the predicted local secondary structures, but A47U/U313A substitutions do not because they still maintain the base pairing. We postulated that these nucleotide substitutions allowed PSTVdNB to traffic from the bundle sheath to mesophyll in all young leaves. To test this hypothesis, we used site-directed mutagenesis to determine the impact of each nucleotide change on this trafficking. First, we changed nucleotide U201 in PSTVdNB to G (PSTVdNBU201G), as is present in PSTVdNT (see Supplemental Figure 4 online). This would allow determination of whether U201 is necessary for the trafficking. Conversely, we mutated G201 in PSTVdNT to U as in PSTVdNB (PSTVdNTG201U; see Supplemental Figure 4 online) to determine whether U201, if necessary, is itself alone sufficient to potentiate the trafficking. We established that both mutants retained the ability to replicate at the cellular level by inoculating them into tobacco BY2 cells and then assaying viroid accumulation by RNA gel blots (Figure 4A). Sequence analyses of RT-PCR products confirmed maintenance of the genomic sequence of each mutant. We then used mechanical inoculation and CoYMV promoter–directed companion cell expression in transgenic tobacco, respectively, to test for the capacity of these mutants to traffic from the phloem to bundle sheath and then to mesophyll. In situ hybridization showed that both PSTVdNBU201G and PSTVdNTG201U remained in the phloem/bundle sheath of young leaves in both transgenic (Figure 4B) and mechanically inoculated plants (data not shown). Both mutants could replicate normally in mesophyll cells, as revealed by in situ hybridization detection of their (−)-strand RNAs in transgenic tobacco expressing their respective (+)-strand cDNAs under the control of the CaMV 35S promoter (see Supplemental Figure 5 online) and by RNA gel blot analysis showing the accumulation of the mature viroid RNA in such plants (see Supplemental Figure 6 online). Thus, U201 in PSTVdNB is necessary but is not sufficient by itself to mediate PSTVd trafficking from the bundle sheath to mesophyll. Sequence analysis of the RT-PCR products confirmed the mutant sequences of the viroid progeny in all transgenic plants analyzed.
Figure 4.
Replication and Cellular Distribution of PSTVdNB- and PSTVdNT-Derived Mutants.
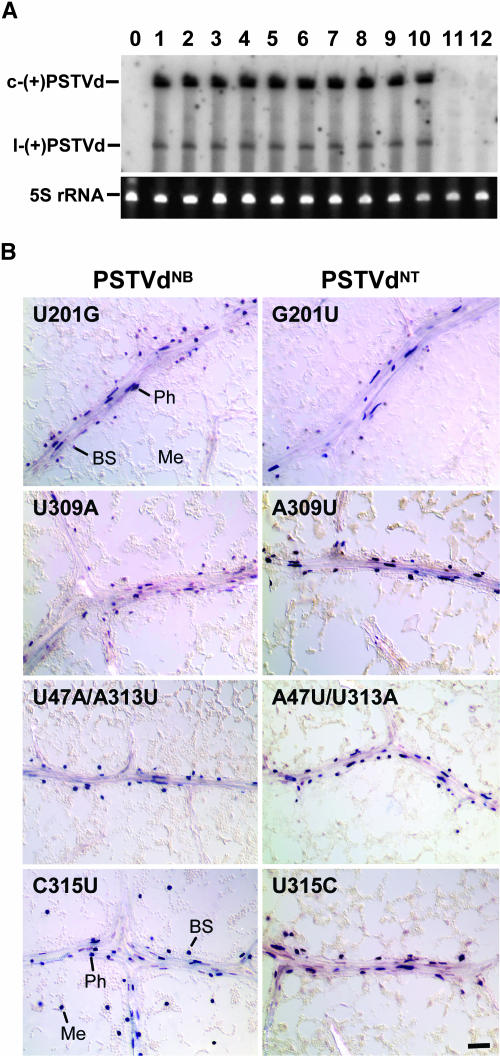
(A) RNA gel blot shows accumulation of the mutants in protoplasts of tobacco BY2 cells. c-(+)PSTVd and l-(+)PSTVd denote circular and linear (+)-PSTVd, respectively. Bands of 5S rRNA show equal loading of RNA in all lanes. Lane 0, mock inoculation; lane 1, PSTVdNT; lanes 2 to 5, PSTVdNT-derived mutants G201U, A309U, U315C, and A47U/U313A; lanes 6 to 12, PSTVdNB-derived mutants U201G, U309A, C315U, U47A/A313U, U47A, and A313U.
(B) In situ hybridization shows cellular distribution of the mutants in the 7th leaves of transgenic tobacco plants expressing the cDNAs under the control of the companion cell-specific CoYMV promoter. Note presence of PSTVdNBC315U in the mesophyll (Me), bundle sheath (BS), and phloem (Ph). All other mutants are present predominantly in the phloem and the associated bundle sheath. Bar = 20 μm.
Because U201 in PSTVdNB is necessary, but is not sufficient for bundle sheath–mesophyll trafficking, the other PSTVdNB-specific nucleotides in the classical pathogenicity domain must also be required. The role of these nucleotides in the trafficking was investigated by mutational analysis as described above. Supplemental Figure 4 online lists all of the mutants that were generated by site-directed mutagenesis. The replication capacity of each mutant was tested in tobacco BY2 protoplasts. Interestingly, two mutants (NBU47A and NBA313U) were defective in replication (Figure 4A) and were not tested further for trafficking functions. The other mutants were fully competent for replication and were tested for trafficking function by mechanical inoculation and the transgenic method as described above. These analyses showed that U309 as well as U47/A313 were each required, but not sufficient, for bundle sheath-to-mesophyll trafficking (Figure 4B). Interestingly, C315 is neither necessary nor sufficient for this trafficking (Figure 4B). As a further support of these conclusions, all mutants defective in this trafficking were fully competent in replication in the mesophyll cells of young leaves from transgenic tobacco expressing the mutant cDNAs under the control of the CaMV 35S promoter, as shown by in situ hybridization (see Supplemental Figure 5 online) and by RNA gel blots (see Supplemental Figure 6 online). Collectively, our analyses demonstrated that U201, U309, and U47/A313 are all necessary and together sufficient to mediate trafficking of PSTVdNB from the bundle sheath to mesophyll. These nucleotides define a bipartite trafficking motif: U201 in a loop in the right-terminal domain and U309/U47/A313 in the classical pathogenicity domain.
Trafficking of PSTVd across the Mesophyll–Bundle Sheath Boundary Can Be Unidirectional
The observation that PSTVdNB- and PSTVdNT-derived mutants that failed to traffic from the bundle sheath into mesophyll in young leaves could initiate systemic infection by mechanical inoculation implies that these mutants could traffic from the mesophyll into bundle sheath for further entry into the phloem in an inoculated leaf. In other words, these mutants seem to be able to traffic unidirectionally from the mesophyll to bundle sheath but not in the reverse direction. However, we could not rule out the possibility, even though remote, that mechanical inoculation introduced the viroid inoculum directly into the bundle sheath or the phloem in an inoculated leaf. To resolve this uncertainty, we conducted an extensive analysis using an alternative inoculation method. We used biolistic bombardment to deliver cDNAs of PSTVdNB- and PSTVdNT-derived variants, under the control of the CaMV 35S promoter, into epidermal cells of a young tobacco leaf that would not allow bundle sheath-to-mesophyll movement of these variants. We established that this method only delivered cDNAs into the epidermis using green fluorescent protein (GFP) expression as a reporter (Figure 5A). Therefore, there was little possibility of directly inoculating the phloem/bundle sheath cells, which are three to four cell layers away from the epidermis. Primary viroid transcripts generated from the CaMV 35S promoter would initiate viroid RNA–RNA replication. If a viroid variant could move from epidermis to mesophyll and bundle sheath and finally into the phloem to embark on systemic infection, we would expect to detect the viroid signal in the systemic leaves by RNA gel blots. As shown in Figure 5B, every PSTVd variant that was defective in bundle sheath-to-mesophyll trafficking could be detected in the systemic leaves at 5 weeks after bombardment. These data provide conclusive evidence that every such PSTVd variant could traffic in the reverse direction: from the mesophyll to bundle sheath. Thus, RNA trafficking across a specific cellular boundary can be unidirectional, and trafficking in both directions involves different regulations.
Figure 5.
Replication/Accumulation of PSTVdNB- and PSTVdNT-Derived Mutants in Systemic Leaves of Biolistically Inoculated Tobacco Plants.
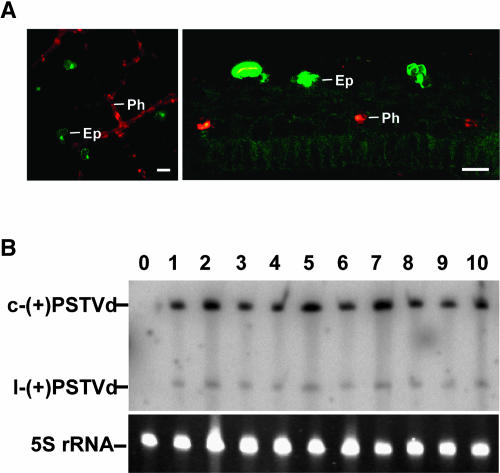
(A) Surface and transverse sectional views of tobacco leaves show transient expression of dimeric GFP, upon biolistic bombardment of plasmids containing CaMV 35S:dGFP, in the epidermis (Ep). The bombarded plants are transgenic for the expression of dimeric dsRed in the phloem (Ph). The images show that biolistic bombardment does not deliver cDNAs into cells below the epidermis. Bars = 40 μm.
(B) RNA gel blot shows accumulation of PSTVdNB, PSTVdNT, and their mutants in systemic leaves of bombarded plants. c-(+)PSTVd and l-(+)PSTVd denote circular and linear (+)-PSTVd, respectively. Bands of 5S rRNA show equal loading of RNA in all lanes. Lane 0, mock inoculation; lane 1, PSTVdNT; lanes 2 to 4, PSTVdNT-derived mutants G201U, U315C, and A309U; lane 5, PSTVdNB; lanes 6 to 8, PSTVdNB-derived mutants U201G, C315U, and U309A; lane 9, PSTVdNTA47U/U313A; lane 10, PSTVdNBU47A/A313U.
DISCUSSION
Proper functioning of all gene products, including RNAs and proteins, requires correct subcellular localization. Extensive studies on the intracellular trafficking of macromolecules in eukaryotic cells have established the general principles that control such trafficking and illuminated how such trafficking serves to regulate gene expression at the cellular level. Systemic RNA trafficking defines a new level of gene regulation that potentially has broad functional implications in coordinating development, physiology, and defense responses (Jorgensen et al., 1998; Ding et al., 1999; Citovsky and Zambryski, 2000; Lucas et al., 2001; Ueki and Citovsky, 2001; Haywood et al., 2002; Jorgensen, 2002; Wu et al., 2002). Elucidating how such trafficking is regulated at the cellular and molecular level should lead to establishment of new paradigms to study gene regulation.
An RNA Motif Mediates Directional RNA Trafficking across a Specific Cellular Boundary
The experimental data presented here provide genetic evidence to support the hypothesis that distinct RNA motifs mediate trafficking between specific cells (Lucas et al., 2001; Ding et al., 2003) and further reveal some basic principles that govern this trafficking: (1) trafficking of an RNA across the boundary interconnecting two types of cells can be unidirectional, (2) distinct RNA motifs may be required to mediate trafficking across such a cellular boundary in opposite directions, and (3) the molecular interactions that regulate such trafficking can be reprogrammed during organ development.
In the case of PSTVdNB, we have identified a bipartite motif that is both necessary and sufficient to mediate trafficking of the viroid RNA from the bundle sheath to the mesophyll in young leaves: one part in the right-terminal domain and the other in the classical pathogenicity domain. Whether this motif functions at the primary nucleotide sequence or secondary structural level requires further analysis. Nonetheless, identification of this motif sets the stage for further investigations on how the motif interacts with cognate cellular factors to confer selectivity and to potentiate trafficking. The finding that this motif is only required for trafficking from the bundle sheath to mesophyll implies that other yet to be identified motifs guide trafficking from the mesophyll to bundle sheath and trafficking between other cells. Consistent with this hypothesis, we previously showed that the intermediate strain of PSTVd can mediate trafficking of a fused RNA that otherwise would not traffic between mesophyll cells, suggesting existence of a motif in the PSTVd that mediates trafficking between such cells (Ding et al., 1996).
The requirement for distinct motifs to mediate trafficking of an RNA across a cellular boundary in opposite directions suggests that the plasmodesmal openings in the two adjoining cells have different molecular compositions to recognize different RNA motifs. Alternatively, each type of cell has unique factors that interact with the RNA motifs to potentiate trafficking. It is also possible that in at least some cases, an RNA contains motifs that are recognized by cellular factors for retention in specific cells. Thus, whether an RNA traffics between two cells in a defined direction at a particular organ developmental stage may depend on the release of the RNA from retention and further recognition for trafficking.
The Bundle Sheath–Mesophyll Boundary Is a Checkpoint for RNA Trafficking between the Phloem and Nonvascular Tissues
Extensive studies on the systemic movement of plant viral RNAs have identified the phloem parenchyma–bundle sheath boundary as a major checkpoint for viral entry into and exit from the phloem (reviewed in Gilbertson and Lucas, 1996; Ghoshroy et al., 1997; Ding, 1998). Phloem-limited viruses usually cannot move from the phloem parenchyma into the bundle sheath (Sanger et al., 1994), and viruses that cannot enter the phloem usually fail in moving from the bundle sheath into the phloem parenchyma (Goodrick et al., 1991; Wintermantel et al., 1997; Wang et al., 1998; Ghoshroy et al., 1998). There are also examples that phloem entry and exit of a virus (Citovsky et al., 1998; Ghoshroy et al., 1998) or viroid (Zhu et al., 2002) use different mechanisms. The precise cellular boundary that is involved in the control in these cases is not clear. It should be noted that viral movement involves complex interactions among the viral genome, viral proteins, and cellular factors (Carrington et al., 1996; Gilbertson and Lucas, 1996; Lazarowitz and Beachy, 1999), complicating analysis of the direct role of cellular factors in recognizing an RNA for trafficking between specific cells. Our current analyses of PSTVd trafficking, which depends solely on the interaction of this RNA with cellular factors, identified the bundle sheath–mesophyll boundary as a novel checkpoint for RNA trafficking between the phloem and nonvascular tissues. The function of the bundle sheath is elusive in C3 plants, including tobacco. Its strategic location between the vascular tissue and the mesophyll suggests a role in regulating transport between the vascular and nonvascular tissues. The results from this study provide one piece of evidence to support this hypothesis.
The Role of an RNA Motif in Mediating Trafficking Is Dependent on Development
The finding that bundle sheath-to-mesophyll trafficking of PSTVdNT does not occur in leaves up to the stage of maturation into a source for photoassimilates but occurs in older leaves reveals a novel aspect of developmental regulation of RNA trafficking. It is interesting to note that although PSTVdNT can invade mesophyll cells in older leaves, such invasion is not as extensive as PSTVdNB invasion. The basis for this difference is unclear. Perhaps the bipartite motif identified as essential for bundle sheath-to-mesophyll trafficking has a supporting, but not decisive, role also in trafficking between mesophyll cells and/or between the mesophyll and epidermis. Lacking this motif, PSTVdNT trafficking between these cells is less efficient. Further experimental studies are required to address this issue.
Developmental regulation of intercellular PSTVd RNA trafficking echos, but does not mirror, the developmental regulation of intercellular protein trafficking. In tobacco leaf epidermis, signal-directed cell–cell trafficking of the Cucumber mosaic virus 3a movement protein occurs in source but not sink leaves (Itaya et al., 1998, 2000). By contrast, cell–cell diffusion of GFP occurs in sink but not source leaves (Imlau et al., 1999; Oparka et al., 1999; Itaya et al., 2000). Altogether, these observations suggest that during leaf development, the symplasmic network continuously remodels its capacity to traffic proteins and RNAs of diverse nature between various cells. A key feature of this remodeling is presumably turnover of the molecular compositions of plasmodesmata as well as dedicated cellular trafficking factors. This dynamic regulation of macromolecular trafficking may be important for development. A critical test of this hypothesis will be identification of the macromolecules that traffic between specific cells and at specific developmental stages and to elucidate the molecular mechanisms that control such trafficking.
Broad Implications of Motif-Mediated RNA Trafficking
Our results suggest that motif-mediated trafficking of an RNA across a specific cellular interface and in a defined direction is a basic mechanism for a plant to sort and target various RNAs to the correct cellular sites for function. This model, however, does not exclude alternative mechanisms and pathways. Pathogens such as viroids and viruses have evolved to adapt the endogenous trafficking control machinery to achieve successful systemic infection. Studies on viral movement have largely focused on the role of viral proteins (reviewed in Carrington et al., 1996; Ghoshroy et al., 1997; Ding, 1998; Lazarowitz and Beachy, 1999). There is indication that some viral RNA may play a direct role in systemic movement (Lauber et al., 1998; Qiu and Scholthof, 2000). Further studies on the role of viral RNAs as well as viroids in trafficking should continue to contribute new insights into the inner workings of the endogenous trafficking machinery in addition to enhancing understanding of the coevolution of pathogen–host interactions that promote pathogen systemic infection or host defense/resistance.
Given that RNA trafficking across different cellular boundaries may be uniquely regulated, understanding systemic RNA trafficking requires detailed analysis of how each cellular boundary functions and how the functions of all cellular boundaries can be integrated. In this regard, the experimental approaches we have developed should be useful. By expressing an RNA in a specific cell type, one can track its movement into other cell types. In conjunction with mutational analysis, the transgenic approach should allow identification of RNA motifs that mediate trafficking between specific cell types.
Systemic RNA trafficking is not unique to plants. There is a strong indication that the mobile signals that traffic intercellularly to potentiate systemic RNA silencing in plants (Palauqui et al., 1997; Voinnet and Baulcombe, 1997; Voinnet et al., 1998), Caenorhabditis elegans (Fire et al., 1998), and Tribolium (Coleoptera) (Bucher et al., 2002) consist of RNAs, at least partially. The occurrence of intercellular RNA trafficking in these diverse organisms supports the hypothesis that such trafficking has an ancient evolutionary origin (Ding et al., 1999). Further studies on this trafficking in various organisms may shed new light on the evolution of intercellular communication mechanisms and RNA functions and help to understand the modern role of such trafficking in organismal development and physiology.
METHODS
Plant Material and Growth Conditions
Tobacco plants (Nicotiana tabacum cv Samsum n/n) were grown in a growth chamber with a 27/22°C day/night temperature regime and a 14-h/10-h light/dark cycle. Suspension cells of tobacco BY2 (N. tabacum var Bright Yellow 2) were cultured in Murashige and Skoog medium (MS salts; Life Technologies, Rockville, MD) supplemented with 30 g/L of sucrose, 256 mg/L of KH2PO4, 100 mg/L of myo-inositol, 1 mg/L of thiamine, and 1 mg/L of 2,4-D with a final pH of 5.5 adjusted with 1 M KOH. Cells were maintained by shaking at 250 rpm in constant light at 24°C and subcultured weekly by 1:25 dilution.
Recovery of the PSTVdNT and PSTVdNB Isolates
Isolation of the PSTVdNT strain was previously described by Wassenegger and coworkers (1996). The PSTVdNB variant was first isolated from N. benthamiana plants that were mechanically inoculated with total RNA from the transgenic tobacco SR1-4(−)/6 line. This plant line contained 4.4 head-to-tail linked copies of the PSTVdKF440-2 cDNA in which the CaMV 35S promoter–driven transcription of the PSTVd transgene resulted in viroid infection (Wassenegger et al., 1994). Although it was demonstrated that in primary SR1-4(−) transformants the original autonomously replicating viroid represented the PSTVdNT strain (Wassenegger et al., 1996), it turned out that in the SR1-4(−)/6 line that was maintained for several years in the greenhouse by propagating cuttings; the PSTVdNT further evolved into PSTVdNB variant. Thus, PSTVdNB was not an adaptation of the PSTVdNT to N. benthamiana but was the result of a long-term selection in tobacco SR1-4(−)/6. Compared with PSTVdKF440-2, the PSTVdNT displayed a C → U nucleotide substitution at position 259 (Wassenegger et al., 1996). The development of the PSTVdNB involves five additional substitutions at positions 47, 201, 309, 313, and 315 (Figure 1A; see Supplemental Figure 1 online). Isolation of RNAs from N. benthamiana and N. tabacum plants, cDNA synthesis, PCR amplification, and cloning and sequencing of the PSTVdNB were performed according to the procedures described by Wassenegger (2001) using the 325-H/326-R and the 175-H/176-R PCR primer pairs.
PSTVd cDNA Constructs
Construction of plasmids pInter(+), pInter(−), and pRZ:NT was described by Qi and Ding (2002). The 294-bp EagI-Eco47III fragment (nucleotides 145 to 359/1 to 79) of PSTVdNB cDNA was transferred to pRZ:NT by replacing the corresponding region of PSTVdNT, giving rise to plasmid pRZ:NB. Plasmids pRZ:NBU201G, pRZ:NBU309A, pRZ:NBU47A, pRZ:NBA313U, pRZ:NBU47A/A313U, pRZ:NBC315U, pRZ:NTG201U, pRZ:NTA309U, pRZ:NTA47U/U313A, and pRZ:NTU315C were generated by site-directed mutagenesis using the Quickchange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions using plasmid pRZ:NB or pRZ:NT as the template and complementary primers with sequences corresponding or complementary to PSTVd sequences with the desired mutations. The introduced mutations were verified by sequencing.
For inoculating plants with PSTVd variants by biolistic bombardment, viroid cDNA fragments flanked by ribozymes in pRZ:NT, pRZ:NB, and pRZ:NT/NB-derived mutants were excised by digestion with SmaI and inserted into the SmaI site, downstream of the CaMV 35S promoter, of vector pRTL2 (Carrington and Freed, 1990; Restrepo et al., 1990).
Binary Vector Construction for Transgenic Expression of PSTVd Variants
For expression of PSTVd variants in transgenic plants, two types of constructs were made. First, the 35S:PSTVd fragments in the pRTL2 constructs were excised by digestion with HindIII and then transferred into the binary vector pGA482. Second, SmaI-SmaI fragments of pRZ:PSTVd were cloned by conventional blunt-end ligation at the SmaI site of binary vector pCOI (provided by Gary Thompson University of Arkansas, Little Rock), downstream of the companion cell-specific CoYMV promoter (Matsuda et al., 2002).
Binary Vector Construction for Transgenic Expression of DsRed2 Protein
To construct the dimeric DsRed2 gene, monomeric DsRed2 gene was obtained by PCR amplification from pDsRed2-C1 (Clontech, Palo Alto, CA) using primers designed to incorporate a BamHI site at the 3′-end without stop codon. The PCR fragment was digested with SacI and cloned into pRTL2 that was linearized by digestion with NcoI (blunted by Pfu DNA polymerase) and SacI, which gave rise to construct pRTL2:mDsRed. Similarly, another monomeric DsRed2 gene with a stop codon was amplified by PCR to incorporate a SacI site at the 5′-end and a BamHI site at the 3′-end and was inserted into pRTL2:mDsRed between the SacI and BamHI sites downstream of the first DsRed2 gene, resulting in pRTL2:dDsRed. Finally, the fusion fragment of TEV leader sequence and dimeric DsRed2 gene was excised from pRTL2:dDsRed by digestion with EcoRI and BamHI, filled in by treatment with Pfu DNA polymerase, and cloned into the SmaI site of pCOI binary vector downstream of the companion cell-specific CoYMV promoter.
Tobacco Transformation
After verification of sequences and insert orientations, the above binary constructs were transferred into Agrobacterium tumefaciens (LBA4404). A standard Agrobacterium-mediated leaf disc transformation method (Horsch et al., 1985) was used to generate transgenic tobacco plants. Positive PSTVd-transgenic lines were identified by RNA gel blots to detect the circular (+)-PSTVd. Positive dimeric DsRed-transgenic plants were selected by DsRed fluorescence using a Nikon E600 epifluorescence microscope (Nikon, Tokyo, Japan) with a filter set consisting of an excitation filter of 540 to 580 nm, a dichroic mirror of 595 nm, and a barrier filter of 600 to 620 nm.
In Vitro Transcription
To prepare in vitro transcripts of the PSTVd variants for plant or protoplast inoculation, plasmids pRZ:NT, pRZ:NB, and pRZ:NT/NB-derived mutants were linearized by HindIII and used as templates for in vitro transcription using the T7 MEGAscript kit (Ambion, Austin, TX) following the manufacturer's directions.
To prepare riboprobes for RNA gel blots or in situ hybridization, [α-32P]- or digoxigenin-UTP-labeled antisense and sense riboprobes were prepared by in vitro transcription using the T7 Maxiscript kit (Ambion) following the methods recommended by the manufacturer, using SpeI-linearized pInter(−) and pInter(+) as the templates, respectively. After in vitro transcription, the DNA templates were removed by digestion with RNase-free DNase I. The RNA transcripts were purified with the MEGAclear kit (Ambion). Nonradioactive RNA transcripts were quantified by UV spectrometry.
Protoplast and Plant Inoculation
Protoplasts were prepared from tobacco BY-2 cultured cells and inoculated with PSTVd transcripts as described (Qi and Ding, 2002). At 3 d postinoculation, the protoplasts were collected for RNA extraction.
Tobacco seedlings were inoculated with PSTVd using two methods: (1) young leaves of tobacco seedlings were dusted with carborundum and mechanically inoculated with PSTVd in vitro transcripts (100 ng/μL) or with water alone as mock inoculum; (2) young leaves of tobacco seedlings were bombarded with gold particles coated with plasmid pRTL2:PSTVd as described (Itaya et al., 1997). Empty vector without PSTVd insert was bombarded as mock inoculum. At 4 to 5 weeks postinoculation, systemic leaves were collected for RNA extraction and processed for in situ hybridization.
Transient Expression of Dimeric GFP
To verify that biolistic bombardment only delivered a plasmid to the epidermis, we bombarded plasmid pRTL2:dGFP (dimeric GFP) (Itaya et al., 2002b), which contains cDNAs to express a dimeric GFP under the control of the CaMV 35S promoter, into leaves of CoYMV:dDsRed-transgenic tobacco. The leaf epidermis and transverse leaf sections were examined under a PCM2000 confocal laser scanning microscope (Nikon) with argon and green HeNe lasers to detect GFP and DsRed fluorescence, respectively. Such analyses allowed determination that GFP fluorescence was only found in individual epidermal cells and was never found in the phloem cells (containing DsRed fluorescence).
Determination of Sink–Source Status of Tobacco Leaves
The sink/source status of a leaf in PSTVd-infected or PSTVd-transgenic tobacco plants was determined by comparison with leaves of comparable developmental stages in two other types of tobacco plants, all growing under identical conditions. First, in transgenic tobacco expressing the Cucumber mosaic virus 3a movement protein fused to GFP under the control of the CaMV35S promoter, the fusion protein is targeted to plasmodesmata in source leaves but not in sink leaves (Itaya et al., 1998, 2000). Second, free GFP transported in the phloem remains in the phloem of a source leaf but diffuses from the phloem into mesophyll in a sink leaf (Oparka et al., 1999). We constructed a graft union between 35S:GFP-transgenic tobacco (Itaya et al., 1998) rootstock and nontransgenic tobacco scion. The GFP from the rootstock moved into the scion via the phloem. In the scion, diffusion of the GFP from the phloem in mesophyll marks a leaf as sink and retention in the phloem marks the leaf as source.
RNA Extraction and RNA Gel Blotting
Total RNAs were isolated from plants or protoplasts using the RNeasy plant mini kit (Qiagen, Valencia, CA) and quantified by UV spectrometry. For RNA gel blot analysis, RNA aliquots were fractionated by electrophoresis at 55°C in 5% polyacrylamide gels containing 1× Tris-borate/EDTA buffer/8 M urea. After electrophoresis, the gels were stained with ethidium bromide and examined under UV light to determine the integrity and equal loading of the RNA samples. The RNAs were transferred to a Hybond-XL nylon membrane using a vacuum blotting system (Amersham, Piscataway, NJ). Hybridizations were performed at 65°C with ULTRAhyb reagent (Ambion) and in vitro transcribed [α-32P]-UTP–labeled antisense riboprobes. The membranes were washed at 68°C and exposed to storage phosphor screen (Kodak, Rochester, NY). Quantification of the radioactivity was performed by Molecular Imager FX using Quantity One-4.1.1 software (Bio-Rad, Hercules, CA).
Sequencing of RNA Progeny
The protocols for preparing cDNAs of the PSTVd progeny isolated from the infected plants or protoplasts were essentially as described by Qi and Ding (2002). Briefly, cDNAs of PSTVd RNA were RT-PCR amplified and sequenced in both directions using the ABI377 DNA sequencer (Perkin-Elmer, Boston, MA) at the DNA Sequencing Facility at Ohio State University.
Dot Blot Hybridization
To test whether (+)- and (−)-PSTVd probes would cross-hybridize to PSTVd RNAs of the same polarity, dot blot hybridization was performed with PSTVd RNAs of a series of dilutions. Aliquots (2 μL) of RNAs were spotted onto nylon membranes, and the membranes were irradiated with 1.2 × 105 μJ of UV light in a Stratalinker UV cross-linker (Stratagene). Prehybridization was performed at 50°C for 1 h with a hybridization solution containing 50% formamide, 5% dextran sulfate, 1% blocking reagent (Boehringer Mannheim, Indianapolis, IN), 0.3 M NaCl, 10 mM Tris-HCl, 1 mM EDTA, and 150 μg/mL of yeast tRNA, pH 7.5. Afterwards, a fresh solution containing 300 ng/mL of digoxigenin-labeled PSTVd RNA probes was added, and the membranes were incubated at 50°C for 19 to 20 h. After two washes with 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) at 50°C (15 min each) and incubation (20 min at room temperature) in 2× NTE buffer (500 mM NaCl, 10 mM Tris, and 1 mM EDTA, pH 8.0) containing 2 μg/mL of RNaseA, the membranes were washed twice (10 min each) with 0.2× SSC containing 0.1% SDS at 65°C and once (3 min) with maleate buffer (100 mM maleic acid and 150 mM NaCl, pH 7.5). The washed membranes were blocked for 30 min in maleate buffer containing 1% blocking reagent (Boehringer Mannheim) and incubated for 30 min with alkaline phosphatase–conjugated anti-digoxigenin antibodies (Boehringer Mannheim; 1:5,000 dilution). After two washes (15 min each) in maleate buffer and one wash (10 min) in AP buffer (100 mM Tris-HCl, 100 mM NaCl, and 50 mM MgCl2, pH 9.5), the membranes were incubated in color substrate solution (100 μL of nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate in 5 mL of AP buffer; Boehringer Mannheim) in the dark. Once color was fully developed, the membranes were washed with TE buffer and water (10 min each) and air dried.
Tissue Processing and In Situ Hybridization
Leaf samples were fixed, frozen, and embedded essentially as described by Itaya et al. (1998). The samples were sectioned to 12-μm thickness using a Microm HM500 cryostat (Walldorf, Germany). In situ hybridization was performed as described previously (Zhu et al., 2001) with modifications, using digoxigenin-labeled sense and antisense PSTVd riboprobes. Sections were incubated in PBS for 5 min and then permeabilized by proteinase K treatment for 30 min. Prehybridization, hybridization with digoxigenin-labeled PSTVd probes, immunodetection, and color development were performed essentially as described above, except that maleate buffer was replaced with buffer I (100 mM Tris-HCl and 150 mM NaCl, pH 8.0). Afterwards, the sections were examined and photographed with the Nikon Eclipse 600 light microscope. Images were captured and processed with a SPOT 2 Slider CCD camera and the associated software (Diagnostics Instruments, Sterling Heights, MI).
Statistical Analysis of the Localization Patterns of PSTVdNT and PSTVdNB
For each PSTVd strain in a leaf, >400 loci (a locus is defined as a vein and associated nonvascular tissues) in 144 in situ hybridization sections from six independent biological repeats were investigated. The maximum distance from phloem (layers of cells) where the viroid was detected was determined. The data were used to categorize five localization patterns: phloem limitation, bundle sheath, one to two layers of mesophyll cells, three to four layers of mesophyll cells, and more than five layers of mesophyll cells. The average percentage of each pattern was calculated and presented with the standard deviation.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AJ634596 (PSTVdNB sequence).
Supplementary Material
Acknowledgments
We thank Richard Nelson and Xuehua Zhong for helpful discussions and comments on the manuscript. We are indebted to the anonymous reviewers for their constructive suggestions. This work was supported by USDA Grant NRICGP 2001-35303-11073 and National Science Foundation Grant IBN-0238412 to B.D. and Deutsche Forschungsgemeinschaft Grants Wa 1019/1-1 to 1-4 to M.W.
Online version contains Web-only data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Biao Ding (ding.35@osu.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.021980.
References
- Branch, A.D., and Robertson, H.D. (1984). A replication cycle for viroids and other small infectious RNA's. Science 223, 450–455. [DOI] [PubMed] [Google Scholar]
- Bucher, G., Scholten, J., and Klingler, M. (2002). Parental RNAi in Tribolium (Coleoptera). Curr. Biol. 12, R85–R86. [DOI] [PubMed] [Google Scholar]
- Carrington, J.C., and Freed, D.D. (1990). Cap-independent enhancement of translation by a plant potyvirus 5′ nontranslated region. J. Virol. 64, 1590–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington, J.C., Kasschau, K.D., Mahajan, S.K., and Schaad, M.C. (1996). Cell-to-cell and long distance transport of viruses in plants. Plant Cell 8, 1669–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky, V., Ghoshroy, S., Tsui, F., and Klessig, D. (1998). Non-toxic concentrations of cadmium inhibit systemic movement of turnip vein clearing virus by a salicylic acid-dependent mechanism. Plant J. 16, 13–20. [DOI] [PubMed] [Google Scholar]
- Citovsky, V., and Zambryski, P. (2000). Systemic transport of RNA in plants. Trends Plant Sci. 5, 52–54. [DOI] [PubMed] [Google Scholar]
- Diener, T.O. (2001). The viroid: Biological oddity or evolutionary fossil? Adv. Virus Res. 57, 137–184. [DOI] [PubMed] [Google Scholar]
- Ding, B. (1998). Intercellular protein trafficking through plasmodesmata. Plant Mol. Biol. 38, 279–310. [PubMed] [Google Scholar]
- Ding, B., Itaya, A., and Woo, Y. (1999). Plasmodesmata and cell-to-cell communication in plants. Int. Rev. Cytol. 190, 251–316. [Google Scholar]
- Ding, B., Itaya, A., and Qi, Y. (2003). Symplasmic protein and RNA traffic: Regulatory points and regulatory factors. Curr. Opin. Plant Biol. 6, 596–602. [DOI] [PubMed] [Google Scholar]
- Ding, B., Kwon, M.O., Hammond, R., and Owens, R. (1996). Cell-to-cell movement of potato spindle tuber viroid. Plant J. 12, 931–936. [DOI] [PubMed] [Google Scholar]
- Ding, B., Li, Q., Nguyen, L., Palukaitis, P., and Lucas, W.J. (1995). Cucumber mosaic virus 3a protein potentiates cell-to-cell trafficking of CMV RNA in tobacco plants. Virology 207, 345–353. [DOI] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Flores, R., Di Serio, F., and Hernández, C. (1997). Viroids: The non-encoding genomes. Semin. Virol. 8, 65–73. [Google Scholar]
- Flores, R., Randles, J.W., Bar-Josef, M., and Diener, T.O. (2000). Subviral agents: Viroids. In Virus Taxonomy, Seventh Report of the International Committee on Taxonomy of Viruses, M.H.V. van Regenmortel, C.M. Fauquet, D.H.L. Bishop, E.B. Carstens, M.K. Estes, S.M. Lemon, J. Manilof, M.A., Mayo, D.J. McGeoch, C.R. Pringle, and R.B. Wickner, eds (San Diego, CA: Academic Press), pp. 1009–1024.
- Fujiwara, T., Giesmann-Cookmeyer, D., Ding, B., Lommel, S.A., and Lucas, W.J. (1993). Cell-to-cell trafficking of macromolecules through plasmodesmata potentiated by the red clover necrotic mosaic virus movement protein. Plant Cell 5, 1783–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoshroy, S., Lartey, R., Sheng, J., and Citovsky, V. (1997). Transport of proteins and nucleic acids through plasmodesmata. Annu. Rev. Plant Physiol. Plant Mol. Biol 48, 27–50. [DOI] [PubMed] [Google Scholar]
- Ghoshroy, S., Freedman, K., Lartey, R., and Citovsky, V. (1998). Inhibition of plant viral systemic infection by non-toxic concentrations of cadmium. Plant J. 13, 591–602. [DOI] [PubMed] [Google Scholar]
- Gilbertson, R.L., and Lucas, W.J. (1996). How do viruses traffic on the ‘vascular highway’? Trends Plant Sci. 1, 260–268. [Google Scholar]
- Goodrick, B.J., Khun, C.W., and Hussey, R.S. (1991). Restricted systemic movement of cowpea chlorotic mottle virus in soybean with nonnecrotic resistance. Phytopathology 81, 1426–1431. [Google Scholar]
- Gross, H.J., Domdey, H., Lossow, C., Jank, P., Raba, M., Alberty, H., and Sanger, H.L. (1978). Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature 273, 203–208. [DOI] [PubMed] [Google Scholar]
- Haywood, V., Kragler, F., and Lucas, W.J. (2002). Plasmodesmata: Pathways for protein and ribonucleoprotein signaling. Plant Cell 14 (suppl.), S303–S325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch, R.B., Fry, J., Hoffmann, N.L., Wallroth, M., Eichholtz, D., Rogers, S.G., and Fraley, R.T. (1985). A simple and general method for transferring genes into plants. Science 227, 1229–1231. [DOI] [PubMed] [Google Scholar]
- Imlau, A., Truernit, E., and Sauer, N. (1999). Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink transport. Plant Cell 11, 309–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itaya, A., Hickman, H., Bao, Y., Nelson, R., and Ding, B. (1997). Cell-to-cell trafficking of cucumber mosaic virus movement protein:green fluorescent protein fusion produced by biolistic bombardment in tobacco. Plant J. 12, 1223–1230. [Google Scholar]
- Itaya, A., Liang, G., Woo, Y.-M., Nelson, R., and Ding, B. (2000). Nonspecific intercellular protein trafficking probed by green-fluorescent protein in plants. Protoplasma 213, 165–175. [Google Scholar]
- Itaya, A., Ma, F., Qi, Y., Matsuda, Y., Zhu, Y., Liang, G., and Ding, B. (2002. b). Plasmodesma-mediated selective protein traffic between “symplasmically isolated” cells probed by a viral movement protein. Plant Cell 14, 2071–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itaya, A., Matsuda, Y., Gonzales, R., Nelson, R.S., and Ding, B. (2002. a). Potato spindle tuber viroid strains of different pathogenicity induced and suppressed expression of common and unique genes in infected tomato. Mol. Plant-Microbe Interact. 15, 990–999. [DOI] [PubMed] [Google Scholar]
- Itaya, A., Woo, Y.M., Masuta, C., Bao, Y., Nelson, R.S., and Ding, B. (1998). Developmental regulation of intercellular protein trafficking through plasmodesmata in tobacco leaf epidermis. Plant Physiol. 118, 373–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, R.A. (2002). RNA traffics information systemically in plants. Proc. Natl. Acad. Sci. USA 99, 11561–11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, R.A., Atkinson, R.G., Forster, R.L.S., and Lucas, W.J. (1998). An RNA-based information superhighway in plants. Science 279, 1486–1487. [DOI] [PubMed] [Google Scholar]
- Kim, M., Canio, W., Kessler, S., and Sinha, N. (2001). Developmental changes due to long-distance movement of a homeobox fusion transcript in tomato. Science 293, 287–289. [DOI] [PubMed] [Google Scholar]
- Kühn, C., Franceschi, V.R., Schulz, A., Lemoine, R., and Frommer, W.B. (1997). Macromolecular trafficking indicated by localization and turnover of sucrose transporters in enucleate sieve elements. Science 275, 1298–1300. [DOI] [PubMed] [Google Scholar]
- Lauber, E., Guilley, H., Tamada, T., Richards, K.E., and Jonard, G. (1998). Vascular movement of beet necrotic yellow vein virus in Beta macrocarpa is probably dependent on an RNA3 sequence domain rather than a gene product. J. Gen. Virol. 79, 385–393. [DOI] [PubMed] [Google Scholar]
- Lazarowitz, S.G., and Beachy, R.N. (1999). Viral movement proteins as probes for intracellular and intercellular trafficking in plants. Plant Cell 11, 535–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas, W.J., Bouche-Pillon, S., Jackson, D.P., Nguyen, L., Baker, L., Ding, B., and Hake, S. (1995). Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 270, 1980–1983. [DOI] [PubMed] [Google Scholar]
- Lucas, W.J., Ding, B., and Van der Schoot, C. (1993). Plasmodesmata and the supracellular nature of plants. New Phytol. 125, 435–476. [DOI] [PubMed] [Google Scholar]
- Lucas, W.J., Yoo, B.C., and Kragler, F. (2001). RNA as a long-distance information macromolecule in plants. Nat. Rev. Mol. Cell Biol. 2, 849–857. [DOI] [PubMed] [Google Scholar]
- Matsuda, Y., Liang, G., Zhu, Y., Ma, F., Nelson, R.S., and Ding, B. (2002). The Commelina yellow mottle virus promoter drives companion cell-specific gene expression in multiple organs of transgenic tobacco. Protoplasma 220, 51–58. [DOI] [PubMed] [Google Scholar]
- Mlotshwa, S., Voinnet, O., Mette, M.F., Matzke, M., Vaucheret, H., Ding, S.W., Pruss, G., and Vance, V.B. (2002). RNA silencing and the mobile silencing signal. Plant Cell 14 (suppl.), S289–S301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, L., Lucas, W.J., Ding, B., and Zaitlin, M. (1996). Viral trafficking is inhibited in replicase-mediated resistant transgenic tobacco plants. Proc. Natl. Acad. Sci. USA 93, 12643–12647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oparka, K.J., Roberts, A.G., Boevink, P., Santa Cruz, S., Roberts, I., Pradel, K.S., Imlau, A., Kotlizky, G., Sauer, N., and Epel, B. (1999). Simple, but not branched, plasmodesmata allow the nonspecific trafficking of proteins in developing tobacco leaves. Cell 97, 743–754. [DOI] [PubMed] [Google Scholar]
- Palauqui, J.C., Elmayan, T., Pollien, J.M., and Vaucheret, H. (1997). Systemic acquired silencing: Transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 16, 4738–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palukaitis, P. (1987). Potato spindle tuber viroid: Investigation of the long-distance, intra-plant transport route. Virology 158, 239–241. [DOI] [PubMed] [Google Scholar]
- Qi, Y., and Ding, B. (2002). Replication of Potato spindle tuber viroid in cultured cells of tobacco and Nicotiana benthamiana: The role of specific nucleotides in determining replication levels for host adaptation. Virology 302, 445–456. [DOI] [PubMed] [Google Scholar]
- Qi, Y., and Ding, B. (2003. a). Inhibition of cell growth and shoot development by a specific nucleotide sequence in a noncoding viroid RNA. Plant Cell 15, 1360–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, Y., and Ding, B. (2003. b). Differential subnuclear localization of RNA strands of opposite polarity derived from an autonomously replicating viroid. Plant Cell 15, 2566–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, W., and Scholthof, K.-B.G. (2000). In vitro and in vivo-generated defective RNAs of satellite panicum mosaic virus defines cis-acting RNA elements required for replication and movement. J. Virol. 74, 2247–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo, M.A., Freed, D.D., and Carrington, J.C. (1990). Nuclear transport of plant potyviral proteins. Plant Cell 2, 987–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Medrano, R., Xoconostle-Cazares, B., and Lucas, W.J. (1999). Phloem long-distance transport of CmNACP mRNA: Implications for supracellular regulation in plants. Development 126, 4405–4419. [DOI] [PubMed] [Google Scholar]
- Sanger, M., Passmore, B., Falk, B.W., Bruening, G., Ding, B., and Lucas, W.J. (1994). Symptom severity of beet western yellows virus strain ST9 is conferred by the ST9-associated RNA and is not associated with virus release from the phloem. Virology 200, 48–55. [DOI] [PubMed] [Google Scholar]
- Ueki, S., and Citovsky, V. (2001). RNA commutes to work: Regulation of plant gene expression by systemically transported RNA molecules. Bioessays 23, 1087–1090. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., and Baulcombe, D.C. (1997). Systemic signalling in gene silencing. Nature 389, 553. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., Vain, P., Angell, S., and Baulcombe, D.C. (1998). Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell 95, 177–187. [DOI] [PubMed] [Google Scholar]
- Wang, H.L., Wang, Y., Giesman-Cookmeyer, D., Lommel, S.A., and Lucas, W.J. (1998). Mutations in viral movement protein alter systemic infection and identify an intercellular barrier to entry into the phloem long-distance transport system. Virology 245, 75–89. [DOI] [PubMed] [Google Scholar]
- Wassenegger, M. (2001). Advantages and disadvantages of using PCR techniques to characterize transgenic plants. Mol. Biotech 17, 73–82. [DOI] [PubMed] [Google Scholar]
- Wassenegger, M., Heimes, S., Riedel, L., and Sänger, H.L. (1994). RNA-directed de novo methylation of genomic sequences in plants. Cell 76, 567–576. [DOI] [PubMed] [Google Scholar]
- Wassenegger, M., Spieker, R.L., Thalmeir, S., Gast, F.U., Riedel, L., and Sanger, H.L. (1996). A single nucleotide substitution converts potato spindle tuber viroid (PSTVd) from a noninfectious to an infectious RNA for nicotiana tabacum. Virology 226, 191–197. [DOI] [PubMed] [Google Scholar]
- Water, A.E., Turner, D.H., Kim, J., Lyttle, M.H., Muller, P., Mathews, D.H., and Zuker, M. (1994). Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc. Natl. Acad. Sci. USA 91, 9218–9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermantel, W.M., Banerjee, N., Oliver, J.C., Paolillo, D.J., and Zaitlin, M. (1997). Cucumber mosaic virus is restricted from entering minor veins in transgenic tobacco exhibiting replicase-mediated resistance. Virology 231, 248–257. [DOI] [PubMed] [Google Scholar]
- Wu, X., Weigel, D., and Wigge, P.A. (2002). Signaling in plants by intercellular RNA and protein movement. Genes Dev. 16, 151–158. [DOI] [PubMed] [Google Scholar]
- Zhu, Y., Green, L., Woo, Y.-M., Owens, R., and Ding, B. (2001). Cellular basis of potato spindle tuber viroid systemic movement. Virology 279, 69–77. [DOI] [PubMed] [Google Scholar]
- Zhu, Y., Qi, Y., Xun, Y., Owens, R., and Ding, B. (2002). Movement of Potato spindle tuber viroid reveals regulatory points of phloem-mediated RNA traffic. Plant Physiol. 130, 138–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.