Abstract
Calcineurin (CN) is a unique calcium/calmodulin (CaM)-activated serine/threonine phosphatase. To perform its diverse biological functions, CN communicates with many substrates and other proteins. In the physiological activation of T cells, CN acts through transcriptional factors belonging to the NFAT family and other transcriptional effectors. The classic immunosuppressive drug cyclosporin A (CsA) can bind to cyclophilin (CyP) and compete with CN for the NFAT LxVP motif. CsA has debilitating side effects, including nephrotoxicity, hypertension and tremor. It is desirable to develop alternative immunosuppressive agents. To this end, we first tested the interactions between CN and the LxVP-type substrates, including endogenous regulators of calcineurin (RCAN1) and NFAT. Interestingly, we found that quercetin, the primary dietary flavonol, can inhibit the activity of CN and significantly disrupt the associations between CN and its LxVP-type substrates. We then validated the inhibitory effects of quercetin on the CN-NFAT interactions in cell-based assays. Further, quercetin also shows dose-dependent suppression of cytokine gene expression in mouse spleen cells. These data raise the possibility that the interactions of CN with its LxVP-type substrates are potential targets for immunosuppressive agents.
Keywords: calcineurin, quercetin, RCAN1, NFAT, LxVP-motif, cyclosporine A, cytokine expression
1. Introduction
Calcineurin (CN), a serine/threonine phosphatase, is highly expressed in the brain. It comprises a catalytic subunit (CNA; 61 kDa) and a regulatory subunit (CNB; 19 kDa). CNA consists of four regions: the catalytic domain includes 20-340 amino acid residues, the CNB-binding domain comprises 349-372 amino acid residues, the calmodulin-binding segment comprises 390-414 amino acid residues, and a C-terminal autoinhibitory domain comprises 469-486 amino acid residues [1]. CN is a key player transducing the information from calcium signals to effectors that control cellular responses and gene transcription [2]. To perform its multiple biological functions, CN interacts with a large number of substrates and other proteins.
In mammals, many CN-dependent processes involve the nuclear factor of activated T cells (NFAT) family of transcription factors. In resting cells, the hyperphosphorylated NFAT proteins mainly reside in the cytoplasm. Upon activation, CN dephosphorylates these proteins, which then translocate to the nucleus. In the nucleus, they regulate gene transcription via interaction with other transcription factors [3,4]. Two domains of an NFAT protein that interact with CN have been defined: the PxIxIT motif, situated near the N terminus of the regulatory region [5,6] of NFAT and the LxVP motif, located near the C terminus [7,8].
The LxVP motif has attracted renewed attention [9,10]. It binds to a hydrophobic pocket at the interface of the CNA and CNB subunits. Binding requires Ca2+ and calmodulin (CaM), which indicates that the interaction occurs only with activated CN. Glutathione S-transferase (GST) pull-down data obtained with mutations in the conserved docking surface on CN, and analyses using the AutoDock software, predict that LxVP-containing sequences have a similar mechanism of action to cyclosporin A (CsA)-cyclophilin (CyP) complexes, which implies that the hydrophobic pocket in CN binds not only NFAT but also CsA-CyP complexes, the latter inhibiting activation of NFAT.
RCAN proteins are endogenous CN regulators, conserved from yeast to humans, which have both positive and negative roles in CN signaling. It has been shown that p38α MAP kinase can phosphorylate RCAN1 at multiple sites in vitro and that phospho-RCAN1 is an efficient substrate for CN [11–14]. Rodriguez et al. also found that a sequence on yeast Rcn1 closely matches the LxVP-type motif of NFAT (KQYLKVPESEKVF, aa 98-110) [9]. This motif in Rcn1 mediates the interaction between CN and Rcn1. Subsequently, Grigoriu et al. predicted that the HLAPP motif of human RCAN1 is an LxVP-type motif [10]. In the present study, we developed cell-free assays for the interactions between CN and LxVP-type substrates, namely RCAN1 and NFAT.
In a previous study, we showed that quercetin inhibits CN in vitro and in Jurkat cells [15]. In this study, we show that quercetin inhibits the interactions of CN with the LxVP-type motifs in its substrates. We also show that quercetin inhibits the CN-NFAT interaction in cell-based assays as well as NFAT nuclear import and NFAT-mediated cytokine gene expression. We conclude that quercetin inhibits CN signaling by interacting with LxVP-type sites on CN substrates. This suggests the possibility that the interactions of CN with LxVP-type substrates may be useful targets for screening immunosuppressive agents.
2. Materials and Methods
2.1 Materials
The RII peptide, a CN substrate, was purchased from Biomol Research Laboratories, Inc. (PA, USA). CsA was purchased from Sigma Chemical Co. (MI, USA). Quercetin was from Melone Pharmaceutical Co. (Dalian, China). Peptides were synthesized by Scilight-Peptide Co. (Beijing, China). Other reagents were of the highest quality obtainable from commercial suppliers.
2.2 Preparation of mouse brain lysates
Male Kunming mice (weight 16 ± 2 g, 4 weeks of age) were obtained from the Experimental Animal Center of Peking University. They were housed in at 20 ± 1°C, and 40–60% humidity, on a12:12-L/D light cycle. The mice were anesthetized with sodium pentobarbital, and all experimental procedures were approved by the Animal Ethics Committee of Beijing Normal University. After the mice were killed, their brains were removed and homogenized by passage via syringe into a solution of 50 mM Tris-HCl, pH 7.5, 0.1 mM EDTA, 0.1 mM EGTA, 1.0 mM dithiothreitol, 0.2% NP-40, 1.0 mM phenylmethylsulfonyl fluoride, 5 μg/ml leupeptin, 5 μg/ml aprotinin, and 2 μg/ml pepstatin at 4°C. After sonication, the homogenate was centrifuged at 16,000 × g and 4°C for 60 min, and the supernatant was used as a source of CN in GST pull-down assays.
2.3 Expression of GST fusion proteins, pull-down assays, and western blotting
Plasmids encoding peptides fused to GST were obtained by cloning overhang-double-stranded annealed oligonucleotides into Eco RI + Xho I-digested pGEX-4T-1 plasmid [9]. The sequences of the oligonucleotides used as GST-peptide fusion proteins were as follows: HLAPP sense, 5′AATTCGGAAGTTCACACCTGGCTCCGCCCAATCCCGACAAACAGTTCCTCTAAC3′; HLAPP antisense, 5′TCGAGTTAGAGGAACTGTTTGTCGGGATTGGGCGGAGCCAGGTGTGGAACTTCCG3′; YLAVP sense, 5′ AATTCGATCAGTACTTGGCCGTACCACAGCATCCGTATCAATGGGCTAAGTAAC3′; YLAVP antisense, 5′TCGAGTTACTTAGCCCATTGATACGGATGCTGTGGTACGGCCAAGTACTGATCG3′.
The GST fusion proteins were expressed in E. coli and proteins were quantified by the Bradford procedure. Unless otherwise specified, all pull-down experiments were performed in 50 mM Tris-HCl, pH 7.5, 1.5 mM CaCl2, 2 μM CaM, 1.0 mM dithiothreitol, and 0.5 mM MnCl2. Glutathione-agarose beads coated with GST or GST peptide were incubated with brain lysates for 1 h at 4°C with end-over shaking. CN was detected by immunoblotting with anti-CNA antibody, designated pan-calcineurin A antibody, at a 1:1000 dilution, or anti-GST antibody.
2.4 Expression and purification of proteins
CNA, CNB and CaM were expressed in Escherichia coli BL21 (DE3) cells and purified as previously described [16–18]. pTrcHis C/CyP was purified with a Ni-nitrilotriacetic acid-agarose column [19].
2.5 Assay of calcineurin activity
The CNA and CNB subunits were expressed and purified. Their purity was assessed by SDS-PAGE and the purified CNA was concentrated with an Amicon Ultra Filter Unit. CN activity was determined by colorimetric assay using the RII peptide as substrate [20].
2.6 Cell culture and transfection
Plasmids encoding LxVP peptides fused to GFP were obtained by direct cloning of overhang-double-stranded annealed oligonucleotides into Eco RI+Xho I-digested pEGFP-C1 plasmid (GFP-YLAVP sense: 5′TCGAGCTGATCAGTTTCTTTCAGTTCCTTCACCCTTTACCTGGAGCAAACCATAAG3′; GFP-YLAVP antisense: 5′AATTCTTATGGTTTGCTCCAGGTAAAGGGTGAAGGAACTGAAAGAAACTGATCAGC3′).
HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum. The cells were grown at 37°C in 5% CO2. GFP-LxVP was transfected into HEK293T cells for 12 h and then incubated with quercetin (50 μM) or DMSO vehicle for an additional 12 h. The cells were then harvested and lysed in lysis buffer: 20 mM Tris-HCl, pH 8.0, 10 mM NaCl, 1 mM EDTA, 0.5% NP-40 and the cell lysates were used in co-immunoprecipitation assays. In these assays we preincubated protein A agarose with 2 μg anti-GFP antibody (rabbit polyclonal antibody), then the cell lysates were incubated with the protein A agarose for 2 h at 4°C with end-over shaking. The GFP-LxVP was immunoblotted with anti-GFP antibody (mouse monoclonal antibody) at 1:5000 dilution and the bound CNA was detected with pan-calcineurin A antibody, at 1:1000 dilution.
2.7 Microscale thermophoresis (MST)
The method of MST has been described in detail elsewhere [21,22]. We used a Monolith NT.115 from NanoTemper Technologies to measure the Kd of binding of FAM-labeled LxVP peptide to CN. A solution of CN (CNA: CNB: CaM, 1: 1: 2) was serially diluted in PBS buffer. The FAM-labeled LxVP peptide (100 nM, 10 μl) was added to each dilution (10 μl), and after incubation at 25°C for 1 h, the samples were loaded into silica capillaries (Polymicro Technologies). We performed the measurements at 20°C using 40% LED power and 20% IR-laser power. Data were analyzed with NanoTemper Analysis software, v.1.2.101.
2.8 RNA extraction and quantitative real-time PCR
To stimulate Ca2+/CN signaling, 1 μM ionomycin and 50 ng/ml phorbol ester (PMA) were added to primary cultured spleen cells, and 10 μM CsA was used as a positive control. Total RNA was extracted with TRIzol (Sigma), and 400 ng RNA was reverse-transcribed to cDNA with MMLV-RT (Invitrogen). 20 ng of cDNA was used in quantitative real-time PCR (qRT-PCR) with TaqMan probes. The qRT-PCR reactions were performed in triplicate in an ABI Prism 7000 Sequence Detection System (Applied Biosystems). The probes used were as follows: IL-2 sense, 5′-CCCAGGATGCTCACCTTCA-3′; IL-2 antisense, 5′-GCAGAGGTCCAAGTTCATCTTC-3′; IL-10 sense, 5′-GCCAGAGCCACATGCTCCTA-3′; IL-10 antisense, 5′-GATAAGGCTTGGCAACCCAAGTAA-3′; IFN-γ sense, 5′-CGGCACAGTCATTGAAAGCCTA-3′; IFN-γ antisense, 5′-GCCAGAGCCACATGCTCCTA-3′ (Takara Bio Company).
2.9 Model building and simulations
Simulations were based on the crystal structure of the A238L-CN complex in the RCSB Protein Data Bank (PDB ID: 4F0Z). The following three peptide/CN systems were built for CN: CN binding to FLCVK from A238L; CN binding to YLAVP from NFAT and CN binding to HLAPP from RCAN1. The other two residues, located at the N and C terminals of the original structure, were included with the neutralized terminals. The initial structures were constructed based on the A238L-CN complex crystal structure by mutating the corresponding residues. The titration states of ionizable residues (glutamate, aspartate, lysine, histidine, arginine, and tyrosine) were assigned based on pKa predictions carried out using PROPKA, a program for estimating the pKa values of the ionizable groups in proteins. This revealed that almost all of the residues were in their default titration states, and only Asp121 was protonated. The system was then solvated in a box of water using the SOLVATE program, with at least 10 Å between the protein and the boundary of the box. Then, 150 mM NaCl was added to neutralize the net charge of the system using the AUTOIONIZE plugin of VMD.
All of the simulations were carried out using NAMD2, the CHARMM36 force field for proteins and ions, and the TIP3P model for explicit water. Periodic boundary conditions and a time step of 2 fs were used throughout and the SHAKE algorithm was used to assign the bond distances involving hydrogen atoms.
After an initial 10,000 steps of energy minimization with all Cα atoms fixed, we equilibrated the system in an NVT ensemble at 310 K for 500 ps, during which all protein Cα atoms were constrained (k = 1 kcal/mol/Å2) to allow relaxation of the side chains and water. All of the following equilibrium simulations were performed in an NVT ensemble for 100 ns, and the active site was always restrained to its configuration in the crystal structure.
For all of the simulations, we used Langevin dynamics with a damping coefficient γ of 0.5 ps−1 to maintain a constant temperature. Also we used a cutoff distance of 12 Å to calculate short-range, non-bonded interactions, and we calculated long-range electrostatic forces by the particle mesh Ewald (PME) method.
2.10 Statistical analysis
In order to quantitatively assess marker protein distributions, we used identical settings and exported to ImageJ (NIH) to take images for imaging analyses. The data were analyzed with GraphPad Prism 5 using the means plus/minus standard error of the mean (means ± SEM).
3. Results
3.1 The classic immunosuppressive drug CsA bound to cyclophilin competes with CN for the conserved HLAPP motif of RCAN1
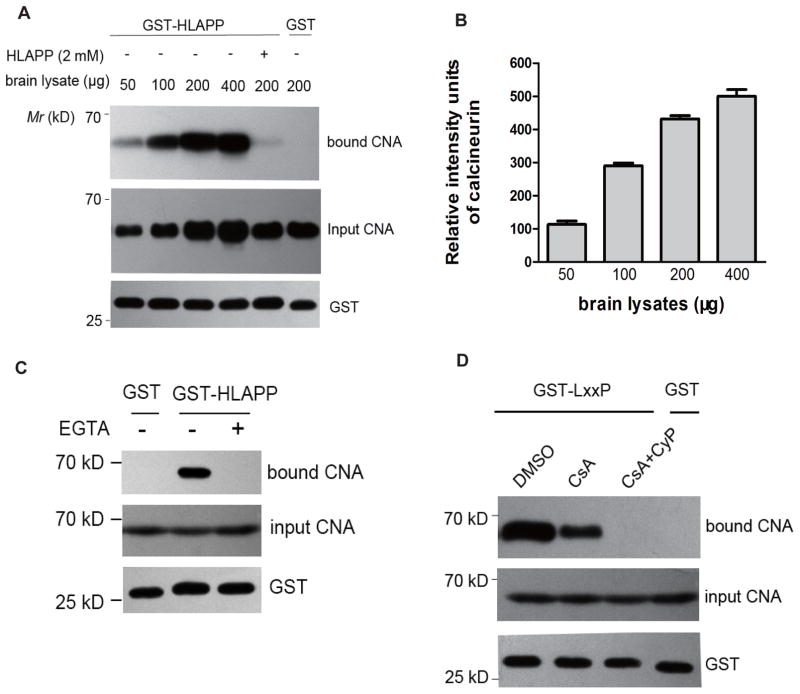
A previous study by Rodriguez [9] demonstrated that Rcn1, one of the conserved substrates of CN in yeast, contains a sequence (KQYLKVPESEKVF) that is a close match to the YLAVP core sequence of NFATc1 (DQYLAVPQHPYQWAK). We tested whether the shorter HLAPP motif of RCAN1 (RCAN1145-159, GSSHLAPPNPDKQFL, also named the LxxP-motif) interacts with CN in lysates of the mouse brain as a source of CN. Densitometric quantification of the bound CNA in these assays indicated that the HLAPP motif bound CN in a dose-dependent manner (Figs. 1A and 1B). CN is activated by the binding of Ca2+ to its B subunit and CaM [23]. The addition of 10 mM EGTA eliminated the interaction between CNA/CNB and CaM (Fig. 1C), indicating that the binding of the HLAPP motif of RCAN1 to CN requires Ca2+ and CaM and thus involves a mechanism similar to binding of the LxVP motif of NFAT to CN.
Figure 1. CsA-CyP competes with CN for the conserved HLAPP motif of RCAN1.
(A) GST-HLAPP (GSSHLAPPNPDKQFL) was incubated with increasing amounts of CN in pull-down assays. The bound CN was visualized by western blot with a monoclonal anti-CNA antibody: the bound CN is shown in the upper blot, and the the input CNA is shown in the middle panel. The bottom blot confirms that equal amounts of GST fusion proteins were used in the reactions. (B) The bound CN was quantified densitometrically and the histograms show the relative intensity units of bound CN. The CN bound in 50 μg of mouse brain lysate was set at 100%. The data are means ± SEM (n = 3). (C) CN binding by GST-HALPP requires Ca2+ and CaM. Pull-down experiments with the GST-HLAPP motif in the absence or presence of 10 mM EGTA. The bound CNA was detected by western blotting. (D) Competition with the binding of CN to GST-LxxP by CsA alone or by the CsA-CyP complex (20 μM and 200 nM, respectively). The bound CN was visualized by western blotting with a monoclonal anti-CNA antibody.
Rodriguez [9] demonstrated that NFAT and the immunosuppressant- immunophilin complex have a common feature in that LxVP-type sites, such as CsA-CyP, inhibit CN by disturbing this mode of substrate recognition. We tested the effect of CsA-CyP on the interaction of GST-LxxP with CN. We found that the amount of CNA that was pulled down with GST-LxxP was reduced following treatment of lysates with the classic immunosuppressant CsA (20 μM) in the absence of exogenous recombinant CyP. Moreover, the further addition of CyP (200 nM) nearly abolished the interaction between CN and the LxxP peptide (Fig. 1D). Thus, these observations indicate that the LxxP motif of RCAN1 interacts with CN in a way similar to the LxVP motif of NFAT.
3.2 The binding between CN and the HLAPP motif of RCAN1 is weaker than that between CN and the YLAVP motif of NFAT
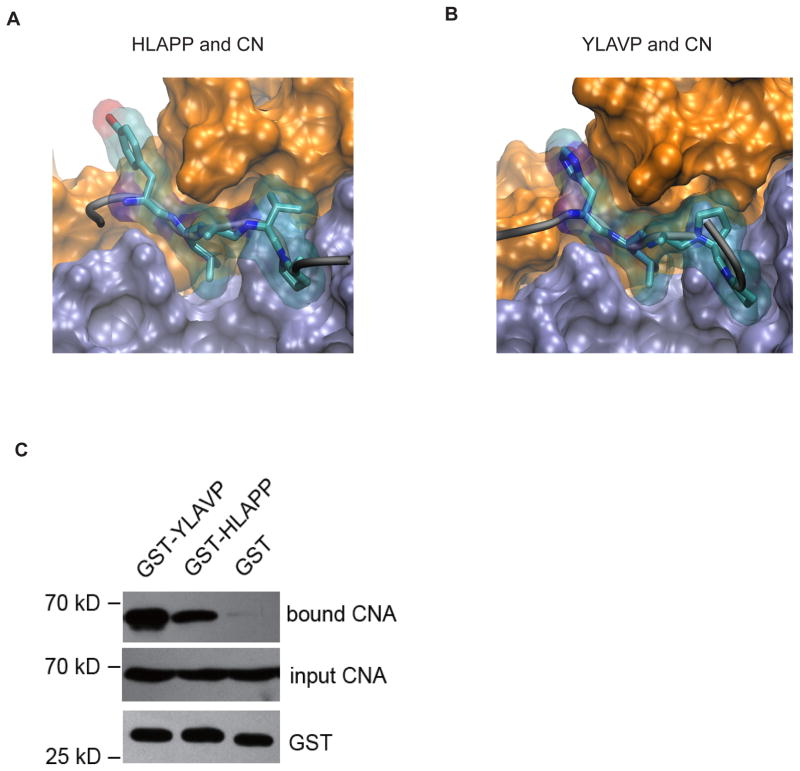
Molecular dynamics simulations were performed to characterize the binding of CN to peptides to HLAPP from RCAN1 and YLAVP from NFAT. The initial structures were constructed based on the crystal structure of the A238L-CN complex (PDB: 4F0Z) by mutating equivalent residues, and each peptide-CN complex was simulated for 100 ns.
Consistent with the crystal structure and simulations of the binding of FLCVK to CN, three residues from peptide YLAVP, namely Y, L and V, contributed to the motif binding to CN (Fig. 2A). Leu and Val were inserted into a hydrophobic groove between CNA and CNB, and both side chains fitted into the available space. In addition, Tyr was found to occupy the crevice between two EF-hand domains of CNB [9].
Figure 2. The binding between CN and the HLAPP motif of RCAN1 is weaker than that between CN and the YLAVP motif of NFAT.
In MD simulations the HLAPP (A) and YLAVP (B) motifs bind to CN in a similar manner. Both CNA (iceblue) and CNB (orange) are shown in surface presentation, while the LxxP motifs are drawn as stick and transparent surface representations. (C) Comparison of the binding between CN-YLAVP and CN-HLAPP in the GST pull-down experiments.
Similarly, His in the HLAPP motif inserts in the same site as Tyr in YLAVP and Phe in FLCVK. All of these findings suggest that an aromatic ring at position “Y” in YLAVP fits into the crevice between the two EF-hand domains of CNB and contributes to binding of the peptide to CN (Fig. 2B). In addition, Leu, as the most conserved residue in these three peptides, binds at the same site, whereas Pro could not fit in the local region like Val in YLAVP and FLCVK. This result is consistent with the finding that the binding between CN and HLAPP motif was weaker than that between CN and the YLAVP motif in the GST pull-down experiments (Fig. 2C).
3.3 Quercetin interferes with the interactions between CN and LxxP-type or LxVP-type motifs
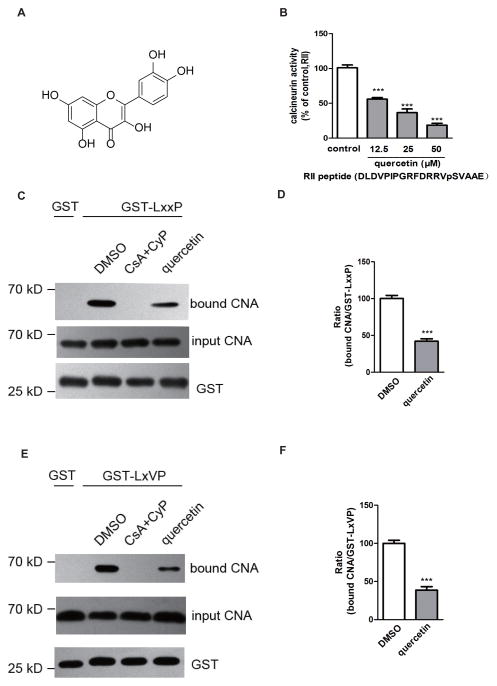
Quercetin (3,3′,4′,5,7-pentahydroxyflavone) is a typical flavonoid ubiquitous in vegetables and fruits (Fig. 3A). In a previous study, we found that quercetin exerts a powerful inhibitory effect on CN [15]. In the present study, we found that quercetin at a concentration of 50 μM competed with LxxP (RCAN1) or the LxVP peptide (NFAT) for binding to CN in brain lysates (Figs. 3B, 3C, 3D and 3E). Quercetin thus interferes with the interactions in the absence of a immunophilin, clearly differentiating it from CsA.
Figure 3. Quercetin interferes with the interactions between CN and the LxxP-type and LxVP-type motifs.
(A) Structure of quercetin. (B) CN is inhibited by quercetin in the presence of 20 mM RII peptide (DLDVPIPGRFDRRVpSVAAE) as substrate. (C) The CsA-CyP complex (20 μM and 200 nM) and quercetin (50 μM) compete with binding of CN to GST-LxxP. The bound CN was visualized by western blot with a monoclonal anti-CNA antibody. (D) The bar graph depicts the ratios of bound CNA and GST-LxxP. The histograms present the values as percentages of the DMSO control binding (100%). The data are means ± SEM. n = 3, *** p < 0.001. (E) The CsA-CyP complex (20 μM and 200 nM) and quercetin (50 μM) compete with binding of CN to GST-LxVP. (F) Other information as in (D)
3.4 Quercetin attenuates the interaction between the LxVP-type motif peptide and purified CN
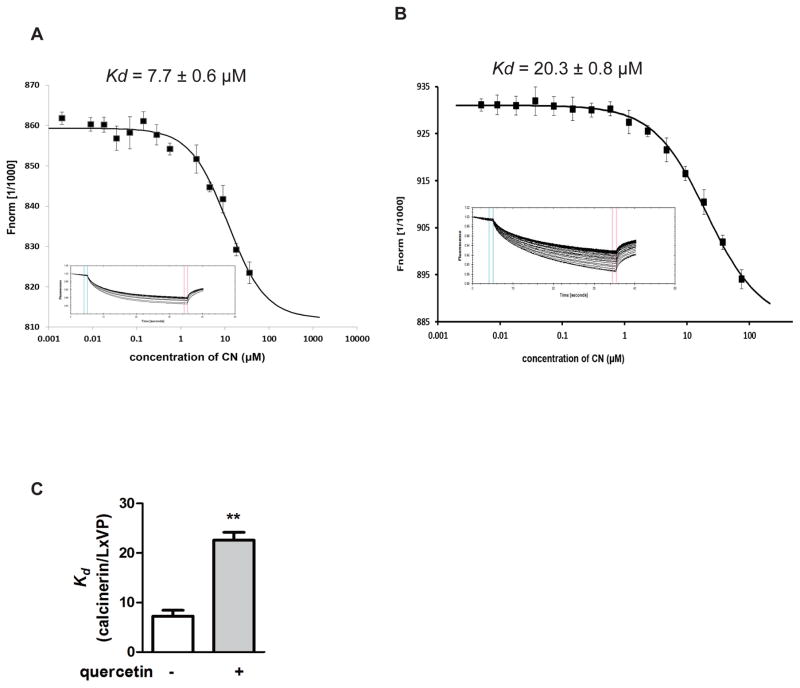
We incubated 100 nM of the FAM-LxVP peptide (NFATc1: DQYLAVPQHPYQWAK) with serial dilutions of CN from 36 μM to 2 nM in PBS buffer. Figure 4A shows that the LxVP peptide bound to CN with a dissociation constant (Kd) of 7.7 ± 0.6 μM. The MST data showed that in the presence of 50μM quercetin, the binding affinity of the FAM-LxVP peptide for CN was lower than in the absence of quercetin, with a dissociation constant of 20.3 ± 0.8 μM (Figure 4B and 4C).
Figure 4. Quercetin attenuates the interactions between the LxVP-type motif peptide and purified CN.
(A) The affinity of the FAM-labeled LxVP peptide for calcineurin was analyzed. The CN protein was titrated against a fixed concentration of labeled LxVP (100 nM). The top panel indicates the isotherm derived from the raw data, fitted to a sigmoidal dose-response curve. The bottom panel displays the raw data for thermophoresis recorded at 20°C using 40% LED and 20% MST power. (B) The affinity of FAM-labeled LxVP peptide to calcineurin in the presence of 50 μM quercetin. The CN protein with 50 μM quercetin was titrated against a fixed concentration of labeled LxVP (100 nM). (C) Comparison of Kd in the absence and presence of 50 μM quercetin. The data are showed as means ± SEM. n = 3, ** p < 0.01.
3.5 Quercetin suppresses CN-NFAT signaling and downstream cytokine gene expression
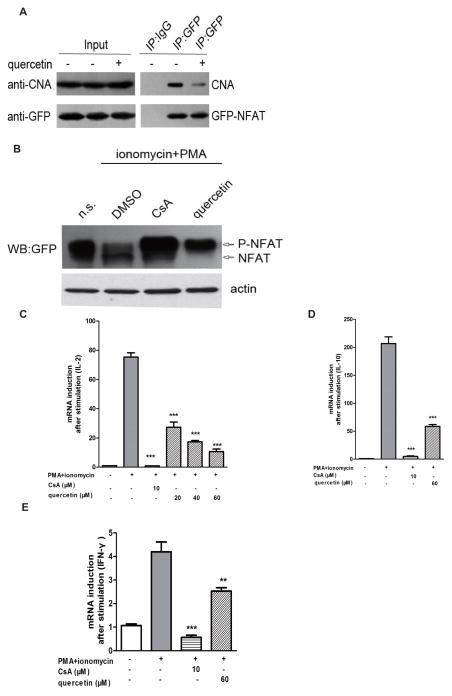
We also tested whether the CN-LxVP interaction is disrupted by quercetin in live cells in coprecipitation experiments in HEK293 cells. Compared with the DMSO control group, precipitation of CNA was clearly decreased in the presence of quercetin (Fig. 5A).
Figure 5. Quercetin suppresses CN-NFAT signaling and the downstream cytokine gene expression.
(A) Empty vector cDNAs or GFP-LxVP cDNAs were transfected into HEK293T cells. The HEK293T cells were pretreated with DMSO or quercetin for 12 h and then subjected to co-precipitation and western blot analysis. (B) Six μg of empty vector cDNAs or NFATc3 (3-407)-GFP were transfected into HEK293T cells. The cells were stimulated with PMA (50 ng/ml) plus ionomycin (1 μM). CsA (10 μM) was used as a positive control. The blot shows the expression of GFP-NFAT in total lysates, and also indicates the positions of phosphorylated and dephosphorylated proteins. (C, D, E) Quercetin inhibits NFAT-driven gene expression. Total mRNA was extracted 5 h after stimulation with ionomycin and PMA. The mRNA levels of IL-2 (C), IL-10 (D) and IFN-γ (E) were quantified by qRT-PCR. The mRNA levels of the control group were set at 100%. The data are shown as means ± SEM of three independent experiments. ** p < 0.01, *** p < 0.001 compared with the PMA + ionomycin group.
We also examined the in vivo effect of quercetin on the CN-NFAT signaling pathway. Pre-treatment with CsA or quercetin is known to prevent the dephosphorylation of NFATc3 caused by ionomycin (1 μM) and PMA (50 ng/ml) in HEK293T cells (Fig. 5B) [24]. We therefore determined NFAT-regulated cytokine expression in mouse spleen cells in the presence and absence of quercetin; quercetin clearly decreased the PMA + ionomycin-induced expression of the NFAT-dependent genes IL-2, IL-10 and IFN-γ (Figs. 5C, 5D, and 5E).
4. Discussion
Our studies have defined the interaction between the LxxP motif of RCAN1 and CN. More importantly, we used the CN-LxxP and CN-LxVP interactions to demonstrate that quercetin can interfere with the interaction between CN and its substrates. The subsequent in vivo experimental results demonstrated that quercetin suppresses the cytokine gene expression resulting from CN-dependent NFAT signaling, which suggests that quercetin prevents the interactions between LxVP-type structures and CN that lead to immunosuppression. These structures could be a starting point at the molecular level for the design of inhibitors of the interactions between CN and its substrates.
Grigoriu has reported that an LxVP-type sequence regulates the interaction of several substrates of A238L, which is a protein inhibitor, and RCAN1 with CN activated by Ca2+/CaM [10]. The LxVP binding motif of CN is composed of residues from both CNA and CNB. RCAN1 is an endogenous regulator of CN. It was first identified as a gene in the Down’s syndrome critical region on human chromosome 21 [25–28]. Rodriguez reported that the changing the conserved YLKVP motif to YaKaa in yeast Rcn1 affected the amount of bound CNA and decreased the inhibition of CNA signaling. Our GST pull-down data demonstrate that the short HLAPP motif can definitely bind to CN in mouse brain lysates. However, compared with the LxVP motif of NFAT, the replacement of the Tyr (Y) and Val (V) with His (H) and Pro (P) in the LxxP motif may reduce CN binding.
It is well known that CsA complexed with its corresponding immunophilin inhibits immune responses by preventing CN-dependent dephosphorylation and the nuclear translocation of NFAT. Moreover in the crystal structure of the CyP/CsA/CN ternary complex residues 3–9 of CsA and Trp-121 of CyP form a compound surface for interaction with CN, and CN goes through a modest closure when it binds to CsA/CyP.
CsA seems to hijack CyP to produce an inhibitory complex in vivo. Neither CsA nor CyP interacts with the active site of CN directly, but the CsA-CyP complex places itself over the active site in such a manner that it prevents substrate from interacting with the catalytic residues [29–31]. Rodriguez reported that CsA-CyP inhibits the immune response by disrupting the interaction between CN and the NFAT LxVP motif, suggesting that disrupting this interaction may be of therapeutic value. Quercetin blocks the interactions between CN and LxVP and between CN and LxxP, and can also inhibit NFAT-induced gene expression in vivo.
Quercetin has a wide range of biological activities, including anti-oxidant, anti-bacterial, anti-viral, and anti-allergic actions. It also can be used as an anti-inflammatory agent [32] and has a direct immunosuppressive effect [33].
In previous work, we reported that quercetin inhibited CN activity in human T-cell leukemia cells (Jurkat cells). Through fluorescence spectroscopy measurements [34], we demonstrated that quercetin binds to CN, and molecular docking data showed that it binds at the junction of CNA and CNB, the rectangle possesses the binding amino acid residues of Asp348, Gln164, Phe346, Ile168, Met347 and Thr351, just like CsA. We also found that this region was important for recognition of the NFAT substrate by CN. In the present study, we showed that quercetin exerts its immunosuppressive activity by suppressing NFAT activation via a mechanism consistent with that of CsA. These findings provide a novel mechanism for quercetin-mediated immunosuppression.
Protein-protein interactions play important regulatory roles in pathological and physiological processes and are closely related to human diseases. Our study offers new insights into the regulation of the interaction of CN with its substrates by small molecules. Our data reveal that quercetin inhibits CN signaling by interfering with the LxVP-type sites of CN substrates. Thus the interactions of CN with its LxVP-type substrates provide potential assays for screening immunosuppressive agents.
Acknowledgments
The present work was supported by the National Nature Science Foundation of China (Project 81373389 and Project 31540011).
References
- 1.Rusnak F, Mertz P. Calcineurin: form and function. Physiol Rev. 2000;80:1483–1521. doi: 10.1152/physrev.2000.80.4.1483. [DOI] [PubMed] [Google Scholar]
- 2.Rao A. Signaling to gene expression: calcium, calcineurin and NFAT. Nat Immunol. 2009;10:3–5. doi: 10.1038/ni0109-3. [DOI] [PubMed] [Google Scholar]
- 3.Müller MR, Rao A. NFAT, immunity and cancer: a transcription factor comes of age. Nat Rev Immunol. 2010;10:645–656. doi: 10.1038/nri2818. [DOI] [PubMed] [Google Scholar]
- 4.Crabtree GR, Schreiber SL. Ca2+-calcineurin-NFAT signaling. Cell. 2009;138:210. doi: 10.1016/j.cell.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li H, Rao A, Hogan PG. Interaction of calcineurin with substrates and targeting proteins. Trends Cell Biol. 2011;21:91–103. doi: 10.1016/j.tcb.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aramuru J, Yaffe MB, Lopez-Rodriguez C, et al. Affinity-driven peptide selection of an NFAT inhibitor more selective than cyclosporin A. Science. 1999;285:2129–2133. doi: 10.1126/science.285.5436.2129. [DOI] [PubMed] [Google Scholar]
- 7.Martínez-Martínez S, Rodríguez A, Lopez-Maderuelo MD, Ortega-Perez I, Vazquez J, Redondo JM. Blockade of NFAT activation by the second calcineurin binding site. J Biol Chem. 2006;281:6227–6235. doi: 10.1074/jbc.M513885200. [DOI] [PubMed] [Google Scholar]
- 8.Liu J, Arai K, Arai N. Inhibition of NFATx activation by an oligopeptide: disrupting the interaction of NFATx with calcineurin. J Immunol. 2001;167:2677–2687. doi: 10.4049/jimmunol.167.5.2677. [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez A, Roy J, Martinez-Martinez S, Lopez-Maderuelo D, Nino-Moreno P, Orti L, Pantoja-Uceda D, Pineda-Lucena A, Cyert MS, Redondo JM. A conserved docking surface on calcineurin mediates interaction with substrates and immunosuppressants. Mol Cell. 2009;33:616–626. doi: 10.1016/j.molcel.2009.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grigoriu S, Bond R, Cossio P, Chen JA, Ly N, Hummer G, Page, Cyert M, Peti W. The molecular mechanism of substrate engagement and immunosuppressant inhibition of calcineurin. Plos Biol. 2013;11:e1001492. doi: 10.1371/journal.pbio.1001492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma L, Tang HP, Ren Y, Deng HT, WUJW, Wang ZX. p38α MAP kinase phosphorylates RCAN1 and regulates its interaction with calcineurin. Sci China Life Sci. 2012;55:559–566. doi: 10.1007/s11427-012-4340-9. [DOI] [PubMed] [Google Scholar]
- 12.Vega RB, Yang J, Rothermel BA, Bassel-Duby R, Williams RS. Multiple domains of MCIP1 contribute to inhibition of calcineurin activity. J Biol Chem. 2002;277:30401–30407. doi: 10.1074/jbc.M200123200. [DOI] [PubMed] [Google Scholar]
- 13.Mehta S, Li H, Hogan PG, Cunningham KW. Domain architecture of the regulators of calcineurin (RCANs) and identification of a divergent RCAN in yeast. Mol Cell Biol. 2009;29:2777–2793. doi: 10.1128/MCB.01197-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aubareda A, Mulero MC, Pérez-Riba M. Functional characterization of the calcipressin 1 motif that suppresses calcineurin-mediated NFAT-dependent cytokine gene expression in human T cells. Cell signal. 2006;18:1430–1438. doi: 10.1016/j.cellsig.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 15.Wang H, Zhou CL, Lei H, Wei Q. Inhibition of calcineurin by quercetin in vitro and in Jurkat cells. J Biochem. 2010;147:185–190. doi: 10.1093/jb/mvp163. [DOI] [PubMed] [Google Scholar]
- 16.Ma Y, Jiang G, Wang Q, Sun Y, Zhao Y, Tong L, Luo J. Enzymatic and thermodynamic analysis of calcineurin inhibition by RCAN1. Int J Biol Macromol. 2015;72:254–260. doi: 10.1016/j.ijbiomac.2014.08.042. [DOI] [PubMed] [Google Scholar]
- 17.Wei Q, Lee EY. Expression and reconstitution of calcineurin A and B subunits. IUBMB life. 1997;41:169–177. doi: 10.1080/15216549700201171. [DOI] [PubMed] [Google Scholar]
- 18.Liu P, Huang C, Wang HL, Zhou K, Xiao FX, Wei Q. The importance of Loop 7 for the activity of calcineurin. FEBS Lett. 2004;577:205–208. doi: 10.1016/j.febslet.2004.09.082. [DOI] [PubMed] [Google Scholar]
- 19.Wang H, Du Y, Xiang B, Lin W, Wei Q. The regulatory domains of CNA have different effects on the inhibition of CN activity by FK506 and CsA. IUBMB life. 2007;59:388–393. doi: 10.1080/15216540701370721. [DOI] [PubMed] [Google Scholar]
- 20.Blumenthal D, Takio K, Hansen RS, Krebs EG. Dephosphorylation of cAMP-dependent protein kinase regulatory subunit (Type II) by calmodulin-dependent protein phosphatase. J Biol Chem. 1986;262:8140–8145. [PubMed] [Google Scholar]
- 21.Jerabek-Willemsen M, Wienken CJ, Braun D, Baaske P, Duhr S. Molecular interaction studies using microscale thermophoresis. Assay Drug Dev Technol. 2011;9:342–353. doi: 10.1089/adt.2011.0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin CC, Melo FA, Ghosh R, Suen KM, Stagg LJ, Kirkpatrick J, Arold ST, Ahmed Z, Ladbury JE. Inhibition of basal FGF receptor signaling by dimeric Grb2. Cell. 2012;149:1514–1524. doi: 10.1016/j.cell.2012.04.033. [DOI] [PubMed] [Google Scholar]
- 23.Watanabe Y, Perrino BA, Chang BH, Soderling TR. Identification in the calcineurin A subunit of the domain that binds the regulatory subunit. J Biol Chem. 1995;270:456–460. [PubMed] [Google Scholar]
- 24.Liu C, Hermann TE. Characterization of ionomycin as a calcium ionophore. J Biol Chem. 1978;253:5892–5894. [PubMed] [Google Scholar]
- 25.Hilioti Z, Cunningham KW. The RCN family of calcineurin regulators. Biochem Biophys Res Commun. 2003;311:1089–1093. doi: 10.1016/s0006-291x(03)01515-8. [DOI] [PubMed] [Google Scholar]
- 26.Fuentes JJ, Genescà L, Kingsbury TJ, Cunningham KW, Pérez-Riba M, Estivill X, Luna S. DSCR1, overexpressed in Down syndrome, is an inhibitor of calcineurin-mediated signaling pathways. Hum Mol Genet. 2000;9:1681–1690. doi: 10.1093/hmg/9.11.1681. [DOI] [PubMed] [Google Scholar]
- 27.Davies KJ, Ermak G, Rothermel BA, Pritchard M, Heitman J, et al. Renaming the DSCR1/Adapt78 gene family as RCAN: regulators of calcineurin. FASEB J. 2007;21:3023–3028. doi: 10.1096/fj.06-7246com. [DOI] [PubMed] [Google Scholar]
- 28.Martínez-Martínez S, Genescà L, Rodríguez A, Raya A, Salichs E, et al. The RCAN carboxyl end mediates calcineurin docking-dependent inhibition via a site that dictates binding to substrates and regulators. Proc Natl Acad Sci USA. 2009;106:6117–6122. doi: 10.1073/pnas.0812544106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J, Farmer JD, Jr, Lane WS, Friedman J, Weissman I, et al. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 30.Jin L, Harrison S. Crystal structure of human calcineurin complexed with cyclosporin A and human cyclophilin. Proc Natl Acad Sci USA. 2002;99:13522–13526. doi: 10.1073/pnas.212504399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huai Q, Kim H-Y, Liu Y, Zhao Y, Mondragon A, et al. Crystal structure of calcineurin-cyclophilin-cyclosporin shows common but distinct recognition of immunophilin-drug complexes. Proc Natl Acad Sci USA. 2002;99:12037–12042. doi: 10.1073/pnas.192206699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miles SL, McFarland, Niles RM. Molecular and physiological actions of quercetin: need for clinical trials to assess its benefits in human disease. Nutr Rev. 2014;72:720–734. doi: 10.1111/nure.12152. [DOI] [PubMed] [Google Scholar]
- 33.Huang RY, Yu YL, Cheng WC, OuYang CN, Fu E, Chu CL. Immunosuppressive effect of quercetin on dendritic cell activation and function. J Immunol. 2010;184:6815–6821. doi: 10.4049/jimmunol.0903991. [DOI] [PubMed] [Google Scholar]
- 34.Lei H, Luo J, Tong L, Peng LQ, Qi Y, Jia ZG, Wei Q. Quercetin binds to a similar region with FKBP-FK506 and CyPA-CsA on calcineurin. Food Chemistry. 2011;127:1169–1174. doi: 10.1016/j.foodchem.2011.01.119. [DOI] [PubMed] [Google Scholar]