Abstract
Anthocyanin biosynthesis is one of the most thoroughly studied enzymatic pathways in biology, but little is known about the molecular mechanisms of its final stage: the transport of the anthocyanin pigment into the vacuole. We have identified a multidrug resistance–associated protein (MRP), ZmMrp3, that is required for this transport process in maize (Zea mays). ZmMrp3 expression is controlled by the regulators of anthocyanin biosynthesis and mirrors the expression of other anthocyanin structural genes. Localization of ZmMRP3 in vivo shows its presence in the tonoplast, the site at which anthocyanin transport occurs. Mutants generated using antisense constructs have a distinct pigmentation phenotype in the adult plant that results from a mislocalization of the pigment as well as significant reduction in anthocyanin content, with no alteration in the anthocyanin species produced. Surprisingly, mutant plants did not show a phenotype in the aleurone. This appears to reflect the presence of a second, highly homologous gene, ZmMrp4, that is also coregulated with the anthocyanin pathway but is expressed exclusively in aleurone tissue. This description of a plant MRP with a role in the transport of a known endogenous substrate provides a new model system for examining the biological and biochemical mechanisms involved in the MRP-mediated transport of plant secondary metabolites.
INTRODUCTION
Sequestration of reactive metabolites is crucial for cell survival. Plant cells in particular produce many secondary metabolites that must be excluded from the cytoplasm to prevent cellular damage. Anthocyanin pigments are a visible example of this type of plant product. Compartmentalization of anthocyanin in the vacuole is required both to limit the mutagenic and oxidative effects of synthesis pathway intermediates (Ahmed et al., 1994; Rueff et al., 1995) and for proper biological function of the final product. The biosynthesis of anthocyanin is one of the most thoroughly studied biochemical pathways in biology. Easily identified phenotypes and complete mutant viability have made anthocyanin production an ideal model for analysis of gene regulation, paramutation, and transposon biology. Recent work on the last genetically defined step in this pathway (Marrs et al., 1995; Alfenito et al., 1998) has also identified anthocyanin biosynthesis as an important model for studying the mechanisms for the detoxification and vacuolar sequestration of heterocyclic organic anions. In addition to anthocyanins, this class of compounds includes a vast array of endogenous and xenobiotic chemicals involved in pest and disease resistance, pigmentation, UV protection, intracellular and extracellular signaling, and weed control (Abell et al., 1993; Shirley, 1996; Hammerschmidt, 1999). Understanding the molecular mechanisms involved in transporting these molecules to the correct cellular locations represents a fundamental, yet poorly understood, problem in biology.
The synthesis of anthocyanin bears a striking similarity to the detoxification of heterocyclic organic xenobiotics. Both processes occur in the cytoplasm and begin with a relatively nonreactive hydrophobic compound that is modified by a series of enzymes to introduce reactive centers and add hydrophilic molecules such as sugars or amino acids. Such modifications not only reduce hydrophobicity but can also act as tags for recognition by the proteins that transport the modified product out of the cytoplasm (Ishikawa et al., 1997). The enzymatic modifications in anthocyanin biosynthesis serve a different purpose: the alteration of the chemical properties of the molecule to fulfill its cellular functions. Anthocyanin pigment acts as UV-B sunscreen when deposited into the vacuole, a process that requires a specific glutathione S-transferase (GST) (Marrs et al., 1995). In their most well characterized role, GSTs catalyze the addition of a glutathione (GSH) molecule to a heterocyclic organic anion (Edwards and Dixon, 2000). In such instances, the GSH conjugate serves the dual purpose of increasing hydrophilicity and marking the molecule for transport by a specific family of ABC proteins, the multidrug resistance–associated proteins (MRP) (Ishikawa et al., 1997).
Also termed GS-X pumps because of their association with the transport of glutathionated compounds, the ∼190-kD MRPs constitute large protein families in many eukaryotes. There are nine MRPs described in Homo sapiens (Dean and Allikmets, 2001), 14 in Arabidopsis thaliana (Sánchez-Fernández et al., 2001; Kolukisaoglu et al., 2002), and at least 12 in Oryza sativa (Jasinski et al., 2003). These MRPs share significant protein sequence homology between species and even across kingdoms. In A. thaliana, 8 of the 15 MRPs are found in homologous pairs that can have >85% sequence identity (Lu et al., 1998; Kolukisaoglu et al., 2002). Interestingly, the highly homologous pair AtMRP1 and AtMRP2 shows both significant overlap of, and fundamental differences in, their in vitro substrate preferences (Lu et al., 1998; Liu et al., 2001). Therefore, both functional redundancy and unique biological roles likely exist for closely related MRPs.
Structurally, MRPs are large, modular proteins made up of three transmembrane domains (TMDs) and two ATP binding cassette domains (nucleotide binding domains [NBDs]) (Borst et al., 1999). MRPs use the energy generated by the hydrolysis of MgATP to facilitate the transmembrane movement of a variety of small molecules. Substrates are not limited to glutathionated compounds but include a range of heterocyclic organic anions and heavy metals (Rea et al., 1998). Such compounds may or may not be conjugated to sugars or amino acids and can be transported alone or in conjunction with free GSH (Liu et al., 2001). In humans, MRP substrates include anticancer drugs and other xenobiotics, including plant flavonoids, and endogenous molecules, such as bile salts and leukotrienes (Borst et al., 1999). Individual MRPs of A. thaliana demonstrate in vitro transport activity with herbicide metabolites, chlorophyll catabolites, and model organic substrates (Lu et al., 1997, 1998; Tommasini et al., 1998; Liu et al., 2001). Isolated plant vacuoles also actively transport chlorophyll catabolites, phytoalexins, and metabolized herbicides (Martinoia et al., 1993; Hinder et al., 1996; Klein et al., 2000). This activity is sensitive to vanadate, a specific inhibitor of ABC transporters, but is unaffected by disruptions in proton and charge gradients, indicating a specific role for ABC transporter activity (Martinoia et al., 1993). To date, however, there is no direct evidence for an in vivo substrate for any plant MRP.
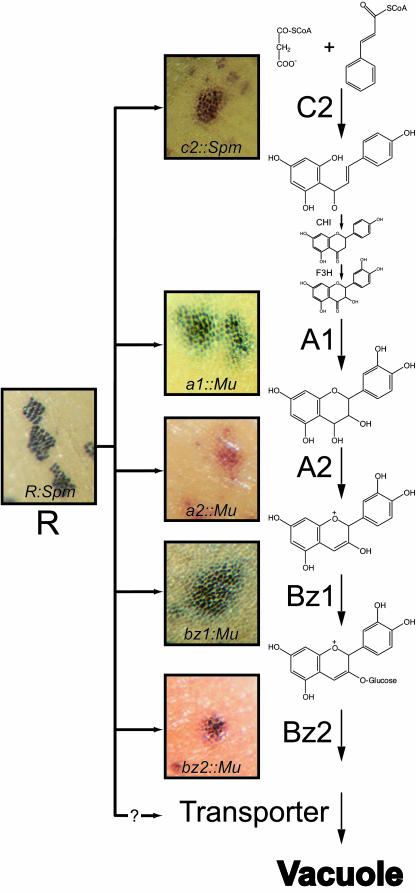
Several lines of indirect evidence implicate an MRP in the anthocyanin biosynthetic pathway of Zea mays. Genetic analysis demonstrates that the actions of the known anthocyanin biosynthetic genes are not cell autonomous. In tissues from mutant plants, all of the anthocyanin pathway genes are present and active, with the exception of the mutated structural gene. Revertant sectors generated by unstable mutants of individual structural genes all have indistinct boundaries resulting from the sequestration of a small amount of anthocyanin in vacuoles of nonrevertant cells surrounding the revertant sector (Figure 1). This indicates that some component produced by the intact anthocyanin pathway in the revertant sector can move to the adjacent cells, partially complementing the mutation in those cells. In tissues from plants with unstable mutations in the anthocyanin regulators, in our lines a complex of the myc and myb-type proteins B and Pl in the plant body and R and C1 in the aleurone, none of the anthocyanin structural genes are active outside of the revertant sectors. If the known structural genes represent all of the anthocyanin biosynthetic genes, we would expect that the components that can diffuse from structural gene revertant sectors would also be able to diffuse out of revertant sectors generated by unstable mutants of the transcription factors and that these revertant sectors would also have diffuse boundaries. However, revertant sectors of the anthocyanin transcription factors have distinct boundaries (Figure 1; Kermicle and Alleman, 1990). This cell autonomy of the anthocyanin regulatory genes argues strongly for the presence of a yet to be described structural gene that is controlled by the anthocyanin transcription factors and converts diffusible anthocyanin into a cell-limited marker. The most obvious candidate for such a gene is a membrane transporter whose expression is regulated by the anthocyanin transcription factors.
Figure 1.
Cell Autonomy Analysis of Anthocyanin Biosynthesis in Z. mays.
The anthocyanin biosynthesis pathway in maize is diagrammed accompanied by the aleurone phenotype of unstable Mutator (Mu) or Suppressor-mutator (Spm) transposon insertions in the structural genes colorless2 (c2::Spm), anthocyaninless1 (a1::Mu), anthocyaninless2 (a2::Mu), bronze1 (bz1::Mu), and bronze2 (bz2::Mu) and the transcription factor R (R::Spm). Transposon excision during aleurone development produces a lineage of cells with normal gene function that form a wild-type (pigmented) sector in an otherwise mutant aleurone.
Further evidence suggests the presence of such a transporter and offers clues to its identity. The ability of vanadate, a specific inhibitor of the ABC family of transmembrane transporters, to block pigment sequestration in maize cells (Marrs et al., 1995) strongly suggests that anthocyanin transport requires the activity of an ABC transporter. Similarities between the anthocyanin biosynthesis and xenobiotic detoxification pathways, particularly the requirement for the activity of a GST to ensure proper anthocyanin localization, suggest that the ABC transporter responsible for anthocyanin sequestration is an MRP. Indeed, in a study of anthocyanin production in maize cell culture, two ABC transporters, one of which is an MRP, were identified as being upregulated in cells overexpressing the anthocyanin transcription factors R and C1 (Bruce et al., 2000). In this article, we identify that MRP, ZmMRP3, as coregulated with the anthocyanin pathway in planta, characterize the ZmMRP3 gene, and demonstrate the involvement of the corresponding protein in the transport of anthocyanin into the vacuole.
RESULTS
MRP Expression and Anthocyanin Biosynthesis
Screening of 30,000 Pioneer Hi-Bred maize EST sequences for those with homology to human, Arabidopsis, and yeast MRPs identified nine candidate MRPs. Full sequencing of the inserts yielded 1.0 to 3.0 kb (20 to 60% of estimated cDNA size) of the 3′ end of seven unique genes. One of these was found to be an ABC transporter of a different class and was not characterized further. Subsequent searches of the publicly available EST database yielded 3′ sequence for four additional MRP-like genes. Two of the genes identified in this screen were recently cloned and named ZmMrp1 and ZmMrp2 (Swarbreck et al., 2003). Fragments of the 3′ end from each of these 10 genes were used as probes on RNA gel blots to compare expression patterns in eight tissue types using samples from plants with either high (B and Pl) or no (b and pl) anthocyanin (Figures 2A and 2B). Six of the ten MRPs were expressed in at least one tissue (Table 1), but only ZmMrp3 showed an expression pattern that correlated with anthocyanin levels and with the expression of Anthocyaninless2 (A2), the gene encoding anthocyanidin synthase (Figure 2C). DNA gel blot analysis using the same ZmMRP3 probe indicates that it hybridizes to a single gene (data not shown).
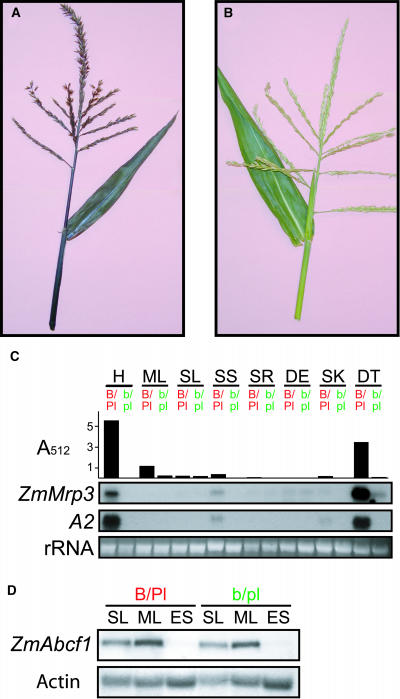
Figure 2.
ZmMrp3 Is Upregulated in Pigmented Tissues.
(A) Fully pigmented maize. Upper leaf and tassel of a maize plant with full anthocyanin pigmentation. In this maize line, anthocyanin pigmentation of the plant body is controlled by the activity of two interacting transcription factors, B and Pl.
(B) Maize lacking anthocyanin. Upper leaf and tassel from plants that are not expressing the anthocyanin transcription factors B and Pl and are, therefore, lacking all anthocyanin pigmentation.
(C) ZmMrp3 gene expression in relation to anthocyanin synthesis. Anthocyanin content, as measured by absorbance at 512 nm, and RNA gel blot of eight maize tissues from the lines shown in (A) and (B). Anthocyanin and total RNA were isolated from the same tissue sample. The blot was probed with the 3′ UTR of ZmMrp3 and the complete A2 cDNA.
(D) ZmAbcf1 gene expression in relation to anthocyanin synthesis. An RNA gel blot of total RNA from three maize tissues from the maize lines shown in (A) and (B). Blots were probed with the 3′ UTR of ZmAbcf1 and with maize actin as a loading control.
H, husk; ML, mature leaf; SL, 14-d-old seedling leaf; SS,14-d-old seedling shoot; SR, 14-d-old seedling root; DE, developing ear first day after silk emergence; SK, silks 3 d after emergence; DT, developing tassel 3 to 5 d before pollen shed; ES, 14-d-old etiolated seedling. B/Pl, tissue containing anthocyanin because of the expression of the anthocyanin transcription factors B and Pl; b/pl, tissue lacking anthocyanin because of the absence of expression of the anthocyanin transcription factors B and Pl.
Table 1.
Relative Expression of Z. mays MRPs
|
ZmMrp Designated by GenBank Accession Number
|
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |||||||||||
|
AY186246
|
AY186247
|
AY609318
|
BG349642
|
BU050629
|
BU050290
|
BU051439
|
BF729302
|
BU098343
|
AW066678
|
|||||||||||
| Tissue | B/Pl | b/pl | B/Pl | b/pl | B/Pl | b/pl | B/Pl | b/pl | B/Pl | b/pl | B/Pl | b/pl | B/Pl | b/pl | B/Pl | b/pl | B/Pl | b/pl | B/Pl | b/pl |
| Etiolated seedling | ++ | ++ | NDa | ND | − | − | − | − | ++ | ++ | + | + | ++ | ++ | ND | ND | ND | ND | ND | ND |
| Seedling root | − | − | − | − | − | − | − | − | ND | ND | ND | ND | ++ | ++ | − | − | − | − | − | − |
| Seedling shoot | ++ | ++ | − | − | +++ | − | − | − | ++ | ++ | ++ | ++ | ++ | ++ | − | − | − | − | − | − |
| Seedling leaf | + | + | − | − | − | − | − | − | ++ | ++ | ++ | ++ | ++ | ++ | − | − | − | − | − | − |
| Developing adult leaf | − | − | ND | ND | ++ | − | − | − | ++ | ++ | +++ | +++ | ++ | ++ | ND | ND | ND | ND | ND | ND |
| Mature adult leaf | ND | ND | + | + | − | − | − | − | ND | ND | ND | ND | ++ | ++ | − | − | − | − | − | − |
| Husk | ND | ND | − | − | ++++ | − | − | − | ND | ND | ND | ND | ++ | ++ | − | − | − | − | − | − |
| Developing ear | − | − | − | − | + | + | − | − | ++ | ++ | ++ | ++ | ++ | ++ | − | − | − | − | − | − |
| Developing tassel | + | + | − | − | ++++ | ++ | − | − | ND | ND | ND | ND | ++ | ++ | − | − | − | − | − | − |
| Silk | − | − | − | − | + | − | − | − | + | + | ++ | ++ | − | − | − | − | − | − | − | − |
| Pollen | − | − | ND | ND | − | − | − | − | − | − | ND | ND | − | − | ND | ND | ND | ND | ND | ND |
ND, not determined.
It was surprising to find that neither ZmMrp3 nor A2 were expressed at detectable levels in fully pigmented, mature adult leaves. To determine if this was a consequence of leaf age, we screened adult leaves of the same anthocyanin genotypes shortly after they emerged and before the development of full pigmentation. Both ZmMrp3 and A2 were highly expressed in expanding adult leaves from pigmented plants (Table 1; data not shown). This indicates that as adult leaves age and accumulate the indelible dark pigmentation, they downregulate expression of the anthocyanin pathway genes. This downregulation appears to eliminate detectable anthocyanin gene expression by the 15th to 20th day after leaf emergence.
In addition to anthocyanin-regulated expression, ZmMRP3 transcript was also found in the unpigmented developing tassel and in the developing ear, where anthocyanin is also absent (Figure 2C, Table 1). This suggests functions for this MRP in addition to anthocyanin transport. Both the developing ear and immature tassels are composed of several tissue types; therefore, the expression data offer few clues to ZmMrp3 functions. The lack of expression of ZmMrp3 in mature pollen and the scarcity of pollen-specific enhancer elements in the ZmMrp3 promoter (Figure 3C) suggest that ZmMrp3 does not have a function within pollen.
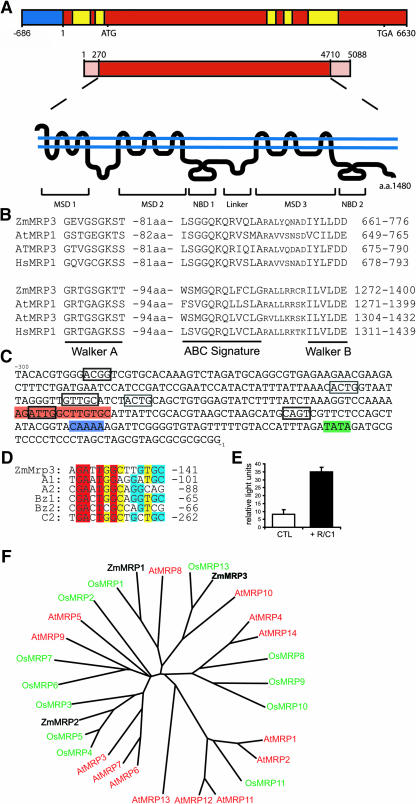
Figure 3.
Molecular Characterization of ZmMrp3.
(A) Structure of ZmMrp3. Diagram of ZmMRP3 representing the known genomic sequence with promoter (blue), intron (yellow)/exon (red) structure, the cDNA sequence with open reading frame (red) and UTRs (orange), and the predicted protein structure indicating the site of putative membrane spanning domains (MSD), NBDs, and the cytoplasmic linker. a.a., amino acid.
(B) ABC motifs in predicted ZmMRP3 protein. Alignment of the NBDs of ZmMRP3, A. thaliana MRP1 and MRP3, and H. sapiens MRP1 showing the Walker A and B boxes and the ABC signature motif.
(C) ZmMrp3 promoter. Partial promoter sequence of ZmMRP3 showing TATA (green), CAAT (blue), overlapping anthocyanin regulatory elements (ARE) (orange), and myb-like motifs (boxed). Note that some of the myb motifs may be encoded on the cDNA strand and may represent degenerate myb core structures that have been found to be sufficient for anthocyanin promoter activation (Tuerck and Fromm, 1994).
(D) The ZmMrp3 promoter contains an anthocyanin regulatory element. Alignment of the AREs, as described by Tuerck and Fromm (1994), from maize anthocyanin structural genes indicating their location relative to transcript start site.
(E) The ZmMrp3 promoter is regulated by the anthocyanin transcription factors R and C1. Luciferase activity derived from the luciferase gene driven by a 687-bp fragment of the ZmMrp3 promoter in the presence or absence of R and C1.
(F) ZmMRP3 is closely related to OsMRP13. Unrooted phylogenetic tree comparing the full-length protein sequences of A. thaliana and O. sativa MRPs and ZmMRP1, ZmMRP2, and ZmMRP3.
Previously, Bruce et al. (2000) reported that the expression of a second ABC protein, originally identified as an MRP, was coregulated with anthocyanin biosynthesis in Black Mexican Sweet culture cells after coexpression of the R and C1 transcription factors. However, in differentiated maize tissues, the expression of this gene is regulated by light and is not affected by the expression of the anthocyanin transcription factors (Figure 2D, Table 1). Sequencing of a full-length cDNA clone of this gene allowed us to determine that it is not an MRP but is instead a member of the ABCF family of ABC proteins (Dean and Allikmets, 2001). Based on its structure, we have named this gene ZmAbcf1. Like the other ABCF genes described to date, the predicted ZmABCF1 protein lacks TMDs and is therefore assumed to be soluble. The other members of the ABCF family described to date are not involved in transmembrane transport. Rather, they appear to play a role in protein translation (Tyzack et al., 2000).
A complete survey of the expression of the available maize MRPs (Table 1) showed that five maize MRPs are expressed in multiple tissues. Of these, ZmMrp3, ZmMrp5, ZmMrp6, and ZmMrp7 are expressed in all of the tissues that synthesize anthocyanin. As noted above, however, only ZmMrp3 shows any change in expression in response to anthocyanin content. ZmMrp7 shows the most consistent levels of expression throughout the plant but is not expressed in silks or in mature pollen. One of the most striking findings was that none of the ZmMrps tested showed detectable expression in mature pollen.
In our RNA gel blot analysis, we found that ZmMrp1 is expressed primarily in seedling tissues, whereas ZmMrp2 expression is only detectable in adult leaves. Within the limits of the differing genetic backgrounds and tissue types examined in these two studies, our results for the expression of ZmMrp1 and ZmMrp2 agree with an RT-PCR based expression analysis reported by Swarbreck et al. (2003). The primary difference between the two studies is the detection of very low levels of ZmMrp2 expression in all tissues tested by RT-PCR that were not shown by RNA gel blot analysis. This leaves open the possibility that genes we report as not being expressed are present but at levels below the limits of detection of RNA gel blotting.
Four of the ZmMrps are not expressed at detectable levels in the tissues tested but may be active in tissue types that were not included in this survey or, as noted above, may be present at levels below the detection limits of RNA gel blots. To shed light on the expression pattern of these MRPs, we examined the tissue of origin for their corresponding ESTs. Three of the ZmMrps were identified in tissues not included in the blot. ZmMrp4 ESTs were recovered from libraries of stressed seedlings and Lesion mimic mutant leaves, ZmMrp9 was found in a tassel primordium library, and ZmMrp10 ESTs were recovered from both the immature embryo and tassel primordia cDNA collections. The ZmMrp8 ESTs were recovered from a library derived from a mixture of adult tassel, silk, kernel, root, and leaf RNA, making it impossible to determine if this MRP is present at levels below the limit of RNA gel blot detection or was derived from one of the library tissues not included in our survey.
Structure of ZmMrp3
The partial clone of the anthocyanin-regulated MRP, ZmMrp3, contained only 1.8 kb of an estimated 5-kb cDNA; the complete transcript was recovered using rapid amplification of cDNA ends (5′ RACE). A full-length cDNA was generated by RT-PCR, cloned into bacteriophage M13, and fully sequenced. The full-length ZmMrp3 transcript is 5088 bp, with a 269-bp 5′ untranslated region (UTR) and 376-bp 3′ UTR bracketing a 4443-bp open reading frame encoding 1480 amino acids (Figure 3A). PCR amplification of the full-length genomic clone from inbred line A188 demonstrated a gene structure with eight exons and seven short introns (Figure 3A).
BLAST searches using the 1480–amino acid predicted protein sequence identified significant homology between ZmMrp3 and MRPs from other organisms, particularly A. thaliana and O. sativa. BLAST CD motif searches identified two ABC TMDs and two ABC transporter ATPase motifs (NBDs) arranged in the TMD-NBD-TMD-NBD sequence characteristic of many full-sized eukaryotic ABC transporters (Theodoulou, 2000). The putative NBDs contain both the Walker A and B boxes (Walker et al., 1982) as well as the ABC signature motif (Higgins et al., 1986) characteristic of all ABC transporters (Figure 3B). The protein sequences in these domains have extensive homology with the NBDs of A. thaliana MRPs 1 and 3 (Figure 3B), both of which have been shown to have ATPase and transporter activity (Lu et al., 1997; Tommasini et al., 1998). Kyte-Doolittle hydropathy analysis confirms the presence and relative location of the two predicted TMDs and also identifies a third likely TMD at the N terminus (Figure 3A). This N-terminal TMD is a unique feature of the MRP family of ABC transporters (Hipfner et al., 1997; Bakos et al., 1998), and in conjunction with the strong sequence homology with the Arabidopsis MRPs and the other structural features, identifies ZmMRP3 as a member of the MRP family of ABC proteins.
A genomic library was screened by PCR to recover 0.69 kb of sequence upstream of the putative transcription start site. Consistent with this being a promoter sequence, the cloned region contains both TATA and CAAT motifs directly upstream of the proposed transcription start site (Figure 3C). The promoter also contains both published types of anthocyanin-responsive element (ARE) (Tuerck and Fromm, 1994; Lesnick and Chandler, 1998), which overlap (Figure 3C). Figure 3D shows the alignment of the anthocyanin-responsive element described by Tuerck and Fromm (1994) and found in all published maize anthocyanin gene promoters. Ten of the fourteen bases in the ZmMrp3 ARE are identical to the consensus site, including all four of the bases that are conserved in this type of maize ARE (Figure 3D). Several degenerate myb-like binding (Sainz et al., 1997) sites were identified both immediately upstream and downstream from the ARE (Figure 3C). This arrangement of ARE and myb-like sites is found in the promoters of many of the maize anthocyanin structural genes and is required for proper B/Pl and R/C1 mediated regulation in both the Bronze1 and A1 genes (Roth et al., 1991; Tuerck and Fromm, 1994; Sainz et al., 1997).
To further test the regulation of ZmMrp3 by the anthocyanin transcription factors, we fused 687 bp of the ZmMrp3 promoter, including the putative ARE site, to a luciferase reporter gene (Bodeau and Walbot, 1996) to produce the ZmMrp3pLuc construct. Luciferase expression from this construct was measured in maize leaf protoplasts both in the presence and absence of anthocyanin synthesis controlled by R/C1 (Figure 3E). The expression of the reporter was regulated by the expression of the anthocyanin transcription factors, increasing approximately fourfold when present in anthocyanin producing cells.
Sequence Relationship between ZmMrp3 and Other MRPs
Based on alignments of the ZmMRP3 protein sequence with sequences of the MRPs from Arabidopsis and rice, ZmMRP3 falls into the Clade II grouping of MRPs (Kolukisaoglu et al., 2002); it is most similar to AtMRP10 and the recently annotated rice MRP with the GenBank accession number AP005828.1, which we term OsMRP13 (Figure 3F). It was interesting to note that the intron/exon structure of ZmMRP3 was not entirely consistent with that found for other members of Clade II. Whereas other Clade II members have between seven and ten introns that are found exclusively in the region between the two NBDs, ZmMRP3 has only five introns in this region but has two introns in the 5′ UTR that are absent in the other members of this clade (Kolukisaoglu et al., 2002).
The most striking finding in the sequence analysis was the similarity between ZmMRP3 and OsMRP13. Over the entire protein, these two genes share >79% amino acid identity. This similarity increases to 85% if the highly variable 5′ TMD is excluded. When compared with the relationships between all MRPs identified from A. thaliana (Kolukisaoglu et al., 2002) and O. sativa (Jasinski et al., 2003), the level of homology between ZmMRP3 and OsMRP13 is found only in duplicate gene pairs from within the same species, such as AtMRP 1 and 2, 4 and 14, 7 and 8, and 12 and 13 (Kolukisaoglu et al., 2002). The closest ZmMRP3 homolog in Arabidopsis is AtMRP10, which shares 60% amino acid identity with both ZmMRP1 and OsMRP13. This level of homology is similar to that found among the Arabidopsis and rice MRPs that are grouped together outside of the duplicate pairs (Figure 3E). Both OsMRP13 and AtMRP10 were identified based on homology searches of sequenced genomes; consequently, nothing is known about their functions.
Although the lack of full-length sequence from most of the maize MRPs precluded useful phylogenetic analysis, ClustalW comparisons of the available EST sequences did identify an extremely close homology between ZmMrp3 and ZmMrp4. The alignment of the entire 1.7-kb EST sequence of ZmMrp4 with the corresponding region of ZmMrp3 shows a nucleotide identity of 77%, which rises to 88% if only the coding regions of the genes are compared. Comparison of the predicted protein sequence shows 90% amino acid identity over the available open reading frame. Additional genomic sequencing of ZmMrp4 identified several differences in intron/exon structure compared with ZmMrp3, confirming that ZmMrp4 is a unique gene (data not shown). As noted in Table 1, ZmMrp4 expression was not detectable by RNA gel blot analysis in the tissues tested.
ZmMRP3 Is Localized to the Tonoplast
To act as an anthocyanin transporter, ZmMRP3 protein must be present at the point at which transmembrane transport occurs: the tonoplast. Subcellular localization of the protein was tested using a green fluorescent protein (GFP) fusion. The vacuole was stained with Lysotracker Red (Molecular Probes), a red fluorescent dye primarily localized to the vacuole (Swanson et al., 1998). The full-length ZmMrp3 cDNA was fused to eGFP (Cormack et al., 1996) under the control of the 35S promoter of Cauliflower mosaic virus (CaMV) in a fusion cassette created in bacteriophage M13. When this construct was electroporated into seedling protoplasts, fluorescence was visible only in a thin membrane outlining the large, red fluorescent central vacuole and around several smaller vacuoles (Figure 4A). Protoplasts electroporated with the empty vector showed GFP fluorescence throughout the cytoplasm (Figure 4B). It was interesting to note that protoplasts stained with Lysotracker Red showed both red and green fluorescence in the chloroplasts. Protoplasts transformed with ZmMRP3:GFP, GFP alone, and no GFP construct all showed a similar level of background green fluorescence in the chloroplast, indicating that this green fluorescence was unrelated to the expression of GFP.
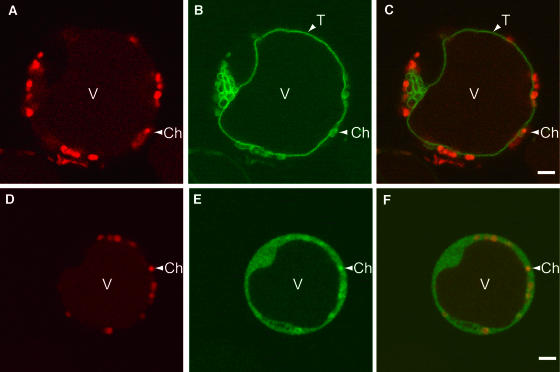
Figure 4.
Subcellular Localization of ZmMRP3.
Laser scanning confocal microscopy images of Z. mays leaf protoplasts. V, vacuole; Ch, chloroplast; T, tonoplast. Bars = 5 μm.
(A) to (C) Protoplasts expressing a ZmMRP3:GFP fusion under the control of the CaMV35S promoter.
(D) to (F) Protoplasts expressing GFP under the control of the CaMV35S promoter.
(A) and (D) Red fluorescence from Lysotracker Red staining and chloroplast autofluorescence.
(B) and (E) Green fluorescence from eGFP.
(C) and (F) Merged images showing both red and green fluorescence.
Antisense Mutants Alter Anthocyanin Biosynthesis
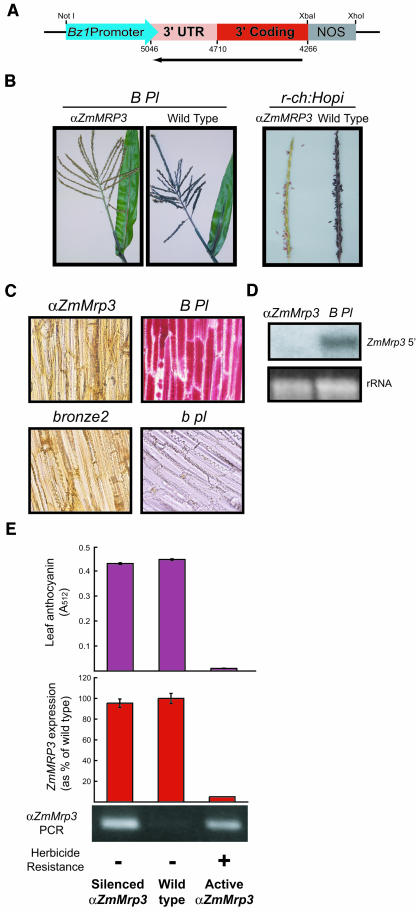
To test the in vivo role of ZmMrp3, we generated transgenic plants expressing an antisense construct of the 3′end of ZmMrp3 driven by the anthocyanin-regulated promoter of the Bronze1 gene (Figure 5A). This promoter was chosen to ensure that the antisense construct would be expressed coordinately and specifically with the anthocyanin pathway; if ZmMrp3 performs essential roles in other cells, expression should be unaffected. Twelve independent transformants were recovered after particle bombardment. The initial transgenic lines contain nonfunctional alleles of the required anthocyanin transcription factors. Consequently, each independent transformant was introgressed into two different stocks in which deep pigmentation is expected in leaves, tassel, anthers, and/or aleurone. In one of the twelve independent transformants screened, plants carrying the transgene and the transcription factors necessary for full pigmentation showed a marked reduction in pigmentation. The observed phenotype varied with genetic background, ranging from a burnished color to an almost complete absence of pigment in anthocyanin containing tissues of the adult plant, particularly the leaves and tassel (Figure 5B). These phenotypes bear a striking resemblance to that seen in the leaves and tassel of plants carrying a mutation in the Bz2 gene (Neuffer et al., 1997).
Figure 5.
Phenotypes of Pigmented Maize Expressing the Antisense Construct αZmMrp3.
(A) The antisense construct αZmMRP3.
(B) Phenotype of αZmMrp3 expression in the adult plant. Upper leaf and tassel phenotype of sibling plants expressing (αZmMrp3) or not expressing (wild type) the αZmMRP3 transgene in the strongly pigmented B/Pl and r-ch:Hopi genetic backgrounds.
(C) Cellular phenotype of αZmMrp3. Microscopic images (400× magnification) of glume face tissue taken from plants lacking anthocyanin (b/pl), with full anthocyanin content (B/Pl), or in a B/Pl genetic background, but showing pigment defects resulting from the presence of either the αZmMRP3 construct or the bronze2 mutation.
(D) αZmMRP3 eliminates the expression of ZmMrp3. RNA gel blot of total RNA from the leaf tissue of the B/Pl plants in (B) showing the loss of ZmMRP3 transcript in the antisense plants. Blots were probed with the 5′ UTR of ZmMrp3. Ethidium bromide–stained rRNA is included as a loading control.
(E) αZmMRP3 expression is required to produce the anthocyanin phenotype. Plants with silenced transgenes were identified by the absence of herbicide resistance combined with the presence of transgene, as shown by genomic PCR. Levels of native ZmMrp3 expression and anthocyanin pigmentation are fully restored to wild-type levels in the presence of silenced αZmMrp3.
To define the cellular nature of this phenotype, we examined the localization of anthocyanin in the face of the glume, a single layer tissue in which pigmentation is regulated by B/Pl expression. Consistent with this regulation, glumes from plants without anthocyanin (b/pl) lacked pigmentation, whereas glumes from plants with high levels of anthocyanin expression (B/Pl) showed high anthocyanin content with the pigment localized to the vacuole (Figure 5C). Glume tissue from the αZmMRP3 plants showed the bronze color characteristic of anthocyanin that cannot enter the vacuole (Marrs et al., 1995). This phenotype is strikingly similar to that seen in glumes from bronze2 mutant plants (Figure 5C). Because of the predominance of the vacuole in glume cells, it is impossible to determine if the mislocalized anthocyanin is accumulating in the thin layer of cytoplasm or is being deposited in the cell wall.
In all cases, reduced pigmentation was correlated with the presence of the transgene and elimination of the full-length ZmMrp3 transcript as tested by RNA gel blot analysis (Figure 5D). To confirm that reduction in ZmMRP3 expression resulted from antisense suppression and was not a product of the insertion of the transgene into an anthocyanin-related gene, we took advantage of the tight genetic linkage between the herbicide resistance marker and the transgene (data not shown) and the natural tendency of transgenes to be transcriptionally silenced over several generations of backcrossing. Silenced plants were identified by the absence of herbicide resistance combined with the presence of the transgene (Figure 5E). This indicates silencing of the entire transgene array, including both the herbicide resistance gene and the antisense construct. In silenced plants, the levels of both native ZmMrp3 expression and anthocyanin content were restored to wild-type levels (Figure 5E), confirming that transgene expression is required to produce the observed phenotype.
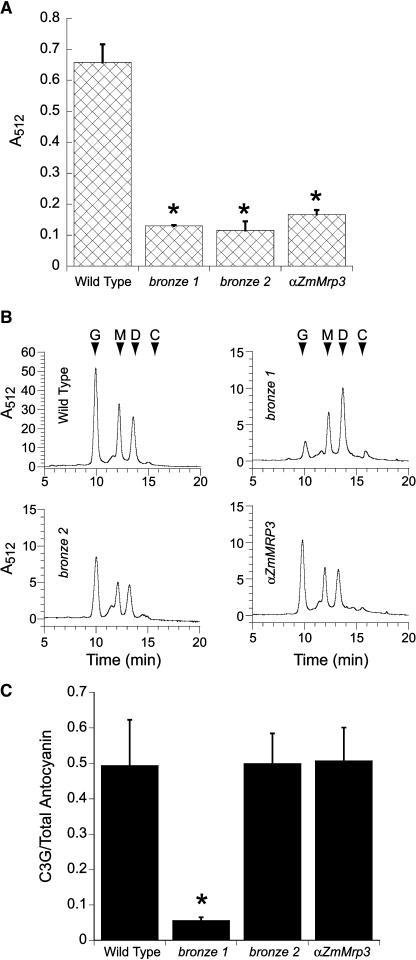
To characterize the biochemical nature of the color phenotype in comparison to mutants in both synthesis and localization, flavonoids were isolated from the adult leaves of sibling plants in families in which the transgene was segregating; siblings expressing the transgene are referred to as αZmMrp3 plants, whereas those not carrying the transgene are referred to as wild-type plants. We also extracted flavonoids from both bronze1 and bronze2 mutants. The bronze1 mutant lacks the glucosyl transferase required to synthesize cyanidin 3-glucoside from cyanidin (Larson and Coe, 1977), and bronze2 mutants lack an anthocyanin-specific GST and, as a result, are unable to localize cyanidin 3-glucoside to the vacuole (Marrs et al., 1995). Quantitatively, the αZmMRP3, bronze1, and bronze2 mutant plants tested all had a similar reduction in anthocyanin pigments compared with the purple control plants (Figure 6A). Expression levels of the anthocyanin biosynthetic genes A1 and A2 remain unchanged in the bronze1, bronze2, and αZmMrp3 plants, whereas these genes are not expressed in b/pl plants, which have no detectable anthocyanin (data not shown).
Figure 6.
Antisense Plants Have Reduced Overall Anthocyanin Content but No Change in the Ratio of Anthocyanin Species Present.
(A) Anthocyanin content of the adult leaves of plants in B/Pl genetic backgrounds. Shown are levels of anthocyanin in plants expressing the αZmMRP3 construct and sibling lacking the transgene as well as plants with both the bronze1 and bronze2 mutations. Results are the means of three separate extractions from the same tissue type. Bars represent standard error.
(B) αZmMRP3 plants have the same anthocyanin profile as the wild type and bronze2 mutant plants. Reverse phase HPLC analysis of anthocyanin pigment extracted from leaf tissue described in (A). Retention times of commercial anthocyanin standards are indicated by arrows.
(C) αZmMRP3 plants have the same ratio of anthocyanin pigments as the wild type and bronze2 mutant plants. Ratio of cyanidin 3-glucoside to total anthocyanin content in adult leaves as measured by reverse phase HPLC. Results for each background represent the mean of anthocyanin extractions from three individual plants. Bars represent standard error.
G, cyanidin 3-glucoside; M, cyandin 3-(6″-malonylglucoside); D, cyanidin 3-(3″,6″dimalonylglucoside); C, cyanidin. The asterisk indicates statistically significant difference from the wild type at P < 0.01.
HPLC analysis of the flavonoids in maize shows that the pigment profile in the bronze1 mutant differs from that seen in the αZmMRP1 and bronze2 plants in terms of anthocyanin species present. Purple maize contains three main anthocyanin pigments: cyanidin 3-glucoside (C3G), cyanidin 3-malonyl glucoside, and cyanidin 3-dimalonyl glucoside as well as a small amount of the C3G precursor cyanidin (Figure 6B; Fossen et al., 2001). The absence of the cyanidin specific UDP:glucosyl transferase activity in the bronze1 mutant (Larson and Coe, 1977) is reflected in the almost complete absence of C3G in bronze1 mutants as compared with wild-type plants (Figures 6B and 6C). By contrast, anthocyanins produced by the αZmMrp3 and bronze2 plants are found in ratios indistinguishable from wild-type plants (Figures 6B and 6C), indicating that all of the enzymatic steps of anthocyanin biosynthesis remain fully functional in both the αZmMrp3 and bronze2 plants. Therefore, the reduction in anthocyanin content in the αZmMRP3 and bronze2 plants appears to stem from the inability of C3G to be transported into the vacuole.
The Lack of an Aleurone Phenotype Suggests Redundancy in Anthocyanin Transport
Observations of kernels from >75 ears segregating for ZmMrp3 antisense expression showed no visible change in aleurone pigmentation. This was true of all plants, including homozygous antisense kernels derived from self-pollination of parents with a strong antisense induced phenotype in the plant body. The absence of an aleurone pigmentation phenotype suggests the presence of an aleurone-specific transporter that is able to compensate for the reduction in ZmMrp3 expression.
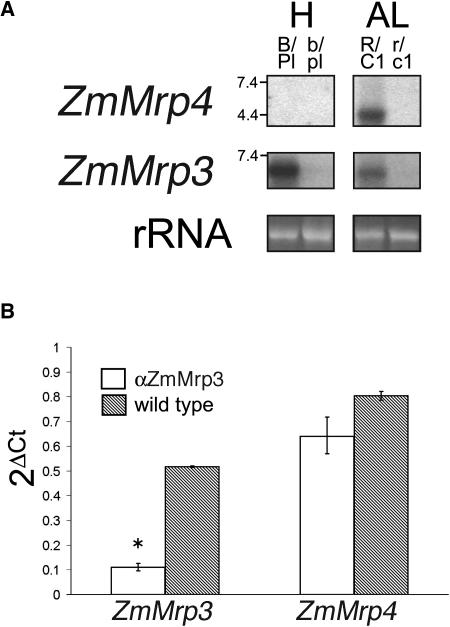
To investigate this possibility, we used the suite of MRP-specific probes described earlier to screen blots containing aleurone RNA. As expected, ZmMrp3 was expressed strongly in pigmented aleurone but was not expressed in near-isogenic unpigmented tissue (Figure 7A). This confirms that ZmMrp3 is coregulated with the anthocyanin structural genes in the aleurone. Surprisingly, ZmMrp4, which is not expressed in the plant body (Table 1), is highly expressed in pigmented aleurone (Figure 7A). ZmMrp4 was the only other MRP to show anthocyanin-regulated activity in the aleurone. It was curious to note that the ZmMRP4 transcript at ∼4.4 kb was significantly shorter than ZmMrp3. The extensive homology between ZmMrp3 and ZmMrp4 would suggest that these two genes have significant overlap in their substrate specificity, but the smaller size of the transcript indicates a large deletion in the ZmMrp4 gene. The predicted open reading frames for ZmMRP2, AtMRP13, and OsMRP1 are also significantly shorter than the other MRPs and hydropathy plots, and sequence alignments of the predicted proteins indicate that all three MRPs lack the 5′ TMD (Sánchez-Fernández et al., 2001; Jasinski et al., 2003; Swarbreck et al., 2003,). The lack of a 5′ TMD domain has also been shown in human Mrp5, a fully active transporter (Bakos et al., 1998). Therefore, it seems likely that the 5′ TMD is also absent in ZmMrp4. Confirmation of this through the cloning and sequencing of a full-length ZmMrp4 gene is ongoing.
Figure 7.
ZmMrp3 and ZmMrp4 Are Expressed in the Aleurone under the Control of the Anthocyanin Transcription Factors R and C1, but Only ZmMrp3 Expression Is Reduced by Antisense Expression.
(A) ZmMRP3 and ZmMRP4 expression is regulated by R and C1 in the aleurone. RNA gel blot of total RNA from the husk and aleurone tissue of the maize lines shown in Figure 2. The same blot was probed with the 3′ UTRs of ZmMRP3 and ZmMRP4. Ethidium bromide–stained rRNA is included as a loading control.
(B) ZmMrp4 expression is not affected by αZmMRP3. Quantitative RT-PCR of total RNA extracted from individual pigmented aleurones taken from three ears segregating for αZmMRP3 expression. The presence of the transgene was detected in 8 of 12 aleurones tested. ZmMRP3 and ZmMRP4 expression reported as 2−ΔCt values, which represents the cycle number at which log phase amplification begins, normalized for the expression of actin (see Methods). All reactions were done in triplicate.
H, husk; A, aleurone; B/Pl, tissue containing anthocyanin because of the expression of B/Pl; b/pl, tissues lacking anthocyanin because of the absence of B/Pl expression; R/C1, tissue containing anthocyanin because of the expression of R/C1; r/c1, tissues lacking anthocyanin because of the absence of R/C1 expression. The asterisk indicates statistically significant difference at P < 0.01.
Given the extensive sequence similarity between ZmMrp3 and ZmMrp4, there was a significant possibility that the αZmMrp3 construct could suppress the expression of both genes. To test the effect that the presence of the αZmMrp3 transgene has on ZmMrp4 expression, we use quantitative RT-PCR to measure the expression of ZmMrp3 and ZmMrp4 in individual aleurones taken from an ear that was segregating for transgene expression. In aleurones that expressed the transgene, there is a fivefold reduction in ZmMrp3 expression (Figure 7B). However, there is no statistically significant change in ZmMrp4 expression in the presence of the transgene (Figure 7B).
DISCUSSION
The MRPs are the most thoroughly studied of the plant ABC transporters, with many in vitro studies detailing the transport kinetics and offering suggestions of possible biological roles. This characterization of ZmMrp3 is however the first to describe the role of a plant MRP in the in vivo transport of a specific endogenous substrate. Our work demonstrates that ZmMrp3 is strongly expressed in tissues actively synthesizing anthocyanin (Figure 2C), and its promoter is regulated by the maize anthocyanin transcription factors (Figure 3E). As well as being expressed in the expected tissue types, we also show that the ZmMRP3 protein is localized to the tonoplast, the site of transport of the anthocyanin pigment into the vacuole (Figure 4). Transgenic antisense plants in which ZmMrp3 expression is reduced below detectable levels are defective in anthocyanin accumulation, but in the presence of a silenced transgene, expression and pigmentation are restored (Figures 5 and 6). Together, these data confirm the role the encoded protein plays in the transport of anthocyanin.
Although ZmMrp3 plays a role in the proper sequestration of anthocyanin, it does not appear to be the only transporter involved in this process. The antisense knockout of ZmMrp3 produces a strong phenotype in the leaves and tassels of pigmented plants, even when it is present as a single transgene array. However, this same construct fails to produce a phenotype in the aleurone when one, two, or three copies of the transgene array are present. In addition, the large number of genetic screens done by our lab and others have failed to identify a transporter gene as a component of the anthocyanin pathway in maize aleurone. This suggests the presence of a redundant transporter and/or an alternative and essential biological role for ZmMrp3.
Expression evidence argues strongly for the presence of an alternative transporter in the aleurone. ZmMrp4, a close homolog of ZmMrp3, is expressed in pigmented aleurone in a manner consistent with regulation by R and C1 (Figure 7A) and is unaffected by the expression of the antisense transgene (Figure 7B). This pattern of regulation is consistent with ZmMrp4 playing a role in anthocyanin sequestration in the aleurone and with genetic evidence suggesting that the transport step confers cell autonomy to the anthocyanin pathway. It is interesting to note that the transcript size of ZmMrp4 is significantly shorter than ZmMrp3, possibly reflecting the absence of the 5′-most TMD as seen in MRPs identified in maize (Swarbreck et al., 2003) and in many other organisms (Bakos et al., 1998; Kolukisaoglu et al., 2002; Jasinski et al., 2003). Studies to confirm the structure of this Zmrp4 and to test its ability to transport anthocyanin are in progress.
Redundancy in anthocyanin structural genes is not uncommon. Both the chalcone synthase and chalcone isomerase genes are present in multiple copies in maize (Coe et al., 1981; Grotewold and Peterson, 1994). The chalcone synthase genes Colorless2 (C2) and White pollen (Whp) present the most striking parallels to the anthocyanin transporters ZmMrp3 and ZmMrp4. C2 is expressed in all tissues that produce anthocyanin, whereas the second copy of chalcone synthase, Whp, is expressed in only a single tissue—the tapetum of the developing tassel (Coe et al., 1981). Either C2 or Whp is sufficient to ensure flavonol synthesis in the tapetum, which is essential for the structural integrity of the pollen tube during subsequent germination (Pollak et al., 1995). This redundancy helps ensure continued male fertility in the event that one of the chalcone synthase genes is lost. ZmMrp3 and ZmMrp4 appear to have a similar expression pattern, with ZmMRP3 expressed in all anthocyanin producing tissues, whereas ZmMrp4 is expressed in a single tissue, the aleurone.
The existence of another possible mechanism of anthocyanin transport has been inferred by data from A. thaliana, indicating that flavonoid transport is not limited to the MRPs. The tt12 mutant described by Debeaujon et al. (2001) yields seeds that show reduced accumulation of proanthocyanidin pigments in endothelial cells. This phenotype results from the loss of a multidrug and toxic compound extrusion (MATE) protein. MATE transporters are a recently described class of drug resistance proteins that use a Na+ antiport mechanism to drive transport (Morita et al., 2000). A similar MATE-type permease has been identified as being upregulated in tomato (Lycopersicon esculentum) plants with high anthocyanin content resulting from overexpressing the myb-type transcription factor Ant1 (Matthews et al., 2003), suggesting that this transport mechanism is conserved in many plant species.
The tt12 mutant has no impact on the anthocyanin content in the Arabidopsis plant body, indicating that another transporter is involved in anthocyanin sequestration in these tissues. Indeed, maize, Arabidopsis, and petunia (Petunia hybrida) all require the activity of a specific GST for correct anthocyanin sequestration (Marrs et al., 1995; Alfenito et al., 1998; Kitamura et al., 2004). This strongly suggests a role for an MRP in this process because GST activity is intimately linked with transport by MRPs in all systems studied to date. This conclusion is further supported by the ability to generate a phenocopy of the bronze2 mutant in maize cell lines through the application of vanadate (Marrs et al., 1995), a compound that specifically blocks ABC transporter activity but does not affect the function of MATE-type transporters (Morita et al., 2000).
The presence of a MATE-type flavonoid transporter needs to be reconciled with the GST-mediated transport of anthocyanin in maize, petunia, and Arabidopsis. The finding that the tt19 gene in Arabidopsis encodes a GST that is required for anthocyanin pigmentation in the plant body (Kitamura et al., 2004) combined with evidence for strong upregulation of a close homolog of the anthocyanin-specific GST from petunia (Alfenito et al., 1998) in Ant1 overexpressing tomato plants (Matthews et al., 2003) argues for the presence of both MATE and MRP transport systems in tomato and Arabidopsis. The simplest explanation is that these transport mechanisms are compound specific. In bronze2 and An9 mutants and in αZmMrp3 plants, anthocyanins are mislocalized (Marrs et al., 1995; Alfenito et al., 1998; Figure 5C), whereas in Arabidopsis tt12 mutants, the substrates in question are proanthocyanidins (Debeaujon et al., 2001). The fact that tt12 mutants do not affect anthocyanin pigmentation argues that the TT12 transporter is not involved in anthocyanin transport in Arabidopsis. Interestingly, bronze2 and αZmMrp3 plants show no alteration in the localization of the flavan-4-ol phlobaphenes (data not shown) confirming the fact that structurally similar substrates can have different transport mechanisms. The upregulation of both a GST and MATE permease in tomato plants that are overexpressing the anthocyanin pathway may reflect the channeling of excess anthocyanin precursors into the synthesis of proanthocyanidins or other related compounds that utilize a MATE permease for correct localization.
Another possibility is that the MRP is not a transporter but acts to modify the activity of MATE permeases. In mammals, a subclass of the MRPs termed sulfonylurea receptors (SUR) form a multimeric complex with a potassium channel, with the SUR proteins regulating the activity of the channel proteins (Babenko et al., 1998). Whereas SUR homologs are not present in plants (Sánchez-Fernández et al., 2001), the possibility exists that an MRP, perhaps in conjunction with an anthocyanin-specific GST, interacts with and regulates the anthocyanin transport activity of a MATE permease. Indeed, the regulation of ion channels by plant MRPs has been suggested as a possible role for AtMRP5 in Arabidopsis (Gaedeke et al., 2001; Klein et al., 2003). Some interplay between the Arabidopsis MATE transporter and the TT19 GST activity is suggested by the inability of tt19 mutant plants to efficiently sequester proanthocyanidins in the vacuole (Kitamura et al., 2004), but the development of an unusual intracellular structure as well as the inability of heterologously expressed petunia AN9 to complement only this aspect of the tt19 phenotype raises some doubts about the involvement of an MRP–MATE interaction. The identification of an anthocyanin-specific MRP will provide a launching point for future experiments to dissect these transport mechanisms.
Indirect evidence suggests that ZmMrp3 may play other, possibly essential roles, in maize. The expression of ZmMrp3 in the developing ear and unpigmented developing tassel (Figure 2C, Table 1) strongly argues for ZmMrp3 functions beyond anthocyanin transport. This would not be surprising given the extensive range of substrates and pleiotropic mutant phenotypes found for the A. thaliana MRPs (Lu et al., 1998; Gaedeke et al., 2001; Liu et al., 2001), which clearly suggest multiple biological roles for this family of transporters. Further, although we were able to generate a mutant phenotype by limiting the expression of the αZmMrp3 construct to tissues that accumulate anthocyanin, we were unable to recover antisense mutants driven by the constitutively expressed CaMV 35S promoter. This suggests that a loss of ZmMrp3 function throughout the plant may negatively impact viability.
Our discovery of an anthocyanin-specific MRP provides an important model system for investigating the biology and biochemistry of both GST and MRP activity. Anthocyanin sequestration, with a known substrate and specific GST and MRP requirements, provides an excellent system for investigating several additional questions relating to the molecular mechanism of GST-MRP transport. Is the GST only involved in binding the substrate or, once bound, does it actively direct the substrate to the MRP? Does transport involve a direct GST–MRP interaction, and is this process selective? Does the GSH bound to the GST act to enhance the rate of transport? In light of the size of the GST and MRP families and the rate at which endogenous GST and MRP substrates are being identified, discovering the role of each component of this transport mechanism will be key to understanding how these molecules are efficiently moved from the cytoplasm to the correct subcellular location.
METHODS
Plant Material
The R-r C1 B Pl and r-g c1 b pl anthocyanin tester lines are in A188/W23 genetic background. Transgenic lines were generated in A188xB73 (Hi Type II) and crossed into the anthocyanin tester lines listed. Inbred line A188 (r-r c1 b pl) seedlings were grown as recommended by Sheen (1993) for protoplast isolation. Genomic and cDNA sequences were generated from RNA and DNA extracted from A188 or W23 inbred lines.
Reagents
Cyanidin and C3G were purchased from Indofine Chemical Company (Somerville, NJ). Cyanidin 3-(6″-malonylglucoside) and cyanidin 3-(3″,6″dimalonylglucoside) were purchased from Polyphenols AS (Sandnes, Norway). Lysotracker Red DND-99 and SyberGreen were purchased from Molecular Probes (Eugene, OR). All other chemicals were purchased from Sigma (St. Louis, MO) except for HPLC grade solvents, which were purchased from Mallinckrodt (Paris, KY).
Gene Expression Analysis
Total RNA was isolated, according to the manufacturer's instructions, using RNAwiz (Ambion, Austin, TX) for all tissues except aleurone. Aleurone RNA was isolated with the Plant RNAeasy kit (Qiagen, Alameda, CA). RNA gel blotting was done as described by Raizada et al. (2001). The probes used are listed in Table 2. Probes were labeled with [32P]dCTP using the DECAprime II kit (Ambion) according to manufacturer's instructions. Probe specificity was confirmed by DNA gel blot analysis (Rudenko and Walbot, 2001).
Table 2.
Tissue-Specific Probes for RNA Gel Blots
| Gene | Probe Location | Probe Construction |
|---|---|---|
| ZmMrp1 | 3′ coding/UTR | 936-bp Product from 3′RACE clone digested with NcoI and NotI |
| ZmMrp2 | 3′ UTR | 368-bp PCR product: D156F 5′-AGCCGCAATGGAATGGTTGT- 3′ and D157R 5′-CGTCAGGCCTTTTAGAACAA-3′ |
| ZmMrp3 | 3′ UTR | 405-bp PCR product: D63F 5′-GTGACGGGAAAGTAGTGGAGTA-3′ and D64R 5′-CTGCCGCACAAGCATTTCTGT-3′ |
| ZmMrp3 | 5′ UTR/coding | 212-bp PCR product: D78F 5′-CATCGATCCGCGCTTTGCTTGATGGGATAA-3′ and D110R 5′-GCAGCTGGAGGAGTCGAGTATGTC-3′ |
| ZmMrp4 | 3′ UTR | 226-bp PCR product: DSP36F 5′-ATATCTAGAAGCCACCTC-3′ and D143R 5′-TGGAAGGGAAGCAATGAC-3′ |
| ZmMrp5 | 3′ coding/UTR | 604-bp Product from EST clone digested with EcoRV and HindIII |
| ZmMrp6 | 3′ coding/UTR | 736-bp Product from EST clone digested with HindIII |
| ZmMrp7 | 3′ UTR | 293-bp PCR product: D58F 5′-AGTGGATCCGCACGAGTTATTCCCTTGGTTCT-3′ and D57R 5′-AGTGGATCCGCACGAGTTATTCCCTTGGTTCT-3′ |
| ZmMrp8 | 3′ UTR | 329-bp PCR product: D150F 5′-TGAAGCGGAGCAAGATCCTC-3′ and D151R 5′-CGTGGAGCCAAAGCAACCTT-3′ |
| ZmMrp9 | 3′ UTR | 356-bp PCR product: D166F 5′-CTCAGATCTCGCTTTGGCATA-3′ and D167R 5′-ATCTGGGTCTGCTTCAGTATG-3′ |
| ZmMrp10 | 3′ UTR | 402-bp PCR product: D168F 5′-CCCAGGTCAGCGTCAACTTGT-3′ and D169R 5′-CACACTTCCTAGGGCGTTTGT-3′ |
| ZmAbcf1 | 3′ coding/UTR | 450-bp Product from EST clone cut with SstI and NotI |
Quantitative RT-PCR was done using an Opticon 2 (MJ Bioworks, South San Francisco, CA). Primers used for amplification were as follows: ZmMrp3, 5′-CATCTCTGTGGAAAGGGTTC-3′ and 5′-CTGTTCGCCCAACTATACCG-3′; ZmMrp4, 5′-TTCCGTGACCTCGTCAAAG-3′ and 5′-TAGTTCTTCGGATGTGCAAG-3′; αZmMrp3, 5′-CGGAATAAAGCGGACACGTT-3′ and 5′-TCTAGTGGAAGGCGCATCAA-3′; and actin, 5′-CGATTGAGCATGGCATTGTCA-3′ and 5′-CCCACTAGCGTACAACGAA-3′. All primer pairs were determined to have equal amplification efficiency, and the PCR products were resolved by electrophoresis and sequenced to confirm the identity of each amplicon. The rate of amplification was monitored using SyberGreen fluorescence at 83°C. The amplification of all samples was normalized to actin amplification to account for variation in RNA levels, and all biological samples were tested in triplicate. Results are reported as 2−ΔCt, where ΔCt is the number of PCR cycles required to the log phase of amplification for the experimental gene minus the same measure for actin (Livak and Schmittgen, 2001), or as 2−ΔCt normalized to wild-type 2−ΔCt. Aleurone samples giving a positive signal for the αZmMrp3 were classified as transgenic. Standard error of the mean was calculated using standard statistical methods.
Cloning and Characterization of ZmMrp3 and ZmAbcf1
The full-length cDNA sequences of these genes were recovered using the 5′RACE system version 2.0 from Invitrogen (Carlsbad, CA) according to manufacturer's instructions with the following modification. Nested PCR was used to amplify the 5′RACE product. The product of the first PCR reaction was separated on agarose gel, and the DNA present in that section of the gel corresponding to the expected size of the 5′RACE fragment was purified. This gel-purified DNA was used as the template for the second PCR reaction.
Full-length cDNA clones were amplified by PCR using Taq HiFi (Invitrogen) with the following primers. For ZmMrp3: D109F, 5′-CCATCGATATCCGCACGCGCCTTTAGCTACTA-3′, and D95R, 5′-TACCGCGGATTAGATGTGTATGACCAGTACTC-3′; for ZmABCF1I: D88F, 5′-GCGGATCCACCCAACCCAACCAAGAAACAGAGG-3′, and D89R, 5′-GGAATTCATAGCATACGGCACAGCACGCAGAGT-3′. ZmMrp3 is toxic in standard Escherichia coli cloning strains so it was cloned into the ClaI/SstII sites of Phagescript M13 (Stratagene, La Jolla, CA). ZmABCF1 was cloned into the EcoR1/BamH1 sites of pBluescript SK− (Stratagene). Both clones were fully sequenced. Genomic sequence of ZmMrp3 was recovered by PCR with Taq HiFi. Primers designed to sequences throughout the gene were used to ensure 3× coverage of the entire gene. Promoter sequence was recovered by digesting genomic DNA with HindIII and ligating it into pBluescript KS+. The resulting library was amplified by nested PCR using Taq HiFi. Amplification was done in two rounds with M13 reverse and T3 sequencing primers and the gene-specific primers D110R, 5′-GCAGCTGGAGGAGTCGAGTATGTC-3′, and D129R, 5′-TCCCCGCGGATGATTCGTGCTTCAACTCCA-3′. The product was TA cloned into the pGEM-T vector (Promega, Madison, WI). Three independent clones were sequenced.
ARE motifs in ZmMRP3 were identified by manual sequence analysis and using the MEME system (Bailey and Elkan, 1994). The proximal 687 bp of the ZmMrp3 promoter were cloned into the luciferase construct described by Bodeau and Walbot (1996). The resulting construct was electroporated into maize (Zea mays) leaf protoplasts following the protocol of Sheen (1993) in the presence or absence of the anthocyanin transcription factor R and C1. Luciferase expression was determined as previously described (Bodeau and Walbot, 1996).
Phylogenetic and Sequence Analysis
ClustalW alignment of DNA and protein sequences was done with the Megalign package (DNAStar, Madison, WI). Protein distance matrix, bootstrap values (1000 replicates), and neighbor-joining consensus trees were calculated using PHYLIP (Felsenstein, 1989). Arabidopsis thaliana and rice (Oryza sativa) sequences were recovered according to Kolukisaoglu et al. (2002) and Jasinski et al. (2003). In one instance where a discrepancy was found between the annotation of OsMRP1 and OsMRP10 in published articles and National Center for Biotechnology Information (NCBI) databases, the NCBI designations were used.
GFP Constructs and Confocal Scanning Microscopy
Because the full-length clone of ZmMrp3 was toxic to E. coli, a GFP construct was constructed in Phagescript M13. The CaMV 35S promoter from pMR51 (Raizada and Walbot, 2000) was cloned into the KpnI/SalI sites of Phagescript M13. The enhanced GFP (Cormack et al., 1996) ocs 3′ cassette from pEZS-NL (D. Ehrhardt, http://deepgreen.stanford.edu) was cloned into the EcoRI/SpeI site of pZERO-2 (Invitrogen). The resulting construct was cut with SstI and SstII and cloned into the Phagescript M13 carrying the CaMV 35S promoter to produce the 35SGFPM13 vector. Full-length ZmMrp3 was cloned into the ClaI/SacII sites of 35SGFPM13, resulting in ZmMrp3:GFP. The ZmMrp3:GFP fusion site was sequenced to ensure that the two genes were in frame.
ZmMrp3:GFP was electroporated into etiolated maize protoplasts as described (Sheen, 1993) with the following modifications. The final electroporation was done in 1 mL of buffer with 50 μg of ZmMrp3:GFP DNA. Protoplasts were incubated overnight in the light. Protoplasts were then incubated for 4 h in protoplast incubation solution containing 10 μM Lysotracker Red DND-99 and washed three times in protoplast incubation solution before viewing.
Scanning confocal imaging was done using a Nikon Diaphot inverted fluorescence microscope (Tokyo, Japan) with a Nikon 60 × 1.2 numerical aperture water immersion objective and a Bio-Rad MRC 1024 confocal head. Images were analyzed with LASERSHARP software (Bio-Rad, Hercules, CA).
Transgenic Plants
Two antisense vectors were constructed for bombardment. The ZmMrp3 cDNA was amplified by PCR using the primers D60F 5′-GGAATTCATCGATCTTATCATGGCTCTTCTTCCTC-3′ and D59R 5′-GGAATTCAGATCTTTAAAGCCTTGGAACAGCCT-3′. To produce the ZmMrp3 antisense construct driven by the CaMV35s promoter, this PCR product was cut with XbaI and BglI and cloned into the BamHI/XbaI sites of pMR51 (Raizada and Walbot, 2000) resulting in clone p35SαZmMrp3. The ZmMrp3 insert was fully sequenced. To create an antisense clone driven by the Bronze1 promoter, the PCR product was cut with XbaI and ClaI and cloned into pBluescript SK to create pBSKcd. This clone was fully sequenced. The promoter region from the Bronze1 genomic clone pMbzR1 (Klein et al., 1989) was cut out with PstI and BamHI and cloned into pZero-2. This construct was then cut with XhoI and HindIII and cloned into pBSKcd. The resultant construct was cut with EcoRI and XhoI, and the nos 3′ region from pMR51 (Raizada and Walbot, 2000) was inserted, producing construct αZmMrp3.
Both constructs αZmMrp1 and p35sαZmMrp1 were submitted to the Iowa State Plant Transformation Facility (Ames, IA), which produced transgenic plants by the particle bombardment method (http://www.agron.iastated.edu/ptf/Web/-mainframe.htm). Twenty-nine independent transformants were received, 10 expressing the p35sαZmMrp1 construct and 19 expressing the pBz1αZmMrp3 construct, as measured by RNA gel blot analysis. PCR screening for the presence of the transgene was done under standard conditions using the same αZmMrp3 primers used in the expression analysis. Further characterization of the transgenic plants is discussed in Results.
Pigment Extraction, Quantification, and HPLC
Anthocyanin pigments for absorbance analysis were extracted by grinding 250 mg of tissue in 1 mL of 0.1 N HCl in methanol. The ground tissue was left in the HCl/methanol overnight and then centrifuged for 5 min at full speed in a microcentrifuge to pellet the solid plant material. Absorbance of a 200-μL volume of the supernatant was read at 512 nm using a SpectraMax 250 plate reader (Molecular Devices, Sunnyvale, CA).
Anthocyanin extractions for HPLC analysis were done as above except the tissue was ground in 0.1 N HCl/methanol for 1 min and immediately spun at full speed in a microcentrifuge for 5 min. The supernatant was removed and centrifuged for a further 5 min. The resulting supernatant was diluted with 5% acetic acid in ratios ranging from 1:1 to 20:1, depending on tissue type, and immediately loaded onto the HPLC.
HPLC was done using a Dionex GP40 gradient pump (Dionex, Sunnyvale, CA) using a Microsorb 100-5 C18 column (Varian, Palo Alto, CA). Data were collected and analyzed using PEAKNET software (Dionex, Sunnyvale, CA). Pigment separation was by gradient elution with a flow rate of 0.75 mL/min. Solvent A, 5% acetic acid; solvent B, acetonitrile, 1 min at 90% A, 10% B; from 90% A, 10% B to 55% A, 45% B in 17.5 min; to 100% B in 2.5 min, at 100% B for 1 min; to 90% A, 10% B in 3 min; at 90% A, 10% B for 3 min. Absorbance was detected at 512 nm using a Dionex AD20 detector.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AP005828.1, AY609318 (ZmMrp3), and AY615521 (ZmAbcf1).
Acknowledgments
We would like to thank R. Meeley and Pioneer Hi-Bred for access to their EST database and the provision of materials, F. Theodoulou and J.V. Dean for providing data before publication, and R. Kurtz and MJ Bioworks for providing the Opticon 2 and for technical advice on quantitative real-time PCR. We would also like to thank K. Brewer, M. Abreu, J. Fernandes, T. DeHoog, and other members of the Walbot Lab for laboratory and field assistance. We would like to acknowledge D. Ehrhardt for the gift of the 35SGFP constructs and assistance with confocal microscopy. We would like to thank S. Long for providing materials and HPLC equipment. L. Mueller, G. Rudenko, C. Schmid, and S. Shah provided helpful suggestions and discussion. This work was supported by a grant from the National Science Foundation (IBN 9603927). C.D.G. was supported in part by a Natural Sciences and Engineering Research Council of Canada PGS-B fellowship.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Virginia Walbot (walbot@stanford.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.022574.
References
- Abell, L.M., Schloss, J.V., and Rendina, A.R. (1993). Target-site directed herbicide design. ACS Symp. Ser. 524, 16–37. [Google Scholar]
- Ahmed, M.S., Ainley, K., Parish, J.H., and Hadi, S.M. (1994). Free radical-induced fragmentation of proteins by quercetin. Carcinogenesis 15, 1627–1630. [DOI] [PubMed] [Google Scholar]
- Alfenito, M.R., Souer, E., Goodman, C.D., Buell, R., Mol, J., Koes, R., and Walbot, V. (1998). Functional complementation of anthocyanin sequestration in the vacuole by widely divergent glutathione S-transferases. Plant Cell 10, 1135–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babenko, A.P., Aguilar-Bryan, L., and Bryan, J. (1998). A view of sur/KIR6.X, KATP channels. Annu. Rev. Physiol. 60, 667–687. [DOI] [PubMed] [Google Scholar]
- Bailey, T.L., and Elkan, C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36. [PubMed] [Google Scholar]
- Bakos, E., Evers, R., Szakács, G., Tusnády, G.E., Welker, E., Szabó, K., de Haas, M., van Deemter, L., Borst, P., Varádi, A., and Sarkadi, B. (1998). Functional multidrug resistance protein (MRP) lacking the N-terminal transmembrane domain. J. Biol. Chem. 273, 32167–32175. [DOI] [PubMed] [Google Scholar]
- Bodeau, J.P., and Walbot, V. (1996). Structure and regulation of the maize Bronze2 promoter. Plant Mol. Biol. 32, 599–609. [DOI] [PubMed] [Google Scholar]
- Borst, P., Evers, R., Kool, M., and Wijnholds, J. (1999). The multidrug resistance protein family. Biochim. Biophys. Acta 1461, 347–357. [DOI] [PubMed] [Google Scholar]
- Bruce, W., Folkerts, O., Garnaat, C., Crasta, O., Roth, B., and Bowen, B. (2000). Expression profiling of the maize flavonoid pathway genes controlled by estradiol-inducible transcription factors CRC and P. Plant Cell 12, 65–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe, E.H., McCormick, S.M., and Modena, S.A. (1981). White pollen in maize. J. Hered. 72, 318–320. [Google Scholar]
- Cormack, B.P., Valdivia, R.H., and Falkow, S. (1996). FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173, 33–38. [DOI] [PubMed] [Google Scholar]
- Dean, M., and Allikmets, R. (2001). Complete characterization of the human ABC gene family. J. Bioenerg. Biomembr. 6, 475–479. [DOI] [PubMed] [Google Scholar]
- Debeaujon, I., Peeters, A.J.M., Léon-Kloosterziel, K.M., and Koornneef, M. (2001). The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell 13, 853–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, R., and Dixon, D.P. (2000). The role of glutathione transferases in herbicide metabolism. In Herbicides and Their Mechanisms of Action, A.H. Cobb and A.C. Kirkwood, eds (Sheffield: Sheffield Academic Press), pp. 33–71.
- Felsenstein, J. (1989). PHYLIP—Phylogeny inference package (version 3.2). Cladistics 5, 164–166. [Google Scholar]
- Fossen, T., Slimestad, R., and Andersen, Ø.M. (2001). Anthocyanins from maize (Zea mays) and reed canarygrass (Phalaris arundinacea). J. Agric. Food Chem. 49, 2318–2321. [DOI] [PubMed] [Google Scholar]
- Gaedeke, N., Klein, M., Kolukisaoglu, U., Forestier, C., Müller, A., Ansorge, M., Becker, D., Mamnun, Y., Kuchler, K., Schulz, B., Mueller-Roeber, B., and Martinoia, E. (2001). The Arabidopsis thaliana ABC transporter AtMRP5 controls root development and stomata movement. EMBO J. 20, 1875–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold, E., and Peterson, T. (1994). Isolation and characterization of a maize gene encoding chalcone flavonone isomerase. Mol. Gen. Genet. 242, 1–8. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt, R. (1999). Phytoalexins: What have we learned after 60 years? Annu. Rev. Phytopathol. 37, 285–306. [DOI] [PubMed] [Google Scholar]
- Higgins, C.F., Hiles, I.D., Salmond, G.P.C., Gill, D.R., Downie, J.A., Evans, I.J., Holland, I.B., Gray, L., Buckel, S.D., Bell, A.W., and Hermodson, M.A. (1986). A family of related ATP-binding subunits coupled to many distinct biological processes in bacteria. Nature 323, 448–450. [DOI] [PubMed] [Google Scholar]
- Hinder, B., Schellenberg, M., Rodoni, S., Ginsburg, S., Vogt, E., Martinoia, E., Matile, P., and Hörtensteiner, S. (1996). How plants dispose of chlorophyll catabolites. Directly energized uptake of tetrapyrrolic breakdown products into isolated vacuoles. J. Biol. Chem. 271, 27233–27236. [DOI] [PubMed] [Google Scholar]
- Hipfner, D.R., Almquist, K.C., Leslie, E.M., Gerlach, J.H., Grant, C.E., Deeley, R.G., and Cole, S.P.C. (1997). Membrane topology of the multidrug resistance protein (MRP). A study of glycosylation-site mutants reveals an extracytosolic NH2 terminus. J. Biol. Chem. 272, 23623–23630. [DOI] [PubMed] [Google Scholar]
- Ishikawa, T., Li, Z.S., Lu, Y.P., and Rea, P.A. (1997). The GS-X pump in plant, yeast, and animal cells: Structure, function, and gene expression. Biosci. Rep. 17, 189–207. [DOI] [PubMed] [Google Scholar]
- Jasinski, M., Ducos, E., Martinoia, E., and Boutry, M. (2003). The ATP-binding cassette transporters: Structure, function, and gene family comparison between rice and Arabidopsis. Plant Physiol. 131, 1169–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermicle, J.L., and Alleman, M. (1990). Gametic imprinting in maize in relation to the angiosperm life cycle. Dev. Suppl. 9–14. [PubMed]
- Kitamura, S., Shikazono, N., and Tanaka, A. (2004). TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J. 37, 104–114. [DOI] [PubMed] [Google Scholar]
- Klein, M., Martinoia, E., Hoffmann-Thoma, G., and Weissenböck, G. (2000). A membrane-potential dependent ABC-like transporter mediates the vacuolar uptake of rye flavone glucuronides: Regulation of glucuronide uptake by glutathione and its conjugates. Plant J. 21, 289–304. [DOI] [PubMed] [Google Scholar]
- Klein, M., Perfus-Barbeoch, L., Frelet, A., Gaedeke, N., Reinhardt, D., Mueller-Roeber, B., Martinoia, E., and Forestier, C. (2003). The plant multidrug resistance ABC transporter AtMRP5 is involved in guard cell hormonal signalling and water use. Plant J. 33, 119–129. [DOI] [PubMed] [Google Scholar]
- Klein, T.M., Roth, B.A., and Fromm, M.E. (1989). Regulation of anthocyanin biosynthetic genes introduced into intact maize tissues by microprojectiles. Proc. Natl. Acad. Sci. USA 86, 6681–6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolukisaoglu, H.Ü., Bovet, L., Klein, M., Eggmann, T., Geisler, M., Wanke, D., Martinoia, E., and Schulz, B. (2002). Family business: The multidrug-resistance related protein (MRP) ABC transporter genes in Arabidopsis thaliana. Planta 216, 107–119. [DOI] [PubMed] [Google Scholar]
- Larson, R.L., and Coe, E.H. (1977). Gene dependent flavonoid glucosyltransferase in maize. Biochem. Genet. 15, 153–156. [DOI] [PubMed] [Google Scholar]
- Lesnick, M.L., and Chandler, V.L. (1998). Activation of the maize anthocyanin gene a2 is mediated by an element conserved in many anthocyanin promoters. Plant Physiol. 117, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, G.S., Sánchez-Fernández, R., Li, Z.S., and Rea, P.A. (2001). Enhanced multispecificity of Arabidopsis vacuolar multidrug resistance-associated protein-type ATP-binding cassette transporter, AtMRP2. J. Biol. Chem. 276, 8648–8656. [DOI] [PubMed] [Google Scholar]
- Livak, K.J., and Schmittgen, T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lu, Y.-P., Li, Z.-S., Drozdowicz, Y.M., Hörtensteiner, S., Martinoia, E., and Rea, P.A. (1998). AtMRP2, an Arabidopsis ATP binding cassette transporter able to transport glutathione S-conjugates and chlorophyll catabolites: Functional comparisons with AtMRP1. Plant Cell 10, 267–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Y.P., Li, Z.S., and Rea, P.A. (1997). AtMRP1 gene of Arabidopsis encodes a glutathione S-conjugate pump: Isolation and functional definition of a plant ATP-binding cassette transporter gene. Proc. Natl. Acad. Sci. USA 94, 8243–8248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs, K.A., Alfenito, M.R., Lloyd, A.M., and Walbot, V. (1995). A glutathione S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2. Nature 375, 397–400. [DOI] [PubMed] [Google Scholar]
- Martinoia, E., Grill, E., Tommasini, R., Kreuz, K., and Amrhein, N. (1993). ATP-dependent glutathione S-conjugate export pump in the vacuolar membrane of plants. Nature 364, 247–249. [Google Scholar]
- Matthews, H., Clendennen, S.K., Caldwell, C.G., Liu, X.L., Connors, K., Matheis, N., Schuster, D.K., Menasco, D.J., Wagoner, W., Lightner, J., and Wagner, D.R. (2003). Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell 15, 1689–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita, Y., Kataoka, A., Shiota, S., Mizushima, T., and Tsuchiya, T. (2000). NorM of vibrio parahaemolyticus is an Na(+)-driven multidrug efflux pump. J. Bacteriol. 182, 6694–6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuffer, M.G., Coe, E.H., and Wessler, S.R. (1997). Mutants of Maize. (Plainview, NY: Cold Spring Harbor Laboratory Press), p. 77.
- Pollak, P.E., Hansen, K., Astwood, J.D., and Taylor, L.P. (1995). Conditional male-fertility in maize. Sex. Plant Reprod. 8, 231–241. [Google Scholar]
- Raizada, M.N., Benito, M.I., and Walbot, V. (2001). The MuDR transposon terminal inverted repeat contains a complex plant promoter directing distinct somatic and germinal programs. Plant J. 25, 79–91. [DOI] [PubMed] [Google Scholar]
- Raizada, M.N., and Walbot, V. (2000). The late developmental pattern of Mu transposon excision is conferred by a cauliflower mosaic virus 35S-driven MURA cDNA in transgenic maize. Plant Cell 12, 5–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea, P.A., Li, Z.S., Lu, Y.P., Drozdowicz, Y.M., and Martinoia, E. (1998). From vacuolar GS-X pumps to multispecific ABC transporters. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 727–760. [DOI] [PubMed] [Google Scholar]
- Roth, B.A., Goff, S.A., Klein, T.M., and Fromm, M.E. (1991). C1- and R-dependent expression of the maize Bz1 gene requires sequences with homology to mammalian myb and myc binding sites. Plant Cell 3, 317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko, G.N., and Walbot, V. (2001). Expression and post-transcriptional regulation of maize transposable element MuDR and its derivatives. Plant Cell 13, 553–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueff, J., Gaspar, J., and Laires, A. (1995). Structural requirements for mutagenicity of flavonoids upon nitrosation. A structure-activity study. Mutagenesis 10, 325–328. [DOI] [PubMed] [Google Scholar]
- Sainz, M.B., Grotewold, E., and Chandler, V.L. (1997). Evidence for direct activation of an anthocyanin promoter by the maize C1 protein and comparison of DNA binding by related Myb domain proteins. Plant Cell 9, 611–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Fernández, R., Davies, T.G.E., Coleman, J.O.D., and Rea, P.A. (2001). The Arabidopsis thaliana ABC protein superfamily, a complete inventory. J. Biol. Chem. 276, 30231–30244. [DOI] [PubMed] [Google Scholar]
- Sheen, J. (1993). Protein phosphatase activity is required for light-inducible gene expression in maize. EMBO J. 12, 3497–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley, B.W. (1996). Flavonoid biosynthesis: ‘New’ functions for an ‘old’ pathway. Trends Plant Sci. 1, 377–382. [Google Scholar]
- Swanson, S.J., Bethke, P.C., and Jones, R.L. (1998). Barley aleurone cells contain two types of vacuoles. Characterization of lytic organelles by use of fluorescent probes. Plant Cell 10, 685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbreck, D., Ripoll, P.-J., Brown, D.A., Edwards, K.J., and Theodoulou, F. (2003). Isolation and characterisation of two multidrug resistance associated protein genes from maize. Gene 315, 153–164. [DOI] [PubMed] [Google Scholar]
- Theodoulou, F.L. (2000). Plant ABC transporters. Biochim. Biophys. Acta 1465, 79–103. [DOI] [PubMed] [Google Scholar]
- Tommasini, R., Vogt, E., Fromenteau, M., Hörtensteiner, S., Matile, P., Amrhein, N., and Martinoia, E. (1998). An ABC-transporter of Arabidopsis thaliana has both glutathione-conjugate and chlorophyll catabolite transport activity. Plant J. 13, 773–780. [DOI] [PubMed] [Google Scholar]
- Tuerck, J.A., and Fromm, M.E. (1994). Elements of the maize A1 promoter required for transactivation by the anthocyanin B/C1 or phlobaphene P regulatory genes. Plant Cell 6, 1655–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyzack, J.K., Wang, X., Belsham, G.J., and Proud, C.G. (2000). ABC50 interacts with eukaryotic initiation factor 2 and associates with the ribosome in an ATP-dependent manner. J. Biol. Chem. 275, 34131–34139. [DOI] [PubMed] [Google Scholar]
- Walker, J.E., Saraste, M., Runswick, M.J., and Gay, N.J. (1982). Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1, 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]