Abstract
Secretion is a fundamental process providing plants with the means for disposal of solutes, improvement of nutrient acquisition, and attraction of other organisms. Specific secretory organs, such as nectaries, hydathodes, and trichomes, use a combination of secretory and retrieval mechanisms, which are poorly understood at present. To study the mechanisms involved, an Arabidopsis thaliana activation tagged mutant, glutamine dumper1 (gdu1), was identified that accumulates salt crystals at the hydathodes. Chemical analysis demonstrated that, in contrast with the amino acid mixture normally present in guttation droplets, the crystals mainly contain Gln. GDU1 was cloned and found to encode a novel 17-kD protein containing a single putative transmembrane span. GDU1 is expressed in the vascular tissues and in hydathodes. Gln content is specifically increased in xylem sap and leaf apoplasm, whereas the content of several amino acids is increased in leaves and phloem sap. Selective secretion of Gln by the leaves may be explained by an enhanced release of this amino acid from cells. GDU1 study may help to shed light on the secretory mechanisms for amino acids in plants.
INTRODUCTION
Although plants store a variety of compounds in the vacuole, which are not currently needed or which may even be potentially harmful, they also excrete substances from roots into the soil and release compounds from the aerial parts. In leaves, excretion, which serves a variety of purposes, is mediated by highly specialized structures called hydathodes. Hydathodes are positioned at the leaf margins, close to the endings of conducting vessels. Hydathodes mediate guttation (i.e., secretion of a sap containing ions, metabolites, and proteins) (Komarnytsky et al., 2000; Mizuno et al., 2002; Grunwald et al., 2003). The guttation process seems to be related to the water status of the plant (i.e., stomata opening and leaf water potential) (Takeda and Glenn, 1989; Enns et al., 2000).
The structure of the hydathodes suggests an involvement not only in the active secretion of solutes but also in the selective absorption and retrieval of both inorganic and organic solutes (Esau, 1977). Little is known about the exact mechanism of hydathode function. A widely accepted hypothesis is that, in analogy to kidney, hydathodes retrieve ions and organic molecules from the apoplasm and the xylem sap and exudate water via the guttation stream. In agreement with this hypothesis, several genes encoding transporters were found to be expressed in hydathodes (e.g., transporters for potassium, sulfate, or N-heterocycles) (Lagarde et al., 1996; Shibagaki et al., 2002; Bürkle et al., 2003; Pilot et al., 2003). Alternatively, secretion may be driven by active export of osmolytes leading to passive efflux of water. Under certain conditions, large quantities of solutes may be excreted by hydathodes (e.g., fertilized rye grass and tea plants secrete amino acids from hydathodes) (Greenhill and Chibnall, 1934; Nagata, 1986), indicating that hydathodes fulfill a specific excretory role. Whether accumulation of these substances is because of active excretion from the cells or lack of retrieval from the guttation sap is currently unclear.
To gain insight into the molecular process of guttation and the specific recovery or secretion of compounds, we searched for mutants with altered hydathode activity. Because no such mutants have been reported by screening conventional ethyl methanesulfonate–mutagenized or knockout libraries of Arabidopsis thaliana, a collection of activation tagged lines was screened. Activation tagging enables unbiased upregulation on a genome-wide scale by the insertion of a T-DNA containing endogenous enhancers or promoters (Weigel et al., 2000). The T-DNA used here contained the BASTA resistance gene (BASTAR) and four repeats of the Cauliflower mosaic virus (CaMV) 35S enhancer sequence, a system successfully used for activation tagging (Borevitz et al., 2000; Weigel et al., 2000; Li et al., 2001; Xia et al., 2004). Mutants were searched that would oversecrete compounds leading to precipitation on the leaf surface. For this purpose, growth conditions were established in which plants were watered from below to prevent the washing out of the precipitate. Two independent activation tagged lines secreting Gln through the hydathodes were identified. In both mutants, hypersecretion is a result of overexpression of a novel gene, GLUTAMINE DUMPER1 (GDU1), with homologs found only in plants. GDU1 contains a single membrane span. Phenotypic and physiological characterization of the mutants suggests that GDU1 is involved in export of amino acids.
RESULTS
Amino Acids Are Present in the Guttation Fluid
Wild-type Arabidopsis plants were grown in growth chambers and exposed to high humidity for one night. Guttation droplets were collected the next morning, lyophilized, and analyzed for amino acid content by HPLC. The guttation contained significant amounts of Asp, Gln, and His (Table 1). For comparison, root xylem exudate from Arabidopsis mainly contained Gln, whereas leaf exudate (phloem sap) composition was more complex (Table 1), consistent with previous reports (Shelp, 1987; Schobert and Komor, 1989, 1990; Lam et al., 1995). Thus, the amino acid composition of the guttation fluid in wild-type plants is different from that of xylem and phloem saps.
Table 1.
Relative Amino Acid Content of Guttation Fluid, Root Exudate, and Leaf Exudate from Wild-Type Plants
| Amino Acid | Guttation (n = 1)a | Root Xylem Exudate (n = 4) | Leaf Exudate (n = 8) |
|---|---|---|---|
| Asp | 19.3 | 0.6 ± 0.4 | 7.2 ± 2.1 |
| Thr | ND | 1.6 ± 0.3 | 2.4 ± 1.2 |
| Ser | ND | 1.3 ± 0.2 | 6.3 ± 2.7 |
| Asn | ND | 2.9 ± 1.4 | 4.3 ± 1.3 |
| Glu | ND | 1.8 ± 0.3 | 11.6 ± 4.9 |
| Gln | 51.9 | 87.4 ± 6.6 | 38.4 ± 8.5 |
| His | 28.8 | 0.7 ± 0.5 | 1.1 ± 0.3 |
| Other AAb | ND | 4.6 ± 2.1 | 35.3 ± 11.6 |
Values are average ± sd of the percentage (molar fraction of all amino acids detected by HPLC) of each amino acid (AA). ND, not detected.
Number of samples.
Namely, Pro, Gly, Ala, Val, Met, Ile, Leu, Tyr, Phe, γ-amino-butyric acid, Orn, Lys, and Arg.
Identification of Two Salt Secreting Mutants
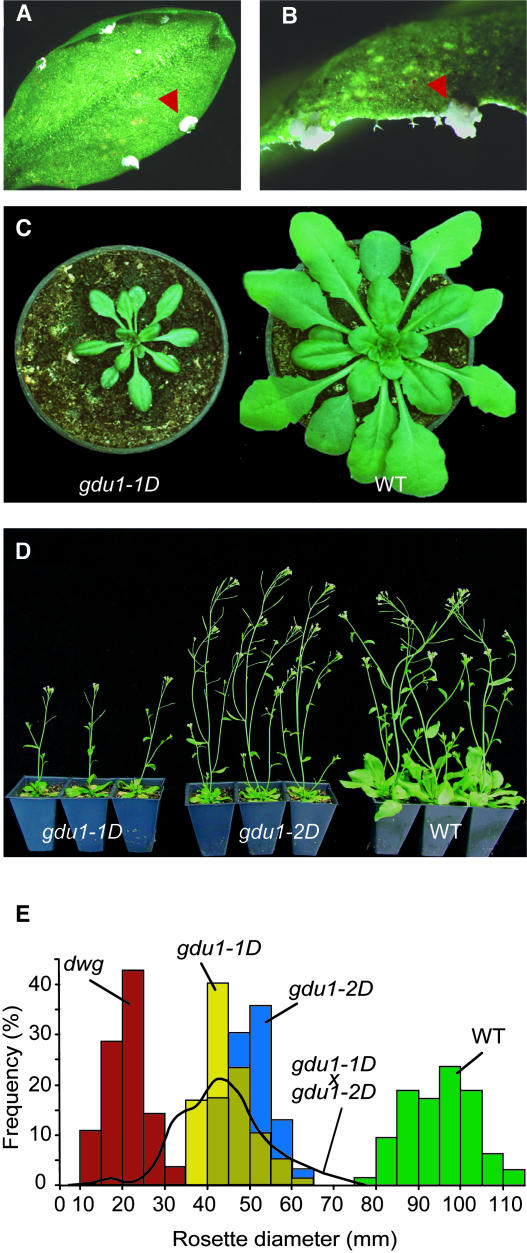
To study the secretion mechanism in Arabidopsis, two independent lines (named gdu1-1D and gdu1-2D, see below) that had white salty deposits at the hydathodes (Figures 1A and 1B) were identified by screening ∼10,000 independent T2 activation tagged lines from Agrinomics (Portland, OR). The white flecks reappeared locally a few days after leaves were washed, showing that the salt deposit is indeed secreted by hydathodes. In young seedlings grown under long-day conditions, deposits often appeared at the tips of cotyledons. Mutant plants were smaller than the wild type when grown in soil in both short- or long-day conditions (Figures 1C to 1E) or in axenic culture (data not shown). Mutant leaves were curled and darker green compared with the wild type. The mutants flowered at the same time as the wild type. All phenotypic alterations observed in gdu1-2D were weaker than those in gdu1-1D: specifically, gdu1-2D secreted less than gdu1-1D and had an intermediate size between gdu1-1D and the wild type (Figures 1D and 1E).
Figure 1.
Phenotype of the gdu1 Mutants.
(A) and (B) Leaves of gdu1-1D mutant displaying the secretion at the hydathodes (arrowheads).
(C) Four-week-old gdu1-1D and wild-type plants grown in short-day conditions.
(D) Five-week-old gdu1-1D, gdu1-2D, and wild-type plants grown in long-day conditions.
(E) Distribution of the rosette diameter of the progeny of gdu1D mutants compared with the wild type. The plants were grown for five weeks in the greenhouse. The diameter of the rosette of 63 (Columbia; green bars), 77 (gdu1-1D; yellow bars), and 92 (gdu1-2D; blue bars) plants was recorded. The black curve corresponds to the size distribution of 454 offspring of five plants produced by crossing gdu1-1D and gdu1-2D (gdu1-1D × gdu1-2D). Red bars correspond to the size distribution of 28 dwg (dwarf gdu1) plants observed in 86 offspring of a heterozygous gdu1-1D plant (group C line; see Table 3).
The Secretion Is Composed of Gln
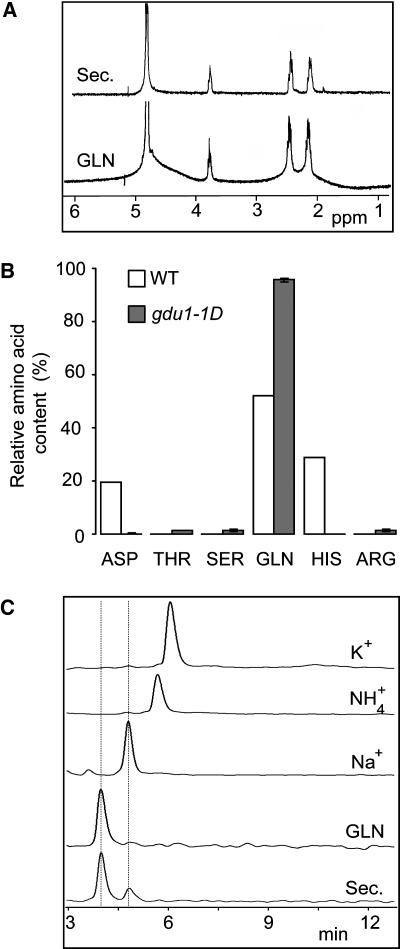
Several samples of the salty deposit were collected and analyzed. Crystals contained few inorganic cations: 18.5% Na+, 0.16% Ca2+, and <0.1% K+ and Mg2+ (w/w) as assessed by flame spectrometry. Combustion analysis revealed the presence of a large proportion of organic matter, with 35.6% carbon, 6.7% hydrogen, and 15.6% nitrogen (w/w). The liquid chromatography–mass spectrometry analysis identified a single compound, which produced a main peak with a mass of 146 atomic units and fragmenting in daughter ions, as detected by the tandem mass spectrometry analysis (data not shown). NMR spectra were identical to those produced by analysis of pure Gln (Figure 2A). The carbon, hydrogen, and nitrogen proportions of the secretion were similar to the expected values for Gln as the sole organic compound (34.4, 5.6, and 15.7%, respectively). HPLC assessed purity of Gln. Gln represented 95% of the detected amino acids (∼90% of secretion weight). Apart from Gln, <2% (1% of secretion weight) each of Asp, Thr, Ser, and Arg were present (Figure 2B). Inorganic anions (NO3−, SO42−, and Cl−) were not detected in the secretion. HPLC cation analysis detected Gln and Na+ but not K+ or NH4+ (Figure 2C). The secretion was thus composed of Gln (>80% [w/w]) and Na+ (between 2 and 20% [w/w]). Based on the composition of the secretion, the mutants were called glutamine dumper. The amino acid composition of the solution produced by the hydathodes is different in gdu1-1D than that of the wild type (Figure 2B). Interestingly, the amino acid composition of the xylem and the secretion of gdu1-1D are very similar: both contain >90% Gln, each of the other amino acids representing <2% of the amino acid content (cf. Table 1 and Figure 2B).
Figure 2.
Chemical Analysis of the Secretion.
(A) The NMR spectra of the secretion (Sec) and d-Gln (GLN) showed identical multiplets centered at 2.1, 2.5, and 3.8 ppm in a 2:2:1 ratio that represents the β-CH2:γ-CH2:α-CH, respectively.
(B) Comparison of amino acid content of the guttation fluid from wild-type plants and of the secretion from gdu1-1D mutant (expressed as a percentage of total amino acid content).
(C) HPLC spectra of the secretion (Sec), d-Gln (GLN), Na+, NH4+, and K+. The two peaks of the secretion spectrum correspond perfectly to the peaks of Gln and Na+.
Free Amino Acid Content of the Leaf Is Increased in gdu1-1D, and Gln Synthetase Activity Is Not Augmented
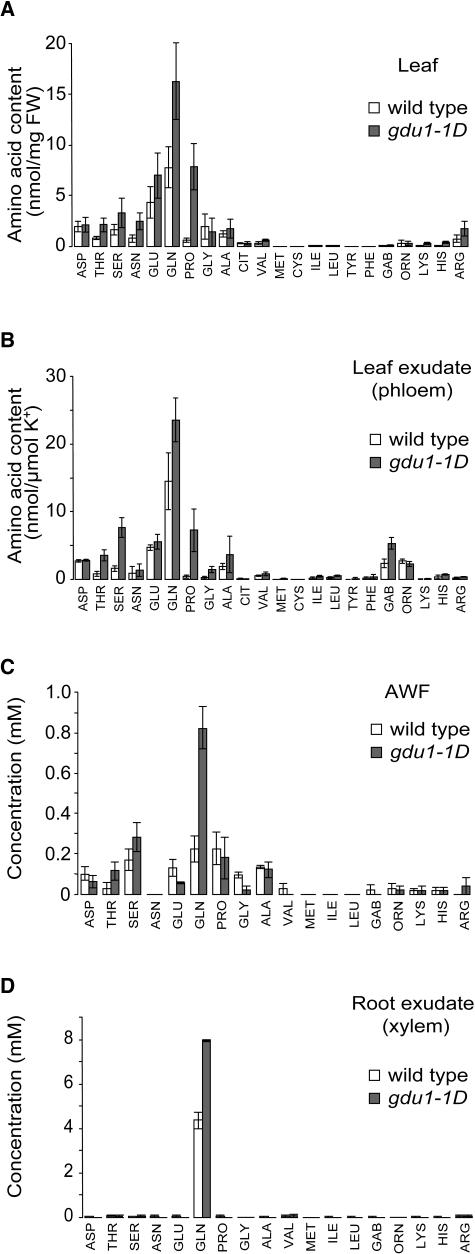
To assess the origin of the Gln secreted by the hydathodes in gdu1-1D, free and total amino acid contents of the leaf were analyzed. Whereas Ala, Gly, and Asp contents were poorly affected, Ser, Gln, Thr, Asn, and Pro levels increased 2, 2.1, 2.5, 3.1, and 12.8 times, respectively (Figure 3A). Interestingly, the leaf content of basic amino acids, such as Arg, Lys, and His, increased 2.3, 2.6, and 3.7 times, respectively. The total free amino acid content of the leaf was increased by 100%. At the same time, the total amino acid content (free plus protein amino acids) of leaf was increased by ∼15% (data not shown). These data show that the content of several amino acids, and not only Gln, is increased in the leaf, showing that the composition of the secretion at the hydathodes is not correlated to amino acid content of the leaf.
Figure 3.
Composition in Free Amino Acids of Whole Leaves, Leaf Exudate, AWF, and Root Xylem Exudate from Wild-Type and gdu1-1D Plants.
(A) Leaf amino acid content. Values and errors bars are average and standard deviation, respectively, of six samples (three from long-day and three from short-day grown plants). FW, fresh weight.
(B) Ratio of amino acid content and potassium content of the leaf exudate. Values are the average of four plants; error bars represent standard deviation.
(C) and (D) Concentration of amino acids in AWF (C) and in root xylem exudate (D). Values are the average of two samples, each collected from four to five plants. Error bars represent the two values.
CIT, citrulline; GAB, γ-amino-butyric acid.
The effects of gdu1-1D mutation on amino acid accumulation are different from those reported for Gln synthetase (GS) overexpression in tobacco (Nicotiana tabacum) plants. In these tobaccos, only Gln levels were significantly augmented (Fuentes et al., 2001). Consistently, total GS activity in leaves of gdu1-1D was not higher than in wild-type plants: when grown in short days, gdu1-1D plants had a 30% lower GS activity than the wild type, and when grown in long days, GS activity was identical in gdu1-1D and the wild type (Table 2). When Gln synthesis was artificially upregulated in wild-type plants by growth on ammonium-rich medium, Gln content increased sixfold, whereas the total amount of the other amino acids was only elevated by ∼25% (data not shown), but no secretion was observed at the hydathodes. In another experiment, gdu1-1D and wild-type plants were grown on media containing variable amounts of ammonium nitrate (0 to 10 mM). The size of the plants increased with the quantity of available nitrogen, but the ratio between the dry weight of the mutant and that of the wild type remained constant (around 0.6; data not shown). On the contrary, it has been shown that transgenic Lotus corniculatus plants overexpressing soybean (Glycine max) GS1 displayed growth enhancement on ammonium-rich media (Vincent et al., 1997). In another report, growth of GS1 overexpressing tobacco showed improvements under nitrogen limiting conditions but was indistinguishable from the wild type in nitrogen-rich conditions (Fuentes et al., 2001).
Table 2.
Total GS Activity in Leaves of Wild-Type and gdu1-1D Plants
| GS Activitya (Transferase Assay)
|
||
|---|---|---|
| Long Days | Short Days | |
| Wild type | 814 ± 48b | 720 ± 75 |
| gdu1-1D | 755 ± 87 | 537 ± 65 |
μmol γ-glutamyl-mono hydroxamate/mg protein/min.
Average ± sd of three replicates of two samples.
These results suggest that the excretion of Gln from the leaves is not the result of an overproduction of Gln or an increased GS activity in the leaves.
Root Xylem Exudates and Apoplasm Wash Fluid of gdu1-1D Contain More Gln
Gln export out of the hydathode seems to be specifically affected in gdu1-1D plants. To analyze whether Gln export is also altered at other sites in the plant, the amino acid composition of leaf exudate, root xylem exudates, and apoplasm wash fluid (AWF) from gdu1-1D and the wild type, corresponding respectively to phloem sap, xylem sap, and apoplasm, were compared.
In the mutant and in the wild type, the free amino acid composition of leaf exudate was comparable to leaf composition, as previously described (Winter et al., 1992; Lohaus et al., 1995; Lohaus and Heldt, 1997). As in the leaf, Gln content was also elevated in phloem sap. However, there was no specific increase in Gln content but an augmentation in several amino acids (Figure 3B), leading to a twofold increase in total amino acid content. Interestingly, Pro levels increased severalfold, suggesting stress responses in the mutant.
In leaves, an apoplasmic-loading step for amino acids requires a cellular export process (Lohaus et al., 1995). Consistent with a general defect in amino acid export of gdu1-1D, the composition of AWF from leaves was also modified compared with the wild type: ∼45% more free amino acids were detected in gdu1-1D (Figure 3C). This augmentation was mainly because of an increase in Gln concentration (more than three times), which led to a modification in the relative composition in amino acids, with an increase of Gln proportion from 18 to 46% (Figure 3C). In the mutant, the qualitative composition in amino acids of the root exudate was largely unaffected; however, the concentration of Gln was increased by ∼80% (Figure 3D).
The selective increase in Gln levels in hydathode secretion, xylem sap, and AWF in gdu1-1D suggests that a common export mechanism for Gln at these different sites in the plant is affected in the mutant.
The gdu1D Mutations Are T-DNA Linked and Lead to a Gain of Function
To genetically characterize the gdu1-1D mutation, the genotype and phenotype of 36 offspring of gdu1-1D were determined. Plants were sorted into four groups based on segregation for BASTAR (harbored by the T-DNA) and Gdu1− phenotype (Table 3). Group B plants segregated for BASTAR with a 3:1 ratio (χ2 test, P = 0.90), suggesting a single insertion locus. Groups A and D plants did not segregate for BASTAR: progeny of group A plants was BASTAR, whereas progeny of group D plants was BASTA susceptible (BASTAS). At the same time, group A plants displayed the Gdu1− phenotype as did their progeny, whereas group D plants and their offspring were indistinguishable from the wild type. Thus, group A plants were homozygous for gdu1-1D and BASTAR, and group D plants were homozygous for the corresponding wild-type alleles. Segregation analysis of group B plants showed that they also segregate for Gdu1− phenotype with a 3:1 ratio (χ2 test, P = 0.88), consistent with a gain-of-function mutation. Furthermore, all Gdu1− plants from this group were BASTAR. Together with the data provided by the analysis of groups A and D, this showed that the gdu1-1D mutation is dominant and genetically linked to the BASTAR locus, strongly indicating that the gdu1-1D mutation was because of activation tagging by insertion of the T-DNA.
Table 3.
Segregation for BASTAR and Gdu1− Phenotype in 36 Offspring of a Heterozygous gdu1-1D Plant
| Progeny (%)
|
||||||
|---|---|---|---|---|---|---|
| In Vitro
|
In Soil
|
|||||
| Groups | Number of Plants | Phenotypea | BASTAR | Gdu1+ | Gdu1− | Dwg− |
| A | 10 | Gdu1− | 100 | 0 | 100 | 0 |
| Bb | 15 | Gdu1− | 74.8 | 24.1 | 74.4 | 1.5 |
| Cc | 1 | Gdu1− | 66.6 | 24.8 | 41.0 | 34.2 |
| D | 10 | Gdu1+ | 0 | 100 | 0 | 0 |
Phenotype of the plants of each group grown in soil.
For BASTAR and Gdu1− segregation, 1074 and 457 plants were analyzed, respectively. All Gdu1− and Dwg− plants were BASTAR (spraying with 0.1% BASTA). All Gdu1+ plants were BASTAS.
For BASTAR and Gdu1− segregation, 419 and 117 plants were analyzed, respectively. All Dwg− and 65% of the Gdu1− plants were BASTAR. All Gdu1+ plants were BASTAS.
Interestingly, the progeny of several heterozygous gdu1-1D plants contained a small number of dwarfed plants (groups B and C, Table 3). These plants were significantly smaller than gdu1-1D (Figure 1E) but showed similar leaf shape and color as gdu1-1D, and all of them were BASTAR, suggesting that the mutation is also T-DNA linked. These plants were called dwarf gdu1 (dwg). Group C contained a single line, which segregated for Dwg− with a higher proportion compared with group B lines and for BASTAR in a non-Mendelian way (Table 3). Moreover, some of the Gdu1− progeny of the group C line were also BASTAS, an attribute never seen in the lines of groups A, B, and D. The abnormal segregation and the generation of a variable percentage of Dwg− plants is likely to come from a genome rearrangement caused by the T-DNA insertion (see below; Nacry et al., 1998; Tax and Vernon, 2001). The Dwg− phenotype segregated in a complex way in the next generation (data not shown) and was not further investigated.
Genetic analysis of gdu1-2D mutant showed that, similar to gdu1-1D, the gdu1-2D mutation is dominant (χ2 test, P = 0.89) and BASTAR linked (data not shown). Unlike gdu1-1D, no Dwg− plants were observed in the progeny of any heterozygous plant. Allelism of the gdu1-1D and gdu1-2D mutation was established from the analysis of the F2 plants obtained from crosses between two homozygous gdu1-1D and gdu1-2D plants: the offspring did not segregate for BASTAR and was always smaller than the wild type, whereas the size of the rosettes was variable (Figure 1E).
GDU1 Overexpression Is the Cause of the Phenotype
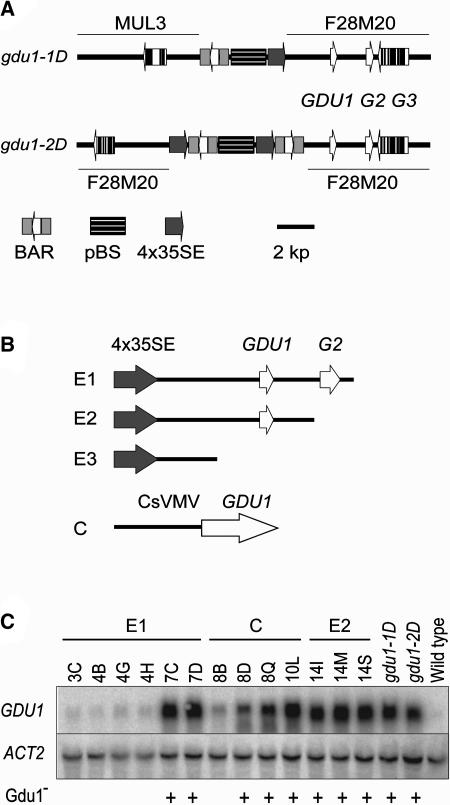
The genomic DNA into which the T-DNAs were inserted was identified by plasmid rescue for both mutant lines. Genomic DNA gel blots were consistent with the restriction patterns obtained from the deduced insertion maps (data not shown). The insertion in gdu1-1D (Figure 4A) is composed of a single T-DNA that bridged segments of chromosomes IV (BAC F28M20) and V (BAC MUL3). Chromosomal rearrangements upon insertion have frequently been observed (Nacry et al., 1998; Tax and Vernon, 2001). This rearrangement may explain the distortion in segregation and the increased frequency of recombination (that may lead to the deletion of an important gene for Arabidopsis growth) observed with the appearance of the Dwg− plants in the progeny of the heterozygous gdu1-1D plants (see above). The gdu1-2D insertion is composed of one full T-DNA flanked by two partial T-DNAs (Figure 4A), consistent with previous observations regarding the complexity of T-DNA insertions (Nacry et al., 1998; Rios et al., 2002). This T-DNA is inserted in BAC F28M20 ∼1250 bp downstream from the T-DNA insertion site in gdu1-1D.
Figure 4.
The Overexpression of GDU1 Is Responsible for the Gdu1− Phenotype.
(A) Maps of the T-DNA insertion in gdu1-1D and gdu1-2D. The name of the BACs is written above (in gdu1-1D) or below (in gdu1-2D) the drawings. GDU1, G2, and G3 correspond to the loci At4g31730, At4g31740, and At4g31750, respectively, according to The Institute for Genomic Research gene index.
(B) Maps of the constructs transferred into wild-type plants for recapitulation of the phenotype. E1, E2, and E3 are drawn to scale.
BAR, BASTA resistance gene; pBS, pBluescript KS+ plasmid; 4x35SE, four repetitions of the CaMV 35S enhancer; CsVMV, promoter of the cassava vein mosaic virus. Plant DNA is drawn with a thick line. Genes are symbolized by open (exons) and closed (introns) boxes; the direction of transcription is given by an arrow.
(C) RNA gel blot analysis of the accumulation of GDU1 and ACT2 transcripts in independent lines containing constructs E1, C, and E2, as indicated above the lanes, in gdu1-1D, gdu1-2D, and Columbia-7 wild type. The lines displaying Gdu1− phenotype are marked with a plus sign below the autoradiograms.
In the 8-kb region flanking the T-DNAs, a segment of BAC F28M20 containing three genes, namely loci At4g31730, At4g31740, and At4g31750, was common between the two mutants (Figure 4A). Real-time RT-PCR showed that the accumulation of At4g31730 mRNA (named thereafter GDU1) increased by ∼20 and 40 times in gdu1-2D and gdu1-1D, respectively, whereas expression levels of the other genes remained unaffected (data not shown). To address whether overexpression of GDU1 is the cause for the Gdu1− phenotype, three DNA fragments derived from gdu1-1D, containing the CaMV 35S enhancer and various lengths of genomic DNA (constructs E1, E2, and E3, Figure 4B; see Methods) and a construction composed of the cassava vein mosaic virus promoter (CsVMV; Verdaguer et al., 1996, 1998) fused to GDU1 coding sequence (construct C, Figure 4B) were introduced into wild-type Arabidopsis. E3 produced transformants indistinguishable from the wild type (∼60 lines analyzed), whereas more than half of the E1 and E2 transformants showed secretion and reduced size. To test the relation between the presence of secretion and overaccumulation of GDU1 mRNA, 21 E1, 12 E2, and 9 C transformants with a single kanamycin resistance locus, irrespective of the phenotype, were selected. Leaf tissues from 8 to 10 plants per line were analyzed for GDU1 mRNA content by RNA gel blot analysis (Figure 4C) and by real-time RT-PCR (data not shown). Twenty-six of these lines displayed the Gdu1− phenotype. In these 26 lines, GDU1 mRNA accumulation was increased more than 10 times compared with the wild type. Secretion was more abundant in the highly expressing plants. In the remaining 16 lines, which did not present the Gdu1− phenotype, GDU1 mRNA accumulated at lower levels only (Figure 4C). Overaccumulation of GDU1 mRNA was thus sufficient and necessary to generate the Gdu1− phenotype.
GDU1 Is a Member of a Novel Multigenic Family of Putative Membrane Proteins
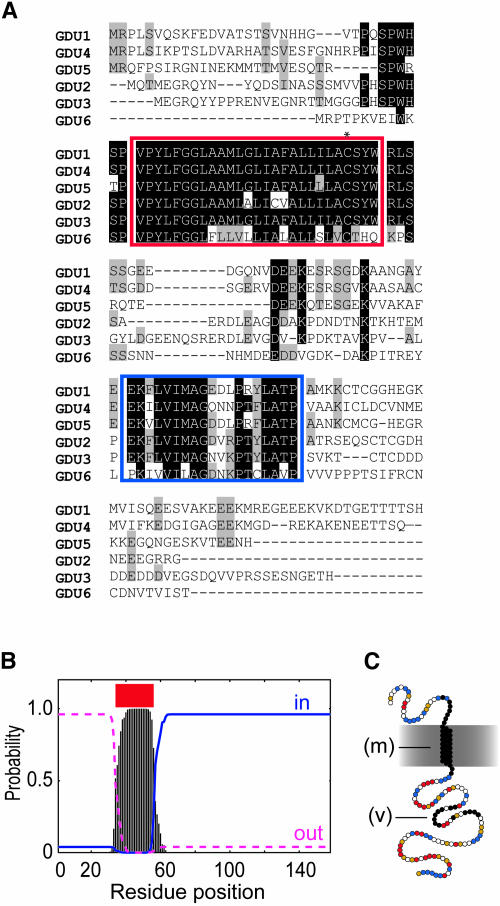
Database searching identified one EST and one Riken Arabidopsis full-length cDNA clone corresponding to GDU1 (see supplemental table online). Comparisons showed that the GDU1 gene contains no intron and encodes a novel 158–amino acid protein (calculated molecular mass of 17.2 kD). Pfam and SMART database searches (http://smart.embl-heidelberg.de/; Letunic et al., 2002) did not identify any known motif in GDU1. BLAST searches (Altschul et al., 1990) of Genbank EST, NR, and HGTS databases (http://www.ncbi.nlm.nih.gov/blast/html/blastcgihelp.html#nucleotide_databases) identified 104 plant DNA sequences encoding proteins similar to GDU1. One entry (AF420333) came from an ectomycorrhizal genome project, potentially corresponding to contamination with a plant cDNA. No significant similarity to bacterial or animal genes was found. The Arabidopsis genome contains five loci, which encode GDU1-related proteins sharing between 32 and 76% protein sequence similarity with GDU1. These proteins were named GDU2 to GDU6. A multiple alignment of GDU1 protein family of Arabidopsis (Figure 5A) and of the 51 GDU1-related proteins (see Supplemental Figure 1 online) identified two conserved regions of ∼36 and ∼20 amino acids, whereas the other parts of the proteins were very divergent. Several transmembrane (TM) domain prediction softwares identified the first conserved region as a membrane-spanning domain (Figure 5B; Schwacke et al., 2003; ARAMEMNON database, http://aramemnon.botanik.uni-koeln.de/). The TM domain does not show amphipathy and contains a single conserved Cys residue (Figure 5A). The second domain contains a highly conserved motif: VIMAG. The C-terminal part, downstream from the putative TM domain, was predicted to be cytoplasmic (Figure 5B; ARAMEMNON) and contains 41 and 22% of charged and polar residues, respectively, often grouped in clusters (Figure 5C).
Figure 5.
GDU1 Gene Encodes a Putative Membrane Protein.
(A) Multiple sequence alignment of GDU1 and the five GDU1-related proteins from Arabidopsis, realized by ClustalX (Thompson et al., 1997). GDU2, GDU3, GDU4, GDU5, and GDU6 correspond to the proteins from the loci At4g25760, Atg57685, At2g24762, At5g24920 (without the intron present in the annotation; data not shown), and At3g30725, respectively. Identical residues at each position are shaded in black; similar residues are shaded in gray. The putative membrane domain is boxed in red; the conserved VIMAG domain is boxed in blue. The asterisk shows the conserved Cys.
(B) TMHMM (Krogh et al., 2001) output from the analysis of GDU1 protein sequence. Similar plots were obtained for the five other Arabidopsis members. Vertical bars, probability of a single residue to be in a membrane spanning region; red line, predicted membrane domain; blue line, probability to be inside the cytoplasm; pink line, probability to be outside.
(C) Putative topology of GDU1. Circles represent amino acids. Black circles, residues conserved within the Arabidopsis family; red circles, negatively charged residues (D and E); yellow circles, positively charged residues (K, R, and H); blue circles, polar residues (Q, N, Y, C, S, and T). m, putative membrane α-helix; V, VIMAG motif-containing domain.
To assess the subcellular localization of GDU1, the green fluorescent protein was fused to the C terminus of GDU1 and expressed transiently in Arabidopsis protoplasts and tobacco epidermis cells. In most of the cases, fluorescence localized only at the plasma membrane in protoplasts (see Supplemental Figure 2 online), whereas several protoplasts also showed staining in endomembranes (data not shown). In tobacco leaves transiently transformed by Agrobacterium tumefaciens, bright green fluorescent protein fluorescence was localized in endomembranes, whereas little fluorescence was detected at the plasma membrane (data not shown). Additional experiments will be required to assess unequivocally the localization of GDU1.
GDU1 Is Expressed in the Vasculature of the Plant and in Hydathodes
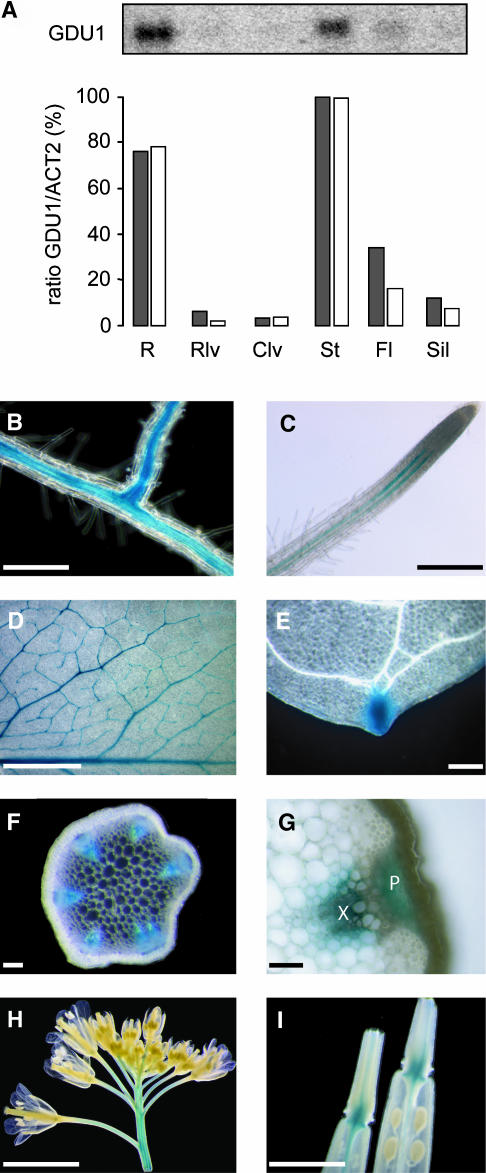
To explore whether expression of GDU1 provides a hint toward function, expression analyses were performed. GDU1 mRNA was detected preferentially in roots and stems by RNA gel blot analysis and real-time RT-PCR (Figure 6A). Expression of the GDU1 gene was investigated in more detail by promoter fusion to the β-glucuronidase (GUS) gene. GDU1 promoter was active in the vasculature throughout the whole plant (i.e., roots, leaves, stems, flowers, and base of the siliques) (Figures 6B to 6I). In the majority of transgenic lines, staining was also visible in hydathodes (Figure 6E). In roots, staining was located along the stele (Figure 6B) and restricted to two stripes close to the tip (Figure 6C). GUS activity was found in both xylem and phloem of the stem (Figure 6G) and the leaf (data not shown).
Figure 6.
GDU1 Gene Is Active in the Vasculature of the Whole Plant.
(A) Quantitation of GDU1 mRNA accumulation in the various organs of 5-week-old plants. Top: RNA gel blot analysis of total RNA (10 μg) hybridized with GDU1 cDNA probe. Bottom: ratios between GDU1 and ACT2 mRNA accumulation as determined by RNA gel blot analysis (gray bars) and by real-time RT-PCR (white bars), expressed as a percentage of the ratio obtained in stems. R, roots; Rlv, rosette leaves; Clv, caulinary leaves; St, stem; Fl, flowers; Sil, siliques.
(B) to (I) Localization of GDU1 promoter activity using GUS reporter gene in root (B), root tip (C), mature leaf (D), hydathode (E), stem cross section (F), detail of the vasculature in a stem cross section (G), inflorescence (H), and base of the siliques (I). X, xylem; P, phloem. Bar in (G) = 20 μm; bars in (B), (C), (E), and (F) = 100 μm; bars in (D) and (I) = 1 mm; bar in (H) = 5 mm.
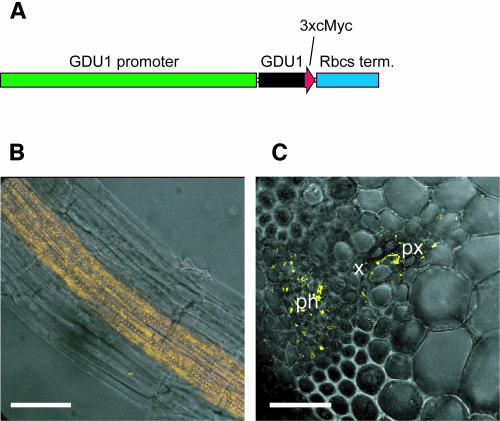
Immunolocalization experiments were performed to assess the localization of GDU1 protein in the plant. Because of the low expression levels of GDU1, a transgenic approach was chosen. Wild-type plants were transformed with a construct in which a triple c-Myc tag was fused to the GDU1 open reading frame, placed under the control of its own promoter (Figure 7A). Localization of the c-Myc tag was determined in roots and stems, where background fluorescence was found to be low. The immunochemical reaction was performed in parallel on wild-type and transformed plants using an anti-c-Myc antibody and a Cy3-labeled secondary antibody. No Cy3 fluorescence was detected in wild-type roots or stems, whereas a strong fluorescence was visible in the transformed plants. In roots, only the stele cells were labeled (Figure 7B), with stronger labeling in pericycle cells (data not shown). In stems, the c-Myc tag was detected in phloem cells, the parenchyma cells of the protoxylem, as well as the cells surrounding xylem vessels (Figure 7C). These results confirm the GUS analysis and show that GDU1 is transcribed in the vascular tissues and that GDU1 protein is present in the phloem and xylem. Root pericycle cells are supposed to play a role in xylem loading because they express transporters known to be involved in xylem loading, namely SKOR (Gaymard et al., 1998), BOR1 (Takano et al., 2002), and SOS1 (Shi et al., 2002). GDU1 expression in the pericycle would be in agreement with a role of this protein in Gln secretion into the xylem.
Figure 7.
GDU1 Protein Is Expressed in the Vascular Tissues of Root and Stem.
(A) Tagged GDU1 construct. The green bar represents the 3 kb upstream from GDU1 ATG (GDU1 promoter), and the blue bar represents the 3′-UTR of Pisum sativum Rbcs gene (Rbcs term.). The arrow represents the GDU1 open reading frame (black) fused to three copies of the c-Myc tag (red).
(B) and (C) Immunolocalization of GDU1-c-Myc in roots and stem, respectively, observed by confocal microscopy. Cy3 fluorescence is shown in yellow. The fluorescence pictures (projection of 10 sections) are merged with one transmission image.
(B) Whole-mount immunolocalization of roots from 2-week-old plants. The c-Myc tag was detected only in the stele of the root, excluding the endodermis.
(C) Immunolocalization of a 50-μm section from Arabidopsis stem. GDU1-c-Myc is detected in phloem and xylem parenchyma cells.
Bars = 20 μm. ph, phloem; px, protoxylem; x, xylem.
Quantitative Relationship between Strength of Phenotype and Accumulation of GDU1 mRNA
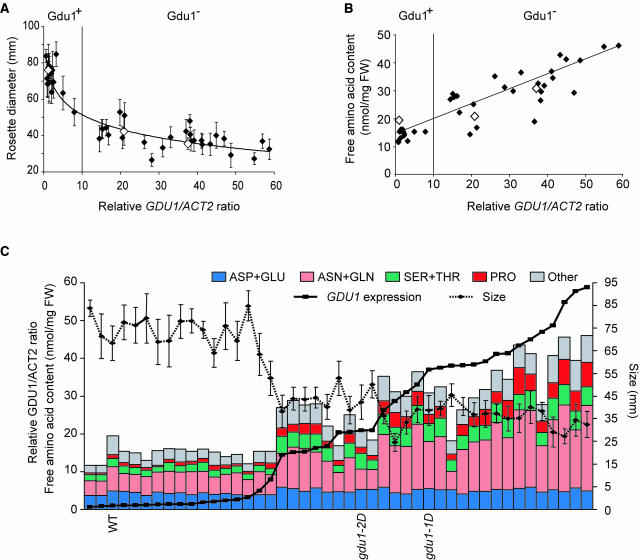
Together with the secretion of Gln, it was noticed that the mutant lines gdu1-1D and gdu1-2D were smaller compared with the wild type (Figure 1). To further characterize the relationship between overexpression of GDU1 and the phenotype, each of the previously selected 42 recapitulation lines, gdu1-1D, gdu1-2D, and wild-type plants, were studied. The size (i.e., diameter of the rosette) of the plants was recorded. A clear correlation existed between size and leaf accumulation of GDU1 mRNA (Figure 8A). The data point distribution could be fitted by a nonlinear, logarithmic, curve (r2 = 0.89; Figure 8A). Although a 10-fold overaccumulation of GDU1 mRNA led to a strong decrease (40%) in the size of the plants, an increase in GDU1 overexpression by 30 and 60 times led to a further decrease in size of 10 and 20%, respectively. The relationship between GDU1 expression and leaf amino acid content was studied on the 42 recapitulation lines, the gdu1 mutants, and wild-type plants. The free amino acid content increased linearly (r2 = 0.81) with GDU1 mRNA accumulation, reaching a threefold increase when the mRNA quantity was augmented 60 times compared with the wild type (from 15 ± 5 to 46 ± 5 nmol·mg−1 fresh weight; Figure 8B). The content of each of the amino acids was either unchanged (Asp, Glu, Ala, citrulline, and γ-amino-butyric acid) or increased (all other amino acids) upon GDU1 overexpression (see Supplemental Figures 3 and 4 online).
Figure 8.
Analysis of Gdu1− Phenotype in the Recapitulation Lines.
(A) Relation between GDU1 mRNA accumulation and the size of the 42 recapitulation lines (closed diamonds), gdu1-1D, gdu1-2D, and wild-type plants (open diamonds). The expression data come from the quantitation of the signals obtained by RNA gel blot analyses presented in Figure 4C. The ratios of the intensities of GDU1 and ACT2 signals were expressed according to the ratio found in wild-type plants, which was set at 1. Size values are the average of the diameter of at least five plants; errors bars are standard deviation. Data point distribution was logarithmically fitted using Excel (Microsoft). Similarly, when using logarithm of mRNA accumulation, the data point distribution can be fitted linearly. The thin vertical line corresponds to the separation between Gdu1+ and Gdu1− plants.
(B) Relation between GDU1 mRNA accumulation and free amino acid content of the leaf. For each line, amino acid content is the sum of the contents of each amino acid determined by HPLC, expressed according to fresh weight (FW) (all plants had a similar relative water content of 88.4 ± 2%). The distribution of data points coming from the recapitulation lines (closed diamonds) and from gdu1-1D, gdu1-2D, and wild-type plants (open diamonds) was fitted linearly. The vertical line corresponds to the separation between Gdu1+ and Gdu1− plants.
(C) Grouping of the amino acid content, size, and GDU1 expression for each of the 45 lines. Lines have been sorted according to the relative GDU1 mRNA accumulation. The bars corresponding to wild-type, gdu1-1D, and gdu1-2D plants are labeled accordingly.
The whole set of data regarding size, mRNA overaccumulation, and amino acid content were grouped in a single graph (Figure 8C). For most of the 45 lines, there is a clear correlation between all three parameters. The correlation between GDU1 mRNA accumulation and size of the plant is thus clearly established, but the reason for this relation is at present unknown.
DISCUSSION
Arabidopsis wild-type plants secrete small amounts of three amino acids from the hydathodes in guttation droplets, namely Gln, Asp, and His. A mutant was identified that deposits salt crystals at the hydathodes. The deposit consists of almost pure Gln (Figure 2). In the mutant, the xylem sap and the AWF also contain elevated levels of Gln. In leaves, Gln content is increased as well, but the content of other amino acids also increases, especially Pro (Figure 2A). The overall elevation of amino acid pools in the whole leaf does not seem to be because of an increase in GS activity and, thus, Gln synthesis. There are three main sites of amino acid export in the plant: at hydathodes, which play a crucial role in solute absorption and possibly excretion; at xylem parenchyma into xylem sap; and from mesophyll into leaf apoplasm. The presented results show that Gln content is specifically increased at these three sites of amino acid export. These data are consistent with a specific increase in net Gln export from hydathodes, export into the xylem, and also potentially export from leaf mesophyll cells. These effects are because of activation tagging of a novel gene, GDU1, encoding a protein with a single putative TM span and a cytosolic domain. GDU1 may thus represent a component of a mechanism responsible for secretion of Gln.
Mechanisms for the Export of Amino Acids
Cellular export of metabolites is one of the least understood phenomena in most organisms (e.g., the mechanism of glucose export from liver is currently subject to debate). Although the GLUT2 transporter had been shown to mediate glucose uniport, recent reports rather suggest a vesicular secretion route for glucose in hepatocytes (Burcelin et al., 2000; Stumpel et al., 2001; Hosokawa and Thorens, 2002). In plants, too, the mechanism of sucrose export from leaf mesophyll cells has not been elucidated. Similarly, amino acid export in animals and plants seems poorly understood, with the exception of neurotransmitter release, which occurs via vesicular secretion. It has been suggested that amino acid export (e.g., from intestinal cells into the blood stream) may also be mediated by amino acid exchangers belonging to the amino acid-polyamine-choline (APC) (SLC7) family (for review, see Wipf et al., 2002). However, net export cannot be achieved by exchange of amino acids. So far, amino acid export systems have been described only in bacteria (e.g., a transporter for Lys excretion) (Vrljic et al., 1996). Conceptually, several export routes can be envisaged: (1) passive efflux by transporters (e.g., uniporters); (2) secondary active transport by antiporters; (3) active export by ABC transporters; or (4) vesicular pathways, such as in neuronal synapses, where specific proton antiporters concentrate the compound in vesicles, which are targeted to the plasma membrane, and then fuse and release their content into the extracellular space (for review, see Wipf et al., 2002). In plants, amino acid export systems are required for a variety of purposes: for the loading of the xylem, for release of amino acids into the apoplasmic space in the leaf as a first step toward phloem loading, and for excretion, especially at secretory organs such as hydathodes or nectaries. It has been suggested that the mechanisms for secretion are similar at the different sites; thus, secretory organs may serve as model systems to study the fundamental processes involved in cellular export of compounds from plant cells (Frey-Wyssling, 1935). Hydathodes are organs located at the leaf edges and are responsible for guttation. Guttation varies between species and depends on environmental conditions. However, the underlying secretion mechanisms are not understood (Frey-Wyssling, 1935 and references therein). The secretion phenomenon is supposed to be composed of both specific export and retrieval mechanisms operating in a way similar to those in kidney. In agreement with this assumption, hydathodes, like nectaries, can also take up amino acids provided from the external medium (Ziegler and Lüttge, 1959) and were shown to express several cellular uptake systems for purines, sulfate, and potassium (Lagarde et al., 1996; Shibagaki et al., 2002; Bürkle et al., 2003; Pilot et al., 2003).
Putative Role of GDU1 in Gln Export
Based on the structure of GDU1, GDU1 may (1) assemble as a homo-oligomer to function as a transporter itself, (2) serve as a β-subunit of a typical polytopic Gln transporter, (3) function as a component of a vesicular export mechanism, or (4) indirectly affect the transport mechanism by altering membrane properties. The small size of the GDU1 protein may make a function as an amino acid sensor per se unlikely. Nevertheless, it cannot be ruled out that GDU1 assembles in a multiprotein complex involved in sensing.
GDU1 was found to be located at the plasma membrane and in intracellular compartments and contains a putative TM domain with a short C terminus probably located in the cytosol. The only homo-oligomeric transporters known so far that are formed from subunits with a single TM are viral channels (Fischer et al., 2000). However, in contrast with viral polypeptides, the TM domain of GDU1 is not amphipathic, arguing against a function as part of the Gln pore in an exporter. However, GDU1 could represent a subunit of a transporter. The existence of β-subunits is a common feature of ion channels. More interestingly, members of the mammalian APC (SLC7) family of amino acid transporters have to associate with heavy subunits belonging to the SLC3 family (4F2hc or rBAT) to form functional transporters (Chillaron et al., 2001). SLC3 auxiliary proteins contain a single TM domain but also have a large extracellular part, which is not present in GDU family members. Similar to SLC3s, GDUs also contain a conserved Cys within the membrane span, which may be relevant for hetero-oligomerization as in the case of heteromeric amino acid transporters (Wagner et al., 2001). SLC3 polypeptides are required for plasma membrane localization of APC transporters. Localization of GDU1 at the cell periphery is consistent with a function in helping a transporter to traffic to the plasma membrane. The Arabidopsis genome contains multiple APC homologs, which may thus represent potential candidates for Gln exporters (Wipf et al., 2002). Alternatively, the results do not exclude that GDU1 is involved in a vesicular export route. Localization close to the plasma membrane may suggest that, in this case, GDU1 could display a specific function in vesicle release. Finally, GDU1 shares structural properties with stomatins, which are polypeptides known to affect raft formation and to modify transport properties of the membrane (Snyers et al., 1999; Schwarz et al., 2001; Zhang et al., 2001). Although the selectivity for Gln suggests that GDU1 plays a specific role in Gln secretion, the data do not preclude other more indirect modes of action (e.g., a role in retrieval or regulatory roles in metabolism or transport). The search for partners interacting with the conserved cytosolic domain using two-hybrid (Bartel and Fields, 1995) or split ubiquitin (Schulze et al., 2003) systems may help to distinguish between the different hypotheses.
Conclusion
During a search for mutants affected in hydathode function, we identified a novel gene that seems to be involved in the regulation of amino acid export out of the cell. The study of Gdu1− mutants could help to gain insights into the mechanism involved in export of metabolites. Furthermore, overexpression of the other members of the GDU family may represent a tool to identify export systems for other compounds.
METHODS
Plant Growth and Transformation
Arabidopsis thaliana plants (ecotype Columbia, Col-7) were grown in soil (Metromix 350) either (1) in a greenhouse (25°C, 75% relative humidity, 16-h light) and watered automatically from below twice a week or (2) in growth chambers (22°C, 50% relative humidity, 16-h light, ∼120 μmol/m2/s) and watered manually from below twice a week. BASTAR segregation was analyzed in vitro on MS medium (Sigma M5524; St. Louis, MO) containing 15 μg/mL of glufosinate-ammonium (BASTA; Bayer CropScience, Frankfurt, Germany), 0.7% agar, and 1% sucrose (16-h light, 22°C, and 100 μmol/m/s).
Binary plasmids were introduced into plants using heat shock–transformed GV3101 (pMP90) Agrobacterium tumefaciens strain (Van Larebeke et al., 1974; Koncz and Schell, 1986) and the floral dip method (Clough and Bent, 1998). Transgenic plant selection and segregation for kanamycin resistance analyses were performed in vitro as described above, but with 50 μg/mL of kanamycin instead of BASTA.
Xylem Exudate, AWF, and Leaf Exudate Collection
Root xylem exudate was collected by cutting the hypocotyl of 4-week-old plants below the rosette. The first drop emerging from the root was discarded to prevent contamination of the xylem exudate by sap from damaged cells. The secretion was then collected with a micropipette during 30 min, and the volume of the samples was measured. The solution was lyophilized and stored at −20°C before amino acid analysis.
AWF was collected using the infiltration-centrifugation method (Lohaus et al., 2001). Briefly, leaves of ∼10 plants grown in short-day conditions for 2 months (to increase the number of leaves of the mutant) were collected and washed in ice-cold milli-Q water (Milipore, Schwalbach, Germany). Leaves were infiltrated with ice-cold milli-Q water by five applications of a pressure of −0.8 bar for 2 min and wiped dry with tissues. AWF was collected by centrifugation for 20 min at 80g. The volume of the collected liquid was measured, and samples were lyophilized and stored at −20°C before amino acid analysis.
Leaf phloem exudate was collected in 5 mM EDTA, pH 7, solution (Corbesier et al., 2001). The samples were split in two and lyophilized. One half was assayed for amino acid content by HPLC, whereas the other half was assayed for cation content (see below).
Analytical Methods
Amino acids were extracted from liquid nitrogen–ground material once in 80% methanol and once in 20% methanol. Both extracts were pooled and lyophilized. Pellets were resuspended in lithium buffer (0.7% lithium acetate and 0.6% LiCl; Pickering Laboratories, Mountain View, CA) spiked with 0.2 mM norleucine. Amino acids were separated by HPLC on a cation-exchange column (high efficiency fluid column, 3 mm × 150 mm; Pickering Laboratories) using lithium buffer as eluant. Amino acids were derivatized with ninhydrine before photometric detection. Total amino acids were analyzed from ground rosette tissues by Ansynth (Roosendaal, The Netherlands).
Crystals secreted by gdu1-1D leaves were solubilized in water with a micropipette, lyophilized, and weighed. A few milligrams were subjected to flame spectrometry for cation analysis and combustion analysis to determine carbon, hydrogen, and nitrogen content (Galbraith Laboratories, Knoxville, TN). Liquid chromatography–mass spectrometry analysis was performed on a Perkin-Elmer Sciex 3000 LC/MS/MS system (Foster City, CA) with an Ion Spray Electrospray Interface to a Shimadzu LC-10AD VP HPLC pumps (Columbia, MD) with 250 μL of high-pressure mixer and a SCL-10A VP pump controller. The columns on the HPLC were two YMC ODS AQ, 2 × 50-mm, 3-μm particle size, 120-Å pore size columns closely coupled in series with a mobile phase of 68% acetonitrile/32% (1.5% acetic acid in water). NMR analyses were performed on a Varian 300 MHz model G300B-2974 NMR system (Palo Alto, CA) with a Sun workstation. The solvent was D2O, frequency of 300.092 Hz, sweep width of 4500.5 Hz, and the number of scans was 16. Cation and anion content of the secretion were respectively analyzed by HPLC with a Hamilton PRP-X200 column (Reno, NV; eluant: 2 mM HNO3 and 15% methanol) and a standard anion chromatography column from Wescan (Gamma Analysen Technik, Bremerhaven, Germany; eluant: 2 mM phtalic acid and 10% methanol, pH 5), and detected by conductivity on a Wescan ion analyzer. Cation content of phloem was analyzed by HPLC (DX120 ion chromatography; Dionex, Idstein, Germany) with an IonPac CS12A column (eluant: 20 mM methanesulfonic acid) and an AS40 automated sampler (Dionex).
Total GS activity was determined using the transferase assay (Shapiro and Stadtman, 1971). Approximately 50 mg of plant tissues were ground and mixed with 200 μL of extraction buffer (50 mM Tris, pH 8, 10 mM imidazole, pH 7, and 0.5% β-mercaptoethanol), centrifuged for 15 min at 20,000g at 4°C. Ten microliters of supernatant were mixed to 500 μL of reaction buffer (100 mM Gln, 50 mM Tris, pH 7.5, 50 mM NH2OH, 20 mM K2HAsO4, 1 mM MnCl2, and 0.5 mM ADP). After 1 h at 37°C, 510 μL of stop buffer (200 mM FeCl3, 200 mM trichloroacetic acid, and 600 mM HCl) were added. OD500 was recorded after removal of the precipitate by centrifugation. Results were compared with a standard curve using γ-glutamyl-monohydroxamate. Total protein content was determined using the Bradford assay.
Plasmid Rescue Experiments
The method was described by Weigel et al. (2000). Briefly, genomic DNA from gdu1-1D and gdu1-2D was extracted using the Nucleon Phytopure kit (Amersham Biosciences, Piscataway, NJ), digested by BamHI, EcoRI, PstI, and SpeI, ligated using the Quick ligation kit (New England Biolabs, Beverly, MA) and introduced in ElectroMAX Escherichia coli DH10β electrocompetent cells (Invitrogen, Carlsbad, CA). Genomic DNA from the plasmids was sequenced using T-DNA–specific primers (Weigel et al., 2000). For gdu1-2D, the complexity of the insertion was resolved using EcoRI partially digested genomic DNA. Fragments of the longest plasmid obtained by plasmid rescue, estimated to contain genomic DNA, were subcloned and sequenced.
Recapitulation Constructs
The PstI fragment obtained by plasmid rescue from gdu1-1D was cleaved with KpnI and partially digested by HindIII. The resulting fragments were cloned into KpnI and HindIII sites of the binary vector pAG2370 (Agrinomics). pAG2370 is a modified pBIN19 plasmid (Bevan, 1984) containing the promoter of CsVMV (Verdaguer et al., 1998), the nopaline synthetase termination sequence, and the neomycin phosphotransferase gene under the control of the RE4 promoter. Cloning in KpnI and HindIII sites led to removal of the CsVMV promoter from pAG2370 and the insertion of enhancer sequences from the T-DNA together with two fragments of the flanking genomic DNA (1804 bp and 6737 bp; constructs E1 and E3, Figure 2B). A fragment of 1594 bp, containing the At4g31740 gene, was removed from the 6737-bp fragment by digestion by BsiWI, leading to a genomic DNA fragment of 5143 bp (E2 construct, Figure 2B). GDU1 coding and 3′-untranslated sequences were amplified by PCR from genomic DNA (there is no intron in this gene; see Results) with Pfu Turbo (Stratagene, Amsterdam, The Netherlands) using primers 5′-TTGGATCCAACAAAAAGAGATTACACAC-3′ and 5′-TTTGGTACCGAGCTCAAAAACCCCAAAATTTTGCTCTC-3′ harboring BamHI and KpnI sites, respectively, cloned into the BamHI and KpnI sites of pAG2370, between the CsVMV promoter and the nopaline synthetase termination sequence. This led to construct C (Figure 2B). E1, E2, E3, and C constructs were introduced in wild-type Arabidopsis.
Expression Analyses
Total RNA was extracted as described previously (Downing et al., 1992). For RNA gel blot analyses, RNA (10 μg) was fractionated on a 1.2% agarose gel in 20 mM Mops [3-(N-morpholino)-propanesulfonic acid]-NaOH, pH 7, 1 mM sodium acetate, 1 mM EDTA, and 6.6% formaldehyde, transferred onto nylon membranes (Amersham Biosciences, Freiburg, Germany), and hybridized with 32P-labeled DNA probes at 45°C in 50% formamide, 10% dextran sulfate, 1% N-laurylsarcosine, 75 mM sodium citrate, 750 mM NaCl, and 0.1 mg/mL of denatured, sheared herring sperm DNA (Sigma). Signals were quantitated with the Storm scanning system and ImageQuant software (Molecular Dynamics, Sunnyvale, CA). For real-time RT-PCR, RNA (3 μg) was reverse transcribed using the TaqMan reverse transcription kit, and real-time PCR was performed using the SYBR Green PCR master mix on the ABI PRISM 7700 sequence detection system (Applied Biosystems, Foster City, CA) with the following primers: GDU1, 5′-ATGGCCGGAGAAGATTTGC-3′ and 5′-CGCCTTCTCTCATCTTCTCTTCC-3′; ACT2 (At3g18780), 5′-GGTAACATTGTGCTCAGTGGTGG-3′ and 5′-AACGACCTTAATCTTCATGCTGC-3′. GDU1 expression was determined relative to ACT2 expression (2ΔCt; ΔCt = CtGDU1 − CtACT2).
The GDU1 promoter region (3 kb) was amplified by PCR from the PstI insert obtained by plasmid rescue (see above) with Pfu Turbo using the primers proF 5′-TTTCTGCAGTTCGTTGGTGATTGGTCGCCTC-3′ and proR 5′-TTTTCTAGATTTTGTTGTTGTTGTGTGTAATCTC-3′. The PCR fragment was digested by PstI and XbaI and cloned into the pPZP212 vector (Hajdukiewicz et al., 1994) containing the GUS gene (Jefferson et al., 1987) inserted between SacI and BamHI restriction sites. Wild-type Arabidopsis was transformed with the resulting construct. GUS staining was performed according to Lagarde et al. (1996). Organs were embedded in 4% agarose and cut in 50-μm sections using a Leica vibratome VT 1000S (Wetzlar, Germany).
GDU1 gene (3-kb promoter and coding sequence) was amplified by PCR with Pfu Turbo and the primer proF and 5′-TTCTCGAGGTGACTTGTAGTAGTTGTCTCGC-3′. The PCR fragment was digested by PstI and XhoI and cloned in frame with a tripled c-Myc tag, previously cloned in pBluescript KS+. The construct was cut out using KpnI and SacI and inserted in front of the 3′ region of the P. sativum Rbcs gene of a modified pPZP212 binary vector, where the 35S-nptII cassette was replaced by a mannopine (mas) promoter-nptII-mas terminator cassette (provided by Ji Hoon Ahn, Seoul National University). Approximately 15 transformed plants were assayed for GDU1-c-Myc expression by immunolocalization. For stems, stalk of 5- to 6-week-old Arabidopsis plants were cut in 5-mm pieces and fixed overnight at 4°C in 0.1% glutaraldehyde, 4% paraformaldehyde in MTSB (50 mM Pipes, 5 mM EGTA, and 5 mM MgSO4, pH 7). The pieces were embedded in 4% agarose and cut in 50-μm sections using a Leica vibratome VT 1000S in PBS, pH 7.2. The sections were blocked for 2 h at 37°C in 3% BSA in PBS. The reaction with the anti-c-Myc antibody (1:600; Santa Cruz Biotechnology, Heidelberg, Germany) was performed overnight at 4°C and 2 h at 37°C in 3% BSA in PBS. Sections were washed six times for 10 min in PBS. Detection was performed using Cy3-conjugated goat anti-mouse antibody (1:200; Jackson ImmunoResearch Laboratories, West Grove, PA) for 4 h at 37°C in 3% BSA in PBS. Sections were washed six times for 10 min in PBS and observed by confocal microscopy (Leica DMRE with TCS SP). Root immunolocalization was performed as described (Friml et al., 2003) using the same antibodies at the same dilution as described above.
Phenotypic Analysis of Recapitulation Lines
Plants were grown in soil under short-day conditions (8-h light, 50% relative humidity, ∼120 μmol/m/s) for 5 weeks. The rosette diameter of four to eight plants per line was recorded, and rosette tissues were collected, pooled, and kept frozen at −80°C until analysis. Tissue was ground in liquid nitrogen, aliquoted, and subjected to RNA and amino acid analyses.
Supplementary Material
Acknowledgments
We would like to thank B. Stadelhofer for all HPLC analyses, C. Brancato and P. Neumann for the perfect technical help, A. Martone for the NMR analyses, R. Seymour for the liquid chromatography tandem mass spectrometry experiments, J. Pilot for the helpful discussions on the real-time PCR experiments, and F. de Courcy for the critical reading of the manuscript. G.P. was supported by an EMBO long-term fellowship. This work was supported by the Koerber Foundation and by the Deutsche Forschungsgemeinschaft (Leibniz-Award to W.B.F.).
Online version contains Web-only data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Wolf B. Frommer (wfrommer@stanford.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.021642.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Bartel, P.L., and Fields, S. (1995). Analyzing protein-protein interactions using two-hybrid system. Methods Enzymol. 254, 241–263. [DOI] [PubMed] [Google Scholar]
- Bevan, M.W. (1984). Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 12, 8711–8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz, J.O., Xia, Y., Blount, J., Dixon, R.A., and Lamb, C.J. (2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12, 2383–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burcelin, R., del Carmen Munoz, M., Guillam, M.T., and Thorens, B. (2000). Liver hyperplasia and paradoxical regulation of glycogen metabolism and glucose-sensitive gene expression in GLUT2-null hepatocytes. Further evidence for the existence of a membrane-based glucose release pathway. J. Biol. Chem. 275, 10930–10936. [DOI] [PubMed] [Google Scholar]
- Bürkle, L., Cedzich, A., Dölpke, C., Stransky, H., Okumoto, S., Gillissen, B., Kühn, K., and Frommer, W.B. (2003). Transport of cytokinins mediated by purine transporters of the PUP family expressed in phloem, hydathodes, and pollen of Arabidopsis. Plant J. 34, 13–26. [DOI] [PubMed] [Google Scholar]
- Chillaron, J., Roca, R., Valencia, A., Zorzano, A., and Palacin, M. (2001). Heteromeric amino acid transporters: Biochemistry, genetics, and physiology. Am. J. Physiol. Renal Physiol. 281, F995–F1018. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Corbesier, L., Havelange, A., Lejeune, P., Bernier, G., and Périlleux, C. (2001). N content of phloem and xylem exudates during the transition to flowering in Sinapsis alba and Arabidopsis thaliana. Plant Cell Environ. 24, 367–375. [Google Scholar]
- Downing, W.L., Mauxion, F., Fauvarque, M.O., Reviron, M.P., de Vienne, D., Vartanian, N., and Giraudat, J. (1992). A Brassica napus transcript encoding a protein related to the Kunitz protease inhibitor family accumulates upon water stress in leaves, not in seeds. Plant J. 2, 685–693. [PubMed] [Google Scholar]
- Enns, L.C., Canny, M.J., and McCully, M.E. (2000). An investigation of the role of solutes in the xylem sap and in the xylem parenchyma as the source of root pressure. Protoplasma 211, 183–197. [Google Scholar]
- Esau, K. (1977). Anatomy of seed plants. (New York: John Wiley and Sons).
- Fischer, W.B., Forrest, L.R., Smith, G.R., and Sansom, M.S. (2000). Transmembrane domains of viral ion channel proteins: A molecular dynamics simulation study. Biochim. Biophys. Acta 53, 529–538. [DOI] [PubMed] [Google Scholar]
- Frey-Wyssling, A. (1935). Stoffausscheidung der höheren Pflanzen. (Berlin, Germany: Von Julius Springer).
- Friml, J., Benkova, E., Mayer, U., Palme, K., and Muster, G. (2003). Automated whole mount localisation techniques for plant seedlings. Plant J. 34, 115–124. [DOI] [PubMed] [Google Scholar]
- Fuentes, S.I., Allen, D.J., Ortiz-Lopez, A., and Hernández, G. (2001). Over-expression of cytosolic glutamine synthetase increases photosynthesis and growth at low nitrogen concentrations. J. Exp. Bot. 52, 1071–1081. [DOI] [PubMed] [Google Scholar]
- Gaymard, F., Pilot, G., Lacombe, B., Bouchez, D., Bruneau, D., Boucherez, J., Michaux-Ferrière, N., Thibaud, J.-B., and Sentenac, H. (1998). Identification and disruption of a plant shaker-like outward channel involved in K+ release into the xylem sap. Cell 94, 647–655. [DOI] [PubMed] [Google Scholar]
- Greenhill, A.W., and Chibnall, A.C. (1934). The exudation of glutamine from perennial rye-grass. Biochem. J. 28, 1422–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald, I., Rupprecht, I., Schuster, G., and Kloppstech, K. (2003). Identification of guttation fluid proteins: The presence of pathogenesis-related proteins in non-infected barley plants. Physiol. Plantarum 119, 192–202. [Google Scholar]
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994. [DOI] [PubMed] [Google Scholar]
- Hosokawa, M., and Thorens, B. (2002). Glucose release from GLUT2-null hepatocytes: Characterization of a major and a minor pathway. Am. J. Physiol. Endocrinol. Metab. 282, E794–E801. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kananagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnytsky, S., Borisjuk, N.V., Borisjuk, L.G., Alam, M.Z., and Raskin, I. (2000). Production of recombinant proteins in tobacco guttation fluid. Plant Physiol. 124, 927–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz, C., and Schell, J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204, 383–396. [Google Scholar]
- Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E.L.L. (2001). Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305, 567–580. [DOI] [PubMed] [Google Scholar]
- Lagarde, D., Basset, M., Lepetit, M., Conejero, G., Gaymard, F., Astruc, S., and Grignon, C. (1996). Tissue-specific expression of Arabidopsis AKT1 gene is consistent with a role in K+ nutrition. Plant J. 9, 195–203. [DOI] [PubMed] [Google Scholar]
- Lam, H.M., Coschigano, K.T., Schultz, C., Melooliveira, R., Tjaden, G., Oliveira, I.C., Ngai, N., Hsieh, M.H., and Coruzzi, G.M. (1995). Use of Arabidopsis mutants and genes to study amide amino acid biosynthesis. Plant Cell 7, 887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic, I., Goodstadt, L., Dickens, N.J., Doerks, T., Schultz, J., Mott, R., Ciccarelli, F., Copley, R.R., Ponting, C.P., and Bork, P. (2002). Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 30, 242–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Lease, K.A., Tax, F.E., and Walker, J.C. (2001). BRS1, a serine carboxypeptidase, regulates BRI1 signaling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 98, 5916–5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohaus, G., and Heldt, H.W. (1997). Assimilation of gaseous ammonia and the transport of its products in barley and spinach leaves. J. Exp. Bot. 48, 1779–1796. [Google Scholar]
- Lohaus, G., Pennewiss, K., Sattelmacher, B., Hussmann, M., and Hermann Muehling, K. (2001). Is the infiltration-centrifugation technique appropriate for the isolation of apoplastic fluid? A critical evaluation with different plant species. Physiol. Plant 111, 457–465. [DOI] [PubMed] [Google Scholar]
- Lohaus, G., Winter, H.C., Riens, B., and Heldt, H.W. (1995). Further studies of the phloem loading process in leaves of barley and spinach. The comparison of metabolite concentrations in the apoplastic compartment with those in the cytosolic compartment and in the sieve tubes. Bot. Acta 108, 270–275. [Google Scholar]
- Mizuno, N., Takahashi, A., Wagatsuma, T., Mizuno, T., and Obata, H. (2002). Chemical composition of guttation fluid and leaves of Petasites japonicus v. giganteus and Polygonum cuspidatum growing on ultramafic soil. Soil Sci. Plant Nutr. 48, 451–453. [Google Scholar]
- Nacry, P., Camilleri, C., Courtial, B., Caboche, M., and Bouchez, D. (1998). Major chromosomal rearrangements induced by T-DNA transformation in Arabidopsis. Genetics 149, 641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata, T. (1986). The exudation of glutamine and theanine from tea leaf. Study of Tea 69, 17–21. [Google Scholar]
- Pilot, G., Gaymard, F., Mouline, K., Chérel, I., and Sentenac, H. (2003). Regulated expression of Arabidopsis Shaker K+ channel genes involved in K+ uptake and distribution in the plant. Plant Mol. Biol. 51, 773–787. [DOI] [PubMed] [Google Scholar]
- Rios, G., et al. (2002). Rapid identification of Arabidopsis insertion mutants by non-radioactive detection of T-DNA tagged genes. Plant J. 32, 243–253. [DOI] [PubMed] [Google Scholar]
- Schobert, C., and Komor, E. (1989). The differential transport of amino acids into the phloem of Ricinus communis L. seedlings as shown by the analysis of sieve-tube sap. Planta 177, 342–349. [DOI] [PubMed] [Google Scholar]
- Schobert, C., and Komor, E. (1990). Transfer of amino acids and nitrate from the roots into the xylem of Ricinus communis seedlings. Planta 181, 85–90. [DOI] [PubMed] [Google Scholar]
- Schulze, W.X., Reinders, A., Ward, J., Lalonde, S., and Frommer, W.B. (2003). Interactions between co-expressed Arabidopsis sucrose transporters in the split-ubiquitin system. BMC Biochem. 4, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke, R., Schneider, A., Van Der Graaff, E., Fischer, K., Catoni, E., Desimone, M., Frommer, W.B., Flugge, U.I., and Kunze, R. (2003). ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 131, 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, K., Simons, M., Reiser, J., Saleem, M.A., Faul, C., Kriz, W., Shaw, A.S., Holzman, L.B., and Mundel, P. (2001). Podocin, a raft-associated component of the glomerular slit diaphragm, interacts with CD2AP and nephrin. J. Clin. Invest. 108, 1621–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro, B.M., and Stadtman, E.R. (1971). Glutamine synthetase (Escherichia coli). Methods Enzymol. 17A, 910–922. [DOI] [PubMed] [Google Scholar]
- Shelp, B.J. (1987). The composition of phloem exudate and xylem sap from Broccoli (Brassica oleracea var. italica) supplied with NH4+, NO3− or NH4NO3. J. Exp. Bot. 38, 1619–1636. [Google Scholar]
- Shi, H., Quintero, F.J., Pardo, J.M., and Zhu, J.K. (2002). The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14, 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibagaki, N., Rose, A., McDermott, J.P., Fujiwara, T., Hayashi, H., Yoneyama, T., and Davies, J.P. (2002). Selenate-resistant mutants of Arabidopsis thaliana identify Sultr1;2, a sulfate transporter required for efficient transport of sulfate into roots. Plant J. 29, 475–486. [DOI] [PubMed] [Google Scholar]
- Snyers, L., Umlauf, E., and Prohaska, R. (1999). Association of stomatin with lipid-protein complexes in the plasma membrane and the endocytic compartment. Eur. J. Cell Biol. 78, 802–812. [DOI] [PubMed] [Google Scholar]
- Stumpel, F., Burcelin, R., Jungermann, K., and Thorens, B. (2001). Normal kinetics of intestinal glucose absorption in the absence of GLUT2: Evidence for a transport pathway requiring glucose phosphorylation and transfer into the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 98, 11330–11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano, J., Noguchi, K., Yasumori, M., Kobayashi, M., Gajdos, Z., Miwa, K., Hayashi, H., Yoneyama, T., and Fujiwara, T. (2002). Arabidopsis boron transporter for xylem loading. Nature 420, 337–340. [DOI] [PubMed] [Google Scholar]
- Takeda, F., and Glenn, M.D. (1989). Hydathode anatomy and the relationship between guttation and plant water status in strawberry (Fragaria x ananassa Duch.). Acta Hortic. 265, 387–392. [Google Scholar]
- Tax, F.E., and Vernon, D.M. (2001). T-DNA-associated duplication/translocation in Arabidopsis. Implications for mutant analysis and functional genomics. Plant Physiol. 126, 1527–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1997). The CLUSTAL X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Larebeke, N., Engler, G., Holsters, M., Van den Elsacker, S., Zaenen, I., Schilperoort, R.A., and Schell, J. (1974). Large plasmid in Agrobacterium tumefaciens essential for crown gall-inducing ability. Nature 252, 169–170. [DOI] [PubMed] [Google Scholar]
- Verdaguer, B., de Kochko, A., Beachy, R.N., and Fauquet, C. (1996). Isolation and expression in transgenic tobacco and rice plants, of the cassava vein mosaic virus (CVMV) promoter. Plant Mol. Biol. 31, 1129–1139. [DOI] [PubMed] [Google Scholar]
- Verdaguer, B., de Kochko, A., Fux, C.I., Beachy, R.N., and Fauquet, C. (1998). Functional organization of the cassava vein mosaic virus (CsVMV) promoter. Plant Mol. Biol. 37, 1055–1067. [DOI] [PubMed] [Google Scholar]
- Vincent, R., Fraisier, V., Chaillou, S., Limami, M.A., Deleens, E., Phillipson, B., Douat, C., Boutin, J.P., and Hirel, B. (1997). Overexpression of a soybean gene encoding cytosolic glutamine synthetase in shoots of transgenic Lotus corniculatus L. plants triggers changes in ammonium assimilation and plant development. Planta 201, 424–433. [DOI] [PubMed] [Google Scholar]
- Vrljic, M., Sahm, H., and Eggeling, L. (1996). A new type of transporter with a new type of cellular function: L-lysine export from Corynebacterium glutamicum. Mol. Microbiol. 22, 815–826. [DOI] [PubMed] [Google Scholar]
- Wagner, C.A., Lang, F., and Broer, S. (2001). Function and structure of heterodimeric amino acid transporters. Am. J. Physiol. Cell Physiol. 281, C1077–C1093. [DOI] [PubMed] [Google Scholar]
- Weigel, D., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122, 1003–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter, H.C., Lohaus, G., and Heldt, H.W. (1992). Phloem transport of amino acids in relation to their cytosolic levels in barley leaves. Plant Physiol. 99, 996–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wipf, D., Ludewig, U., Tegeder, M., Rentsch, D., Koch, W., and Frommer, W.B. (2002). Conservation of amino acid transporters in fungi, plants and animals. Trends Biochem. Sci. 27, 139–147. [DOI] [PubMed] [Google Scholar]
- Xia, Y., Suzuki, H., Borevitz, J., Blount, J., Guo, Z., Patel, K., Dixon, R.A., and Lamb, C. (2004). An extracellular aspartic protease functions in Arabidopsis disease resistance signaling. EMBO J. 23, 980–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J.Z., Abbud, W., Prohaska, R., and Ismail-Beigi, F. (2001). Overexpression of stomatin depresses GLUT-1 glucose transporter activity. Am. J. Physiol. Cell Physiol. 280, C1277–C1283. [DOI] [PubMed] [Google Scholar]
- Ziegler, H., and Lüttge, U. (1959). Uber die Resorption von C14-Glutaminsaüre durch sezernierende Nektarien. Naturwissenschaften 5, 176–177. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.