Abstract
Reversible protein phosphorylation regulates many cellular processes, including the dynamics and organization of the microtubule cytoskeleton, but the events mediating it are poorly understood. A semidominant phs1-1 allele of the Arabidopsis thaliana PROPYZAMIDE-HYPERSENSITIVE 1 locus exhibits phenotypes indicative of compromised cortical microtubule functions, such as left-handed helical growth of seedling roots, defective anisotropic growth at low doses of microtubule-destabilizing drugs, enhancement of the temperature-sensitive microtubule organization1-1 phenotype, and less ordered and more fragmented cortical microtubule arrays compared with the wild type. PHS1 encodes a novel protein similar to mitogen-activated protein kinase (MAPK) phosphatases. In phs1-1, a conserved Arg residue in the noncatalytic N-terminal region is exchanged with Cys, and the mutant PHS1 retained considerable phosphatase activity in vitro. In mammalian MAPK phosphatases, the corresponding region serves as a docking motif for MAPKs, and analogous Arg substitutions severely inhibit the kinase–phosphatase association. Transgenic studies indicate that the phs1-1 mutation acts dominant negatively, whereas the null phs1-2 allele is recessive embryonic lethal. We propose that the PHS1 phosphatase regulates more than one MAPK and that a subset of its target kinases is involved in the organization of cortical microtubules.
INTRODUCTION
Microtubules are cylindrical polymers built from heterodimers of highly conserved α- and β-tubulin. Microtubules are nucleated at and grow from distinct subcellular sites containing γ-tubulin, after which the polymer ends exhibit alternate periods of growth and rapid shortening, a behavior called dynamic instability (Desai and Mitchison, 1997; Job et al., 2003). The dynamic assembly and disassembly of microtubules is essential for a variety of cellular functions, such as the maintenance or establishment of cell morphology and polarity, cell division, and intracellular trafficking (Kirschner and Mitchison, 1986; Vega and Solomon, 1997). Remarkably, the stability and spatial organization of microtubules vary extensively during the cell cycle and between different cell types, but the physiological determinants for such diversity are not fully understood (Joshi, 1998; Gundersen and Cook, 1999).
In plant cells, microtubules are organized into several distinct structures during the cell cycle, including preprophase bands, mitotic spindles, phragmoplasts, and cortical microtubules (Wasteneys, 2002). In postcytokinetic epidermal and cultured cells of Arabidopsis thaliana, cortical microtubules actively migrate across the cell cortex by a hybrid treadmilling mechanism and form highly dynamic arrays (Chan et al., 2003; Shaw et al., 2003). In Arabidopsis root epidermal cells, the orientation of cortical microtubules changes during cell elongation and differentiation. In these cells, microtubules adopt consistently parallel arrays that are transverse to the long axis of the cell while the growth rate is increasing but change to form shallow helices that are consistently right-handed when growth is starting to decline (Sugimoto et al., 2000). In the Arabidopsis lefty1 and lefty2 mutants, microtubule stability is compromised by the tubulin missense mutations, resulting in right-handed or less ordered helical arrays of cortical microtubules in the rapidly elongating root epidermal cells (Thitamadee et al., 2002; Abe et al., 2004). The lefty mutant cells or wild-type cells treated with microtubule-targeted drugs (Baskin et al., 1994; Furutani et al., 2000) show left-handed or grossly isotropic growth. These and other studies (for a review, see Lloyd and Chan, 2003) indicate that proper control of microtubule stability is important for the organization of cortical microtubule arrays and for strict anisotropic growth in plants.
Microtubule dynamics and organization are regulated by distinct groups of proteins that directly bind or act on microtubules. Microtubule-associated proteins (MAPs) bind microtubules on the lateral walls or preferentially at the polymer ends (Mandelkow and Mandelkow, 1995; Howard and Hyman, 2003). Other microtubule regulators include tubulin sequestrating proteins, microtubule-depolymerizing unconventional kinesins, and microtubule-severing katanin (Heald and Nogales, 2002). These proteins can promote stabilization, polymerization, and depolymerization of microtubles, cross-linking of adjacent microtubules, association of microtubules with motor protein complexes, and targeting of microtubule ends to a specific area of the cell cortex or restricted subcellular regions. Several microtubule regulators have been recently identified from plants through biochemical purification, mutant isolation, and homology to animal and fungal MAPs (for reviews, see Lloyd and Hussey, 2001; Hashimoto, 2003). One such example is the XMAP215 family MAP (Ohkura et al., 2001). The Arabidopsis XMAP215 homolog has been identified as missense mutations in the MOR1 gene (Whittington et al., 2001). Temperature-sensitive mor1 alleles show fragmentation and disorganization of cortical microtubules at restrictive temperatures while the mutant plants grow normally at permissive temperatures. Null alleles of MOR1 (called gem1) are also defective in the cytokinetic phragmoplast (Twell et al., 2002).
Environmental, developmental, and cell cycle cues may be transduced to MAPs and other regulators of microtubule dynamics to reorganize the microtubule cytoskeleton. A dominant mechanism of information transfer in cells is protein phosphorylation, and the activities of animal microtubule regulators are modified posttranslationally through reversible phosphorylation and dephosphorylation (Cassimeris, 1999). A well-known example of a phosphorylation-responsive microtubule regulator is animal stathmin, which was initially identified as a protein phosphorylated in response to several extracellular signals, and was later rediscovered as a potent microtubule destabilizer (Belmont and Mitchison, 1996). Phosphorylation of distinct Ser residues decreases the binding affinity of stathmin for tubulin and consequently turns off its microtubule destabilizing activity (Cassimeris, 2002).
Protein phosphorylation has been suggested to play an important role in the stability and organization of plant microtubules as well, but the molecular components involved remain unknown (Cyr and Palevitz, 1995; Hepler and Hush, 1996; Azimzadeh et al., 2001). Protein kinase inhibitors and inhibitors for types 1 and 2A protein phosphatases are reported to disorganize cortical microtubules (Baskin and Wilson, 1997, and references therein). Addition of protein kinase inhibitors to suspension-cultured tobacco (Nicotiana tabacum) cells rendered cortical microtubules more resistant to cold-induced depolymerization (Mizuno, 1992), whereas an inhibitor of types 1 and 2A protein phoshatases promoted microtubule depolymerization in maize (Zea mays) root cells (Tian et al., 2004), indicating that cortical microtubule stability is regulated by protein phosphorylation. A recent genetic approach identified a putative protein phosphatase 2A regulatory subunit to be involved in the control of cortical microtubule structures (Camilleri et al., 2002). A genetic screen for mutants defective in microtubule functions should provide further information on the control mechanisms of plant microtubules. In this study, we isolated many Arabidopsis mutants that are more sensitive to microtubule-destabilizing drugs than the wild type. We show that a novel phosphatase is defective in one of the drug-hypersensitive mutants and may be involved in phosphorylation cascades that control the dynamics of cortical microtubules in plant cells.
RESULTS
Screening of Propyzamide-Hypersensitive Mutants
We have previously shown that the two left-handed helical growth Arabidopsis mutants lefty1 and lefty2 are caused by dominant negative mutations at the tubulin intradimer interface of α-tubulins 4 and 6 and that the cortical microtubules in these mutants had increased sensitivity to microtubule-specific drugs, including a microtubule destabilizing drug, propyzamide (Thitamadee et al., 2002). To further explore the genetic factors affecting the microtubule organization, we screened Arabidopsis mutants with increased sensitivity to propyzamide. T-DNA insertion lines and M2 seedlings mutagenized by ethyl methanesulfonate were grown on hard agar plates containing 3 μM propyzamide and screened for seedlings whose primary roots were stunted and swollen. Candidate seedlings were transferred to drug-free agar plates, and the recovery of root growth was tested. We obtained 39 propyzamide-hypersensitive mutants that were grouped into three types. Twenty eight mutants were mapped to TUBULIN loci, in which missense mutations were found in the corresponding tubulin genes. These tubulin mutants will be reported elsewhere. Ten mutants were recessive and showed morphologies reminiscent of the katanin p60 mutant fra2/bot1/erh3/lue1 (Bichet et al., 2001; Burk et al., 2001; Webb et al., 2002; Bouquin et al., 2003). Indeed, these mutants were mapped to the AtKTN1 katanin p60 locus and were found to be fra2 alleles by complementation tests. One remaining mutant was named propyzamide-hypersensitive 1-1 (phs1-1). Unless stated otherwise, homozygous phs1-1 plants were used in the subsequent experiments.
Plant Morphology and Sensitivity to the Microtubule-Interacting Drugs
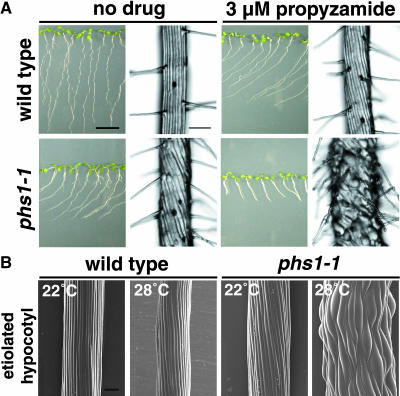
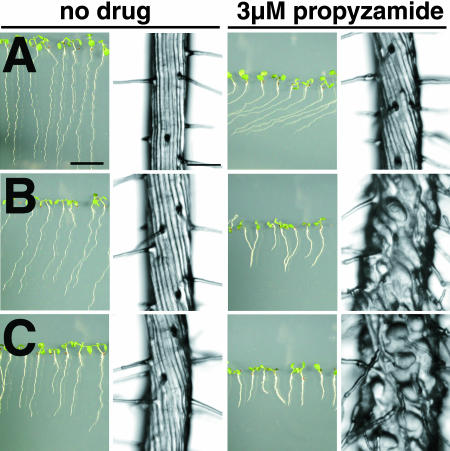
Without drug treatment, the epidermal cell files of phs1-1 roots formed left-handed (S-form) helices, in contrast with straight wild-type Columbia (Col) cell files (Figure 1A). When seedlings were cultured vertically against an impenetrable agar surface, phs1-1 primary roots skewed toward the left side of the plate, as viewed from above, whereas wild-type roots grew downward in the direction of gravity (Figure 1A). Addition of 3 μM propyzamide to the culture medium caused the epidermal cells of phs1-1 roots to swell, and the elongation of phs1-1 roots was severely inhibited (Figure 1A). At the same drug concentration, the wild-type root epidermal cell files formed left-handed helices, but no swelling was apparent in the epidermal cells (Figure 1A).
Figure 1.
Morphological Phenotypes of phs1-1.
(A) Wild-type (Col) and phs1-1 seedlings grown for 7 d on a vertically placed agar plate containing nutrient medium with or without 3 μM propyzamide. Root epidermal cells are shown in an enlarged magnification at the right side of each seedling picture. Bar in seedling view = 1 cm; bar in root view = 100 μm.
(B) Scanning electron micrographs of upper hypocotyl regions of wild-type and phs1-1 seedlings grown in the dark for 5 d at 22 or 28°C. Bar = 100 μm.
The epidermal cell files of phs1-1 etiolated hypocotyls were slightly skewed in the direction of S-form at 22°C, whereas the anisotropic growth was moderately impaired and the epidermal cells were somewhat swollen when the mutant seedlings were grown in the dark at 28°C (Figure 1B). The epidermal cells of wild-type etiolated hypocotyls grew straight at either growth temperature (Figure 1B). Other cell types of phs1-1 plants appeared to be indistinguishable morphologically from wild-type cells.
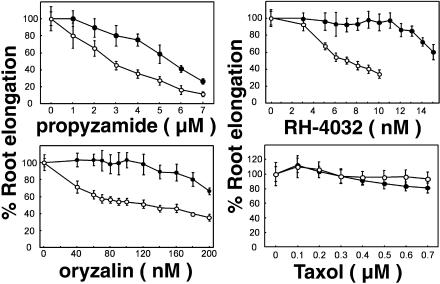
To test the specificity of phs1-1 sensitivity to microtubule-targeted drugs, we examined the inhibition of primary root elongation by several drugs that affect microtubule stability. phs1-1 roots were more sensitive than wild-type roots to propyzamide, RH-4032 (a potent propyzamide analog; Young and Lewandowski, 2000), and another class of microtubule-destabilizing drug, oryzalin (Figure 2). For example, RH-4032 at 10 nM and oryzalin at 120 nM did not affect the growth of wild-type roots but reduced the length of phs1-1 roots by 70 and 50%, respectively. By contrast, phs1-1 roots displayed wild-type sensitivity to the microtubule-stabilizing drug Taxol (Figure 2) and to the actin filament disrupting drug cytochlasin D (data not shown).
Figure 2.
phs1-1 Responses to Microtubule-Interacting Drugs.
After 7 d of growth on medium supplemented with the indicated concentrations of propyzamide, RH-4032, oryzalin, and Taxol, Col (closed circles) and phs1-1 (open circles) seedlings were removed from the agar, and the length of the primary root was measured. Results were standardized against growth on unsupplemented medium. Error bars represent standard deviations of the means (n = 30).
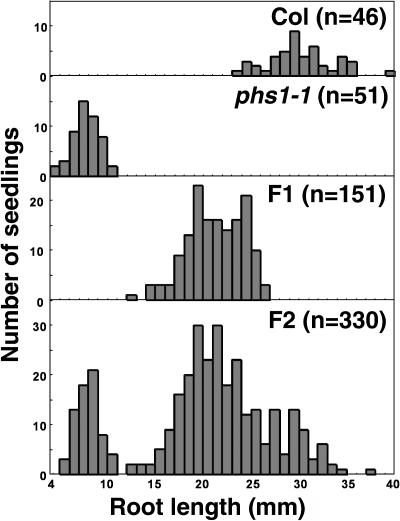
The response of phs1-1 heterozygotes to propyzamide was intermediate between those of phs1-1 homozygotes and the wild type (Figure 3). In the F2 segregating population made from a cross between phs1-1 and the Col parental ecotype, the wild-type phenotype, the intermediate phenotype, and the phs1-1 hypersensitive phenotype were found roughly in a 1:2:1 ratio. Therefore, the phs1-1 mutation is semidominant.
Figure 3.
phs1-1 Is a Semidominant Allele.
After 7 d of growth on medium supplemented with 3 μM propyzamide, seedlings were removed from the agar, and the length of the primary root was measured. Results are displayed as distribution histograms. The number of plants analyzed is indicated in parentheses.
Microtubule Organization in phs1-1 Cells
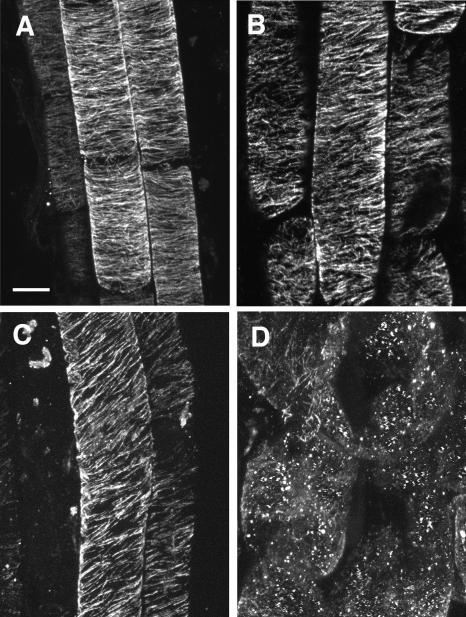
The increased sensitivity of phs1-1 roots to microtubule-destabilizing drugs suggests that the microtubule organization is impaired in the mutant cells. To assess this possibility, we examined cortical microtubules underneath the outer tangential wall of epidermal cells in the root elongation zone by immunofluorescent staining with an anti-tubulin antibody. In the root epidermal cells without drug treatment, cortical microtubule arrays appeared to be less ordered and slightly more fragmented in phs1-1 than in the wild type (Figures 4A and 4B). We quantified microtubule orientations from immunofluorescent images of fixed cells. Most of the wild-type cortical microtubules deviated by <20° from the transverse axis, with an average angle of 89.4°, whereas the angular deviation in phs1-1 cells increased significantly (see supplemental data online). It is noted that the average microtubule angle in phs1-1 cells was shifted to 97.2°, which indicates that the mutant microtubule arrays on average formed shallow right-handed helices.
Figure 4.
Cortical Microtubule Arrays in Root Epidermal Cells.
Confocal micrographs of immunofluorescently labeled microtubules in root epidermal cells from 4-d-old seedlings. Bar = 10 μm.
(A) Col (wild type) without drug treatment.
(B) phs1-1 without drug treatment.
(C) Col grown in the medium supplemented with 150 nM oryzalin.
(D) phs1-1 grown in the medium supplemented with 150 nM oryzalin.
When seedlings were grown in the presence of 150 nM oryzalin, cortical microtubule arrays of wild-type root epidermal cells were skewed slightly to form shallow right-handed helices but were not visibly fragmented (Figure 4C). By contrast, cortical microtubule arrays in swollen phs1-1 cells were heavily fragmented and distributed highly randomly (Figure 4D). Many fluorescent dots that presumably represent aggregated tubulin oligomers were also observed. Similar results were also obtained with 3 μM propyzamide (data not shown). We conclude that cortical microtubules in phs1-1 cells are more destabilized than wild-type microtubules.
phs1-1 Mutation Exaggerates the Temperature-Sensitive mor1 Phenotypes
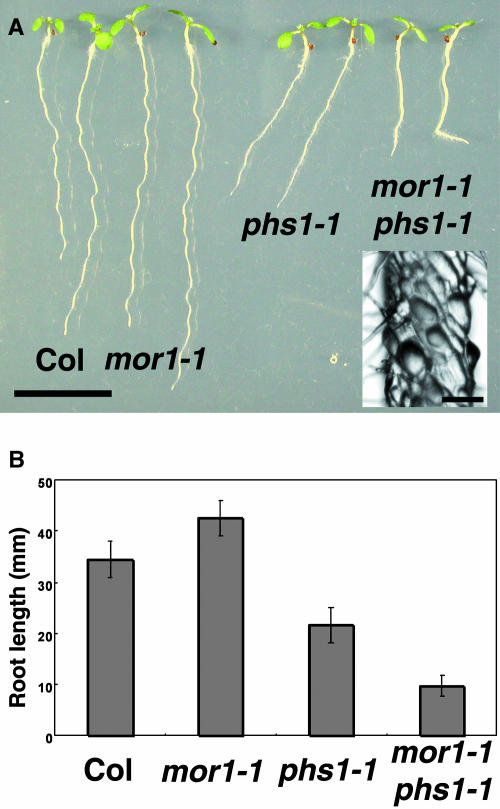
To test genetic interactions, we generated double mutants of phs1-1 with several microtubule-related mutants, including lefty1, lefty2 (Thitamadee et al., 2002), spiral1, spiral2 (Furutani et al., 2000), and mor1 (Whittington et al., 2001). Among the double mutants, the most striking phenotype was observed in the phs1-1 microtubule organization1-1 (mor1-1) plants. mor1-1 is a temperature-sensitive mutant allele of XMAP215-class MAP and at 22°C does not show any obvious phenotypes (Whittington et al., 2001) but showed a slightly enhanced sensitivity to propyzamide that was intermediate between the wild type and phs1-1 (our unpublished results). At this permissive temperature, the morphological phenotypes of phs1-1 mor1-1 were much more severe than those of either single mutant (Figure 5A). The exaggerated defects were observed not only in the impaired growth of primary roots (Figure 5B) but also in the overall stunted growth statute, short petiole, stubby trichome stalks at maturity, and male sterility (data not shown). These phenotypes were at least partly attributable to a defect in anisotropic cell expansion (Figure 5A, inset).
Figure 5.
Genetic Interaction between phs1-1 and mor1-1.
(A) Seedlings grown for 7 d on a vertically placed agar plate. Genotypes are shown below the seedlings. Inset shows a higher magnification image of the mor1-1 phs1-1 root epidermal cells. Bar = 1 cm; bar in inset = 100 μm.
(B) Root growth of 7-d-old seedlings. Lengths of primary roots were measured in 30 seedlings for each genotype.
phs1-1 Has a Missense Mutation in a Putative Phosphatase
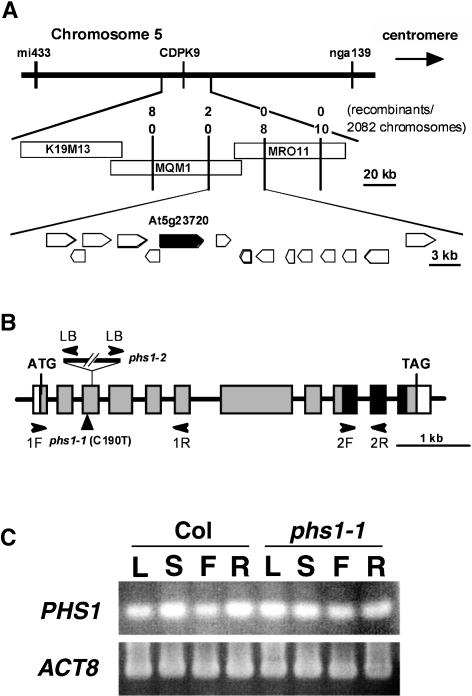
We identified the PHS1 gene using map-based positional cloning (see Methods). PHS1 was mapped to the top of chromosome 5 between the markers mi433 and nga139. Further fine mapping narrowed down the PHS1 position to a 63-kb region in the two BACs MQM1 and MRO11 (Figure 6A). We then sequenced all the potential open reading frames predicted in this region and found a C-to-T point mutation in the third exon of a putative phosphatase, At5g23720 (Figure 6B). A cDNA clone representing this transcription unit was assembled from an incomplete EST (AV546458) and RT-PCR fragments. Sequencing analysis of our full-length cDNA clone revealed that the nucleotide sequence deposited at the National Center for Biotechnology Information (AY099681) contains two deletions of 5 and 18 bp in the predicted coding sequence, which results in a wrong protein prediction of the N-terminal region and a deletion of six amino acid residues at the C-terminal region. The corrected protein sequence, which was corroborated by the BAC sequence, shows that an Arg at position 64 was changed to a Cys codon in phs1-1.
Figure 6.
Map-Based Positional Cloning of PHS1.
(A) Recombination mapping of phs1-1 localized the PHS1 locus within the 60-kb region spanned by BACs MQM1 and MRO11. Potential open reading frames are indicated by pentagons, and the gene At5g23720 (PHS1) is indicated by a closed pentagon.
(B) PHS1 contains 11 exons (thick boxes) separated by 10 introns (lines). Protein coding region is indicated by gray boxes, and the dual-specificity phosphatase catalytic domain is indicated by closed boxes. phs1-1 has a C-to-T mutation at position 190 (where 1 is the A of the initiator ATG), whereas phs1-2 contains a T-DNA insertion in the third exon. The position of PCR primers used for genotyping the phs1-2 allele (LB, 1F, and 1R) and RT-PCR analysis (2F and 2R) is indicated by arrowheads.
(C) RT-PCR analysis of PHS1 gene expression in Arabidopsis organs of Col and phs1-1. Total RNA was extracted from roots (R) of 14-d-old seedlings and from leaves (L), stems (S), and flowers (F) of 40-d-old plants. Expression of an actin gene (ACT8) was also analyzed as a control.
To confirm that this gene was responsible for the phs1-1 phenotypes and to gain an insight into the semidominant nature of the phs1-1 allele, we performed two transformation experiments. First, we transformed the phs1-1 plants with a 6.1-kb wild-type genomic region surrounding the At5g23720 gene. In five independent homozygous T3 lines among a total of 20 transgenic lines with single-locus T-DNA insertions, wild-type phenotypes were restored, including straight root growth and the normal leftward skewing response of the primary root on the propyzamide-supplemented agar medium (Figure 7A). Second, we also transformed the wild-type Col plants with the same 6.1-kb genomic region as above, except that the C-to-T point mutation found in phs1-1 was introduced in the third exon of At5g23720. When 26 independent transgenic lines with single-locus T-DNA insertions were examined, six homozygous T3 lines displayed clear phs1-1 phenotypes, such as leftward root bending on drug-free agar plates and stunted root growth in the presence of 3 μM propyzamide (Figure 7B). Similar propyzamide-hypersensitive phenotypes were also observed when the PHS1 cDNA with the phs1-1 mutation was expressed under the control of the 35S promoter of Cauliflower mosaic virus (CaMV35S) in the wild-type plants (Figure 7C). However, when wild-type PHS1 cDNA was expressed with the CaMV35S promoter in the wild type, none of the 31 transgenic lines showed either reduced anisotropic growth or increased sensitivity to propyzamide (data not shown). These transgenic experiments indicate that phs1-1 phenotypes are caused by a dominant-negative mutation in At5g23720.
Figure 7.
Morphological Phenotypes of Transgenic Plants Expressing Wild-Type or Mutant PHS1 Transgenes.
Seedlings were grown for 7 d on a vertically placed agar plate with or without 3 μM propyzamide in the medium. Root epidermal cells are shown on the right side of each seedling picture. Bar in seedling view = 1 cm; bar in magnified root view = 100 μm.
(A) phs1-1 seedlings transformed with a wild-type genomic clone of PHS1.
(B) Wild-type seedlings transformed with a PHS1 genomic clone containing the R64C mutation.
(C) Wild-type seedlings transformed with the R64C mutant PHS1 cDNA under the control of the CaMV35S promoter.
Because the steady state levels of PHS1 mRNA were undetectable by RNA gel blot analysis, the expression was examined by RT-PCR. PHS1 was expressed in all wild-type organs tested, including leaves, stems, flowers, and roots (Figure 6C). In phs1-1 mutants, the PHS1 gene was expressed in all organs at similar levels to the wild type.
PHS1 Is a MAP Kinase Phosphatase-Like Protein
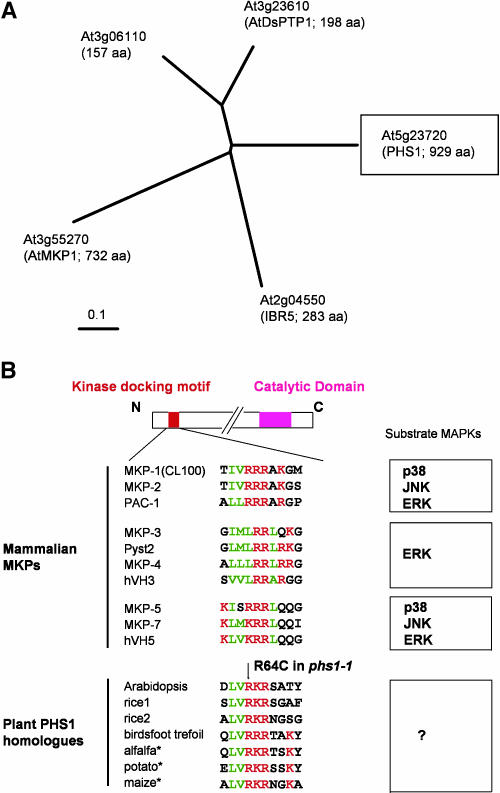
PHS1 encodes a 929–amino acid protein that possesses a dual-specificity phosphatase catalytic domain at the C terminus, but its long N-terminal extension shows no significant homology with other proteins encoded in the Arabidopsis genome (Figure 6B). Two bipartite nuclear targeting sequences are predicted at positions 351 to 367 and 811 to 827 by PROSITE (http://kr.expasy.org/cgi-bin/scanprosite). Dual-specificity phosphatases are evolutionarily related to protein Tyr phosphatases but act at both Tyr and Ser/Thr residues. The Arabidopsis genome contains 18 potential dual-specificity phosphatases, among which five are classified as the mitogen-activaged protein kinase (MAPK) phosphatase subclass, based on the amino acid sequence similarity of the phosphatase catalytic domain to established animal MAPK phosphatases (Kerk et al., 2002). As shown in Figure 9A, PHS1 (At5g23720) belongs to this MAPK phosphatase subclass.
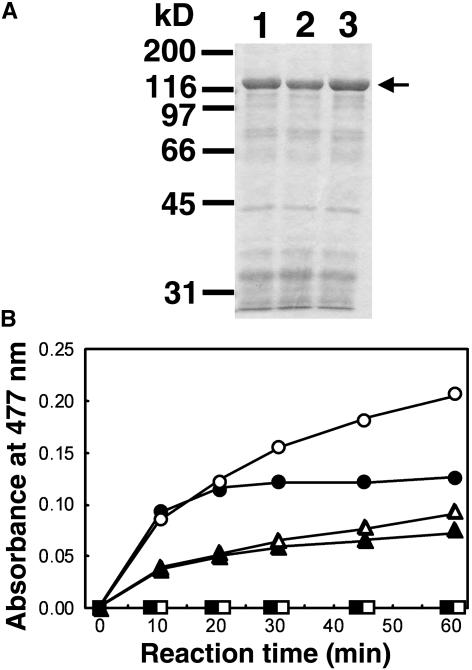
Figure 9.
Phosphatase Activity of Recombinant PHS1 Proteins.
(A) SDS-polyacrylamide gel electrophoresis of the partially purified recombinant GST-PHS1 fusion proteins. Proteins were separated on 10% gels and visualized with Coomassie blue. Positions of molecular weight markers are indicated at the left, and the arrow at the right shows the PHS1 proteins. Lane 1, partially purified GST-PHS1; lane 2, partially purified PHS1R64C; lane 3, partially purified PHS1C792S.
(B) Phosphatase activity of GST-PHS1 proteins. Reaction mixtures contained an artificial substrate OMFP and 60 μg of partially purified PHS1 fusion proteins of the wild type (circles), R64C mutant (triangles), or C792S mutant (squares). The reaction was performed at either 20°C (open symbols) or 30°C (closed symbols) for the period indicated.
Typical mammalian MAPK phosphatases contain a catalytic phosphatase domain at the C terminus and an N-terminal noncatalytic domain that are divergent among MAPK phosphatases but are important for MAPK binding specificity (Theodosiou and Ashworth, 2002). A well-characterized MAPK-interacting motif is composed of two or three consecutive basic amino acids (mostly Arg) preceded by hydophobic amino acids (Figure 9B; Tanoue et al., 2002). We found a similar motif in the N terminus of PHS1 and its putative orthologs in other plants (Figure 8B). Interestingly, the phs1-1 mutation R64C destroys one of the strictly conserved basic residues in this putative MAPK-interacting motif.
Figure 8.
PHS1 Is a MAPK Phosphatase-Like Protein Containing a Putative Kinase Docking Motif.
(A) Phylogenetic tree of five Arabidopsis dual-specificity phosphatases belonging to the MAPK phosphatase subclass. The portions of the protein corresponding to the catalytic domain (residues 703 to 845 in PHS1) were aligned with the ClustalW method (http://www.ebi.ac.uk/clustalw/#), and the unrooted phylogram was generated using TreeView (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html). aa, amino acids.
(B) Alignment of the kinase docking motif found in 10 members of mammalian MAPK phosphatases and also of the putative MAPK docking motif in plant PHS1 homologs. The red characters indicate negatively charged amino acids, whereas the green characters indicate hydrophobic amino acids. Possible substrate MAPKs are shown at the right. In phs1-1, the conserved Arg residue in this motif is changed to Cys. The GenBank accession numbers of plant PHS1 homologs are TC137403 for rice1, TC157626 for rice2, AP006392.1 for birdsfoot trefoil (Lotus corniculatus var japonicus), TC83892 for alfalfa (Medicago sativa), TC82139 for potato (Solanum tuberosum), and CF244510 for maize. Asterisks indicate partial N-terminal sequences available from EST sequences. Modified from Tanoue et al. (2002).
The phs1-1 Mutation Does Not Abolish the Phosphatase Activity
To test whether PHS1 possesses phosphatase activity, we fused PHS1 to glutathione S-transferase (GST), expressed recombinant GST-PHS1 proteins in Escherichia coli, and purified the fusion proteins (Figure 9A). We tested the phosphatase activities of wild-type PHS1, PHS1R64C (a mutant phosphatase expressed in phs1-1), and PHS1C792S as a negative control. The Cys792 in PHS1 is located in the active site signature motif, and the Cys residues in the equivalent position of MAPK phosphatases have been shown to be one of the three invariant amino acids that are absolutely essential for catalysis (Stewart et al., 1999; Theodosiou and Ashworth, 2002, and references therein).
First, we tried to measure the phosphatase activity using pyronitrophenyl phosphate (pNPP) as an artificial substrate but found no significant activity in the three GST-PHS1 fusion proteins. Dual-specificity phosphatases prefer bulky polycyclic aryl phosphates and show weak enzyme activity toward simple aryl phosphates, such as pNPP (Chen et al., 1996; Gottlin et al., 1996), probably because of their shallower active site pockets (Yuvaniyama et al., 1996; Stewart et al., 1999). Therefore, we next tested a bulky polycyclic substrate, 3-O-methylfluorescein phosphate (OMFP), which is known to boost the second order rate constant kcat/Km of the mammalian MAPK phosphatase MKP-3 over 280-fold compared with pNPP (Zhou and Zhang, 1999). Wild-type PHS1 indeed dephosphorylated OMFP at a considerable rate, whereas PHS1C792S was totally inactive (Figure 9B). The phosphatase activity was markedly inactivated during the reaction, and the inactivation was somewhat ameliorated when the reaction was performed at 20°C. Interestingly, the PHS1R64C mutant showed clear phosphatase activity that was roughly half that of the wild-type enzyme at either 20 or 30°C under the reaction conditions used. These results demonstrate that PHS1 is an active phosphatase and that the phs1-1 mutation compromises but does not abolish the enzyme activity in vitro.
A Knockout Allele of PHS1 Is Recessive Lethal
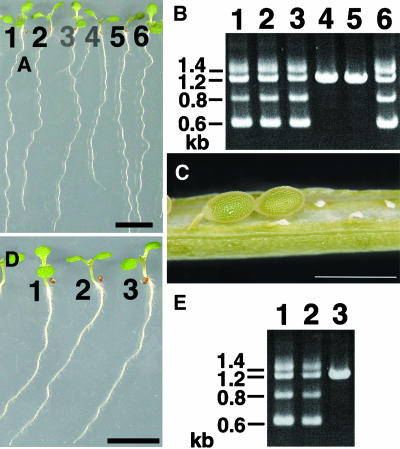
Because the phs1-1 is a semidominant missense mutation, we sought knockout alleles of PHS1 in the T-DNA insertion mutants. The phs1-2 mutant has a T-DNA insertion within the third exon of the PHS1 gene (Figure 6B) and is expected to be a nonfunctional null allele. When F2 seedlings from a cross between Col wild type and heterozygous phs1-2 were analyzed by genomic PCR for the presence of T-DNA at the PHS1 locus, either the wild-type allele (21 plants out of 96 plants analyzed) or the phs1-2 heterozygous allele (remaining 75 plants) was recovered, but no homozygous phs1-2 plants were found (Figure 10B). The phs1-2 heterozygous plants were indistinguishable from wild-type plants in growth and morphology (Figure 10A), indicating that phs1-2 is a fully recessive allele. In the immature siliques of the Col × phs1-2 F1 plants, a high percentage (∼60%) of embryos was aborted early in seed development (Figure 10C). These results suggest that the phs1 null mutant is embryonic lethal. Our attempts to generate Arabidopsis plants with a highly reduced but residual PHS1 expression using antisense RNA technology have not yet recovered transgenic lines with clear phenotypes.
Figure 10.
T-DNA Insertion Allele of PHS1 Is Recessive Embryonic Lethal.
(A) Selfed progeny of phs1-2 heterozygous plants were grown for 7 d on a vertically placed agar plate. Individual seedlings were numbered from 1 to 6. Bar = 1 cm.
(B) Genomic PCR analysis of the PHS1 locus. The numbers indicate the seedlings in (A) from which genomic DNA was extracted for analysis. Seedlings 1, 2, 3, and 6 were heterozygous phs1-2 plants, whereas seedlings 4 and 5 were wild-type plants.
(C) Siliques of phs1-2 heterozygous plants contained normal seeds and small aborted seeds. Bar = 1 mm.
(D) Selfed progeny of phs1-1/phs1-2 trans-heterozygous plants were grown for 7 d on a vertically placed agar plate. Individual seedlings were numbered from 1 to 3. Bar = 1 cm.
(E) Genomic PCR analysis of the PHS1 locus. The numbers indicate the seedlings in (D) from which genomic DNA was extracted for analysis. Seedlings 1 and 2 were phs1-1/phs1-2 trans-heterozygotes, whereas seedling 3 was a phs1-1 homozygote.
We also analyzed phs1-1/phs1-2 trans-heterozygotes. In the selfed progeny of a plant that possessed one copy each of phs1-1 and phs1-2 mutations, phs1-1/phs1-2 trans-heterozygotes and phs1-1 homozygotes were identified by genomic PCR, but no phs1-2 homozygotes were found (Figure 10E). The phs1-1 homozygotes and the phs1-1/phs1-2 trans-heterozygotes were indistinguishable in growth and morphology (Figure 10D). This result further supports that phs1-1 is a semidominant allele and that the T-DNA insertion in At5g23720 is responsible for the recessive lethal phenotype of the phs1-2 allele.
DISCUSSION
Semidominant phs1-1 Mutation Disrupts Microtubule Functions
Most Arabidopsis mutants that showed stunted and swollen root phenotypes in the presence of propyzamide in our screen turned out to have mutations in either tubulin genes (our unpublished results) or the AtKTN1 gene (Burk et al., 2001). Only one propyzamide-hypersensitive mutant, phs1-1, was isolated whose mutation was mapped to a novel locus distinct from tubulin and katanin genes. Several observations suggest that this semidominant mutation disrupts normal microtuble functions. First, in the absence of microtubule-targeted drugs, the epidermal cell files of phs1-1 roots were twisted in a left-handed helix, which presumably resulted in skewing toward the left side of the plate, as viewed from above. This left-handed helical growth phenotype is reminiscent of lefty1 and lefty2 tubulin mutants (Thitamadee et al., 2002). In the lefty roots, mutant α-tubulins are incorporated into microtubule polymers, producing moderately destabilized and right-handed obliquely oriented cortical microtubule arrays. Anisotropic growth was also impaired in the epidermal cells of etiolated phs1-1 hypocotyls. Second, cortical microtubule arrays in phs1-1 root epidermal cells were somewhat more disorganized and fragmented than wild-type arrays. The angular distribution of microtubules in phs1-1 was shifted slightly toward the direction of right-handed helices, possibly explaining the leftward skewing of the root epidermal cell files. Third, phs1-1 root growth was hypersensitive to different types of microtubule-destabilizing drugs but appeared not to differ from wild-type root growth in response to the microtuble-stabilizing drug taxol and the actin-disrupting drug cytochalasin D. The cortical microtubules in the mutant root cells were readily depolymerized with low doses of microtubule-destabilizing drugs, indicative of moderately destabilized microtubules. Fourth, the phs1-1 mutation significantly enhanced the temperature-sensitive mor1-1 phenotype even at the permissive temperature. Taken together, these results strongly suggest that cortical microtubules are destabilized in phs1-1 cells of roots (and probably of etiolated hypocotyls as well).
PHS1 Is a MAPK Phosphatase-Like Protein
Positional cloning revealed that PHS1 encodes a 104-kD protein with a dual-specificity phosphatase domain at the C terminus. Dual-specificity phosphatases act at both phospho-Tyr and phospho-Ser/Thr residues in protein substrates, and a subclass of dual-specificity phosphatases dephosphorylates MAP kinases and as such are termed MAPK phosphatases (Camps et al., 1999). PHS1 (At5g23720) is one of the five potential MAPK phosphatases encoded in the Arabidopsis genome and is loosely grouped with At3g23610 (AtDSPTP1) and At3g06110 in a phylogenetic tree (Figure 8A; Kerk et al., 2002). AtDSPTP1 has been shown to dephosphorylate and inactivate an Arabidopsis MAPK (AtMPK4) in vitro (Gupta et al., 1998). AtDSPTP1 and At3g06110 are relatively small proteins (18 to 22 kD) and consist mainly of the phosphatase domain, whereas PHS1 differs from these two with respect to its long N-terminal extension. The other two Arabidopsis proteins in the MAPK phosphatase family have been recently characterized. Disruption of AtMKP1 (At3g55270) causes hypersensitivity to genotoxic stress and increased resistance to salinity (Ulm et al., 2001, 2002). AtMKP1 interacts with an Arabidopsis MAP kinase, MPK6, in yeast two-hybrid assays and in vitro, and MPK6 activity and the level of MKP1 in plants are inversely correlated (Ulm et al., 2002). Arabidopsis ibr5 mutants are disrupted in another relatively small MAPK phosphatase-like protein (At2g04550) and are less responsive to auxin and abscisic acid than the wild type (Monroe-Augustus et al., 2003). Although we have yet to provide experimental evidence that PHS1 acts on MAPKs in vivo, the following discussions are based on such an assumption.
phs1-1 Mutation Alters a Putative MAPK Docking Motif
Recently, interaction with MAPK targets has been extensively studied in several mammalian MAPK phosphatases. MKP-3, a prototypical MAPK phosphatase, binds to the MAPK extracellular signal-regulated kinase 2 (ERK2) through its N-terminal domain, resulting in the activation of its C-terminal phosphatase domain (Camps et al., 1998). In the absence of the substrate ERK2, the N-terminal MAPK binding domain associates with the C-terminal phosphatase domain, which is in a low-activity state because of disengagement of the active site residues. Interaction of the MAPK binding domain with ERK2 allosterically affects the active site conformation in the phosphatase domain, leading to enzymatic activation of MKP-3 (Stewart et al., 1999; Farooq et al., 2001). The N-terminal MAPK docking motif consisting of a cluster of positively charged amino acids, surrounded by a loosely positioned cluster of hydrophobic amino acids, plays a major role in the high affinity ERK2 binding of MKP-3 (Zhou et al., 2001). This MAPK docking motif is conserved in other human MAPK phosphatases and, interestingly, in PHS1 and its putative orthologs in other plant species (Figure 8B).
Typical MAPKs contain a common docking domain in the C terminus that is used in the docking interactions with substrates, activators (MAPK kinases), and inactivators (MAPK phosphatases) (Tanoue et al., 2000). Two Asp residues in this common domain of MAPK are essential for electrostatic interactions with clustered basic amino acid residues in their interacting partners. When either one of the clustered Arg residues in the MAPK-docking motif of MKP-2 and MKP-3 is substituted with Ala, the mutant phosphatases scarcely bind with, and are activated by, ERK2 (Chen et al., 2001; Farooq et al., 2001; Zhou et al., 2001). The mutant PHS1 phosphatase expressed in the phs1-1 allele has a substitution of Arg64 with Cys in this putative MAPK-docking motif (Figure 8B) and might be unable to bind efficiently its target MAPKs.
Mammalian MAPKs are grouped into three major classes based on their preferential activation by extracellular stimuli and amino acid sequence homology: ERK, c-Jun N-terminal kinase (JNK), and p38 (Cano and Mahadevan, 1995). On the other hand, human MAPK phosphatases are classified into three groups on the basis of MAPK recognition sites (Tanoue et al., 2002). Whereas the MKP-3 group members act mainly on ERK, the other two MAPK phosphatase groups have been shown to interact with all three MAPK classes (Figure 8B). In the MKP-1 (CL100) and MKP-5 groups, multiple interaction modules are proposed to be used in combination to bind specific MAPKs. This means that the contribution of the N-terminal basic motif to MAPK interaction is different depending on particular MAPK targets. For example, although the Arg residues of MKP-1 and MKP-2 (Figure 8B) are critical for docking to ERK2 and p38, their contribution is much less important for JNK (Chen et al., 2001; Slack et al., 2001; Tanoue et al., 2002). In MKP-7, these Arg residues are essential for interaction with p38 but not with ERK2 or JNK1 (Masuda et al., 2003). Likewise, if PHS1 interacts with multiple Arabidopsis MAPKs, the R64C mutation in phs1-1 might impair its binding to particular MAPKs but not to other target MAPKs.
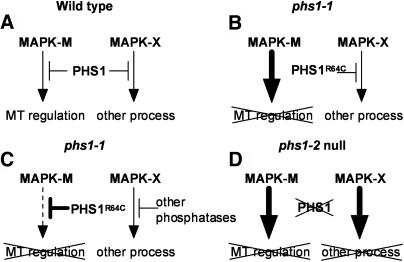
Model for PHS1–MAPK Interaction
We speculate that the PHS1R64C mutant is defective in binding and thus deactivation of a subset of its target MAPKs. In the Arabidopsis genome sequence, 20 genes encoding possible MAPKs were identified and classified into four groups and nine subgroups (Ichimura et al., 2002). Among them, 12 putative MAPKs have the C-terminal common docking domain-like region, which, in analogy to mammalian and yeast MAPKs, might interact with the basic motif in MAPK regulators and substrates. On the other hand, there are five potential MAPK phosphatases, including PHS1, in the Arabidopsis genome. Therefore, at least some Arabidopsis MAPK phosphatases should interact with more than one MAPK subgroup. Although the phosphatase activity of the recombinant PHS1R64C was reduced to about half of the wild-type activity in vitro, the compromised phosphatase activity presumed in phs1-1 is unlikely to be the major cause of the phs1-1 phenotype. This is because the heterozygous phs1-2 T-DNA insertion plants, in which the phosphatase activity should be half the level of the wild-type activity, showed no obvious mutant phenotypes.
In our model, wild-type PHS1 phosphatase binds and inactivates at least two MAPKs, arbitrarily called MAPK-M and MAPK-X (Figure 11A). MAPK-M regulates microtubule-related processes, whereas MAPK-X is involved in other cellular functions. In the phs1-1 mutant, we propose that the mutant PHS1R64C cannot bind and thus is incapable of inactivating MAPK-M but it can still regulate MAPK-X as in the wild type (Figure 11B). A sustained and deregulated activation of MAPK-M may lead to microtubule defects. The MAPK deregulation caused by defective interaction with a phosphatase is best illustrated by a dominant gain-of-function sevenmaker allele of a Drosophila melanogaster MAPK rolled (Brunner et al., 1994). The negatively charged Asp residue in the common docking domain of this MAPK is replaced with Asn in the sevenmaker mutation, which results in greatly reduced binding with its inactivating phosphatases and keeps the mutant MAPK in an activated state (Chu et al., 1996; Camps et al., 1998; Chen et al., 2001; Slack et al., 2001).
Figure 11.
Model for PHS1 Interaction with MAPK.
For details, see Discussion. MT, microtubule.
The semidominant nature of the phs1-1 mutation is more difficult to explain. One possibility is that the PHS1R64C mutant still interacts with the MAP kinase module complex containing MAPK-M and other components, despite its defective interaction with MAPK-M. The long N-terminal extension of PHS1 provides an ample opportunity to interact with multiple targets. The assembly of MAP kinase modules with scaffold proteins is known to dictate pathway specificity and subcellular localization (van Drogen and Peter, 2002; Park et al., 2003). Another possibility comes from the fact that MAPK phosphatases, MAPK kinases, and MAPK substrates, through their Arg-rich basic docking sites, compete with each other for binding to their cognate MAPKs in a competitive and perhaps mutually exclusive fashion (Tanoue et al., 2000; Kusari et al., 2004). In such a case, even a mild perturbation of this competition might result in a profound effect on the MAPK-cascade regulation.
Alternatively, the PHS1R64C mutation may impair its association with MAPK-X, which may lead to an increased proportion of phosphatase activity directed toward MAPK-M rather than MAPK-X (Figure 11C). Enhanced inactivation of MAPK-M could attenuate its function, leading to microtubule defects. Loss of PHS1 inactivation of MAPK-X may be compensated by other phosphatases that redundantly act on MAPK-X. Mammalian MAPKs may be inactivated by multiple specific phosphatases of different classes (Keyse, 2000; Zhou et al., 2002, and references therein). As a variation of this model, the R64C mutation may fortuitously increase the binding affinity of PHS1 for MAPK-M, which the wild-type PHS1 does not normally inactivate efficiently. In this case, wild-type PHS1 may not primarily function in a microtubule-related process. Model C requires the catalytic activity of PHS1 to exert its semidominant phenotype. Expression of the PHS1R64C protein mutated simultaneously at the catalytic Cys792 in transgenic plants should test this model.
Finally, when PHS1 is deleted in the phs1-2 knockout mutant, MAPK-M and MAPK-X are both deregulated (Figure 11D). Activation of the MAPK-X pathway or simultaneous activation of the two MAPK pathways may cause cellular defects much more severe than microtubule defects alone, resulting in embryonic lethality. Our working model on the PHS1 function and specificity should be testable when a full spectrum of PHS1 interactions with its putative MAPK substrates and other signaling components is identified.
MAP Kinase Control of Microtubule Functions
Pharmacological and genetic evidence suggests involvement of type 2A phosphatases in the plant microtubule organization (Baskin and Wilson, 1997; Camilleri et al., 2002; Tian et al., 2004). The paucity of effective and specific inhibitors may have eluded studies on other types of protein phosphatases, but our study expands the list of phosphatases to include a putative MAPK phosphatase as a potential regulator of plant microtubule functions. MAPK phosphatase activity is proposed to be critical for a flexible response of the MAPK signaling network (Bhalla et al., 2002) and is expected to be an important control point for the MAPK cascades.
Transcriptional factors may be cellular targets for MAPKs. To phosphorylate these proteins, activated MAPKs must translocate from the cytoplasm to the nucleus (Keyse, 2000). Some MAPK phosphatases are known to localize in the nucleus or shuttle between the two cellular compartments (Theodosiou and Ashworth, 2002). The resulting transcriptional activation or suppression of MAPs may lead to modulation of the microtubule organization and stability. Two putative bipartite nuclear targeting sequences are found in PHS1. Alternatively, activated MAPKs may phosphorylate target proteins in the cytoplasm. Animal MAPs have been long known to be efficient in vitro substrates for MAPKs and even contributed to the MAPK term (Ray and Sturgill, 1987; Cobb et al., 1991). In mouse fibroblasts, approximately one-third of the total MAPK is associated with the microtubule cytoskeleton (Reszka et al., 1995). Recently, a member of the JNK MAPKs was shown to associate in vivo with two classic stabilizing MAPs, MAP2 and MAP1B, and to be required for maintaining the microtubule integrity of neuronal cells (Chang et al., 2003). In mouse oocytes, a MAPK substrate, DOC1R, localizes to microtubules and controls the microtubule organization (Terret et al., 2003). Genetic interaction between phs1-1 and mor1-1 raises the interesting possibility that one of the cellular targets of a PHS1-regulated MAPK might be the XMAP215-class MAP, MOR1 (Whittington et al., 2001).
Plant MAPK cascades are vital to fundamental physiological functions involved in hormonal responses, cell cycle regulation, abiotic stress signaling, and defense mechanisms. Expansion of the cell plate in plant cytokinesis is regulated by a MAPK cascade, which involves a kinesin-like protein/MAPK kinase kinase complex and requires intact phragmoplast microtubules (Nishihama et al., 2002; Soyano et al., 2003). The complexity and redundancy of the signaling components, the antagonistic nature of distinct pathways, and the use of both positive and negative regulatory mechanisms have been documented (Tena et al., 2001). The semidominant phs1-1 mutation may help to dissect the MAPK signaling cascades and to unravel the hitherto unknown involvement of MAPKs in the regulation of plant microtubule functions. Identification of in vivo substrates for the PHS1 phosphatase is the next essential step toward this goal.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana plants (ecotypes Col and Wassilewskija) mutagenized using ethyl methanesulfonate or T-DNA insertions were obtained from Lehle Seeds (Round Rock, TX) and the ABRC (Columbus, OH), respectively. Seedlings were grown aseptically on a nutrient medium solidified with 1.5% (w/v) agar at 22°C with a 16-h-light/8-h-dark cycle (Furutani et al., 2000), unless otherwise noted. For the screening of mutants, propyzamide (Wako, Kyoto, Japan) was added to the solid medium at a final concentration of 3 μM. On this selection medium, wild-type seedlings displayed left-handed helical growth in roots and petioles. Putative mutants that showed a swollen root morphology were removed after 7 d of germination and transferred onto fresh medium without propyzamide. Only the plants that recovered the radial expansion phenotype were then grown in soil and analyzed. The phs1-1 allele was isolated from the ethyl methanesulfonate–mutagenized seeds of the Col ecotype and backcrossed to the wild type at least three times before analysis. The mor1-1 allele has been described (Whittington et al., 2001). A fat root allele of the fra2 mutant was used for complementation.
Oryzalin (AccuStandard, New Haven, CT), propyzamide, RH-4032 [3,5-dichloro-N-(3-chloro-1-ethyl-1-methyl-2-oxopropyl)-benzamide], Taxol (Nacalai Tesque, Kyoto, Japan), and cytochlasin D (Nacalai Tesque) were dissolved in dimethyl sulfoxide and used so that the final concentration of dimethyl sulfoxide in each medium was <0.1%.
Scanning Electron Microscopy
Replica images of epidermal cell files were obtained from etiolated hypocotyls and analyzed with a N3200 scaning electron microscope (Hitachi, Tokyo, Japan) as described (Furutani et al., 2000).
Immunofluorescent Staining
Whole seedlings were used for immunostaining as described previously (Sugimoto et al., 2000). Primary mouse antibody against α-tubulin (DM1A; Oncogene Research Products, Boston, MA) was used at a dilution of 1:1000 and fluorescein-isothiocyanate–conjugated antimouse IgG (Kirkegaard and Perry Laboratories, Gaithersburg, MD) was used at 1:100 as a secondary antibody. Fluorescent images were collected with an LSM510 confocal laser-scanning microscope (Zeiss, Jena, Germany).
Positional Cloning
phs1-1 was outcrossed to Landsberg erecta, and genomic DNA was isolated from propyzamide-hypersensitive F2 plants. The mutant was mapped using DNA markers available from the Arabidopsis Information Resource database (http://www.arabidopsis.org) and newly developed PCR-based markers. Marker names based on BAC names, primer sequences, restriction enzymes used, and the ecotype cut by the enzyme are as follows: MQM1.1, 5′- GAGCGGAATTACTGTCACTG-3′, 5′- CTGCGCATCTTATCACAGTC-3′, MseI, and Col; MQM1.2, 5′-ACGATGGCACTCTTAAGCAG-3′, 5′-TACACATACGCACCAAGCAC-3′, and no restriction (simple sequence length polymorphism marker); MRO11.1, 5′-ATTTATGTCGTGGGCCAACG-3′, 5′-TCCTCCTCCTAATGAGTGAC-3′, ClaI, and Landsberg erecta; MRO11.2, 5′-GCTGAGAAAACACATCACAC-3′, 5′-CGATAGATAGCCAATTCCGG-3′, EcoT14I, and Col.
PHS1 was mapped to a 63-kb region that spanned the two BAC clones MQM1 and MRO11 on chromosome 5. Fifteen open reading frames predicted in this region (http://www.arabidopsis.org/servlets/sv) were amplified by PCR from the phs1-1 genome and sequenced.
The phs1-2 allele (Garlic 1293 C2) was identified in the SAIL T-DNA insertion lines of Syngenta Biotechnology (Research Triangle Park, NC) (Sessions et al., 2002) and backcrossed to the Col parental wild type three times before analysis. Segregating plants were genotyped by amplifying genomic DNA with a mixture of three PCR primers; a T-DNA left border primer (LB; 5′-CATCTGAATTTCATAACCAACTCGATACAC-3′) and two PHS1-specific primers (PHS1-1F, 5′-GAGAAGAAGCGAGATCAACC-3′; PHS1-1R, 5′-TTACTCTAGCCTGTCGCATG-3′). Because both ends of the T-DNA insert were left borders, this PCR amplification yielded a 1.2-kb fragment from the wild-type allele and two fragments of 0.6 and 0.8 kb from the phs1-2 allele. From genomic DNA of heterozygous phs1-2 plants and phs1-1/phs1-2 trans-heterozygous plants, these three fragments of 0.6, 0.8, and 1.2 kb (plus a 1.4-kb fragment of unknown origin) were amplified. Direct sequencing of PCR fragments determined the precise location of the T-DNA insert.
To create a wild-type genomic transformation clone, DNA from BAC MQM1 was isolated and first cloned at the XmaI site of pUC19 after digestion with MroI and XmaI to give pUC-gPHS1. To create a mutant genomic clone, phs1-1 genomic DNA was amplified by PCR with the primers 5′-GTTGTTGATTTCGACCG-3′ and 5′-TTACTCTAGCCTGTCGCATG-3′. The resulting PCR fragment containing the phs1-1 mutation (R64C) was digested with PacI and MscI and cloned into pUC-gPHS1 that had been digested with the same enzymes to give pUC-gPHS1R64C. The wild-type and mutant PHS1 genes (containing 1.0-kb 5′ and 0.8-kb 3′ flanking sequences) in pUC19 plasmids were then excised with FspI and MunI and subcloned into pBIN19 that had been digested with EcoRI and SmaI to give pBIN-gPHS1 and pBIN-gPHS1R64C, respectively. These binary vectors were electroporated into Agrobacterium tumefaciens strain MP90 and transformed into wild-type and phs1-1 plants using the floral dip method (Clough and Bent, 1998). Transformants were selected on the agar medium supplemented with 40 μg/mL of kanamycin. Homozygous progeny of kanamycin-resistant transformants were used for the phenotypic analysis.
cDNA Isolation and Overexpression
Because publicly available cDNA clones lacked a part of the 5′ region, the missing N-terminal part of PHS1 was obtained by the method of 5′-rapid amplification of cDNA ends (5′-RACE) using the 5′-RACE system, version 2.0 (Gibco BRL, Cleveland, OH). An NaeI site was added in front of the first ATG after reamplification of the 5′-RACE fragment with the primers 5′-GCCGGCATGGCGGAACCTGAG-3′ and 5′-GAAGATTCTCCCTAGAGCTG-3′ and cloned into pGEM-T (Promega, Madison, WI) to give pGEM-PHS1N. A partial PHS1 cDNA clone (AV546458) in pBluescript II SK− (Stratagene, La Jolla, CA) was excised with Bpu1102 I and XhoI and subcloned into pSP72 (Boehringer Mannheim, Mannheim, Germany) to give pSP-PHS1C. A full-length PHS1 cDNA was constructed by ligating the NotI/PstI insert of pGEM-PHS1N into PvuII/PstI-digested pSP-PHS1C to give pSP-PHS1X. Next, an XbaI site was introduced right after the stop codon of the PHS1 cDNA. To this end, a PCR fragment was amplified from pSP-PHS1C with the primers 5′-TTCGAAGGTCGGAGTAGAAG-3′ and 5′-GCTCTAGACTAACCAGGATGAG-3′, cloned in pGEM-T, excised with XcmI and PvuII, and then ligated into XcmI/SmaI-cut pSP-PHS1X to give pSP-PHS1. The phs1-1 mutation was introduced into the wild-type PHS1 cDNA by a PCR-based mutagenesis strategy to give pSP-PHS1R64C. The XhoI/XbaI inserts of the PHS1 cDNA in pSP-PHS1 and pSP-PHS1R64C were ligated into XhoI/XbaI-digested pAVA121 (von Arnim et al., 1998), a plant transformation vector with CaMV35S, and the CaMV35S terminator to yield p35S-PHS1 and p35S-PHS1R64C, respectively. These binary vectors were used to transform wild-type Col plants by the Agrobacterium-mediated method, and the transformants were selected and analyzed as described for transformation of the genomic clones.
Gene Expression Analysis
The level of PHS1 mRNA in various organs was analyzed with RT-PCR. Total RNA of 40-d-old plants was extracted with the RNeasy plant mini kit (Qiagen, Valencia, CA) and used for the synthesis of the first-strand cDNA with Superscript II (Invitrogen, Carlsbad, CA), which was further PCR amplified for 35 cycles with PHS1-specific primers PHS1-2F (5′-TCAAAAGGCTTTGGAGGCTC-3′) and PHS1-2R (5′-GACTTTAACCGGGACAAACC-3′) or with ACTIN8 primers (5′-ATTAAGGTCGTGGCA-3′ and 5′-TCCGAGTTTGAAGAGGCTAC-3′). The PCR products were run on an agarose gel and stained with ethidium bromide.
Protein Expression and Purification
Full-length PHS1 proteins were expressed in Escherichia coli as GST fusion proteins. A MunI site was introduced by a PCR-based method at the first ATG of wild-type and R64C-mutant PHS1 cDNA inserts in pSP-PHS1 and pSP-PHS1R64C, which were subsequently excised with MunI and SalI and ligated into EcoRI/SalI-digested pGEX 6P-1 (Amersham Biosciences, Piscataway, NJ) to give pGST-PHS1 and pGST-PHS1R64C. A catalytically inactivating mutation C792S was introduced to pSP-PHS1 by PCR-mediated mutagenesis using as primers 5′-GAGTAGAAGCGCAACAGTAG-3′ and 5′-CGACCTTCGAAGCTATGAAC-3′ and processed as above to give pGST-PHS1C792S. These three plasmids were used to transform Escherichia coli BL21 (DE3). Single colonies were selected and grown in LB medium supplemented with 100 μg/mL of ampicillin overnight with shaking at 37°C. An overnight preculture was transferred to new LB medium supplemented with 100 μg/mL of ampicillin and shaken at 37°C until the absorbance at 600 nm was between 0.6 and 0.8. After isopropyl-1-thio-β-d-galactopyranoside was added to a final concentration of 1 mM, the culture was incubated at 18°C for an additional 18 h. Bacterial cultures were pelleted and resuspended in a buffer containing 100 mM NaCl, 50 mM Tris-HCl, pH 8.0, 2 μM phenylmethylsulfonylfluoride, 1 mM benzamidine, and 2 mM EDTA before lysis by sonication in the same buffer except that 0.1% (v/v) Triton X-100 and 10 mg/L of lysozyme were added. The cell lysate was pelleted at 12,000g to collect the supernatant containing the fusion protein, which subsequently was purified on a glutathione Sepharose 4B column as recommended by the manufacturer (Pharmacia).
Phosphatase Assay
Phosphatase activity was assayed at either 20 or 30°C in a reaction buffer containing 50 mM 3,3-dimethylglutaric acid, pH 7.0, 1 mM EDTA, 0.15 M NaCl, and 500 μM OMFP in a 0.8-mL volume. The reaction was quenched by addition of 0.2 mL of 5 N NaOH. The amount of product, 3-O-methylfluorescein, was determined from the absorbance at 477 nm using the Ultrospec 3000 spectrophotometer (Amersham Biosciences).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AB161693 (PHS1). PHS1 homologs from other species are rice (TC137403 and TC157626), birdsfoot trefoil (AP006392.1), alfalfa (TC83892), potato (TC82139), and maize (CF244510).
Supplementary Material
Acknowledgments
We thank G. Wasteneys for mor1-1 seeds, V. Lewandowski for RH-4032, A.G. von Arnim for pAVA121, K. Matsui and K. Okada for complementation tests of our putative fra2 alleles with a fat root allele of the fra2 mutant, M. Yoshimura for mutant screening, and T. Yasuhara for plant transformation. The ABRC is acknowledged for supplying EST AV546458, BAC MQM1, and T-DNA insertion lines and Syngenta Biotechnology for phs1-2 seeds. This study was in part supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology (14036225, 15031219, and 15370023) to T.H.
Online version contains Web-only data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Takashi Hashimoto (hasimoto@bs.naist.jp).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.021865.
References
- Abe, T., Thitamadee, S., and Hashimoto, T. (2004). Microtubule defects and cell morphogenesis in the lefty1lefty2 tubulin mutant of Arabidopsis thaliana. Plant Cell Physiol. 45, 211–220. [DOI] [PubMed] [Google Scholar]
- Azimzadeh, J., Traas, J., and Pastuglia, M. (2001). Molecular aspects of microtubule dynamics in plants. Curr. Opin. Plant Biol. 4, 513–519. [DOI] [PubMed] [Google Scholar]
- Baskin, T.I., and Wilson, J.E. (1997). Inhibitors of protein kinases and phosphatases alter root morphology and disorganize cortical microtubules. Plant Physiol. 113, 493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin, T.I., Wilson, J.E., Cork, A., and Williamson, R.E. (1994). Morphology and microtubule organization in Arabidopsis roots exposed to oryzalin and taxol. Plant Cell Physiol. 35, 935–942. [PubMed] [Google Scholar]
- Belmont, L.D., and Mitchison, T.J. (1996). Identification of a protein that interacts with tubulin dimers and increases the catastrophe rate of microtubules. Cell 84, 623–631. [DOI] [PubMed] [Google Scholar]
- Bhalla, U.S., Ram, P.T., and Lyengar, R. (2002). MAP kinase phosphatase as a locus of flexibility in a mitogen-activated protein kinase signaling network. Science 297, 1018–1023. [DOI] [PubMed] [Google Scholar]
- Bichet, A., Desnos, T., Turner, S., Grandjean, O., and Höfte, H. (2001). BOTERO1 is required for normal orientation of cortical microtubules and anisotropic cell expansion in Arabidopsis. Plant J. 25, 137–148. [DOI] [PubMed] [Google Scholar]
- Bouquin, T., Mattsson, O., Naestead, H., Foster, R., and Mundy, J. (2003). The Arabidopsis lue1 mutant defines a katanin p60 ortholog involved in hormonal control of microtubule orientation during cell growth. J. Cell Sci. 116, 791–801. [DOI] [PubMed] [Google Scholar]
- Brunner, D., Oellers, N., Szabad, J., Biggs III, W.H., Zipursky, S.L., and Hafen, E. (1994). A gain-of-function mutation in Drosophila MAP kinase activates multiple receptor tyrosine kinase signaling pathways. Cell 76, 875–888. [DOI] [PubMed] [Google Scholar]
- Burk, D.H., Liu, B., Zhong, R., Morrison, W.H., and Ye, Z.-H. (2001). A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 13, 807–827. [PMC free article] [PubMed] [Google Scholar]
- Camilleri, C., Azimzadeh, J., Pastuglia, M., Bellini, C., Grandjean, O., and Bouchez, D. (2002). The Arabidopsis TONNEAU2 gene encodes a putative novel protein phosphatase 2A regulatory subunit essential for the control of the cortical cytoskeleton. Plant Cell 14, 833–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps, M., Nichols, A., and Arkinstall, S. (1999). Dual specificity phosphatases: A gene family for control of MAP kinase function. FASEB J. 14, 6–16. [PubMed] [Google Scholar]
- Camps, M., Nichols, A., Gillieron, C., Antonsson, B., Muda, M., Chabert, C., Boschert, U., and Arkinstall, S. (1998). Catalytic activation of the phosphatase MKP-3 by ERK2 mitogen-activated protein kinase. Science 280, 1262–1265. [DOI] [PubMed] [Google Scholar]
- Cano, E., and Mahadevan, L. (1995). Parallel signal processing among mammalian MAPKs. Trends Biochem. Sci. 20, 117–122. [DOI] [PubMed] [Google Scholar]
- Cassimeris, L. (1999). Accessory protein regulation of microtubule dynamics throughout the cell cycle. Curr. Opin. Cell Biol. 11, 134–141. [DOI] [PubMed] [Google Scholar]
- Cassimeris, L. (2002). The oncoprotein 18/stathmin family of microtubule destabilizers. Curr. Opin. Cell Biol. 14, 18–24. [DOI] [PubMed] [Google Scholar]
- Chan, J., Calder, G., Doonan, J.H., and Lloyd, C.W. (2003). EB1 reveals mobile microtubule nucleation sites in Arabidopsis. Nat. Cell Biol. 5, 967–971. [DOI] [PubMed] [Google Scholar]
- Chang, L., Jones, Y., Ellisman, M.H., Goldstein, L.S.B., and Karin, M. (2003). JNK1 is required for maintenance of neuronal microtubules and controls phosphorylation of microtubule-associated proteins. Dev. Cell 4, 521–533. [DOI] [PubMed] [Google Scholar]
- Chen, L., Montserat, J., Lawrence, D.S., and Zhang, Z.-Y. (1996). VHR and PTP1 protein phosphatases exhibit remarkably different active site specificities toward low molecular weight nonpeptidic substrates. Biochemistry 35, 9349–9354. [DOI] [PubMed] [Google Scholar]
- Chen, P., Hutter, D., Yang, X., Gorospe, M., Davis, R.J., and Liu, Y. (2001). Discordance between the binding affinity of mitogen-activated protein kinase subfamily members for MAP kinase phosphatase-2 and their ability to activate the phosphatase catalytically. J. Biol. Chem. 276, 29440–29449. [DOI] [PubMed] [Google Scholar]
- Chu, Y., Solski, P.A., Khosravi-Far, R., Der, C.J., and Kelly, K. (1996). The mitogen-activated protein kinase phosphatases PAC1, MKP-1, and MKP-2 have unique substrate specificities and reduced activity in vivo toward the ERK2 sevenmaker mutation. J. Biol. Chem. 271, 6497–6501. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cobb, M.H., Robbins, D.J., and Boulton, T.G. (1991). ERKs, extracellular signal-regulated MAP-2 kinases. Curr. Opin. Cell Biol. 3, 1025–1032. [DOI] [PubMed] [Google Scholar]
- Cyr, R.J., and Palevitz, B.A. (1995). Organization of cortical microtubules in plant cells. Curr. Opin. Cell Biol. 7, 65–71. [DOI] [PubMed] [Google Scholar]
- Desai, A., and Mitchison, T.J. (1997). Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13, 83–117. [DOI] [PubMed] [Google Scholar]
- Farooq, A., Chaturvedi, G., Mujtaba, S., Plotnikova, O., Zeng, L., Dhalluin, C., Ashton, R., and Zhou, M.-M. (2001). Solution structure of ERK2 binding domain of MAPK phosphatase MKP-3: Structural insights into MKP-3 activation by ERK-2. Mol. Cell 7, 387–399. [DOI] [PubMed] [Google Scholar]
- Furutani, I., Watanabe, Y., Prieto, R., Masukawa, M., Suzuki, K., Naoi, K., Thitamadee, S., Shikanai, T., and Hashimoto, T. (2000). The SPIRAL genes are required for directional control of cell elongation in Arabidopsis thaliana. Development 127, 4443–4453. [DOI] [PubMed] [Google Scholar]
- Gottlin, E.B., Xu, X., Epstein, D.M., Burke, S.P., Eckstein, J.W., Ballou, D.P., and Dixon, J.E. (1996). Kinetic analysis of the catalytic domain of human Cdc25B. J. Biol. Chem. 271, 27445–27449. [DOI] [PubMed] [Google Scholar]
- Gundersen, G.G., and Cook, T.A. (1999). Microtubules and signal transduction. Curr. Opin. Cell Biol. 11, 81–94. [DOI] [PubMed] [Google Scholar]
- Gupta, R., Huang, Y., Kieber, J., and Luan, S. (1998). Identification of a dual-specificity protein phosphatase that inactivates a MAP kinase from Arabidopsis. Plant J. 16, 581–589. [DOI] [PubMed] [Google Scholar]
- Hashimoto, T. (2003). Dynamics and regulation of plant interphase microtubules: A comparative view. Curr. Opin. Plant Biol. 6, 568–576. [DOI] [PubMed] [Google Scholar]
- Heald, R., and Nogales, E. (2002). Microtubule dynamics. J. Cell Sci. 115, 3–4. [DOI] [PubMed] [Google Scholar]
- Hepler, P.K., and Hush, J.M. (1996). Behavior of microtubules in living plant cells. Plant Physiol. 112, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, J., and Hyman, A.A. (2003). Dynamics and mechanics of the microtubule plus end. Nature 422, 753–758. [DOI] [PubMed] [Google Scholar]
- Ichimura, K., et al. (2002). Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 7, 301–308. [DOI] [PubMed] [Google Scholar]
- Job, D., Valiron, O., and Oakley, B. (2003). Microtubule nucleation. Curr. Opin. Cell Biol. 15, 111–117. [DOI] [PubMed] [Google Scholar]
- Joshi, H.C. (1998). Microtubule dynamics in living cells. Curr. Opin. Cell Biol. 10, 35–44. [DOI] [PubMed] [Google Scholar]
- Kerk, D., Bulgrien, J., Smith, D.W., Barsam, B., Veretnik, S., and Gribskov, M. (2002). The complement of protein phosphatase catalytic subunits encoded in the genome of Arabidopsis. Plant Physiol. 129, 908–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyse, S.M. (2000). Protein phosphatases and the regulation of mitogen-activated protein kinase signaling. [DOI] [PubMed]
- Kirschner, M.W., and Mitchison, T. (1986). Beyond self-assembly: From microtubules to morphogenesis. Cell 45, 329–342. [DOI] [PubMed] [Google Scholar]
- Kusari, A.B., Molina, D.M., Sabbagh, W., Jr., Lau, C.S., and Bardwell, L. (2004). A conserved protein interaction network involving the yeast MAP kinases Fus3 and Kss1. J. Cell Biol. 164, 267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, C., and Chan, J. (2003). Microtubules and the shape of plants to come. Nat. Rev. Mol. Cell Biol. 5, 13–22. [DOI] [PubMed] [Google Scholar]
- Lloyd, C., and Hussey, P. (2001). Microtubule-associated proteins in plants—Why we need a MAP. Nat. Rev. Mol. Cell Biol. 2, 40–47. [DOI] [PubMed] [Google Scholar]
- Mandelkow, E., and Mandelkow, E. (1995). Microtubules and microtubule-associated proteins. Curr. Opin. Cell Biol. 7, 72–81. [DOI] [PubMed] [Google Scholar]
- Masuda, K., Shima, H., Katagiri, C., and Kikuchi, K. (2003). Activation of ERK induces phosphorylation of MAPK phosphatase-7, a JNK specific phosphatase, at Ser-446. J. Biol. Chem. 278, 32448–32456. [DOI] [PubMed] [Google Scholar]
- Mizuno, K. (1992). Induction of cold stability of microtubles in cultured tobacco cells. Plant Physiol. 100, 740–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe-Augustus, M., Zolman, B.K., and Bartel, B. (2003). IBR5, a dual-specificity phosphatase-like protein modulating auxin and abscisic acid responsiveness in Arabidopsis. Plant Cell 15, 2979–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihama, R., Soyano, T., Ishikawa, M., Araki, S., Tanaka, H., Asada, T., Irie, K., Ito, M., Terada, M., Banno, H., Yamazaki, Y., and Machida, Y. (2002). Expansion of the cell plate in plant cytokinesis requires a kinesis-like protein/MAPKKK complex. Cell 109, 87–99. [DOI] [PubMed] [Google Scholar]
- Ohkura, H., Garcia, M.A., and Toda, T. (2001). Dis1/TOG universal microtubule adaptors—One MAP for all? J. Cell Sci. 114, 3805–3812. [DOI] [PubMed] [Google Scholar]
- Park, S.-H., Zarrinpar, A., and Lim, W.A. (2003). Rewiring MAP kinase pathways using alternative scaffold assembly mechanisms. Science 299, 1061–1064. [DOI] [PubMed] [Google Scholar]
- Ray, L.B., and Sturgill, T.W. (1987). Rapid stimulation by insulin of a serine/threonine kinase in 3T3-L1 adipocytes that phosphorylates microtubule-associated protein 2 in vitro. Proc. Natl. Acad. Sci. USA 84, 1502–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reszka, A.A., Seger, R., Diltz, C.D., Drebs, E.G., and Fischer, E.H. (1995). Association of mitogen-activated protein kinase with the microtubule cytoskeleton. Proc. Natl. Acad. Sci. USA 92, 8881–8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions, A., et al. (2002). A high-throughput Arabidopsis reverse genetic system. Plant Cell 14, 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, S.L., Kamyar, R., and Ehrhardt, D.W. (2003). Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science 300, 1715–1718. [DOI] [PubMed] [Google Scholar]
- Slack, D.N., Seternes, O.-M., Gabrielsen, M., and Keyse, S.M. (2001). Distinct binding determinants for ERK2/p38α and JNK MAP kinases mediate catalytic activation and substrate selectivity of MAP kinase phosphatase-1. J. Biol. Chem. 276, 16491–16500. [DOI] [PubMed] [Google Scholar]
- Soyano, T., Nishihama, R., Morikiyo, K., Ishikawa, M., and Machida, Y. (2003). NQK1/NtMEK1 is a MAPKK that acts in the NPK1 MAPKKK-mediated MAPK cascade and is required for plant cytokinesis. Genes Dev. 17, 1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, A.E., Dowd, S., Keyse, S.M., and McDonald, N.Q. (1999). Crystal structure of the MAPK phosphatase Pyst1 catalytic domain and implications for regulated activation. Nat. Struct. Biol. 6, 174–181. [DOI] [PubMed] [Google Scholar]
- Sugimoto, K., Williamson, R.E., and Wasteneys, G.O. (2000). New techniques enable comparative analysis of microtubule orientation, wall texture, and growth rate in intact roots of Arabidopsis. Plant Physiol. 124, 1493–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanoue, T., Adachi, M., Moriguchi, T., and Nishida, E. (2000). A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat. Cell Biol. 2, 110–116. [DOI] [PubMed] [Google Scholar]
- Tanoue, T., Yamamoto, T., and Nishida, E. (2002). Modular structure of a docking surface on MAPK phosphatases. J. Biol. Chem. 277, 22942–22949. [DOI] [PubMed] [Google Scholar]
- Tena, G., Asai, T., Chiu, W.-L., and Sheen, J. (2001). Plant mitogen-activated protein kinase signaling cascades. Curr. Opin. Plant Biol. 4, 392–400. [DOI] [PubMed] [Google Scholar]
- Terret, M.E., Lefebvre, C., Djiane, A., Rassinier, P., Moreau, J., Maro, B., and Verlhac, M.-H. (2003). DOC1R: A MAP kinase substrate that control microtubule organization of metaphase II mouse oocytes. Development 130, 5169–5177. [DOI] [PubMed] [Google Scholar]
- Theodosiou, A., and Ashworth, A. (2002). MAP kinase phosphatases. Genome Biol. 3, 3009.1–3009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thitamadee, S., Tuchihara, K., and Hashimoto, T. (2002). Microtubule basis for left-handed helical growth in Arabidopsis. Nature 417, 193–196. [DOI] [PubMed] [Google Scholar]
- Tian, G.-W., Smith, D., Glück, S., and Baskin, T.I. (2004). Higher plant cortical microtubule array analyzed in vitro in the presence of the cell wall. Cell Motil. Cytoskeleton 57, 26–36. [DOI] [PubMed] [Google Scholar]
- Twell, D., Park, S.K., Hawkins, T.J., Schuber, D., Schmidt, R., Smertenko, A., and Hussey, P.J. (2002). MOR1/GEM1 has an essential role in the plant-specific cytokinetic phragmoplast. Nat. Cell Biol. 4, 711–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm, R., Ichimura, K., Mizoguchi, T., Peck, S.C., Zhu, T., Wang, X., Shinozaki, K., and Paszkowski, J. (2002). Distinct regulation of salinity and genotoxic stress responses by Arabidopsis MAP kinase phosphatase 1. EMBO J. 21, 6483–6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm, R., Revenkova, E., di Sansebastiano, G., Bechtold, N., and Paszkowski, J. (2001). Mitogen-activated protein kinase phosphatase is required for genotoxic stress relief in Arabidopsis. Genes Dev. 15, 699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Drogen, F., and Peter, M. (2002). MAP kinase cascades: Scaffolding signal specificity. Curr. Biol. 12, R53–R55. [DOI] [PubMed] [Google Scholar]
- Vega, L.R., and Solomon, F. (1997). Microtubule function in morphological differentiation: Growth zones and growth cones. Cell 89, 825–828. [DOI] [PubMed] [Google Scholar]
- Von Arnim, A.G., Deng, X.-W., and Stacey, M.G. (1998). Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene 221, 35–43. [DOI] [PubMed] [Google Scholar]
- Wasteneys, G.O. (2002). Microtubule organization in the green kingdom: Chaos or self-order? J. Cell Sci. 115, 1345–1354. [DOI] [PubMed] [Google Scholar]
- Webb, M., Jouannic, S., Foreman, J., Linstead, P., and Dolan, L. (2002). Cell specification in the Arabidopsis root epidermis requires the activity of ECTOPIC ROOT HAIR 3—A katanin-p60 protein. Development 129, 123–131. [DOI] [PubMed] [Google Scholar]
- Whittington, A.T., Vugrek, O., Wei, K.J., Hasenbein, N.G., Sugimoto, K., Rashbrooke, M.C., and Wasteneys, G.O. (2001). MOR1 is essential for organizing cortical microtubules in plants. Nature 411, 610–613. [DOI] [PubMed] [Google Scholar]
- Young, D.H., and Lewandowski, V.T. (2000). Covalent binding of the benzamide RH-4032 to tubulin in suspension-cultured tobacco cells and its application in a cell-based competitive-binding assay. Plant Physiol. 124, 115–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuvaniyama, J., Denu, J.M., Dixon, J.E., and Saper, M.A. (1996). Crystal structure of the dual specificity protein phosphatase VHR. Science 272, 1328–1331. [DOI] [PubMed] [Google Scholar]
- Zhou, B., Wang, Z.-X., Zhao, Y., Brautigan, D.L., and Zhang, Z.-Y. (2002). The specificity of extracellular signal-regulated kinase 2 dephosphorylation by protein phosphatases. J. Biol. Chem. 277, 31818–31825. [DOI] [PubMed] [Google Scholar]
- Zhou, B., Wu, L., Shen, K., Zhang, J., Lawrence, D., and Zhang, Z.-Y. (2001). Multiple regions of MAP kinase phosphatase 3 are involved in its recognition and activation by ERK2. J. Biol. Chem. 276, 6506–6515. [DOI] [PubMed] [Google Scholar]
- Zhou, B., and Zhang, Z.-Y. (1999). Mechanism of mitogen-activated protein kinase phosphatase-3 activation by ERK2. J. Biol. Chem. 274, 35526–35534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.