Abstract
Testicular germ cell tumor (TGCT) is a highly heritable cancer primarily affecting young white men. Genome-wide association studies (GWAS) have been particularly effective in identifying multiple common variants with strong contribution to TGCT risk. These loci identified through association studies have implicated multiple genes as associated with TGCT predisposition, many of which are unique among cancer types, and regulate processes such as pluripotency, sex specification and microtubule assembly. Together the identification of these biological plausible genes converges upon pathways involved in male germ cell development and maturation, and suggests that perturbation of them confers susceptibility to TGCT, as a developmental defect of germ cell differentiation.
1. Introduction
Testicular germ cell tumor (TGCT) is the most common cancer affecting white men aged 15-45. Although TGCT is relatively rare (lifetime risk 0.4%), rates have doubled over the last 30-40 years, and over 230,000 men in the US are living with the diseases [1]. Incidence of TGCT varies by geography and ethnic group: highest in Nordic populations (11.5/100,000), and lowest in African and Asian countries [2]. In the United States, there is a greater than fivefold incidence difference between non-Hispanic white men (6.2/100,000) and black men (1.2/100,000) [3]. However, the rate of TGCT globally among non-white men is rising, hypothesized to be secondary to changing environmental exposures; non-white men are more likely to present with more advanced disease due to diagnostic delay [3]. TGCT has been described as the model of a curable cancer, is generally exquisite sensitivity to chemotherapy, and has survival rates over 95% [1]. Unfortunately, there is long term morbidity associated with the use of the chemotherapeutics in treatment for TGCT, including cardiovascular disease, metabolic syndrome, and infertility [4].
2. Histopathologic classification
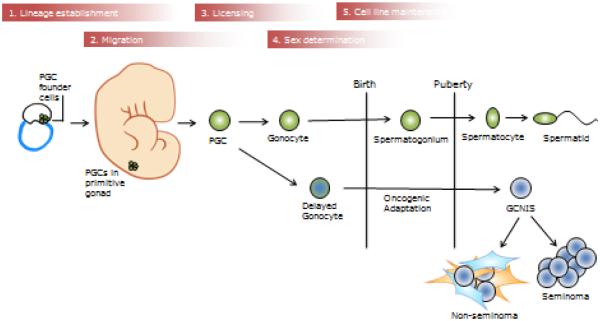
TGCT is a histologically heterogeneous disease, and historically has presented a challenge to classify. The high level of heterogeneity can be attributed to pluripotency of the originating germ cell line, and extended time period during which oncogenic mutations accumulate before rapid invasive growth during or after puberty. TGCT derives from aberrantly arrested fetal gonocytes, which do not develop properly after birth into spermatogonium (Figure 1). Arrested gonocytes accumulate oncogenic genetic adaptations through childhood and puberty, becoming germ cell neoplasia in situ (GCNIS) in childhood and young adulthood, and emerging as invasive TGCT in the young adult. GCNIS can be detected histologically early in childhood, but is challenging to distinguish from normal germ cells in the young child. TGCT is histologically divided into two general subtypes: seminoma and non-seminoma. Seminomas are homogenous tumors that resemble undifferentiated gonocytes, accounting for ~55% of TGCTs, with a peak incidence at ages 35-39. Non-seminomatous germ cell tumors (NSGCT) make up ~44% of TGCTs, are generally more aggressive, and have a younger age of diagnosis at 25-29 years. NSGCT are heterogeneous in composition, reflecting their dysregulated differentiation into embryonal carcinoma, teratatoma, choriocarcinoma, and yolk-sac tumor. Tumors containing both NSGCT and seminoma, known as mixed or combined tumors, are classified as a subtype of NSGCT [5].
Figure 1.
TGCT pathogenesis in relation to germ cell development. The stages of germ cell development are shown above in red. Normal germ cell development stages are shown in green, and aberrant TGCT precursors are shown with blue. PGC=primordial germ cell. GCNIS=germ cell neoplasia in situ.
TGCTs constitute ~98% of testicular cancers. There are two other types of primary testicular cancers that do not arise from GCNIS: 1) spermatocytic seminomas, which generally present at 50-55 years of age, and arise from a distinct pathway involving clonal expansion of the spermatogonium; and 2) childhood tumors, which appear to arise from the primary germ cell (PGC), the precursor of the gonocyte. These tumors are rare and not the focus of this review.
3. Risk Factors
Family history is one of the strongest known risk factors for TGCT, and relatively high as compared to other cancer types. As documented across multiple populations, sons of men with TGCT have a 4-6-fold risk of TGCT (versus generally three-fold or below in other cancer types), and brothers an 8-10-fold risk of TGCT (versus six-fold or below in other types) [6]. The higher rate in brother versus father-son may reflect the complex genetic/shared environmental risk, or an X-linked or autosomal recessive component of complex inheritance. The Nordic twin cohort study determined an estimated hereditary effect of 37%, higher than breast cancer, ranked seventh out of the 15 cancers they reported [7]. Overall risk for TGCT in this cohort was 0.5%, and risk for a man whose co-twin had been diagnosed was 6% for dizigotic and 14% for monozygotic (the total familial effect). In addition to the 37% heredity effect, the strong TGCT familial effect included 24% attributable to shared environment, which was as high as lung cancer [7]. The heritability of TGCT recently was estimated to be 1) ~48% using the Swedish population family-cancer database (over 15 million individuals born in Sweden after 1937) and 2) ~38% using genomic estimates drawn from ~1000 U.K. patients previously included in GWAS studies [8]. Altogether the heritability of TGCT is estimated to be 35-50%, with the higher population-based estimate reflecting multiple components beyond the genetic, or the “missing heritability”, be that shared unmeasured environmental factors, epigenetic effects, or other factors such as imperfect linkage disequilibrium between genotypes, SNPs and casual variants.
Multiple other specific risk factors for TGCT have been evaluated, including various environmental exposures and morphologic differences. Most studies have been negative (non-genitourinary organ malformations and dysmorphology), equivocal (marijuana use), or not consistently repeatable (history of orchitis). Cryptorchidism, subfertility, testicular microlithiasis, and increased adult height have all been consistently associated with TGCT risk in more than one study [9,10]. Cryptorchidism confers a similar level of risk as family history. However, the directionality of the relationship between cryptorchidism and TGCT (both components of the hypothesized Testicular Dysgenesis Syndrome) has not been definitely established, as cryptorchidism may be associated with TGCT through exposure of the developing gonad to an abnormal environment, or as part of a single pathologic process with shared origins. Currently, the former hypothesis is favored.
4. APPROACHES TO THE GENETIC STUDY OF TGCT PREDISPOSITION
4.1 Initial Linkage Studies
Initial studies attempting to identify a genetic etiology for TGCT focused on multiple case families. It was expected, that they would be explained by rare germline mutations in highly penetrant genes, which was not the case. The only locus identified through linkage analysis was at Xq27, founding through linkage of 134 families with family history compatible with an X-linked inheritance pattern [11]. Unfortunately, a follow-up larger independent analysis (n = 237 families) did not confirm the association [12], and it has not been further pursued.
4.2 Candidate Gene Studies
Multiple candidate gene and locus studies failed to identify genetic variation associated with TGCT, with genes studied including DND1, RLN1, ESR1, ESR2, LHCCGR, DICER1, AKT1, PTEN, AR, and 8q24 (reviewed by Greene MH et al.) [13]. The first independently validated candidate locus analysis was the Y-deletion known as “gr/gr”. Based on the co-occurrence of TGCT and subfertility, Nathanson and colleagues investigated a Y-chromosome deletion associated with an increased risk of infertility, and found the gr/gr deletion present in 3.0% of familial TGCT cases (13/431), versus 2% of TGCT without a family history (28/1376), and 1.3% of unaffected males (33/2599) [14]. The identification of gr/gr deletions provided both the first clear evidence of genetic predisposition to TGCT. The gr/gr region, within the AZFc (azoospermia factor) contains genes of the BPY2, CDY1, and DAZ families, all of which are relevant to germ cell maturation and development, foreshadowing the common genetic links tying TGCT to germ cell development. The only other candidate gene identified has been PDE11A, in which inactivating mutations were found in association with TGCT predisposition, similar to those found in other hormonal neoplasms (including adrenal tumors) [15].
4.3 Genome-Wide Association Studies
Genome-wide association studies (GWAS) have revolutionized our understanding of the role of genetic variation in TGCT predisposition. GWAS leverages cohorts of hundreds or thousands of patients to agnostically search for single nucleotide polymorphisms (SNPs) to define regions of the genome associated with the phenotype. The SNP within the region with the strongest association (sentinel variant) may either be the causal variant or a genetic marker for the causal variant that is in linkage disequilibrium, whose mechanistic relationship with the phenotype (TGCT) can then be functionally evaluated.
Ten genome-wide association studies of TGCT have been published, including meta-analyses of previously published and unpublished populations [16–25]. These studies have identified 27 independent loci or genomic regions with specific alleles associated with TGCT. The strength of these associations is greater than other cancers, with all odds ratios over 1.2 to date, including the strongest GWAS signal thus far reported in a cancer (KITLG locus, per allele OR >2.5). Additionally, essentially all of the alleles associated with TGCT are significantly more frequent in non-Hispanic-white than black populations, consistent with disease incidence patterns.
GWAS of TGCT has revealed multiple sentinel variants, many of which are in the introns of, or close proximity to, genes with strong biological plausibility as being associated with disease. As in GWAS generally, TGCT loci are predominantly in non-coding regions of the genome. Detailed biological mechanism is difficult to elucidate from variants in non-coding regions, but close proximity to biologically plausible genes allows inference of potential function. In particular, findings from TGCT GWAS have highlighted the benefit of association studies to deepen our understanding of disease mechanism. Genes implicated in TGCT GWAS fall into multiple pathways. Some of the genes and pathways implicated have been associated with other cancer types (e. g. DNA damage response and telomere length), whereas other genes and pathways are unique to germ cell tumors (e.g. germ cell development, sex determination, and microtubule assembly). All of these pathways also regulate important components of male germ cell development, and so can be organized within that framework.
5. TESTICULAR CANCER IS A DISEASE OF MALE GERM CELL DEVELOPMENT
Male germ cell development is a highly complex process requiring alignment spatially, temporally, and genetically. It begins at the earliest stage of embryogenesis, and continues after birth into puberty, which can be divided into multiple phases which genetically and temporally overlap (Table 1; Figure 1). Genetic variation in genes at each one of these steps has been found to play a role in TGCT predisposition (Table 2). Below, we review the implicated genes and their role in male germ cell development and maturation.
Table 1.
Stages of germcell development.
| 1. | Establishment of the germline lineage from the inner cell mass, with suppression of somatic genes and dynamic activation of germ cell specific genes. |
| 2. | Migration of primordial germ cells (PGCs) to the genital ridge, while maintaining on-going somatic gene repression. |
| 3. | Epigenetic reprogramming, with global demethylation and paternal re-imprinting. |
| 4. | Initial meiosis and sex-specific determination of germ cell and gonadal tissue. |
| 5. | Maintenance of gametes, followed by preparation for regulation of early zygotic processes after fertilization. |
Table 2.
Published GWAS Loci for TGCT Predisposition
| CYTOBAND | SNP* | LOCATION | GENE NEIGHBORHOOD | GERM CELL DEVELOPMENT STAGE |
|---|---|---|---|---|
| lq22 | rs2072499 | 156169610 | PMF1 | 5 |
| lq24.1 | rs3790672 | 165873392 | UCK2 | 5 |
| 3p24.2 | rsl0510452 | 16625048 | DAZL | 3 |
| 3q23 | rsll705932 | 141818850 | TFDP2 | 4 |
| 3q25.3 | rsl510272 | 156300724 |
SSR3
TIPARP |
6 4 |
| 4q22.3 | rsl7021463 | 95224812 | HPGDS | 4 |
| 4q24 | rs2720460 | 104054686 |
CENPE
BDH2 |
5 5 |
| 5pl5.3 | rs2736100 | 1286516 | TERT | 5 |
| 5pl5.3 | rs4635969 | 1308552 | TERT | 5 |
| 5q31.1 | rs3805663 | 134366200 | PITX1 | 5 |
| 5q31.3 | rs4624820 | 141681788 | SPRY4 | 2 |
| 6p21.3 | rs210138 | 33542538 | BAK1 | 2 |
| 7p22.3 | rsl2699477 | 1968953 | MAD1L1 | 5 |
| 8ql3.3 | rs7010162 | 70976505 | PRDM14 | 1 |
| 9p24.3 | rs7040024 | 845516 | DMRT1 | 4 |
| 9p24.3 | rs755383 | 863635 | DMRT1 | 4 |
| llql4.1 | rs7107174 | 77997936 |
GAB2
USP35 |
6 6 |
| 12pl3.1 | rs2900333 | 14653867 | ATF7IP | 3 |
| 12q21.3 | rs995030/rsl 508595 | 88953561 | KITLG | 2 |
| 16pl3.1 | rs4561483 | 11920037 |
GSPT1
RSL1D1 |
5 6 |
| 16ql2.1 | rs8046148 | 50142944 | HEATR3 | 6 |
| 16q23.1 | rs4888262 | 94670458 |
RFWD3
MLKL |
5 5 |
| 16q24.2 | rs55637647 | 88549264 | ZFPM1 | 4 |
| 17ql2 | rs7501939 | 36101156 | HNF1B | 5 |
| 17q22 | rs9905704 | 56632543 |
TEX 14
SEPT4 RAD51C TRIM 37 |
6 6 5 6 |
| 19pl2 | rs2195987 | 24149545 | ||
| 21q22.3 | rs2839186 | 47690068 | MCM3AP | 5 |
First published sentinel variant for this locus
6. undefined impact on germ cell development
4.1 Establishment of the Germline Lineage
Human germ cell development beings with specification of the PGCs, with is thought to happen around the initiation of gastrulation (developmental week 2). PRDM14 at 8q13.3 is thus far the only gene implicated in TGCT predisposition that is known to direct germline lineage determination. Prdm14 is one of the first two primordial germ cell markers in mice [26], and complete Prdm14 knockout mice are sterile (MGI #3588194). PRDM14 is not the primary PGC determinant in humans, where it has been evolutionarily supplanted by BLIMP1-SOX17 [27]. The impact of PRDM14 on TGCT development may involve a recapitulation of its pluripotency role in mice.
4.2 Migration of PGCs to the Genital Ridge
The specified PGCs migrate to the genital ridge developmental weeks three through six, and then colonize the genital ridge. KIT-KITLG-associated MAPK signaling dominates migration, and is tightly integrated with apoptosis signaling to clear malmigrated/misplaced PGCs [28]. The KITLG region contains the haplotype most strongly associated with TGCT, with per allele odds ratio of 2.5-fold [16,20], the strongest association reported for any cancer to-date [29]. The directionality of the TGCT-associated KITLG risk allele is as-yet unclear (whether up- or down-regulation of expression). Increased rates of spontaneous TGCTs occur in the murine model with germ cell-specific loss of the transmembrane (but not soluble) Kitlg isoform, yet other Kitlg deficient murine models are infertile through PGC loss earlier, during migration (MGI #96974) [30]. A potential causal variant within the KITLG locus results in KITLG upregulation via increased p53 binding [31], in contrast with tumor growth associated with Kitlg loss in mice. Although the directionality of effect has not been definitively established, the sensitivity of the KIT-KITLG system to perturbation and its relevance to TGCT development is clear. However, most human evidence suggests that upregulation of the KITLG-KIT-MAPK signaling pathway is associated with TGCT.
Further reinforcing the relevance of variation within the KITLG/MAPK signaling pathway in TGCT susceptibility, two addition downstream effectors have also been identified within TGCT GWAS loci, BAK1 and SPRY4. BAK1 is a pro-apoptotic protein thought to control the death of mislocalized PGCs during migration [32]. SPRY4 is an inhibitor of the KITLG-KIT-MAPK pathway. Relevance of SPRY4 to TGCT is also implicated by increased methylation of the maternal allele promoter in TGCT patients [33]. In vitro evidence suggests that decreased expression of SPRY4 may result in increased cell survival of abnormal PGCs [34], and that the oncogenic signaling may also involve the long-noncoding RNA (lncRNA) SPRY4 intronic transcript (SPRY4-IT1), a negative prognostic indicator in bladder cancer [35]. Involvement of several components of the KITLG-MAPK signaling pathway suggests that variation of signaling within the pathway in the PGC may drive TGCT risk.
4.3 Epigenetic Reprogramming
The epigenome undergoes extensive reprogramming in PGCs, erasing much of the epigenetic memory transmitted from each parent. The progressive demethylation is called “licensing”, as the cell prepares for new methylation programming appropriate for the future sperm or oocyte. A 3p24.3 sentinel variant lies within an intron of DAZL, which is requisite for licensing; deletion of Dazl in mice prevents the PGCs from progressing forward as either type of fetal gonad [36]. Another locus at 12p13 contains a single protein, ATF7IP (alternately known as MCAF1). ATF7IP has multiple roles including as transcription factor (including for TERT: section 4.5.2), and in formation of heterochromatin [37]. In the context of licensing, ATF7IP is a key factor in proviral silencing, the system whereby endogenous retrovirus transcription remains inhibited throughout licensing and gametogenesis to maintain overall sequence integrity [38]. Variation at the DAZL locus may suggest an intersection of global licensing control and TGCT development, and through ATF7IP, variation in the finer control of retaining methylation may also contribute.
4.4 Meiosis Initiation and Sex Determination
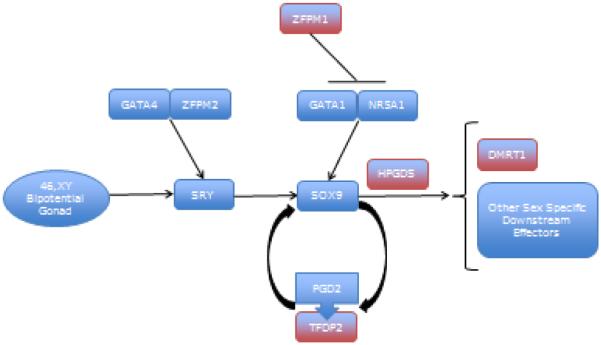
Sex determination is the process of definitively assigning each bipotential gonad as future testicle or ovary. Neither SRY nor SOX9, the lynchpin early sex determination genes have been implicated through TGCT GWAS, but several other members of the pathway have been. Of particular note is DMRT1 (9p24.3), which contains at least two independent intronic risk variants. DMRT1 is a key sex determination gene downstream of SOX9 (Figure 2), with homologues in species ranging from chickens to frogs [39]. DMRT1 is regulated by NR5A1 via SOX9. A TGCT sentinel variant lies with an intron of ZFPM1, which down regulates NR5A1 through direct binding [40]. Given the known relationship of both DMRT1 and NR5A1 loss to sex ambiguity, it is tempting to hypothesize that the pro-oncogenic changes in DMRT1 and ZFPM1 also drive in that direction, toward incomplete or mildly ambiguous PGC determination (decrease in DMRT1 function and increase in ZFPM1 function).
Figure 2.
Sex Determination Signaling. GWAS hits for TGCT predisposition are in red, other components in blue. Most components linking the initially bipotential gonad to DMRT1 and other downstream effectors are transcription factors. Exceptions include HPGDS, required for translocation of SOX9 into the nucleus, and the ligand/receptor pair PGD2 and TFDP2. TIPARP is not included, which influences the overall hormonal milieu.
Two associated genes, HPGDS and TIPARP, were first discovered as responsive elements to environmental dioxin exposure at levels sufficient to cause reproductive toxicity. HPGDS is expressed early in both male and female gonads, and is required for normal translocation of Sox9 [41], upstream of DMRT1. TIPARP influences the overall hormonal milieu or “sex-chromosome specific hormone niche development” through PDGF signaling [42]. HPGDS and TIPARP may represent yet another category of TGCT predisposition loci, those that directly connect the environmental and heritable components of familial predisposition to the cancer.
4.5 Maintenance of Gametes
4.5.1 Microtubule and Kinetochore Assembly
Microtubule and kinetochore assembly is the last germ cell development stage identified in TGCT predisposition, but not other cancers. The sentinel variant at 7p23.2 lies within an intron of MAD1L1, a spindle assembly checkpoint protein, loss of which is implicated in cancer progression related chromosome instability [43]. The 1q22 locus is one in which interpretation of the sentinel variant is more nuanced. The 1q22 sentinel variant lies within an intron of SLC25A44, but in close linkage disequilibrium is an exonic missense variant in the nearby kinetochore assembly protein, PMF1 [44] . Given the biological plausibility and high impact of such a variant, PMF1 becomes the more likely effector of the 1q22 TGCT locus.
4.5.2 Telomerase Function
The remaining pathways of gamete maintenance implicated by TGCT GWAS are observed as loci in other cancer types, and their broad relevance to oncogenic potential through control of cell proliferation is well described. Telomerase function is a hallmark for self-renewal potential in cancer, iPS, and male germ cells [45]. Dysregulated DNA damage repair contributes to oncogenic adaptation accumulation, and change in metabolic maintenance of the cell aid in rapid growth (Warburg effect).
Two independent loci have been identified in the ‘cancer hub’ region at 5p13, in close proximity to TERT. TERT encodes the catalytic subunit of the telomerase complex, which extends cell life through replacement of (TTAGGG)n repeats lost during incomplete DNA replication. Loss of TERT in animal models is associated with slow progressive loss of male germ cells over time (MGI #1202709), suggesting that TERT upregulation may be the culprit in TGCT. However, like ATF7IP, the TERT locus may be one where the relevant gene is clear, but the mechanism may be multifactorial. Non-canonical activities of TERT include a role in RNA synthesis, and heterochromatin maintenance [46]. Another sentinel variant sits within an intron of PITX1, a homeobox gene involved in early limb patterning which PITX1 suppresses TERT through binding to its promoter [47].
4.5.3 DNA Damage Response
Some variant findings in TGCT present an opportunity to elucidate the genetics of complex loci. A sentinel variant lies within an intron of MCM3AP, which is also included as a domain within the larger protein known as GANP. MCM3AP influences DNA damage response through the well-appreciated ATM pathway [48], but the larger protein, GANP, is a member of the THSC complex involved in transcription elongation, mRNA processing, and export (reviewed in Wichramasinghe et al.) [49]. Both genes are potential candidates for TGCT predisposition, and the causal variant could influence regulation of either or both transcripts.
4.5.4 Metabolic Maintenance
The most recent pathway to be associated with TGCT predisposition is metabolic maintenance. UCK2 is the clearest example, a uridine-cytidine kinase important in production of pyrimidine nucleoside triphosphates for RNA/DNA production. UCK2 was originally described through differential mRNA seeking testis-specific genes [50], highlighting its relevance to testes-specific cell maintenance.
4.6 Loci with Multiple Genes
Some loci contain multiple candidate genes, highlighting the need for refined mapping and demonstration of a causative relationship. These sites lack one significant single gene with an overwhelming weight of biological plausibility, and therefore represent an opportunity to perhaps further elucidate novel gene function. The locus at 17q22 contains several genes: TEX14, SEPT4, RAD51C, and TRIM37. Of the four, TEX14 has the greatest biological plausibility, given that murine knockouts have male (but not female) infertility (MGI #1933227), and it regulates kinetochore-microtubule assembly in testicular germ cells [51]. However, the sentinel SNP falls within an intron of the long-coding antisense RNA SEPT4-AS1, thought to regulate the protein SEPT4, a gene required in structural integrity of sperm in mice [52]. RAD51C is a DNA damage response gene defective in autosomal recessive Fanconi Anemia (complementation group O) (OMIM #602774), and mice hypomorphic for RAD51C have both male and female infertility (MGI #2150020). Finally, TRIM37 is a ubiquitin ligase overexpressed in some cancers [53].
Other loci are in linkage disequilibrium and close physical proximity to single genes, but the biological plausibility and mechanistic relationship of that gene to TGCT is unclear. Variants nears HEATR3 (at 16q12.1) also have been associated with both TGCT and esophageal squamous-cell carcinoma [54], but little is known about HEATR3, other than that the homologous gene in yeast is involved in nuclear RNA transport [55]. These new genes present an opportunity for hypothesis-driven exploration of these genes and their function through the TGCT phenotype.
4.7 GWAS Summary
The experience of GWAS in TGCT provides an excellent model for GWAS in cancer predisposition, and indeed in GWAS overall. Effect sizes in TGCT are quite large, and predisposition pathways have been identified and fleshed out. These pathways are expected to be relevant to both TGCT predisposition and later progression, and highlight both the sensitivity of the PGC development system to perturbation, and potentially reveal the more robust and sensitive components of that system. The next phase of challenge is the same as that for all GWAS findings, to demonstrate a causal relationship through functional evidence beyond biological plausibility.
5. Future Areas of Exploration
5.1 Future of GWAS
As collected patient population sizes increase, so increases the power of GWAS studies. As described above, genome-wide complex trait analysis (GCTA) shows that ~37% of TGCT heritability is within common variants. The first 27 GWAS loci for TGCT account for ~10% of this 37%. Statistical analysis suggest the remaining ~27% leaves in excess of 50 variants left yet be identified [8]. Further findings may be limited by statistical strength and number of subjects available, and additional loci can be expected to have lower effect sizes.
5.2 Small Non-coding RNAs
Small non-coding RNA (snRNA) molecules, such as microRNAs (miRNA), regulate translation, are highly tissue specific, and play a key role in cellular differentiation. For example, miRNAs are essential for spermatogenesis through targeted gene regulation [56]. Deletion of Dicer, an endonuclease essential for global normal miRNA biosynthesis, results in smaller testes, disruption of spermatogenesis, and infertility in mice (MGI #2177178). Other miRNAs have been found to regulate germ cell differentiation by targeting NOTCH1 and DAZL. Closer to the clinical realm, several studies suggest that serum levels of particular miRNA clusters (miR-371-2-3 and miR-302/367) are predictive of malignant GCT (reviewed in Rijlaarsdam, et al.) [57]. Causal GWAS variants may influence oncogenic potential through regulation of snRNAs, and provide an opportunity for pre-invasive disease risk stratification through sperm cell or peripheral blood measure.
6. Assessing TGCT Risk – Current and Future Perspectives
GWAS has been extremely valuable in expending our understanding of TGCT predisposition and pathogenesis. An obvious hope was to employ risk loci clinically for patient risk stratification. Using a polygenic risk score combining 23 risk loci, men in the top 1% of genetic risk has a 10.4-fold relative, and 5.2% lifetime, risk of TCGT [24]. The 10.4-fold elevation of TGCT risk is greater than other similar calculations for other cancers (prostate as next-highest at 4.7x risk, aggregating 77 alleles), reflecting the large per-locus ORs in TGCT. Litchfield et al. evaluated a screening model employing various combinations of stratification including sperm analysis, genetic screening, and testicular biopsy. Using semen analysis alone, together with the 19 GWAS risk loci known at that time, their model achieved a 60% positive predictive value, over the ~0.4% population risk [58]. The positive predictive power of a polygenic risk score would increase with the addition of further stratification features, including the over 50 potential additional GWAS loci, family history, and history of cryptorchidism, but the clinical utility and economics would be prohibitive. Although screening is not currently recommended for TGCT, a polygenic risk score might be useful in populations at highest risk, such as those with cryptorchidism. In a future where anticipatory genome sequencing may be done routinely, identification of common risk variants such as those underlying TGCT may play a more practical clinical role. As advances are made toward technologies including peripheral blood miRNA screening, or sperm cell genetic screening, and non-invasive labeled imaging of testicular tissue, screening of these individuals at higher risk for TGCT can become a reality.
Table 3.
TGCT GWAS Loci Associated with Gamete Maintainence
| MICROTUBULE ASSEMBLY |
TELOMERASE FUNCTION |
DNA DAMAGE REPAIR |
METABOLIC MAINTAINENCE |
|---|---|---|---|
| PMF | TERT | MCM3AP | BDH2 |
| MAD1L1 | PITX1 | RFWD3 | HNF1B |
| CENPE | RAD51C | UCK2 |
Acknowledgments
Funding: This work is supported by National Institutes of Health grants CA164947 (KLN), CA114478 (KLN), and T32GM008638 (LCP) and the Abramson Cancer Center at the University of Pennsylvania.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no conflicts of interest or financial relationships to disclose.
References
- [1].Howlader N, Noone AM, Krapcho M, Garshell J, Miller D, Altekruse SF, et al., editors. SEER Cancer Statistics Review, 1975-2012, National Cancer Institute. Bethesda, MD: http://seer.cancer.gov/csr/1975_2012/, based on November 2014 SEER data submission, poste. n.d.; [Google Scholar]
- [2].Chia VM, Quraishi SM, Devesa SS, Purdue MP, Cook MB, McGlynn KA. International trends in the incidence of testicular cancer, 1973-2002. Cancer Epidemiol Biomarkers Prev. 2010;19:1151–9. doi: 10.1158/1055-9965.EPI-10-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ghazarian AA, Trabert B, Devesa SS, McGlynn KA. Recent trends in the incidence of testicular germ cell tumors in the United States. Andrology. 2015;3:13–8. doi: 10.1111/andr.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Travis LB, Beard C, Allan JM, Dahl AA, Feldman DR, Oldenburg J, et al. Testicular cancer survivorship: research strategies and recommendations. J Natl Cancer Inst. 2010;102:1114–30. doi: 10.1093/jnci/djq216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Moch H, Cubilla AL, Humphrey PA, Reuter VE, Ulbright TM. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs-Part A: Renal, Penile, and Testicular Tumours. Eur Urol. 2016 doi: 10.1016/j.eururo.2016.02.029. [DOI] [PubMed] [Google Scholar]
- [6].Hemminki K, Rawal R, Chen B, Bermejo JL. Genetic epidemiology of cancer: from families to heritable genes. Int J Cancer. 2004;111:944–50. doi: 10.1002/ijc.20355. [DOI] [PubMed] [Google Scholar]
- [7].Mucci LA, Hjelmborg JB, Harris JR, Czene K, Havelick DJ, Scheike T, et al. Familial Risk and Heritability of Cancer Among Twins in Nordic Countries. JAMA. 2016;315:68–76. doi: 10.1001/jama.2015.17703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Litchfield K, Thomsen H, Mitchell JS, Sundquist J, Houlston RS, Hemminki K, et al. Quantifying the heritability of testicular germ cell tumour using both population-based and genomic approaches. Sci Rep. 2015;5:13889. doi: 10.1038/srep13889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tan IB, Ang KK, Ching BC, Mohan C, Toh CK, Tan MH. Testicular microlithiasis predicts concurrent testicular germ cell tumors and intratubular germ cell neoplasia of unclassified type in adults: a meta-analysis and systematic review. Cancer. 2010;116:4520–32. doi: 10.1002/cncr.25231. [DOI] [PubMed] [Google Scholar]
- [10].McGlynn KA, Trabert B. Adolescent and adult risk factors for testicular cancer. Nat Rev Urol. 2012;9:339–49. doi: 10.1038/nrurol.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Rapley EA, Crockford GP, Teare D, Biggs P, Seal S, Barfoot R, et al. Localization to Xq27 of a susceptibility gene for testicular germ-cell tumours. Nat Genet. 2000;24:197–200. doi: 10.1038/72877. [DOI] [PubMed] [Google Scholar]
- [12].Crockford GP. Genome-wide linkage screen for testicular germ cell tumour susceptibility loci. Hum Mol Genet. 2005;15:443–451. doi: 10.1093/hmg/ddi459. [DOI] [PubMed] [Google Scholar]
- [13].Greene MH, Mai PL, Loud JT, Pathak A, Peters JA, Mirabello L, et al. Familial testicular germ cell tumors (FTGCT) - overview of a multidisciplinary etiologic study. Andrology. 2015;3:47–58. doi: 10.1111/andr.294. [DOI] [PubMed] [Google Scholar]
- [14].Nathanson KL, Kanetsky PA, Hawes R, Vaughn DJ, Letrero R, Tucker K, et al. The Y deletion gr/gr and susceptibility to testicular germ cell tumor. Am J Hum Genet. 2005;77:1034–43. doi: 10.1086/498455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Azevedo MF, Horvath A, Bornstein ER, Almeida MQ, Xekouki P, Faucz FR, et al. Cyclic AMP and c-KIT signaling in familial testicular germ cell tumor predisposition. J Clin Endocrinol Metab. 2013;98:E1393–400. doi: 10.1210/jc.2012-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Rapley EA, Turnbull C, Al Olama AA, Dermitzakis ET, Linger R, Huddart RA, et al. A genome-wide association study of testicular germ cell tumor. Nat Genet. 2009;41:807–10. doi: 10.1038/ng.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ruark E, Seal S, McDonald H, Zhang F, Elliot A, Lau K, et al. Identification of nine new susceptibility loci for testicular cancer, including variants near DAZL and PRDM14. Nat Genet. 2013;45:686–9. doi: 10.1038/ng.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kanetsky PA, Mitra N, Vardhanabhuti S, Vaughn DJ, Li M, Ciosek SL, et al. A second independent locus within DMRT1 is associated with testicular germ cell tumor susceptibility. Hum Mol Genet. 2011;20:3109–17. doi: 10.1093/hmg/ddr207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Schumacher FR, Wang Z, Skotheim RI, Koster R, Chung CC, Hildebrandt MAT, et al. Testicular germ cell tumor susceptibility associated with the UCK2 locus on chromosome 1q23. Hum Mol Genet. 2013;22:2748–53. doi: 10.1093/hmg/ddt109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kanetsky PA, Mitra N, Vardhanabhuti S, Li M, Vaughn DJ, Letrero R, et al. Common variation in KITLG and at 5q31.3 predisposes to testicular germ cell cancer. Nat Genet. 2009;41:811–5. doi: 10.1038/ng.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Turnbull C, Rapley EA, Seal S, Pernet D, Renwick A, Hughes D, et al. Variants near DMRT1, TERT and ATF7IP are associated with testicular germ cell cancer. Nat Genet. 2010;42:604–7. doi: 10.1038/ng.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chung CC, Kanetsky PA, Wang Z, Hildebrandt MAT, Koster R, Skotheim RI, et al. Meta-analysis identifies four new loci associated with testicular germ cell tumor. Nat Genet. 2013;45:680–5. doi: 10.1038/ng.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Litchfield K, Sultana R, Renwick A, Dudakia D, Seal S, Ramsay E, et al. Multi-stage genome-wide association study identifies new susceptibility locus for testicular germ cell tumour on chromosome 3q25. Hum Mol Genet. 2015;24:1169–76. doi: 10.1093/hmg/ddu511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Litchfield K, Holroyd A, Lloyd A, Broderick P, Nsengimana J, Eeles R, et al. Identification of four new susceptibility loci for testicular germ cell tumour. Nat Commun. 2015;6:8690. doi: 10.1038/ncomms9690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kristiansen W, Karlsson R, Rounge TB, Whitington T, Andreassen BK, Magnusson PK, et al. Two new loci and gene sets related to sex determination and cancer progression are associated with susceptibility to testicular germ cell tumor. Hum Mol Genet. 2015;24:4138–46. doi: 10.1093/hmg/ddv129. [DOI] [PubMed] [Google Scholar]
- [26].Grabole N, Tischler J, Hackett JA, Kim S, Tang F, Leitch HG, et al. Prdm14 promotes germline fate and naive pluripotency by repressing FGF signalling and DNA methylation. EMBO Rep. 2013;14:629–37. doi: 10.1038/embor.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Irie N, Weinberger L, Tang WWC, Kobayashi T, Viukov S, Manor YS, et al. SOX17 is a critical specifier of human primordial germ cell fate. Cell. 2015;160:253–68. doi: 10.1016/j.cell.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Farini D, La Sala G, Tedesco M, De Felici M. Chemoattractant action and molecular signaling pathways of Kit ligand on mouse primordial germ cells. Dev Biol. 2007;306:572–83. doi: 10.1016/j.ydbio.2007.03.031. [DOI] [PubMed] [Google Scholar]
- [29].Chung CC, Magalhaes WCS, Gonzalez-Bosquet J, Chanock SJ. Genome-wide association studies in cancer--current and future directions. Carcinogenesis. 2010;31:111–20. doi: 10.1093/carcin/bgp273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Heaney JD, Lam M-YJ, Michelson MV, Nadeau JH. Loss of the transmembrane but not the soluble kit ligand isoform increases testicular germ cell tumor susceptibility in mice. Cancer Res. 2008;68:5193–7. doi: 10.1158/0008-5472.CAN-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zeron-Medina J, Wang X, Repapi E, Campbell MR, Su D, Castro-Giner F, et al. A polymorphic p53 response element in KIT ligand influences cancer risk and has undergone natural selection. Cell. 2013;155:410–22. doi: 10.1016/j.cell.2013.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Yan W, Samson M, Jégou B, Toppari J. Bcl-w forms complexes with Bax and Bak, and elevated ratios of Bax/Bcl-w and Bak/Bcl-w correspond to spermatogonial and spermatocyte apoptosis in the testis. Mol Endocrinol. 2000;14:682–99. doi: 10.1210/mend.14.5.0443. [DOI] [PubMed] [Google Scholar]
- [33].Karlsson R, Andreassen KE, Kristiansen W, Aschim EL, Bremnes RM, Dahl O, et al. Investigation of six testicular germ cell tumor susceptibility genes suggests a parent-of-origin effect in SPRY4. Hum Mol Genet. 2013;22:3373–80. doi: 10.1093/hmg/ddt188. [DOI] [PubMed] [Google Scholar]
- [34].Felfly H, Klein OD. Sprouty genes regulate proliferation and survival of human embryonic stem cells. Sci Rep. 2013;3:2277. doi: 10.1038/srep02277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhao X-L, Zhao Z-H, Xu W-C, Hou J-Q, Du X-Y. Increased expression of SPRY4-IT1 predicts poor prognosis and promotes tumor growth and metastasis in bladder cancer. Int J Clin Exp Pathol. 2015;8:1954–60. [PMC free article] [PubMed] [Google Scholar]
- [36].Gill ME, Hu Y-C, Lin Y, Page DC. Licensing of gametogenesis, dependent on RNA binding protein DAZL, as a gateway to sexual differentiation of fetal germ cells. Proc Natl Acad Sci U S A. 2011;108:7443–8. doi: 10.1073/pnas.1104501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sasai N, Saitoh N, Saitoh H, Nakao M. The transcriptional cofactor MCAF1/ATF7IP is involved in histone gene expression and cellular senescence. PLoS One. 2013;8:e68478. doi: 10.1371/journal.pone.0068478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Yang BX, EL Farran CA, Guo HC, Yu T, Fang HT, Wang HF, et al. Systematic Identification of Factors for Provirus Silencing in Embryonic Stem Cells. Cell. 2015;163:230–45. doi: 10.1016/j.cell.2015.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Mawaribuchi S, Yoshimoto S, Ohashi S, Takamatsu N, Ito M. Molecular evolution of vertebrate sex-determining genes. Chromosome Res. 2012;20:139–51. doi: 10.1007/s10577-011-9265-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Robert NM, Tremblay JJ, Viger RS. Friend of GATA (FOG)-1 and FOG-2 differentially repress the GATA-dependent activity of multiple gonadal promoters. Endocrinology. 2002;143:3963–73. doi: 10.1210/en.2002-220280. [DOI] [PubMed] [Google Scholar]
- [41].Moniot B, Farhat A, Aritake K, Declosmenil F, Nef S, Eguchi N, et al. Hematopoietic prostaglandin D synthase (H-Pgds) is expressed in the early embryonic gonad and participates to the initial nuclear translocation of the SOX9 protein. Dev Dyn. 2011;240:2335–43. doi: 10.1002/dvdy.22726. [DOI] [PubMed] [Google Scholar]
- [42].Schmahl J, Rizzolo K, Soriano P. The PDGF signaling pathway controls multiple steroid-producing lineages. Genes Dev. 2008;22:3255–67. doi: 10.1101/gad.1723908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Minton K. Mitosis: a safety net for successful mitosis. Nat Rev Mol Cell Biol. 2014;15:220–1. doi: 10.1038/nrm3777. [DOI] [PubMed] [Google Scholar]
- [44].Petrovic A, Pasqualato S, Dube P, Krenn V, Santaguida S, Cittaro D, et al. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J Cell Biol. 2010;190:835–52. doi: 10.1083/jcb.201002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Pech MF, Garbuzov A, Hasegawa K, Sukhwani M, Zhang RJ, Benayoun BA, et al. High telomerase is a hallmark of undifferentiated spermatogonia and is required for maintenance of male germline stem cells. Genes Dev. 2015;29:2420–34. doi: 10.1101/gad.271783.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Maida Y, Masutomi K. Telomerase reverse transcriptase moonlights: Therapeutic targets beyond telomerase. Cancer Sci. 2015;106:1486–92. doi: 10.1111/cas.12806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Qi D-L, Ohhira T, Fujisaki C, Inoue T, Ohta T, Osaki M, et al. Identification of PITX1 as a TERT suppressor gene located on human chromosome 5. Mol Cell Biol. 2011;31:1624–36. doi: 10.1128/MCB.00470-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, Hurov KE, Luo J, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–6. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- [49].Wickramasinghe VO, Laskey RA. Control of mammalian gene expression by selective mRNA export. Nat Rev Mol Cell Biol. 2015;16:431–42. doi: 10.1038/nrm4010. [DOI] [PubMed] [Google Scholar]
- [50].Ozaki K, Kuroki T, Hayashi S, Nakamura Y. Isolation of three testis-specific genes (TSA303, TSA806, TSA903) by a differential mRNA display method. Genomics. 1996;36:316–9. doi: 10.1006/geno.1996.0467. [DOI] [PubMed] [Google Scholar]
- [51].Mondal G, Ohashi A, Yang L, Rowley M, Couch FJ. Tex14, a Plk1-regulated protein, is required for kinetochore-microtubule attachment and regulation of the spindle assembly checkpoint. Mol Cell. 2012;45:680–95. doi: 10.1016/j.molcel.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kissel H, Georgescu M-M, Larisch S, Manova K, Hunnicutt GR, Steller H. The Sept4 septin locus is required for sperm terminal differentiation in mice. Dev Cell. 2005;8:353–64. doi: 10.1016/j.devcel.2005.01.021. [DOI] [PubMed] [Google Scholar]
- [53].Bhatnagar S, Gazin C, Chamberlain L, Ou J, Zhu X, Tushir JS, et al. TRIM37 is a new histone H2A ubiquitin ligase and breast cancer oncoprotein. Nature. 2014;516:116–20. doi: 10.1038/nature13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Wu C, Kraft P, Zhai K, Chang J, Wang Z, Li Y, et al. Genome-wide association analyses of esophageal squamous cell carcinoma in Chinese identify multiple susceptibility loci and gene-environment interactions. Nat Genet. 2012;44:1090–7. doi: 10.1038/ng.2411. [DOI] [PubMed] [Google Scholar]
- [55].Kressler D, Bange G, Ogawa Y, Stjepanovic G, Bradatsch B, Pratte D, et al. Synchronizing nuclear import of ribosomal proteins with ribosome assembly. Science. 2012;338:666–71. doi: 10.1126/science.1226960. [DOI] [PubMed] [Google Scholar]
- [56].Kotaja N. MicroRNAs and spermatogenesis. Fertil Steril. 2014;101:1552–1562. doi: 10.1016/j.fertnstert.2014.04.025. [DOI] [PubMed] [Google Scholar]
- [57].Rijlaarsdam MA, van Agthoven T, Gillis AJM, Patel S, Hayashibara K, Lee KY, et al. Identification of known and novel germ cell cancer-specific (embryonic) miRs in serum by high-throughput profiling. Andrology. 2015;3:85–91. doi: 10.1111/andr.298. [DOI] [PubMed] [Google Scholar]
- [58].Litchfield K, Mitchell JS, Shipley J, Huddart R, Rajpert-De Meyts E, Skakkebæk NE, et al. Polygenic susceptibility to testicular cancer: implications for personalised health care. Br J Cancer. 2015;113:1512–8. doi: 10.1038/bjc.2015.334. [DOI] [PMC free article] [PubMed] [Google Scholar]