Abstract
Fanconi anemia (FA) is a multigenic autosomal recessive cancer susceptibility syndrome. The FA pathway regulates the monoubiquitination of FANCD2 and the assembly of damage-associated FANCD2 nuclear foci. How FANCD2 monoubiquitination is coupled to the DNA-damage response has remained undetermined. Here, we demonstrate that the ATR checkpoint kinase and RPA1 are required for efficient FANCD2 monoubiquitination. Deficiency of ATR function, either in Seckel syndrome, which clinically resembles Fanconi anemia, or by siRNA silencing, results in the formation of radial chromosomes in response to the DNA cross-linker, mitomycin C (MMC), thus mimicking the chromosome instability of FA cells.
Keywords: Fanconi anemia, FANCD2, cancer susceptibility, checkpoint, replication stress
Fanconi anemia (FA) is an autosomal recessive disease associated with cancer susceptibility (D'Andrea and Grompe 2003). FA cells are hypersensitive to DNA cross-linking agents, such as mitomycin C (MMC) and cisplatin, and display chromosome instability characterized by the formation of radial chromosomes (D'Andrea and Grompe 2003). These cellular phenotypes indicate that FA cells have a defect in the DNA-damage response.
The genes for eight FA subtypes (A, C, BRCA2/D1, D2, E, F, G, and L) have been cloned, and the encoded FA proteins cooperate in a common pathway. Six of the FA proteins (A, C, E, F, G, and L) assemble into a nuclear complex that is required for the monoubiquitination (activation) of FANCD2 (Garcia-Higuera et al. 2001; D'Andrea and Grompe 2003; Meetei et al. 2003a). Monoubiquitinated FANCD2 is subsequently targeted into DNA repair foci containing BRCA1 (Garcia-Higuera et al. 2001), BRCA2/FANCD1 (Wang et al. 2004), Rad51 (Taniguchi et al. 2002a), and NBS1 (Nakanishi et al. 2002). Deficiencies of FANCD2 monoubiquitination and foci formation, resulting either from mutations in the genes for upstream FA proteins or nonubiquitinable mutants of FANCD2, are associated with cellular hypersensitivity to cross-linking agents and with chromosome instability (Garcia-Higuera et al. 2001).
FANCD2 is monoubiquitinated both during S phase (Taniguchi et al. 2002a) and in response to various DNA damaging agents, including ionizing radiation (IR) and MMC (Garcia-Higuera et al. 2001). The monoubiquitination of FANCD2 is required for its localization to DNA-damage foci (Garcia-Higuera et al. 2001). Although it is clear from these observations that FANCD2 monoubiquitination is activated by DNA damage, it has not been determined how this process is coupled to the DNA-damage response or to S-phase progression.
At the center of the DNA-damage response, the checkpoint protein kinases ATM (ataxia telangiectasia mutated) and ATR (ATM and Rad3-related) phosphorylate numerous proteins involved in checkpoint function and DNA repair and thereby coordinate these processes (Abraham 2001; Osborn et al. 2002). It is generally believed that ATM and ATR act in parallel pathways that respond to different DNA stresses, including double-strand breaks (DSBs) and collapse of replication forks (treatment with IR or hydroxyurea [HU], respectively). Whereas ATM phosphorylates FANCD2 in response to IR, we previously demonstrated that ATM-deficient cells are proficient for FANCD2 monoubiquitination and foci formation (Taniguchi et al. 2002b). Thus, ATM is not required for activation of FANCD2 in the FA pathway.
ATR is mutated in one complementation group of Seckel syndrome (SCKL1; O'Driscoll et al. 2003), an autosomal recessive disorder resembling FA, Nijmegen Breakage syndrome, and other diseases involving impaired DNA-damage responses. ATR is present at the replication fork (Lupardus et al. 2002; Tercero et al. 2003). It is activated when the replication fork encounters DNA damage and then phosphorylates several substrates, including its partner kinase, Chk1 (Liu et al. 2000; Zhao and Piwnica-Worms 2001). The RPA complex recruits ATR to newly synthesized, single-strand DNA (ssDNA), generated at sites of damage (Zou and Elledge 2003).
In the present study, we demonstrate using RNA silencing and Seckel syndrome cells that ATR is required for efficient FANCD2 monoubiquitination and FANCD2 foci assembly in response to various genotoxic stresses, including IR and MMC. Further, ATR-deficient Seckel syndrome cells form radial chromosomes in response to MMC, thus establishing a genetic relationship between Seckel syndrome and Fanconi anemia. FANCD2 monoubiquitination also requires the DNA-damage sensor RPA1. This is the first demonstration of a DNA-damage response in which ATR is required for the response to either replication stress or IR without any role for ATM.
Results and Discussion
Inhibition of FANCD2 monoubiquitination by ATR silencing
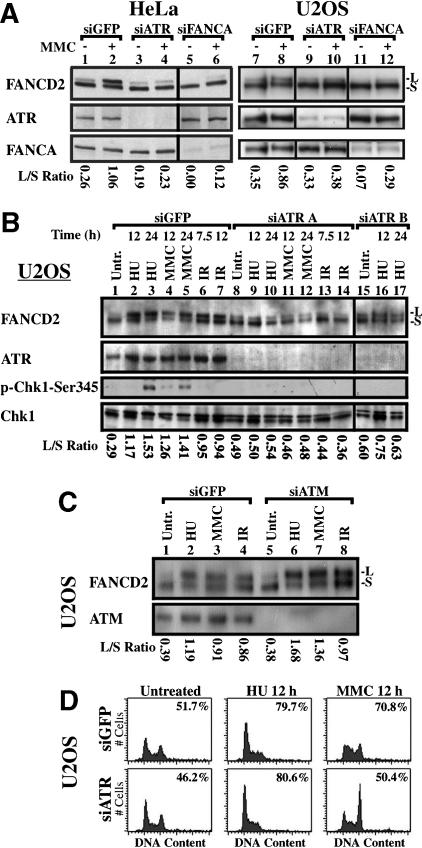
Given the central role of the ATR kinase in surveillance of DNA damage (Abraham 2001; Osborn et al. 2002), we suppressed ATR in HeLa (Fig. 1A, lanes 1-6) and U2OS cells (Fig. 1A, lanes 7-12) with siRNA (Fig. 1A). ATR protein was suppressed by siATR, but not siGFP (green fluorescent protein). ATR suppression resulted in loss of MMC-inducible FANCD2 monoubiquitination. By contrast, silencing of FANCA with siRNA inhibited FANCD2 monoubiquitination both without treatment and following exposure to MMC.
Figure 1.
ATR is required for damage-inducible monoubiquitination of FANCD2. (A) ATR siRNA A strongly inhibits MMC-induced monoubiquitination of FANCD2 in HeLa and U2OS cells. Silencing of FANCA suppresses FANCD2 monoubiquitination either with or without exposure to MMC. (B) Silencing of ATR with siRNA in U2OS cells suppresses FANCD2 monoubiquitination following exposure to HU, MMC, or IR for the indicated times. Suppression of ATR activity is demonstrated by decreased phosphorylation of Chk1 on S345 in response to silencing. (C) Silencing of ATM does not decrease FANCD2 monoubiquitination. (D) Flow cytometric analysis demonstrates that inhibition of damage-induced monoubiquitination of FANCD2 by suppression of ATR is not due to an alteration of cell-cycle progression. Histograms of DNA content are displayed, with S-phase index measured by BrdU incorporation as described (Andreassen et al. 2001), inset in each histogram.
We next tested the effect of ATR silencing on FANCD2 monoubiquitination in response to various DNA damaging agents (Fig. 1B). In untreated control U2OS cells, transfected with GFP siRNA, and in untreated cells transfected with two different sets of ATR siRNA (A or B), FANCD2 was present mainly as a nonubiquitinated isoform (FANCD2-S). Treatment of control cells with HU, MMC, or IR activated the monoubiquitination of FANCD2 (FANCD2-L). Suppression of ATR with siRNA A strongly inhibited FANCD2 monoubiquitination in response to HU, MMC, or IR (Fig. 1B, lanes 8-14). The ratio of monoubiquitinated to nonubiquitinated FANCD2 (L/S ratio) under each condition is shown below the lanes in Figure 1B. ATR siRNA B similarly suppressed damage-induced monoubiquitination of FANCD2 following treatment with HU (Fig. 1B, lanes 15-17). FANCD2 monoubiquitination in response to HU, MMC, and IR was similarly suppressed by siATR in HeLa cells (data not shown).
Chk1 protein kinase is a substrate of ATR and is activated by phosphorylation at S345 (Liu et al. 2000). Chk1 was strongly phosphorylated at S345 following exposure to the replication inhibitor HU, and less strongly following exposure to MMC (Fig. 1B, lanes 3,5). Silencing of ATR suppressed Chk1 phosphorylation.
It has been reported that ATR is required for FANCD2 phosphorylation in response to psorelen/UV-A (Pichierri and Rosselli 2004), but this was not associated with a failure to up-regulate FANCD2 monoubiquitination. This previous study (Pichierri and Rosselli 2004) used an inducible system for the expression of dominant-negative ATR and this may not have inhibited endogenous ATR sufficiently to suppress FANCD2 monoubiquitination. Unlike our observation that ATR is required for FANCD2 monoubiquitination in response to multiple DNA-damage stresses, it is not clear whether this previously reported role for ATR in FANCD2 phosphorylation (Pichierri and Rosselli 2004) is stress-specific. By its dependence upon ATR, the monoubiquitination of FANCD2 is coupled to the DNA-damage response and integrated with other DNA repair and checkpoint processes.
Whereas ATR is required for FANCD2 monoubiquitination in response to many types of DNA damage, including IR, ATM is not required for FANCD2 monoubiquitination (Taniguchi et al. 2002b). We tested an ATM siRNA for the ability to inhibit FANCD2 monoubiquitination in response to DNA damage (Fig. 1C). Silencing of ATM did not inhibit FANCD2 monoubiquitination in response to HU, MMC, or IR, and instead yielded an increased FANCD2 L/S ratio. Exposure of HeLa cells to wortmannin, an inhibitor of PI3-kinase activity, also indicates a role for ATR in the regulation of FANCD2 monoubiquitination (Supplementary Fig. 1).
To exclude the possibility that effects of ATR silencing on FANCD2 monoubiquitination are due to changes in cell-cycle distribution, we analyzed the effects of ATR siRNA on the cell-cycle distribution of U2OS cells (Fig. 1D). Histograms of DNA content, comparing cells treated with GFP siRNA or ATR siRNA, were similar in untreated cells and cells treated with HU (Fig. 1D). ATR deficiency resulted in an increased G2 accumulation following exposure to MMC, but only a modest decrease in the percentage of cells actively synthesizing DNA in S phase, relative to control cells treated with MMC, as quantitated by bromodeoxyuridine (BrdU) incorporation (Fig. 1D, inset in histograms). S phase is the phase when FANCD2 is monoubiquitinated (Taniguchi et al. 2002a; Rothfuss and Grompe 2004). Thus, the effect of ATR silencing on FANCD2 monoubiquitination did not result from an indirect effect on cell-cycle progression.
Inhibition of FANCD2 foci assembly and induction of radial chromosomes by ATR silencing
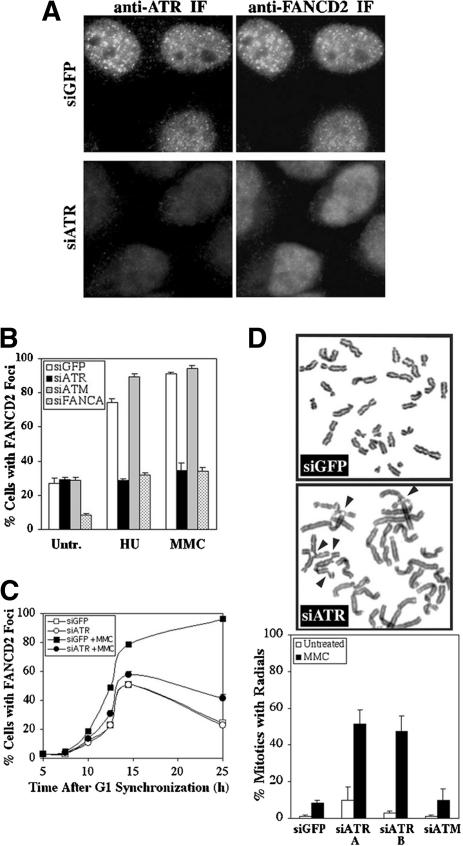
Because monoubiquitination is required for the assembly of FANCD2 foci, we examined the effect of ATR silencing on FANCD2 foci formation in HeLa cells, in which S phase and damage-induced FANCD2 foci have been well characterized (Garcia-Higuera et al. 2001; Taniguchi et al. 2002a). ATR and FANCD2 colocalized in HeLa cells following exposure to MMC (Fig. 2A). This is in accord with Pichierri and Rosselli (2004), who observed the colocalization of FANCD2 and ATR following DNA cross-linking by psoralen/UV-A. Further, transfection with ATR siRNA eliminated ATR foci in >95% of cells (Fig. 2A) and suppressed FANCD2 monoubiquitination (Figs. 1A, 4A, below) in HeLa. Silencing of ATR, but not ATM, suppressed FANCD2 foci formation following treatment with HU or MMC, but did not alter the formation of FANCD2 foci in untreated HeLa cells (Fig. 2B). Using synchronization in G1 by selective detachment of mitotic cells and release from nocodazole treatment, we found that silencing of ATR decreased MMC-induced FANCD2 foci assembly but had no effect on FANCD2 foci assembly during S phase in the absence of exogenous DNA damage (Fig. 2C). S-phase cells, identified by incorporation of BrdU, increased at 10 h following release from synchronization (23%) and peaked at 14.5 h (73%; data not shown).
Figure 2.
ATR is required for FANCD2 function following DNA damage. (A) ATR colocalizes with FANCD2 at DNA-damage foci formed by treatment of HeLa cells with 0.5 μM MMC for 24 h. Silencing of ATR with siATR A can be detected by immunofluorescence microscopy, and such cells are deficient for FANCD2 foci. (B) Silencing of ATR, but not ATM, leads to a strong decrease in the percentage of cells with FANCD2 foci following treatment with HU or MMC, relative to cells transfected with GFP siRNA. (C) HeLa cells treated with siGFP or siATR have similar levels of FANCD2 foci assembly during S phase in the absence of exogenous DNA damage. Cells were treated with 0.04 μg/mL nocodazole for 12 h, 4 d after transfection, and were synchronized in G1 by release in drug-free medium. MMC was added at 5 h release, where appropriate. Values represent three counts of 150 cells each. Standard deviation was <10% of the ordinate value. (D) Transformed human diploid fibroblasts (IMR90) deficient for ATR, following transfection with two different ATR siRNAs (A and B), have elevated formation of radial chromosomes in response to 48 h treatment with 0.032 μM MMC, relative to IMR90 cells transfected with GFP or ATM siRNA.
Figure 4.
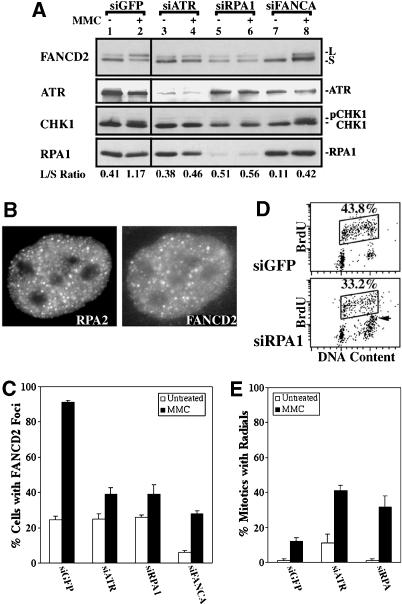
RPA1 is required for FANCD2 monoubiquitination following MMC treatment. (A) Immunoblots demonstrate that attenuation of RPA1 with siRNA strongly inhibits FANCD2 monoubiquitination in HeLa cells following treatment with 0.5 μM MMC for 24 h. (B) RPA2 colocalizes with FANCD2 in DNA-damage foci following 24 h treatment of HeLa cells with 0.5 μM MMC. (C) The formation of FANCD2 foci at 24 h treatment with 0.5 μM MMC is decreased strongly in HeLa cells in which either ATR or RPA1 had been attenuated with siRNA. (D) Dot plots of BrdU incorporation and DNA content demonstrate that siRPA does not suppress normal DNA replication during S phase (box). (E) Transformed human lung fibroblasts (IMR90) in which RPA1 has been silenced with siRNA form radial chromosomes in response to 48 h treatment with 0.032 μM MMC.
The development of radial chromosomes is a measure of cellular sensitivity to MMC, and FA cells deficient for FANCD2 monoubiquitination display elevated levels of radial chromosomes (D'Andrea and Grompe 2003). Accordingly, we found that silencing of ATR in transformed IMR90 cells, a cell line with a normal human karyotype and baseline normal MMC sensitivity, induced the formation of radial chromosomes following exposure to MMC (Fig. 2D). In agreement with Casper et al. (2002), inactivation of ATR did not induce significant karyotypic abnormalities in untreated cells. In contrast, transformed IMR90 cells transfected with GFP or ATM siRNA exhibited only low levels of chromosome damage in response to MMC. Interestingly, ATR is also required for resistance to cisplatin (Yazlovitskaya and Persons 2003), which, like MMC, is a DNA cross-linker.
ATR-deficient Seckel cells are deficient in FANCD2 monoubiquitination and FANCD2 function
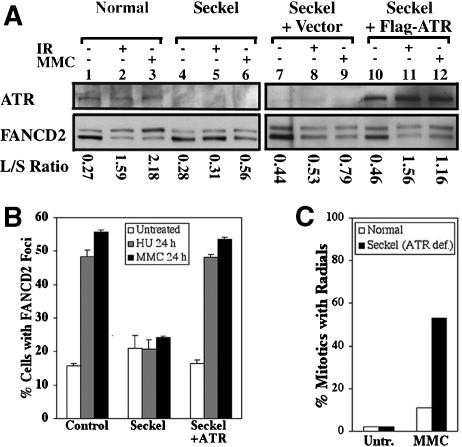
Seckel syndrome and FA share several clinical features, including varying degrees of microcephaly, dwarfism, thumb abnormalities, aplastic anemia, and cancer susceptibility (Chanan-Khan et al. 2003), suggesting the possibility of a mechanistic link between these diseases. We examined FANCD2 monoubiquitination in a Seckel syndrome cell line with biallelic mutation in ATR (Fig. 3A; O'Driscoll et al. 2003). Although untreated Seckel cells displayed monoubiquitinated FANCD2, the L/S ratio did not increase following DNA damage by IR or MMC (Fig. 3A, lanes 4-6,7-9). Control TERT immortalized normal fibroblasts (PD846F) showed a normal damage-inducible increase in FANCD2 monoubiquitination (Fig. 3A, lanes 1-3). The deficient monoubiquitination of FANCD2 in Seckel cells in response to treatment with IR or MMC was corrected by stable retroviral transduction of ATR (Fig. 3A, lanes 10-12). Whereas ATR is required for efficient damage-inducible FANCD2 monoubiquitination, FA subtypes A, C, E, F, G are required for FANCD2 monoubiquitination both during S phase of an unperturbed cell cycle and in response to DNA damage (Taniguchi et al. 2002a). Thus, the control of FANCD2 monoubiquitination by ATR is an additional regulatory mechanism specific to DNA damage.
Figure 3.
Cells derived from a Seckel syndrome patient are defective in the activation of the FA pathway. (A) Immunoblots with TERT immortalized fibroblasts, either wild type (PD846), ATR-deficient Seckel cells (FO2-98), Seckel cells with empty vector, or Seckel cells corrected by the expression of ATR were prepared from untreated specimens or cells exposed to 15 Gy IR for 12 h or 0.5 μM MMC for 24 h. (B) Damage-induced formation of FANCD2 foci occurs in control cells (PD846), but not in Seckel cells treated with 2 mM HU or 0.5 μM MMC for 24 h. (C) F02-98 Seckel cells, but not PD846 normal controls, show a highly elevated level of radial chromosomes in MMC.
FANCD2 foci formed in untreated Seckel cells (Fig. 3B). However, although an increased percentage of control fibroblasts (PD846) and corrected Seckel cells (Seckel-ATR) displayed FANCD2 foci in response to HU or MMC, Seckel cells did not. Conversely, Seckel cells displayed hypersensitivity to MMC, as determined by cytogenetic analysis, whereas the normal cells did not (Fig. 3C). This is in agreement with the finding of O'Driscoll et al. (2003) that these Seckel cells have elevated sensitivity to MMC. These results further suggest that the deficiency in ATR function in Seckel cells disrupts the FA pathway.
The FANCA, C, E, F, G, and L proteins form a nuclear complex that is required for monoubiquitination of FANCD2 (Garcia-Higuera et al. 2001; D'Andrea and Grompe 2003; Meetei et al. 2003a). Because FANCL has ubiquitin ligase activity, this complex may be the specific multi-subunit ubiquitin ligase for FANCD2 (Meetei et al. 2003a). As possible mechanisms of ATR-dependent control of FANCD2 monoubiquitination, (1) ATR may directly phosphorylate FANCD2 and lead to increased FANCD2 monoubiquitination, (2) ATR may phosphorylate members of the FA complex and thereby increase the monoubiquitin ligase activity of the FA complex, or (3) known substrates of ATR, such as BRCA1 or Chk1, may mediate the regulation of FANCD2 monoubiquitination by ATR. Because FANCD2 is a direct substrate of ATM (Taniguchi et al. 2002b), and because ATM and ATR share common substrates (Tibbetts et al. 2000), we speculate that FANCD2 may be a direct substrate of ATR. Accordingly, FANCD2 can be phosphorylated in vitro by ATR at sites distinct from those phosphorylated by ATM (Supplementary Fig. 2). FANCA and FANCG are also phosphoproteins (Kupfer et al. 1999; Qiao et al. 2001), but the responsible kinase(s) and consequences for FANCD2 monoubiquitination have not been determined. The precise mechanism of ATR-dependent activation of FANCD2 by monoubiquitination has not yet been determined.
RPA1 acts as a sensor of DNA damage required for efficient FANCD2 monoubiquitination
RPA1 binds to newly generated ssDNA at sites of DNA damage or replication stress, leading to the activation of ATR (Zou and Elledge 2003). Given the role of ATR in FANCD2 monoubiquitination, we next tested whether RPA1 is also required in this process (Fig. 4). Decreasing the levels of RPA1 protein (RPA70) with siRNA inhibited ATR activation following treatment with MMC, as measured by the suppression of a slower migrating form of Chk1 that is detected in control cells (HeLa-siGFP; Fig. 4A, cf. lanes 2 and 6). Attenuation of RPA1 protein levels also inhibited FANCD2 monoubiquitination following exposure to MMC. These results suggest that RPA1 is a sensor of DNA cross-links and is required for efficient FANCD2 monoubiquitination. RPA1 copurifies with the FA complex (Meetei et al. 2003b), suggesting that the sensing of DNA damage and FA complex-dependent monoubiquitination are physically linked.
Attenuation of FANCA with siRNA inhibited FANCD2 monoubiquitination in untreated cells and cells exposed to MMC (Figs. 1A, 4A). However, deficiency for FANCA was not associated with a decrease in Chk1 phosphorylation, as measured by the appearance of a slower migrating form (Fig. 4A, lane 8). Further, FA cells are proficient for Chk1 phosphorylation in response to DNA damage, as measured with phospho-345 antibodies (Pichierri and Rosselli 2004; data not shown). These results suggest that monoubiquitination of FANCD2 is not required for Chk1 phosphorylation and activation.
RPA2 colocalized with FANCD2 DNA-damage foci following MMC treatment (Fig. 4B). Consistent with a role in activating ATR, the assembly of FANCD2 foci in MMC was strongly decreased by siRNA for RPA1 (Fig. 4C). FANCD2 foci were present, however, in untreated HeLa cells transfected with siRPA1. Inactivation of RPA1 did not affect the ability of untreated HeLa cells (Fig. 4D), or those treated with MMC (data not shown), to efficiently replicate DNA during S phase as measured by BrdU incorporation. RPA1 siRNA induced some G2 accumulation (Fig. 4D, indicated by arrow). Because FANCD2 is monoubiquitinated during S phase (Rothfuss and Grompe 2004), this demonstrates that the effects of siRNA for RPA1 on FANCD2 monoubiquitination and foci formation are not due to blocks to DNA replication. Similarly, incomplete depletion of RPA from Xenopus extracts does not interfere with DNA replication (Walter and Newport 2000). Attenuation of RPA1, like suppression of ATR, generated an MMC-inducible elevation in radial chromosomes in transformed IMR90 cells (Fig. 4E).
Because FANCD2 monoubiquitination following exposure to MMC requires RPA1, and given that the RPA complex recruits ATR to ssDNA (Zou and Elledge 2003), these results suggest that RPA1 acts as a sensor of DNA cross-links and activates ATR, leading to FANCD2 monoubiquitination. The involvement of the ssDNA-binding protein, RPA1, suggests that the signaling of FANCD2 monoubiquitination in response to DNA cross-links is based upon processing to generate ssDNA. FANCD2 monoubiquitination is coincident with the appearance of DSBs recognized by phosphorylated H2AX protein (Rothfuss and Grompe 2004), and these DSBs may be resected to yield ssDNA.
Because ATR is present at the replication fork during S phase (Lupardus et al. 2002; Tercero et al. 2003), this may allow it to mediate processes, such as FANCD2 monoubiquitination, which occur regardless of the type of DNA damage incurred. The positioning of ATR at the advancing replication fork may also provide an explanation for the coupling of damage-induced FANCD2 monoubiquitination to S phase (Rothfuss and Grompe 2004). Further, the Fanconi anemia pathway may be integrated with other checkpoint and repair processes dependent on ATR. Given that monoubiquitinated FANCD2 binds BRCA2 in chromatin (Wang et al. 2004), and the recently determined role for BRCA2, one of the roles of the FA pathway may be to stabilize stalled DNA replication forks (Lomonosov et al. 2003).
Materials and methods
Cell culture
U2OS, HeLa, and transformed IMR90 cells (Garcia-Higuera et al. 2001), Seckel cells (F02-98; O'Driscoll et al. 2003), and PD846 fibroblasts immortalized with pBABE-hTERT-neo (Nakanishi et al. 2002) were grown as described.
Irradiation was delivered using a Gammacell-40 irradiator. Hydroxyurea was added at 2 mM, and mitomycin C was utilized at 0.5 μM.
Retroviral expression of ATR
Human ATR cDNA N-terminally tagged with FLAG, a kind gift from Dr. Karlene Cimprich (Stanford University, Stanford, CA), was inserted into pMMP-puro. Production of pMMP retroviral supernatants and infection of fibroblasts was performed as described (Garcia-Higuera et al. 2001). After 48 h, cells were selected in puromycin (1 μg/mL).
Immunoblotting
Lysates were electrophoresed by SDS-PAGE (6% polyacrylamide, bisacrylamide gels for ATR, ATM, FANCA, and FANCD2; 10% for Chk1 and RPA1). Proteins were then transferred to nitrocellulose, blocked, and incubated with primary antibodies as described (Taniguchi et al. 2002b). Antibodies included anti-FANCA (1:1000) and anti-FANCD2 (E35, 1: 1000; Garcia-Higuera et al. 2001), anti-ATR (1:1000 Santa Cruz [N19]), anti-ATM (1:500, Novus), anti-RPA1 (1:200 Oncogene Research), anti-phospho-S345-Chk1 (1:1000, Cell Signaling), and anti-Chk1 (1:500 Santa Cruz [G4]). Membranes were washed, incubated with HRP-linked secondary antibodies (Amersham), and detected by chemiluminescence (Amersham) as described (Taniguchi et al. 2002b).
siRNA and transfection
Expression of targeted genes was knocked down by transient expression of siRNA directed against ATR (5′-AACCTCCGTGATGTTGCTTGA-3′, A, and 5′-AAGCCAAGACAAATTCTGTGT-3′, B; Casper et al. 2002), GFP (5′-CGGCAAGCTGACCCTGAAGTTCAT-3′), RPA1 (5′-AACACTCTATCCTCTTTCATG-3′; Zou and Elledge 2003), FANCA (5′-AAGGGTCAAGAGGGAAAAATA-3′; Bruun et al. 2003), and ATM (5′-AACATACTACTCAAAGACATT-3′). Cells were plated at least 16 h prior to transfection in six-well plates to yield ∼80% confluency at the time of transfection in 1 mL of media devoid of antibiotics. SiRNA oligos were diluted in 100 μL Opti-MEM I to yield a final concentration of 100 nM siRNA, and were mixed with 6 μL of Lipofectamine 2000 (Invitrogen) diluted in 100 μL of OptiMEM I. Complex formation was allowed to proceed for 20 min at room temperature prior to dropwise addition to cells. SiRNA complexes were incubated with cells for 6 h, at which time cells were changed to normal growth media. At 84-96 h posttransfection, cells were treated with MMC (0.5 μM, 12 or 24 h), HU (2 mM, 12 or 24 h), or IR (10 Gy, 7.5 or 12 h).
Immunofluorescence microscopy
Immunofluorescence microscopy on adherent cells was performed as described (Taniguchi et al. 2002a). Primary antibodies included FANCD2 (E35, 1:200), ATR (N19, 1:100; Santa Cruz), and RPA2 (1:200, Oncogene). Labeling with BrdU, fixation, and permeabilization for detection with a cell proliferation kit (Amersham Pharmacia) was as described (Wu et al. 2000).
Flow cytometry
Incorporation of bromodeoxyuridine (BrdU) and measurement of DNA content with propidium iodide were as described (Andreassen et al. 2001).
Chromosome breakage analysis
Chromosome breakage analysis was performed by the Cytogenetics Core Facility of the Dana-Farber Cancer Institute. For the analysis of IMR90 transfected with various siRNA, cells were plated into T25 flasks at 24 h following transfection. Beginning the next day, cells were treated with 0.063 μM MMC for 2 d. After treatment, cells were exposed to colcemid for 12 h, swollen using 0.075 M KCl, and fixed with 3:1 methanol:acetic acid. Slides were stained with Wrights's stain, and 50 metaphases were scored for radials in two independent experiments using a blinded approach. For hTERT-immortalized fibroblasts, cells were plated in T75 flasks and treated with 0.2 μM MMC for 24 h and colcemid for the last 20 h.
Acknowledgments
We thank P. Jeggo and M. O'Driscoll for the primary Seckel fibroblasts, F02-98. The pBABE-hTERTneo plasmid and human ATR cDNA were generously provided by D. Weaver and K. Cimprich, respectively. We thank L. Moreau for chromosome breakage analysis. This work was supported by NIH grants RO1HL52725, RO1DK43889, and PO1HL54785 (A.D.D.). R.P.A. is a Special Fellow of the Leukemia and Lymphoma Society (LLS). T.T. is a Scholar Fellow of the American Society of Hematology.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1196104.
References
- Abraham R.T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes & Dev. 15: 2177-2196. [DOI] [PubMed] [Google Scholar]
- Andreassen P.R., Lohez, O.D., Lacroix, F.B., and Margolis, R.L. 2001. Tetraploid state induces p53-dependent arrest of nontransformed mammalian cells in G1. Mol. Biol. Cell 12: 1315-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruun D., Folias, A., Akkari, Y., Cox, Y., Olson, S., and Moses, R. 2003. siRNA depletion of BRCA1, but not BRCA2, causes increased genome instability in Fanconi anemia cells. DNA Repair (Amst) 2: 1007-1013. [DOI] [PubMed] [Google Scholar]
- Casper A.M., Nghiem, P., Arlt, M.F., and Glover, T.W. 2002. ATR regulates fragile site stability. Cell 111: 779-789. [DOI] [PubMed] [Google Scholar]
- Chanan-Khan A., Holkova, B., Perle, M.A., Reich, E., Wu, C.D., Inghirami, G., and Takeshita, K. 2003. T-cell clonality and myelodysplasia without chromosomal fragility in a patient with features of Seckel syndrome. Haematologica 88: ECR14. [PubMed] [Google Scholar]
- D'Andrea A.D. and Grompe, M. 2003. The Fanconi anaemia/BRCA pathway. Nat. Rev. Cancer 3: 23-34. [DOI] [PubMed] [Google Scholar]
- Garcia-Higuera I., Taniguchi, T., Ganesan, S., Meyn, M.S., Timmers, C., Hejna, J., Grompe, M., and D'Andrea, A.D. 2001. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell 7: 249-262. [DOI] [PubMed] [Google Scholar]
- Kupfer G., Naf, D., Garcia-Higuera, I., Wasik, J., Cheng, A., Yamashita, T., Tipping, A., Morgan, N., Mathew, C.G., and D'Andrea, A.D. 1999. A patient-derived mutant form of the Fanconi anemia protein, FANCA, is defective in nuclear accumulation. Exp. Hematol. 27: 587-593. [DOI] [PubMed] [Google Scholar]
- Liu Q., Guntuku, S., Cui, X.S., Matsuoka, S., Cortez, D., Tamai, K., Luo, G., Carattini-Rivera, S., DeMayo, F., Bradley, A., et al. 2000. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes & Dev. 14: 1448-1459. [PMC free article] [PubMed] [Google Scholar]
- Lomonosov M., Anand, S., Sangrithi, M., Davies, R., and Venkitaraman, A.R. 2003. Stabilization of stalled DNA replication forks by the BRCA2 breast cancer susceptibility protein. Genes & Dev. 17: 3017-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupardus P.J., Byun, T., Yee, M.C., Hekmat-Nejad, M., and Cimprich, K.A. 2002. A requirement for replication in activation of the ATR-dependent DNA damage checkpoint. Genes & Dev. 16: 2327-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meetei A.R., De Winter, J.P., Medhurst, A.L., Wallisch, M., Waisfisz, Q., Van De Vrugt, H.J., Oostra, A.B., Yan, Z., Ling, C., Bishop, C.E., et al. 2003a. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat. Genet. 35: 165-170. [DOI] [PubMed] [Google Scholar]
- Meetei A.R., Sechi, S., Wallisch, M., Yang, D., Young, M.K., Joenje, H., Hoatlin, M.E., and Wang, W. 2003b. A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol. Cell. Biol. 23: 3417-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K., Taniguchi, T., Ranganathan, V., New, H.V., Moreau, L.A., Stotsky, M., Mathew, C.G., Kastan, M.B., Weaver, D.T., and D'Andrea, A.D. 2002. Interaction of FANCD2 and NBS1 in the DNA damage response. Nat. Cell Biol. 4: 913-920. [DOI] [PubMed] [Google Scholar]
- O'Driscoll M., Ruiz-Perez, V.L., Woods, C.G., Jeggo, P.A., and Goodship, J.A. 2003. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat. Genet. 33: 497-501. [DOI] [PubMed] [Google Scholar]
- Osborn A.J., Elledge, S.J., and Zou, L. 2002. Checking on the fork: The DNA-replication stress-response pathway. Trends Cell Biol. 12: 509-516. [DOI] [PubMed] [Google Scholar]
- Pichierri P. and Rosselli, F. 2004. The DNA crosslink-induced S-phase checkpoint depends on ATR-CHK1 and ATR-NBS1-FANCD2 pathways. EMBO J. 23: 1178-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao F., Moss, A., and Kupfer, G.M. 2001. Fanconi anemia proteins localize to chromatin and the nuclear matrix in a DNA damage- and cell cycle-regulated manner. J. Biol. Chem. 276: 23391-23396. [DOI] [PubMed] [Google Scholar]
- Rothfuss A. and Grompe, M. 2004. Repair kinetics of genomic inter-strand DNA cross-links: Evidence for DNA double-strand break-dependent activation of the Fanconi anemia/BRCA pathway. Mol. Cell. Biol. 24: 123-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T., Garcia-Higuera, I., Andreassen, P.R., Gregory, R.C., Grompe, M., and D'Andrea, A.D. 2002a. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood 100: 2414-2420. [DOI] [PubMed] [Google Scholar]
- Taniguchi T., Garcia-Higuera, I., Xu, B., Andreassen, P.R., Gregory, R.C., Kim, S.T., Lane, W.S., Kastan, M.B., and D'Andrea, A.D. 2002b. Convergence of the fanconi anemia and ataxia telangiectasia signaling pathways. Cell 109: 459-472. [DOI] [PubMed] [Google Scholar]
- Tercero J.A., Longhese, M.P., and Diffley, J.F. 2003. A central role for DNA replication forks in checkpoint activation and response. Mol. Cell 11: 1323-1336. [DOI] [PubMed] [Google Scholar]
- Tibbetts R.S., Cortez, D., Brumbaugh, K.M., Scully, R., Livingston, D., Elledge, S.J., and Abraham, R.T. 2000. Functional interactions between BRCA1 and the checkpoint kinase ATR during genotoxic stress. Genes & Dev. 14: 2989-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J. and Newport, J. 2000. Initiation of eukaryotic DNA replication: Origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase α. Mol. Cell 5: 617-627. [DOI] [PubMed] [Google Scholar]
- Wang X.Z., Andreassen, P.R., and D'Andrea, A.D. 2004. Functional Interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin. Mol. Cell. Biol. 24: 5850-5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Lee, W.H., and Chen, P.L. 2000. NBS1 and TRF1 colocalize at promyelocytic leukemia bodies during late S/G2 phases in immortalized telomerase-negative cells. Implication of NBS1 in alternative lengthening of telomeres. J. Biol. Chem. 275: 30618-30622. [DOI] [PubMed] [Google Scholar]
- Yazlovitskaya E.M. and Persons, D.L. 2003. Inhibition of cisplatin-induced ATR activity and enhanced sensitivity to cisplatin. Anticancer Res. 23: 2275-2279. [PubMed] [Google Scholar]
- Zhao H. and Piwnica-Worms, H. 2001. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol. Cell. Biol. 21: 4129-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L. and Elledge, S.J. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542-1548. [DOI] [PubMed] [Google Scholar]