Abstract
Although most mRNAs initiate translation by 5′ ribosome scanning, some small fraction of mammalian and viral mRNAs utilize either of two alternate mechanisms, known as internal ribosome entry and ribosome shunting. Ribosome shunting is a poorly understood form of initiation in which 40S ribosome subunits are loaded onto mRNA through interactions with the m7GTP cap, but then bypass large segments of the mRNA as directed by cis-acting RNA shunting elements and trans-acting protein factors. Here, we describe the molecular mechanism by which ribosome shunting occurs with high efficiency on adenovirus late mRNAs. We show that the viral 100k protein possesses a selective binding element for the 5′ noncoding region (5′NCR) of viral late mRNAs (known as the tripartite leader), forms a complex with initiation factor eIF4G and poly(A)-binding protein (PABP), and strongly and selectively enhances the level of both factors and 40S ribosome subunits on viral mRNAs in polysomes. Mutational and biochemical studies demonstrate that the ability of 100k protein to bind both the tripartite leader and eIF4G are critical to promote a high level of ribosome shunting. A molecular mechanism for ribosome shunting is described by which enhanced binding of eIF4G and possibly PABP with 100k protein, and simultaneous interaction with the tripartite leader 5′NCR, drives 40S ribosome recruitment and initiation on mRNAs.
Keywords: Translation, ribosome shunting, eIF4G, poly(A)-binding protein, adenovirus
Unorthodox mechanisms of translational control have often been discovered first in viruses and found later in animal cells, generally corresponding to mRNAs of specialized cellular function. Internal initiation of translation through an internal ribosome entry site (IRES) in the mRNA, and the process of nonlinear ribosome scanning known as ribosome shunting, are cases in point (for review, see Schneider and Mohr 2003). Initiation of translation on most eukaryotic mRNAs is the rate-limiting step and involves the recruitment of mRNAs to 40S ribosome subunits (Hershey and Merrick 2000). Eukaryotic mRNAs typically possess a 5′ m7GpppN cap structure that is specifically bound by the cap-initiation complex (eIF4F), which plays a pivotal role in associating capped mRNAs with 40S ribosome subunits. eIF4F functions as a cap-dependent RNA helicase that stimulates protein synthesis by unwinding the 5′ end of mRNAs, recruiting 40S ribosomes, and promoting a 5′-to-3′ search for the initiation codon through a process known as ribosome scanning (Hershey and Merrick 2000; Jackson 2000). Many of the molecular details for IRES-mediated translation initiation are beginning to be understood (Stoneley and Willis 2004), whereas ribosome shunting remains a poorly characterized alternate means of initiation (for review, see Cuesta et al. 2001; Ryabova et al. 2002).
Ribosome shunting was first described for the cauliflower mosaic virus (CaMV) 35S mRNA (Futterer et al. 1993; Ryabova et al. 2000; Pooggin et al. 2001), and remains best understood in that system (Hemmings-Mieszczak et al. 2000). In CaMV, ribosome shunting is mediated by a short upstream open reading frame (ORF) and a downstream stable hairpin structure that together act to accumulate and reinitiate 40S ribosome subunits at a downstream ORF. Ribosome shunting has also been described for adenovirus (Ad) late mRNAs (Yueh and Schneider 1996, 2000), the Sendai virus Y mRNAs (Curran and Kolakofsky 1988; Latorre et al. 1998; de Breyne et al. 2003), papillomavirus E1 mRNA (Remm et al. 1999), and possibly for several mammalian mRNAs (Yueh and Schneider 2000; Rogers et al. 2004). The general mechanism for ribosome shunting in Ad and CaMV involves loading of 40S ribosome subunits onto the 5′ end of the capped mRNA, entry of ribosome subunits a short distance into the mRNA, possibly by limited 5′ scanning, followed by direct translocation of 40S subunits to a downstream initiation codon, directed by cis-acting RNA shunting elements.
In adenovirus, cis-acting RNA shunting elements were identified in the 200 nucleotide 5′NCR known as the tripartite leader, which is found on viral late mRNAs and is essential for viral translation. Structural and mutational studies of the tripartite leader showed that it contains an extensive unstructured 5′ end, followed by a group of stable hairpin structures that form large single-stranded loops (Zhang et al. 1989; Dolph et al. 1990). Several of the hairpin structures possess remarkable complementarities to the 3′ end of 18S rRNA. Whether these sequences function to bind 18S rRNA or act structurally to promote ribosome shunting is unresolved (Yueh and Schneider 2000). In uninfected cells, the tripartite leader directs translation by conventional 5′ scanning of 40S ribosome subunits and by 40S ribosome shunting at roughly equal levels (Yueh and Schneider 1996). However, in late Ad-infected cells, the tripartite leader directs translation solely by ribosome shunting in a manner that is strongly stimulated by one or more late viral gene products (Yueh and Schneider 1996, 2000).
Ad mRNAs are capped and utilize the cap-initiation complex (eIF4F), which consists of a core group of three polypeptides. eIF4F contains a 24-kDa cap-binding protein (eIF4E), a 45-kDa ATP-dependent RNA helicase (eIF4A), and eIF4G, a large molecular adapter protein upon which the complex assembles at the cap (Hentze 1997). Also bound to eIF4G are either of the two eIF4E kinases known as Mnk1 and Mnk2 (Fukunaga and Hunter 1997; Waskiewicz et al. 1997, 1999; Pyronnet et al. 1999), which efficiently phosphorylate eIF4E in vivo only when both are bound to eIF4G (Pyronnet et al. 1999; Waskiewicz et al. 1999; Cuesta et al. 2000). eIF4E phosphorylation positively correlates with increased translation initiation in many, but not all systems (Wang et al. 1998; Fraser et al. 1999; Gingras et al. 1999) and its function remains unknown. eIF4G binds PABP (Tarun et al. 1997; Gradi et al. 1998), which promotes initiation and potentially circularizes capped and polyadenylated mRNAs (Wells et al. 1998), possibly providing surveil-lance for mRNA integrity.
Human adenovirus perturbs the cap-initiation complex to establish two novel translation programs in infected cells. The virus disrupts the cap-initiation complex, blocking host protein synthesis during the late phase of infection (during viral replication), while simultaneously usurping the cap-initiation complex for translation of its own mRNAs by ribosome shunting. Expression of one or more Ad late gene products is associated with both events (Zhang et al. 1994). The viral 100-kDa (100k) nonstructural protein is a nuclear and cytoplasmic protein that was shown to bind the C terminus of translation initiation factor eIF4G, at or near the site occupied by Mnk1. This implicates 100k protein in direct competitive displacement of Mnk1, which is associated with impaired translation of cellular mRNAs (Cuesta et al. 2000, 2004). Nuclear 100k protein is involved in morphogenesis of the viral particle (Oosterom-Dragon and Ginsberg 1980), whereas cytoplasmic 100k protein is associated with several distinct activities, including regulation of granzyme B (Andrade et al. 2001, 2003) and inhibition of cellular protein synthesis (for review, see Cuesta et al. 2001). The disparate activities of 100k protein represent a conundrum, made more puzzling by the early observation that it can be photo-UV cross-linked to both cellular and viral mRNAs in the cytoplasm, but not to other types of RNAs (Adam and Dreyfuss 1987).
To understand the molecular mechanism of ribosome shunting on Ad tripartite leader mRNAs, we sought to identify the viral gene product that strongly and selectively promotes it. Here, we identify the viral gene product as the Ad late 100k protein, and we describe its unusual molecular mechanism of action in facilitating ribosome shunting on Ad late mRNAs. We show that 100k protein selectively binds and recruits eIF4G and PABP to translationally active viral mRNAs through a simultaneous interaction with these factors and the viral tripartite leader 5′NCR. The interaction of 100k with both eIF4G and tripartite leader mRNAs in polysomes are shown to be critical to promote ribosome shunting. Thus, the inhibition of cellular mRNA translation and promotion of ribosome shunting on late Ad mRNAs typically occurs as an integrated translational program that is coordinated by selective 100k binding to late Ad mRNAs, selective binding to eIF4G, and selective recruitment of eIF4G and PABP to these mRNAs. A molecular mechanism for ribosome shunting is described, by which enhanced binding of eIF4G and PABP to 100k protein drives 40S ribosome subunit recruitment and utilization on tripartite leader mRNAs.
Results
Ad 100k protein specifically promotes ribosome shunting on viral late mRNAs
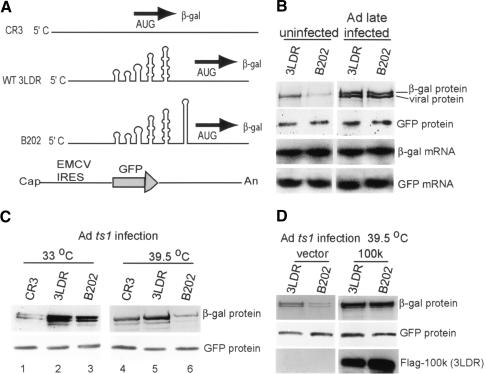
Earlier studies showed that late Ad gene expression strongly facilitates ribosome shunting directed by the tripartite leader (Yueh and Schneider 1996, 2000). To identify the effector of ribosome shunting, we utilized a stable hairpin structure (ΔG = -70 kcal/mole) inserted at the 3′ end of the tripartite leader (Fig. 1A, mutant B202) shown previously to block ribosome scanning and direct translation initiation solely by ribosome shunting (Yueh and Schneider 1996, 2000). The 293 cells were cotransfected with plasmids expressing reporter mRNAs encoding green fluorescent protein (GFP) translated by the EMCV IRES (pIRES-EGFP) and β-galactosidase (β-gal) translated by the wild-type or B202 tripartite leader. Cells were then mock infected or infected with wild-type Ad (dl309), labeled in late infection with [35S]methionine and β-gal recovered to measure translation rates by SDS-PAGE and autoradiography. GFP protein was detected by immunoblot analysis from the same amount of lysates. Reporter mRNA levels were assessed by Northern blot analysis and densitometry, and were found to be unchanged by Ad infection (Fig. 1B). In the absence of Ad infection, there was a threefold reduction in translation from the B202 mRNA when compared with the wild-type tripartite leader (3LDR), reflecting its inability to direct scanning-dependent initiation. During Ad late infection, translation of both the wild-type and the B202 mRNAs was enhanced by fivefold to 10-fold. The enhancement was due exclusively to ribosome shunting, as shown by the B202 mutant and determined previously (Yueh and Schneider 1996; Fig. 1B). Ad infection had no effect on translation of the GFP mRNA directed by the EMCV IRES. These data suggest that one or more late viral protein(s) strongly promote ribosome shunting on tripartite leader mRNAs during late Ad infection. Ad 100k is an abundant late viral protein that is expressed with the onset of the late phase of virus infection, which coincides with the initiation of viral DNA replication. The 100k protein was previously implicated by genetic analysis in Ad translational control (Hayes et al. 1990; Riley and Flint 1993; Zhang 1994). The Ad 100k ts1 mutant contains a temperature-sensitive (ts) mutation in 100k protein, which renders 100k protein nonfunctional at the restrictive temperature (39.5°C) for inhibition of cellular protein synthesis (Hayes et al. 1990; Cuesta et al. 2000). We therefore utilized the ts1 100k mutant to investigate whether 100k protein facilitates tripartite leader translation by ribosome shunting during the late phase of viral infection. The 293 cells were cotransfected with plasmids expressing GFP translated from the EMCV IRES and β-gal translated from the wild-type tripartite leader (3 LDR), the B202 leader mutant, or a 5′NCR that is strongly dependent on eIF4F (CR3; Feigenblum and Schneider 1996). CR3 is comprised of a 5′NCR with an extensive segment of a polylinker region to provide secondary structure and greater eIF4F dependence. Cells were then infected with Adts1 virus and labeled with [35S]methionine during late times of infection at 39.5°C (restrictive temperature) or 33°C (nonrestrictive temperature). Translation of β-gal from eIF4F-dependent CR3 mRNA was impaired fourfold to fivefold at 33°C, but not at 39.5°C, as compared with the wild-type tripartite leader (Fig. 1C, lanes 1,4). Translation of the ribosome shunting-dependent B202 mutant reporter mRNA was reduced by fivefold to 10-fold at the restrictive temperature for 100k protein (39.5°C) with only a threefold decrease for the 3LDR, which can translate by ribosome scanning in the absence of 100k protein (Yueh and Schneider 1996, 2000; Fig. 1C, cf. lanes 2,3 and 5,6). The strong stimulation by 100k protein of translation on the B202 mRNA indicates an ability to promote ribosome shunting. There was no change in EMCV IRES-directed reporter GFP levels (Fig. 1C), and reporter mRNA levels at the different temperatures were unchanged (data not shown). To confirm that the enhanced translation by ribosome shunting at 33°C was mediated by 100k protein and not by some other event of Ad infection, a complementation analysis was carried out to determine whether overexpression of wild-type 100k protein rescues ribosome shunting at the restrictive temperature (39.5°C) in cells infected by Adts1 virus. The 293 cells were cotransfected at 39.5°C with plasmids expressing GFP (EMCV IRES) and β-gal containing the wild-type or B202 tripartite leader, and Flag-100k protein (also translated by the tripartite leader) or empty vector. Cells were then infected with Adts1 virus and labeled with [35S]methionine, β-gal was recovered from equal amounts of lysates by immunoprecipitation, and translation activity was determined by SDS-PAGE and fluorography. At 39.5°C in the absence of wild-type 100k protein, translation of the mutant tripartite leader mRNA (B202) was reduced by approximately fivefold compared with the wild-type tripartite leader, but was restored to wild-type levels by expression of 100k protein (Fig. 1D). Overexpression of ts1 100k in trans instead of wild-type 100k protein did not recover translation (data not shown). There was no change in EMCV IRES-driven GFP levels under any conditions. These data confirm that 100k protein facilitates ribosome shunting on mRNAs containing the Ad tripartite leader.
Figure 1.
Ad late 100k protein specifically facilitates translation by ribosome shunting on the tripartite leader. (A) Diagram of 5′NCR elements used in this study. Shown are the CR3 5′NCR (strongly eIF4F dependent), the wild-type tripartite leader (wt 3LDR), the B202 tripartite leader that translates exclusively by ribosome shunting, and the EMCV IRES-GFP construct. (B) The 293 cells were cotransfected with pIRES-EGFP and plasmids expressing β-galactosidase (β-gal) mRNA containing 3LDR or B202 mutant 5′NCRs. At 18 h posttransfection, cells were mock infected or infected with Addl309 for 30 h, labeled with [35S]methionine for 30 min, β-gal was recovered by immunoprecipitation, resolved by SDS-PAGE and fluorographed. GFP protein was detected by immunoblot using anti-GFP antibodies. mRNA levels were analyzed by Northern blot. A contaminating viral protein is indicated. (C) Cells were transfected as above and infected with 100k mutant Adts1 at 39.5°C (restrictive temperature) for 30 h (late infection), or at 33°C (nonrestrictive temperature) for 60 h (late infection), labeled, and proteins processed as above. (D) The 293 cells were transfected with wild-type or B202 tripartite leader reporters as above, with or without cotransfection of Flag-100k plasmid (controlled by the tripartite leader), infected with Adts1 at 39.5°C for 30 h, and proteins analyzed as described above. Autoradiograms were quantified by densitometry, and typical results of at least three independent experiments were used to calculate mean values and standard deviations reported in the text.
100k protein is sufficient to specifically promote ribosome shunting on the Ad tripartite leader
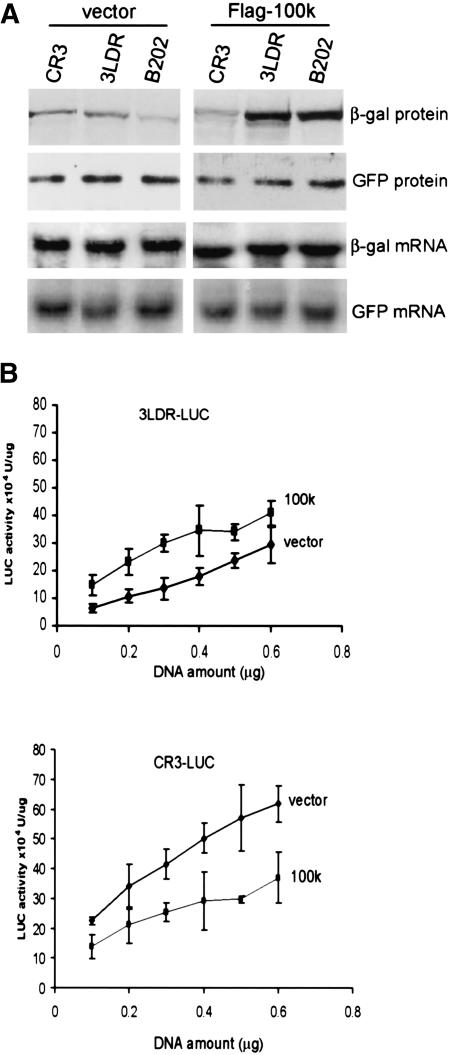
We next investigated whether 100k protein is sufficient to facilitate ribosome shunting on Ad mRNAs, or whether it requires the participation of other late viral gene products. Reporter mRNAs were cotransfected into cells with Flag-100k protein or Flag (empty) vector. Cells were labeled with [35S]methionine for analysis of reporter mRNA translation rates, mRNAs were isolated for Northern blot analysis, and β-gal was recovered by immunoprecipitation (Fig. 2A). Coexpression of Ad 100k protein stimulated tripartite leader specific β-gal translation by fivefold to 10-fold, which resulted entirely from increased ribosome shunting, as shown by an equivalent increase in translation of the B202 mRNA. Importantly, expression of 100k protein strongly decreased the eIF4F dependence of CR3 mRNA translation, but had no effect on that of the EMCV IRES-GFP mRNA, consistent with its role in shutoff of host cell eIF4F-dependent mRNA translation (Cuesta et al. 2000). To exclude that 100k protein promotes ribosome shunting, only on the β-gal tripartite leader mRNA, another set of tripartite leader reporter mRNAs containing luciferase, were examined (Fig. 2B). The 293 cells were cotransfected as above with vectors expressing Flag-100k protein and luciferase reporters controlled by the wild-type tripartite leader (3LDR) or the eIF4F-dependent 5′NCR (CR3). Luciferase activity was measured and normalized to mRNA levels as determined by Northern blot analysis (data not shown). Approximately threefold to fourfold higher levels of tripartite leader-specific translation was observed with expression of 100k protein over a sixfold range in concentration of transfected reporter plasmid. eIF4F-dependent mRNA translation (CR3) decreased by threefold when 100k was expressed (Fig. 2B). The more modest stimulation of tripartite leader-luciferase translation by 100k probably reflects the measurement of steady-state translation levels rather than translation rates, in addition to the fact that luciferase protein is less stable. Because 100k protein actually inhibits scanning-dependent translation on the wild-type tripartite leader, these data indicate that independent of the reporter, 100k protein is sufficient to specifically facilitate ribosome shunting on tripartite leader mRNAs.
Figure 2.
Ad 100K protein is sufficient to facilitate ribosome shunting on tripartite leader mRNAs. The 293 cells were cotransfected with plasmids expressing β-gal mRNA containing wild-type (3LDR) or shunting-only (B202) Ad tripartite leader 5′ UTRs or a 5′ UTR (CR3) that is strongly dependent on eIF4F, with Flag, or Flag-100k plasmids (using the tripartite leader) and pIRES-EGFP. (A) At 36 h posttransfection, β-gal mRNA levels were determined by Northern blot hybridization, cells were labeled with [35S]methionine, normalized to GFP and mRNA levels, and β-gal levels determined by immunoprecipitation, SDS-PAGE, and fluorography. (B) The 293 cells were cotransfected with plasmid pIRES-EGFP, vector alone (pFLAG-CM2), or a Flag-100k expression vector (pTL-FLAG100), and increasing amounts of plasmids pCMV-Ad LUC (3LDR) or pCMV-LUC (CR3). At 36 h posttransfection, luciferase activity was measured and normalized to mRNA levels as determined by Northern blotting. Data were quantified as described in the legend for Figure 1.
100k-eIF4G interaction is necessary, but not sufficient to promote ribosome shunting
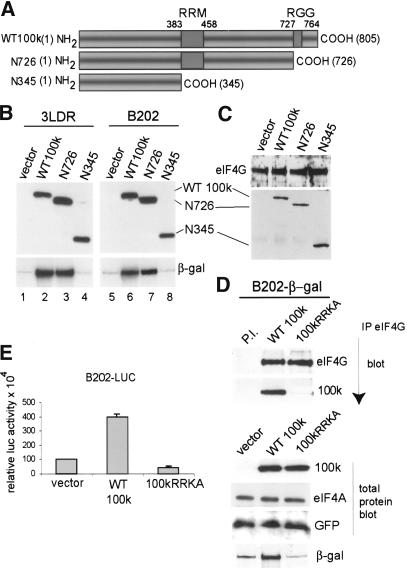
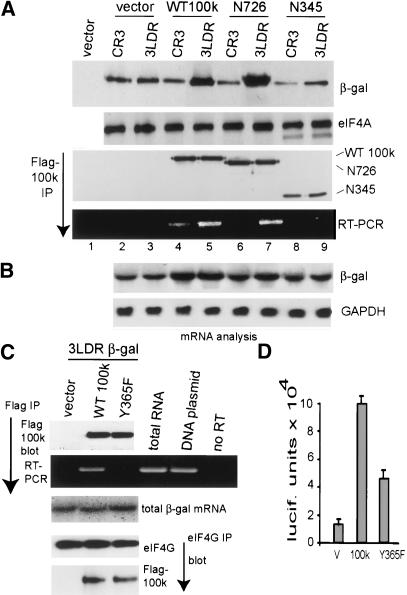
The direct interaction of 100k protein with eIF4G (Cuesta et al. 2000, 2004) implicates eIF4G in 100k protein-directed ribosome shunting. To determine whether eIF4G association with 100k is required and/or sufficient for 100k protein to promote ribosome shunting, we used two 100k protein mutants (100k N345 and 100k N726). These mutants retain eIF4G association, but are C-terminally truncated, progressively deleting at least two distinct RNA-binding domains (Fig. 3A). 293 cells were transfected with vectors expressing wild-type or mutant 100k proteins and plasmids expressing β-gal directed by the wild-type 3LDR or mutant B202 tripartite leader. Wild-type and mutant 100k proteins were expressed at similar levels (Fig. 3B, top). Wild-type 100k protein enhanced ribosome shunting by ∼10-fold on both wild-type and B202 mRNAs (Fig. 3B, bottom, lanes 2,6). The 100k N726 mutant also enhanced ribosome shunting by ∼10-fold on tripartite leader mRNAs (Fig. 3B, lanes 3,7), whereas the 100k N345 mutant had no effect (Fig. 3B, lanes 4,8). The 100k mutants bound eIF4G, as established in cells transfected with Flag-tagged wild-type or mutant 100k proteins (Fig. 3C). Immunoprecipitation of endogenous eIF4G, and immunoblot analysis, showed strong binding with wild-type Flag-100k, as well as with both mutant 100k proteins. These data indicate that 100k protein binding to eIF4G is not sufficient to promote ribosome shunting.
Figure 3.
The 100k protein interaction with eIF4G is necessary but not sufficient for ribosome shunting. (A) Diagrammatic representation of wild-type and mutant 100k proteins used in this study. The general RNA-binding domain (RGG box) and the tripartite leader RNA-recognition motif (RRM) are shown. Numbers refer to amino acid positions of 100k protein. (B) The 293 cells were transfected with vectors expressing wild-type Flag-100k protein, Flag-100k N345 mutant protein, or Flag-100k N726 mutant proteins and β-gal mRNAs containing either the wild-type Ad tripartite leader (3LDR) or tripartite leader mutant B202. At 36 h posttransfection, cells were labeled with [35S]methionine, β-gal protein was recovered by immunoprecipitation, resolved by SDS-PAGE, and fluorographed. Wild-type 100k and mutant 100k proteins were detected by immunoblotting with anti-Flag antibody. (C) The 293 cells were cotransfected with vector alone or wild-type Flag-100k protein, Flag-100k N345 mutant protein, or Flag-100k N726 mutant protein expression vector. Endogenous eIF4GI was recovered from equal amounts of lysate by immunoprecipitation, and the associated wild-type or mutant 100k proteins were detected by immunoblotting with anti-Flag antibody. (D) The 293 cells were cotransfected with pIRES-EGFP and plasmids expressing Flag-tagged wild-type (WT) or mutant 100k RRKA protein and β-gal mRNA. (Top) Immunoprecipitation of endogenous eIF4G and immunoblot of eIF4G and associated Flag-100k proteins, or preimmune [PI] sera. (Bottom) Immunoblot analysis of levels of proteins in total cell lysates are shown for Flag-100k proteins and controls (eIF4A and GFP). (β-gal) Cells were labeled with [35S]methionine for 30 min and β-gal translation rates were determined by immunoprecipitation of β-gal protein (normalized to GFP and RNA levels) and SDS-PAGE/fluorography. (E) Cells were transfected with wild-type or 100kRRKA protein expression vectors expressing luciferase (luc) mRNA containing the B202 Ad tripartite leader. Luciferase activity was measured and translation rates were normalized to GFP protein and luciferase mRNA levels.
Previous studies have shown that both the mouse and human Mnk1 N-terminal polybasic region is required for eIF4G binding (Waskiewicz et al. 1999; Parra-Palau et al. 2003; Cuesta et al. 2004). We recently found that the N-terminal 325-331 amino acid segment of 100k protein is homologous to both the mouse and human Mnk1 N termini (Cuesta et al. 2004). Mutation of Ad 100k protein sequence RRK (amino acids 329-331) to AAA in a truncated form of 100k protein blocks its binding to eIF4G (Cuesta et al. 2004). We therefore constructed the RRK (329-331) to AAA mutation into the full-length 100k protein (100kRRKA), and tested its ability to bind endogenous eIF4GI in cells transfected with vectors expressing Flag-100k or Flag-100kRRKA proteins. The importance of 100k-eIF4G association in ribosome shunting was determined by including vectors expressing β-gal reporters directed by the B202 tripartite leader. Equal amounts of eIF4GI were recovered by immunoprecipitation, and eIF4G or Flag-100k proteins were detected by immunoblot analysis. The 100kRRKA protein bound very weakly to eIF4G compared with wild-type 100k protein (Fig. 3D, top). Expression levels for wild-type and 100kRRKA mutant proteins were the same, as shown by immunoblot analysis of total protein lysates and comparison with endogenous eIF4A and cotransfected GFP (Fig. 3D, bottom). Translation analysis of β-gal by [35S]methionine labeling showed that wild-type 100k protein promoted ribosome shunting on the B202 β-gal mRNA by approximately sixfold, whereas the 100kRRKA mutant was inactive for shunting (Fig. 3D, bottom). Similar results were obtained with the luciferase reporter (Fig. 3E, right). These data indicate that 100k protein association with eIF4G is necessary but not sufficient to facilitate ribosome shunting on tripartite leader mRNAs. Of particular note, the 100kRRKA mutant actually appears to function as a mild suppressor of tripartite leader-directed ribosome shunting. As shown later, it is necessary for 100k to bind to both eIF4G and the tripartite leader to promote ribosome shunting, which likely accounts for the suppressive effect of this mutant.
100k protein selectively recruits eIF4G/PABP to polysomes containing tripartite leader mRNAs
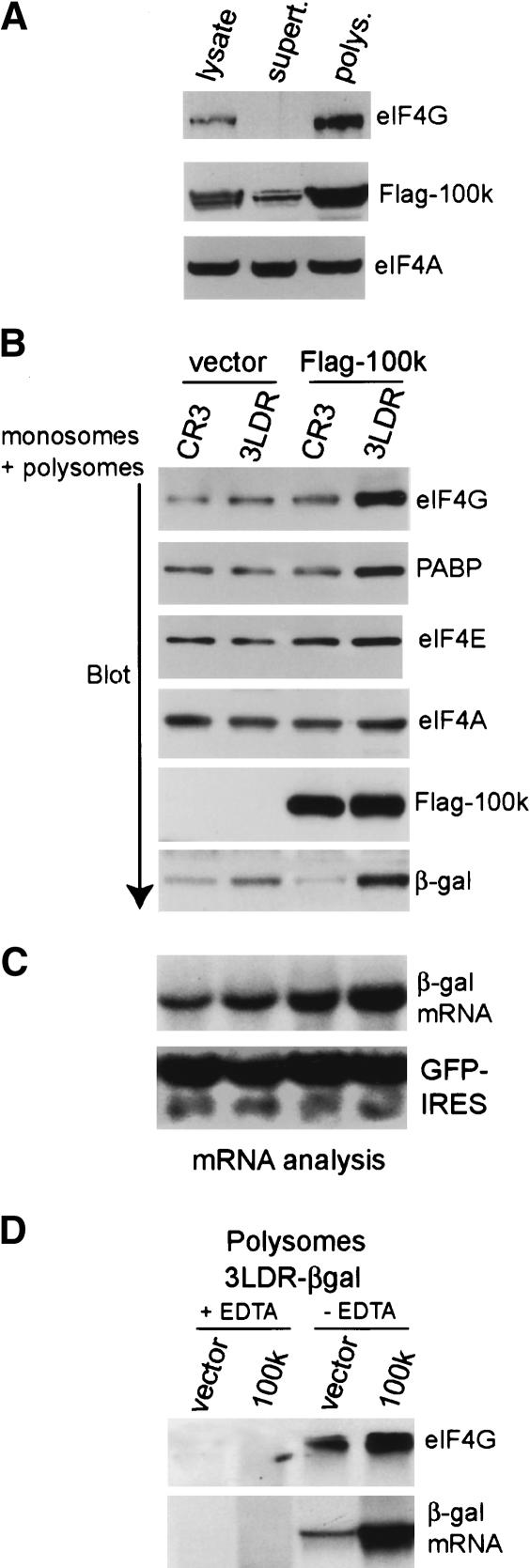
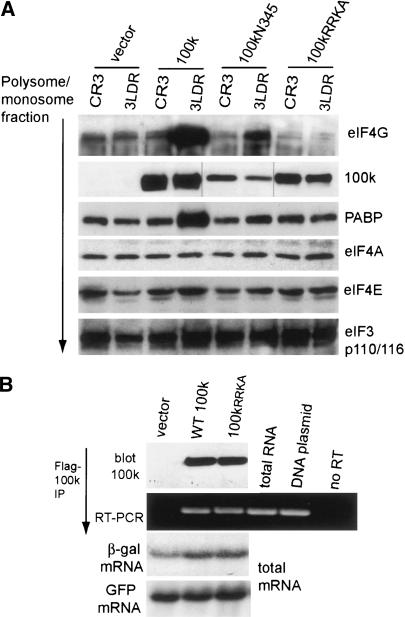
Previous studies showed that 100k protein associates with eIF4G (Cuesta et al. 2000) and can be cross-linked to both cellular and viral poly(A)+ RNAs (Adam and Dreyfuss 1987). Whether 100k binds more selectively or strongly to the tripartite leader was not addressed in these studies. We hypothesized that 100k protein might facilitate ribosome shunting on tripartite leader mRNAs by binding with higher affinity to the tripartite leader and by selectively recruiting certain initiation factors via its interaction with eIF4G. To explore this possibility, 293 cells were transfected with plasmids expressing Flag-100k protein or vector alone, and plasmids expressing β-gal controlled by the CR3 5′NCR or the tripartite leader. The polysome fraction was isolated, equal amounts of protein were resolved by SDS-PAGE, and associated translation initiation factors were identified by immunoblot analysis with specific antisera. Cytoplasmic 100k protein was found predominantly in the monosome/polysome fraction along with eIF4G, with little 100k in the postpolysome supernatant fraction (Fig. 4A). As shown in Figure 4B, in the absence of 100k protein, similar levels of eIF4G, eIF4E, eIF4A, PABP, and eIF3 (Fig. 7A [below] 110/116 kDa subunits) were found in the polysome/monosome fraction, regardless of the presence of different reporter mRNAs in polysomes (CR3 or Ad 3LDR). Surprisingly, in the presence of both 100k protein and the Ad 3LDR β-gal mRNA, there was a significant increase (fivefold to eightfold) of polysome-associated eIF4G and PABP, a small increase in eIF4E (less than twofold), and no change in eIF4A (Fig. 4B) or eIF3 (Fig. 7A, below) compared with polysomes containing the CR3 β-gal mRNA. The presence of 100k protein enhanced tripartite leader β-gal mRNA translation and decreased CR3 mRNA translation, as shown earlier. Northern blot analysis of polysomes, with and without coexpression of 100k protein (Fig. 4C) showed that in the absence of 100k, tripartite leader-β-gal mRNAs in polysomes were represented at levels similar to that of CR3 mRNAs. With expression of 100k protein, the abundance of CR3 mRNA in polysomes increased slightly (by approximately twofold), whereas the tripartite leader mRNA increased approximately fivefold. Notably, despite the slight increase of the CR3 (eIF4F-dependent) mRNA in polysomes, translation of the CR3 mRNA (and native cellular mRNAs) was strongly decreased. These data suggest that 100k protein blocks translation initiation on CR3 mRNAs at an early step that is subsequent to the interaction with a 40S ribosome subunit, as CR3 mRNAs were found at high levels in the monosome/polysome fraction, but were not translated. Earlier studies have excluded the possibility of a block in elongation (for review, see Cuesta et al. 2001). In addition, although it may seem surprising that a measurable increase can be detected in eIF4G and PABP associated with polysomal tripartite leader mRNA, it reflects the significant fraction of polysomal mRNA occupied by the Ad tripartite leader reporter as a result of 100k expression. Control studies were carried out in which the monosome/polysome fraction was prepared in the absence or presence of 25 mM EDTA, which disrupts ribosome-mRNA association (Fig. 4D). EDTA disruption of polysomes eliminated the 100k-directed increase in eIF4G in this fraction, as well as the presence of β-gal reporter mRNAs. The fact that neither eIF4A nor eIF3 are increased on tripartite leader mRNAs by 100k, whereas eIF4G is increased despite its ability to form a complex with these factors, suggests that eIF4G and PABP are likely targeted selectively by 100k protein. Thus, these data indicate that 100k protein preferentially recruits translation initiation factors eIF4G and PABP to polysomes containing Ad tripartite leader mRNAs.
Figure 4.
The 100k protein enhances eIF4G/PABP levels on polysomes containing Ad tripartite leader mRNAs. (A) The 293 cells were transfected with plasmids expressing β-gal mRNA containing the wild-type Ad tripartite leader 5′NCR (3LDR) or the eIF4F-dependent 5′NCR (CR3) and vector or plasmid expressing Flag-100k protein (pFlag-100k). At 36 h posttransfection, cells were fractionated and total lysate, monosome/polysome complexes, and postpolysome supernatants were isolated as described (Morley and Hershey 1990). (A) Equal amounts of total cell lysates, the monosome/polysome fraction, and the postpolysome supernatants were resolved by SDS-PAGE. Proteins were detected by immunoblot using specific antibodies as shown. (B) Equal amounts of monosome/polysome-associated proteins were resolved by SDS-PAGE. Proteins were detected by immunoblot analysis using specific antibodies as shown. (C) RNA was isolated from the monosome/polysome complexes isolated above and detected by Northern blot analysis. (D) Monosome/polysome fractions were prepared in the absence or presence of 25 mM EDTA to disrupt ribosome-mRNA interactions. Fractions were subjected to immunoblot analysis for eIF4G. Northern blot analysis was used to detect reporter β-gal mRNA.
Figure 7.
eIF4G and tripartite leader-specific binding are required for 100k recruitment of eIF4G and PABP into polysomes containing Ad tripartite leader mRNAs. (A) The 293 cells were transfected with plasmids expressing β-gal mRNA containing the wild-type tripartite leader (3LDR) or the eIF4F-dependent 5′NCR (CR3), and plasmids expressing wild-type or mutant 100k proteins (pFlag-100k, pFlag-100kN345, pFlag-100kRRKA). At 36 h posttransfection, monosome/polysomes complexes were isolated, equal amounts of polysome-associated proteins were resolved by SDS-PAGE, and detected by immunoblot analysis using specific antibodies as shown. (B) The 293 cells were transfected with vector alone, plasmids expressing wild-type Flag-100k, or the Flag-100kRRKA mutant, pIRES-EGFP, and tripartite leader β-gal mRNA. Flag-100k association with tripartite leader β-gal mRNA was determined by immunoprecipitation and quantitative RT-PCR. Total mRNA levels were determined by Northern blot analysis.
100k protein preferentially associates with viral tripartite leader mRNAs to promote ribosome shunting
Early studies found an interaction between 100k protein and mRNA (both cellular and Ad), but did not observe preferential binding to late Ad mRNAs (Adam and Dreyfuss 1987; Riley and Flint 1993). In agreement, we found that recombinant or purified 100k protein displays no preferential binding to tripartite leader or other RNAs (data not shown). However, these studies were either conducted using nonquantitative methods (in vivo photo-UV cross-linking) or with purified 100k protein that might alter the binding characteristics of the protein. Studies were therefore carried out to determine whether 100k in association with initiation factors in vivo (i.e., eIF4G) preferentially binds to viral tripartite leader mRNA. The 293 cells were cotransfected with vector alone or a plasmid expressing full-length Flag-100k protein, or truncated 100k protein mutants N726 or N345, and β-gal reporter mRNA containing the tripartite leader or the CR3 5′NCR. 100k proteins were recovered from equal amounts of cell lysates by immunoprecipitation with anti-Flag antibody under conditions that preserve its interaction with eIF4G (Cuesta et al. 2004), mRNA was extracted from the immunoprecipitated pellet and quantified by reverse transcription-quantitative polymerase chain reaction (RT-PCR). Control studies (Fig. 5B) demonstrated similar levels of CR3 and Ad 3LDR mRNAs in transfected cells (within twofold). Steady-state analysis of β-gal reporter mRNA translation by immunoblot of β-gal protein showed a strong increase in tripartite leader, but not CR3 mRNA translation, promoted by either wild-type 100k or the N726 mutant, but not by the N345 mutant. The CR3 mRNA bound to wild-type 100k-eIF4F complexes about threefold to fourfold less well than the tripartite leader (Fig. 5A). Importantly, the truncated N726 100k protein mutant that lacks the general RNA-binding RGG box, does not bind the CR3 mRNA. However, the N726 100k protein still bound the tripartite leader and promoted translation by ribosome shunting. The 100k protein mutant N345 did not stimulate translation by ribosome shunting and was not associated with either CR3 or tripartite leader mRNAs. These data indicate that 100k protein contains a tripartite leader-specific RNA-binding domain that may function to promote ribosome shunting. In addition, the inability of Adts1 virus to promote ribosome shunting on B202 mRNA at the restrictive temperature was complemented by both wild-type 100k protein and the 100k N726 mutant (Fig. 6), again indicating that the general RGG RNA-binding motif is not required for ribosome shunting.
Figure 5.
The 100k protein specifically associates with the viral tripartite leader mRNA in vivo. (A) The 293 cells were cotransfected with either vector alone, plasmids expressing Flag-100k protein, truncated 100k protein mutants N726 or N345, and plasmids expressing β-gal mRNA containing either the wild-type tripartite leader 5′NCR (3LDR) or an eIF4F-dependent 5′ NCR (CR3). Wild-type or mutant 100k proteins were recovered from equal amounts of lysate by immunoprecipitation with anti-Flag antibody, mRNA was extracted from the extensively washed immunoprecipitates and identified by quantitative RT-PCR and ethidium bromide agarose gel electrophoresis. Equal amounts of protein lysates were resolved by SDS-PAGE and immunoblotted with specific antisera as shown. (B) Total RNA was extracted from cells and analyzed by Northern blot hybridization analysis. (C) The 293 cells were transfected with vector alone, plasmids expressing wild-type Flag-100k protein, or 100k point mutant Y36F and tripartite leader β-gal mRNA. Flag-100k proteins were immunoprecipitated and analyzed by quantitative RT-PCR as above. eIF4G was immunoprecipitated and immunoblot analysis carried out for eIF4G and Flag-100k protein. Total β-gal mRNA was analyzed by Northern blot. (D) The 293 cells were cotransfected with a plasmid expressing the tripartite leader B202 β-gal mRNA and vector alone, wild-type Flag-100k, or the Y365F 100k mutant and luciferase activity determined. Standard deviations were calculated from at least three independent experiments.
Figure 6.
The 100k mutant N726 rescues the Adts1 virus to promote ribosome shunting at restrictive temperature. (A) The 293 cells were cotransfected with pIRESEGFP and plasmids expressing β-gal mRNA containing the B202 tripartite leader, Flag-tagged 100k protein or Flag-tagged 100kN726 protein (with the tripartite leader as the 5′NCR). At 18 h posttransfection, cells were infected with Adts1. After 30 h of infection at 39.5°C, cells were labeled with [35S]methionine, β-gal was recovered from equal amounts of lysate by immunoprecipitation, resolved by SDS-PAGE, and fluorographed. Autoradiograms were quantified by densitometry. Other proteins were detected by immunoblot analysis using specific antibodies as shown. Luciferase activity was determined from equal amounts of cell lysates and averaged for three independent experiments.
The N345 mutation represents a large truncation of 100k protein, which would alter activities other than RNA binding. We therefore conducted a point-mutation screen of 100k protein to introduce single site mutations that alter activity. Whereas analysis of this panel of mutants will be published elsewhere, one mutant contained a single site alteration at position Y365F (converted from tyrosine to phenylalanine) within the region that is thought to bind strongly to the tripartite leader. The Flag-100k Y365F mutant protein was expressed at the same levels in transfected cells as the wild-type protein, and bound eIF4G to the same extent, as shown by immunoprecipitation of endogenous eIF4G and immunoblot analysis (Fig. 5C, bottom). The ability of the Y365F 100k mutant to bind the tripartite leader β-gal mRNA was analyzed by immunoprecipitation of equal amounts of wild-type or mutant 100k proteins, followed by quantitative RT-PCR as described above. The Y365F 100k protein mutant bound the tripartite leader reporter mRNA approximately threefold less well than wild-type 100k protein (Fig. 5C, top). The ability of this mutant to promote ribosome shunting was assessed in cells cotransfected with the B202 tripartite leader luciferase reporter mRNA, which translates solely by ribosome shunting (Fig. 5D). Wild-type 100k stimulated ribosome shunting by fivefold to sixfold compared with vector control, whereas the Y365F mutant stimulated shunting by twofold. The reduced capacity to stimulate translation by ribosome shunting is similar to the reduced ability to bind the tripartite leader mRNA. These data therefore support the requirement for 100k interaction with tripartite leader mRNAs to promote ribosome shunting.
We next determined whether the ability to bind both eIF4G and the tripartite leader are critical for 100k recruitment of PABP and eIF4G to polysomes containing the Ad tripartite leader. The 293 cells were transfected with plasmids expressing β-gal directed by the tripartite leader or CR3 5′NCR, and wild-type or mutant 100k proteins (at a higher level than in Fig. 5). Monosome/polysome complexes were isolated, equal amounts of protein were resolved by SDS-PAGE, and associated translation initiation factors were identified by immunoblot analysis with specific antisera (Fig. 7A). Consistent with previous studies, the expression of 100k protein in the presence of tripartite leader mRNA enhanced the polysome accumulation of eIF4G and PABP. Importantly, 100k mutant (100kRRKA) failed to increase recruitment of eIF4G and PABP to tripartite leader containing polysomes. The 100k N345 mutant, which still binds eIF4G, slightly increased eIF4G and PABP levels in tripartite leader polysomes, consistent with its inability to specifically bind the tripartite leader. The 100kRRKA mutant binds the tripartite leader, but not eIF4G, and is associated with polysomal mRNAs, but cannot promote ribosome shunting. The ability of the 100kRRKA mutant to bind tripartite leader β-gal reporter mRNAs was identical to that of wild-type 100k, as shown by immunoprecipitation and RT-PCR analysis (Fig. 7B). These data therefore indicate that the ability of 100k protein to bind both eIF4G and the tripartite leader are critical for selective recruitment of eIF4G and PABP to tripartite leader mRNAs and for promotion of ribosome shunting.
Discussion
It was previously shown that Ad mRNA translation during late times of viral infection occurs exclusively by ribosome shunting, which is directed by the viral tripartite leader 5′NCR (Yueh and Schneider 1996). In this report, we characterized the molecular mechanism by which ribosome shunting occurs and demonstrate that the viral 100k protein is a potent effector that selectively promotes ribosome shunting on late viral mRNAs. The 100k protein was found to contain an RNA-binding element that interacts strongly with the tripartite leader and is essential for ribosome shunting. Another element that functions as a general RNA-binding domain in 100k protein (Cuesta et al. 2004) was found to be dispensable for ribosome shunting. An eIF4G-binding element in 100k protein, which is critical for inhibition of host-cell protein synthesis, was also found to be crucial for promoting ribosome shunting.
Prior to this investigation, only a very general understanding of ribosome shunting on Ad mRNAs was known. Ribosome shunting directed by the tripartite leader was previously known to involve loading of 40S ribosome subunits to the 5′ end of the mRNA and entry into the mRNA of only a short distance (∼40 nucleotides), possibly by limited 5′-to-3′ scanning, followed by direct translocation of 40S subunits to a downstream initiation codon, directed by cis-acting RNA shunting elements (Cuesta et al. 2001). The loading of 40S ribosomes onto the tripartite leader was also known to be a cap-dependent process (Yueh and Schneider 1996, 2000). Here, we have shown that 100k protein is largely in the cytosol in association with eIF4G, PABP, and polysome complexes, and is strongly enriched in polysomes (Fig. 4). Importantly, 100k protein was found to have the ability to strikingly increase and selectively recruit factors to polysomes containing tripartite leader mRNAs. The 100k protein increased the levels of eIF4G and PABP in polysomes containing tripartite leader mRNAs by fivefold to eightfold, and eIF4E by twofold to fourfold (Fig. 4B), to a level considerably greater than the increase in tripartite leader mRNA in polysomes. These data suggested that 100k protein promotes ribosome shunting by either selectively recruiting these factors to tripartite leader mRNAs, or that 100k/eIF4G complexes have greater affinity for tripartite leader mRNAs. The results of in vivo RNA-binding assays makes it clear that 100k protein contains a tripartite leader-specific RNA-binding domain within the middle region of the protein, which confers on 100k protein an ability to bind more specifically to late viral mRNAs (Fig. 5). The 100k protein also contains a general RNA-binding domain located in the C terminus of the protein, which likely directs its binding at some level to most mRNAs (Cuesta et al. 2004). It is possible that 100k interacts with all mRNAs via association with eIF4G and its general RNA-binding motif, but is more strongly retained on tripartite leader mRNAs through stronger or more specific RNA-100k protein interactions. In support of this possibility is the finding that although the 100k protein-eIF4G complex can bind both viral and cellular mRNAs, viral mRNAs appear to interact to an approximately fourfold higher level (Fig. 5).
How does 100k protein discriminate mRNAs from other RNAs in the cell? The 100k protein binds tightly to eIF4G (Cuesta et al. 2004) through a motif shared with the Mnk1 protein, resulting in competitive displacement of Mnk1 from eIF4G and shutoff of cellular protein synthesis. In fact, most cytoplasmic 100k protein was found in association with eIF4G and polysomes, even in the absence of tripartite leader mRNAs (Fig. 4). Thus, it is most likely that 100k protein gains access to mRNAs through its almost quantitative interaction with eIF4G, despite the fact that it possesses a RGG-type general RNA-binding motif. The modified eIF4F complex that contains 100k (Cuesta et al. 2000) associates somewhat preferentially with late viral (tripartite leader) mRNAs in polysomes (Fig. 4). Although the eIF4F/100k complex is nevertheless associated with both viral and cellular mRNAs, our results indicate that only late viral mRNAs can utilize this complex and can promote translation by ribosome shunting. The 100k interaction with eIF4G is therefore important for ribosome shunting, but it is not sufficient (Figs. 3, 4). A second key feature required for ribosome shunting is the ability of 100k protein to bind specifically to tripartite leader mRNAs. This was shown through the use of 100k mutants deleted in the general RGG-type RNA-binding motif and the tripartite leader specific RNA-binding motif, and a point mutant with reduced ability to bind tripartite leader mRNAs. Elimination of the general RNA binding motif did not impair ribosome shunting on the tripartite leader, whereas a more severe truncation of the protein that eliminated the tripartite leader-binding domain was fully impaired in shunting (Fig. 5). The 100k mutant lacking the tripartite leader-binding domain failed to recruit greater amounts of eIF4G and PABP to monosome/polysome complexes containing tripartite leader mRNAs compared with the wild-type 100k protein (Fig. 7). The reduced ability of a point mutant in 100k protein to bind tripartite leader mRNAs, in conjunction with its reduced ability to promote ribosome shunting, particularly underscores the importance of tripartite leader RNA interaction. However, we cannot exclude that other unknown activities of 100k protein, apart from tripartite leader RNA and eIF4G binding, might also be involved in promoting ribosome shunting.
There are a number of features in common between the better-studied CaMV ribosome shunt and that of Ad. The surprising degree of functional similarity between these two systems aids in understanding the mechanism of ribosome shunting. In both CaMV and Ad, capped mRNAs undergo ribosome shunting by binding the cap-initiation complex to load 40S ribosome subunits. Both 5′NCRs appear to undergo a 5′-to-3′ entry of 40S ribosome subunits, which for the Ad tripartite leader is fairly minor, involving the first 40-60 nucleotides, which may or may not constitute actual scanning (Yueh and Schneider 1996, 2000). In CaMV, a short ORF in the 5′NCR must be translated, and must be followed by a stable hairpin, ostensibly to accumulate 40S ribosome subunits for shunting. In Ad, there is no ORF in the 5′NCR, but there is a cluster of stable hairpins possessing a repeated complementarity to the 3′ end of 18S rRNA. It is thought that the 18S rRNA complementarity either binds directly to and captures 40S ribosomes or provides a conformation that is critical for an early event in shunting that is functionally equivalent (Yueh and Schneider 2000). Whether the regions of 18S rRNA complementarity are important for 100k protein stimulation of ribosome shunting is not known, but will be explored in future studies to better understand the mechanistic process. The 100k protein of Ad, by binding with high affinity to eIF4G and the tripartite leader, recruits high levels of PABP and 40S ribosome subunits, the latter probably via eIF3-eIF4F interactions directly to Ad mRNAs. Thus, 100k protein enhances the local abundance of 40S ribosomes on tripartite leader mRNAs, which then enter the shunting pathway through sequence-specific or structural interactions with the tripartite leader.
How does the increased concentration of eIF4G and PABP on tripartite leader mRNAs function to strongly enhance translation by ribosome shunting? An important conceptual distinction between translation initiation by ribosome scanning and ribosome shunting is that in the latter, the act of translation occurs as a consequence of direct translocation of the 40S ribosome from a “take-off” site in the 5′NCR to a “landing site” considerably downstream. Translation initiation is therefore uncoupled from the enzymatic events at the cap in which cap-initiation complexes are thought to unwind the mRNA and facilitate 40S ribosome subunits in search of a downstream initiation codon. However, our data suggests that for each 100k protein associated with the tripartite leader RNA, there is likely only one eIF4G, because the interaction was stoichiometric, and therefore likely only one 40S ribosome subunit that can be involved. Consequently, ribosome shunting on late Ad mRNAs is likely a distributive process in which each initiation event requires an eIF4F-100k complex and a 40S subunit. If the shunting initiation complex directs or accompanies the ribosome to the “landing site” at the downstream AUG, this would also explain the high level of PABP that might be involved in ribosome shunting. The poly(A) tail interaction with the shunting initiation complex would also have to be maintained to provide the signals that are necessary for translation initiation, as suggested for the 5′-to-3′ closed loop model (Wells et al. 1998; Gray et al. 2000). Thus, a modest increase in eIF4E would accompany much greater recruitment of eIF4G/100k complexes to Ad mRNAs, but a high level of eIF4G and PABP would be expected due to continuous interaction with 100k protein, which is essential to direct shunting of 40S subunits downstream and to stimulate translation initiation.
Materials and methods
Virus, antisera, and cells
Ad5dl309 is a phenotypically wild-type Ad5 strain with altered restriction enzyme sites (Jones and Shenk 1979). Ad5ts1 is a mutant that contains a temperature-sensitive mutation in 100k protein (Hayes et al. 1990). Virus stocks were grown and titers determined on 293 cells, a human embryonic kidney cell line transformed with the left 11% of Ad5. Infection of cells with viruses was typically carried out for 1 h with 25 plaque-forming units per cell (PFU/cell). Rabbit polyclonal antisera were raised against full-length recombinant human eIF4E protein, and against the C-terminal protein fragment of human eIF4GI (amino acids 1045-1560). Mouse monoclonal anti-human PABP antibody was provided by G. Dreyfuss (University of Pennsylvania, Philadelphia), and mouse monoclonal anti-rabbit eIF4A antibody was provided by W. Merrick (Case Western Reserve University, Cleveland, OH). Other antisera were from commercial sources and include rabbit polyclonal anti-β-galactosidase (β-gal) antibody (Cortex Biochem), rabbit polyclonal anti-GFP antibody (Molecular Probe), horseradish peroxidase (HRP)-conjugated donkey anti-rabbit or sheep anti-mouse secondary antibodies (Amersham), and mouse mononclonal anti-FLAG antibody (Sigma); enhanced chemiluminescence system (Amersham) was used for detection. The 293 cells were cultured in Dulbecco's modified Eagle medium (DMEM) containing 10% calf serum and 50 μg/mL gentamycin.
Plasmids
Plasmids pCMV-Ad, pCMV-Ad β-gal (Feigenblum and Schneider 1996), pFlag-100kRRKA (Cuesta et al. 2004), pHA-eIF4GI, and pFLAG-100kDa (Cuesta et al. 2000) were described previously. pFLAG-CM2 was from Sigma; pIRES-EGFP was from Clonetech. Plasmid pCMV-CR3 β-gal was generated by digestion of plasmid pCR3 β-gal (Feigenblum and Schneider 1996) with HindIII and SalI, and the fragment was inserted into a HindIII partially-digested and SalI-digested pCMV-Ad β-gal vector. pTLFLAG-CM2 was constructed as follows. The tripartite leader was amplified by PCR using the primers 5′-GCAGAGCTCTCTGGCTAACTAG-3′ and 5′-GCCGAGCTC CCGCTGGAAACTTGC-3′. The resultant PCR product was digested with SacI and subcloned into SacI-digested pFLAG-CM2. Inserting the 2.4-kb ClaI-SalI fragment from pFLAG-100k into the ClaI-SalI-digested pTL-FLAG-CM2 created plasmid pTL-FLAG-100k. Plasmid pTLFlagN726, pTLFlagN345, and pCMVAd LUC were constructed as follows. The C-terminal 726 amino acids of 100k gene DNA and the C-terminal 345 amino acids of 100k DNA were amplified by PCR using the primers 5′-GACTATCGATGGAGTCAGTCGAG-3′, 5′-CAGTGTCG ACTATCCGTGGGTGGCGG-3′ for N726, and primers 5′-GACTATCGATGGAGTCAGTCGAG-3′, 5′-CAGTGTCGAC TAGCACTCAAGCTCC-3′ for N345. The resultant PCR products were digested with ClaI and SalI and subcloned into ClaI-SalI-digested pTLFLAG-CM2. The full length of luciferase-coding region was amplified by PCR using the primers 5′-CGG TACTGTCGACAAATGG-3′ and 5′-CGCTAGATCTAGTTA CATTTTAC-3′. The resultant PCR product was digested with SalI-BglII and subcloned into SalI-BglII-digested pCMV-Ad. Inserting 5× BamHI hairpin into the SalI site of pCMV-AdLUC or pCMV-Ad β-gal created plasmids pCMV-Ad B202 LUC and pCMV-AdB202β-gal. The 100k mutant Y365F was constructed by introducing a point mutation in a fragment of 100k protein using PCR-directed mutagenesis, which was confirmed by DNA sequence analysis and reconstructed into the full-length Flag-100k gene.
Mammalian cell transfection and luciferase assay
The 293 cells were transfected with plasmid constructs using lipofectamine plus (Invitrogen) according to the manufacturer's protocols. Cells were harvested 36 h posttransfection and subjected to detergent lysis 0.5% NP40, 50 mM HEPES at pH 7.0, 250 mM NaCl, 2 mM EDTA, 2 mM sodium orthovanadate, 25 mM glycerophosphate, 1 tablet of protease inhibitor [Roche] per 10 mL) at 4°C for 20 min, nuclei were pelleted by centrifugation, and supernatant lysates were collected. Luciferase assays were performed as per the manufacturer's protocol (Promega).
Labeling of cells and analysis of polypeptides
Cells were labeled with 100 μCi of [35S]methionine (Amersham Pharmacia Biotech) per milliliter in DMEM without methionine for 1 h. Cells were lysed in 0.5% NP-40 lysis buffer (0.5% NP-40, 50 mM HEPES at pH 7.0, 250 mM NaCl, 2 mM EDTA, 2 mM sodium orthovanadate, 25 mM glycerophosphate, 1 tablet of protease inhibitor [Roche] per 10 mL) at 4°C and cleared of debris by centrifugation at 13,000 × g. Immunoprecipitation analysis was performed with normalized equal amounts of protein in cell extracts. Extracts were incubated with antisera to β-galactosidase (10 μg/mL) for 2 h at 4°C, and protein A-Sepharose (Santa Cruz Biotech.) was added to the lysates. After 1 h incubation at 4°C, the beads were recovered, washed four times with lysis buffer, and boiled in SDS sample buffer. Proteins were analyzed by SDS-PAGE and visualized by autoradiography.
Isolation of polysomes
Polysomes were isolated as described by Morley and Hershey (1990) with minor modifications. Cells were treated with 100 μg/mL of cycloheximide 5 min before harvesting. One 150-mm plate of 60% confluent-transfected 293 cells was scraped into 0.5 mL of lysis buffer (20 mM Hepes KOH at pH 7.2, 10 mM NaCl, 3 mM MgCl2, 0.5% NP40, 100 μg/mL cycloheximide, 200 U Rnasin [Promega], and 1 tablet protease inhibitor per 10 mL). After 15-min incubation on ice, lysates were transferred to a 1-mL dounce homogenizer on ice and cells lysed with 10 strokes. The nuclei were pelleted in a microcentrifuge at 3000 × g for 2 min. The supernatant was transferred to a fresh tube and 500 μg/mL heparin was added. Ribosome components were separated from the soluble fraction by centrifugation at 100,000 rpm (∼430,000 × g) for 25 min at 4°C using a TL-120.2 rotor in a Beckmann TL-120 ultracentrifuge. The ribosome pellet was resuspended in lysis buffer. The ribosome pellet and the postribosomal supernatant were resolved on SDS-10%PAGE. Control studies included 25 mM EDTA in the lysate and sucrose gradients.
Northern blotting
Transfected 293 cells were lysed directly into Trizol extraction reagent (Invitrogen). RNA samples were resolved by denaturing formaldehyde gel electrophoresis and transferred to a nylon membrane (Perkin Elmer Life Science, Inc). Membranes were probed overnight at 68°C with [α32P]dCTP probes directed to the β-gal and EGFP-coding regions. Following washes, the membrane was exposed to film for 1 h at -70°C.
In vivo RNA-binding assay
Wild-type 100k protein or mutant protein was recovered from equal amounts of lysate by immunoprecipitation with anti-Flag antibody. mRNAs were extracted from the immunoprecipitated protein using Trizol extraction reagent (Invitrogen). Semiquantitative RT-PCR amplification of immunoprecipitated β-gal mRNA was carried out as described previously (Sarkar et al. 2003).
Acknowledgments
Special thanks to G. Dreyfuss for antibodies as noted in the text. We thank lab members for critical review of the manuscript and helpful discussions. This work was supported by grant CA42357 to R.J.S. from the National Institutes of Health.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1212504.
References
- Adam S.A. and Dreyfuss, G. 1987. Adenovirus proteins associated with mRNA and hnRNA in infected Hela cells. J. Virol. 61: 3276-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade F., Bull, H.G., Thornberry, N.A., Ketner, G.W., Casciola-Rosen, L.A., and Rosen, A. 2001. Adenovirus L4-100K assembly protein is a granzyme B substrate that potently inhibits granzyme B-mediated cell death. Immunity 14: 751-761. [DOI] [PubMed] [Google Scholar]
- Andrade F., Casciola-Rosen, L.A., and Rosen, A. 2003. A novel domain in adenovirus L4-100K is required for stable binding and efficient inhibition of human granzyme B: Possible interaction with a species-specific exosite. Mol. Cell. Biol. 23: 6315-6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta R., Xi, Q., and Schneider, R.J. 2000. Adenovirus specific translation by selective disassembly of cap-initiation complex. EMBO J. 19: 3465-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2001. Preferential translation of adenovirus mRNAs in infected cells. Cold Spring Harb. Symp. Quant. Biol. 66: 259-267. [DOI] [PubMed] [Google Scholar]
- ____. 2004. Structural basis for competitive inhibition of eIF4G-Mnk1 interaction by adenovirus 100k protein. J. Virol. 78: 7707-7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran J. and Kolakofsky, D. 1988. Scanning independent ribosomal initiation of the Sendai virus X protein. EMBO J. 7: 2869-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Breyne S., Simonet, V., Pelet, T., and Curran, J. 2003. Identification of a cis-acting element required for shunt-mediated translational initiation of the Sendai virus Y proteins. Nucleic Acids Res. 31: 608-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitriev S.E., Terenin, I.M., Dunaevsky, Y.E., Merrick, W.C., and Shatsky, I.N. 2003. Assembly of 48S translation initiation complexes from purified components with mRNAs that have some base pairing within their 5′ untranslated regions. Mol. Cell. Biol. 23: 8925-8933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolph P.J., Huang, J., and Schneider, R.J. 1990. Translation by the adenovirus tripartite leader: Elements which determine independence from cap-binding protein complex. J. Virol. 64: 2669-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigenblum D. and Schneider, R.J. 1996. Cap-binding protein (eukaryotic initiation factor 4E) and 4E-inactivating protein BP-1 independently regulate cap-dependent translation. Mol. Cell. Biol. 16: 5450-5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser C.S., Pain, V.M., and Morley, S.J. 1999. Cellular stress in Xenopus kidney cells enhances the phosphorylation of eukaryotic translation initiation factor (eIF)4E and the association of eIF4F with poly(A)-binding protein. Biochem. J. 342 Pt 3: 519-526. [PMC free article] [PubMed] [Google Scholar]
- Fukunaga R. and Hunter, T. 1997. MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 16: 1921-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futterer J., Kiss-Laszlo, Z., and Hohn, T. 1993. Nonlinear ribosome migration on cauliflower mosaic virus 35S RNA. Cell 73: 789-802. [DOI] [PubMed] [Google Scholar]
- Gingras A.-C., Raught, B., and Sonenberg, N. 1999. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68: 913-963. [DOI] [PubMed] [Google Scholar]
- Gradi A., Imataka, H., Svitkin, Y.V., Rom, E., Raught, B., Morino, S., and Sonenberg, N. 1998. A novel functional human eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 18: 334-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray N.K., Coller, J.M., Dickson, K.S., and Wickens, M. 2000. Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J. 19: 4723-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes B.W., Telling, G.C., Myat, M.M., Williams, J.F., and Flint, S.J. 1990. The adenovirus L4 100 kilodalton protein is necessary for efficient translation of viral late mRNA species. J. Virol. 64: 2732-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmings-Mieszczak M., Hohn, T., and Preiss, T. 2000. Termination and peptide release at the upstream open reading frame are required for downstream translation on synthetic shunt-competent mRNA leaders. Mol. Cell. Biol. 20: 6212-6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze M.W. 1997. eIF4G: A multipurpose ribosome adapter? Science 275: 500-501. [DOI] [PubMed] [Google Scholar]
- Hershey J.W.B. and Merrick, W.C. 2000. Pathway and mechanism of initiation of protein synthesis. In Translational regulation of gene expression (eds. J.W.B. Hershey, M.B. Mathews, and N. Sonenberg), pp. 33-88. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Jackson R.J. 2000. A comparative view of initiation site selection mechanisms. In Translational regulation of gene expression (eds. J.W.B. Hershey, M.B. Mathews, and N. Sonenberg), pp. 127-183. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Jones N. and Shenk, T. 1979. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell 17: 683-689. [DOI] [PubMed] [Google Scholar]
- Latorre P., Kolakofsky, D., and Curran, J. 1998. Sendai virus Y proteins are initiated by a ribosomal shunt. Mol. Cell. Biol. 18: 5021-5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley S.J. and Hershey, J.W.B. 1990. A fractionated reticulocyte lysate retains high efficiency for protein synthesis. J. Biol. Chem. 72: 259-264. [DOI] [PubMed] [Google Scholar]
- Oosterom-Dragon E.A. and Ginsberg, H.S. 1980. Purification and preliminary immunological characterization of the type 5 adenovirus nonstructural 100,000 dalton protein. J. Virol. 33: 1203-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra-Palau J.L., Scheper, G.C., Wilson, M.L., and Proud, C.G. 2003. Features in the N and C termini of the MAPK-interacting kinase Mnk1 mediate its nucleocytoplasmic shuttling. J. Biol. Chem. 278: 44197-44204. [DOI] [PubMed] [Google Scholar]
- Pooggin M.M., Futterer, J., Skryabin, K.G., and Hohn, T. 2001. Ribosome shunt is essential for infectivity of cauliflower mosaic virus. Proc. Natl. Acad. Sci. 98: 886-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyronnet S., Imataka, H., Gingras, A.C., Fukunaga, R., Hunter, T., and Sonenberg, N. 1999. Human eukaryotic translation initiation factor 4G (eIF4G) recruits Mnk1 to phosphorylate eIF4E. EMBO J. 18: 270-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remm M., Remm, A., and Ustav, M. 1999. Human papilloma-virus type 18 E1 protein is translated from polycistronic mRNA by a discontinuous scanning mechanism. J. Virol. 73: 3062-3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley D. and Flint, S.J. 1993. RNA-binding properties of a translational activitor, the adenovirus L4 100-kilodalton protein. J. Virol. 67: 3586-3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers G.W., Edelman, G.M., and Mauro, V.P. 2004. Differential utilization of upstream AUGs in the β-secretase mRNA suggests that a shunting mechanism regulates translation. Proc. Natl. Acad. Sci. 101: 2794-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryabova L.A., Pooggin, M.M., Dominguez, D.I., and Hohn, T. 2000. Continuous and discontinuous ribosome scanning on the cauliflower mosaic virus 35 S RNA leader is controlled by short open reading frames. J. Biol. Chem. 275: 37278-37284. [DOI] [PubMed] [Google Scholar]
- Ryabova L.A., Pooggin, M.M., and Hohn, T. 2002. Viral strategies of translation initiation: Ribosomal shunt and reinitiation. Prog. Nucleic Acid Res. Mol. Biol. 72: 1-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar B., Xi, Q., Cheng He, and Schneider, R.J. 2003. Selective degradation of AU-rich mRNAs promoted by the p37 AUF1 protein isoform. Mol. Cell. Biol. 23: 6685-6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R.J. and Mohr, I. 2003. Translation initiation and viral tricks. Trends Biochem. Sci. 28: 130-136. [DOI] [PubMed] [Google Scholar]
- Stoneley M. and Willis, A.E. 2004. Cellular internal ribosome entry segments: Structures, trans-acting factors and regulation of gene expression. Oncogene 23: 3200-3207. [DOI] [PubMed] [Google Scholar]
- Tarun S.Z., Wells, S.E., and Sachs, A.B. 1997. Translation initiation factor eIF-4G mediates in vitro polyA tail dependent translation. Proc. Natl. Acad. Sci. 94: 9046-9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Flynn, A., Waskiewicz, A.J., Webb, B.L.J., Vries, R.G., Baines, I.A., Cooper, J.A., and Proud, C.G. 1998. The phosphorylation of eukaryotic initiation factor eIF-4E in response to phorbol esters, cell stresses and cytokines is mediated by distinct MAP kinase pathways. J. Biol. Chem. 273: 9373-9377. [DOI] [PubMed] [Google Scholar]
- Waskiewicz A.J., Flynn, A., Proud, C.G., and Cooper, J.A. 1997. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 16: 1909-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskiewicz A.J., Johnson, J.C., Penn, B., Mahalingham, M., Kimball, S.R., and Cooper, J.A. 1999. Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol. Cell. Biol. 19: 1871-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells S.E., Hillner, P.E., Vale, R.D., and Sachs, A.B. 1998. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 2: 135-140. [DOI] [PubMed] [Google Scholar]
- Yueh A. and Schneider, R.J. 1996. Selective translation by ribosome jumping in adenovirus infected and heat shocked cells. Genes & Dev. 10: 1557-1567. [DOI] [PubMed] [Google Scholar]
- ____. 2000. Translation by ribosome shunting on adenovirus and Hsp70 mRNAs facilitated by complementarity to 18S rRNA. Genes & Dev. 14: 414-421. [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Dolph, P.J., and Schneider, R.J. 1989. Secondary structure analysis of adenovirus tripartite leader. J. Biol. Chem. 264: 10679-10684. [PubMed] [Google Scholar]
- Zhang Y., Feigenblum, D., and Schneider, R.J. 1994. A late adenovirus factor induces eIF-4E dephosphorylation and inhibition of cell protein synthesis. J. Virol. 68: 7040-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]