Abstract
Messenger RNA decay mediated by the c-fos major protein coding-region determinant of instability (mCRD) is a useful system for studying translationally coupled mRNA turnover. Among the five mCRD-associated proteins identified previously, UNR was found to be an mCRD-binding protein and also a PABP-interacting protein. Interaction between UNR and PABP is necessary for the full destabilization function of the mCRD. By testing different classes of mammalian poly(A) nucleases, we identified CCR4 as a poly(A) nuclease involved in the mCRD-mediated rapid deadenylation in vivo and also associated with UNR. Blocking either translation initiation or elongation greatly impeded poly(A) shortening and mRNA decay mediated by the mCRD, demonstrating that the deadenylation step is coupled to ongoing translation of the message. These findings suggest a model in which the mCRD/UNR complex serves as a “landing/assembly” platform for formation of a deadenylation/decay mRNA-protein complex on an mCRD-containing transcript. The complex is dormant prior to translation. Accelerated deadenylation and decay of the transcript follows ribosome transit through the mCRD. This study provides new insights into a mechanism by which interplay between mRNA turnover and translation determines the lifespan of an mCRD-containing mRNA in the cytoplasm.
Keywords: CCR4, UNR, PABP, translational control, mRNA turnover, poly(A) nuclease
Regulation of mRNA turnover rate is an important control point, and the mRNA lifetimes in eukaryotic cells can vary over several orders of magnitude. These differences in mRNA stability provide a cell with flexibility in effecting a rapid change in mRNA abundance and thus in gene expression (for review, see Wilusz et al. 2001; Parker and Song 2004).
Messenger RNA (mRNA) decay mediated by the c-fos major protein-coding determinant of instability (mCRD) is unique in that it represents a “suicide” mechanism in which translation of the mCRD-containing mRNA results in rapid degradation of the message (Schiavi et al. 1994; Grosset et al. 2000). However, the participating trans-acting factors and underlying mechanism remain to be elucidated. Previous studies of c-fos mCRD-mediated decay have shown that (1) deadenylation precedes the decay of the RNA body (Shyu et al. 1991; Schiavi et al. 1994); (2) decay is tightly coupled to translation (Schiavi et al. 1994; Chen et al. 1995); (3) the mCRD RNA sequence per se, not the corresponding translated peptide, is recognized during the decay process (Wellington et al. 1993); (4) the minimal functional sequence of the mCRD is mapped to an 87-nucleotide (nt) purine-rich region (Grosset et al. 2000); and (5) the mRNA-destabilizing function of mCRD depends on its distance from the poly(A) tail (Grosset et al. 2000). Five mCRD-associated proteins were identified recently (Grosset et al. 2000): UNR, a cold-shock domain (CSD)-containing RNA-binding protein involved in internal ribosomal entry site (IRES)-mediated translation initiation (Hunt et al. 1999; Boussadia et al. 2003); PABP, the major cytoplasmic poly(A)-binding protein (Mangus et al. 2003); Paip1, a PABP-interacting protein that can enhance translation initiation (Craig et al. 1998); hnRNP D, an AU-rich element binding protein with multiple roles in mRNA turnover (Shyu and Wilkinson 2000); and NSAP1 (Harris et al. 1999), also termed hnRNP Q, suggested to play a role in nuclear pre-mRNA splicing (Mourelatos et al. 2001). Ectopic expression of individual NSAP1, Paip1, or UNR stabilized mCRD-containing mRNA by impeding deadenylation (Grosset et al. 2000).
In yeast and in mammalian cells, many RNA destabilizing elements found throughout the messages mediate mRNA decay by triggering removal of the 3′ poly(A) tail (Beelman and Parker 1995; Jacobson and Peltz 1996), which plays an important role in eukaryotic mRNA metabolism, affecting virtually every step in an mRNA's life in the nucleus and in the cytoplasm (for review, see Kahvejian et al. 2001; Mangus et al. 2003). The cytoplasmic actions of the poly(A) tail are mediated by PABP, which interacts with many proteins involved in translation initiation and termination (Hoshino et al. 1999; Kahvejian et al. 2001; Mangus et al. 2003). Although the 3′ poly(A) tail complexed with PABPs may fulfill its role in determining a transcript's fate through dynamic interactions with other cis-acting elements complexed with their cognate binding proteins, it remains unclear how individual RNA stability determinants, along with their cognate binding proteins, trigger rapid removal of the poly(A) tail. Recent studies using mammalian cytoplasmic extracts identified a poly(A)-specific nuclease, PARN, as a major nuclease activity responsible for the poly(A) shortening in vitro (Korner and Wahle 1997; Dehlin et al. 2000; Gao et al. 2000; Martinez et al. 2000). In addition, mammalian orthologs of yeast major poly(A) nucleases, CCR4 and PAN2, have been cloned, and their poly(A) nuclease activities have been characterized in vitro (Albert et al. 2000; Chen et al. 2002; Uchida et al. 2004). However, the in vivo roles of these various mammalian poly(A) nucleases in cytoplasmic mRNA turnover remain unclear.
In this study, we identify UNR as a critical factor in mCRD-mediated mRNA turnover. It functions both as an mCRD-binding protein and as a PABP-interacting protein. In vitro and in vivo evidence indicates a physical interaction between the poly(A)/PABP complex and the mCRD/UNR complex, and interference with this interaction diminishes the mCRD-destabilizing function. Moreover, we identify human CCR4 as a poly(A) nuclease involved in mCRD-mediated mRNA decay and show that it also associates with UNR. These findings support a model in which the mCRD/UNR complex serves as a “landing/assembly” platform for formation of a deadenylation/decay mRNP complex involving PABP and CCR4 poly(A) nuclease.
Results
Identification of c-fos mCRD-binding component in the mCRD-associated protein complex
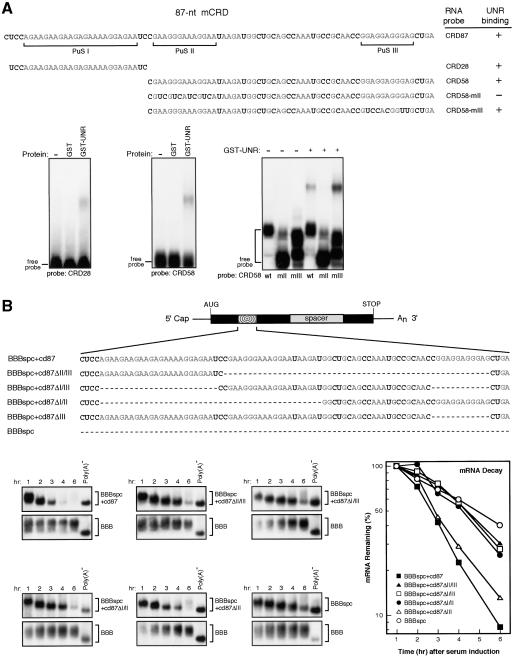
Sucrose-gradient fractionation and coimmunoprecipitation (co-IP) experiments suggested that the five mCRD-associated proteins form a multiprotein complex in vivo (Grosset et al. 2000). We first sought to identify which of these proteins directly bind mCRD. GST-tagged UNR, PABP, NSAP1, Paip1, and hnRNP D recombinant proteins were affinity-purified in a soluble form from Escherichia coli lysates (Fig. 1A, left). Their ability to bind the 87-nt minimal functional sequence of the c-fos mCRD (CRD87) was examined by gel electrophoretic mobility shift assay (GEMSA). As shown in Figure 1A, right, the [32P]-labeled CRD87 RNA was shifted by UNR and PABP and not by NSAP1, Paip1, hnRNP D, or the control protein, chloramphenicol acetyl transferase (CAT). Binding of both UNR and PABP to CRD87 was specific because there was clear competition for binding by UNR or PABP from nonlabeled CRD87 RNA but not from nonspecific RNA (Fig. 1B). We tested whether UNR also binds poly(A) as PABP does. UNR was unable to shift poly(A) RNA, whereas PABP resulted in a profound shift (Fig. 1C).
Figure 1.
Identification of UNR as the c-fos mCRD-binding protein in mCRD-associated protein complex. (A, left) The GST fusion proteins tested were shown by 10% SDS-PAGE and Coomassie blue staining. (Right) The ability of each mCRD-associated protein to bind c-fos mCRD was tested by gel electrophoretic mobility shift assay (GEMSA). (B) Specificity of binding of GST-UNR or GST-PABP to c-fos mCRD was demonstrated by competition GEMSA. (C) Binding ability of GST-UNR or GST-PABP to poly(A) RNA was tested by GEMSA. (D) Competition GEMSA showing that mCRD RNA cannot compete with poly(A) RNA for PABP binding. (E) Competition GEMSA showing that poly(A)25 RNA outcompetes mCRD RNA for PABP binding.
Because the poly(A) tails of cytoplasmic mRNAs are coated with PABPs in vivo, we then tested whether association of PABP with poly(A) of mRNA allows it to bind the mCRD. Nonlabeled poly(A) competed effectively with [32P]-labeled poly(A) for PABP binding, whereas CRD87 RNA had no effect, even at 1000-fold molar excess (Fig. 1D). Conversely, poly(A) easily out-competed [32P]-labeled CRD87 for PABP binding (Fig. 1E). Collectively, our data support the notion that PABP does not bind the mCRD while associating with poly(A), and that UNR, among the five mCRD-associated proteins, is the one that directly binds mCRD.
Functional characterization of UNR-binding motifs in c-fos mCRD
UNR is known to recognize purine-rich sequences (Triqueneaux et al. 1999), and the 87-nt mCRD contains three such stretches (Fig. 2A, PuS I-III). A [32P]-labeled RNA probe (CRD28) containing PuS I supported formation of an mRNP complex with purified GST-UNR (Fig. 2A). A probe (CRD58) containing the other two purine stretches also interacted with UNR (Fig. 2A). The UNR binding was reduced by decreasing the purine residues in PuS II (CRD58-mII), but similar mutation in PuS III (CRD58-mIII) did not have such effect (Fig. 2A), indicating that PuS I and PuS II contained the specific UNR-binding sites.
Figure 2.
UNR-binding motifs in the c-fos mCRD are involved in mRNA destabilization. (A) Mapping of UNR-binding motifs in the 87-nt minimal functional element of c-fos mCRD. (Top) Sequences of the 87-nt mCRD and its derivatives. Purine residues are shown in gray, and the three purine stretches (PuS I, PuS II, and PuS III) are indicated. (Bottom) Different portions of mCRD RNA were [32P]-labeled and tested for their ability to bind GST-UNR (0.5 pmole) by GEMSA. The results are summarized on the right of the top panel (+ indicates detection of band-shifted complexes; - indicates band-shifted complex is not readily detectable). (B, top) Physical map of β-globin mRNA carrying the 87-nt mCRD (BBBspc + cd87). Solid lines indicate 5′ and 3′ UTRs from β-globin mRNA. (Rectangle) Protein coding region from β-globin. (Radial box) The 87-nt c-fos mCRD. A 314-nt protein coding sequence from rat GAPDH (gray box-marked spacer) is referred to as “spc” in the names for hybrid mRNAs. (Bottom) In vivo functional tests of individual or combination of UNR-binding motifs in mRNA decay by Northern blot analysis. BBB mRNAs were expressed constitutively and served as an internal control. The times given at the top correspond to hours after serum stimulation. Poly(A)- RNA was prepared in vitro by treating RNA sample from early time point with oligo(dT) and RNase H. The amount of tested RNAs on Northern blots was quantitated by phosphorimaging, normalized to the amount of BBB RNA, and plotted semilogarithmically.
A series of in-frame deletion constructs were then derived from plasmid pBBBspc + cd87, which carries the sequences encoding the 87-nt minimal functional sequence of mCRD (Grosset et al. 2000). Time-course experiments (Fig. 2B) showed that β-globin mRNA carrying PuS I (BBBspc + cd87ΔII/III), PuS II (BBBspc + cd87ΔI/III), or PuS III (BBBspc + cd87ΔI/II) was stable, with a decay rate similar to that of the control (BBBspc, a β-globin mRNA derivative). However, mRNA carrying both PuS I and PuS II (BBBspc + cd87ΔIII) decayed rapidly, with a rate similar to the BBBspc + cd87 message (Fig. 2B). These findings indicate that none of the purine stretches alone is sufficient to destabilize the message and that the two UNR-binding sites, PuS I and PuS II, can work together to confer the mRNA destabilization capacity.
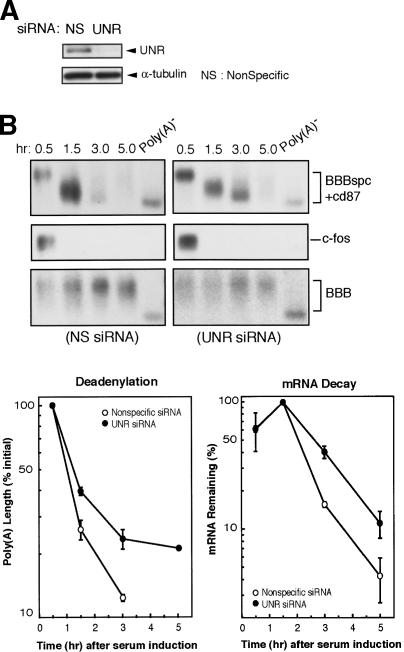
Knocking down UNR expression slows down the mRNA decay mediated by the c-fos mCRD
Decay of the BBBspc + cd87 message was monitored in NIH3T3 cells transfected with either UNR-specific or nonspecific siRNAs. As shown in Figure 3A, UNR protein expression was significantly decreased by UNR-specific siRNA, accompanied by a decreased rate of BBBspc + cd87 mRNA deadenylation and decay (Fig. 3B). By contrast, these effects were not observed in cells treated with the nonspecific siRNAs. Importantly, decreasing UNR expression also had no stabilizing effect on rapid decay of c-fos mRNA (Fig. 3B), which carries other destabilizing elements, for example, the AU-rich element in the 3′ untranslated region, besides the mCRD. This result indicated that the mRNA stabilization effect by knocking down UNR is specific to the c-fos mCRD. Taken together, these results show that UNR plays a role in the mRNA deadenylation and decay mediated by the c-fos mCRD.
Figure 3.
Knocking down UNR expression slows down the deadenylation and decay of c-fos mCRD-containing mRNA. (A) Western blots of whole-cell lysate showing knock-down of UNR protein expression by the specific (UNR) siRNA and not by the nonspecific (NS) siRNA. (B) Northern blots showing decay and deadenylation of BBBspc + cd87 mRNA isolated from NIH 3T3 cells transfected with either UNR-specific or nonspecific siRNA. BBB mRNA was expressed constitutively and served as an internal standard. Poly(A) lengths were compared by calculating from the difference in electrophoretic mobility between each message and cognate Poly(A)- RNA. Deadenylation curves were plotted as described previously (Shyu et al. 1991).
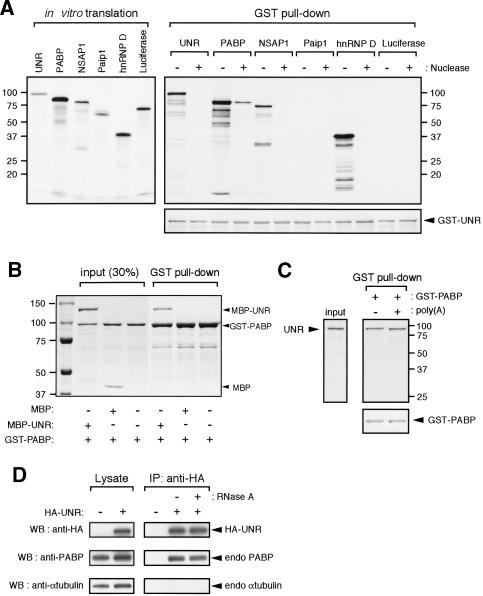
UNR is a new PABP-interacting protein
Knowing that UNR binds the mCRD and PABP associates with the poly(A) tail, we therefore addressed whether the UNR/mCRD complex and PABP/poly(A) complex connect with each other via direct UNR-PABP interaction or through association with other protein(s) in the multiprotein complex. In vitro GST pull-down assays were performed to evaluate the binding of [35S]methionine-labeled UNR, PABP, NSAP1, Paip1, and hnRNP D (Fig. 4A, left) to the GST-UNR fusion protein. Labeled luciferase served as a control. As shown in Figure 4A (right), without RNase A treatment, UNR exhibited interactions with itself, PABP, NSAP1, and hnRNP D, but not with Paip1 or luciferase. After RNase A treatment, only PABP interacted with UNR. Similar amounts of recombinant GST-UNR were used in each case for GST pull-down assays (Fig. 4A, right, bottom). To further substantiate the above findings, purified recombinant GST-PABP and maltose-binding protein (MBP)-UNR fusion proteins were mixed together and applied to glutathione column. The result (Fig. 4B) showed that significant MBP-UNR, but not MBP, was retained with GST-PABP on the column. Western blot analysis was performed and confirmed the identities of fusion proteins (data not shown). These experiments demonstrated that only PABP displays direct interaction with UNR in a RNA-independent manner.
Figure 4.
Identification and characterization of the PABP-binding activity of UNR. (A) GST pull-down assay examining the interactions between UNR and other mCRD-associated proteins. Each of the in vitro translated [35S]methionine-labeled proteins with (+) or without (-) RNase A treatment was incubated with immobilized GST-UNR on glutathione column. The bound protein complexes were eluted and analyzed by SDS-PAGE (10%) followed by Coomassie blue staining for GST-UNR (lower right) and then autoradiography for labeled proteins (top right). (Left) Coomassie blue-stained SDS-PAGE gel showing 20% of the in vitro translated proteins used for pull-down assay. (B) MBP-UNR but not MBP alone interacts with GST-PABP. Equal moles of GST-PABP and MBP-UNR fusion proteins were mixed together and applied to glutathione-column. Bound proteins were eluted from the column and analyzed by SDS-PAGE (7.5%) followed by Coomassie blue staining. (C) UNR can interact with PABP associated with poly(A). In vitro translated [35S]methionine-labeled UNR was incubated with GST-PABP immobilized glutathione-Sepharose that had been preincubated with (+) or without (-) poly(A) RNA as described in Materials and Methods Bound protein complexes were eluted and analyzed by SDS-PAGE (10%) followed by Coomassie blue staining for GST-PABP (lower right) and then autoradiography (top right) for labeled UNR. (Left) Twenty percent of the in vitro translated proteins used for pull-down assay. (D) Co-IP of UNR and PABP. COS7 cells were transfected with vector alone or the vector expressing HA-tagged UNR. Cytoplasmic lysates were prepared and incubated with anti-HA agarose in the absence (-) or presence (+) of RNase A. Following precipitation, bound proteins were analyzed by Western blotting using various antibodies as indicated. The lysate panel shows 6% of the input.
To determine if UNR is able to interact with PABP associated with poly(A) sequence, [35S]-labeled UNR was applied to a GST-PABP column that was presaturated with poly(A). Similar amounts of UNR were pulled down by GST-PABP in the presence and absence of poly(A) (Fig. 4C), indicating that association of PABP with poly(A) does not disrupt its interaction with UNR.
Co-IP/Western blot analyses were performed to determine if interaction between UNR and PABP exists in vivo. Cytoplasmic extracts prepared from COS7 cells expressing HA-epitope tagged UNR were immunoprecipitated with anti-HA antibody. Western blot analysis of the immunoprecipitates showed that endogenous PABP, and not endogenous α-tubulin, coprecipitated with HA-UNR (Fig. 4D). Importantly, RNase A treatment did not abolish the co-IP of UNR and PABP (Fig. 4D). These results establish UNR as a PABP-interacting protein and raise the possibility that the PABP/poly(A) complex and the UNR/mCRD complex are brought together through a direct interaction between UNR and PABP.
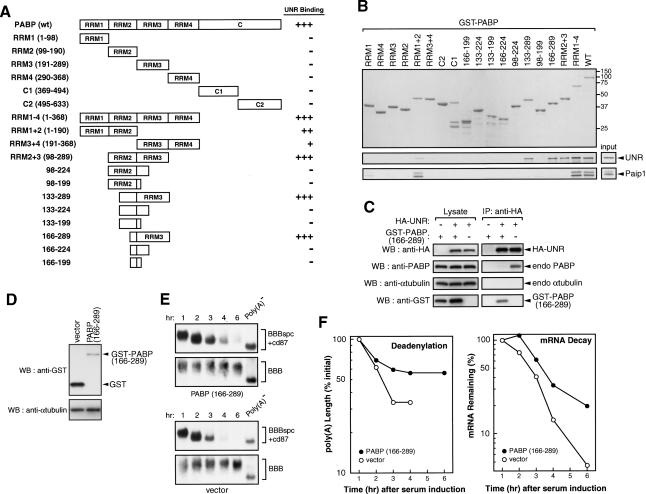
UNR-PABP interaction is required for full destabilization function of the c-fos mCRD
To address the functional significance of the UNR-PABP interaction in the mCRD-mediated RNA decay, we first used GST pull-down analysis to map the interaction site(s) of UNR on PABP. GST-PABP and a series of truncated derivatives (Fig. 5A) were purified and used in glutathione affinity chromatography to test their ability to retain [35S]-labeled UNR. The results (Fig. 5A) localized the UNR-binding site to the end of RRM2 and entire RRM3 of PABP (amino acids 166-289). As a control, Paip1-binding site was mapped to RRM1 and RRM2 (RRM1 + RRM2; Fig. 5B, bottom), consistent with the previous report (Roy et al. 2002). The PABP region recognized by UNR is distinct from the strong poly(A)-binding domains (RRM1 + RRM2; Kuhn and Pieler 1996; Deardorff and Sachs 1997; Mangus et al. 2003), substantiating the observation that UNR interaction with PABP is not impaired by poly(A) association with PABP (Fig. 4C).
Figure 5.
Identification and functional analysis of UNR-binding sites in PABP. (A) Schematic diagram of GST-PABP and its derivatives. +++, ++; and + indicate relative levels of labeled proteins pulled down, and - denotes not detectable. (B) GST pull-down assay mapping UNR-binding sites in PABP. GST-PABP or its derivatives (Imataka et al. 1998; Khaleghpour et al. 2001) was immobilized on glutathione-Sepharose beads. [35S]-methionine-labeled UNR or Paip1 was incubated with GST-tagged PABP, or its derivative proteins immobilized glutathione-Sepharose as indicated. The bound protein complexes were eluted and analyzed by SDS-PAGE (11%) followed by Coomassie blue staining (top) for GST-fusion proteins and then autoradiography (bottom) for labeled proteins. Twenty percent of the in vitro translated proteins used for pull-down assay were shown on the bottom right. The results of UNR binding to PABP and its derivatives are summarized in A. (C) Co-IP experiments showing that overexpression of PABP(166-289) interferes with the interaction between UNR and endogenous PABP. HA-UNR and GST-PABP (166-289) are expressed individually or in combination as indicated (+ indicates present in lysate; - indicates absent in lysate). (D) Western blot analysis showing the expression of GST-tagged PABP (166-289) in the cytoplasm of NIH 3T3 cells. The blot was probed with an antibody against the GST-tag and a control antibody against α-tubulin. (E) Northern blot analysis showing deadenylation and decay of BBBspc + cd87 mRNA in the absence (vector) or presence of ectopically expressed PABP (166-289). BBB mRNA was served as an internal standard. Poly(A)- RNA preparation was carried out as described in the legend for Figure 2. (F) Comparison of deadenylation (top) and decay (bottom) kinetics of BBBspc + cd87 mRNA in the absence (open circle) or presence (filled circle) of ectopically expressed PABP (166-289). Data quantitation and plotting were as described in the legend for Figure 3.
We then asked whether a competition for interacting with UNR exists in intact cells between PABP and the PABP (166-289). The open reading frame (ORF) of GSTPABP (166-289) was subcloned into a mammalian expression vector and was transfected into COS7 cells. Ectopic expression of HA-UNR led to co-IP of endogenous PABP in a RNA-independent manner, as the experiment was performed by using RNase A-treated lysates (Fig. 5C). However, coexpression of GST-PABP (166-289) along with HA-UNR essentially eliminated the co-IP of endogenous PABP with HA-UNR; instead, a significant amount of GST-PABP (166-289) protein was coimmunoprecipitated. Taken together, these experiments further substantiated the findings that UNR is a PABP-interacting protein and amino acids 166-289 of PABP is sufficient to support association with UNR.
The functional significance of UNR-PABP interaction in the mCRD-mediated mRNA decay was tested by monitoring the decay of BBBspc + cd87 mRNA in cells expressing GST-PABP (166-289) or GST alone. Although the expression level of PABP(166-289) protein was modest in NIH3T3 cells (Fig. 5D), it slowed down BBBspc + cd87 mRNA decay and also reduced the extent of poly(A) shortening (Fig. 5E,F). By contrast, robust expression of recombinant GST (Fig. 5D) did not affect mRNA decay (Fig. 5E,F). These results are consistent with the notion that in vivo UNR-PABP interaction plays a role in the mCRD-mediated mRNA turnover.
CCR4 poly(A) nuclease is involved in the c-fos mCRD-mediated mRNA turnover
We then set out to identify the poly(A) nuclease(s) involved in mCRD-mediated mRNA decay and examine their interaction with UNR. In various reconstituted decay systems, PARN, a mammalian poly(A) nuclease not found in yeast, has been shown to account for the major poly(A) nuclease activity in vitro (see Korner and Wahle 1997; Dehlin et al. 2000; Gao et al. 2000; Martinez et al. 2000). Recently, human orthologs of major yeast poly(A) nuclease complexes, CCR4/CAF1 and PAN2/3, have been cloned and their poly(A) nuclease activities have been characterized (Albert et al. 2000; Chen et al. 2002; Baggs and Green 2003; Uchida et al. 2004). Although CCR4 belongs to the super-family of exoIII nuclease, PARN and PAN2 are members of the DEDD nuclease super-family (Dupressoir et al. 2001; Zuo and Deutscher 2001). Importantly, catalytically inactive mutants that remain able to bind their substrates have been described for members of these two super-families, including PARN (Ren et al. 2002; Baggs and Green 2003; Uchida et al. 2004). We took the dominant-negative approach of interfering with individual endogenous poly(A) nuclease activity by ectopically overexpressing catalytically inactive mutants of PARN, CCR4, or PAN2 in NIH3T3 cells, and the decay of BBBspc + cd87 mRNA was then monitored.
The results of time-course experiments (Fig. 6A) showed that ectopic expression of mutant CCR4, but not mutant PARN or mutant PAN2, impeded rapid deadenylation and decay of BBBspc + cd87 mRNA. Moreover, although ectopic expression of wild-type PARN or PAN2 had little effect on deadenylation kinetics of BBBspc + cd87 mRNA, overexpression of wild-type CCR4 increased the rate and extent of deadenylation of the same message (Fig. 7A). Western blot analysis of cytoplasmic lysates showed that all recombinant poly(A) nucleases were expressed at readily detectable levels (Fig. 6B). It is worth noting that the level of CCR4 expression in NIH3T3 cells is the least among the three poly(A) nucleases. We conclude that CCR4 is involved in mediating the rapid deadenylation step directed by the mCRD.
Figure 6.
Identification of a poly(A) nuclease involved in mCRD-directed mRNA turnover. (A) Northern blots showing mRNA deadenylation and decay of the BBBspc + cd87 mRNA in absence (vector) or presence of ectopically expressed poly(A) nucleases, CCR4, CCR4-mt, PAN2, PAN2-mt, PARN, and PARN-mt. Corresponding deadenylation rate of the BBBspc + cd87 mRNA in each case was plotted. Plates of cells for the same time-course experiment were mixed and reseeded 10 h after transfection, which allows endogenous GAPDH mRNA to be served as a control for sample handling and loading. (B) Western blot analysis of cytoplasmic lysates showing that all ectopically expressed poly(A) nucleases can be readily detected by anti-HA (for CCR4 and PAN2) or anti-V5 tag (for PARN) antibody. (C) Immunoprecipitation experiment showing co-IP of UNR and CCR4.
Figure 7.
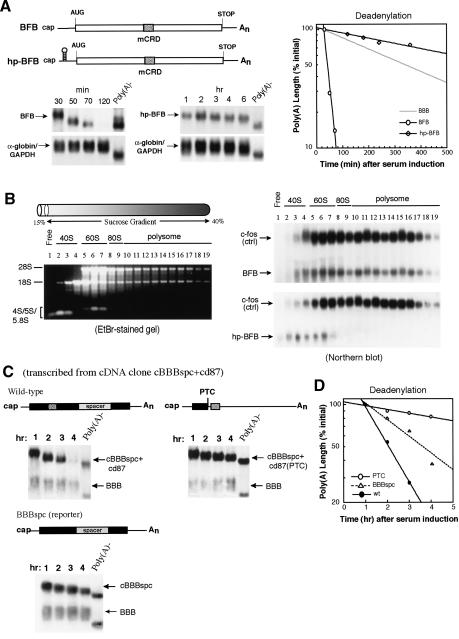
Ribosome transit is required to trigger accelerated deadenylation for mCRD-mediated mRNA turnover. (A, left) Physical maps and Northern blots showing deadenylation and decay of BFB and hp-BFB hybrid mRNAs. Solid lines indicate 5′ and 3′ UTRs from β-globin mRNA. (Rectangles) Protein-coding region from c-fos mRNA. (Radial box) c-fos mCRD. (hp) Hairpin structure. Northern blots showing deadenylation and decay of BFB and hp-BFB mRNA transcribed from the c-fos promoter in serum-induced NIH3T3 cells. (Right) Semi-log plot showing the deadenylation kinetics of BFB, hp-BFB, and BBB mRNA. α-globin/GAPDH (control) mRNAs were expressed constitutively and served as an internal standard. The times given at the top correspond to minutes or hours after serum stimulation. Deadenylation curves were plotted as described previously (Shyu et al. 1991). (B) Polysome profiles showing that the hairpin inserted in the 5′ UTR of BFB mRNA efficiently and specifically blocked the ongoing translation of the message. (Left) Evaluation of positions and distributions of the 40S, 60S, 80S, and polysome by running RNA samples from each fraction on a 1% nondenaturing agarose gel. (Right) Distribution of BFB, hp-BFB, and c-fos mRNAs as identified by Northern blotting. (C) Physical maps and Northern blots showing deadenylation and decay of BBBspc, cBBBspc + cd87, and cBBBspc + cd87(PTC) mRNAs. Note that cBBBspc + cd87 and cBBBspc + cd87(PTC) mRNAs are expressed from intronless cDNA constructs. The schematic drawings are as described in the legend for Figure 2. (PTC) Premature termination codon. (D) Semi-log plot showing deadenylation kinetics of the three transcripts: cBBBspc + cd87(PTC; open circles), BBBspc (open triangles), and cBBBspc + cd87 (filled circles).
We carried out co-IP/Western blot analysis to determine whether CCR4 associates with the mCRD-binding protein, UNR. V5-tagged CCR4 and HA-tagged UNR were ectopically expressed in COS7 cells, and UNR was then immunoprecipitated from RNase A-treated lysates by antibody against HA-tag. Western blot analysis of the immunoprecipitates (Fig. 6C) showed that expression of HA-UNR led to co-IP of V5-CCR4, but not an abundant cytoplasmic protein, α-tubulin, indicating an interaction between CCR4 and UNR.
Ribosome transit is required for accelerated deadenylation in mCRD-mediated mRNA turnover
c-fos mCRD-mediated decay is tightly coupled to translation. Taken with results from the experiments above, this suggests a model in which mCRD/UNR interacts with poly(A) tail/PABP before translation initiation to help form a deadenylation/decay complex that is kept in a dormant state prior to translation. We hypothesized that ribosome transit alters the interaction, allowing CCR4 to attack the poly(A) tail. This model predicts that preventing ribosomes from transiting the mCRD would slow down deadenylation of the mCRD-containing mRNA due to the dormant conformation of a protein/RNA complex formed between the mCRD/UNR and poly(A)/PABP. We took two approaches to test this hypothesis: blocking translation initiation of the mCRD-containing mRNA and prematurely terminating ribosome transit upstream of the mCRD.
Rapid deadenylation of BFB, a highly unstable hybrid message consisting of the β-globin 5′ and 3′ UTRs and the entire c-fos ORF, is mediated by the mCRD and precedes decay of the RNA body (Shyu et al. 1991; Chen et al. 1992). Blocking translation initiation of the BFB mRNA by introducing a hairpin (hp) in its 5′ UTR stabilized the transcript (Chen et al. 1995), but it was not determined whether the deadenylation kinetics was affected. As shown in Figure 7A, the hp-BFB mRNA exhibited little deadenylation over the 6-h time course, retaining >75% of its poly(A) tail. In contrast, it took <50 min (compared with 30- and 70-min time points) to shorten the poly(A) tail of the labile BFB message by ∼90%. Significantly, deadenylation of hp-BFB was even slower than for the stable β-globin (BBB) mRNA (Fig. 7A, gray line of the plot; Chen and Shyu 2003). Deadenylation of all three transcripts exhibited first-order kinetics (Fig. 7A). Polysome profile analysis by sucrose gradient fractionation (Fig. 7B) confirmed that the hp-BFB mRNA cosedimented with free ribosomal subunits, whereas much of the BFB mRNA was associated with polysomes, supporting an efficient blockade of translation initiation by the hp insertion. In each case, endogenous c-fos mRNA profiles were unchanged (Fig. 7B).
As a further test, we prevented ribosomes from transiting the mCRD while allowing translation initiation and elongation of the transcript. This was accomplished by creating a premature termination codon (PTC) upstream of the mCRD (Fig. 7C). To avoid nonsense-mediated decay, the PTC was introduced into an intronless cDNA construct derived from the BBBspc + cd87 genomic construct (cBBBspc + cd87). As shown in Figure 7C, cBBBspc + cd87 mRNA expressed from the intronless cDNA construct underwent deadenylation and decay at the same rate as the mRNA transcribed from the genomic construct (BBBspc + cd87 in Fig. 2B; see also Grosset et al. 2000). The results (Fig. 7C) showed that the cBBBspc + cd87(PTC) transcript exhibited deadenylation even slower than the control message, BBBspc, indicating that deadenylation is retarded when the ribosome does not transit the mCRD, thus presumably preserving the interaction between the mCRD and the poly(A) tail.
Discussion
UNR, a new PABP partner, is a key factor with multiple functions in the c-fos mCRD-mediated mRNA turnover
In this study we identified UNR as the mCRD-binding protein among the five proteins that associate with the mCRD. Several lines of evidence we obtained support that UNR is a novel PABP-interacting protein. First, in vitro GST-pull down assays demonstrated that UNR exhibits RNA-independent and direct interaction with PABP, whereas interactions of hnRNP D and NSAP1 (hnRNP Q) with UNR are RNA-dependent (Fig. 4A). Second, we were able to map the UNR-binding site on PABP to the end of RRM2 and the entire RRM3 of PABP, which is immediately downstream of the poly(A)-binding site on PABP (Fig. 5). The mapping data are consistent with the observation that association of poly(A) with PABP does not interfere with interaction between UNR and PABP/poly(A) complex. Third, we showed that in intact cells UNR and PABP can be coimmunoprecipitated and that this co-IP is abolished when the UNR-interacting domain of PABP (166-289) is ectopically overexpressed. The functional significance of UNR-PABP interaction in the mCRD-mediated decay was further addressed by showing slow deadenylation and decay kinetics of mCRD-containing mRNA in cells with diminished UNR-PABP interaction when PABP (166-289) is ectopically expressed (Fig. 5). Collectively, these data support the existence of direct interaction between the mCRD/UNR and the poly(A)/PABP in the mCRD-mediated decay in vivo.
Mammalian UNR belongs to a large family of proteins consisting of prokaryotic cold shock proteins (CSPs) and eukaryotic CSD-containing proteins (Graumann and Marahiel 1998). UNR has five CSDs and all are required for proper functioning of several viral IRESs (internal ribosomal entry sites; Hunt et al. 1999; Boussadia et al. 2003). The modular structure of UNR allows it to interact both with RNA (Jacquemin-Sablon et al. 1994) and also with the transcription factor ALL-1 (Leshkowitz et al. 1996). It has recently become clear that prokaryotic CSP function as RNA chaperones (Graumann and Marahiel 1998). Binding of CSPs to RNA is cooperative and is believed to prevent the formation of secondary structure. Consistent with the functions of prokaryotic CSPs, a recent study (Mitchell et al. 2003) suggests that UNR acts as a RNA chaperone, changing the structure of the Apaf-1 IRES to permit translation initiation in mammalian neuronal-derived cells. Taking those studies with our observation that UNR has at least two binding sites in the c-fos mCRD necessary for mRNA destabilization, it is possible that multiple UNRs bind the mCRD and then elicit interactions with individual PABPs associated with poly(A), establishing strong interactions between the mCRD/UNR complex and the poly(A)/PABP complex, and that the interactions are altered by ribosome transit.
CCR4 functions in accelerated deadenylation directed by the c-fos mCRD
Here, our in vivo evidence showed that CCR4 poly(A) nuclease is involved in mCRD-mediated mRNA decay in the cytoplasm. In yeast, the CCR4/CAF1 complex is known to be the major deadenylase in cytoplasmic mRNA decay (Tucker et al. 2001, 2002). However, the PAN2/3 complex might also participate in cytoplasmic deadenylation, as the largest retardation effect on cytoplasmic deadenylation was seen when both complexes were disrupted (Tucker et al. 2001). Recently, mammalian orthologs of yeast CCR4/CAF1 and Pan2/Pan3 with poly(A) nuclease activities have been described (Albert et al. 2000; Chen et al. 2002; Uchida et al. 2004). In addition, several studies using in vitro reconstituted systems have identified a different poly(A) nuclease, PARN, as the major poly(A) nuclease responsible for accelerated deadenylation that precedes decay of the RNA body in cytoplasmic extracts (Korner and Wahle 1997; Dehlin et al. 2000; Gao et al. 2000; Martinez et al. 2000).
The potential functional redundancy among the several classes of poly(A) nuclease present in mammalian cells (Dupressoir et al. 2001) makes it difficult to employ the siRNA knock-down approach to assign in vivo deadenylation function to a particular poly(A) nuclease. This study circumvented these potential problems by combining transcription pulsing to monitor mRNA deadenylation and decay kinetics and a dominant-negative approach to specifically inactivate individual poly(A) nucleases (Fig. 6). This combined approach allowed identification of CCR4 as a poly(A) nuclease involved in mCRD-mediated mRNA turnover. The affected mCRD-containing mRNA showed slow deadenylation kinetics, reminiscent of the phenotype seen with the yeast CCR4 mutant (Tucker et al. 2001). This result could mean that the residual CCR4 activity is responsible for the slow deadenylation or a different poly(A) nuclease, such as PAN2 or PARN, is responsible for the default deadenylation. To our knowledge, these experiments provide the first in vivo kinetic evidence for a role of CCR4 in accelerated deadenylation directed by the mCRD. The present data suggest that the poly(A) nuclease function of CCR4 in cytoplasmic deadenylation is evolutionarily conserved.
Accelerated deadenylation directed by the mCRD is coupled to translation
The present results indicate that deadenylation is coupled to translation in the process of mCRD-mediated decay. Our data provide mechanistic insights into how the coupling may be accomplished. Blocking translation initiation or halting translation elongation leads to profound impairment of deadenylation and stabilization of the message (Fig. 7). Two observations argue that this retarded deadenylation is specific to nontranslated mCRD-containing mRNA and not a general nonspecific effect of nontranslated mRNA. First, when the same kind of hp structure was introduced into the 5′ UTR of β-globin mRNA or β-globin mRNA containing an ARE to block translation initiation, there was essentially no change in deadenylation or decay kinetics (Chen et al. 1995; Chen and Shyu 2003). Second, prematurely terminating translation upstream of the mCRD had the same outcome as preventing translation initiation by a strong stem-loop structure in the 5′ UTR (Fig. 7). Moreover, the c-fos ARE introduced into the 3′ UTR of the nontranslated hp-BFB mRNA still destabilizes the resulting transcript (data not shown), indicating that the nontranslated hp-BFB mRNA is not present as an aberrant and nondegradable mRNP. Taken together, these observations argue for the notion that interaction between the mCRD/UNR complex and the poly(A)/PABP complex locks the mRNP in a form resistant to poly(A) shortening until ribosome transit occurs.
A model for translationally coupled mRNA decay
The accumulating information suggests a model in which UNR binding to the mCRD initiates assembly of the mRNP complex involving interaction with the poly(A)/PABP complex. The observations that UNR associates with CCR4 and that decreasing UNR expression slows deadenylation suggest that the CCR4 nuclease is recruited to the mCRD-containing mRNP complex through its interaction with UNR. Prior to translation initiation, the UNR-PABP interaction may protect the poly(A) tail from attack by CCR4. However, after the ribosome transits the mCRD, disruption of the complex allows CCR4 access to the poly(A) tail, leading to rapid deadenylation. At present, it is not clear when CCR4 joins the mRNP complex. An important aspect of the model is that the mCRD serves as a landing/assembly platform for formation of a deadenylation/decay complex that is kept latent prior to translation, consistent with observation of little deadenylation in the absence of ribosome transit of the mCRD.
In summary, the present results provide important new insights into the mechanism of translationally coupled mRNA turnover directed by the mCRD in the c-fos proto-oncogene transcript. The mechanism is distinct from other modes of translationally coupled mRNA decay that appear not to be triggered by deadenylation, including nonsense-mediated decay (Lejeune et al. 2003), nonstop decay (Frischmeyer et al. 2002; van Hoof et al. 2002), c-myc coding determinant-mediated decay (Lemm and Ross, 2002), histone RNA decay (Dominski et al. 2003), and tubulin mRNA decay (Yen et al. 1988). The c-fos gene is one of a large group of so-called early-response genes (ERGs) with transcripts that typically decay rapidly (Herschman 1991). It will thus be interesting to assess the generality of mRNA decay mediated by destabilizing coding-region determinants in other ERG mRNAs. A critical pending issue concerning mammalian mRNA degradation is which poly(A) nuclease(s) is responsible in vivo for the accelerated poly(A) shortening directed by mRNA destabilizing elements, such as AREs. In addition, future experimentation to elucidate the regulation of the multiple functions of UNR in mCRD-mediated decay and the roles of other factors, such as Paip1, hnRNP D, and NSAP1, will help to elucidate the mechanisms that control the fate of cytoplasmic mRNA by coupling message decay to translation.
Materials and methods
Plasmid construction
Construction of plasmids pSVα1/GAPDH (Chen and Shyu 1994), pBFB, pBBB, pBBB + ARE (Shyu et al. 1989), php-BFB (Chen et al. 1995), pBBBspc, pBBBspc + cd87 (Grosset et al. 2000), pSVB10 (Shyu et al. 1989), pSV-myc37AUF1 (Loflin et al. 1999a), pSG-UNRFlag (Grosset et al. 2000), and pTetMycAUF1 (Xu et al. 2001) has been described previously. Please refer to the Supplemental Material for details about construction of the following plasmids: pBBBspc + cd87ΔI/II, pBBBspc + cd87ΔIII, pBBBspc + cd87ΔIII, pcDNA6-hnRNP D, pcDNA6-NSAP1, pcDNA6-Paip1, pcDNA6/V5-His-CCR4, pcDNA3.1/GS-PARN-mt, pGST-hnRNP D, pGST-NSAP1, pGST-Paip1, pGST-UNR, pMBP-UNR, pMyc-BBB, pMyc-cBBB + spc + cd87, pSR-GST, pSR-GST-PABP(166-284), pSR-HA-CCR4, pSR-HA-CCR4-mt, pSR-HA-PAN2, pSR-HA-PAN2-mt, and pSR-HA-UNR.
Cell culture and transfection
NIH3T3 cells were cultured in DMEM medium containing 10% calf serum, 2 mM glutamine, 100 U/mL penicillin, and 100 mg/ml streptomycin at 37°C/8% CO2. COS7 cells were cultured as described above except in 10% fetal calf serum. DNA transfection using calcium phosphate technique and serum induction of quiescent cells to induce transient transcription from the c-fos promoter was conducted as described previously (Loflin et al. 1999b). For DNA transfections using liposome-based techniques, 2 × 106 NIH3T3 cells were plated per 10-cm plate and cultured 24 h. Thereafter, test plasmid DNAs (7 μg total) were transfected by using PolyFect (QIAGEN) according to manufacturer's protocol. Analyses were performed 36-48 h after transfection. For transfection involving both siRNA and DNA, NIH3T3 cells were split to a density of 1.8 × 106/100-mm dish 24 h before transfection. Thirty micrograms of siRNAs and 90 μL RNAiFect (QIAGEN) mixture and 7 μg DNA (1.5 μg of test plasmid, 3.0-4.5 μg of internal control plasmid, and 1.0-2.5 μg of carrier plasmid) and 40 μL PolyFect (QIAGEN) mixture were prepared separately according to manufacturer's protocols. Both mixtures were then added simultaneously to the culture dish. After 21 h, cells were serum-starved in DMEM/0.5% calf serum for 25 h and then stimulated with DMEM/20% calf serum. siRNAs targeting mouse UNR (accession. no. NM_144901) and nonspecific control siRNAs were purchased from Dharmacon SMARTpool.
RNA isolation and analyses
Isolation of total cytoplasmic RNA and Northern blot analysis were conducted as described previously (Shyu et al. 1996). Analyses of polysome profile by sucrose gradient fractionation and subsequent analyses of specific distribution of tested mRNAs (exogenous BFB and hp-BFB mRNAs) and a control mRNA (endogenous c-fos mRNA) in the fractions were performed as described previously (Chen et al. 1995). Gene-specific DNA probes were prepared by the method of random oligo-nucleotide priming for Northern blot analysis. Because of its relatively long size (1.5 kb), RNase H-directed cleavage was performed to cleave the hp-BFB mRNA into a 5′ 0.93-kb fragment and a 3′ fragment containing poly(A) tail, which ranges in size from 0.38 to 0.56 kb (Shyu et al. 1991). The 3′ fragments were detected by a probe corresponding to the β-globin 3′ UTR. A control plasmid encoding a stable message with a size of ∼1.4 kb, termed α-globin/GAPDH, served as an internal standard for normalization of the test messages (Chen and Shyu 1994). The [32P]-labeled probes were produced by inclusion of [α-32P] dCTP (>6000 Ci/mmole; DuPont). The quantitation of data was obtained by PhosphorImager (Bio-Rad). RNase H treatment of cytoplasmic mRNA to generate poly(A)- RNA was carried out as described previously (Shyu et al. 1991). Deadenylation and decay curves were plotted as described previously (Shyu et al. 1991).
Western blot analysis
For lysate preparations, plates of transfected cells (100-mm culture dish) from experiments parallel to the time course experiment were harvested for preparing total or cytoplasmic extracts as described previously (Peng et al. 1998). Protein concentration was analyzed by the BCA protein assay reagent (Pierce). For Western blot analysis, cytoplasmic lysates (5-40 μg) were resolved on a SDS-polyacrylamide gel, and the PVDF blots were probed with specific antibodies as described in the figure legends. SuperSignal West Pico Chemiluminescent Substrate (Pierce) was used for detection. The mAb against the V5 epitopetag (Invitrogen) was used at 1:2000 dilution. The mAb for HA-tag was used at 1:2000 dilution. The goat polyclonal antibody against GST (Amersham) was used at 1:2000 dilution. The mAb against α-tubulin (DM1A, Sigma) was used at 1:5000 dilution as a positive control for cytoplasmic protein preparations. To detect endogenous UNR in NIH3T3 cells, a polyclonal antibody was generated in rabbits immunized with GST-UNR fusion protein purified from E. coli, and the antibody was used at 1:2000 dilution. Endogenous PABP was detected by a rabbit polyclonal antibody (1:5000 dilution) against the RRM4 of human PABP (kindly provided by R. Lloyd, Baylor College of Medicine, Houston, TX). The rabbit polyclonal antibody against HA-tag (Bethyl) was used at 1:2000 dilution.
Immunoprecipitation
For immunoprecipitation, cytoplasmic lysates were incubated with anti-HA agarose (Roche) with rotation for 2 h at 4°C. When necessary, RNase A (0.1 mg/ml) was included during incubation. Following precipitation, the resin was washed five times with 1 mL lysis buffer (20 mM Tris-HCl at pH 7.4, 150 mM NaCl, 0.4% NP-40, 1 mM sodium orthovanadate, 1 mM sodium pyrophosphate, 1 mM NaF, 100 nM okadaic acid, and protease inhibitor cocktail). Elution was carried out for 15 min at 37°C in lysis buffer supplemented with 1 mg/mL HA peptide (Roche). The eluted fraction was resolved by SDS-PAGE. Western blotting was performed with indicated antibodies as described above.
Recombinant protein expression and purification
Clones for GST-fused proteins, GST alone, MBP-fused proteins, and MBP alone were expressed in E. coli BL21 (Amersham) and then induced with 0.4 mM isopropyl β-D-thiogalactoside for 2.5 h at 30°C. The cells were harvested and resuspended in ice-cold phosphate-buffered saline (PBS) containing 1 mM dithiothreitol (DTT) and protease inhibitor cocktail (Roche). Following incubation with 0.1 mg/mL lysozyme on ice for 30 min, DNase I was added to a final concentration of 10 μg/mL, and the lysates were subjected to sonication for three cycles on ice. The lysates were clarified by centrifugation at 14,000 × g for 10 min. The expressed proteins in the clear supernatant were purified by using either MicroSpin GST Purification Modules (Amersham) or amylose resin chromatographic column (New England BioLab) according to the manufacture's instructions.
Preparation of RNA substrates
In vitro transcription was performed by using T3 or T7 RNA polymerase (Promega) according to the manufacture's instruction. Labeled RNA transcripts were produced by inclusion of [γ-32P]UTP (800 Ci/mmole; PerkinElmer) in the reaction mixtures.
The DNA template CRD28 used for in vitro transcription was generated by annealing pairs of DNA oligomers carrying T7 promoter and desired CRD sequences (Fig. 2A). All others were generated by PCR amplification of desired region by using pT18-cdi2, a pT7/T3α-18 (Bethesda Research Laboratory [BRL]) derivative with a fragment spanning the 320-nt c-fos mCRD (Grosset et al. 2000) inserted between the HincII and BamHI sites, as templates. For nonspecific probe (NS), a 38-nt sense transcript was synthesized from HincII-linearized pT7/T3α-18. Except probe NS from T3 promoter, all the above probes were transcribed from T7 promoter. For poly(A) probe, a 25-nt poly(A) RNA was purchased from Dharmacon Research, Inc. Poly(A) RNA (10 pmole) was 5′ end labeled with 150 mCi of [γ-32P]ATP and 30 U polynucleotide kinase in 30 μL for 1 h at 30°C. Labeled poly(A) was purified on CHROMA spin columns (Clontech).
Electrophoretic mobility shift assay
Purified recombinant proteins (0.5 pmole) and [32P]-labeled RNA probes (0.5 pmole) were incubated at room temperature for 15 min in a binding buffer (10 μL) containing 10 mM HEPES (pH 7.6), 3 mM MgCl2, 100 mM KCl, 5 mM EDTA, 2 mM DTT, 5% glycerol, 0.5% Nonidet P-40 (NP-40), 0.2 μg/μL yeast RNA, and 1.5 μg/μL heparin. For experiments using [32P]-labeled poly(A)25 RNA, 0.01 pmole was used. RNA-protein complexes were resolved on a 5%-6% native polyacrylamide gel at 4°C. The results were analyzed by drying down the gel and subjecting the gel to autoradiography. For competition experiment, the [32P]-labeled RNA probe and indicated amounts of unlabeled competitor RNA were first mixed in the same binding buffer as mentioned above and then incubated with the recombinant proteins at room temperature for 15 min. RNA-protein complexes were resolved, and the results were analyzed as described above.
GST pull-down assay
GST fusion proteins were immobilized on glutathione-Sepharose by using MicroSpin GST Purification Modules (Amersham) according to the manufacture's instruction, and then incubated with 5 μL of the in vitro translated protein in 250 μL of binding buffer (10 mM HEPES at pH 7.6, 3 mM MgCl2, 100 mM KCl, 5 mM EDTA, 5% glycerol, 0.5% NP-40, and protease inhibitor cocktail). After incubation with rotation for 1.5 h at 4°C, the resin was washed two times with 250 μL RIPA buffer (10 mM sodium phosphate at pH 7.2, 150 mM NaCl, 1% NP-40, and 0.5% sodium deoxycholate) and once with 500 μL of PBS. The bound fraction was eluted in 100 μL of elution buffer (10 mM glutathione, 50 mM Tris-HCl at pH 8.0) and analyzed by SDS-PAGE. The gels were stained by Coomassie blue to show equal loading of GST fusion protein and then dried for autoradiography to detect pull-down proteins labeled with [35S]methionine.
The [35S]methionine-labeled proteins were produced in the rabbit reticulocyte lysate by using an in vitro coupled transcription/translation system (Promega) in the presence of [35S]methionine (Amersham). When necessary, the in vitro translated products were treated with DNase I (0.5 U/μL) and RNase A (0.1 mg/mL) for 15 min at 30°C.
For GST-PABP/poly(A) pull-down experiment, GST-PABP was first immobilized on glutathione-Sepharose as described above and incubated with 100 pmole of 25-nt poly(A) RNA in 200 μL of binding buffer (described in Electrophoretic Mobility Shift Assay). After incubation with rotation for 30 min at 4°C, the resin was washed two times with 250 μL of DEPC-treated water. The in vitro translated [35S]methionine-labeled UNR was then incubated with immobilized GST-PABP/poly(A). The subsequent procedures were as described above.
Acknowledgments
We thank Dr. R. Kulmacz for critical reading of the manuscript and his valuable comments, Dr. T. Nagase (Kazusa DNA Research Institute) for providing KIAA1194 clone, Dr. S. Ohno (Yokohama City University) for the expression vectors, SRD4 and SRHisHA, Dr. S. Hoshino (University of Tokyo) for the pGST-PABP plasmid, Dr. H. Imataka (McGill University) for the pGST-CAT plasmid, and Dr. R. Lloyd (Baylor College of Medicine) for the anti-PABP antibody. Special thanks go to N. Xu for technical assistance. This work was supported by grants from National Institutes of Health (GM 46454 and GM 59211) to A.-B.S.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1219104.
References
- Albert T.K., Lemaire, M., van Berkum, N.L., Gentz, R., Collart, M.A., and Timmers, H.T.M. 2000. Isolation and characterization of human orthologs of yeast CCR4-NOT complex subunits. Nucl. Acids Res. 28: 809-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggs J.E. and Green, C.B. 2003. Nocturnin, a deadenylase in Xenopus laevis retina: A mechanism for posttranscriptional control of circadian-related mRNA. Curr. Biol. 13: 189-198. [DOI] [PubMed] [Google Scholar]
- Beelman C.A. and Parker, R. 1995. Degradation of mRNA in eukaryotes. Cell 81: 179-183. [DOI] [PubMed] [Google Scholar]
- Boussadia O., Niepmann, M., Creancier, L., Prats, A.-C., Dautry, F., and Jacquemin-Sablon, H. 2003. Unr is required in vivo for efficient initiation of translation from the internal ribosome entry sites of both rhinovirus and poliovirus. J. Virol. 77: 3353-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y. and Shyu, A.B. 1994. Selective degradation of early-response-gene mRNAs: Functional analyses of sequence features of the AU-rich elements. Mol. Cell. Biol. 14: 8471-8482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2003. Rapid deadenylation triggered by a nonsense codon precedes decay of the RNA body in a mammalian cytoplasmic nonsense-mediated decay pathway. Mol. Cell. Biol. 23: 4805-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y., You, Y., and Shyu, A.B. 1992. Two cellular proteins bind specifically to a purine-rich sequence necessary for the destabilization function of a c-fos protein-coding region determinant of mRNA instability. Mol. Cell. Biol. 12: 5748-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y., Xu, N., and Shyu, A.B. 1995. mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: Different deadenylation kinetics and uncoupling from translation. Mol. Cell. Biol. 15: 5777-5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Chiang, Y.-C., and Denis, C.L. 2002. CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 21: 1414-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig A.W., Haghighat, A., Yu, A.T., and Sonenberg, N. 1998. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature 392: 520-523. [DOI] [PubMed] [Google Scholar]
- Deardorff J.A. and Sachs, A.B. 1997. Differential effects of aromatic and charged residue substitutions in the RNA binding domains of the yeast Poly(A)-binding protein1. J. Mol. Biol. 269: 67-81. [DOI] [PubMed] [Google Scholar]
- Dehlin E., Wormington, M., Korner, C.G., and Wahle, E. 2000. Cap-dependent deadenylation of mRNA. EMBO J. 19: 1079-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z., Yang, X.C., Kaygun, H., Dadlez, M., and Marzluff, W.F. 2003. A 3′ exonuclease that specifically interacts with the 3′ end of histone mRNA. Mol. Cell 12: 295-305. [DOI] [PubMed] [Google Scholar]
- Dupressoir A., Morel, A.-P., Barbot, W., Loireau, M.-P., Corbo, L., and Heidmann, T. 2001. Identification of four families of yCCR4- and Mg2+-dependent endonuclease-related proteins in higher eukaryotes, and characterization of orthologs of yCCR4 with a conserved leucine-rich repeat essential for hCAF1/hPOP2 binding. BMC Genomics 2: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischmeyer P.A., van Hoof, A., O'Donnell, K., Guerrerio, A.L., Parker, R., and Dietz, H.C. 2002. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295: 2258-2261. [DOI] [PubMed] [Google Scholar]
- Gao M., Fritz, D.T., Ford, L.P., and Wilusz, J. 2000. Interaction between a poly(A)-specific ribonuclease and the 5′ cap influences mRNA deadenylation rates in vitro. Mol. Cell 5: 479-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann P.L. and Marahiel, M.A. 1998. A superfamily of proteins that contain the cold-shock domain. Trends Biochem. Sci. 23: 286-290. [DOI] [PubMed] [Google Scholar]
- Grosset C., Chen, C.-Y.A., Xu, N., Sonenberg, N., Jacquemin-Sablon, H., and Shyu, A.-B. 2000. A mechanism for translationally coupled mRNA turnover: Interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell 103: 29-40. [DOI] [PubMed] [Google Scholar]
- Harris C.E., Boden, R.A., and Astell, C.R. 1999. A novel heterogeneous nuclear ribonucleoprotein-like protein interacts with NS1 of the minute virus of mice. J. Virol. 73: 72-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschman H.R. 1991. Primary response genes induced by growth factors and tumor promoters. Annu. Rev. Biochem. 60: 281-319. [DOI] [PubMed] [Google Scholar]
- Hoshino S., Imai, M., Kobayashi, T., Uchida, N., and Katada, T. 1999. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-poly(A) tail of mRNA: Direct association of erf3/GSPT with polyadenylate-binding protein. J. Biol. Chem. 274: 16677-16680. [DOI] [PubMed] [Google Scholar]
- Hunt S.L., Hsuan, J.J., Totty, N., and Jackson, R.J. 1999. unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes & Dev. 15: 437-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imataka H., Gradi, A., and Sonenberg, N. 1998. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 17: 7480-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson A. and Peltz, S.W. 1996. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Ann. Rev. Biochem. 65: 693-739. [DOI] [PubMed] [Google Scholar]
- Jacquemin-Sablon H., Triqueneaux, G., Deschamps, S., le Maire, M., Doniger, J., and Dautry, F. 1994. Nucleic acid binding and intracellular localization of unr, a protein with five cold shock domains. Nucl. Acids Res. 22: 2643-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahvejian A., Roy, G., and Sonenberg, N. 2001. The mRNA closed-loop model: The function of PABP and PABP-interacting proteins in mRNA translation. Cold Spring Harb. Symp. Quant. Biol. 66: 293-300. [DOI] [PubMed] [Google Scholar]
- Khaleghpour K., Kahvejian, A., De Crescenzo, G., Roy, G., Svitkin, Y.V., Imataka, H., O'Connor-McCourt, M., and Sonenberg, N. 2001. Dual Interactions of the translational repressor Paip2 with Poly(A) binding protein. Mol. Cell. Biol. 21: 5200-5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korner C.G. and Wahle, E. 1997. Poly(A) tail shortening by a mammalian poly(A)-specific 3′-exoribonuclease. J. Biol. Chem. 272: 10448-10456. [DOI] [PubMed] [Google Scholar]
- Kuhn U. and Pieler, T. 1996. Xenopus poly(A) binding protein: Functional domains in RNA binding and protein-protein interaction. J. Mol. Biol. 256: 20-30. [DOI] [PubMed] [Google Scholar]
- Lejeune F., Li, X., and Maquat, L. 2003. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell 12: 675-687. [DOI] [PubMed] [Google Scholar]
- Lemm I. and Ross, J. 2002. Regulation of c-myc mRNA decay by translational pausing in a coding region instability determinant. Mol. Cell. Biol. 22: 3959-3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshkowitz D., Rozenblatt, O., Nakamura, T., Yano, T., Dautry, F., Croce, C.M., and Canaani, E. 1996. ALL-1 interacts with unr, a protein containing multiple cold shock domains. Oncogene 13: 2027-2031. [PubMed] [Google Scholar]
- Loflin P.A., Chen, C.-Y.A., and Shyu, A.-B. 1999a. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes & Dev. 13: 1884-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loflin T.L., Chen, C.-Y.A., Xu, N., and Shyu, A.-B. 1999b. Transcriptional pulsing approaches for analysis of mRNA turnover in mammalian cells. Methods 17: 11-20. [DOI] [PubMed] [Google Scholar]
- Mangus D.A., Evans, M.C., and Jacobson, A. 2003. Poly(A)-binding proteins: Multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 4: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J., Ren, Y., Thuresson, A., Hellman, U., Astrom, J., and Virtanen, A. 2000. A 54-kDa fragment of the poly(A)-specific ribonuclease is an oligomeric, processive, and cap-interacting poly(A)-specific 3′ exonuclease. J. Biol. Chem. 275: 24222-24230. [DOI] [PubMed] [Google Scholar]
- Mitchell S.A., Spriggs, K.A., Cloldwell, M.J., Jackson, R.J., and Willis, A.E. 2003. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol. Cell 11: 757-771. [DOI] [PubMed] [Google Scholar]
- Mourelatos Z., Abel, L., Yong, J., Kataoka, N., and Dreyfuss, G. 2001. SMN interacts with a novel family of hnRNP and spliceosomal proteins. EMBO J. 20: 5443-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R. and Song, H. 2004. The enzymes and control of eukaryotic mRNa turnover. Nat. Strut. Mol. Biol. 11: 121-127. [DOI] [PubMed] [Google Scholar]
- Peng S.-P., Chen, C.-Y., Xu, N., and Shyu, A.-B. 1998. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 17: 3461-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y.-G., Martinez, J., and Virtanen, A. 2002. Identification of the active site of poly(A)-specific ribonuclease by site-directed mutagenesis and Fe2+-mediated cleavage. J. Biol. Chem. 277: 5982-5987. [DOI] [PubMed] [Google Scholar]
- Roy G., De Crescenzo, G., Khaleghpour, K., Kahvejian, A., O'Connor-McCourt, M., and Sonenberg, N. 2002. Paip1 interacts with Poly(A) binding protein through two independent binding motifs. Mol. Cell. Biol. 22: 3769-3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavi S.C., Wellington, C.L., Shyu, A.B., Chen, C.Y., Greenberg, M.E., and Belasco, J.G. 1994. Multiple elements in the c-fos protein-coding region facilitate mRNA deadenylation and decay by a mechanism coupled to translation. J. Biol. Chem. 269: 3441-3448. [PubMed] [Google Scholar]
- Shyu A.B. and Wilkinson, M.F. 2000. The double lives of shuttling mRNA binding proteins. Cell 102: 135-138. [DOI] [PubMed] [Google Scholar]
- Shyu A.B., Greenberg, M.E., and Belasco, J.G. 1989. The c-fos mRNA is targeted for rapid decay by two distinct mRNA degradation pathways. Genes & Dev. 3: 60-72. [DOI] [PubMed] [Google Scholar]
- Shyu A.-B., Belasco, J.G., and Greenberg, M.G. 1991. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes & Dev. 5: 221-232. [DOI] [PubMed] [Google Scholar]
- Shyu A.B., Garcia-Sanz, J.A., and Mullner, E. 1996. Analysis of mRNA decay in mammalian cells. In: The immunology methods manual (ed., I. Lefkovits), pp. 450-462. Academic Press, London.
- Triqueneaux G., Velten, M., Franzon, P., Dautry, F., and Jacquemin-Sablon, H. 1999. RNA binding specificity of Unr, a protein with five cold shock domains. Nucl. Acids Res. 27: 1926-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M., Valencia-Sanchez, M.A., Staples, R.R., Chen, J., Denis, C.L., and Parker, R. 2001. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104: 377-386. [DOI] [PubMed] [Google Scholar]
- Tucker M., Staples, R.R., Valencia-Sanchez, M.A., Muhlrad, D., and Parker, R. 2002. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 21: 1427-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N., Hoshino, S.-i., and Katada, T. 2004. Identification of a human cytoplasmic Poly(A) nuclease complex stimulated by Poly(A)-binding protein. J. Biol. Chem. 279: 1383-1391. [DOI] [PubMed] [Google Scholar]
- van Hoof A., Frischmeyer, P.A., Dietz, H.C., and Parker, R. 2002. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295: 2262-2264. [DOI] [PubMed] [Google Scholar]
- Wellington C.L., Greenberg, M.E., and Belasco, J.G. 1993. The destabilizing elements in the coding region of c-fos mRNA are recognized as RNA. Mol. Cell. Biol. 13: 5034-5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz C.W., Wormington, M., and Peltz, S.W. 2001. The capto-tail guide to mRNA turnover. Nat. Rev. Mol. Cell. Biol. 2: 237-246. [DOI] [PubMed] [Google Scholar]
- Xu N., Chen, C., and Shyu, A. 2001. Versatile role for hnRNP D isoforms in the differential regulation of cytoplasmic mRNA turnover. Mol. Cell. Biol. 21: 6960-6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen T.J., Machlin, P.S., and Cleveland, D.W. 1988. Autoregulated instability of β-tubulin mRNAs by recognition of the nascent amino terminus of β-tubulin. Nature 334: 580-585. [DOI] [PubMed] [Google Scholar]
- Zuo Y. and Deutscher, M.P. 2001. Exoribonuclease superfamilies: Structural analysis and phylogenetic distribution. Nucl. Acids Res. 29: 1017-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]