Abstract
Enhancers are cis-acting DNA elements that play critical roles in distal regulation of gene expression. Identifying enhancers is an important step for understanding distinct gene expression programs that may reflect normal and pathogenic cellular conditions. Experimental identification of enhancers is constrained by the set of conditions used in the experiment. This requires multiple experiments to identify enhancers, as they can be active under specific cellular conditions but not in different cell types/tissues or cellular states. This has opened prospects for computational prediction methods that can be used for high-throughput identification of putative enhancers to complement experimental approaches. Potential functions and properties of predicted enhancers have been catalogued and summarized in several enhancer-oriented databases. Because the current methods for the computational prediction of enhancers produce significantly different enhancer predictions, it will be beneficial for the research community to have an overview of the strategies and solutions developed in this field. In this review, we focus on the identification and analysis of enhancers by bioinformatics approaches. First, we describe a general framework for computational identification of enhancers, present relevant data types and discuss possible computational solutions. Next, we cover over 30 existing computational enhancer identification methods that were developed since 2000. Our review highlights advantages, limitations and potentials, while suggesting pragmatic guidelines for development of more efficient computational enhancer prediction methods. Finally, we discuss challenges and open problems of this topic, which require further consideration.
Keywords: gene regulation, enhancers, chromatin signatures, histone modification marks, genome annotation, machine learning, bioinformatics, computer science
Introduction
Gene expression in eukaryotes is governed by complex processes orchestrated by the interplay of various elements located in DNA regulatory regions [1–4]. Enhancers represent one of the better-characterized regulatory elements. Enhancers increase the transcriptional output in cells manifesting distinct properties, which are summarized as follows [5–8]: (a) enhancers reside thousands of base pairs upstream or downstream from the transcription start sites (TSSs) of their target genes or they can even be on different chromosomes relative to their targets, (b) they may exhibit tissue-specific properties and (c) they may initiate RNA polymerase II transcription, producing a new class of non-coding RNAs called enhancer RNAs (eRNAs).
Previous gene regulation studies have emphasized the role of enhancers in transcription initiation [9]. Analysis of enhancer properties has also raised key questions about mechanisms that govern the fate of temporal and tissue-specific gene expression. In addition, several studies [10, 11] have linked variations in enhancer sequences to cancer and other diseases. In particular, identifying enhancers and understanding their mechanisms of functioning is an area of great interest that may enrich our current knowledge about diseases and therapeutic strategies [12, 13].
So far, some review articles have focused on different aspects of enhancer functions that characterize cell identity or pathogenic states [14, 15]. In addition, the enhancer mechanistic properties aimed at identifying active enhancers are well documented in several studies and reviews, including advances in high-throughput experimental technologies [16–18]. However, because active enhancers are characterized by specific cellular properties and because there are numerous cellular conditions, experimental identification of enhancers faces certain limitations [17]. For this reason, computational identification of enhancers has been well studied in recent years and has resulted in a number of computational methods that complement the experimental techniques [19, 20]. Moreover, the generation of new types of high-throughput data helped to improve prediction models for enhancers. However, despite the efforts to develop accurate enhancer prediction methods [21–55], the current solutions generate significantly different enhancer predictions. In Table 1, we present the pairwise intersection of enhancer predictions as obtained in [50] by five state-of-the-art methods across six ENCODE (Encyclopedia of DNA Elements) cell lines. It is apparent that the overlap of computationally predicted sets of enhancers is relatively small. Consequently, it will be beneficial for the research community to have an overview of the strategies and solutions developed in this field.
Table 1.
Comparison analysis of enhancer predictions obtained by different methods across six ENCODE cell lines
| Method 1 versus Method 2 | Gm12878 | H1hesc | K562 | HeLa | HepG2 | HUVEC | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Coverage 1 versus. Coverage 2 (million bases) | Overlap (million bases) | Jaccard inde× (%) | Coverage 1 versus Coverage 2 (million bases) | Overlap (million bases) | Jaccard inde× (%) | Coverage 1 versus Coverage 2 (million bases) | Overlap (million bases) | Jaccard inde× (%) | Coverage 1 versus Coverage 2 (million bases) | Overlap (million bases) | Jaccard index (%) | Coverage 1 versus Coverage 2 (million bases) | Overlap (million bases) | Jaccard inde× (%) | Coverage 1 versus Coverage 2 (million bases) | Overlap (million bases) | Jaccard index (%) | |
| CSI-ANN versus ENCODE annotation | 10.7 versus 42.69 | 5.3 | 11.0 | 19.1 versus 56.5 | 2.2 | 3.0 | 34.5 versus 28.1 | 8.0 | 14.8 | 26.6. versus 42.9 | 5.3 | 8.3 | 40.8 versus 24 | 6.4 | 11.0 | 49.4 versus 47.1 | 21.3 | 28.3 |
| RFECS versus ENCODE annotation | 344.1 versus 42.69 | 31.6 | 8.9 | 124.7 versus 56.5 | 29.5 | 19.4 | 130.6 versus 28.1 | 18.5 | 13.1 | 87.4 versus 42.9 | 26.4 | 25.4 | 253.1 versus 24 | 12 | 4.5 | 191.2 versus 47.1 | 33.6 | 16.4 |
| ChromHMM versus ENCODE annotation | 82.7 versus 42.69 | 37.7 | 43.0 | 80.5 versus 56.5 | 36.9 | 36.8 | 111.4 versus. 28.1 | 24.8 | 21.6 | 70.9 versus 42.9 | 36.0 | 46.2 | 72.88 versus 24 | 10.8 | 12.6 | 107.2 versus 47.1 | 40.0 | 35.0 |
| Segway versus ENCODE annotation | 119.5 versus 42.69 | 39.1 | 31.7 | 404.9 versus 56.5 | 20.3 | 4.6 | 282.5 versus 28.1 | 27.6 | 9.7 | 124.8 versus 42.9 | 41.1 | 32.7 | 230.3 versus 24 | 11.6 | 4.5 | 189.6 s. 47.1 | 39.1 | 19.7 |
| CSI-ANN versusRFECS | 10.7 versus 344.1 | 6.1 | 1.7 | 19.1 versus 124.7 | 2.2 | 1.5 | 34.5 versus 130.6 | 11.4 | 7.4 | 26.6 versus 87.4 | 5.71 | 5.2 | 40.8 versus 253.1 | 12.6 | 4.4 | 49.4 versus 191.2 | 21.9 | 10.0 |
| RFECS versus ChromHMM | 344.1 versus 82.7 | 55.9 | 15.0 | 124.7 versus 80.5 | 40.3 | 24.4 | 34.5 versus 111.4 | 51.1 | 26.7 | 87.4 versus 70.9 | 42.1 | 36.1 | 253.1 versus 72.88 | 45.2 | 16.2 | 191.2 versus 107.2 | 73.7 | 32.7 |
| RFECS versus Segway | 344.1 versus 119.5 | 71.6 | 18.2 | 124.7 versus 404.9 | 52.3 | 10.9 | 130.6 versus. 282.5 | 80.5 | 24.2 | 87.4 versus 124.8 | 51.1 | 31.7 | 253.1 versus 230.3 | 77.8 | 19.1 | 191.2 versus 189.6 | 92.4 | 32.0 |
| CSI-ANN versus ChromHMM | 10.7 versus 82.7 | 6.0 | 6.8 | 19.1 versus 80.5 | 1.6 | 1.6 | 34.5 versus 111.4 | 12.0 | 8.9 | 26.6 versus 70.9 | 5.8 | 6.3 | 40.8 versus 72.88 | 10.9 | 10.6 | 49.4 versus 107.2 | 25.8 | 19.7 |
| ChromHMM versus Segway | 82.7 versus 119.5 | 63.8 | 46.1 | 80.5 versus 404.9 | 52.5 | 12.1 | 111.4 versus 282.5 | 100.6 | 34.2 | 70.9 versus 124.8 | 61.4 | 45.7 | 72.88 versus 230.3 | 56.3 | 22.8 | 107.2 versus 189.6 | 99.0 | 50.0 |
| CSI-ANN versus Segway | 10.7 versus 119.5 | 8.0 | 6.5 | 19.1 versus 404.9 | 1.3 | 0.3 | 34.5 versus 282.5 | 19.74 | 6.6 | 26.6. versus 124.8 | 10.0 | 7.1 | 40.8 versus 230.3 | 18.1 | 7.1 | 49.4 versus 189.6 | 30.4 | 14.5 |
We report the total number of bases in millions predicted as belonging to enhancers. Coverage 1 corresponds to enhancers predicted by Method 1, while Coverage 2 corresponds to enhancers predicted by Method 2. The overlap column corresponds to the same enhancer predictions in million bases as obtained by Method 1 and Method 2. In the third column, we report similarity of predictions of Method 1 and Method 2 based on the Jaccard similarity index (as percentage)
With this issue in mind, we focused our efforts on bioinformatics approaches for enhancer identification published from 2000 to 2015, characterized by the use of data from high-throughput experiments for the development of enhancer prediction models. First, we present the basic principles of a general framework for enhancer identification. Next, we cover a comprehensive list of over 30 existing enhancer recognition tools and methods that have been developed in the considered period. Our aim is to analyse the existing approaches to provide useful comments regarding the data sets used and the prevalent computational solutions. In a separate section, we comment on obstacles that the existing methods face, address challenges and open questions related to enhancer identification and hint on promising directions for future research. Finally, we summarize available enhancer resources and suggest pragmatic guidelines for using the available computational solutions and relevant enhancer data.
Computational identification of enhancers: the framework
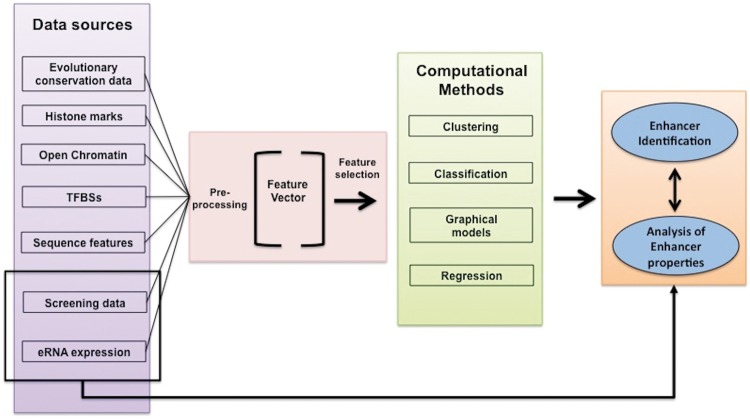
The problem of computational identification of enhancers can be formulated as follows: ‘Given a DNA region described by multiple data types, determine if it can function as an enhancer'. Figure 1 depicts an overview of a general enhancer identification process.
Figure 1.
This figure shows basic components of a general enhancer identification system. The first block on the left (lille colour) handles integration and preprocessing of different data types. These data types (summarized in Table 2) can be combined in different ways to generate feature vectors that describe DNA regions. The feature values can be normalized or rescaled (second block-red colour). Then, FS techniques can be applied to reduce the number of features and select smaller sets of features with higher discriminative capabilities. The feature vectors feed computational models that make decisions using unsupervised and/or supervised algorithms (third block-green colour). Outcome is a list of identified enhancer regions (fourth block-orange colour), which can be analysed further using computational techniques.
The first step concerns integration of different data types coming from different data sources and preprocessing to generate feature vectors that serve as input for the enhancer identification and analysis system. The feature vectors contain information that describes data instances. Typically, these feature vectors capture information about evolutionary conservation [20] (e.g. regions or motifs that are highly conserved across different species), and/or chromatin profiles of histone marks as derived from ChIP-seq (chromatin immunoprecipitation with massively parallel DNA sequencing) data [28] and/or chromatin accessibility information as derived from DNase I hypersensitivity sites (DHS). The previous data types are frequently combined with transcription factor-binding sites (TFBSs) for identifying different classes of regulatory elements (e.g. enhancers, promoters, etc.) [25]. Note that with the acronym TFBSs, we refer to both the actual and the predicted DNA-binding sites of DNA-binding proteins that facilitate transcription, including transcription factors (TFs) and additional binding proteins or protein complexes such as the nucleosome remodelling complex (e.g. SWI/SNF), or histone acetyltransferases (HATs; e.g. P300 from HATs) and histone methyltransferases (HMTs; e.g. ASH1L from HMTs). Recently, enhancer-screening data, as well as expression of eRNAs, can serve as input for identifying enhancers and analysing their properties. In Table 2, we present an overview of the features used by different computational methods for enhancers’ identification. The process of generating feature vectors may include additional steps of normalization or rescaling of the feature values.
Table 2.
Overview of data and features used for enhancer identification
| Data sources | Feature example | Advantage | Disadvantage | Representative methods |
|---|---|---|---|---|
| Evolutionary conservation | Conserved motifs across species | Easy to compute | Insufficient information for predicting enhancer's tissue-specific activity | [20] |
| Histone marks | ChIP-seq from H3K4me1 | Provides cell-line-/tissue-specific information that characterize enhancers and also different categories of enhancers (e.g. poised versus active) | Different cell lines/tissues are associated with different combination of histone marks | [21, 28, 33, 34] |
| TFBSs | ChIP-seq from P300 | Provides cell-line-/tissue-specific information that characterize enhancers. High-resolution data for testing activity of enhancer-related TFs | Not available for many cell lines/tissues | [23, 29] |
| Open chromatin | DHS | High discriminative capacity when combined with other data types, e.g. P300-binding sites | Regions with enriched DHS activity do not necessarily correspond to enhancers | [25] |
| Sequence characteristics | Kmers of size 5 | Easy to compute | Insufficient information for predicting enhancers’ activity across different tissues | [39, 51] |
| eRNA expression | CAGE data | High accuracy | eRNA regulation mechanisms are unknown, and not all of the enhancers are known to produce eRNAs | [40] |
| Enhancer-screening data | STARR-seq | High accuracy for testing enhancer activity | Not useful for ab initio discovery of enhancers | [42, 43, 52] |
In the second step, different computational models use feature vectors to annotate DNA regions. The computational models are developed by computational methods, unsupervised or supervised, using the same feature vectors to describe the data. The methods used include state-of-the-art clustering algorithms such as K-means [21] or bi-clustering [24], probabilistic graphical models (PGMs) such as Hidden Markov Models (HMMs) [30] or Dynamic Bayesian Networks (DBNs) [31], regression models such as least absolute shrinkage and selection operator [53] and more advanced supervised classification systems, such as support vector machines (SVMs) [34], artificial neural networks (ANNs) [33], decision trees (DTs) [38] and random forests (RFs) [37]. The most important difference between supervised and unsupervised techniques is the fact that supervised methods require prior knowledge (e.g. some representative enhancers and when available, non-enhancer examples) for training. In contrast, this is not the case for unsupervised methods, where enhancer regions (and other regulatory elements in general) can be identified ab initio and without any prior knowledge. Unsupervised techniques rely strongly on some ad hoc rules for assigning regions to the class of enhancers, and thus their predictive abilities have some limitations. An example is identification of enhancers using only H3K4me1 profiles, which of course is correct, but is insufficient because there is no guarantee that they can characterize in the same way enhancers from different cell lines and tissues can.
The main outcome of an enhancer identification system is a catalogue of predicted enhancers. The identified enhancers can be further analysed computationally for their properties, deciphering their regulatory roles and associating them with target genes and eRNAs.
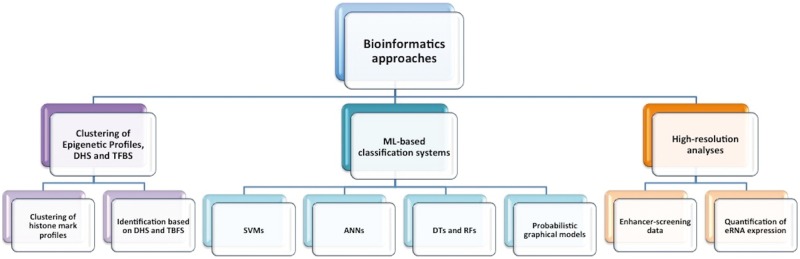
A conceptually simple way to classify enhancer identification methods can be based on the available data sources (e.g. grouping together all methods that rely on evolutionary conservation). However, this is not readily applicable because different methods rely on a mixture of different data sets/features, and frequently the deployed algorithms combine supervised and unsupervised components. In this review, we group the available methods into three categories. The first category includes computational methods that identify DNA regulatory elements (including enhancers) using epigenetic signatures such as ChiP-seq of histone marks, DHS peaks and/or TFBSs mainly through unsupervised learning and clustering techniques [21–29]. The second category represents systems based on supervised machine learning (ML) classification that use mainly ChIP-seq data of histone marks frequently combined with sequence motifs, to distinguish enhancers from non-enhancers and identify features that characterize enhancers in an optimized way [33–39]. In this category, we also cover methods based on PGMs that are in the group of supervised learning methods [30–32]. As the third category, we consider recent bioinformatics methods that identify enhancers using as input experimental enhancer-screening data and data from some more targeted experiments. Although these methods are in principle experimental, the analysis of the results relies strongly on advanced bioinformatics methods combined with ML algorithms for deciphering the enhancer context [42–49]. Figure 2 gives the outline of existing bioinformatics approaches for enhancer identification. In Table 3, we further highlight the most popular approaches and mark those that are accessible and functional.
Figure 2.
The figure presents the roadmap of existing approaches for enhancer identification. We have categorized the methods into three basic streams, which we partitioned further into subcategories based on the underlying computational solutions and the combination of relevant enhancer data.
Table 3.
Summary of the most popular bioinformatics approaches for enhancer identification
| Name | Computational method | Highlight | Link | Reference |
|---|---|---|---|---|
| Heintzman et al. | Clustering and correlation of histone marks profiles | High-recognition performance in HeLa | – | [21] |
| ChromaSig (*) | Identification of specific histone mark motifs and clustering | The method is sensitive enough to capture patterns characterizing different classes of enhancers. | http://bioinformatics-renlab.ucsd.edu/rentrac/ wiki/ChromaSig | [22] |
| Rye et al. | Clustering of profiles | The results indicate that selection of relevant TFs may be sufficient to identify regulatory elements | – | [23] |
| Won et. al. | HMMs | State-of-the-art method suggesting that HMMs are capable of integrating information from multiple histone marks for predicting regulatory elements | http://http/nash.ucsd.edu/chromatin.tar.gz | [54] |
| Boyle et al. | Combination of DHS with TFBSs | Active enhancers usually overlap with open chromatin regions, but not all of the DNA accessible regions correspond to enhancers | – | [25] |
| ChromHMM (*) | HMMs | State-of-the-art genome annotation method by ENCODE | http://compbio.mit.edu/ChromHMM | [30] |
| Segway (*) | DBNs | State-of-the-art genome annotation method by ENCODE | http://www.pmgenomics.ca/hoffmanlab/ proj/segway/ | [31] |
| ChroModule | HMMs | Annotated human genome for eight cell lines and improved the AUC compared with state-of-the-art HMM based methods | – | [32] |
| CSI-ANN (*) | ANNs | Effective combination of ANNs with FDA for FS | http://www.healthcare.uiowa.edu/labs/ tan/CSIANNWebpage.html | [33] |
| ChromaGenSVM (*) | SVMs | Effective combination of SVMs with GA for optimization and FS | http://sysimm.ifrec.osaka-u.ac.jp/download/ Diego/ | [34] |
| EnhancerFinder | MKL | Functional genomics combined with sequence motifs can accurately identify developmental enhancers | – | [35] |
| RFECS (*) | RFs | Method less prone to overfitting, which introduces additional novelties on the way enhancer predictions are validated | http://enhancer.ucsd.edu/renlab/RFECS_ enhancer_prediction/ | [37] |
| DEEP (*) | SVMs and ANNs | Novel ensemble-learning-based algorithm with good generalization capabilities in unknown cell lines.. | http://cbrc.kaust.edu.sa/deep/ | [36] |
| kmer-SVM (*) | SVMs | Study extensively the enhancer sequence context | http://kmersvm.beerlab.org/ | [39] |
| dREG (*) | SVR | Usage of GRO-seq data combined with regression analysis | https://github.com/Danko-Lab/dREG/ | [41] |
| DELTA (*) | AdaBoost | Introduces the concept of shape features from ChiP-seq data | https://github.com/drlu/delta | [38] |
| Andersson et al. (*) | eRNA expression analysis | Introduces one of the most accurate features for enhancer identification | http://enhancer.binf.ku.dk/enhancers.php | [40] |
| CoSBI (*) | Bi-clustering | Reports combination of histone marks with high discriminative power for the category of enhancers | http://www.healthcare.uiowa.edu/labs/tan/ CoSBIWebpage.html | [24] |
Note. With (*), are marked the methods that provide source codes or executable files.
Identification of enhancers based on clustering of epigenetic profiles, DHS and TFBSs
Over the past years, advances in high-throughput experiments such as ChIP-seq have generated vast amounts of data describing the epigenetic landscape of different human and non-human cells and tissues [55–57]. The produced data characterize profiles of different epigenetic marks, identify or estimate many TFBSs and describe the chromatin accessibility of DNA. Systematic analysis of these data generated global epigenetic maps for different cell lines and tissues and enabled inference of the core principles that characterize different categories of DNA regulatory elements [58]. For example, based on data from ChIP-seq experiments, it is found that active enhancers are frequently associated with H3K27ac, while active and poised enhancers are associated with H3K4me1 [17]. Such information made space for the development of several computational methods for identification of enhancers and other regulatory elements in a cell-line-/tissue-specific context. Essentially, all of the methods that fall into this category initially estimate the profiles (therefore called epigenetic signatures) of histone marks and/or the profile of DHS from different genomic regions. In a later step, these genomic regions are assigned into different regulatory classes via unsupervised learning techniques (e.g. grouping of similar epigenetic profiles) or by the binding fingerprint of enhancer-related TFBSs [59–61].
Methods based on Clustering of Chromatin Profiles
Typical example of this subcategory is the bioinformatics analysis presented in Heintzman et al. [21], which studied the chromatin landscape of promoters and enhancers in HeLa cell line from ENCODE experiments [62]. In the first stage, the analysis revealed that promoters are characterized by H3K4me3, while enhancers are characterized by H3K4me1, but not H3K4me3. In the second stage, the outcome of this analysis served as a basis for developing a two-step algorithm that scans genomic regions from new cell lines and classifies genomic segments as promoters and enhancers based on the similarity of chromatin profiles with existing annotated segments. Although the reported enhancers [21] were derived from a single data set, the main findings have served as a baseline for many subsequent studies for enhancers characterized by the presence of P300-binding sites. Another example is ChromaSig [22] that uses signatures of nine core chromatin marks to generate groups of distinct histone modification profiles that can be further assigned to different classes of regulatory elements. Analysis over HeLa and CD4T cells identified 8 and 16 clusters of chromatin profiles, respectively, that were enriched in enhancer- and promoter-related TFBSs. Overall, ChromaSig is sensitive enough to distinguish different classes of enhancers, and the results are in agreement with the enhancer lists reported by previous studies [21].
Following the above-mentioned concepts, several other methods [23, 24] used diverse data sets and different clustering techniques to identify enhancers. As an example, clustering of TFBS profiles from 67 binding factors and 9 histone marks from ENCODE Gm12878 and K562 cell lines revealed that between those two cell lines, H3K4me1 marker is more frequent in enhancer clusters compared with P300 or H3K27ac [23]. The main outcome of this study indicates that an adequate selection of TFs may be used to identify different regulatory elements in the genome. In another study, the problem of describing more effectively combinatorial histone modification patterns is tackled using a novel algorithm for clustering called CoSBI (Coherent and Shifted Bicluster Identification) [24]. CoSBI follows the concept of coherent bi-clustering applied to 39 chromatin modification maps from CD4T cells [63]. The algorithm reported 843 patterns of core chromatin modification marks that effectively distinguish different regulatory elements, including the category of enhancers.
Methods based on Chromatin Accessibility and TFBSs
There are several other studies for enhancer recognition that rely mainly on the effective combination of DHS footprints with TFBSs of enhancer-related binding factors like P300 or CREBBP (therefore called CBP) [64, 65]. Here, we highlight the high-resolution identification of DNA regulatory elements in seven lymphoblastoid cell lines and other five human cells/cell lines with diverse characteristics [K562, HeLa, HUVEC, NHEK and embryonic stem cells (ESCs)] [25]. Active enhancers were found to overlap with DHS. Note that not all highly accessible DNA regions correspond to enhancers. To mitigate the above-mentioned limitation, DHS information can also be combined with more advanced algorithms such as CENTIPEDE [26] and Wellington [27] for identifying binding sites of enhancer-related binding factors. We note that TFBSs and ChIP-seq data from histone marks, combined with PGMs and clustering techniques, have been successfully applied to studies of the mouse genome [28, 29]. Finally, an algorithm called Prestige [66] uses histone H3K4me1 profiles from ChIP-seq data, combined with gene expression from RNA-seq, to identify enhancers and associate variations of the enhancer region sequences with diseases through genome-wide association studies.
Identification based on ML classification methods
Methods of this category reformulate the enhancer identification problem as a binary classification task for predicting enhancer regions as being different from non-enhancer (negative control) regions. So far, SVMs, ANNs, DTs, RFs, PGMs and ensemble techniques have successfully been applied. All these methods have found use in bioinformatics [67–69] and could be applied to enhancer prediction problems [30–38]. We also note that ensemble-learning methods have documented advantages for the class-imbalance problem, which is also present in enhancer identification [69]. Briefly, the class-imbalance problem occurs when the number of samples from the class of interest (e.g. enhancers) differs significantly from the number of samples from other classes (e.g. non-enhancers).
Typically, supervised ML classification systems are combined with feature selection (FS) techniques to extract small sets of features (in our case, histone modification marks and/or sequence characteristics and/or TFBS/binding motifs), which, all together, are capable of maximizing the separation between enhancers and non-enhancers [70, 71]. In addition, a combination of supervised classification systems with global optimization techniques such as Genetic Algorithms (GA) or Simulated Annealing can be used for tuning the model parameters and optimizing several steps of the enhancer recognition process [72].
Solutions that use PGMs
The methods we survey here are used for genome-wide annotation purposes. In principle, some of these tools [30, 31] segment genomes into intervals and develop PGMs from large numbers of chromatin modifications coming from multiple cell lines and tissues. The identified chromatin states are then grouped and annotated as enhancers, promoters, repressed regions or transcribed regions based on the known functional sites.
The most popular genome-wide annotation tool for genome segmentation in the above-mentioned manner proposed by the ENCODE consortium is ChromHMM [30]. ChromHMM uses a probabilistic model based on a multivariate HMMs. ChromHMM segments the genome into 200 bp intervals, and a single model is trained on data from six available cell lines. Segway [31], on the other hand, is an alternative genome annotation tool based on DBNs. Segway offers a higher-resolution analysis because it annotates the genome for every single base (e.g. has 1 bp resolution). In addition, it trains cell-specific models and is more computationally demanding than ChromHMM.
Although ChromHMM and Segway were developed independently, the ENCODE consortium combined these programs to annotate the human genome in a more comprehensive way. The annotation proposed by Hoffman et al. [73] combines the results produced by ChromHMM and Segway with other relevant experimental data such as DHS, FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) assays and several ChIP-seq data sets for transcription regulators (e.g. CTCF, POL II, P300) to generate annotation maps for Gm12878, K562, H1, HeLa, HepG2 and HUVEC cell lines. Note that this annotation serves as the baseline annotation proposed by the ENCODE consortium. Specifically, the integrative annotation categorizes enhancers into three states, Enh, EnhF and EnhWF, with Enh representing the class of enhancers with the strongest enrichment of TFBS (therefore called strong enhancers) [73]. Finally, other probabilistic graphical methods for enhancer identification exist, as well as many independent genome annotation tools [32, 54, 74, 75]. Here, we highlight ChroModule [32], which annotated human genome characteristics for eight cell lines and reported higher recognition performance compared with [30] as indicated by the area under curve (AUC).
Solutions that use ANNs
In particular, CSI-ANN [33] is one of the first enhancer classification systems that rely on an ANN using chromatin signatures as input. Putative enhancers derived from human CD4T cell data from Wang et al. [63] based on P300 ChIP-seq peak distal to TSS overlapping with computationally predicted enhancers from PreMod database [76]. The FS component of CSI-ANN, based on Fisher Discriminant Analysis (FDA), reported several histone marks such as H3K4me3, H4Ac and H3, which separate enhancers from background sequences in an optimized way. In terms of recognition performance, CSI-ANN reported higher Positive Predictive Value (PPV) on untreated HeLa cells (maximum PPV of 66.3% based on the overlap of predictions with P300- or DHS- or TRAP220-binding sites) as compared with [21] and [54].
Solutions that use SVMs
ChromaGenSVM [34] is a typical enhancer classification system that uses SVMs. ChromaGenSVM is trained on HeLa enhancer data (the authors also developed a second model on CD4+T cells from [63]) from Heintzman et al. [21] using core ChIP-seq histone modification markers. For FS and SVM parameter optimization, ChromaGenSVM uses a global optimization technique based on GA. The optimal ChromaGenSVM model identified histones H3, H3K4me1 and H3K4me3 as the most prominent features for describing enhancers versus the background sequences. In terms of recognition performance, ChromaGenSVM reported PPV ∼90% on CD4+T and on untreated HeLa cells achieved comparable PPV with [21], [33] and [54] (maximum PPV of ∼57% based on the overlap of predictions with P300- or DHS- or TRAP220-binding sites).
The idea of integrating diverse data sets from multiple sources to accurately identify developmental enhancers is the main contribution introduced by EnhancerFinder [35]. EnhancerFinder’s underlying classification method is based on the use of Multiple Kernel Learning (MKL), with the training data sets derived from VISTA database [77]. EnhancerFinder also investigates the discriminative power of different data sets and features, concluding that sequence motifs, combined with functional genomics data (e.g. H3K4me1 or P300), are capable of identifying enhancers. This, of course, relates only to a subset of enhancers. In terms of recognition performance, when applied to the entire genome, EnhancerFinder predicted 84 031 developmental enhancers and achieved much higher recognition performance compared with [30] and [31].
To achieve better generalization capabilities in unknown tissues and cell lines, DEEP (Dragon Ensemble Enhancer Predictor) [36] introduces a two-layer classification algorithm based on SVMs and ANNs and training based on data from multiple cell lines and tissues. In its first step, DEEP trains multiple SVM models on data from different cell lines and tissues, which are combined in a second step via an ANN for finally distinguishing enhancers from non-enhancers. DEEP uses putative enhancers from the ENCODE annotation proposed by Hoffman et al. [73], actively transcribed enhancers from FANTOM5 (Functional Annotation of the Mammalian Genome) Atlas [40], and a small set of developmental enhancers achieved in VISTA database [77]. An exhaustive search technique applied on the set of 11 core histone modification markers revealed that different ENCODE cell lines are characterized by different optimized sets of histone marks. In these sets, only H3K4me1 characterizes enhancer regions from different cell lines studied in DEEP. In terms of performance, DEEP reported higher PPV compared with [30, 31, 33 and 37] on HeLa and K562 cell lines (PPV was computed based on the overlap of predictions with P300-binding sites or DHS). When considering the number of predicted enhancers that overlap with promoters, DEEP achieved lower or comparable overlap with the competitor methods.
Solutions that use DTs and RFs
For reducing the effects of class-imbalance between enhancer/non-enhancer samples and eliminating limitations coming from the small size of the training data, RFECS (Random Forest-based Enhancer identification from Chromatin States) [37] introduces a RF-based classification system trained on H1 and IMR90 data from the NIH Epigenome Roadmap project [78]. RFECS introduces additional novelties in the way putative enhancer regions are selected and in the way genome-wide predictions are validated. Overall, RFECS tested on CD4+T and H1-hESC cell lines achieves higher true-positive rate and lower false-positive rate compared with state-of-the-art enhancer recognition systems [33, 34, 54] (RFECS achieved true-positive rate of ∼70% and ∼82.5% and false-positive rate of ∼7% and ∼4.9%, respectively). We note that the true-positive rate was measured by the overlap of predictions with DHS-, P300- and CBP-binding sites and the false-positive rate was measured by the overlap of predictions with TSSs as annotated by UCSC Genome Browser. In addition, an out-of-bag FS technique reported histone marks H3K4me3, H3K4me1 and H3K4me2 as the most important features for the enhancer’s recognition problem by this approach. DTs have been successfully applied in another method called DELTA (Distal Enhancer Locating Tool based on AdaBoost) [38]. DELTA is based on the AdaBoost algorithm applied to a set of features characterizing the shape of ChIP-seq peaks of core chromatin markers. In terms of performance, DELTA further improved the prediction accuracy on CD4+T and H1-hESC cell lines, achieving a misclassification rate of 2% and 1.6%, respectively.
Solutions that use classification algorithms to study the enhancer DNA sequence context
The problem of identifying enhancers based solely on sequence characteristics (e.g. motifs or kmers) is tackled in [79]. In another study [51], sequence features capable of discriminating mammalian enhancer sequences from random genomic loci are systematically identified. The proposed ‘kmer-frequency vector' [39], which captures the full set of kmers of varying length (3–10 nucleotides), and its refined version called ‘gapped kmer-vector' [80] were used in SVM models to predict enhancers.
Identification of enhancers using high-resolution data
The presence of deep sequence data has enabled development of a variety of bioinformatics methods to detect active enhancers and test directly their ability to trigger transcription in messenger RNA (mRNA) promoters. Nowadays several enhancer-testing and in vivo-screening methods exist for human, mouse, flies and yeast, such as STARR-seq [44], CRE-seq [45], FIREWACh [46] and several others [47–49], which are surveyed comprehensively in [17].
Methods based on Enhancer Screening Data
This subcategory of methods describes bioinformatics analyses for investigating mechanisms that trigger regulation activities related to enhancers and promoters, combining several high-throughput data sets, sequence characteristics or TFBSs and more targeted mutation experiments [81, 82]. A typical example is an analysis based on MPRA-derived data (massively parallel reporter assay) from K562 and Hep cell lines that reconfirmed previously published results for cell type specificity of enhancer chromatin states [42]. In a similar fashion, functional testing of computationally predicted enhancers with CRE-seq data in K562 cell line revealed that previously reported chromatin states can distinguish active enhancers from negative samples, but TFBS motifs also have high discriminative power and characterize in a better way the most active enhancer regions [43]. Note that an analysis based on STARR-seq data from Drosophila cells reported interesting mechanistic properties of enhancers and can serve as a paradigm for similar studies in humans [52].
Identification based on Quantification Analysis of RNA
A popular subcategory identifies enhancer regions using high-throughput techniques that measure the production of RNA based on Cap Analysis of Gene Expression (CAGE) or calculation of transcription rate using Genomic Run-on (GRO-seq). In particular, using bidirectional CAGE tags, over 135 tissues and 241 cell lines were analysed in FANTOM experiments [83]. A total of 43 011 putative enhancer regions that were depleted in CpG islands were reported [40]. The so-called ‘Atlas of actively transcribed enhancers' also reported core differences between enhancers and mRNA promoters, whereas the results complement findings reported by the ENCODE consortium. Note that another CAGE analysis from FANTOM5 data revealed that transcription in enhancer regions is the earliest event that leads to many subsequent transcriptional changes during cellular differentiation [84]. Finally, a high-throughput recognition system called dREG [41] uses GRO-seq data [85] and Support Vector Regression (SVR) to identify and characterize effectively active transcriptional regulatory elements, including the category of enhancers.
Challenges and obstacles in computational identification of enhancers
Here, we address several challenges and open questions related to the enhancer identification.
Challenges and open questions
Computational prediction of enhancers does not guarantee that the identified enhancers are real. Because there exists no large, sufficiently comprehensive and experimentally validated enhancer set for humans (or other species), one of the major issues related to enhancer identification is how to assess the correctness of predictions. One possible way of validation is to link the predicted enhancers to their target genes. This, complementary to computational prediction of enhancers, is without a doubt the most difficult challenge. Below, we summarize the most important streams for enhancer target identification, and we discuss relevant sub-problems:
Enhancers can be located relatively close (e.g. few thousands of bases) or much further away (e.g. hundred thousands of bases) to the genes they affect [86]. Consequently, some methods identify enhancer targets based on their relative location to enhancers (e.g. an enhancer interacts with its neighbouring mRNA promoter). These models are oversimplified because there are no clear distance boundaries for the enhancer–promoter interactions. Some of the existing approaches [87] have defined arbitrary thresholds for the relative location of enhancers and mRNA promoters (e.g. minimum distance 5000 bases and maximum 125 000 bases). Although these approaches are easy to implement, they generate a trade-off between distance threshold and number of true and false positives.
More sophisticated approaches for identifying enhancer targets can be based on correlated activity of enhancers and mRNA promoters. This category is promising because it is based on cell-line-/tissue-specific information. However, the largest obstacle stems from the limited knowledge about enhancer and mRNA promoter co-activity [40, 65]. One possible solution can be based on the identification of all possible pairs of enhancers and promoters within a predefined distance threshold combined with correlation analysis and representative data sets and markers (e.g. correlated expression activity between eRNAs and target genes or correlated DHS activity) [84]. However, this is also challenging because enhancers and mRNA promoters have many-to-many relationships, meaning that one promoter can be associated with multiple enhancers, and one enhancer can be associated with different promoters. Thus, the problem becomes computationally expensive, and efficient pruning techniques are required to restrict the number of candidate associations between enhancers and promoters.
The most promising direction for identifying enhancer–promoter associations can be based on chromatin conformation data as captured by 3C/5C [88] or ChIA-PET (Chromatin Interaction Analysis by Paired-End Tag Sequencing) [89]. These data sets can be used to identify associations of enhancers with known mRNA promoters in the three-dimensional space. A typical example of this category is the method introduced in [86], which combines ChIA-PET data with supervised learning based on RFs for linking enhancers to their target genes. With all these methods, there are still areas for improvements, such as noise and bias removal in chromatin conformation data sets or utilization of additional features to link enhancer–promoter associations with regulatory functions with much higher confidence.
Except for the enhancer target identification, identifying the tissue-specific activity of enhancers is another promising area of research. For example, histone modification mark data, DHSs, different TFBSs as derived from ChIP-seq experiments and expression of eRNAs can characterize enhancers in a cell-line-/tissue-specific context. In contrast, sequence characteristics or evolutionary-conserved motifs do not contain sufficient information to describe enhancer activity in different tissues. Consequently, methods that rely solely on ChiP-seq data from histone marks, DHS and/or TFBSs may maximize the enhancer recognition performance in specific cell lines and tissues, but frequently the developed models achieve lower generalization capabilities in unknown cell lines [21, 33–36]. To mitigate this trade-off, mixtures of cell-specific features and sequence characteristics appear to be a promising direction [35, 36].
Another important challenge related to the enhancer identification problem concerns the role of eRNAs in transcription regulation. Recent evidence [90] indicates that many TSSs of eRNAs and protein-coding genes present similar architecture that is differentiated only at the post-transcriptional regulatory layer. Consequently, understanding the functional mechanisms of eRNAs and inferring rules that link eRNA transcription with transcription initiation through mRNA promoters [84] is a question warranting further exploration.
Obstacles of existing approaches
Many obstacles derive from the input data sets that existing methods use and the fact that an optimal combination of features for describing enhancers across different cell-lines and tissues does not exist. [36]. There are also specific technical limitations introduced by the existing computational solutions.
Regarding the used data sets and features, it is documented that information on evolutionary conservation cannot help much [91] in the prediction of enhancers’ activity because few non-coding elements and motifs appear to be well conserved in other species, and because enhancers are largely tissue specific. On the other hand, ChIP-seq data for histone marks and TFBSs capture cell-line-/tissue-specific information. Using these ChIP-seq data, however, requires a demanding data preprocessing phase. This preprocessing phase usually segments genome into small intervals (e.g. 100or 200 bp), but a clear answer to the optimal way of selecting this interval size does not exist. The step of identifying significant ChIP-seq peaks (therefore called the peak-calling step), as derived from programs like MACS (Model-based Analysis of ChIP-Seq) [92] or SICER (Spatial clustering approach for the identification of ChIP-enriched regions) [93], is sensitive to the selection of parameters, which are usually data set dependent and different among different cellular conditions (e.g. HeLa versus K562). Guidelines about the optimal selection of publicly available peak-calling programs for ChIP-seq data can be found in [94] and [95]. Note that some of the existing approaches for enhancer prediction recommend use of specific ChIP-seq peak-calling programs [34, 37], which represent a limitation because different and possibly better solutions for peak calling could be available in future. Furthermore, ChIP-seq data are not available for many of the existing cell lines and tissues. This represents a real obstacle, as it limits the scope of potential studies that rely on such information. To mitigate this problem, data imputation techniques for histone modification marks have been proposed [96].
Moreover, methods that rely on DHS footprints for finding regulatory elements usually lack specificity between different functional categories (e.g. promoters versus enhancers versus insulators) [97]. In other words, DNA regions with enriched DHS activation are not necessarily enhancers. Also, the identification step of TFBSs is also problematic because not all of the enhancers are marked by the same combination of regulatory proteins or present similar histone modification patterns. This simply means that genomic regions with enrichment in specific histone marks (e.g. H3K4me1) or binding factors (e.g. P300) are not necessarily enhancers. To complicate the problem even more, even the antibodies that are used by ChIP-related experiments may not be always available because enhancers are characterized by different (and maybe unknown) combinations of enhancer co-activators [4]. On the other hand, identification of binding sites based on the Positional Weight Matrices (PWMs) prediction models faces limitations and frequently achieves poor recognition performance [98, 99].
Further, supervised and unsupervised ML methods also face limitations. For the unsupervised clustering of histone mark profiles, rules that have been applied for identifying enhancers are not general enough because different combinations of histone markers and enhancer-related TFBSs characterize enhancers in different cell lines and tissues. This argumentation raises several questions that need to be addressed. For example, to what extent chromatin-defined enhancers in multiple cell lines/tissues have exactly the same chromatin states? Or which cell lines and tissues have exactly the same sets of active enhancers?
In addition, the main challenge that all of the ML-based classification methods face is the selection of high-quality samples to represent adequately the positive (enhancers) and negative classes (non-enhancers). In the absence of a ‘ground truth enhancer' data set, the first ML-based classification systems introduced rules to select enhancer regions for training [33, 34, 37]. The most prominent rule is the selection of DNA segments distal to protein-coding TSSs, characterized by open chromatin as indicated by DHS data that are also enriched in enhancer-related TFBSs (e.g. P300 and/or CBP). For the selection of negative samples, random sequences not annotated as enhancers or promoters are frequently used. An alternative way to generate negative control samples is to shuffle the genomic content of existing enhancer regions (e.g. scrambled enhancers). However, with the recent advances on computational and experimental techniques, the ENCODE integrative annotation [70], the Atlas of actively transcribed enhancers [40], the VISTA enhancer browser [75] and the outcome of individual studies based on enhancer-screening data (similar to those we summarized before) can serve as baseline sources for implementing more reliable ML-based recognition systems [35, 36].
Finally, the class-imbalance problem [36, 37], tuning of classification model parameters (e.g. number of neurons or hidden layers for ANNs or parameter C and gamma for SVMs) [34], overfitting issues, poor generalization capabilities of the developed models in unknown cell lines/tissues and ad hoc rules for validating genome-wide predictions of enhancers are some technical problems related to enhancer recognition via ML-based classification systems.
Enhancer-related resources
In this section, we report available online resources related to enhancers, which include databases, repositories of experimental data, computational tools and other material useful for subsequent enhancer identification studies.
Regarding the enhancer databases, PReMod [76] (http://genomequebec.mcgill.ca/PReMod/) and PEDB (Mammalian Promoter/Enhancer DataBase) [100] (http://promoter.cdb.riken.jp/) are two of the first resources that archived computationally predicted enhancers in human and mouse. Currently, the state-of-the-art database for enhancers is the ‘Human Transcribed Enhancer Atlas' that contains actively transcribed enhancers based on the analysis of eRNA expression [40] (http://enhancer.binf.ku.dk/enhancers.php). Except for the list of human enhancers in multiple tissues and organs, the Atlas contains utilities for downstream analysis, such as TF motif enrichment in enhancer sequences, as well as a selection of enhancers based on expression levels. In addition, all the results are publicly available as flat files or can be visualized in the Genome Browser. On the other hand, VISTA enhancer browser (http://enhancer.lbl.gov/) contains a set of developmental enhancers extremely conserved in mouse and human [77]. This list of developmental enhancers is experimentally validated in mouse [77]. There are also some other enhancer sources that archive enhancers in an integrative way. Examples are dbSUPER (http://bioinfo.au.tsinghua.edu.cn/dbsuper/index.php), which contains 66 033 super enhancer regions predicted [101] from 96 human and 5 mouse tissues, and DENdb (Dragon Enhancer DataBase) [50] (http://www.cbrc.kaust.edu.sa/dendb/), which is the first online repository of putative enhancers, from 15 ENCODE cell lines computationally predicted by five state-of-the-art ML enhancer recognition systems. DENdb also incorporates utilities such as overlap of enhancers with TFBS from ChIP-seq data or predictions of TFBSs obtained by PWM from HOCOMOCO (Homo Sapiens Comprehensive Model Collection) database [102], interactions of enhancers with other genomic loci as captured by chromatin conformation technologies such as 3C/5C or ChIA-PET archived in 4DGenome database [103] (http://4dgenome.int-med.uiowa.edu/) and overlap of enhancers with open chromatin regions via DHS.
Conclusion
Bioinformatics approaches for enhancer identification are valuable for validating hypotheses and assumptions in gene regulation studies. Here, we went through >30 bioinformatics approaches that have been developed over the past few years. We covered three basic streams of computational methods including: (a) methods that identify DNA regulatory elements via clustering of histone marks profiles, open chromatin information and TFBSs; (b) ML-based classification systems; and (c) bioinformatics analyses based on high-resolution enhancer-screening data sets.
During our review process, we identified and reported limitations and advantages of the existing computational methods. In addition, we summarized a comprehensive list of enhancer resources that include databases for enhancers, data repositories and open-source programs useful for further analyses. A large-scale comparison analysis of the performance of the existing methods may provide meaningful insights about the discriminative capacity of different genomic and epigenetic data sets that feed different computational solutions.
We also commented on some promising areas of research, and we reported challenges that require further investigation. Among them, linking enhancers with their in vivo target genes and understanding the role of eRNAs for transcription regulation are among the most challenging topics for future research.
To conclude, we anticipate that our review will complement subsequent gene regulation studies aimed at resolving questions regarding the role of enhancers into cellular transcriptional activities.
Key Points
Interplay between histone modification profiles, open chromatin information and TFBSs can characterize enhancer regions with increased accuracy in a cell-line-/tissue-specific content.
Developed models based on SVMs, ANNs, RFs, use with various level of success, features such as binding sites of P300, CBP, TRAP220 proteins, sequence compositional properties, DHS, different chromatin marks.
The effectiveness of ML models critically depend on the selected set of features, and the most promising solutions use combinations of features deriving from genomic and epigenomic data.
Acknowledgements
We thank Haitham Ashoor from the Computational Bioscience Research Center of King Abdullah University of Science and Technology for his helpful comments.
Biographies
Dimitrios Kleftogiannis is a PhD candidate at King Abdullah University of Science and Technology (KAUST). His research interests include Gene Regulation, Computational Biology and Bioinformatics.
Panos Kalnis is professor of Computer Science at King Abdullah University of Science and Technology (KAUST). His current research interests include Databases, Information Management and Bioinformatics.
Vladimir B. Bajic is professor of Applied Mathematics and Computational Science at King Abdullah University of Science and Technology (KAUST) and Director of KAUST’s Computational Bioscience Research Center (CBRC). His current research interests include Data, Text and Knowledge Mining, Data Integration, Gene Regulation, Drug Repositioning and Bioinformatics.
Funding
This study is supported by the KAUST Research Funds via AEA KAUST-Stanford Round 3 Global Collaborative Research Program and by KAUST Base Research Funds to PK and VBB.
References
- 1.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature 2001;409:860–921. [DOI] [PubMed] [Google Scholar]
- 2.Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science 2001;291:1304–51. [DOI] [PubMed] [Google Scholar]
- 3.Maston GA, Evans SK, Green MR. Transcriptional regulatory elements in the human genome. Annu Rev Genomics Hum Genet 2006;7:29–59. [DOI] [PubMed] [Google Scholar]
- 4.Heintzman ND, Ren B. Finding distal regulatory elements in the human genome. Curr Opin Genet Dev 2009;19:541–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell 1981;27:299–308. [DOI] [PubMed] [Google Scholar]
- 6.Visel A, Rubin EM, Pennacchio LA. Genomic views of distant-acting enhancers. Nature 2009;461:199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bulger M, Groudine M. Enhancers: the abundance and function of regulatory sequences beyond promoters. Dev Biol 2010;339:250–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim TK, Hemberg M, Gray JM, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 2010;465:182–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnone MI, Davidson EH. The hardwiring of development: organization and function of genomic regulatory systems. Development 1997;124:1851–64. [DOI] [PubMed] [Google Scholar]
- 10.Altshuler DM, Gibbs RA, Peltonen L, et al. Integrating common and rare genetic variation in diverse human populations. Nature 2010;467:52–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell 2012;150:12–27. [DOI] [PubMed] [Google Scholar]
- 12.Herz HM, Hu D, Shilatifard A. Enhancer malfunction in cancer. Mol Cell 2014;53:859–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith E, Shilatifard A. Enhancer biology and enhanceropathies, Nat Struct Mol Biol 2014;21:210–9. [DOI] [PubMed] [Google Scholar]
- 14.Heinz S, Romanoski CE, Benner C, et al. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol 2015;16:144–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatzis P, Talianidis I. Dynamics of enhancer-promoter communication during differentiation-induced gene activation. Mol Cell 2002;10:1467–77. [DOI] [PubMed] [Google Scholar]
- 16.Plank JL, Dean A. Enhancer function: mechanistic and genome-wide insights come together. Mol Cell 2014;55:5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions, Nat Rev Genet 2014;15:272–86. [DOI] [PubMed] [Google Scholar]
- 18.Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why?, Mol Cell 2013;49:825–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yip KY, Cheng C, Gerstein M. Machine learning and genome annotation: a match meant to be? Genome Biol 2013;14:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Visel A, Bristow J, Pennacchio LA. Enhancer identification through comparative genomics. Semin Cell Dev Biol 2007;18:140–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heintzman ND, Stuart RK, Hon G, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 2007;39:311–8. [DOI] [PubMed] [Google Scholar]
- 22.Hon G, Ren B, Wang W. ChromaSig: a probabilistic approach to finding common chromatin signatures in the human genome. PLoS Comput Biol 2008;4:e1000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rye M, Saetrom P, Handstad T, et al. Clustered ChIP-Seq-defined transcription factor binding sites and histone modifications map distinct classes of regulatory elements. BMC Biol 2011;9:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ucar D, Hu Q, Tan K. Combinatorial chromatin modification patterns in the human genome revealed by subspace clustering. Nucleic Acids Res 2011;39:4063–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyle AP, Song L, Lee BK, et al. High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Res 2011;21:456–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pique-Regi R, Degner JF, Pai AA, et al. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res 2011;21:447–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piper J, Elze MC, Cauchy P, et al. Wellington: a novel method for the accurate identification of digital genomic footprints from DNase-seq data. Nucleic Acids Res 2013;41:e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Visel A, Blow MJ, Li Z, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 2009;457:854–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Won KJ, Ren B, Wang W. Genome-wide prediction of transcription factor binding sites using an integrated model. Genome Biol 2010;11:R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods 2012;9:215–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoffman MM, Buske OJ, Wang J, et al. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat Methods 2012;9:473–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Won KJ, Zhang X, Wang T, et al. Comparative annotation of functional regions in the human genome using epigenomic data. Nucleic Acids Res 2013;41:4423–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Firpi HA, Ucar D, Tan K. Discover regulatory DNA elements using chromatin signatures and artificial neural network. Bioinformatics 2010;26:1579–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernandez M, Miranda-Saavedra D. Genome-wide enhancer prediction from epigenetic signatures using genetic algorithm-optimized support vector machines. Nucleic Acids Res 2012;40:e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Erwin GD, Oksenberg N, Truty RM, et al. Integrating diverse datasets improves developmental enhancer prediction. PLoS Comput Biol 2014;10:e1003677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kleftogiannis D, Kalnis P, Bajic VB. DEEP: a general computational framework for predicting enhancers. Nucleic Acids Res 2015;43:e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rajagopal N, Xie W, Li Y, et al. RFECS: a random-forest based algorithm for enhancer identification from chromatin state. PLoS Comput Biol 2013;9:e1002968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu Y, Qu W, Shan G, et al. DELTA: a distal enhancer locating tool based on AdaBoost algorithm and shape features of chromatin modifications. PLoS One 2015;10:e0130622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fletez-Brant C, Lee D, McCallion AS, et al. kmer-SVM: a web server for identifying predictive regulatory sequence features in genomic data sets. Nucleic Acids Res 2013;41:W544–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andersson R, Gebhard C, Miguel-Escalada I, et al. An atlas of active enhancers across human cell types and tissues. Nature 2014;507:455–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Danko CG, Hyland SL, Core LJ, et al. Identification of active transcriptional regulatory elements from GRO-seq data. Nat Methods 2015;12:433–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kheradpour P, Ernst J, Melnikov A, et al. Systematic dissection of regulatory motifs in 2000 predicted human enhancers using a massively parallel reporter assay. Genome Res 2013;23:800–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kwasnieski JC, Fiore C, Chaudhari HG, et al. High-throughput functional testing of ENCODE segmentation predictions. Genome Res 2014;24:1595–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arnold CD, Gerlach D, Stelzer C, et al. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science 2013;339:1074–7. [DOI] [PubMed] [Google Scholar]
- 45.Mogno I, Kwasnieski JC, Cohen BA. Massively parallel synthetic promoter assays reveal the in vivo effects of binding site variants. Genome Res 2013;23:1908–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murtha M, Tokcaer-Keskin Z, Tang Z, et al. FIREWACh: high-throughput functional detection of transcriptional regulatory modules in mammalian cells. Nat Methods 2014;11:559–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gisselbrecht SS, Barrera LA, Porsch M, et al. Highly parallel assays of tissue-specific enhancers in whole Drosophila embryos. Nat Methods 2013;10:774–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Melnikov A, Murugan A, Zhang X, et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat Biotechnol 2012;30:271–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patwardhan RP, Hiatt JB, Witten DM, et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nat Biotechnol 2012;30:265–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ashoor H, Kleftogiannis D, Radovanovic A, et al. DENdb: database of integrated human enhancers. Database (Oxford) 2015;2015:bav085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee D, Karchin R, Beer MA. Discriminative prediction of mammalian enhancers from DNA sequence. Genome Res 2011;21:2167–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yanez-Cuna JO, Arnold CD, Stampfel G, et al. Dissection of thousands of cell type-specific enhancers identifies dinucleotide repeat motifs as general enhancer features. Genome Res 2014;24:1147–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Narlikar L, Sakabe NJ, Blanski AA, et al. Genome-wide discovery of human heart enhancers. Genome Res 2010;20:381–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Won KJ, Chepelev I, Ren B, et al. Prediction of regulatory elements in mammalian genomes using chromatin signatures. BMC Bioinformatics 2008;9:547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heintzman ND, Hon GC, Hawkins RD, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 2009;459:108–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rada-Iglesias A, Bajpai R, Swigut T, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 2011;470:279–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ram O, Goren A, Amit I, et al. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell 2011;147:1628–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ernst J, Kellis M. Interplay between chromatin state, regulator binding, and regulatory motifs in six human cell types. Genome Res 2013;23:1142–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hallikas O, Palin K, Sinjushina N, et al. Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell 2006;124:47–59. [DOI] [PubMed] [Google Scholar]
- 60.Jolma A, Yan J, Whitington T, et al. DNA-binding specificities of human transcription factors. Cell 2013;152:327–39. [DOI] [PubMed] [Google Scholar]
- 61.Yip KY, Cheng C, Bhardwaj N, et al. Classification of human genomic regions based on experimentally determined binding sites of more than 100 transcription-related factors. Genome Biol 2012;13:R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Skipper M, Dhand R, Campbell P. Presenting ENCODE. Nature 2012;489:45. [DOI] [PubMed] [Google Scholar]
- 63.Wang Z, Zang C, Cui K, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 2009;138:1019–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lickwar CR, Mueller F, Hanlon SE, et al. Genome-wide protein-DNA binding dynamics suggest a molecular clutch for transcription factor function. Nature 2012;484:251–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thurman RE, Rynes E, Humbert R, et al. The accessible chromatin landscape of the human genome. Nature 2012;489:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Corradin O, Saiakhova A, Akhtar-Zaidi B, et al. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res 2014;24:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang ZR. Biological applications of support vector machines. Brief Bioinform 2004;5:328–38. [DOI] [PubMed] [Google Scholar]
- 68.Lancashire LJ, Lemetre C, Ball GR. An introduction to artificial neural networks in bioinformatics–application to complex microarray and mass spectrometry datasets in cancer studies. Brief Bioinform 2009;10:315–29. [DOI] [PubMed] [Google Scholar]
- 69.Lin WJ, Chen JJ. Class-imbalanced classifiers for high-dimensional data. Brief Bioinform 2013;14:13–26. [DOI] [PubMed] [Google Scholar]
- 70.Gola D, Mahachie John JM, van Steen K, et al. A roadmap to multifactor dimensionality reduction methods. Brief Bioinform 2015. pii: bbv038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu C, Ma S. A selective review of robust variable selection with applications in bioinformatics. Brief Bioinform 2015;16:873–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Larranaga P, Calvo B, Santana R, et al. Machine learning in bioinformatics. Brief Bioinform 2006;7:86–112. [DOI] [PubMed] [Google Scholar]
- 73.Hoffman MM, Ernst J, Wilder SP, et al. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res 2013;41:827–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sinha S, van Nimwegen E, Siggia ED. A probabilistic method to detect regulatory modules. Bioinformatics 2003;19 (Suppl 1):i292–301. [DOI] [PubMed] [Google Scholar]
- 75.Mammana A, Chung HR. Chromatin segmentation based on a probabilistic model for read counts explains a large portion of the epigenome. Genome Biol 2015;16:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ferretti V, Poitras C, Bergeron D, et al. PReMod: a database of genome-wide mammalian cis-regulatory module predictions. Nucleic Acids Res 2007;35:D122–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Visel A, Minovitsky S, Dubchak I, et al. VISTA Enhancer Browser–a database of tissue-specific human enhancers. Nucleic Acids Res 2007;35:D88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bernstein BE, Stamatoyannopoulos JA, Costello JF, et al. The NIH roadmap epigenomics mapping consortium. Nat Biotechnol 2010;28:1045–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Leung G, Eisen MB. Identifying cis-regulatory sequences by word profile similarity. PLoS One 2009;4:e6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ghandi M, Lee D, Mohammad-Noori M, et al. Enhanced regulatory sequence prediction using gapped k-mer features. PLoS Comput Biol 2014;10:e1003711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sharon E, Kalma Y, Sharp A, et al. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nat Biotechnol 2012;30:521–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pengelly AR, Copur O, Jackle H, et al. A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science 2013;339:698–9. [DOI] [PubMed] [Google Scholar]
- 83.Ravasi T, Suzuki H, Cannistraci CV, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell 2010;140:744–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Arner E, Daub CO, Vitting-Seerup K, et al. Gene regulation. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science 2015;347:1010–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Core LJ, Martins AL, Danko CG, et al. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat Genet 2014;46:1311–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.He B, Chen C, Teng L, et al. Global view of enhancer-promoter interactome in human cells. Proc Natl Acad Sci USA 2014;111:E2191–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ernst J, Kheradpour P, Mikkelsen TS, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 2011;473:43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dostie J, Zhan Y, Dekker J. Chromosome conformation capture carbon copy technology. Curr Protoc Mol Biol 2007;Chapter 21:Unit 21.14. [DOI] [PubMed] [Google Scholar]
- 89.Fullwood MJ, Liu MH, Pan YF, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature 2009;462:58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Weingarten-Gabbay S, Segal E. A shared architecture for promoters and enhancers. Nat Genet 2014;46:1253–4. [DOI] [PubMed] [Google Scholar]
- 91.Meireles-Filho AC, Stark A. Comparative genomics of gene regulation-conservation and divergence of cis-regulatory information. Curr Opin Genet Dev 2009;19:565–70. [DOI] [PubMed] [Google Scholar]
- 92.Zhang Y, Liu T, Meyer CA, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008;9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zang C, Schones DE, Zeng C, et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics 2009;25:1952–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Koohy H, Down TA, Spivakov M, et al. A comparison of peak callers used for DNase-Seq data. PLoS One 2014;9:e96303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wilbanks EG, Facciotti MT. Evaluation of algorithm performance in ChIP-seq peak detection. PLoS One 2010;5:e11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ernst J, Kellis M. Large-scale imputation of epigenomic datasets for systematic annotation of diverse human tissues. Nat Biotechnol 2015;33:364–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Teytelman L, Thurtle DM, Rine J, et al. Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc Natl Acad Sci USA 2013;110:18602–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bajic VB. Comparing the success of different prediction software in sequence analysis: a review. Brief Bioinform 2000;1:214–28. [DOI] [PubMed] [Google Scholar]
- 99.Budden DM, Hurley DG, Crampin EJ. Predictive modelling of gene expression from transcriptional regulatory elements. Brief Bioinform 2015;16:616–28. [DOI] [PubMed] [Google Scholar]
- 100.Kumaki Y, Ukai-Tadenuma M, Uno KD, et al. Analysis and synthesis of high-amplitude Cis-elements in the mammalian circadian clock. Proc Natl Acad Sci USA 2008;105:14946–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Loven J, Hoke HA, Lin CY, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 2013;153:320–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kulakovskiy IV, Medvedeva YA, Schaefer U, et al. HOCOMOCO: a comprehensive collection of human transcription factor binding sites models. Nucleic Acids Res 2013;41:D195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Teng L, He B, Wang J, et al. 4DGenome: a comprehensive database of chromatin interactions. Bioinformatics 2015;31:2560–64. [DOI] [PMC free article] [PubMed] [Google Scholar]