Abstract
Cytosine methylation regulates many biological processes such as gene expression, chromatin structure and chromosome stability. The whole genome bisulfite sequencing (WGBS) technique measures the methylation level at each cytosine throughout the genome. There are an increasing number of publicly available pipelines for analyzing WGBS data, reflecting many choices of read mapping algorithms as well as preprocessing and postprocessing methods. We simulated single-end and paired-end reads based on three experimental data sets, and comprehensively evaluated 192 combinations of three preprocessing, five postprocessing and five widely used read mapping algorithms. We also compared paired-end data with single-end data at the same sequencing depth for performance of read mapping and methylation level estimation. Bismark and LAST were the most robust mapping algorithms. We found that Mott trimming and quality filtering individually improved the performance of both read mapping and methylation level estimation, but combining them did not lead to further improvement. Furthermore, we confirmed that paired-end sequencing reduced error rate and enhanced sensitivity for both read mapping and methylation level estimation, especially for short reads and in repetitive regions of the human genome.
Keywords: whole genome bisulfite sequencing, DNA methylation, WGBS analysis step evaluation, read quality trimming, WGBS mapping software
Introduction
DNA methylation is an important epigenetic modification that is used to regulate many biological processes such as gene expression, chromatin structure, imprinting and chromosome stability [1-3]. In mammals, DNA methylation occurs mostly for cytosines in the CG context (∼80% of CG dinucleotides in both genomic strands) and rarely in the CHG or CHH contexts (∼3%; H = A, T, or C) [4]. Because abnormality of DNA methylation is observed in various diseases, especially cancers, there is a growing interest in developing novel medical interventions that target aberrant DNA methylation [5].
Improvements in high-throughput sequencing technologies have enabled cytosine methylation to be monitored at single-nucleotide resolution throughout the genome, using a method called whole genome bisulfite sequencing (WGBS) [6], even in a single cell [7]. Bisulfite treatment converts unmethylated cytosines to uracils and leaves methylated cytosines intact [8]. When the sequencing reads are mapped back to the reference genome, methylated and unmethylated cytosines can be identified computationally. Because bisulfite alters ∼90% of cytosines in the genome [9], mapping bisulfite converted reads to the genome poses major computational challenges. Furthermore, it can be difficult to distinguish a converted nucleotide (nt) from a sequencing error. Thus, it is important to assess the accuracy of the methods that map reads and estimate methylation levels.
As reviewed by Adusumalli et al. [10], the entire WGBS analysis workflow is composed of many steps, including adapter removal, read quality trimming, read mapping, postalignment read filtering, sample heterogeneity assessment and identification of differentially methylated regions between two conditions. In this study, we focused on three analysis steps that most directly impact the performance of methylation level estimation—read quality trimming (henceforth preprocessing), read mapping and postalignment read filtering (henceforth postprocessing).
Many algorithms have been developed to map bisulfite-converted reads to the reference genome (Table 1) [11-28]. These algorithms are classified into two groups, wild-card and three-letter aligners [29]. Wild-card aligners treat Cs and Ts in the reads as matches for Cs in the reference genome, or they use a modified nucleotide similarity score matrix that contains a positive match score between a T in a read and a C in the reference genome. Three-letter aligners first reduce the four-nucleotide lexicon of DNA (A, C, G, T) into three nucleotides (A, G, T) by converting all Cs in the reads and both strands of the reference genome into Ts. After this computational nucleotide conversion, the reads are mapped to the reference genome using a standard read mapping algorithm such as BWA and Bowtie [29].
Table 1.
Publicly available tools for mapping bisulfite converted reads
| Program | Version | Aligner type | Aligner description | Language | Alignment engine | Paired end | Color space | Non directional | Multithread | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| BatMeth | 1.04b | Three letter | FM index of the C-to-T converted genome. Filtering step for low complexity reads with Shannon's entropy. | C, Perl | None | Yes | Yes | Yes | Yes | [11] |
| Bismark | v0.14.2 | Three letter | FM index of the C-to-T converted genome. Mapping step taken into account basecall qualities. | Perl | Bowtie, Bowtie2 | Yes | No | Yes | Yes | [12] |
| Bisulfighter | 1.3 | Wild-card | Spaced suffix array index of the original (i.e. unconverted) reference genome. | C/C++, Python, R | LAST | Yes | No | Yes | No | [13] |
| BRAT-BW | 2.0.1 | Three letter | FM index of the C-to-T converted genome. Multi-seeding starting from different positions within reads. | C++ | None | Yes | No | Yes | Yes | [14] |
| BSMAP | 2.74 | Wild-card | Hash table of the original genome. Based on SOAP alignment algorithm. | C++, Python | None | Yes | No | Yes | Yes | [15] |
| BSmooth | 0.8.1 | Wild-card | FM-index, nucleotide base Y for C and T matches. Support for color-space read mapping. | Perl | Bowtie 2, Merman | Yes (with Bowtie2) | Yes (with Merman) | Yes | Yes | [16] |
| BS-Seeker | – | Three letter | FM index of the C-to-T converted genome. Only accepts reads in fixed length in FASTQ. | Python | Bowtie | No | No | Yes | No | [17] |
| BS-Seeker2 | v2.0.9 | Three letter | FM index of the C-to-T converted genome. Filtering step for the reads with incomplete bisulfite conversion. | Python | Bowtie, Bowtie2, SOAP, RMAP | Yes | No | Yes | Yes | [18] |
| B-SOLANA | 1.0 | Color-space | FM-index of the C-to-T converted genome. Support for color-space read mapping. | Python | Bowtie | No | Yes | No | Yes | [19] |
| GSNAP | 2014‐01‐21 | Wild-card | Hash table of the C-to-T converted genome. Uses wild-card letter Y for Cs and Ts in reads. | C, Perl | None | Yes | No | Yes | Yes | [20] |
| ERNE-BS5 | 2.1 | Wild-card | Hashing the reference genome with 5-letter, A, T, G, Cu, Cm. | C++ | None | Yes | Yes | No | Yes | [21] |
| LAST | 548 | Wild-card | Spaced suffix array index of the original genome. Modified score matrix for C-to-T and G-to-A matches. | C/C++, Python | None | Yes | No | Yes | Yes (requires GNU parallel) | [22] |
| MAQ | v0.6.6 | Wild-card | Multiple hash tables of reads. Nonunique reads assigned randomly to one of the best-matching positions. | C/C++, Perl | None | Yes | Yes | No | No | [23] |
| MethylCoder | 0.3.8 | Three letter | Remapping step for unmapped reads to the original genome. Support for color-space reads. | Python | Bowtie, GSNAP | Yes | Yes | Yes | Yes | [24] |
| Novoalign | V3.02.13 | Wild card | Hash table of the C-to-T converted genome. Commercial software package. | C/C++ | None | Yes | Yes | Yes | No | http://www.no vocraft.com/ |
| Pash | 3.0 | Wild card | Multi-positional hash tables containing all possible C-to-T conversion combinations of reads. | C | None | No | No | No | Yes | [25] |
| RMAP | v2.05 | Wild card | Hash table based on layered bit-mask seeds of reads. | C++ | None | Yes | No | Yes | Yes | [26] |
| Segemehl | 0.2.0 | Wild card | Enhanced suffix array index for the converted genome. Based on Myers bit-vector algorithm for matching reads. | C | None | Yes | No | Yes | Yes | [27] |
| SOCS-B | 2.1 | Color space | Iterative Rabin-Karp hashing for the C-to-T converted genome and reads. Support for color-space reads. | Perl | None | No | Yes | Yes | No | [28] |
Many preprocessing tools also exist for read quality trimming to eliminate nucleotides of low sequencing qualities (Table 2). These tools can be grouped into two main classes, running sum and window based [30]. Running-sum methods globally scan a read and use a cumulative quality score to determine the position for trimming the 3′-end of the read [30-32]. Window-based methods apply sliding or nonoverlapping windows to locally scan a read and clip off the low-quality regions [32-38].
Table 2.
Publicly available read quality trimming tools
| Program | Version | Trimmer type | Trimmer description | Language | Adapter trimming | Paired end | Color space | Reference |
|---|---|---|---|---|---|---|---|---|
| Adapter removal | 2.1.0 | Window based | Longest stretches of high-quality bases in a read. Scans bases from both ends of a read. | C++ | 5′ and 3′ | Yes | No | [33] |
| Btrim | 0.3.0 | Window based | Trims a read and the following 3′ bases at the point where an average quality score within a window drops below a cutoff. | C | 5′ and 3′ | No | No | [34] |
| ConDeTri | v2.3 | Window based | Trims a read and the following 3′ bases at the position before a certain number of consecutive high-quality bases. Rescues a certain number of consecutive low-quality bases if high-quality bases surround the low-quality base block. | Perl | No | Yes | No | [35] |
| Cutadapt | 1.8 | Running-sum | Subtracts a preset cutoff quality score from the quality score at each position and adds the remainder to a cumulative score at the position. Trims at the 3′ portion of a read starting from the position with the minimum cumulative score. | Python, C | 5′ and 3′ | Yes | Yes | [31] |
| ERNE-FILTER | 2.1 | Running-sum | Subtracts a preset cutoff quality score from the quality score at each position and adds the remainder to a cumulative score at the position. Trims at the 3′ portion of a read starting from the position with the minimum cumulative score. | C++ | No | Yes | No | [30] |
| FASTX toolkit | 0.0.13 | Window based | Longest stretch of high-quality bases in a read. | C++ | 3´ | No | No | http://hannon lab.cshl.edu/ fastx_toolkit/ |
| Kraken | 13‐274 | Window based | Trims a read and the following 3′ bases at the point where a median quality score within a window drops below a cutoff. | C | 5′ and 3′ | Yes | No | [36] |
| PRINSEQ | 0.20.4 | Window based | Longest stretch of high-quality bases in a read. Two cutoffs for both ends of a read, Qleft and Qright. | Perl | No | Yes | No | [37] |
| SolexaQA | v3.1.3 | Window based, running sum | Longest stretch of high-quality bases in a read. The option—BWA for read trimming with running sum algorithm. | Perl | No | Yes | No | [32] |
| Trimmo-matic | 0.33 | Window based | Trims a read and the following 3′ bases at the point where an average quality score within a window drops below a cutoff. | Java | 5′ and 3′ (Illumina-specific sequences only) | Yes | No | [38] |
As another approach for quality control, a postprocessing step is often integrated after read mapping and before computing methylation levels. There are two kinds of postprocessing methods. Coverage filtering methods discard the cytosines covered by fewer reads than a preset cutoff. Quality filtering methods exclude a read from the estimation of the methylation level of a cytosine if the sequencing quality of the read at the corresponding position is lower than a preset cutoff.
Several studies compared the sensitivity and accuracy of a panel of read-mapping algorithms using experimental WGBS data and simulated reads, mostly in the context of assessing a newly developed algorithm; however, only a few studies evaluated the accuracy of computing methylation levels after read mapping [13, 39]. So far, no study has evaluated the combination of preprocessing, mapping and postprocessing steps, especially with regard to estimation of methylation levels.
Paired-end WGBS data are becoming increasingly available, with a pair of reads sequenced from both ends of each DNA fragment, as opposed to single-end data with only one end of each DNA fragment sequenced. Because the length distribution of DNA fragments is known, paired-end reads are constrained spatially in genomic coordinates, which allows them to be more accurately mapped to the reference genome than single-end reads, especially in repetitive regions [40, 41]. However, no study has systematically compared the performance of paired-end and single-end WGBS data sets at an equivalent sequencing depth.
Compared with earlier studies, which were mainly focused on the performance of read mapping algorithms, we further benchmarked various combinations of pre- and postprocessing methods with read mapping algorithms [42, 43]. Another novel aspect of our study is that we compared single-end and paired-end reads at an equal sequencing depth. We simulated single-end and paired-end reads that incorporated single nucleotide polymorphisms (SNPs), sequencing errors and DNA methylation derived from experimental WGBS libraries. We tested five widely used WGBS mapping algorithms, Bismark [12], BSMAP [15], GSNAP [20], BRAT-BW [14] and LAST [22], in combination with three preprocessing methods and two postprocessing methods each with several thresholds. For Bismark, the most widely used algorithm, we tested two alignment engines, Bowtie and Bowtie2, each with two different seed lengths. In total, we tested 192 combinations.
We found that Mott trimming and quality filtering individually improved the results of both read mapping and methylation level estimation, but combining them did not lead to further improvements. Furthermore, we confirmed that at equivalent sequencing depth, paired-end sequencing reduced the error rate and enhanced the sensitivity on read mapping and methylation level estimation, especially for short reads and repetitive regions. Bismark and LAST were the most robust mapping algorithms throughout most simulated data sets, and constituted the best WGBS analysis pipelines when combined with Mott trimming and no filtering.
Material and methods
Genome annotations and WGBS data sets
We downloaded the genomic sequence of human chromosome 21 (chr21; version hg19), the allele frequencies of SNPs (snp138Common.txt) [44], annotated repeats (rmsk.txt) [45] and annotated segmental duplications (genomicSuperdup.txt) [46] from the UCSC genome database [47]. To simulate bisulfite-converted reads, we retrieved three WGBS libraries from the Gene Expression Omnibus: SRR901864 [48] for 101 nt single-end reads, SRR568015 [49] for 45 nt and 50 nt paired-end reads (paired-end data set A) and SRR771408 [39] for 100 nt paired-end reads (paired-end data set B).
Benchmark data
We simulated bisulfite converted reads by randomly drawing from the human chr21 genome sequence. Taking into account cytosine contexts (CG, CHG and CHH), we randomly assigned a methylation level to every cytosine in both strands of chr21. We used the methylation levels in Table 2 of a previous publication [22]. The CG context was given higher probability of being methylated than CHG and CHH. For each allele, we inserted random polymorphisms including SNPs and indels based on annotated allele frequencies in the chr21 sequence.
We used the DNemulator algorithm [22] to simulate reads. We randomly extracted 50 million (50M) fragments, of the same lengths in the experimental libraries, from the chr21 sequence with simulated methylation levels and alleles. These correspond to 140-, 130- and 280-fold genome coverage for the single-end data set and paired-end data sets A and B, respectively. To simulate paired-end reads, we used a normal distribution of insert lengths with the following mean and standard deviation: 400 nt and 25 nt for the paired-end data set A, and 350 nt and 40 nt for the paired-end data set B. We also simulated a single-end data set (i.e. all reads were randomly drawn from chr21 independently) equivalent to each paired-end data set in terms of read length, total read depth and sequence quality scores to compare the benefits of paired-end versus single-end libraries at equivalent sequencing depth (i.e. equal cost).
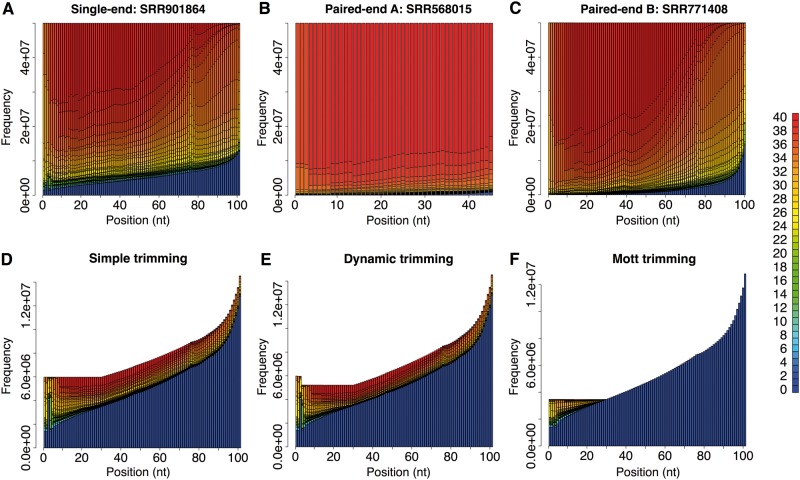
We then simulated bisulfite conversion with the efficiency of 99% on all the reads. After that, we mutated nucleotides in the simulated reads to account for sequencing errors, according to the per-base quality scores of the first 50M reads in the experimental data sets. The quality score distribution of the first 50M reads in each data set is shown in Figure 1A–C. To test data sets with different sequencing depths, we down-sampled the data sets from 50M reads (or read pairs) to 10M and 20M reads (read pairs). Note that the sequencing depth of 10M for chr21 corresponds to 26–56 folds of coverage, which is achieved by most present-day WGBS experimental data sets.
Figure 1.
Quality score distribution of experimental data used for simulation and after trimming. The horizontal axis shows read positions, and colors represent quality scores. Panels DEF are for the single-end data set. (A) Quality score distribution in the single-end data set. (B) Quality score distribution in the paired-end data set A. (C) Quality score distribution in the paired-end data set B. (D) Quality score distribution of trimmed regions and discarded reads by simple trimming. (E) Quality score distribution of trimmed regions and discarded reads by dynamic trimming. (F) Quality score distribution of trimmed regions and discarded reads by Mott trimming. A colour version of this figure is available online at BIB online: https://academic.oup.com/bib.
Preprocessing: trimming methods
To test the performance of preprocessing methods, we implemented the following three trimming algorithms:
Mott trimming (running sum): The method starts from the 3′-end of each read, subtracts a preset cutoff quality score from the quality score at each position and adds the remainder to a cumulative score at the position. The 3′ portion of the read starting from the position with the minimum cumulative score is trimmed [50].
Dynamic trimming (window based): The method searches for the longest stretch of positions (window) in each read such that the quality scores of each position in the window exceed a preset threshold [32].
Simple trimming: The method scans from the 5′ end of each read. As soon as it detects a position with quality scores below a preset threshold, it discards this position and the remaining positions at the 3′-end of the read.
We used the Phred+33 score as the quality score for all three trimming algorithms. We set a cutoff of 3 for all three trimming methods based on an earlier study [51]. The Python scripts implemented for this study can be downloaded from https://github.com/jnktsj/trim-suppl_BIB100115.
Mapping: bisulfite-seq mapping algorithms
We compared five bisulfite-seq mapping algorithms: Bismark (v0.14.2) [12], BSMAP (2.74) [15], GSNAP (the 21 January 2014 version) [20], BRAT-BW (2.0.1) [14] and LAST (version 548) [22]. We tested two different alignment engines for Bismark, Bowtie (1.1.1) [52] and Bowtie2 (2.2.5) [53], each with two different seed lengths: l = 28 or 50 for Bowtie and L = 20 or 22 for Bowtie2. The command lines of each software package used in this study are summarized in Supplementary Materials.
Postprocessing: filtering methods
Before computing methylation levels, we tested two filtering methods as postprocessing quality control:
Coverage filtering: We only computed methylation levels for cytosines covered by at least n reads. We tested n = 3, 5 and 10.
Quality filtering: To compute the methylation level for a cytosine, we only used the reads that covered this cytosine and had quality scores greater than or equal to q at this position. We tested q = 10 and 20.
Evaluation of performance
We evaluated the performance of each mapping algorithm in isolation for read mapping accuracy, also in combination with the preprocessing and postprocessing steps for methylation level estimation accuracy.
To evaluate read mapping accuracy, we took a read-centric approach and simply counted the fraction of correctly mapped reads. We defined mapping sensitivity as the fraction of correctly mapped reads out of all simulated reads and error rate as the fraction of incorrectly mapped reads out of all simulated reads. For those mapping algorithms that perform local alignments (LAST, Bismark with Bowtie2 and GSNAP), if at least one base in a read is correctly mapped, the read is considered correctly mapped.
To evaluate the accuracy of methylation level estimation, we separated cytosines into two groups and computed their fractions: cytosines that had one or more simulated reads and were also covered by one or more mapped reads (henceforth, ‘Cs correctly covered'), and cytosines that did not have simulated reads but were covered by one or more mapped reads (henceforth, ‘Cs falsely covered'). For Cs correctly covered, we further computed two metrics: the fraction of Cs with correctly estimated methylation levels, i.e. zero error (henceforth, ‘Cs perfectly estimated') and ‘the error of estimated methylation levels', determined using the gold standard methylation levels assigned during simulation. We plot the cumulative fraction of Cs correctly estimated as a function of the allowed error of estimated methylation levels. We calculated the average error of methylation level estimation for all Cs correctly covered.
Performance in repetitive regions of the genome
In addition to genome-wide performance, we also compared the performance of read mapping and methylation level estimation in repetitive regions. For Alu elements, we focused on the AluY subfamily that is the youngest and the least diverged. Similarly, we focused on the LINE-1P subfamily of the LINE elements. The fractions of simulated reads in all data sets that fell in the repetitive regions within chromosome 21 were 0.48–1.21% in AluY, 2.54–5.34% in LINE-1P, 0.05–0.20% in AT-rich low complexity regions (LCR), 0.05–0.15% in GC-rich LCR, 0.02–0.10% in other LCR and 1.70–3.41% in segmental duplications.
Runtime and memory usage
To measure the runtime and memory usage of the mapping algorithms, we used the paired-end data set A. We mapped those reads in both single-end and paired-end modes with different coverages: 10M, 20M and 50M reads. The runtime measurement covered the entire process including reading and outputting files. Runtime was measured for one CPU (Dell R815 AMD Opteron 6380, 2.50 GHz) with 512 GB RAM (random access memory).
Results
Mott trimming improves mapping accuracy
We tested three read trimming algorithms in preprocessing step: Mott, dynamic and simple trimming (described in ‘Methods’ section). For all data sets, Mott trimming retained the most reads and simple trimming retained the fewest reads, and the differences among them were greater for the single-end data set than for the two pair-end data sets (Supplementary Table S1) because the single-end data set had poorer sequencing quality (Figure 1A).
The impact on read mapping accuracy with preprocessing is shown in Figure 2A for the single-end data set (with 10M reads) and in Figure 3A for the two paired-end data sets (each data set with 10M read pairs). Trimming tended to increase sensitivity, but sometimes slightly increased error rate (note the different scales for sensitivity and error rate in Figures 2A and 3A). Mott trimming showed better performance than simple trimming and dynamic trimming. In contrast, simple and dynamic trimming algorithms locally trim reads based on a single base with a quality score below threshold. As shown in Figure 1D–F for the single-end data set, when there was a single nucleotide with a poor quality score in the middle of a read that was composed of mostly high-quality bases, simple and dynamic trimming methods discarded the downstream bases with high-quality scores. Thus, we will focus our discussion on Mott trimming.
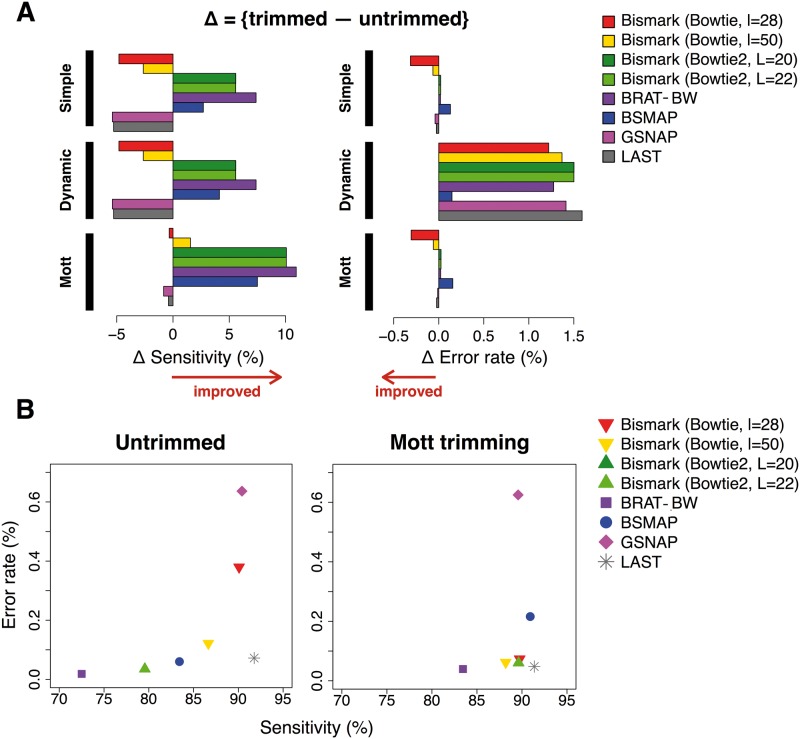
Figure 2.
Evaluation of preprocessing and mapping performance on the single-end data set. (A) The differences in sensitivity and error rate with preprocessing and those without preprocessing. The order of the bars in each preprocessing method corresponds to the order of alignment algorithms shown in the legend. (B) Mapping accuracy without trimming (left panel) and with Mott trimming (right panel). A colour version of this figure is available online at BIB online: https://academic.oup.com/bib.
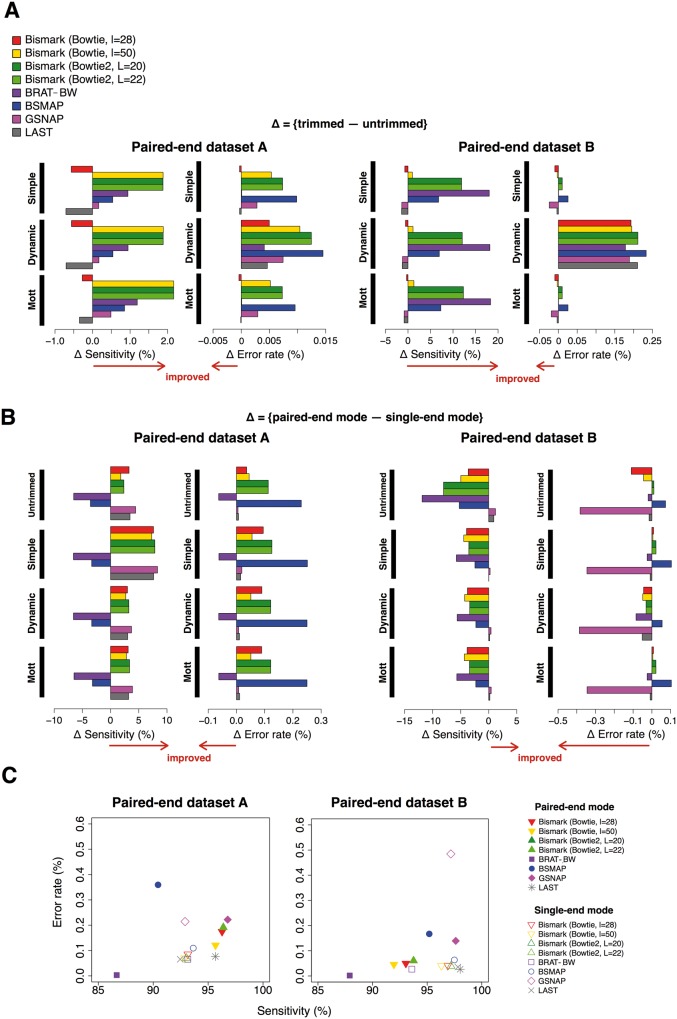
Figure 3.
Evaluation of preprocessing and mapping performance on paired-end data sets. (A) Differential sensitivity and error rate by preprocessing. (B) Differential sensitivity and error rate by paired-end information. (C) Comparison of the mapping accuracy of reads after Mott trimming between the paired-end data sets and their corresponding single-end data sets at equal sequencing depth. A colour version of this figure is available online at BIB online: https://academic.oup.com/bib.
Mott trimming substantially increased the mapping sensitivity for most alignment algorithms with minor impact on their error rates; the sensitivity improvement ranged 1.54–10.9% for the single-end data set (Figure 2A), 1.19–2.16% for the paired-end data set A and 1.32–18.3% for the paired-end data set B (Figure 3A). Read trimming is especially effective in increasing the sensitivity of mapping longer reads (the 101 nt single-end data set and the 100 nt paired-end data set B). Read trimming differed drastically in how it impacted Bismark with the two search engines (Bowtie and Bowtie2). Large improvement was seen on Mott trimming for Bismark with Bowtie2, but for Bismark with Bowtie (especially with seed length l = 28) Mott trimming had small and mixed impacts. The two seed lengths for Bowtie2 produced nearly identical results for all aspects of our study. For LAST and GSNAP, Mott trimming had little impact on mapping quality. These conclusions hold for 20M and 50M simulated reads (Supplementary Figure S1–S3).
Comparison of the mapping performance of the mapping algorithms
Among the mapping tools we tested, LAST exhibits the highest sensitivity (91.8% and 98.9%) and the lowest error rate (0.04% and 0.07%) in the single-end data set and the paired-end data set B, likely because these data sets have longer read length than the paired-end data set A (Figures 2B and 3C). As mentioned in the previous section, LAST shows equally good performance on mapping untrimmed reads.
Aided by Mott trimming, Bismark also showed high sensitivity and low error rate. The four settings of Bismark performed similarly on Mott trimmed single-end reads, with slightly lower sensitivities than LAST (Figure 2B). Bismark with Bowtie2 achieved slightly higher sensitivity but slightly increased error than Bismark with Bowtie on paired-end data sets, and higher sensitivity than LAST for data set A but lower for data set B (Figure 3C).
GSNAP achieves nearly as high sensitivity as LAST and Bismark. Especially for paired-end data set A, GSNAP shows the highest sensitivity; yet its error rate remains higher than the other algorithms. Among the five mapping algorithms with Mott trimming, GSNAP and BSMAP had the highest error rates and BRAT-BW showed the lowest sensitivity for all three data sets (Figures 2B and 3C).
Paired-end information helps accurate mapping of short reads
To investigate how much paired-end information increases mapping accuracy, we simulated a single-end data set (called single-end mode) that matched of each the two paired-end data sets in read length, total read depth and sequencing quality scores. For example, the paired-end data set A had 10M pairs of reads while the matched single-end data set A had 20M reads, with all the reads randomly drawn from chr21 and then assigned matching sequencing quality scores, SNPs, etc.
For shorter reads (the 50 nt and 45 nt paired-end data set A, Figure 3B, left panel), paired-end information improved the sensitivity for Bismark, GSNAP and LAST, at a small cost of error rate. For longer reads (the 100 nt paired-end data set B; Figure 3C, right panel), LAST showed a slight improvement and GSNAP showed a noticeable improvement in the paired-end mode; however, the other algorithms, including Bismark with all four options, performed worse in the paired-end mode than in the single-end mode. Similar results were seen for 20M and 50M read pairs (Supplementary Figure S2–S3).
BSMAP performed much worse on both paired-end data sets than on their matching single-end data sets. BRAT-BW achieved slightly lower error rate at a large cost of 5.68–6.48% sensitivity in the paired-end mode for both data sets (Figure 3C). Thus, the mapping algorithms differ in their efficiencies of using paired-end information.
Mott trimming improves the accuracy of methylation level estimation consistently
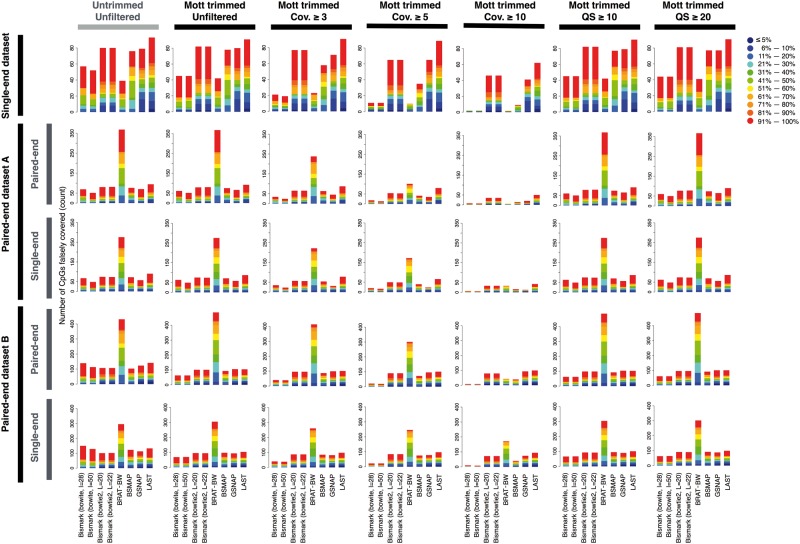
We evaluated the accuracy of methylation level estimation using three metrics: (i) the fraction of Cs correctly covered, i.e. cytosines that have one or more simulated reads and are also covered by one or more mapped reads (Supplementary Table S2–S4), and the fraction of Cs perfectly estimated, i.e. cytosines with perfectly estimated methylation levels (Figure 4); (ii) the average error in estimating methylation levels for the Cs correctly covered (Figure 4); and (iii) the number of Cs falsely covered, i.e. cytosines that do not have simulated reads but are covered by one or more mapped reads (Figure 5).
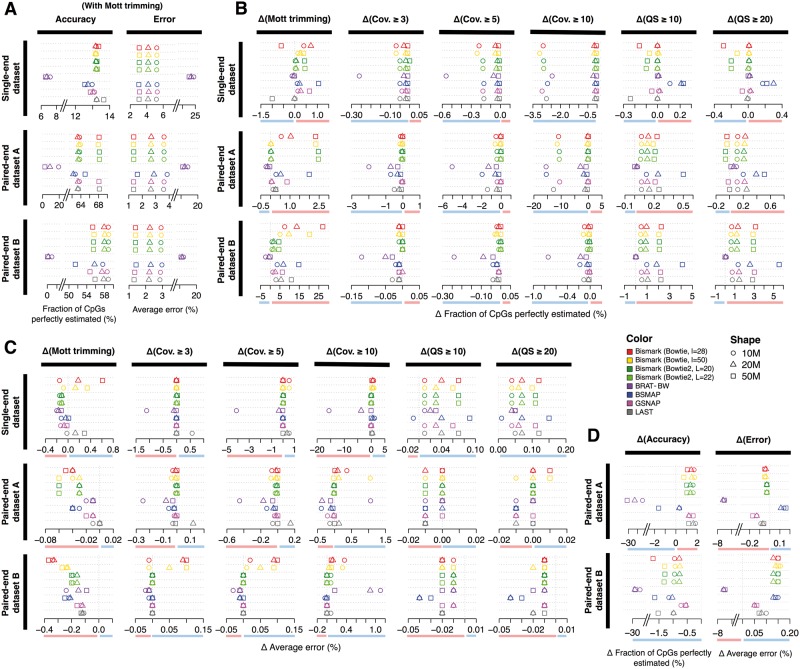
Figure 4.
Performance of methylation level estimation and improvements by pre- and postprocessing steps. (A) Fractions of CpGs perfectly estimated (left panel) and average errors (right panel) are plotted for each alignment algorithm (rows, corresponding to the order of alignment algorithms in the legend) in three simulated benchmark data sets at three sequencing depths (indicated by shapes of the symbols), after Mott trimming and no postprocessing. (B) Difference in the fraction of CpGs perfectly estimated after Mott trimming compared with untrimmed data sets (the left most column), and after Mott trimming and five options of postprocessing compared with the data sets just with Mott trimming (remaining columns). Cov. indicates coverage and QS indicates quality score. (C) Like B, but for difference in the average errors of methylation level estimation. (D) Difference in the fraction of CpGs perfectly estimated (left panels) and difference in average error (right panels) between the paired-end data sets and their matching single-end data sets at equal sequencing depth. For panels BCD, improved and worsened performance is highlighted with a red and a blue bar, respectively, below the X-axis. A colour version of this figure is available online at BIB online: https://academic.oup.com/bib.
Figure 5.
Number of CpGs falsely covered. The number of CpGs falsely covered is shown. Upper segments of each bar show higher methylation levels assigned during simulation of the reads. The trimming and filtering options are indicated on the top of each column of panels. We used the simulated data sets with 10M reads. A CpG was deemed falsely covered if it was not covered by any simulated reads but was covered by one or more mapped reads. Out of the 760 888 CpGs in chr21, in total 193 CpGs were falsely covered in the single-end data set. In the paired-end data sets A and B, in total 716 and 840 CpGs were falsely covered respectively, and 706 and 783 CpGs were falsely covered in their corresponding single-end data sets with equivalent sequencing depth. Cov. indicates coverage and QS indicates quality score. A colour version of this figure is available online at BIB online: https://academic.oup.com/bib.
Although Mott trimming improved the mapping accuracy of Bismark with Bowtie2, BRAT-BW and BSMAP on the single-end data set, it only slightly improved the accuracy of methylation level estimation using reads mapped by these three algorithms (Figure 4B, the first column labeled ‘Δ(Mott trimming)’). Take Bismark with Bowtie2 at 50M reads as an example, the fraction of CpGs with perfectly estimated methylation levels was 12.82% for untrimmed reads and increased to 13.25% for Mott trimmed reads (green rectangles in Figure 4A and B and Supplementary Table S2), and the average error in estimating methylation levels for CpGs correctly covered was 3.21% for untrimmed reads and decreased to 3.09% for Mott-trimmed reads (Figure 4C and Supplementary Table S2). Mott trimming did not improve the mapping accuracies for LAST on the single-end data set, and accordingly slightly worsened the methylation level estimation accuracy. Mott trimming decreased the number of Cs falsely covered for most mapping algorithms on the single-end data set, with the largest decreases on the number of the CpGs falsely covered for Bismark with Bowtie (Figure 5) and for cytosines in the CHG or CHH contexts (Supplementary Figure S4).
The benefit of read trimming on methylation level estimation is more noticeable for the two paired-end data sets. Mott trimming consistently improved all algorithms, even for LAST, which did not show obvious improvement on mapping accuracy (Figure 4B and Supplementary Tables S3–S4). For Bismark with Bowtie2 on the paired-end data set B (50M reads), the fraction of CpGs perfectly estimated increased from 51.9% to 55.53% and the average error decreased from 1.59% to 1.39% (Figure 4A–C and Supplementary Table S4). Although Mott trimming did not alter the mapping accuracy of LAST for this data set, the fraction of CpGs perfectly estimated improved from 44.95% to 55.85% and the average error decreased from 1.43% to 1.30% (gray rectangles in Figure 4A–C and Supplementary Table S4). Moreover, Mott trimming decreased the number of Cs falsely covered for most algorithms on the two paired-end data sets (Figure 5 and Supplementary Figures S5 and S6). These results indicate that trimming the low-quality nucleotides can decrease the error of methylation level estimation.
Quality filtering improves the accuracy of methylation level estimation slightly for paired-end data sets
We tested six postprocessing options: filtering out cytosine position covered by fewer reads <3, 5 or 10 reads, or discarding the reads covering a cytosine but with quality scores <10 or 20 at that position. We then evaluated the accuracy of estimating methylation levels for each combination of four trimming, eight mapping and six postprocessing options based on four metrics: the fraction of Cs correctly covered and the fraction of Cs perfectly estimated, the average error and the number of Cs falsely covered. Supplementary Tables S2–S4 detail the absolute performance of these combinations. Because we have established in the previous section that Mott trimming generally improves methylation level estimation, in Figure 4B and C we plot the relative performance (Δ) of each postprocessing option combined with Mott trimming in comparison with Mott trimming alone.
Coverage or quality filtering worsened the performance for the single-end data set judged by the fraction of Cs correctly covered, the fraction of Cs perfectly estimated or the average error (Figure 4B and C and Supplementary Tables S2–S4), but decreased the number of Cs falsely covered (Figure 5 and Supplementary Figures S4–S6). Rather, filtering improved the results for the two paired-end data sets, especially data set B. Thus, the remaining discussion of this section will pertain to the paired-end data sets.
Coverage filtering with the cutoffs 3 or 5 had little impact on the fraction of Cs correctly covered and the fraction of Cs perfectly estimated, or the average error for most alignment algorithms (Figure 4B and C); however, the stricter threshold of 10 reads decreased the fraction of Cs perfectly estimated and increased the average error compared with the pipeline without any postprocessing, especially for the paired-end data set A at the 10M sequencing depth. As expected, coverage filtering reduced the number of Cs falsely covered for all alignment algorithms in all data sets (Figure 5). Based on those observations, we concluded that gentle coverage filtering with the cutoff of 3 reads slightly decreased falsely covered Cs with little impact on the accuracy of estimating correctly covered Cs.
Quality filtering increased the fraction of Cs perfectly estimated and decreased the average error for most alignment algorithms on the paired-end data sets, especially data set B (Figure 4B and Supplementary Tables S3–S4). The results for the two quality filtering cutoffs (10 or 20) led to highly similar results. As expected, quality filtering also reduced the number of Cs falsely covered for all alignment algorithms in all data sets without read trimming (Figure 5).
When combined with Mott trimming, quality filtering did not further improve the results for the single-end data set, and only improved the results slightly for the paired-end data sets (the last two columns in Figure 4B and C; note the small ranges of the axes), suggesting that Mott trimming and quality filtering targeted the same set of low-quality reads. Because Mott trimming led to overall a slightly better and more stable improvement than quality filtering, we suggest Mott trimming without any filtering, which will be the focus of our discussion for the rest of the ‘Results’ section.
Comparison of the methylation level estimation performance of the mapping algorithms
LAST achieved the highest fraction of Cs perfectly estimated and the lowest average error on the two longer read data sets—the 101 nt single-end data set and the 100 nt paired-end data set B (Figure 4A and Supplementary Tables S2 and S4). After Mott trimming and no filtering, Bismark with Bowtie2 was a close second to LAST on these data sets. On the paired-end data set A, LAST and Bismark with Bowtie2 performed equally well (Figure 4A and Supplementary Table S3). GSNP and BSMAP performed slightly worse than LAST and Bismark for the three data sets.
Paired-end sequencing led to more accurate methylation level estimation for shorter reads
LAST, Bismark and GSNAP performed better on the shorter, 45 nt and 50 nt, paired-end data set A than its single-end data set with matching sequencing depth (Figure 4D). In contrast, the performance of all algorithms was worse on the 100 nt paired-end data set B than its matching single-end data set (Figure 4D). This is consistent with the higher mapping rate in the single-end mode for data set B (Figure 3B). Thus, with sufficiently long sequencing length (e.g. the 100 nt paired-end data set B), it is more cost-effective to perform single-end sequencing than paired-end sequencing if the overall methylation level estimation is the goal. In the next section, we show that paired-end sequencing achieves more accurate methylation level estimation than single-end sequencing in repetitive regions, even for data set B.
Read mapping and methylation level estimation in repetitive regions
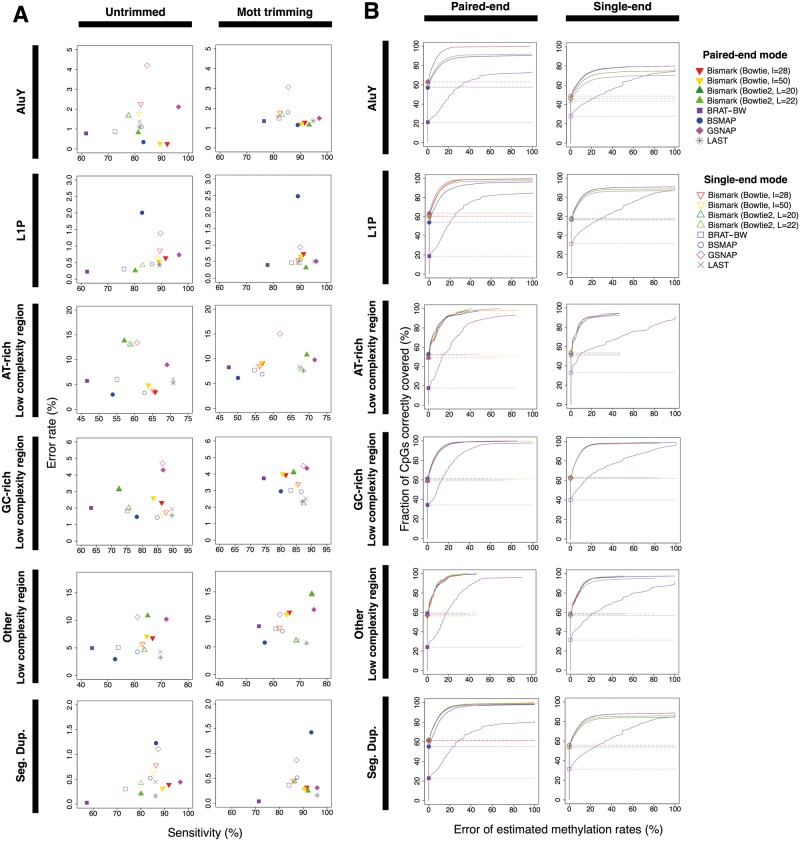
We also compared the performance of read mapping and methylation level estimation in repetitive regions. Consistent with the overall results mentioned above, Mott trimming substantially increased mapping sensitivity for most mapping algorithms in repetitive regions, at the cost of increased error rates for LCR (Supplementary Figure S7–S9). In particular, there were consistent improvement in AluY, LINE-1P and segmental duplications (sensitivity increase = 1.33–13.28%; error rate decrease = 0.06–0.55%; Figure 6A).
Figure 6.
Performance of mapping and CpG methylation estimation in repetitive regions. (A) Mapping accuracy in repetitive regions for the paired-end data set B. (B) Methylation level estimation performance in repetitive regions on the paired-end data set A. The cumulative fraction of Cs correctly covered is plotted against the error of estimated methylation levels, i.e. the difference in predicted and true methylation levels. A colour version of this figure is available online at BIB online: https://academic.oup.com/bib.
Paired-end information improved mapping accuracy in repetitive regions. Even though paired-end information did not improve the overall mapping accuracy for the paired-end data set B, it improved mapping sensitivity in repetitive regions by 13.28% with slightly worse error rates for LCR (Figure 6A; filled symbols versus open symbols). Paired-end information also improved methylation level estimation, indicated by the much higher fraction of Cs correctly covered across the entire range of estimated methylation levels (Figure 6B and Supplementary Tables S5–S7). In particular, we observed the greatest improvement in estimated methylation levels for AluY, LINE-1P and segmental duplication, concomitant with the improved mapping accuracy for these families of repeats.
For the single-end data set, LAST and Bismark with Bowtie2 showed higher sensitivities yet lower error rates than the other mapping algorithms for most repeat families (Supplementary Figure S7). Accordingly, these two algorithms performed the best for methylation level estimation, especially for AT-rich repeats (Supplementary Table S5).
For the paired-end data set A, Bismark with Bowtie2 performed the best in both mapping and methylation level estimation in repetitive regions, with the exception of LCR. Bismark with Bowtie, LAST and GSNAP showed nearly the same performance on both mapping and methylation level estimation in segmental duplications (Supplementary Figure S8 and Table S6). Bismark and GSNAP exhibited better mapping and methylation level estimation accuracies in AluY and LINE-1P, respectively.
For the paired-end data set B, LAST showed the most robust performance on mapping of all repeats (Figure 6A and Supplementary Figure S9). For methylation level estimation, LAST and Bismark performed better than the other alignment algorithms (Supplementary Table S7).
Running time and memory usage of mapping algorithms
We measured running time and peak memory usage for each mapping algorithm in single-end and paired-end modes on the paired-end data set A (Table 3). The runtime measurement includes the entire process such as reading and outputting files.
Table 3.
Running time and peak memory usage on the paired-end data set A in 10M pairs
| Methods | Memory usage (MB) | CPU time (min) | ||
|---|---|---|---|---|
| Single end | Paired end | Single end | Paired end | |
| Bismark (Bowtie, l = 28) | 104.12 | 105.06 | 100.51 | 105.12 |
| Bismark (Bowtie, l = 50) | 104.14 | 105.63 | 87.61 | 90.58 |
| Bismark (Bowtie2, L = 20) | 104.14 | 104.13 | 146.00 | 160.54 |
| Bismark (Bowtie2, L = 22) | 104.14 | 105.33 | 130.90 | 149.74 |
| BRAT_BW | 538.72 | 538.74 | 17.34 | 16.20 |
| BSMAP | 943.11 | 942.82 | 6.33 | 7.51 |
| GSNAP | 1724.90 | 1752.40 | 75.40 | 207.86 |
| LAST | 131.31 | 129.05 | 141.92 | 134.82 |
BSMAP was the fastest on both single-end and paired-end mapping but used a large amount of memory. Bismark with Bowtie ran faster than with Bowtie2, 1.67 and 1.77 times faster in the single-end and paired-end mode, respectively. Bismark used the least memory through the entire mapping process. The runtime and the memory usage of LAST were approximately 20% and 30% worse than those of Bismark with Bowtie.
We tested the latest version of Bismark (v0.14.2), which added the functionality of running Bowtie using multiple CPUs. (This functionality existed for Bowtie2 in the previous version of Bismark already.) However, as pointed out in Bismark’s manual, Bowtie running in parallel consumes more memory than Bowtie2 running in parallel, with the difference proportional to the total number of CPUs. Because most WGBS data sets are very large (hundreds of millions of reads for each human data set), Bowtie2 is a more practical option for most users.
Discussion
Overall recommendation
It would be desirable to analyze experimental WGBS data sets directly; however, the true methylation levels are not known. Therefore, we simulated single-end and paired-end benchmark data sets by incorporating error profiles in experimental data sets and experimentally measured frequencies of SNPs and indels. Judging by the performance on the three simulated data sets, we recommend Mott trimming for preprocessing combined with Bismark or LAST for mapping without any further filtering as the best approach for accurate methylation level estimation. However, the data sets we used did not completely simulate experimental data as discussed in the next section, “Limitation of benchmark data sets”. Thus, the other alignment algorithms may still be useful in practice. For paired-end data sets, Bismark requires the user to input the mean and standard deviation of fragment lengths. One advantage of LAST is that it does not require such user input because it can directly estimate fragment length distribution after mapping reads. Because the fragment length distribution may be unavailable or may not be measured accurately, this feature of LAST can be desirable. Paired-end sequencing achieves better performance than single-end sequencing for short read lengths (e.g. 50 nt). For long reads (e.g. 100 nt), single-end sequencing yields slightly higher overall accuracy for methylation level estimation; yet, paired-end sequencing achieves higher accuracy at repetitive regions.
Limitation of benchmark data sets
Although our benchmark data sets were based on experimental data, we did not accounted for all sources of experimental noise. For example, our simulated data sets did not contain any contaminant DNA sequences, nor did they contain entire or parts of adapter sequences at the end of reads that experimental data sets usually contain. Another factor that may need to be considered during the simulation is the degree of DNA degradation, because low-quality DNA samples can adversely impact the entire WGBS analysis. In the simulated paired-end data sets, we assumed that fragment lengths followed a uniform distribution while extracting reads from the reference genome; however, the distributions of fragment lengths in experimental data sets may vary depending on experimental protocols and sequencers. Another aspect that may improve the simulated data sets is to consider differential methylation levels of CpGs in different locations of the genome, specifically the low levels in promoters and CpG islands. One possible approach is to first estimate the methylation levels of an experimental data set and use these as the gold standard methylation levels for simulating reads.
Preprocessing methods
We observed that Mott trimming weeded out sequencing errors and improved the performance of both mapping and methylation level estimation. The Mott trimming algorithm determines a trimming position in a read by computing the global minimum of a cumulative quality score. This global evaluation is more effective than other trimming algorithms because it preserves the largest possible portion of the read with high quality (Figure 1D–F). Although Mott trimming did not improve the mapping accuracy for LAST, it improved the subsequent methylation level estimation for both paired-end data sets. This indicates that although low-quality reads can be mapped to the correct locations of the genome, their errors affect methylation level estimation, and trimming these erroneous nucleotides can lead to better results.
Mapping algorithms
Among the five algorithms for mapping bisulfite converted reads that we tested, LAST, BSMAP and GSNAP belong to the wild-card class of aligners while Bismark and BRAT-BW belong to the three-letter class. We found LAST to achieve the best performance over all. LAST uses a modified scoring matrix with a positive score between a C and a T (match = 6; C/T match = 3; mismatch = –18) [22]. The unique aspect of LAST is that it constructs adaptive seeds that are of variable lengths, determined after taking into account their rareness in the reference genome [54]. In comparison, BSMAP and GSNAP use fixed-length seeds and evaluate an alignment based on the number of mismatches between any types of nucleotides [15, 20]. The mismatch-oriented evaluation of BSMAP and GSNAP tend to discard or incorrectly align T-rich sequences in bisulfite-converted reads against CT-rich regions in the reference genome. Furthermore, BSMAP has a stringent scheme that checks a seed match only at the beginning of each read, which may also impact its accuracy. Bismark showed comparable performance with LAST when combined with read trimming. Although Bismark also assesses the best alignment for each read by tallying up mismatches, the evaluation is based on the number of mismatches between the nucleotides that are not bisulfite-converted [12, 55]. Overall, these penalty and alignment evaluation schemes in LAST and Bismark for bisulfite-converted reads may contribute to their better performance.
Postprocessing methods
We observed that the stricter thresholds of coverage filtering with the cutoff 10 decreased sensitivity and increased the average error. Coverage filtering with a gentle threshold such as the cutoff 3 can eliminate inaccurate methylation estimation originating from a few wrongly mapped reads. Quality filtering with quality score cutoff 10 is the most effective postprocessing method, when not combined with Mott trimming.
Possible enhancement for methylation level estimation at low sequencing depth
In this study, we simulated three data sets with ∼30-fold or greater genome coverage, following the recommendations of the ENCODE and IHEC consortia [56, 57]. Despite the rapid developments of sequencing technologies, it remains challenging for WGBS libraries to achieve 30-fold coverage of mammalian genomes. In addition to sequencing cost, the amount of DNA sample can also be limiting because bisulfite treatment can lead to fragmentation of DNA and substantial loss of sample DNA [58].
For the WGBS libraries that have low genomic coverage, a possible enhancement for methylation level estimation is integrating methods like BSmooth [16] as a postprocessing step. BSmooth assumes that cytosines in genomic blocks (e.g. CpG islands and CpG island shores) have similar methylation levels, and conducts a local likelihood smoothing to rescue the cytosines with low read coverage. Although the smoothing may miss individual cytosines that exhibit sharp changes in methylation levels within the genomic block, it can greatly aid the detection of differentially methylated regions in low-coverage samples.
Summary
In summary, we created three benchmark data sets based on three experimental WGBS data sets. Using these data sets, we compared choices of preprocessing methods, mapping algorithm and postprocessing methods on accurate mapping and methylation level estimation. We also showed how the paired-end information improved the performance of WGBS analysis, especially in repetitive regions. This comprehensive evaluation of WGBS pipelines should provide a practical guide for researchers to choose the most suitable methods with the optimal parameters.
Key Points
Whole genome bisulfite sequencing (WGBS) analysis steps that included 192 combinations of preprocessing, mapping and postprocessing methods were evaluated using simulated single-end and paired-end data sets that closely matched experimental WGBS data sets.
Mott trimming for preprocessing combined with Bismark or LAST for mapping without any further postprocessing showed the best accuracy on methylation level estimation.
Paired-end sequencing reduced error rate and enhanced sensitivity for both read mapping and methylation level estimation, especially for short reads and in repetitive regions of the human genome.
Supplementary Data
Supplementary data are available online at https://academic.oup.com/bib.
Acknowledgements
We thank Dr Benjamin Hitz and Dr Martin C. Frith for valuable discussions on the implementation of a uniform processing pipeline of WGBS data for the ENCODE consortium. We also thank Michael Purcaro for proofreading our manuscript.
Biographies
Junko Tsuji is a postdoctoral research associate in the Program in Bioinformatics and Integrative Biology at University of Massachusetts Medical School. Her research interests include genome sequence analysis and epigenetic regulation of gene expression.
Zhiping Weng is a professor and the director of the Program in Bioinformatics and Integrative Biology at University of Massachusetts Medical School. She develops and applies computational algorithms for studying a wide range of biological problems in genomics, epigenomics and gene regulation.
Funding
National Institutes of Health (grant no. U41 HG007000 to Z.W.).
References
- 1.Hawkins RD, Hon GC, Lee LK, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell 2010;6:479–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cantone I, Fisher AG. Epigenetic programming and reprogramming during development. Nat Struct Mol Biol 2013;20:282–9. [DOI] [PubMed] [Google Scholar]
- 3.Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 2009;41:178–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 2010;11:204–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol 2005;2:S4–11. [DOI] [PubMed] [Google Scholar]
- 6.Laird PW. Principles and challenges of genome-wide DNA methylation analysis. Nat Rev Genet 2010;11:191. [DOI] [PubMed] [Google Scholar]
- 7.Smallwood SA, Lee HJ, Angermueller C, et al. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat Methods 2014;11:817–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frommer M, McDonald LE, Millar DS, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci 1992;89:1827–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Capuano F, Mülleder M, Kok R, et al. Cytosine DNA Methylation is found in Drosophila melanogaster but absent in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and other yeast species. Anal Chem 2014;86:3697–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adusumalli S, Omar MFM, Soong R, et al. Methodological aspects of whole-genome bisulfite sequencing analysis. Brief Bioinform 2015;16:369–79. [DOI] [PubMed] [Google Scholar]
- 11.Lim JQ, Tennakoon C, Li G, et al. BatMeth: improved mapper for bisulfite sequencing reads on DNA methylation. Genome Biol 2012;13:R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011;27:1571–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saito Y, Tsuji J, Mituyama T. Bisulfighter: accurate detection of methylated cytosines and differentially methylated regions. Nucleic Acids Res 2014;42:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris EY, Ponts N, Le Roch KG, et al. BRAT-BW: efficient and accurate mapping of bisulfite-treated reads. Bioinformatics 2012;28:1795–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xi Y, Li W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics 2009;10:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen KD, Langmead B, Irizarry RA, et al. BSmooth: from whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biol 2012;13:R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen PY, Cokus SJ, Pellegrini M. BS seeker: precise mapping for bisulfite sequencing. BMC Bioinformatics 2010;11:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo W, Fiziev P, Yan W, et al. BS-Seeker2: a versatile aligning pipeline for bisulfite sequencing data. BMC Genomics 2013;14:774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kreck B, Marnellos G, Richter J, et al. B-SOLANA: an approach for the analysis of two-base encoding bisulfite sequencing data. Bioinformatics 2012;28:428–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 2010;26:873–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prezza N, Del Fabbro C, Vezzi F, et al. ERNE-BS5: aligning BS-treated sequences by multiple hits on a 5-letters alphabet. Proceedings of the ACM Conference on Bioinformatics, Computational Biology and Biomedicine, 2012, Orlando, Florida, USA, pp. 12–19. [Google Scholar]
- 22.Frith MC, Mori R, Asai K. A mostly traditional approach improves alignment of bisulfite-converted DNA. Nucleic Acids Res 2012;40:e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 2008;18:1851–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pedersen B, Hsieh TF, Ibarra C, et al. MethylCoder: software pipeline for bisulfite-treated sequences. Bioinformatics 2011;27:2435–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coarfa C, Yu F, Miller CA, et al. Pash 3.0: a versatile software package for read mapping and integrative analysis of genomic and epigenomic variation using massively parallel DNA sequencing. BMC Bioinformatics 2010;11:572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith AD, Chung WY, Hodges E, et al. Updates to the RMAP short-read mapping software. Bioinformatics 2009;25:2841–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otto C, Stadler PF, Hoffmann S. Fast and sensitive mapping of bisulfite-treated sequencing data. Bioinformatics 2012;28:1698–704. [DOI] [PubMed] [Google Scholar]
- 28.Ondov BD, Cochran C, Landers M, et al. An alignment algorithm for bisulfite sequencing using the Applied Biosystems SOLiD System. Bioinformatics 2010;26:1901–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bock C. Analysing and interpreting DNA methylation data. Nat Rev Genet 2012;13:705–19. [DOI] [PubMed] [Google Scholar]
- 30.Del Fabbro C, Scalabrin S, Morgante M, et al. An extensive evaluation of read trimming effects on illumina NGS data analysis. PLoS ONE 2013;8:e85024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.J 2011;17:10. [Google Scholar]
- 32.Cox MP, Peterson DA, Biggs PJ. SolexaQA: at-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics 2010;11:485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindgreen S. AdapterRemoval: easy cleaning of next-generation sequencing reads. BMC Res Notes 2012;5:337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kong Y. Btrim: a fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics 2011;98:152–3. [DOI] [PubMed] [Google Scholar]
- 35.Smeds L, Künstner A. ConDeTri–a content dependent read trimmer for Illumina data. PloS One 2011;6:e26314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis MPA, van Dongen S, Abreu-Goodger C, et al. Kraken: a set of tools for quality control and analysis of high-throughput sequence data. Methods 2013;63:41–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011;27:863–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kunde-Ramamoorthy G, Coarfa C, Laritsky E, et al. Comparison and quantitative verification of mapping algorithms for whole-genome bisulfite sequencing. Nucleic Acids Res 2014;42:e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hormozdiari F, Hajirasouliha I, McPherson A, et al. Simultaneous structural variation discovery among multiple paired-end sequenced genomes. Genome Res 2011;21:2203–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shrestha AMS, Frith MC. An approximate Bayesian approach for mapping paired-end DNA reads to a reference genome. Bioinformatics 2013;29:965–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chatterjee A, Stockwell PA, Rodger EJ, et al. Comparison of alignment software for genome-wide bisulphite sequence data. Nucleic Acids Res 2012;40:e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tran H, Porter J, Sun M, et al. Objective and comprehensive evaluation of bisulfite short read mapping tools. Adv Bioinforma 2014;2014:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 2001;29:308–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smit AF, Hubly R, Green P. RepeatMasker Open-4.0. 2013. http://www.repeatmasker.org/ (9 March 2014, date last accessed). [Google Scholar]
- 46.Bailey JA, Gu Z, Clark RA, et al. Recent segmental duplications in the human genome. Science 2002;297:1003–7. [DOI] [PubMed] [Google Scholar]
- 47.Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Res 2002;12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lister R, Mukamel EA, Nery JR, et al. Global Epigenomic reconfiguration during Mammalian Brain development. Science 2013;341:1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Myers R, Pauli F. Whole Genome Bisulfite Sequencing by ENCODE/HAIB (GSE40832). 2012. , http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE40832 (11 November 2014, date last accessed). [Google Scholar]
- 50.Ewing B, Hillier L, Wendl MC, et al. Base-calling of automated sequencer traces usingPhred. I. Accuracy assessment. Genome Res 1998;8:175–85. [DOI] [PubMed] [Google Scholar]
- 51.MacManes MD. On the optimal trimming of high-throughput mRNA sequence data. Front Genet 2014;5:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langmead B, Trapnell C, Pop M, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009;10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kiełbasa SM, Wan R, Sato K, et al. Adaptive seeds tame genomic sequence comparison. Genome Res 2011;21:487–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krueger F, Kreck B, Franke A, et al. DNA methylome analysis using short bisulfite sequencing data. Nat Methods 2012;9:145–51. [DOI] [PubMed] [Google Scholar]
- 56.ENCODE consortium. ENCODE Standards and Guidelines for Whole Genome Shotgun Bisulfite Sequencing (WGBS). https://www.encodeproject.org/about/experiment-guidelines/. 2015 [Google Scholar]
- 57.International Human Epigenome Consortium (IHEC). Reference Epigenome Standards Whole Genome Shotgun Bisulfite Sequencing. http://ihec-epigenomes.org/research/reference-epigenome-standards/. 2013 [Google Scholar]
- 58.Miura F, Enomoto Y, Dairiki R, et al. Amplification-free whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res 2012;40:e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.