Abstract
ChIP-seq has become a widely adopted genomic assay in recent years to determine binding sites for transcription factors or enrichments for specific histone modifications. Beside detection of enriched or bound regions, an important question is to determine differences between conditions. While this is a common analysis for gene expression, for which a large number of computational approaches have been validated, the same question for ChIP-seq is particularly challenging owing to the complexity of ChIP-seq data in terms of noisiness and variability. Many different tools have been developed and published in recent years. However, a comprehensive comparison and review of these tools is still missing. Here, we have reviewed 14 tools, which have been developed to determine differential enrichment between two conditions. They differ in their algorithmic setups, and also in the range of applicability. Hence, we have benchmarked these tools on real data sets for transcription factors and histone modifications, as well as on simulated data sets to quantitatively evaluate their performance. Overall, there is a great variety in the type of signal detected by these tools with a surprisingly low level of agreement. Depending on the type of analysis performed, the choice of method will crucially impact the outcome.
Keywords: ChIP-seq, differential analysis, software
Introduction
High-throughput sequencing (HTS) has become a standard method in genomics research and has almost completely superseded array-based technologies, owing to the ever-decreasing costs and the variety of different assays that are based on short read sequencing. Most array-based assays have now a counterpart based on HTS, with a generally improved dynamic range in the signal. Genome sequence (whole genome or exome), DNA-methylation (whole genome bisulfite sequencing), gene expression (RNA-seq, CAGE-seq), chromatin accessibility (DNAse1-seq, ATAC-seq, FAIRE-seq) or chromatin interaction (ChIP-seq) all belong to the standard repertoire of genomic studies, and follow standardized protocols. However, the broad availability of these approaches should not hide the fact that they are still highly complex, requiring a number of experimental steps that can lead to considerable differences in the readout for a same assay performed by different groups [1, 2]. Large-scale consortia such as ENCODE or Roadmap, Epigenomics, which rely on different sequencing centers for the data collection, have faced the problem of harmonizing the results obtained by different centers, which require systematic bias correction before data integration can be achieved in a meaningful way. Clearly, the more complex the experimental setup is, the more it is subject to biases, which can be introduced in the different steps of the experimental protocol or the downstream analysis [3, 4]. Among the approaches listed previously, those based on immunoprecipitation are the more complex ones, as the antibody-based precipitation usually represents a critical step, and leads to variations in the precipitation efficiency, the cross-reaction probability, conditioned by the quality of the antibody. Hence, the reproducibility of the assays is often limited, especially in cases with additional constraints, for example low input material. The amount of noise in the data can be substantial: a standard measure of the signal-to-noise ratio is the FRiP (fraction of reads in peaks), which measures how many sequencing reads are located in enriched regions, compared with the total amount [1]. In the ENCODE project, this ratio was in the range of a few percent, indicating that the amount of noise is >90%. To detect consistent signal between replicates, special statistical methods have been developed such as the Irreproducible Discovery Rate [5], which allow to detect consistent signal between replicates.
Detecting differential gene expression between several conditions is one of the most common analysis steps since the advent of genome-wide expression measurements based on microarrays, and a considerable literature has been dedicated to the development of solid statistical procedures. Expression measurements based on HTS has also lead to the development of new tools or the adjustment of existing procedures. Differential analysis are however not restricted to gene expression but can in principle be extended to any quantitative assay, such as the measurement of DNA methylation levels or the enrichment of ChIP signal. Accordingly, many tools are available either to detect differential gene expression or to delimit differentially methylated regions (DMRs). Similarly, a number of tools and methods have been published to detect differences in ChIP signal between several conditions [6–19]. If the underlying question is similar for ChIP-seq experiments (where are the regions showing significant differential signal between two conditions?), there are a number of particularities that make this simple question particularly challenging in the case of ChIP-seq data sets, as compared with RNA-seq or whole-genome bisulfite sequencing. In the case of RNA-seq, most of the signal is concentrated in regions that are either annotated as genes or easily recognized as enriched regions, representing unannotated transcripts. Hence, the search space for differential signal is well defined. In the case of differential DNA methylation, the search space is extended to the complete genome, as CpGs occur virtually everywhere. However, the signal range is constrained to a finite interval (between 0% and 100% methylation) and the amount of noise is low, two properties which make the search for DMRs a tractable problem. In the case of ChIP-seq, we are facing multiple challenges: (i) the search space is not limited to a particular region of the genome, as differential binding can occur everywhere; this implies that regions of interest, in which differential signal should be looked for, need to be defined first; (ii) the range of the signal is not constrained to a finite interval and requires transformation such that standard statistical tools can be applied; (iii) the amount of noise in considerable, making variations in the signal challenging to detect, especially when these differences are subtle, and (iv) the properties of the enriched regions (in particular their length) differ substantially depending on the protein or epigenetic modification targeted by the immunoprecipitation.
In this article, we have performed a comprehensive comparison of a large number of tools, which have been published to detect differential signals in ChIP-seq assays. Our criterion for tool selection was the availability of a working software that could be implemented without the need for extensive efforts for porting the code. These tools differ in many ways, and this diversity reflects the diverse challenges listed previously: some require preliminary detection of enriched regions by external peak-calling algorithms, while others implement their own detection method or work using sliding windows; others differ in the underlying statistical modeling of the signal distribution, based either on Poisson distribution or on a more flexible negative binomial distribution. Finally, some tools work in the absence of replicates for each condition, and others require replicates to provide differential analysis. Importantly, some tools have been specifically designed for particular ChIP-seq data, such as histone modifications or transcription factor (TF) binding. We have indicated this in the tool description (Supplementary Table S1); however, we have tried to apply these tools to all types of data sets to verify their behavior. For the evaluation, and given the absence of a gold standard for differential enrichment in ChIP signal, we have adopted a multistep strategy. First, the tools have been compared using various published ChIP-seq data sets, to compare global statistics on the sets of differential regions (DR) detect by each tool, such as the number of DR, their length distribution and the pairwise overlap between the outputs. We have made the distinction between tools that do not require replicates for the definition of DR (single-replicate) and those that only work when replicates are available (multi-replicates), and performed all analysis for both categories separately. To estimate rates of true positives and false positives, we have performed a simulation of synthetic data sets, but made sure that these simulated data sets are as close as possible to real data sets and represent realistic and fair test sets. Using these data sets, we have compared the sensitivity and specificity of each tool. In particular, we have investigated the threshold in differential signal that is required for each tool to start detecting a significant difference. Finally, we have conducted a functional annotation of the sets of DR and related the DR to differentially expressed genes (DEG). This last analysis is meant to address typical biological questions that are mostly focused at difference in gene expression driven by differential enrichments.
Overall, we have seen considerable differences between the tools, which can be traced back to the underlying statistical assumptions or the way the initial search space is defined, either based on peak calling or using a windowing approach. Researchers that are interested in finding DR in their study should be aware of these differences, and should adapt the choice of the tool to their particular experimental question. We believe that our extensive study can help in making a motivated choice and avoid obvious pitfalls.
Material and methods
Tools
We have selected 14 available tools for differential peak calling, based on a variety of algorithmic approaches [6–19]. A detailed description of each tool is given in Supplementary Text 1, whereas the parameters used are described in Supplementary Table S1.
Data set sources
A summary of the data sets used and statistics on the number of reads is given in Supplementary Table S2.
Transcription factor data
We obtained FoxA1 ChIP-seq data from a previously published study for two experimental conditions including estradiol (E2)- and vehicle (Veh)-treated MCF7 cells each in two biological replicates (GEO: GSE59530). Additionally, expression data as already aligned reads for both conditions derived from global run-on sequencing (GRO-seq) were taken from this study (GEO:GSE59531) [20].
Sharp histone post translational modification (PTM) data
Two biological replicates of H3K27ac ChIP-seq were retrieved from a study that differentiates embryonic stem cells (hESC-H1) to mesenchymal stem cells (GEO: GSE16256) [21].
Broad histone PTM data
Moreover, we collected data sets in two biological replicates for H3K36me3 in an MMSET (multiple myeloma SET domain) overexpression (TKO) against physiological expression (NTKO) setup in myeloma cells (GEO: GSE57632) [22].
Simulated sharp signal data
As an example of data with localized signal giving rise to sharp peaks, we used the merged replicates of the previously described FoxA1 ChIP in E2-treated MCF7 cells as a source for our simulation of a sharp signal data set (GEO: GSE59530) [20].
Simulated broad signal data
Moreover, two biological replicates of H3K36me3 in MCF7 cells were downloaded and pooled, which was used as source for the simulation of a broad signal data set (GEO: GSE31755) [22].
Input data for background simulation
As a control data set, we used DNA, which is not precipitated (‘input DNA'), which represents the standard control in ChIP-seq experiments. Five input experiments from different MCF7 studies were collected and further used to generate an additive background model (GEO: GSM1059389, GSM1089816, GSM1143667, GSM1241757 and GSM1383864). We performed a genome-wide correlation of the input signals to verify their similarities and pooled them together as source for our background signal simulation. Data simulation was then performed as described in the section ‘Simulation of a differential dataset'.
ChIP-seq preprocessing and analysis
The data sets were downloaded as sra files from the Sequence Read Archive (SRA) and converted to fastq with the SRA Toolkit. Reads were aligned to the hg19 reference genome using BWA aln with default settings [23]. Subsequently, duplicated reads and reads with bad mapping quality (-q 1) were removed from alignments.
We performed peak calling on all ChIP-seq data sets using MACS2 with additional parameters according to underlying signal type as followed: for sharp peaks ‘-g hs -q 0.1 –call-summits' and for broad peaks ‘-g hs -q 0.1 –broad' [24]. Resulting peak sets were used as input for tools that require peaks/regions as input for differential enrichment analysis.
Differential peak calling
We performed differential peak calling with each tool according to the settings recommended by the developers of the tools or, if not indicated explicitly, with default settings as listed in Supplementary Table S1.
Moreover, tools were classified into two categories according to their ability to use biological replicates or only single samples (Supplementary Table S1).
For the tools that do not consider replicates, the replicates of each ChIP-seq experiment were pooled to create a single data set for each condition. Because each of these tools calculates different significant measures, a general significant threshold for all of them could not be defined (Supplementary Table S1). However, for tools using an false-discovery rate (FDR) or P-value measure, we decided to set the thresholds as FDR ≤ = 0.01 or P-value ≤ = 0.05.
GRO-seq analysis
We obtained already aligned GRO-seq reads for the FoxA1 data set as an input for groHMM, which is an R pipeline for the analysis of GRO-seq data [25]. Differential GRO-seq analysis was performed according to the manual of groHMM using edgeR with a significance threshold of P ≤ = 0.05 [25, 26]. Resulting list of DEGs between E2- and Veh-treated MCF7 cells was used for further integration with differential peak data sets.
DEG enrichment analysis
Lists of DEGs for the H3K27ac and H3K36me3 data sets were taken from the corresponding studies to integrate them with computed differential peak sets. For FoxA1, we used the results of the GRO-seq analysis described above. Additionally, a list of house-keeping genes (HKGs) was obtained (http://www.wikicell.org/index.php/HK_Gene) and used as a control to compare enrichment of DEGs with HKGs.
First, DR were ranked for each tool according to the associated significance measure, and the top 1000 unique nearby genes assigned to DR were kept for this analysis. We performed a cumulative recovery analysis of DEGs as well as HKGs in each gene ranking. The resulting DEG (respectively HKG) ranks were used to compute the area under curve (AUC) for each tool. Moreover, a background model was computed for each ranking by randomly sampling 10,000 times a new gene ranking from all human genes (UCSC Known Gene Annotation) followed by the same cumulative recovery analysis. The resulting background AUC distributions were used to compute the normalized enrichment score (NES) with following formula:
where corresponds to the mean and to the standard deviation of the background model.
In addition, we introduced for each ChIP-seq data set another control experiment: we ranked all genes using the coverage fold-change within gene bodies (H3K36me3) and promoters (FoxA1 / H3K27Ac). We used these rankings to perform the same recovery analysis.
Gene ontology enrichment analysis
As large variations in terms of ‘number of differential peaks' were observable for all data sets, we decided to use a ‘gene centric' approach, focusing on the top 1000 genes that were close to a differential region. Therefore, each peak was annotated with its nearest gene using the R-package ChIPseeker [27]. We ranked these genes according to corresponding peak significance measures and obtained the top 1000 unique genes for each differential peak set. Regions ±1.5 kb around the consensus transcription start side (TSS) were used as input for the annotation tool GREAT with the setting 'single nearest gene' [28].
As the transcription factors or histone modifications studied here are considered to be positively correlated with gene expression, we used DEGs as a positive control. Therefore, regions ±1.5 kb around the consensus TSS of these genes were used as an input for a GREAT analysis with the setting ‘single nearest gene'.
Differential peak calling data presentation
Signal tracks were generated for each replicate using deepTools [29]. For sharp signal data, scaling factors were estimated with the signal extraction scaling (SES) option, and further log2 ratios of ChIP over input signal were computed [30]. Broad signal data were scaled according to the total number of reads and again log2 ratios were calculated.
Additionally, all differential peaks can be displayed in a single genome-wide coverage track, which was generated using the bedtools genomecov function [31].
All differential peak sets, signal tracks and genome-wide coverage tracks can be displayed using following UCSC genome browser trackhubs:
http://goo.gl/5WI5w3 (FoxA1); http://goo.gl/HyfooK (H3K36me3); http://goo.gl/5uYXsn (H3K27ac).
Simulation of a differential data sets
To simulate a realistic ‘gold standard' data set for differential ChIP enrichment, we started from real ChIP data sets: one data set targeting FoxA1, as an example of a data set with sharp signal, and a H3K36me3 data set as an example of a broad enrichment. In each case (FoxA1 and H3K36me3), we simulated two ‘treatment' data sets (T1 and T2) representing the reference condition and the comparison condition, as well as ‘control' data sets (C1 and C2), corresponding to input. The treatment data sets (T) were obtained by merging regions corresponding to real signal (S) with background noise (B) (hence, T = S + B) The signal data set was obtained as follows: we performed peak calling using MACS2, using the provided input data set as a control. We selected the top 20,000 peaks identified by the peak caller, as they are most likely to represent truly enriched regions. In a first step, only the reads located in these regions were considered, and represent the reference signal data set (S1). To simulate the second condition, these peak regions were split into two groups: 10,000 peaks were kept as is (S20), and represent nondifferential peaks. The remaining 10,000 peaks were divided into 10 groups of 1000 peaks each, which were downsampled by random read sampling by 10%, 20%, etc. (S210,S220,…). These represent sets of differential peaks in which the level of differential enrichment is variable. Next, we simulated realistic background signal B: several public input data sets for the MCF7 cell line were downloaded, aligned and compared. The input data set showing the highest correlation with the original input was used to sample reads, until the library size of the real ChIP-seq experiment was reached. If not enough reads were available in one input, it was merged with a second similar one. This is done twice, to simulate a background for the reference and the second condition (B1 and B2). In summary, our two data sets are composed of T1 = S1 + B1 and T2 = S2 + B2, where S2 = S20 + Σi = 10 → 100 S2i. Comparing these two data sets should ideally yield 10,000 differential peaks, which represents our gold standard.
The Bioconductor packages IRanges and GenomicRanges [32] were used to detect overlapping peak regions. For the simulated data set, a GenomicRanges object comprising all true differential binding events (n = 10,000) was implemented as a reference to find overlaps with DR called by the respective tools. We considered a called DR to be a true event if at least 25% of its length overlapped with a real DR.
Using this data set, we computed two different measures to infer sensitivity and specificity of the analyzed tools. For differential ChIP-seq analysis, the number of negatives outnumbers by far the number of positives, as most of the genome is not differentially enriched. Hence, the specificity is not a good measure, as all tools will have artificially high specificity, given the large number of true negatives. We therefore prefer using a ‘precision'-like measure. To take into account the fact that some tools call large DR, we defined the ‘Jaccard index precision' as as a more stringent precision measure, penalizing imprecise peak calling (either over- or under-calling). Here, Ai represents a DR called by a tool and Bj(i) represents the true DR that intersects Ai. The norm is meant as the genomic length of the intersection or the union, and N indicates the number of DR called by the tool considered. We define the recall as the proportion of true DR that intersects a called DR with a 25% overlap.
Results
Comparison using real data sets
We applied the single and multi-replicates pipelines to selected data sets, using the default or recommended settings for the tools (see Supplementary Table S1 for methods and parameters used). As mentioned in the Introduction, a major distinction can be made between tools that require a predefined set of regions of interest (for example, peaks) and those that implement an internal procedure to define these regions of interest. We call the first set of tools EROI (for external regions of interest), and the second IROI (for internal regions of interest), and distinguish these two groups (in addition to the requirement for replicates or not) in the subsequent analysis.
Our first question was whether the tools would report consistent sets of differential peaks.
For both types of ChIP-seq data sets (sharp TF binding and broad histone modification enrichment), we observed considerable differences in the number of reported DR (Figure 1A and B). The sets of DR ranged from no detected differential peak (QChIPat) to about 150,000 DR for ODIN-poisson for the FoxA1 data set. Five of the methods had a relatively consistent number of DR, in the range of 25,000–35,000 peaks. Among the tools that were given the same set of predetermined regions using an external peak caller (MACS2 in this case), the number of DR does not show particular consistency, with MAnorm having about 23,000 DR and QChIPat only a handful, indicating that the number of input regions is not the primary determinant of the number of detected DR. Obviously, the number of DR depends on the choice of parameters, which we have chosen in a consistent way across tools. We were interested in determining how sensitive the number of peaks is on this choice. Hence, we have varied the threshold on the P-value or FDR, and recorded the number of DR (Figures 1C and 2C). Surprisingly, the IROI tools from the single replicate pipeline show a step-like behavior, with a pronounced jump at small P-values, and a quasi saturation beyond this threshold. Other tools show a more progressive increase. Hence, the choice of P-value threshold is crucial for the first class of tools, as this number can vary abruptly by several orders of magnitude. As for the tools with a small number of DR (MMDiff, DBChIP), this number indeed increases when relaxing the threshold, but remains at a low level, much lower than other tools from the same class, indicating an intrinsic difference in the internal statistical procedure to call DR.
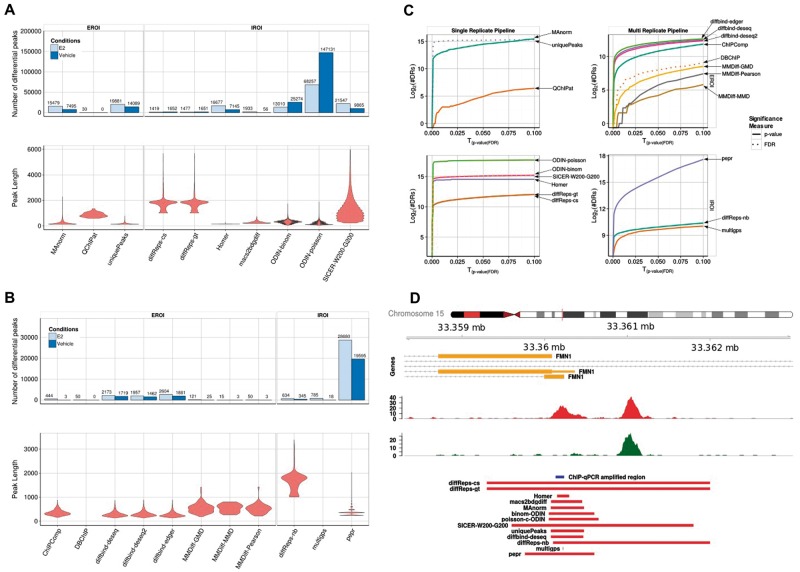
Figure 1.
Overview of the DR called by the single and multi-replicate pipelines for the FoxA1 data set: (A) Number (barplots) and size distribution (violin plots) of detected DR in each condition and size distribution for the single replicate pipeline. (B) Same as shown in A for the multi-replicate pipeline. (C) Plots showing the number of DR returned by each method as a function of the P-value threshold (plain lines) or FDR (dotted lines). (D) An example of a region, highlighting the difference between the different tools in terms of length of DR. A colour version of this figure is available online at BIB online: https://academic.oup.com/bib.
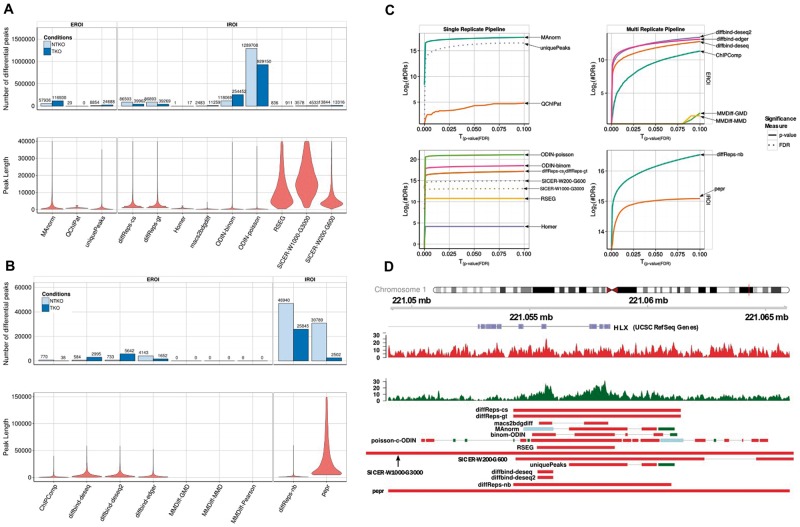
Figure 2.
Same as Figure 1 for the H3K36me3 data set. A colour version of this figure is available online at BIB online: https://academic.oup.com/bib.
Whereas the total number of DR is subject to the choice of parameters, we expected that, beyond the amount of DR, the proportion in each condition should be consistent between the tools, showing, for example, systematically more DR in one direction. To our surprise, there was a lack of consistency in this respect, with four tools predicting a higher number of DR enriched in the vehicle condition compared with the estrogen-treated condition, while five tools predicted more DR enriched in the estrogen-treated condition. The naive approach based on the number of unique peaks predicts indeed more specific peaks in the treated condition, and hence, we tend to have greater confidence in the prediction of the five tools that confirm this tendency. Additionally, this is in accordance with the biological expectation, as it was previously reported that FoxA1 shows increased binding on estrogen treatment [20].
The number of DR is related to the size of the reported regions: some tools might call a large number of small regions, while others would aggregate them into larger domains. Here also, we observe huge differences, related to the underlying method (Figure 1A and B, lower panel). Some tools report small regions, while others like diffReps or SICER identify large regions of up to 2 kb as being differentially enriched. While this might be realistic for histone modifications, it appears to be a clear overestimation of the real size in the case of TFs (Figure 1C). In particular, this overestimation implies that any subsequent analysis of enriched DNA motifs in the DR will be obscured by noise. Tools based on fixed windows approaches (diffReps, SICER,…) generally call broader regions, compared with the tools based on an initial peak calling step.
The tools included in the multi-replicate pipeline consistently report a much lower number of DR. diffReps, which can handle either single or multiple replicates, reports less than half of the DR in the multiple replicates setting, compared with the single replicate in which the data sets were pooled. Hence, the tools including the replicates appear to be much more conservative, at least with the default parameters we applied. Interestingly, all tools from the multi-replicate pipeline agree in predicting more peaks enriched in the estrogen-treated condition.
Despite the difference in the type of regions, the same observations hold true for the data sets for H3K36me3 (Figure 2), a histone modification characterized by broad domains of enrichment, especially in the gene body of transcribed genes, as well as for H3K27ac (Supplementary Figure S1), a mark associated with active regulatory regions. The length distribution is variable among tools, and for H3K36me3 does not reflect the size distribution of transcripts, as could have been anticipated for this histone mark. Figure 2D shows how different the lengths of DR are on the example of the HLX gene, indicating that the differential signal does not cover the whole transcript. The proportion of DR between the two conditions is not consistent either for both histone data sets, with some tools predicting more DR in one direction while other predict the opposite (Figure 2, Supplementary Figure S1). PePr, for example, predicts a 12-fold higher number of peaks enriched in the NTKO condition, while diffBind predicts an 8-fold enrichment in the other direction.
Unlike the FoxA1 data set, in the H3K36me3 data set, the tools in the replicate pipeline do not agree regarding the number of enriched peaks in each condition. Both IROI tools (diffreps-nb and PePr) do agree, while the discrepancy is greater among the EROI tools, despite the fact that they were supplied the same set of regions. Surprisingly, the three algorithms implemented in diffBind yield contradictory results, which might be related to the different normalization approaches used by DESeq and edgeR. They do, however, agree for the H3K27ac data set, indicating that the broad signal of H3K36me3 enrichment represents a particular challenge to these tools.
Next, we asked to what extent the sets of DR were overlapping between tools. Given the broad range of DR sizes between tools described previously, it is not obvious to determine whether two peaks agree. Instead, we considered the genomic range covered by the peak sets of each algorithm.
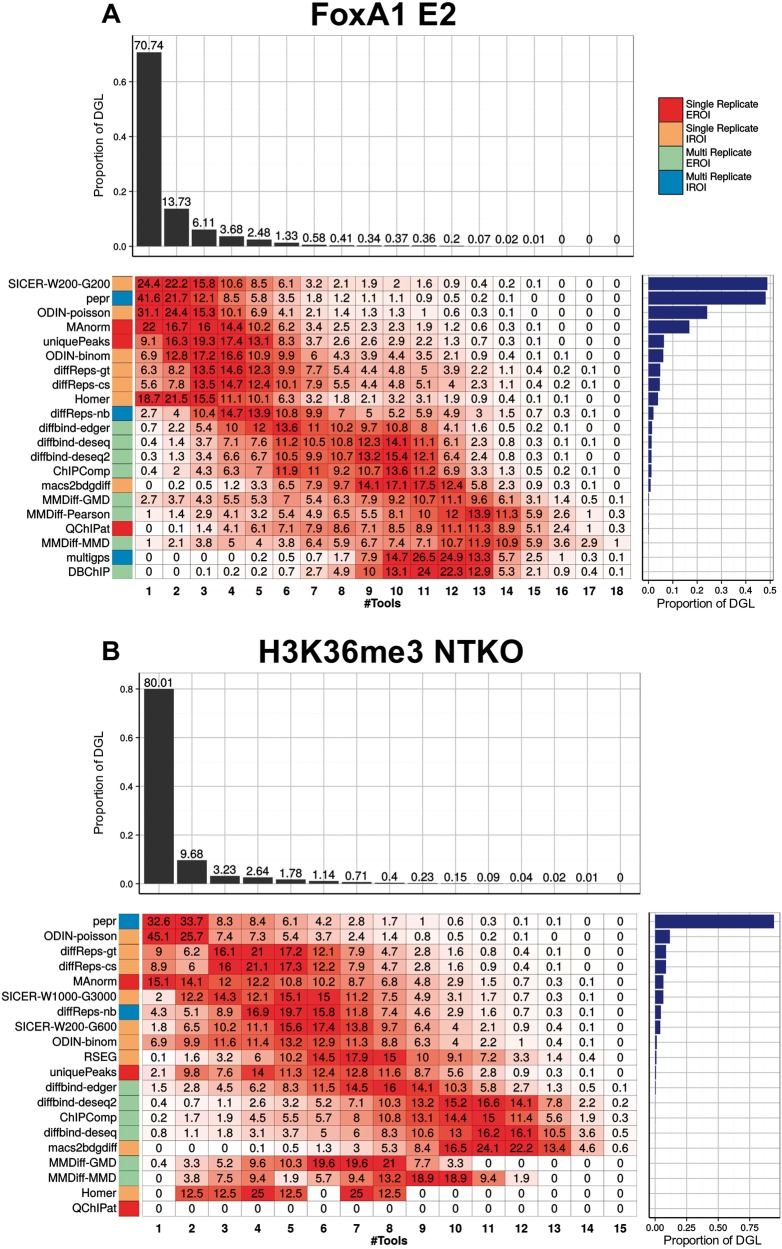
We first determined the genomic regions covered by the union of all DR from all tools (differential genomic loci or DGL), and determined the coverage of these regions, i.e. the proportion that is covered by peaks from n tools. For FoxA1, 71% of the regions that have been called as differential by any tool arise from one single tool (Figure 3A). We call these regions ‘private DR', as they are specific to one single tool. Less than 14% of the total DGL is covered by two or more tools. These proportions are more or less consistent between the data sets, with between 71% and 80% of the DGL being private.
Figure 3.
Comparison of the sets of DR between tools for the FoxA1 E2 treatment condition (A) and H3K36me3 NTKO data sets (B). We consider the union of all regions determined as being differential by any of the tools, and determine which proportion is called by 1,2,… tools (upper barplot). For each tool, we determine which proportion of the differential genomic locus (DGL) is called by this tool alone, or corresponds to regions called by several tools (heatmap). We indicate the total size of the DGL as a barplot on the right. A colour version of this figure is available online at BIB online: https://academic.oup.com/bib.
Next, we wanted to determine whether a tool has a propensity to call private DR, or rather has a higher agreement with other tools. We therefore determined what proportion of the DGL of a given tool has coverage n (Figure 3B and D). We expect that tools that call many more DR than others will have a tendency to call more private peaks. Hence, we ordered the tools according to the size of the DGL. As anticipated, we observed that the tools with the largest DGL tend to call more private DR (e.g. for PePr, 41.6% of the DGL are specific to this tool). ODIN-binom calls few private DR, and shows a good agreement with other tools. On the other hand, more than two-thirds of the DR called by SICER on the FoxA1 Vehicle data set are private DR, which is related to the large size of the regions, and indicates that the fixed windows settings of SICER, even using parameters indicated for sharp transcription factors, are not adapted to detect accurately small-scale variations.
As a conclusion, we observe striking differences in the number and sizes of DR detected by the different tools, and inconsistencies in the direction of the differential enrichment. Tools based on multiple replicates call less DR, which seems to indicate that the proportion of false positives might be lower, however, possibly at the expense of the sensitivity. Also, these tools show a more progressive increase in the number of DR when relaxing the threshold, compared with the single-replicate tools, which have a sharp increase followed by a saturation. For data sets with broader signal, we observe a clear difference between tools calling a large number of small peaks (e.g. ODIN), and others that aggregate these into broader domains (e.g. RSEG or SICER), which better reflects the true signal.
Comparison using simulated data sets
As no golden standard is available apart from a limited number of qPCR-validated regions, no assessment of the rate of true/false positives can be made using real data sets. Hence, we simulated differential data sets. Several other studies have addressed the question of simulating ChIP-seq data sets [33, 34]. These procedures have focused on simulating ChIP-seq data sets for transcription factors only, and they rely on a theoretical statistical model to describe the expected distribution of reads from a ChIP-seq experiment. To avoid biasing the benchmark toward one or the other tool, our simulation does no rely on a particular theoretical model of read distribution. Instead, we used real data sets as a basis for this simulation, and reasoned that the best ranking peaks identified by MACS2 most probably represent true binding/enriched regions. We therefore considered the 20,000 best ranking peaks, of which we downscaled 10,000 in a stepwise manner, while maintaining a set of 10,000 nondifferential binding events (see ‘Methods' section). These true binding events were merged with real input data sets, to ensure that no binding event occurs outside of the selected regions, while providing a realistic background (Supplementary Figures S2 and S3). Hence, we can evaluate for each tool how many true/false positives/negatives have been detected.
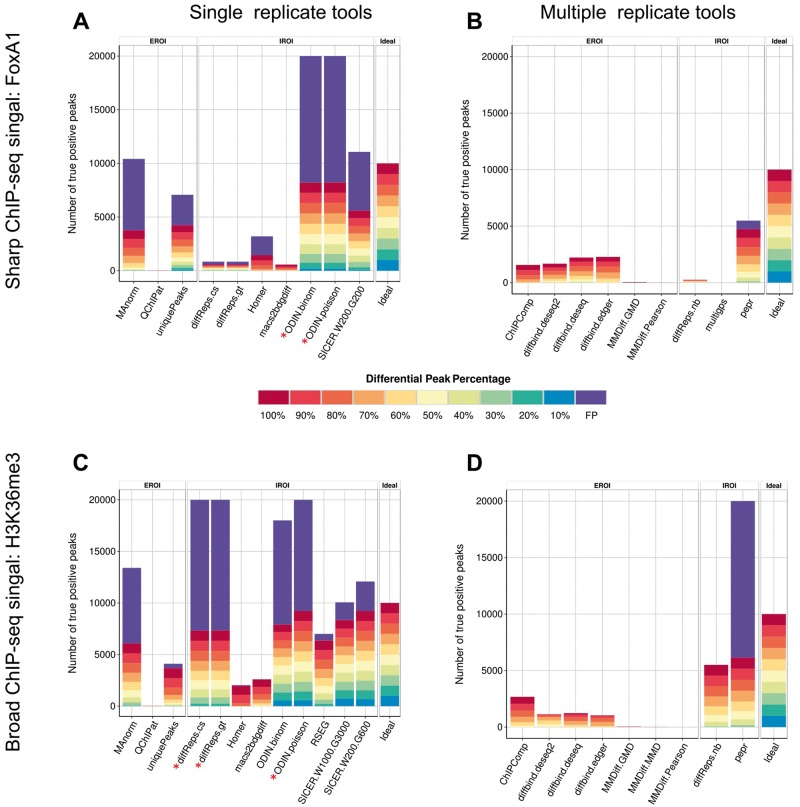
In the ideal case, the tools should detect 10,000 DR. For the single replicate pipeline, ODIN-binom and SICER returned the largest number of DR for the FoxA1 simulated data set (Figure 4A). Given that ODIN outnumbered by far the other tools in the number of DR detected in the real FoxA1 data set, it is likely that the high recall rate of ODIN is at the expense of a high number of false positives. Indeed, both versions of ODIN had an important proportion of false positives. Both tools called >300,000 DR, of which more than 95% are false positives. We believe that the HMM approach taken by ODIN renders it extremely sensitive to small fluctuations in the background, hence leading to many false-positive hits. On the other end of the scale, QChIPAT only returned a limited number of DR. As expected, most tools were able to detect at least partially the strongest differential peaks (100% down-sampling). HOMER only detected DR, which were down-sampled by ≥80%. This is likely owing to the additional filtering parameter implemented by HOMER, which requires a minimum fold-change (by default 4) in addition to a P-value cutoff to call differential peaks.
Figure 4.
Proportion of true and false positives for each tool on the simulated FoxA1 data set (A, B) and H3K36me3 data set (C, D). The percentages in the legend indicate the level of downsampling performed. A colour version of this figure is available online at BIB online: https://academic.oup.com/bib.
For the multi-replicate pipeline, the ranking in the number of detected DR follows what has been observed in the real data set, with PePr detecting the largest number of regions, of which most are true positives, with a small fraction of false positives (Figure 4B). The different variants of the diffBind tool detected around 30% of the true DR, with no false positives, while all other tools had a high proportion of false negatives. This confirms our assumption of the first section, that the much lower number of DR from the tools in the replicate pipeline comes at the expense of a much reduced sensitivity. Considering the H3K36me3 simulation, the overall number of detected DR agreed with the real data set (Figures 4C, D and 2), with ODIN-poisson showing the highest number of regions, followed by the two variants of diffReps. As expected previously, this comes at the expense of a high proportion of false positives, at least with our parameter settings. On the other hand, RSEG and SICER with the various settings show excellent performances with a high recall rate and a limited number of false positives. In the multi-replicate pipeline, the tools call a much lower number of peaks, corresponding to a high rate of false negatives, with two exceptions: diffReps-nb performs well on this simulation with a decent recall, whereas PePr calls many false positives.
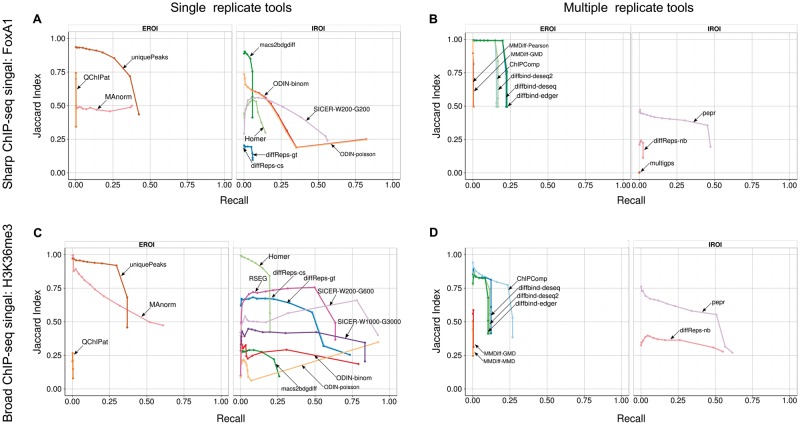
As the number of regions returned by each tool depends on the parameter settings, we decided to compare the precision and recall of each tool at various stringency levels: for each tool, we selected the top 10, 50, 100, 250, 500, 750, 1000, 1500, 2000, 3000, 5000, 10,000 and all peaks, and computed the recall (i.e. the proportion of true differential peaks detected) and an adapted version of the precision, for which we computed the proportion of the total length of predicted DR that coincides with a true differential region. The rational of this Jaccard index (abbreviated JI in the following) is to avoid providing a biased advantage to those tools that tend to call large regions, which automatically increases the probability to hit a true differential region. For the single replicate tools on the simulated FoxA1 data set, we see that ODIN shows a good overall recall of above 75% when taking into account all peaks, but a low JI of 25% (Figure 5A). The best performing approach appears to be the naive approach based on the set of unique peaks between the two conditions. However, this results from a bias of our approach, which used MACS2 to define the initial set of 20,000 binding events, and used MACS2 again to determine the sets of peaks in both conditions in the simulated data set, hence giving an obvious advantage to this approach. Overall, SICER and MAnorm show a good trade-off between recall and JI; however, MAnorm also relies on MACS2 peaks and hence has an advantage in this simulation. Strikingly, most tools show a rapid drop in the JI with increasing sets of DR, indicating that even tools with a limited number of DR tend to overcall DR. The difference is striking for the multi-replicate pipeline (Figure 5B), where diffBind shows a perfect precision of 100% up to 2000 DR, before dropping as more peaks are included. Together with PePr, these are the only tools that achieve decent performance, and a relatively stable precision rate.
Figure 5.
JI (y-axis) and recall (x-axis) for the tools in the single replicate (left column) and multiple-replicate pipeline (right column) for the FoxA1 (A, B) and H3K36me3 (C, D) simulated data sets. We make the distinction between tools that require external regions of interest (EROI) and those that determine them internally (IROI). A colour version of this figure is available online at BIB online: https://academic.oup.com/bib.
The overall performances are better on the H3K36me3 data set (Figure 5C and D), with a high recall rate here and a reasonable JI. The higher average values of JI comes from the fact that the simulated regions are larger, and hence, they intersect a higher proportion of the predicted DR. Note that despite the higher number of false positives in PePr, the final JI is similar between diffReps-nb and PePr, when considering all DR, whereas PePr has a much higher JI when considering only the top 5000 peaks. This indicates that the false-positive peaks called by PePr are among the lower-ranking peaks, while the top peaks contain a high proportion of true positives.
In summary, our analysis on simulated data sets confirms that (1) the replicate tools have a higher rate of false negatives, but (2) fewer false positives, and that (3) most tool seem to perform better on the broad histone mark than on the sharp TF binding peaks.
Functional annotation
When comparing two biological conditions, one often aims at relating the observed changes to genes to obtain a functional interpretation of the conditions under study, in terms of enriched functional categories. This is particularly the case when performing differential expression analysis based on transcriptome data, and a plethora of tools have been developed to turn lists of DEGs into functional categories or pathways mostly affected by the changes. In the case of ChIP-seq, similar questions can be asked, by looking at genes that are potentially affected by differential binding sites or enriched regions, thus generating biological hypothesis about affected biological processes. Assigning a gene to a genomic region, however, introduces an additional uncertainty, as regulatory regions, for example, can act over large distance [35]. Various approaches have been proposed to make the assignment more accurate, based e.g. on published interaction data sets or looking at the correlation between the ChIP signal and the expression of surrounding genes [36–38]. However, we still expect that for most of the cases, relating a differential peak to the closest gene(s) will be a reasonable proxy, especially if the differential region lies close to gene promoters. If expression data are available for the same condition, one can compare the two differential data sets (expression and ChIP binding/enrichment) and validate to what extent the biological interpretation based on differential expression and differential enrichment are compatible. In this section, we used the DR obtained in the first section, and performed functional enrichment of the detected regions for each of the tools. This adds an additional layer of validation. Indeed, irrespective of whether DR overlap, if they lie in proximity to the same genes, the biological interpretation will be similar. We made the assumption that FoxA1 represents a transcriptional activator, hence the direction of the differential ChIP enrichment and the differential expression should be the same.
First, we checked if the DEGs can be recovered from the DR. As a negative control, we used housekeeping genes (HKG) that are not differentially expressed. For each tool, we ranked the DR according to their significance, and assigned to each DR the closest gene, up to 1000 genes. Using these rankings, we computed the enrichment of DEG versus HKG using a NES, which represents a normalized version of the AUC (see ‘Methods' section). We compared the performance of the tools with a naive approach, consisting in ranking the genes according to their log-fold enrichment on the gene body (H3K36me3) or on the promoter (FoxA1 and H3K27ac). The results are shown in Supplementary Figure S4 for the FoxA1,H3K36me3 and H3K27ac data sets. As noticed previously, some tools have a low number of DR and therefore associate to a small number of genes. Overall, most of the tools show a good performance in recovering DEG from DR for FoxA1, better than the naive approach based on promoter enrichment alone, possibly reflecting the fact that FoxA1 also binds outside of promoters and acts over longer distances. Somewhat unexpectedly, the results are worse for the H3K36me3 data set, especially for DR enriched in the TKO condition. Here, the tools from the single-replicate pipeline show particularly poor performance, as can be seen from the small difference in NES score between the DEG and the HKGs. Most tools, however, perform better than the naive approach, which seems to indicate that small differences in the enrichment, possibly restricted to a portion of the transcript, are better captured by the tools. For H3K27ac, the performance of most tools is good, with some unexpected behaviors: for one of the conditions, we note again the difference between the DESeq, DESeq2 and edgeR algorithms implemented in diffBind, possibly reflecting the different normalizations applied and the greater stringency of the original DESeq algorithm.
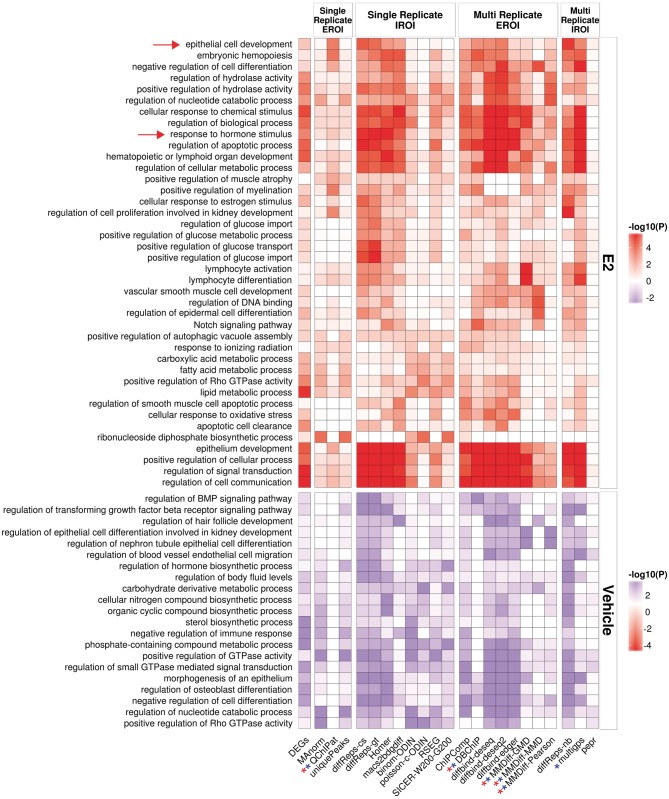
Next, we asked whether the sets of DR would lead to similar biological interpretations. For each method, as previously, we selected the first 1000 genes that were nearby a DR (DR were ordered by decreasing significance). We submitted the promoters of these genes to GREAT, and collected for each tool the top 3 enriched functional terms, and displayed the union of these terms over all tools in a heatmap. As a gold standard, we also performed the same analysis using the set of DEGs, by submitting their promoter to GREAT.
Comparing the output for each set of tool (Figure 6A and B), we observe a good consistency between the tools for some terms such as ‘response to hormone stimulus', which is also found among the DEG. Most tools indeed show enrichment for these terms, except the tools from the single-replicate/EROI group (Manorm, QChIPat) and PePr.For QChIPat, and this can be explained by the low number of DR and hence the small set of affected genes. The functional enrichments obtained from the DR do not necessarily correspond to the terms obtained from the DEG, with, for example, ‘epithelial cell development' being highly enriched among the DR of many tools, but showing only a minor enrichment among the DEG. Given that FoxA1 has been described as being involved in epithelial lung cell differentiation [39], the presence of this term makes sense in this context. It is striking that the approaches from the single replicate with external regions of interest show a clear depletion in enrichments; while this can be explained by the small number of DR returned by QChIPat, this is more surprising for MAnorm. It also highlights that the naive approach based on taking the unique peaks in each condition also fails to find most of the interesting functional enrichments.
Figure 6.
Functional enrichment obtained by a GREAT analysis on the sets of DR and using the promoters of the DEG as a baseline. Shown heatmaps are based on DR derived from FoxA1 E2- and Veh-treated data sets. Stars indicate tools for which <1000 genes were available in either condition. The arrows highlight terms discussed in the text. A colour version of this figure is available online at BIB online: https://academic.oup.com/bib.
Discussion and conclusion
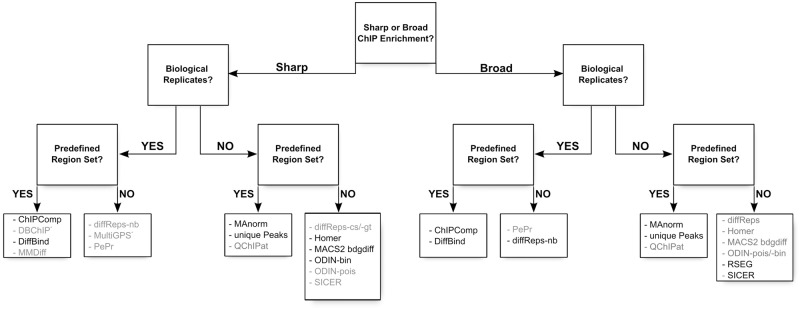
We have performed a comprehensive comparison of a large number of tools developed for the detection of differentially enriched regions from ChIP-seq experiments. As ChIP-seq is a widely applicable method and can target many transcription factors or epigenetic modifications, the type of signal is diverse, and makes the detection of DR a challenging task. Hence, we would not expect that these tools be universally applicable to any type of ChIP-seq, from transcription factor ChIP to broad histone modifications such as H3K36me3 or H3K27me3. Indeed, several of the tools we have included in this study have been specifically designed for either of these data sets; MultiGPS, for example, is specifically designed for transcription factors, while tools such as diffReps or RSEG have been developed to detect differential histone modifications and indeed show poor performances on the transcription factor data set. Hence, these tools cannot be used universally. On the other hand, most other tools appear to be applicable in both cases, in particular if an external set of regions of interest is provided. We have summarized in Figure 7 and Supplemental Table S1 the main characteristics of the tools, and this can be used as a guide for selection of a tool specifically suited for a particular application.
Figure 7.
Decision tree indicating the proper choice of tool depending on the data set: shape of the signal (sharp peaks or broad enrichments), presence of replicates and presence of an external set of regions of interest. We have indicated in dark the name of the tools that give good results using default settings, and in gray the tools that would require parameter tuning to achieve optimal results: some tools suffer from an excessive number of DR (PePr, ODIN-pois), an insufficient number of DR (QChIPat, MMDiff, DBChIP) or from an imprecise definition of the DR for sharp signal (SICER, diffReps-nb). *MultiGPS has been explicitly developed for transcription factor ChIP-seq.
The proper choice of the algorithm also depends on the types of questions: if the study is primarily gene centric, i.e. aims at identifying genes in the vicinity of the DR to derive a biological interpretation based on the annotation of the genes, the tools offer rather consistent results, as is shown from the recovery analysis of DEG and the analysis of term enrichments using tools such as GREAT. On the other hand, if one is more interested in chromatin organization and dynamics on treatment, and focuses on the properties of the differential enriched regions (such as size, exact localization, etc…), then the differences in the tools will have a huge impact on the results. Similarly, any analysis that focuses on enriched motifs within the DR is also sensitive to the proper definition of the DR, as any over-estimation of the size of the DR will decrease the signal-to-noise ratio and result in less accurate motif inference. For example, SICER, even with the parameters recommended for TF binding, yields large regions, which might introduce a high level of noise for motif analysis. A similar problem arises in single-condition peak calling for ChIP-seq, for which differences in peak properties impact the accuracy of motif discovery.
Each tool involves several ad hoc filtering steps, with the number of tunable parameters being often substantial. In this analysis, scanning the full parameter space for all tools over all data sets was beyond our scope. Of course, we cannot exclude that some results would have differed if, for example, the developers of the individual tools would have optimized the parameters using their insight knowledge of their own tool. Figures 1C and 2C show that the number of DR is more or less sensitive to the choice of threshold, which should be taken into account when using one or the other tool. This type of comparative studies is for example applied in the DREAM challenges or in a motif discovery benchmark [40]. As we had no bias toward any of the analyzed tools, we reasoned that a moderately expert user would most likely rely on the recommendations and default parameters suggested by the developers in the documentation of the tool. Hence, we took this approach to determine the parameters that should be applied, and believe that this represents a fair option for an unbiased comparison. For example, some tools did report a limited number of DR. This is probably a consequence of our parameter choice, and different results would probably have been obtained using other values. However, this also indicates that these tools do require optimization, and cannot be used with the standard settings, whereas others give reasonable results ‘out-of-the-box' (Figure 7).
In the workflow from bam files down to list of DR, several steps have a strong impact on the results: first, the tools differ in their approach to define the regions to test for differential enrichment. We can broadly categorize them into three categories, with the exception of MultiGPS: (1) the tools that rely on an external set of regions provided by the user, generally based on the output of a peak caller algorithm, (2) the tools that use a fixed window-based approach to compare enrichments, and (3) the tools that implement a hidden-Markov approach. Another crucial point is the normalization of the data sets, either treatment versus control or both treated data set against each other. As indicated in Supplementary Table S1, the tools use different normalization strategies, from the simple library size normalization to more sophisticated approaches such as the SES method. Recently, a publication has compared the impact of normalization schemes on different data sets and shown differences especially when data sets are based on genomes with chromosomal aberrations [41]. Most tools presented in this study are based on normalization methods initially developed for gene expression analysis with the underlying assumption that only a small number of regions are truly differential. However, this is not suitable in case of global epigenome changes, e.g. owing to inhibition of epigenetic enzymes. Therefore, a number of ChIP-seq protocols are now being published using as internal experimental control a spiking-in of a known amount of chromatin from a different species (Drosophila melanogaster or Mus musculus) [42, 43]. This internal reference allows to precisely determine a sample-specific normalization factor and therefore enables a more robust genome-wide quantitative comparison across samples.
However, in our comparison, the largest difference comes from the availability of replicates. Tools that handle replicates call a much smaller number of DR, and achieve a much better precision, yet at the expense of the recall. Importantly, if multiple replicates are available, they should be considered independently using one of the multiple replicate tools, rather than pooled, as the level of false positives is generally much lower for this category, owing to the implementation of robust statistical tests. In the absence of replicates, the user should be aware that most tools in the single replicate category require extensive parameter fine-tuning to achieve a good trade-off between precision and recall.
According to our results, it is crucial to generate replicate ChIP-seq data sets when looking for differential enrichment, as is now an established procedure when looking for DEGs from RNA-seq data, to achieve a sufficient specificity.
Note that we have not here addressed the question of comparing multiple conditions, as none of the tools presented here allow this type of analysis. For that, novel methods need to be developed, for example, based on an ANOVA testing of variable regions [44].
In conclusion, our study highlights the general lack of consistency between the tools considered here, in terms of number and location of DR. Depending on the biological focus, this might yield substantially different interpretations. We therefore recommend to use and compare several of these tools to obtain a confident consensus set of DR, but most importantly our study highlights the importance of generating biological replicates for ChIP-seq, like what has become standard practice for RNA-seq.
Key Points
Tools for differential ChIP-seq analysis show important differences in the number and size of detected differential regions (DR).
Methods taking into account replicates appear to be more robust than those handling single replicate data sets.
Inconsistent sets of DR will affect results based on sequence analysis, like detection of enriched transcription factor binding sites.
However, analysis of functional enrichments based on neighboring genes appears to be more robust.
Some tools give good results with default parameters, like ChIPComp or diffBind when replicates are available, or MAnorm, Homer, macs2bdgdiff and RSEG with single replicates. The other tools would require more extensive fine-tuning of parameters to achieve satisfactory results.
Supplementary Data
Supplementary data are available online at https://academic.oup.com/bib.
Acknowledgments
We thank Karsten Rippe and Jan-Phillip Mallm for discussions, and Morgane Thomas-Chollier for comments on the manuscript.
Biographies
Sebastian Steinhauser is a master's student in the Cancer Regulatory Genomics group headed by Carl Herrmann in the Division of Theoretical Bioinformatics, DKFZ, Heidelberg, Germany.
Nils Kurzawa is a master's student in the Cancer Regulatory Genomics group headed by Carl Herrmann in the Division of Theoretical Bioinformatics, DKFZ, Heidelberg, Germany.
Roland Eils is Professor at the Institute for Pharmacy and Molecular Biotechnologies, University Heidelberg, and Head of the Division of Theoretical Bioinformatics at the DKFZ, Heidelberg.
Carl Herrmann is assistant professor at the Institute for Pharmacy and Molecular Biotechnologies, University Heidelberg, and heads the Cancer Regulatory Genomics group in the Division of Theoretical Bioinformatics, DKFZ, Heidelberg.
Funding
Funding for open access charge: DKFZ internal funding.
References
- 1.Landt SG, Marinov GK, Kundaje A, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res 2012;22:1813–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Y, Negre N, Li Q, et al. Systematic evaluation of factors influencing ChIP-seq fidelity. Nat Methods 2012;9:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diaz A, Park K, Lim DA, et al. Normalization, bias correction, and peak calling for ChIP-seq. Stat Appl Genet Mol Biol 2012;11:Article 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang K, Keles S. Normalization of ChIP-seq data with control. BMC Bioinformatics 2012;13:199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Q, Brown JB, Huang H, et al. Measuring reproducibility of high-throughput experiments. Ann Appl Stat 2006;5:1752–79. [Google Scholar]
- 6.Zang C, Schones DE, Zeng C, et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics 2009;25:1952–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L, Wang C, Qin ZS, et al. A novel statistical method for quantitative comparison of multiple ChIP-seq datasets. Bioinformatics 2015;31:1889–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahony S, Edwards MD, Mazzoni EO, et al. An Integrated model of multiple-condition ChIP-seq data reveals predeterminants of Cdx2 binding. PLoS Comput Biol 2014;10:e1003501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang K, Keles S. Detecting differential binding of transcription factors with chiP-seq. Bioinformatics 2012;28:121–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allhoff M, Sere K, Chauvistre H, et al. Detecting differential peaks in ChIP-seq signals with ODIN. Bioinformatics 2014;30:3467–75. [DOI] [PubMed] [Google Scholar]
- 11.Ross-Innes CS, Stark R, Teschendorff AE, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 2012;481:389–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen L, Shao N-YY, Liu X, et al. diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS One 2013;8:e65598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song Q, Smith AD. Identifying dispersed epigenomic domains from ChIP-Seq data. Bioinformatics 2011;27:870–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shao Z, Zhang Y, Yuan G-C, et al. MAnorm: a robust model for quantitative comparison of ChIP-Seq data sets. Genome Biol 2012;13:R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schweikert G, Cseke B, Clouaire T, et al. MMDiff: quantitative testing for shape changes in ChIP-Seq data sets. BMC Genomics 2013;14:826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Liu T, Meyer CA, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008;9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Lin Y-H, Johnson TD, et al. PePr: a peak-calling prioritization pipeline to identify consistent or differential peaks from replicated ChIP-Seq data. Bioinformatics 2014;30:2568–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu B, Yi J, Sv A, et al. QChIPat: a quantitative method to identify distinct binding patterns for two biological ChIP-seq samples in different experimental conditions. BMC Genomics 2013;14 (Suppl 8):S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010;38:576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franco HL, Nagari A, Kraus WL. TNFα signaling exposes latent estrogen receptor binding sites to alter the breast cancer cell transcriptome. Mol Cell 2015;58:21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie W, Schultz MD, Lister R, et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell 2013;153:1134–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Popovic R, Martinez-Garcia E, Giannopoulou EG, et al. Histone Methyltransferase MMSET/NSD2 alters EZH2 binding and reprograms the myeloma epigenome through global and focal changes in H3K36 and H3K27 methylation. PLoS Genet 2014;10:e1004566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Liu T, Meyer CA, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008;9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hah N, Danko CG, Core L, et al. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell 2011;145:622–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu G, Wang L-G, He Q-Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 2015;31:2382–3. [DOI] [PubMed] [Google Scholar]
- 28.McLean CY, Bristor D, Hiller M, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 2010;28:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramírez F, Dündar F, Diehl S, et al. DeepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 2014;42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonhoure N, Bounova G, Bernasconi D, et al. Quantifying ChIP-seq data: A spiking method providing an internal reference for sample-to-sample normalization. Genome Res 2014;24:1157–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010;26:841–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lawrence M, Huber W, Pagès H, et al. Software for computing and annotating genomic ranges. PLoS Comput Biol 2013;9:e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang ZD, Rozowsky J, Snyder M, et al. Modeling ChIP sequencing in silico with applications. PLoS Comput Biol 2008;4;e1000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Humburg P. ChIPsim: Simulation of ChIP-seq experiments. 2011, R package version 1.24.0. [Google Scholar]
- 35.Li G, Ruan X, Auerbach RK, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 2012;148:84–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dixon JR, Selvaraj S, Yue F, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012;485:376–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He B, Chen C, Teng L, et al. Global view of enhancer-promoter interactome in human cells. Proc Natl Acad Sci USA 2014;111:E2191–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thurman RE, Rynes E, Humbert R, et al. The accessible chromatin landscape of the human genome. Nature 2012;489:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshimi T, Nakamura N, Shimada S, et al. Homeobox B3, FoxA1 and FoxA2 interactions in epithelial lung cell differentiation of the multipotent M3E3/C3 cell line. Eur J Cell Biol 2005;84:555–66. [DOI] [PubMed] [Google Scholar]
- 40.Tompa M, Li N, Bailey TL, et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat Biotechnol 2005;23:137–44. [DOI] [PubMed] [Google Scholar]
- 41.Wu D-Y, Bittencourt D, Stallcup MR, et al. Identifying differential transcription factor binding in ChIP-seq. Front Genet 2015;6:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonhoure N, Bounova G, Bernasconi D, et al. Quantifying ChIP-seq data: a spiking method providing an internal reference for sample-to-sample normalization. Genome Res 2014;24:1157–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Orlando DA, Chen MW, Brown VE, et al. Quantitative ChIP-Seq normalization reveals global modulation of the epigenome. Cell Rep 2014;9:1163–70. [DOI] [PubMed] [Google Scholar]
- 44.Kasowski M, Kyriazopoulou-Panagiotopoulou S, Grubert F, et al. Extensive variation in chromatin states across humans. Science 2013;342:750–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.