Abstract
Respiratory viruses alter the nasopharyngeal microbiome and may be associated with a distinct microbial signature. To test this hypothesis, we compared the nasopharyngeal microbiome of 135 previously healthy infants with acute respiratory infection due to human rhinovirus (HRV; n = 52) or respiratory syncytial virus (RSV; n = 83). The nasopharyngeal microbiome was assessed by sequencing the V4 region of the 16S ribosomal RNA. Respiratory viruses were identified by quantitative reverse-transcription polymerase chain reaction. We found significant differences in the overall taxonomic composition and abundance of certain bacterial genera between infants infected with HRV and those infected with RSV. Our results suggest that respiratory tract viral infections are associated with different nasopharyngeal microbial profiles.
Keywords: microbiome, nasopharynx, respiratory syncytial virus, human rhinovirus, children
Human rhinovirus (HRV) and respiratory syncytial virus (RSV) are the most common etiologies of acute respiratory tract infections (ARIs) in early life [1]. HRV is the leading cause of upper respiratory tract infections and usually produces a mild disease in previously healthy infants, whereas RSV is the number one cause of lower respiratory tract infections and is the most common cause of hospitalization in young children [1, 2]. Infants with ARIs due to HRV and RSV, particularly those with lower respiratory tract infections, also have an increased risk of childhood asthma [2], but the mechanisms underlying these associations are largely unknown.
There is increasing evidence to suggest that the nasopharyngeal microbiome plays an important role in the pathogenesis of viral ARIs [1, 3–5]. Certain respiratory viruses have been shown to alter bacterial adherence and colonization, thus increasing the risk of secondary bacterial infections [1]. The host immune response to respiratory viruses may also depend on the prevalence of certain bacteria [4, 6]. Likewise, the presence of certain bacteria has been associated with a more clinically severe presentation of viral ARIs in some studies and with the development of childhood asthma in others [3, 4]. In view of all these, it has been previously suggested that a virus-specific bacterial profile in the respiratory tract could potentially be characterized [7].
We hypothesized that individual viral ARIs are associated with a distinct nasopharyngeal microbiome signature. To test this hypothesis, we assessed the nasopharyngeal microbiome among infants enrolled in the Infant Susceptibility to Pulmonary Infections and Asthma Following RSV Exposure (INSPIRE) study who met prespecified criteria for ARIs [8]. In these infants, we used next-generation sequencing of the 16S ribosomal RNA (rRNA) to compare the nasopharyngeal microbiome of infants with viral ARIs due to HRV and RSV.
METHODS
INSPIRE is a population-based, prospective cohort of previously healthy, term infants enrolled near birth with biweekly surveillance of respiratory illnesses during their first winter viral season. The detailed methods for INSPIRE have been previously reported [8]. One parent of each infant provided informed consent for participation. The Institutional Review Board of Vanderbilt University approved this study.
For the current study, we included 135 infants enrolled in the INSPIRE cohort who met prespecified criteria for an ARI. These infants underwent an in-person visit, which included the administration of a respiratory illness questionnaire, a nasal wash to identify respiratory viruses and characterize the nasopharyngeal microbiome, and a physical examination. The severity of the ARI was measured with the respiratory severity score (RSS) [9]. The diagnosis of HRV or RSV infection was made by quantitative reverse-transcription polymerase chain reaction [8]. We compared 52 infants with HRV ARI to 83 infants with RSV ARI. Infants with HRV/RSV coinfection were excluded from this study. Full details can be found in the Supplementary Data.
We have previously described the methods for nasopharyngeal sampling, viral detection, and characterization of the nasopharyngeal microbiome in detail [5, 8, 10]. In brief, after cell lysis and extraction of the microbial genomic DNA from the nasal wash samples, the V4 region of the 16S rRNA was amplified with 515F/806R primers. Sequencing was performed on an Illumina MiSeq sequencer with 2 × 300 base pair reads. A mothur-based automated annotation pipeline, YAP (available at: https://github.com/andreyto/YAP), was used to perform initial processing of the 16S rRNA data sets. Operational taxonomic units (OTUs) were clustered at 97% sequence identity. Statistical analyses were done in R, using the MGSAT package (Supplementary Data).
RESULTS
The baseline characteristics of the 135 infants with ARIs due to HRV or RSV can be found in Table 1. The majority of infants in our study (approximately 80%) were <6 months of age at the time of the ARI and did not require an emergency department visit or hospitalization (approximately 74%). Infants with RSV ARI had a significantly higher RSS and were more likely to have been hospitalized when compared to those with HRV ARI. There were no significant differences between these 2 groups in regard to age at the time of illness, sex, race or ethnicity, gestational age, birth weight, mode of delivery, exposure to antibiotics in utero or after birth, breastfeeding, maternal smoking, maternal asthma, or type of insurance.
Table 1.
Baseline Characteristics of Infants With Acute Respiratory Tract (ARI) Infections Due to Respiratory Syncytial Virus (RSV) or Human Rhinovirus (HRV)
| Characteristic | Infants With HRV ARI (n = 52) | Infants With RSV ARI (n = 83) |
|---|---|---|
| Age at time of illness, wk | 17.5 (10.5–23.1) | 19.1 (10.4–25.9) |
| Female sex | 22 (42.3) | 35 (42.2) |
| Race or ethnicity | ||
| Black, non-Hispanic | 10 (19.2) | 15 (18.1) |
| White, non-Hispanic | 32 (61.5) | 50 (60.2) |
| Hispanic | 4 (7.7) | 8 (9.6) |
| Othera | 6 (11.5) | 10 (12.1) |
| Gestational age, wk | 39.0 (38.5–40.0) | 39.0 (38.0–40.0) |
| Birth weight, gb | 3377.0 (2894.0–3859.0) | 3377.0 (2894.0–3859.0) |
| Birth by cesarean section | 20 (38.5) | 28 (33.7) |
| Exposure to antibiotics in utero | 27 (51.9) | 36 (43.4) |
| Exposure to antibiotics between birth and enrollment | 7 (13.5) | 9 (10.8) |
| Exposure to antibiotics for current ARI | 11 (21.2) | 19 (22.9) |
| Any breastfeeding | 43 (82.7) | 58 (69.9) |
| Maternal smoking at enrollment | 9 (17.3) | 20 (24.1) |
| Maternal asthma | 12 (23.1) | 14 (16.9) |
| Level of care required for this ARIc | ||
| Emergency department | 5 (9.5) | 6 (7.2) |
| Hospitalization | 2 (3.9) | 22 (26.5) |
| Otherd | 45 (86.6) | 55 (66.3) |
| Respiratory severity scorec | 2.0 (1.5–3.0) | 3.0 (2.0–6.0) |
| Insurance type | ||
| Medicaid | 27 (51.9) | 45 (54.2) |
| Private | 24 (46.2) | 36 (43.4) |
| Other | 1 (1.9) | 2 (2.4) |
Data are median values (interquartile ranges) or no. (%) of subjects. Percentages are calculated for children with complete data.
a Includes mixed race and unknown.
b It is a coincidence that the value and medians are the same. The means were slightly different.
c P < .05 for the comparison between groups for any of the variables presented in the table, using the Mann–Whitney U test or the Pearson χ2 test, as appropriate.
d Includes well-child care visits, outpatient sick visits, and research visits.
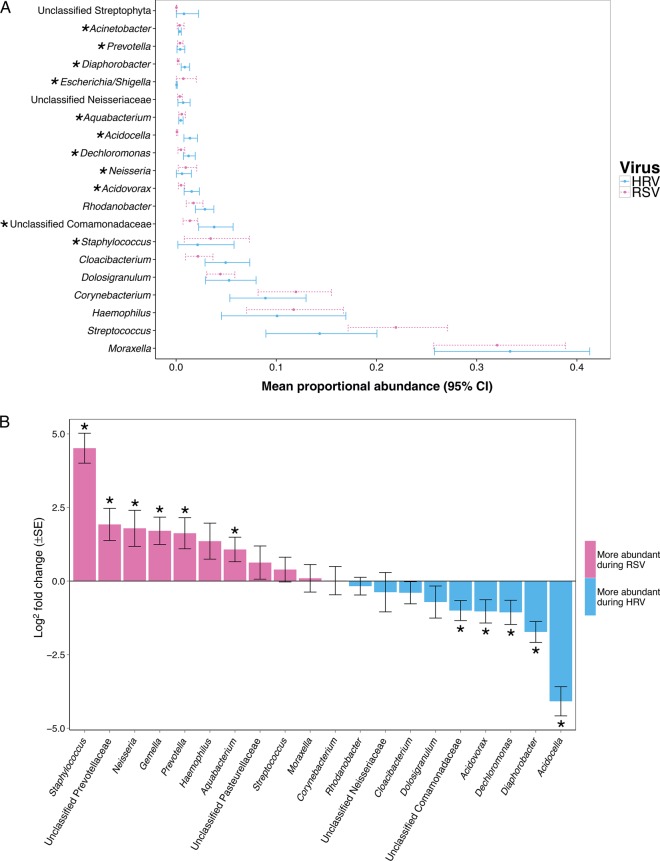
The most abundant bacterial genera in these 135 infants included Moraxella (32.9%), Streptococcus (18.4%), Corynebacterium (10.7%), Haemophilus (10.2%), and Dolosigranulum (4.6%). The sample means of the relative abundance of the top most abundant bacterial genera with bootstrap confidence intervals at the 95% confidence levels are shown in Figure 1A. Among the HRV-positive and RSV-positive samples retained for analysis, the median sequence count per sample was 20 000 (range, 5302–80 070). In the DESeq2 analysis called from the MGSAT pipeline (using a Wald test with Benjamini and Hochberg correction for multiple comparisons), the abundances of 19 bacterial genera were detected to be significantly different between the 2 groups when the statistical significance threshold was set at 0.05 (Supplementary Table 1). For instance, Staphylococcus (adjusted base mean, 264.32; log2 fold change, 4.51; q<0.001) was significantly higher during RSV ARI when compared to HRV ARI. In addition, there was a trend toward a higher abundance of Haemophilus during RSV ARI, although this was not significant (adjusted base mean, 3907.14; log2 fold change, 1.36; q = 0.06; Figure 1B).
Figure 1.
Comparison of the abundance of nasopharyngeal bacterial genera between infants with acute respiratory tract infection due to human rhinovirus (HRV) or respiratory syncytial virus (RSV). Bacterial genera that are significantly different between groups after adjusting for multiple comparisons by using the DESeq2 package (see “Methods” section) are indicated by an asterisk. A, Mean proportional abundance and 95% bootstrap confidence intervals (CIs) of nasopharyngeal bacterial genera according to viral etiology. Within each sample, counts were normalized to simple proportions. Overlapping confidence intervals do not imply a lack of a significant difference in the DESeq2 analysis. The relative abundance of the 20 most abundant bacterial genera by simple proportions is shown. B, Log2 fold change and log2 fold change standard error of nasopharyngeal bacterial genera according to viral etiology as calculated with the DESeq2 analysis. The log2 fold changes of the 20 most abundant bacterial genera by the DESeq2 adjusted base mean are shown. A log2 fold change of >0 (pink bars) indicates that abundance was detected to be higher during RSV ARI as compared to HRV ARI, while a log2 fold change of <0 (blue bars) indicates that abundance was detected to be higher during HRV ARI as compared to RSV ARI. Abbreviation: SE, standard error.
The overall taxonomic composition of the nasopharyngeal microbiome also differed between HRV-positive and RSV-positive infants at both the genus level (P = .01) and OTU level (P = .01) in permutational multivariate analysis of variance (Adonis), using the Bray-Curtis dissimilarity index, after normalizing counts to simple proportions. There were no significant differences in richness at either the genus or OTU levels between the 2 groups, using 2 abundance-based estimators, Chao1 and observed taxa counts, when testing for a nonzero coefficient of a normal linear model that used group membership as predictor of richness (P ≥ .05 for all estimates). At the OTU level, both the Shannon index (P = .008) and the inverse Simpson index (P = .006) were significantly higher during HRV ARI (mean Hill number N1, 44.78; mean Hill number N2, 12.70), compared with RSV ARI (mean Hill number N1, 33.90; mean Hill Number N2, 9.30), indicating an increase in α-diversity.
To control for the potential confounding effect of the severity of the ARI, we (1) examined the association between observed nasopharyngeal microbial patterns and RSS and (2) conducted exploratory analyses in a subset of HRV-positive and RSV-positive infants with similar RSSs. There was no strong association between the nasopharyngeal microbiome and RSS in regard to the overall taxonomic composition, richness, or abundance of bacterial genera in HRV-positive infants, RSV-positive infants, or the full sample set (Supplementary Data). Within the subset of infants restricted to those with similar disease severity (n = 109), the median RSSs for infants with HRV ARI (n = 52) and those with RSV ARI (n = 57) were 2 (interquartile range, 1.8–3) and 2 (interquartile range, 2–3), respectively (P = .4). When comparing the bacterial genera abundance between infants with HRV ARI and infants with RSV ARI among this subgroup, we observed that 10 of the initial 19 identified bacterial genera identified in the full sample set remained statistically significant (Supplementary Table 2). The overall taxonomic composition and α-diversity indices also remained statistically different in this subgroup analysis (Supplementary Data).
We conducted similar exploratory analyses to control for the potential confounding effect of postnatal antibiotic exposure, obtaining similar results to our main analysis (data not shown).
DISCUSSION
Early-life exposure to respiratory viruses and bacteria is important in the development of an infant's immune response [11]. The respiratory microbiome and how acute illnesses such as ubiquitous infant viral ARIs alter it may help us to better understand the predisposition to secondary bacterial infections and long-term respiratory outcomes, including childhood asthma. Furthermore, modifications of the infant respiratory microbiome could be an approach to respiratory morbidity prevention [2]. Because these modifications may be virus specific, better characterizing the changes in the respiratory microbiome after the most common viral ARIs in infancy is important.
In our study, which mostly included children <6 months of age, we found that the nasopharyngeal microbiome of infants during HRV and RSV ARIs is largely dominated by Moraxella, Streptococcus, Corynebacterium, Haemophilus, and Dolosigranulum. This pattern contrasts with the nasopharyngeal microbiome in healthy infants <6 months of age that we and others have previously described, in which Haemophilus tends to be less common and Staphylococcus is one of the most predominant genera [3, 10, 12, 13]. Interestingly, early-life colonization with Haemophilus influenzae has been associated with a proinflammatory immune response and the development of childhood asthma [4, 6, 14]. In addition, we found differences in the abundance of various bacterial genera, including a higher abundance of Staphylococcus and a trend toward a higher abundance of Haemophilus in RSV-positive infants, and in the overall bacterial composition between infants with HRV and RSV ARIs. Thus, it is possible that virus-specific compositional shifts in the nasopharyngeal microbiome contribute to worse outcomes after early-life ARIs.
Most studies attempting to characterize a virus-specific bacterial profile in the respiratory tract have used conventional culture techniques and/or have been done in adults. To our knowledge, this is the first study comparing the nasopharyngeal microbiome profile of infants with RSV and HRV ARIs, the most common respiratory viral pathogens in early life. Only 1 other study has directly compared the nasopharyngeal microbiome between children with respiratory viruses by using next-generation sequencing [15]. In their study, Hyde et al demonstrated that, in children up to 2 years of age, an increased detection of H. influenzae and Moraxella catarrhalis distinguished children with HRV/RSV coinfection and those with only one of these two respiratory viruses [15]. In line with our findings, they also found that H. influenzae was commonly detected in children with RSV-only ARIs but not in those with HRV-only ARIs. Their study population importantly differs from ours in including older children who were hospitalized with LRTIs, who had prior wheezing, and/or who had a history of prematurity.
Our study has considerable strengths, including the population-based design of the parent study, the inclusion of infants with different degrees of ARI severity, the use of predefined criteria for ARIs, and the close surveillance during the first winter viral season to try to capture the infant's initial RSV ARI. We also acknowledge several limitations. First, the changes in the overall taxonomic composition or in the abundance of bacterial genera may have preceded the viral ARIs. However, other studies have shown that respiratory viruses are indeed capable of causing alterations in the composition of the nasopharyngeal microbiome [1, 3]. Second, the changes we observed could be the result of residual confounding from measured (eg, postnatal exposure to antibiotics or ARI severity) or unmeasured (eg, coinfection with other respiratory viruses or certain immunizations) variables. We conducted several exploratory analyses to assess the potential confounding effect of measured variables, obtaining similar results to those of our main analysis, which makes this unlikely. Last, we were unable to analyze the different bacteria below the genus level and therefore were unable to identify specific species associated with HRV or RSV ARIs. Despite these limitations, our study adds to the small but increasing literature on the interactions between respiratory viruses and the development and modification of the infant respiratory microbiome.
In conclusion, we found significant differences in the nasopharyngeal microbiome profile between infants with HRV and RSV ARIs. Future studies in this area are needed to better understand the interactions between respiratory viruses and commensal bacteria and how these affect the development of the immune response in early life, the severity of the ARI, and the development of childhood asthma.
Supplementary Data
Supplementary materials are available at http://jid.oxfordjournals.org. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Notes
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health (NIH).
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases, NIH (grants U19 AI095227 and K24 AI77930; grant U19 AI110819 via the J. Craig Venter Institute Genomic Centers for Infectious Diseases Program); the National Center for Advancing Transitional Sciences, NIH (grants UL1 TR000445 and U54 RR24975 via the Vanderbilt Institute for Clinical and Translational Research); and the NIH (contract HHSN272200900007C).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Tregoning JS, Schwarze J. Respiratory viral infections in infants: causes, clinical symptoms, virology, and immunology. Clin Microbiol Rev 2010; 23:74–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feldman AS, He Y, Moore ML, Hershenson MB, Hartert TV. Toward primary prevention of asthma. Reviewing the evidence for early-life respiratory viral infections as modifiable risk factors to prevent childhood asthma. Am J Respir Crit Care Med 2015; 191:34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teo SM, Mok D, Pham K et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 2015; 17:704–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Steenhuijsen Piters WA, Heinonen S, Hasrat R et al. Nasopharyngeal microbiota, host transcriptome and disease severity in children with respiratory syncytial virus infection. Am J Respir Crit Care Med 2016; 194:1104–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosas-Salazar C, Shilts MH, Tovchigrechko A et al. Nasopharyngeal microbiome in respiratory syncytial virus resembles profile associated with increased childhood asthma risk. Am J Respir Crit Care Med 2016; 193:1180–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Folsgaard NV, Schjorring S, Chawes BL et al. Pathogenic bacteria colonizing the airways in asymptomatic neonates stimulates topical inflammatory mediator release. Am J Respir Crit Care Med 2013; 187:589–95. [DOI] [PubMed] [Google Scholar]
- 7.Yi H, Yong D, Lee K, Cho YJ, Chun J. Profiling bacterial community in upper respiratory tracts. BMC Infect Dis 2014; 14:583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larkin EK, Gebretsadik T, Moore ML et al. Objectives, design and enrollment results from the Infant Susceptibility to Pulmonary Infections and Asthma Following RSV Exposure Study (INSPIRE). BMC Pulm Med 2015; 15:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodriguez H, Hartert TV, Gebretsadik T, Carroll KN, Larkin EK. A simple respiratory severity score that may be used in evaluation of acute respiratory infection. BMC Res Notes 2016; 9:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shilts MH, Rosas-Salazar C, Tovchigrechko A et al. Minimally invasive sampling method identifies differences in taxonomic richness of nasal microbiomes in young infants associated with mode of delivery. Microb Ecol 2016; 71:233–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daley D. The evolution of the hygiene hypothesis: the role of early-life exposures to viruses and microbes and their relationship to asthma and allergic diseases. Curr Opin Allergy Clin Immunol 2014; 14:390–6. [DOI] [PubMed] [Google Scholar]
- 12.Bosch AA, Levin E, van Houten MA et al. Development of upper respiratory tract microbiota in infancy is affected by mode of delivery. EBioMedicine 2016; 9:336–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biesbroek G, Tsivtsivadze E, Sanders EA et al. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med 2014; 190:1283–92. [DOI] [PubMed] [Google Scholar]
- 14.Bisgaard H, Hermansen MN, Buchvald F et al. Childhood asthma after bacterial colonization of the airway in neonates. N Engl J Med 2007; 357:1487–95. [DOI] [PubMed] [Google Scholar]
- 15.Hyde ER, Petrosino JF, Piedra PA, Camargo CA Jr, Espinola JA, Mansbach JM. Nasopharyngeal Proteobacteria are associated with viral etiology and acute wheezing in children with severe bronchiolitis. J Allergy Clin Immunol 2014; 133:1220–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.