Abstract
Background. Many countries worldwide have reported increasing numbers of emm89 group A Streptococcus (GAS) infections during last decade. Pathogen genetic factors linked to this increase need assessment.
Methods. We investigated epidemiological characteristics of emm89 GAS bacteremic infections, including 7-day and 30-day case-fatality rates, in Finland during 2004–2014 and linked them to whole-genome sequencing data obtained from corresponding strains. The Fisher exact test and exact logistic regression were used to compare differences between bacteremic infections due to emm89 GAS belonging to different genetic clades and subclades.
Results. Out of 1928 cases of GAS bacteremic infection, 278 were caused by emm89 GAS. We identified 2 genetically distinct clades, arbitrarily designated clade 2 and clade 3. Both clades were present during 2004–2008, but clade 3 increased rapidly from 2009 onward. Six subclades (designated subclades A–F) were identified within clade 3, based on phylogenetic core genome analysis. The case-fatality rate differed significantly between subclades (P < .05), with subclade D having the highest 30-day estimated case-fatality rate (19% vs 3%–14%).
Conclusions. A new emm89 clone, clade 3, emerged in 2009 and spread rapidly in Finland. Patients infected with certain subclades of clade 3 were significantly more likely to die. A specific polymerase chain reaction assay was developed to follow the spread of subclade D in 2015.
Keywords: Streptococcus pyogenes, bacteremia, whole genome sequencing, surveillance, molecular epidemiology, bacterial genome, emm type
Group A Streptococcus (GAS) is a gram-positive human-adapted pathogen with the ability to cause diseases with a wide clinical range, from mild infections such as pharyngitis to severe infections such as bacteremia and necrotizing fasciitis. When the isolation of GAS from a normally sterile body site is associated with disease, it is referred as an invasive GAS (iGAS) infection. Worldwide, GAS infections are responsible for >600 million cases of mild infections and for 663 000 new cases of iGAS per year, with incidence rates ranging from 2.45 to 46.00 cases per 100 000 population and case-fatality rates (CFRs) of up to 25% [1, 2].
The emm gene, which encodes M protein, one of the most important GAS virulence factors, is a target for genotyping. To date, >240 emm types have been reported, and genetic diversity can exist among strains of the same emm type [3, 4]. Dynamic changes and fluctuations in emm type frequency in iGAS infection occur over time and space [4], including emergence and replacement of new clones or subtypes [5–7]. Some emm types, such as emm1, have been linked to more severe disease manifestations and higher CFRs [4, 8–10].
Our recent study on iGAS infection in Finland showed that although the overall incidence of iGAS infections remained relatively stable during last few years, the incidence of cases caused by emm89 strains increased [11]. Similar findings have been reported from other countries in Europe [4, 6, 12–15], North America [7, 9, 16], and elsewhere [17–19].
The emergence of emm89 GAS strains has led investigators to perform bacterial genomic studies to understand reasons behind it. Thus far, the study of the largest sample of 1125 emm89 isolates, covering 2003–2013 from 3 countries, including Finland, revealed that emm89 strains underwent recombinational replacement events previously reported for the successful intercontinental epidemic emm1 clone [5, 7].
The goal of this study was to analyze, in detail, temporal, spatial, genomic, and clinical aspects of emm89 bacteremic infections in Finland during 2004–2014. We linked the epidemiological information with whole-genome sequencing (WGS) data for the corresponding 272 strains. We found that progeny of a new genetic clone (designated clade 3) emerged in 2009, spread rapidly in Finland, and caused a significantly increased number of iGAS infections throughout the country. Patients infected with certain subclades of this new clone were significantly more likely to die from the infection. A specific polymerase chain reaction (PCR) assay was developed to permit rapid identification of a distinct subclade for epidemiologic purposes.
MATERIALS AND METHODS
Surveillance of Bacteremic Infections Due to GAS
The Finnish healthcare system consists of 20 healthcare districts that are organized in 5 tertiary healthcare districts (population range, 740 000–1 900 000), for this study designated as southern, eastern, northern, central, and western Finland. Since 1995, all clinical microbiological laboratories electronically report each GAS isolate cultured from blood and/or cerebrospinal fluid to the National Infectious Disease Register (NIDR) maintained by the National Institute for Health and Welfare (THL). Each notification includes patient demographic data (date of birth, sex, and age), information regarding the specimen (type and date of sampling), name of the laboratory, date of notification, place of treatment, and, since 2004, the national personal identity code. Through the personal identity code, NIDR data are linked to the National Population Register (NPR) to obtain place of residence and date of death. In case of multiple notifications from the same patient within 90-day interval, notifications are merged and reported as a single case. In addition, clinical microbiological laboratories submit the corresponding GAS isolate to the National Reference Laboratory (NRL) for further analysis such as emm typing [20].
Case Definition and Outcome
A case of bacteremic GAS disease was defined as isolation of Streptococcus pyogenes (GAS) from a blood culture in Finland during 2004–2014 and as an emm89 GAS bacteremic infection when emm89 was specifically detected. All bacteremic GAS cases reported to the NIDR (n = 1977) and all available corresponding isolates (n = 1928) submitted to NRL from January 2004 to December 2014 were included in this study. Data about deaths of bacteremic GAS cases with appropriate personal identity codes (n = 1915) within 7-day and 30-day follow-up periods after GAS blood isolation were obtained from the National Population Register and used to assess 7-day and 30-day CFRs, respectively. In case of suspected epidemiological link among emm89 bacteremic GAS cases, medical records were reviewed by infectious diseases physicians at healthcare districts to assess common exposure or contacts between the patients and to identify underlying conditions known as risk factors for iGAS.
emm Typing
All isolates sent to the NRL were analyzed for emm type and subtype according to the guidelines of the Centers for Disease Prevention and Control (Atlanta, GA) [21].
WGS
Methods used for genome sequencing and analysis were recently described [5, 7, 22]. Briefly, after chromosomal DNA extraction from overnight cultures using the DNeasy 96 Blood and Tissue Kit (Qiagen), sequencing libraries were prepared using Nextera XT DNA Sample and Nextera XT Index V2 Prep kits (Illumina) and sequenced using Illumina instruments (HiSeq2500, NextSeq, and MiSeq). Raw reads were quality filtered using Trimmomatic [23], and errors were corrected using Musket [24]. Reads were aligned to the emm89 clade 3 reference genome MGAS27601 [23], using SMALT software (Wellcome Trust Sanger Institute), while genetic variant calls compared to the reference genome were detected using FreeBayes software. Single-nucleotide polymorphism (SNP) multiple sequence alignments were generated using Prephix and Phrecon scripts (available at: https://github.com/codinghedgehog). Genetic distances among strains and branches were calculated using MEGA6 [25] and R statistical packages (available at: https://www.r-project.org/). De novo assemblies of large contigs containing the ndrI to fabG chromosomal RB-15 region were generated using SPAdes software [26]. The nature of each SNP and INDEL category (coding/noncoding and synonymous/nonsynonymous) was interrogated using the in-house–developed script SNPfx.pl. All genome sequence data were deposited under accession number SRP059971 and SUB1086963 in the Sequence Read Archive (National Center for Biotechnology Information).
PCR to Identify an Emerging emm89 Subclade
A specific PCR targeting the nrdl to fabG chromosomal region was developed to rapidly identify emm89 bacteremic GAS strains belonging to clade 3 subclade D. Primer 1 (5′-GACTAAAACAGCTAAGAAGAAGGAC -3′) was designed to detect all subclades, primer 2 (5′-CATACAAGGTACTATCTTCCTCAAG-3′) to be specific for A-B-C-E-F subclades, and primer 3 (5′-GTCTCTTTTTGATATCACCTCC-3′) to be specific for subclade D. The expected size of amplified products was 626 bp for subclade D and ≥1034 bp for all other subclades. Strains were cultivated overnight on blood agar at 35°C with 5% CO2, and DNA was extracted from bacterial colonies after suspension in 150 µL of Milli-Q water, incubation at 99°C for 10 minutes, and centrifugation at high speed for 5 minutes. Each reaction contained 15.5 µL of nuclease-free water, 25 µL of GoTaq Master Mix (G2 Hot Start polymerase, Promega), 2.5 µL of each primer (10 µM), and 2 µL (50–100 ng) of DNA template. PCR was performed using T100 Thermal Cycler (Biorad, Singapore) with initial denaturation at 95°C for 2 minutes; 30 cycles at 95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 1 minute and 10 seconds; and final extension for 2 minutes at 72°C. ATCC 700294 was used as positive control.
Data Analysis and Statistics
NIDR data for notified cases and emm typing results for corresponding isolates were linked using personal identity code and date of specimen. Annual incidence rates for all bacteremic GAS and emm89 bacteremic GAS in the entire country and in each of the 5 tertiary healthcare districts were calculated using population data for the corresponding year as reported by Statistics Finland. CFRs within 7-day and 30-day follow-up periods after emm89 bacteremic GAS isolation was calculated for clades and subclades within emm89 bacteremic GAS cases. Fisher exact test and exact logistic regression were used to compare differences in 7-day and 30-day CFRs. Yearly overall and district relative increases in emm89 proportion were estimated by binary regression with log-link. Differences were considered significant when P values were < .05. Data were analyzed with Stata 13 (Statacorp, College Station, Texas).
Ethical Study Approval
Ethical committee clearance was not required as all aspects of this study fall within THL legal mandate defined by the Finnish Communicable Diseases Act [27]. No personal information concerning cases was shared with non-THL investigators.
RESULTS
GAS Cases
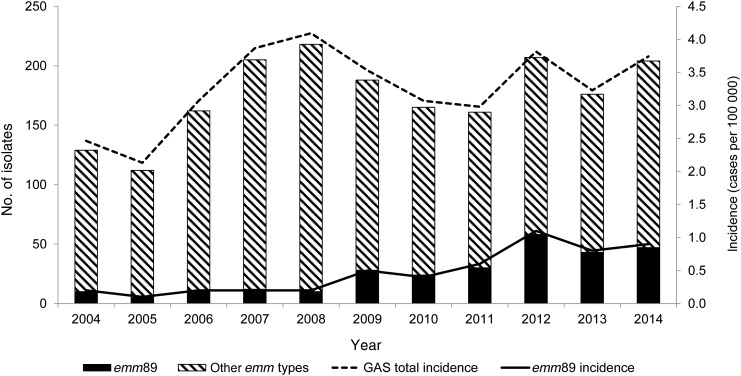
During 2004–2014, 1928 bacteremic GAS cases were identified (range by year, 112–218 cases). The median age of cases was 54 years (range, 0–100 years), and 54% were males. The average annual incidence rate during 2004–2014 was 3.3 cases per 100 000 population (range by year, 2.1–4.1 cases per 100 000 population), with a peak of 4.1 cases per 100 000 population in 2008 (Figure 1). The average incidence rate was highest in the age groups of 55–65 years (4.5 cases per 100 000 population) and >65 years (5.7 cases per 100 000 population). The 7-day CFR was 7% (127 of 1915), and the 30-day CFR was 9% (176 of 1915).
Figure 1.
Number of bacteremic group A Streptococcus (GAS) isolates (left axis) typed at the national reference laboratory and incidence of GAS bacteremia (right axis) in Finland, 2004–2014.
emm89 GAS Cases
During 2004–2014, 278 bacteremic GAS cases with available corresponding strains were caused by emm89 (range by year, 5–58 cases). The median age of emm89 bacteremic GAS cases was 55 years (range, 0–97 years), equally distributed between males and females. The 7-day and 30-day CFRs of emm89 bacteremic GAS were 4% (12 of 278) and 7% (20 of 278), respectively, which were not significantly different from those caused by all other emm types combined (7-day CFR, P = .12; 30-day CFR, P = .26).
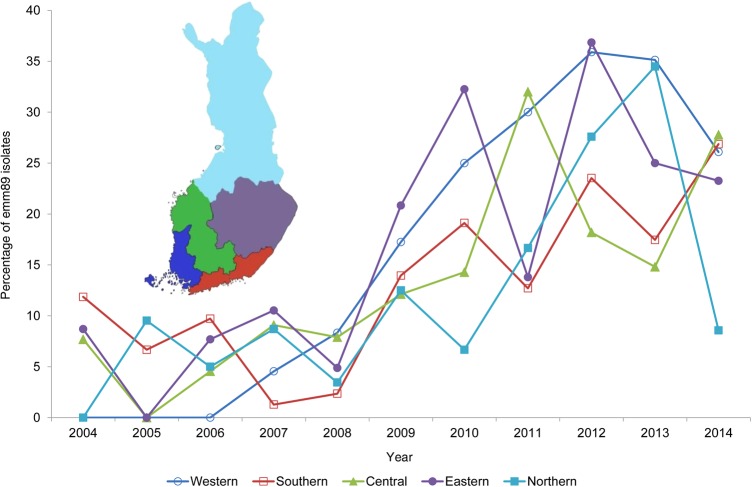
The proportion of emm89 cases among bacteremic GAS cases varied from 4% (5 of 112) in 2005 to 28% (58 of 207) in 2012, corresponding to incidence rates of 0.1 and 1.1 cases per 100 000 population, respectively (Figure 1). In 2004, emm89 bacteremic cases were only notified in southern, central, and western Finland, but within 3 years it caused bacteremic cases throughout all country (Figure 2). The proportion of emm89 increased significantly in all tertiary healthcare districts (mean relative increase, 19%; range, 17.8%–27.9%; P < .005). The majority of emm89 bacteremic GAS cases (276 of 278) were caused by subtype emm89.0. One strain of emm89.0b and one of emm89.1 subtypes were also identified.
Figure 2.
Emergence of emm89 group A Streptococcus (GAS) isolates among all bacteremic GAS isolates by tertiary care district in Finland, 2004–2014. emm89 proportions were calculated for each tertiary care district and for each year. The inset shows a map of Finland, with the 5 tertiary care districts in color.
WGS Analysis
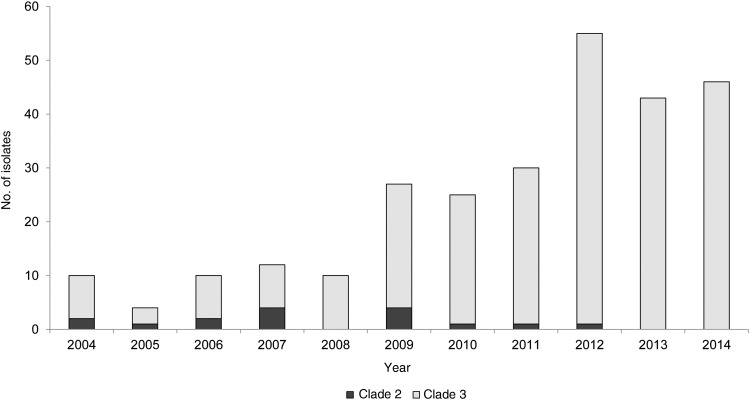
A total of 272 emm89 bacteremic GAS strains isolated in Finland during 2004–2014 were analyzed by WGS, and 2 genetically distinct clades, designated clade 2 and clade 3, as recently described [28], were identified. Clade 2 included 6% (16 of 272), while clade 3 accounted for the remaining 94% of emm89 isolates (256 of 272). A total of 56% of clade 2 cases were in males, whereas clade 3 cases were equally distributed between males and females. Cases with clade 3 strains were significantly older than those with clade 2 (median age, 56 vs 44 years; P < .05). Both clades caused bacteremic GAS cases during 2004–2008, but thereafter clade 3 rapidly increased in frequency, causing virtually all cases from 2009 onward. By 2012, all emm89 bacteremic GAS strains were of clade 3 (Figure 3).
Figure 3.
emm89 bacteremic group A Streptococcus (GAS) isolates by clade, in Finland, 2004–2014.
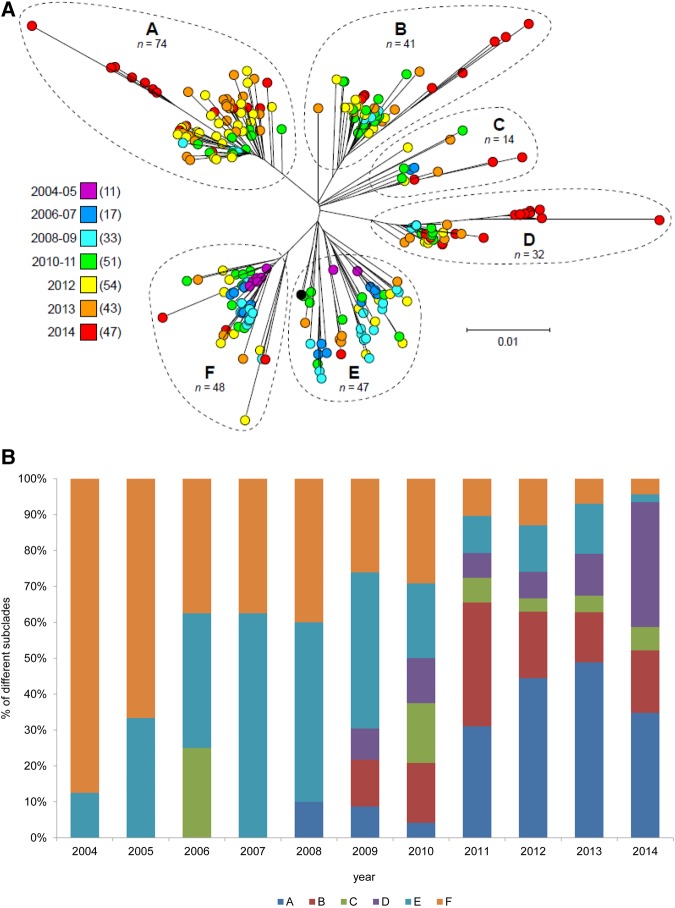
Inasmuch as clade 3 strains increased rapidly in frequency and have showed enhanced virulence in animal models of invasive infection [28], we tested the hypothesis that patients infected with strains of the 2 different clades differed significantly in outcome. Within the 7-day or 30-day follow-up periods, none of the clade 2 cases died, while among clade 3 cases, the 7-day CFR was 5% (12 of 255), and the 30-day CFR was 8% (20 of 255). However, these observations were not statistically significant. On the basis of SNP variation among the core genomes of clade 3 emm89 bacteremic GAS strains, 6 distinct subclades were identified, arbitrarily designated as subclades A through F (Figure 4A and 4B). Subclade A included 74, subclade B 41, subclade C 14, subclade D 32, subclade E 46, and subclade F 49 of the 256 isolates. We identified variations in the temporal distribution of the 6 subclades, and a noteworthy increase of cases caused by subclade D strains occurred during 2013–2014 (Figure 4A and 4B).
Figure 4.
emm89 bacteremic group A Streptococcus clade 3 subclades by year (A) and proportion (B), 2004–2014.
CFRs differed significantly between subclades, both with respect to the 7-day CFR (P = .001) and the 30-day CFR (P = .04; Table 1). Subclade C had the highest 7-day CFR (14%), followed by subclade D, B, E, and both A and F. For the 30-day CFR, subclade D showed the highest rate (19%), followed by subclades C, B, E, F, and A. Subclade A was assigned as a reference when we calculated the estimates of age-adjusted 7-day and 30-day CFR odds (Table 1). Subclade C had the highest age-adjusted 7-day odds of death due to GAS, followed by subclade D, B, E, and F. Subclade D had the highest age-adjusted 30-day odds of death due to GAS, followed by subclade B, C, E, and F.
Table 1.
Number of Deaths, Case-Fatality Rates, and Estimated Age-Adjusted Odds of Death Due to emm89 Clade 3 Subclades for 7-Day and 30-Day Follow-up Periods
| Subclade | Cases, No. | 7-day Follow-up |
30-day Follow-up |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Deaths, No. | CFR, % | OR (95% CI) | P Value | Deaths, No. | CFR, % | OR (95% CI) | P Value | ||
| A | 74 | 0 | 0 | Reference | 2 | 3 | Reference | ||
| B | 41 | 4 | 10 | 10.5 (1.28–∞) | .03 | 5 | 12 | 5.5 (.83–61.0) | .08 |
| C | 14 | 2 | 14 | 12.8 (.97–∞) | .05 | 2 | 14 | 5.2 (.34–80.0) | .28 |
| D | 32 | 4 | 13 | 12.4 (1.49–∞) | .02 | 6 | 19 | 7.2 (1.2–77.7) | .03 |
| E | 46 | 2 | 4 | 4.0 (.32–∞) | .30 | 3 | 7 | 2.7 (.30–34.0) | .51 |
| F | 48 | 0 | 0 | 1.0 (0–∞) | … | 2 | 4 | 1.8 (.12–25.9) | .92 |
Abbreviations: CFR, case-fatality rate; CI, confidence interval; OR, odds ratio.
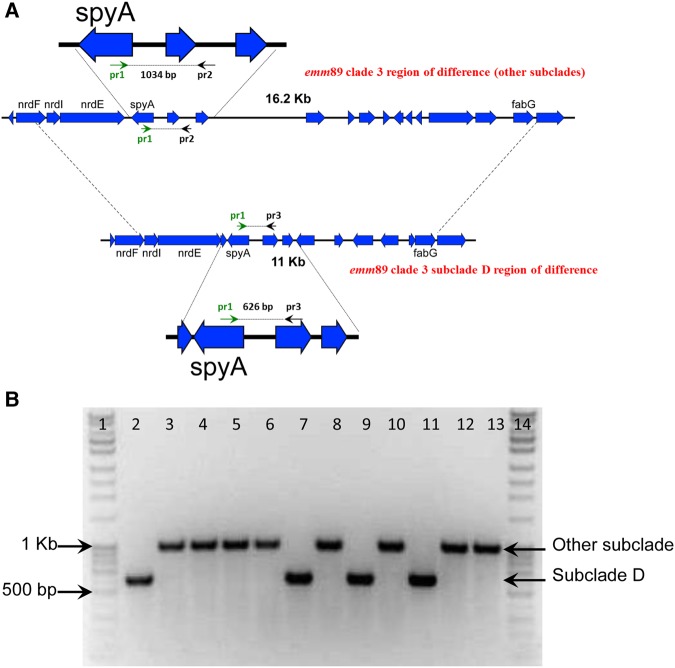
Subclade D strains differed from other clade 3 strains in the ndrI to fabG chromosomal region RB-15 (Figure 5A). In addition, 6 subclade D strains had a nonsynonymous SNP (Lys214Arg) in the 3-component regulatory system liaS gene. Of note, 11 subclade D strains had also 2 unique SNPs [22], resulting in amino acid replacements in CovR (Ser130Asn) and ParC (Asp83Gly) genes, which are part of regulatory systems involved in virulence and DNA topoisomerase IV chromosome partitioning, respectively. Five of these 11 subclade D strains were isolated from cases in eastern Finland, and 4 were isolated from cases in southern Finland, at the border with eastern Finland. Nine cases were identified within a 5-month period in 2014, and 2 cases died within 7 days after the detection of bacteremia. Based on medical records, no epidemiological link could be observed among cases, but they all possessed ≥1 known risk factor for bacteremic GAS infection, such as old age, delivery, alcoholism, liver chronic disease, and malignancy.
Figure 5.
Specific polymerase chain reaction (PCR) assay to detect clade 3 subclade D strains. A, Schematic showing the emm89 clade 3 region of difference. B, Agarose gel elecrophoresis of findings of the specific PCR assay to detect clade 3 subclade D strains. Lane 1, 100-bp DNA ladder; lane 2, subclade D; lanes 3–6, other subclades; lane 7, subclade D; lane 8, other subclade; lane 9, subclade D; lane 10, other subclade; lane 11, subclade D; lanes 12–13, other subclades; lane 14, 100-bp DNA ladder.
Rapid PCR Identification of Recently Emerged Subclade D Strains
The PCR specific for subclade D was validated using 69 emm89 bacteremic GAS strains representing the different subclades isolated during 2009–2014. A band of 600 bp was obtained from all 33 subclade D strains as expected, whereas bands of ≥1000 bp were obtained from 36 A-B-C-E-F subclade strains (Figure 5). After validation, this PCR was used to screen all 33 emm89 bacteremic GAS strains isolated in 2015, and 9 subclade D strains (27%) were identified.
DISCUSSION
GAS has been shown to spread in epidemic waves through the emergence and dissemination of certain strains [5, 7, 29]. Genomic alterations and molecular events leading to events such as upregulated expression of virulence factors contributed to the spread of certain successful GAS lineages, including emm1 [5]. Recently, reports of similar events taking place in emm89 strains have been published [7, 30]. In our present study, we investigated the emergence and spread of emm89 GAS among bacteremic cases in Finland during 2004–2014, using nationwide comprehensive, population-based infectious disease surveillance data and the cognate strain collection. We show how emm89 increase is particularly linked to the recent emergence of clade 3 strains, a genetically distinct clade recently described [7, 22, 28]. The current emergence of 1 subclade among the 6 in clade 3, namely subclade D, was studied in more depth. As we discovered that cases caused by this subclade rapidly increased in frequency among all emm89 cases and had a significantly higher 30-day CFR, targeted surveillance by PCR specific for subclade D was performed during 2015, which revealed its sustained circulation during this period.
The increase of emm89 GAS strains in prevalence and incidence has been reported in many countries [4, 6, 7, 9, 12–17, 19, 31, 32]. The first 2 emm89 bacteremic GAS strains in Finland were reported in 1996 [20], and since then this emm type has progressively increased in number [11]. emm89 GAS strains among bacteremic cases were already reported by all Finnish tertiary healthcare districts in 2007, but its proportion increased nationally, becoming the second most prevalent emm type from 2009 onward. In addition, already during 2008–2009, emm89 isolates were common among a various different infections in Finland, including pharyngitis and deep-tissue infections [18]. Interestingly, the highest number and corresponding proportion of emm89 strains (58 of 207 [28%]) among all bacteremic GAS strains occurred in 2012, a year when the annual incidence of bacteremic GAS cases also peaked at 3.9 cases per 100 000 population. The proportion of bacteremic GAS cases due to emm89 remained constant after 2012, representing at least one fourth of all bacteremic GAS cases annually, although yearly fluctuations were detected on tertiary healthcare district level.
Previous studies of annual incidence and emm type distribution among iGAS strains showed fluctuations over years with emergence and reemergence of specific emm types [13, 14]. In Finland, during past decades, in addition to emm89 strains, these periodic fluctuations were associated with peaks of disease activity caused by emm1 [8, 33, 34], emm28 [8, 11], emm84 [35], and emm33 [11, 36] strains. For decades, the reasons behind fluctuations in emm type distribution were considered to be primarily associated with the spread of distinct emm types in immunological naive populations. Recent studies based on WGS have shown that genetic changes in the organism due to acquisition or upregulation of virulence factors [5, 6, 28, 29] also participate. We show that the change in emm type distribution overlapped changes in emm89 population genomics, and the increasing number of emm89 bacteremic GAS cases during 2009–2012 coincided with the clade 2 to clade 3 shift. Turner et al [6, 37] described a similar event with the emergence of new ST101 clade in England and Wales during 2005–2009, and Friães et al [38] suggested, although without the support of WGS data, that this also happened in Portugal. Based on available information, it thus seems that emm89 ST101 emerged in many countries in a closely similar time frame during the first decade of the 2000s, accompanied by a clade swift from 1 to 2 and subsequent emergence of clade 3. However, clade 1 strains have not been identified in Finland [7, 22].
So far, few studies have been conducted to address molecular mechanisms responsible for the increasing number of emm89 GAS cases [5–7, 28, 37]. Zhu et al [7] analyzed a set of emm89 isogenic mutant strains for biochemical, pathogenesis, and ex vivo studies and demonstrated that the molecular trigger underlying the current emm89 epidemic is associated with the upregulation in the expression of 2 secreted cytotoxins, known as S. pyogenes NADase and streptolysin O. Recently, we showed that a distinct genetic subpopulation of Finnish emm89 bacteremic GAS isolates had an enhanced ability to survive in human saliva ex vivo [22], and this could promote person-to-person spread. In addition, GAS can persist inside host cells [39] and thus evade action of antibiotics and perhaps even reach distant body sites from the point of entry though a Trojan horse mechanism.
emm89 bacteremic GAS cases did not differ from bacteremic GAS cases caused by all other emm types when compared by age. The sex distribution and mean age of these 2 groups were similar. However, when emm89 cases were analyzed at the clade level, clade 3 cases were significantly older than clade 2 cases. Tamayo et al [13] also described an increased number of emm89 cases, especially in the older age group, in Spain during 2009–2011.
The 7-day (4%) and 30-day (7%) CFRs of emm89 bacteremic GAS were lower, compared with values from a previous report [6]. No statistically significant differences were found when comparing 7-day and 30-day CFRs between overall bacteremic GAS cases and emm89 bacteremic GAS cases or between clades 2 and 3, as also previously described [6]. On the contrary, statistically significant differences were found within clade 3 subclades, using exact logistic regression adjusted by age. However, the small numbers among subclades represents a limitation, and, thus, the interpretation needs precaution. The eventual concomitant role of underlying conditions and risk factors could not be assessed because this information is not collected by the surveillance system. Subclade C and subclade D had the highest 7-day and 30-day CFRs, with values of 14% (2 of 14) and 19% (6 of 32), respectively. Although subclade C had the highest 7-day CFR, we focused particularly on subclade D because of its increase in frequency among all emm89 cases during 2013–2014 and additional deaths reported after the 7-day and within the 30-day follow-up periods among cases of this subclade, while no additional deaths after 7-day follow-up and no increase in subclade C cases were observed during our study period. Finnish emm89 bacteremic GAS clade 3 subclade D strains differed from other clade 3 strains in ndrI-to-fabG chromosomal region RB-15, as recently described [22]. A specific PCR based on WGS data analysis was developed to detect subclade D among emm89 bacteremic GAS strains during 2015, and, not surprisingly, this subclade was still circulating in the country. However, no obvious clustering of cases could be observed as the 9 subclade D cases occurred in all 5 tertiary healthcare districts (data not shown). The circulation of subclade D strains still warrant further surveillance of this subclade.
Our study describes the characteristics of the spread of a newly emerged emm89 GAS clone causing bacteremia in Finland during 2004–2014. WGS identified potentially more-virulent clones, such as clade 3 and its subclade D. The spread of subclade D strains was further followed by the use of a specific PCR during 2015. To our best knowledge, this is the first study that systematically combines comprehensive, population-based surveillance data and full-genome analyses of corresponding isolates to highlight the importance of their combined use to better understand the evolution and spread of emm89 GAS. The need for targeted public health strategies to control these life-threatening infections necessitates the establishment of comprehensive iGAS infection surveillance systems, including those for collecting clinical data, and research to develop an effective GAS vaccine should continue.
Notes
Acknowledgments. We thank Teemu Möttönen, for technical expertise in retrieving and combining data from several database sources; Aino Makkonen, Tuula Rantasalo, and Toni Huovinen, for excellent technical assistance; Tiina Sorvari, Pekka Suomalainen, Jukka Heikkinen, and Katariina Kainulainen, for assistance in collecting patient background data; Aftab Jasir, for critical comments on the manuscript; and the clinical microbiology laboratories that submitted notifications and isolates.
Financial support. This work was partially supported by the Academy of Finland (grant 255636), the Hospital District of Southwest Finland, and the Fondren Foundation.
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect Dis 2005; 5:685–94. [DOI] [PubMed] [Google Scholar]
- 2.Sims Sanyahumbi A, Colquhoun S, Wyber R, Carapetis JR. Global disease burden of group A Streptococcus. In: Ferretti JJ, Stevens DL, Fischetti VA, eds. Streptococcus pyogenes: basic biology to clinical manifestations. Oklahoma City, OK: University of Oklahoma Health Sciences Center, 2016. [PubMed] [Google Scholar]
- 3.ftp://ftp.cdc.gov/pub/infectious_diseases/biotech/tsemm/.
- 4.Luca-Harari B, Darenberg J, Neal S et al. . Clinical and microbiological characteristics of severe Streptococcus pyogenes disease in Europe. J Clin Microbiol 2009; 47:1155–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nasser W, Beres SB, Olsen RJ et al. . Evolutionary pathway to increased virulence and epidemic group A Streptococcus disease derived from 3,615 genome sequences. Proc Natl Acad Sci U S A. 2014; 111:E1768–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turner CE, Abbott J, Lamagni T et al. . Emergence of a new highly successful acapsular group A Streptococcus clade of genotype emm89 in the United Kingdom. MBio 2015; 6:e00622–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu L, Olsen RJ, Nasser W et al. . A molecular trigger for intercontinental epidemics of group A Streptococcus. J Clin Invest 2015; 125:3545–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siljander T, Toropainen M, Muotiala A, Hoe NP, Musser JM, Vuopio-Varkila J. emm typing of invasive T28 group A streptococci, 1995–2004, Finland. J Med Microbiol 2006; 55(Pt 12):1701–6. [DOI] [PubMed] [Google Scholar]
- 9.O'Loughlin RE, Roberson A, Cieslak PR et al. . The epidemiology of invasive group A streptococcal infection and potential vaccine implications: United States, 2000–2004. Clin Infect Dis 2007; 45:853–62. [DOI] [PubMed] [Google Scholar]
- 10.Nelson GE, Pondo T, Toews KA et al. . Epidemiology of Invasive Group A Streptococcal Infections in the United States, 2005–2012. Clin Infect Dis 2016; 63:478–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smit PW, Lindholm L, Lyytikainen O, Jalava J, Patari-Sampo A, Vuopio J. Epidemiology and emm types of invasive group A streptococcal infections in Finland, 2008–2013. Eur J Clin Microbiol Infect Dis 2015; 34:2131–6. [DOI] [PubMed] [Google Scholar]
- 12.Friaes A, Pinto FR, Silva-Costa C, Ramirez M, Melo-Cristino J, Portuguese Group for the Study of Streptococcal Infections. Group A streptococci clones associated with invasive infections and pharyngitis in Portugal present differences in emm types, superantigen gene content and antimicrobial resistance. BMC Microbiol 2012; 12:280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamayo E, Montes M, Garcia-Arenzana JM, Perez-Trallero E. Streptococcus pyogenes emm-types in northern Spain; population dynamics over a 7-year period. J Infect 2014; 68:50–7. [DOI] [PubMed] [Google Scholar]
- 14.Creti R, Imperi M, Baldassarri L et al. . emm Types, virulence factors, and antibiotic resistance of invasive Streptococcus pyogenes isolates from Italy: What has changed in 11 years? J Clin Microbiol 2007; 45:2249–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plainvert C, Doloy A, Loubinoux J et al. . Invasive group A streptococcal infections in adults, France (2006–2010). Clin Microbiol Infect 2012; 18:702–10. [DOI] [PubMed] [Google Scholar]
- 16.Shea PR, Ewbank AL, Gonzalez-Lugo JH et al. . Group A Streptococcus emm gene types in pharyngeal isolates, Ontario, Canada, 2002–2010. Emerg Infect Dis 2011; 17:2010–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karaky NM, Araj GF, Tokajian ST. Molecular characterization of Streptococcus pyogenes group A isolates from a tertiary hospital in Lebanon. J Med Microbiol 2014; 63(Pt 9):1197–204. [DOI] [PubMed] [Google Scholar]
- 18.Vahakuopus S, Vuento R, Siljander T, Syrjanen J, Vuopio J. Distribution of emm types in invasive and non-invasive group A and G streptococci. Eur J Clin Microbiol Infect Dis 2012; 31:1251–6. [DOI] [PubMed] [Google Scholar]
- 19.Ikebe T, Tominaga K, Shima T et al. . Increased prevalence of group A streptococcus isolates in streptococcal toxic shock syndrome cases in Japan from 2010 to 2012. Epidemiol Infect 2015; 143:864–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rantala S, Vahakuopus S, Siljander T et al. . Streptococcus pyogenes bacteraemia, emm types and superantigen profiles. Eur J Clin Microbiol Infect Dis 2012; 31:859–65. [DOI] [PubMed] [Google Scholar]
- 21.http://www.cdc.gov/streplab/protocol-emm-type.html.
- 22.Beres SB, Kachroo P, Nasser W et al. . Transcriptome Remodeling Contributes to Epidemic Disease Caused by the Human Pathogen Streptococcus pyogenes. MBio 2016; 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Schroder J, Schmidt B. Musket: a multistage k-mer spectrum-based error corrector for Illumina sequence data. Bioinformatics 2013; 29:308–15. [DOI] [PubMed] [Google Scholar]
- 25.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 2013; 30:2725–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bankevich A, Nurk S, Antipov D et al. . SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012; 19:455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.http://www.finlex.fi/en/laki/kaannokset/1986/en19860583.pdf.
- 28.Zhu L, Olsen RJ, Nasser W, de la Riva Morales I, Musser JM. Trading capsule for increased cytotoxin production: contribution to virulence of a newly emerged clade of emm89 Streptococcus pyogenes. MBio 2015; 6:e01378–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fittipaldi N, Beres SB, Olsen RJ et al. . Full-genome dissection of an epidemic of severe invasive disease caused by a hypervirulent, recently emerged clone of group A Streptococcus. Am J Pathol 2012; 180:1522–34. [DOI] [PubMed] [Google Scholar]
- 30.Musser JM, Zhu L, Olsen RJ, Nasser W. Musser et al. Reply to “Emergence of the Same Successful Clade among Distinct Populations of emm89 Streptococcus pyogenes in Multiple Geographic Regions”. MBio 2015; 6:e01838–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darenberg J, Luca-Harari B, Jasir A et al. . Molecular and clinical characteristics of invasive group A streptococcal infection in Sweden. Clin Infect Dis 2007; 45:450–8. [DOI] [PubMed] [Google Scholar]
- 32.Williamson DA, Morgan J, Hope V et al. . Increasing incidence of invasive group A streptococcus disease in New Zealand, 2002–2012: a national population-based study. J Infect 2015; 70:127–34. [DOI] [PubMed] [Google Scholar]
- 33.Hoe NP, Nakashima K, Lukomski S et al. . Rapid selection of complement-inhibiting protein variants in group A Streptococcus epidemic waves. Nat Med 1999; 5:924–9. [DOI] [PubMed] [Google Scholar]
- 34.Siljander T, Lyytikainen O, Vahakuopus S, Snellman M, Jalava J, Vuopio J. Epidemiology, outcome and emm types of invasive group A streptococcal infections in Finland. Eur J Clin Microbiol Infect Dis 2010; 29:1229–35. [DOI] [PubMed] [Google Scholar]
- 35.Siljander T, Lyytikainen O, Vahakuopus S, Saila P, Jalava J, Vuopio-Varkila J. Rapid emergence of emm84 among invasive Streptococcus pyogenes infections in Finland. J Clin Microbiol 2009; 47:477–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pesola AK, Sihvonen R, Lindholm L, Patari-Sampo A. Clindamycin resistant emm33 Streptococcus pyogenes emerged among invasive infections in Helsinki metropolitan area, Finland, 2012 to 2013. Euro Surveill 2015; 20:21117. [DOI] [PubMed] [Google Scholar]
- 37.Turner CE, Lamagni T, Holden MT et al. . Turner et al. Reply to “Emergence of the Same Successful Clade among Distinct Populations of emm89 Streptococcus pyogenes in Multiple Geographic Regions”. MBio 2015; 6:e01883–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Friaes A, Machado MP, Pato C, Carrico J, Melo-Cristino J, Ramirez M. Emergence of the same successful clade among distinct populations of emm89 Streptococcus pyogenes in multiple geographic regions. MBio 2015; 6:e01780–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Neill AM, Thurston TL, Holden DW. Cytosolic replication of group A Streptococcus in human macrophages. MBio 2016; 7:e00020–16. [DOI] [PMC free article] [PubMed] [Google Scholar]