Abstract
Background. Natural infection induces partial immunity to Chlamydia trachomatis. Identification of chlamydial antigens that induce interferon γ (IFN-) secretion by T cells from immune women could advance vaccine development.
Methods. IFN-γ production by CD4+ and CD8+ peripheral blood T cells from 58 high-risk women was measured after coculture with antigen-presenting cells preincubated with recombinant Escherichia coli expressing a panel of 275 chlamydial antigens. Quantile median regression analysis was used to compare frequencies of IFN-γ responses in women with only cervical infection to those in women with endometrial infection and frequencies in women who remained uninfected for over 1 year to those in women who developed incident infection. Statistical methods were then used to identify proteins associated with protection.
Results. A higher median frequency of CD8+ T-cell responses was detected in women with lower genital tract chlamydial infection, compared with those with upper genital tract chlamydial infection (13.8% vs 9.5%; P = .04), but the CD4+ T-cell response frequencies were not different. Women who remained uninfected displayed a greater frequency of positive CD4+ T-cell responses (29% vs 18%; P < .0001), compared with women who had incident infection, while the frequencies of CD8+ T-cell responses did not differ. A subset of proteins involved in central metabolism, type III secretion, and protein synthesis were associated with protection.
Conclusions. Investigations in naturally exposed women reveal protective responses and identify chlamydial vaccine candidate antigens.
Keywords: Chlamydia trachomatis, genital, women, vaccine, antigen
Chlamydia trachomatis causes the world's most prevalent bacterial sexually transmitted infection, with approximately 3.0 million new infections in the United States [1] and 90 million new infections globally each year [2]. Adolescents and young adults are at greatest risk for infection [3]. Despite accurate diagnostic tests and effective antibiotic therapies, it remains an important global health problem. Most infections are asymptomatic, leading to undetected and untreated infections and sustained high prevalence. Although only a small proportion of infected women develop severe morbidities, including chronic pelvic pain, ectopic pregnancy, and tubal factor infertility, their absolute number is great because infection prevalence is high, driving annual healthcare costs in the United States to an estimated $517 million [4]. Although antibiotics clear infection, destructive pathology is not ameliorated. Thus, a vaccine is urgently needed.
Evidence for naturally acquired immunity to chlamydia in humans is accumulating, including a documented decreased prevalence with increasing age despite continued exposure [5], decreased infection concordance between sex partners with increasing age of the partners, and lower bacterial loads among individuals with a history of infection [6]. Furthermore, in a prospective study of 200 women, those whose chlamydial infections cleared spontaneously between testing and treatment were less likely to become reinfected during the follow-up period [7]. Human vaccine data are derived solely from trials of live and inactivated whole-cell vaccines administered to children living in trachoma-endemic areas in the 1960s, when short-term and serovar-specific protection against reinfection was observed [8–10]. Animal models have illuminated the importance of T cells in control of infection [11]. Resolution of infection with attenuated chlamydial strains leads to partial immunity from reinfection and to significant protection from disease in mice [12] and nonhuman primates [13]. Data from the mouse model indicate that IFN-γ–producing CD4+ T-helper type 1 cells play a central role in infection clearance [14–16], while antichlamydia antibody contributes to resistance to reinfection [17]. Although CD8+ T cells are dispensable for immunity in the mouse genital tract model [18], a role for CD8+ T cells in vaccine-induced protection from ocular infection was demonstrated in macaques [19] and C. trachomatis–specific human CD8+ T-cell clones inhibit chlamydial replication [20], indicating a need to further explore their contribution to protection.
In this study, we build on previous investigations [21, 22] by determining the relative contribution of IFN-γ–producing CD4+ and CD8+ T cells to the control of infection and to protection against incident infection in highly exposed women [23]. We selected a panel of 275 chlamydial proteins to probe antigen-specific CD4+ and CD8+ T-cell IFN-γ responses in women whose extent of infection was defined at enrollment and who were followed longitudinally for reinfection. We hypothesized that women who had infection limited to the cervix would exhibit a greater frequency of positive responses to a specific subset of proteins as compared to women in whom infection ascended to their upper genital tract. Additionally, we hypothesized that women who remained uninfected over 1 year would recognize a specific subset of antigens with increased frequency, compared with women who developed incident infection. Our overall goal was to use a minimally biased screening assay to probe peripheral blood samples available from women with evidence of natural immune protection to identify candidate vaccine T-cell antigens and thereby advance vaccine development.
METHODS
Patient Populations
Blood from 97 women recruited into a previously described T-Cell Response Against Chlamydia (TRAC) cohort was processed for analysis by a T-cell IFN-γ assay [23]. The institutional review boards for human research of the University of Pittsburgh and the University of North Carolina approved the study protocol. All participants provided written informed consent at the time of enrollment and agreed to be contacted to return for follow-up visits 1, 4, 8, and 12 months after enrollment.
TRAC was a longitudinal cohort study investigating cellular responses important for protection against incident C. trachomatis genital tract infection. A total of 225 women aged 15–35 years with lower genital tract sexually transmitted infection (STI) or who were at risk for STI were enrolled in the study from 3 urban sites in Pittsburgh, Pennsylvania, during 2011–2015. Eligibility criteria included clinical evidence of mucopurulent cervicitis, diagnosis of chlamydial infection prior to treatment, or reported sexual contact with an individual with a recent diagnosis of chlamydial urethritis or nongonococcal urethritis. Women with a diagnosis of pelvic inflammatory disease (PID), according to the Centers for Disease Control and Prevention guidelines [24], were excluded from this study and offered enrollment in a parallel study comparing PID antibiotic regimens.
At enrollment, study personnel obtained demographic data and a standardized medical history. Each participant completed a questionnaire, and cervical and endometrial swab samples were obtained for microbiologic testing [23]. Peripheral blood mononuclear cells (PBMCs) were obtained for T-cell assays. Participants returned for cervical infection screening and PBMC collection 1, 4, 8, and 12 months after enrollment. A standardized questionnaire was administered at each follow-up visit, addressing the participant′s interim medical and sexual history. PBMCs available from the first 97 women enrolled in the cohort were processed for assay; assay failure occurred for 39 women, owing to poor cell viability or inadequate cell numbers after expansion, leaving findings from 58 women for comparative analysis.
Definition of Clinical and Microbiological Subgroups
Women were assigned to groups according to the extent of their infection at enrollment. Women with a cervical swab positive for chlamydia by nucleic acid amplification testing (NAAT) but with a negative chlamydia NAAT result for the endometrial sample were defined as Cervix+. Women who tested positive for chlamydia at the cervix and in the endometrium were defined as Endo+. Among 58 women analyzed, 16 (28%) were uninfected, 27 (47%) had lower genital tract infection (the Cervix+ group), and 15 (26%) had upper genital tract infection (the Endo+ group). No participants tested positive solely in the endometrium. The chlamydial cervical burden was estimated via quantitative NAAT, using genomic DNA extracted from reserved cervical swab eluates as a template [23].
Women were also grouped on the basis of their infection status throughout follow-up. The susceptible group contained women who tested positive for chlamydial infection at any follow-up visit or who reported having an infection between visits that was diagnosed at an outside clinic. Women who tested negative at 4 study follow-up visits and never reported having an infection between visits composed the follow-up–negative group. Within these groups, 31 women (53%) were follow-up negative, and 27 (47%) were susceptible.
Selection and Preparation of Antigen Panel
An initial ATLAS screen (see protocol details below) was conducted on a group of 141 subjects by using the entire proteome of C. trachomatis D/UW-3/Cx, which includes approximately 960 candidate chlamydial antigens [21]. Chlamydial proteins were selected for inclusion in further assays if they elicited an IFN-γ response in 15% of all individuals tested and were thus considered immunogenic or if they induced a significantly greater frequency of IFN-γ responses in persons who exhibited an effective immune response versus persons with an ineffective immune response and were thus potentially associated with protection (Supplementary Table 1). An “effective” immune response was defined as one that spontaneously resolved infection or one that kept an individual uninfected despite sexual exposure to an infected person. An immune response was deemed “ineffective” in women with acute chlamydial PID or individuals with a diagnosis of chlamydia who did not clear infection by the time they returned for treatment >2 weeks later [21].
ATLAS T-Cell Screen
PBMCs were isolated from heparinized whole blood by density gradient centrifugation and stored in liquid nitrogen. CD14+ monocytes, CD4+ T cells, and CD8+ T cells were isolated by magnetic bead sorting [21]. Monocyte-derived dendritic cells (MDDCs) were generated to serve as antigen-presenting cells (APCs), while CD8+ and CD4+ T cells were nonspecifically expanded with anti-CD3/CD28 Dynabeads (ThermoFisher) and 20 U/mL recombinant human interleukin 2 for 6 to 7 days. The cells were rested for 48 hours with no cytokine, and the expansion beads were removed ≥4 hours before the cell-based assay. MDDCs were added to the Escherichia coli expression panel in duplicate at a target cell ratio of 1:100 MDDCs to bacteria. After 2 hours of incubation at 37°C, the MDDC-bacterium layer was fixed with 0.1% paraformaldehyde and washed; 4 × 104 T cells were added to each well of pulsed MDDCs and cocultured for 24 hours. This method results in antigen presentation within <2 hours [25]. Cell-free supernatants were harvested in duplicate and assayed via the OptEIA human IFN-γ enzyme-linked immunosorbent assay (Becton Dickinson, Franklin Lakes, NJ). Positive-control wells contained T cells stimulated with phytohemagglutinin (PHA) at 5 μg/mL. Negative-control wells contained E. coli expressing green fluorescent protein (GFP) [21]. Each candidate protein was expressed in 2 separate E. coli strains for optimal antigen presentation to CD4+ and CD8+ T cells.
Statistical Analysis
Four-parameter logistic curve fitting was used to calculate the concentration of IFN-γ. A positive response to antigen was defined as a mean IFN-γ response to antigen greater than the sum of the mean response to GFP (+2 SDs). To compare T-cell responses in the Endo+ group to those in the Cervix+ group, as well as T-cell responses in the susceptible group to those in the follow-up–negative group, we used quantile (median) regression analyses. Quantile regression provides more-robust estimations against response measurement outliers and unbiased estimates even when equal variance assumptions are violated. To determine specific proteins that were significantly more frequently recognized in Cervix+ women, we used the Fisher exact test of cross-sectional enrollment data. To identify specific proteins associated with resistance to incident infection over time, we used mixed-effects logistic regression for repeated measures. The covariate “sex with a new partner since last visit” was adjusted in the model. All analyses were conducted using R (version 3.1.1). Immunodominant proteins were defined as the 20 proteins that elicited the highest CD4+ or CD8+ T-cell response frequencies among the 58 evaluable participants in the cohort. For clinical and bacterial burden data, if outcomes were continuous with a normal distribution, the Student t test was conducted; for ordinal or interval outcomes, the Wilcoxon signed ranks test was used; and for categorical outcomes, the χ2 test was used unless cell sizes were <5, in which case the Fisher exact test was used. A P value of .05 was used to determine significance for all comparisons except for identification of potentially important protein antigens, for which we used a P value of .10.
RESULTS
Baseline Patient Characteristics
We processed PBMCs from 97 women from the TRAC cohort for T-cell analyses; data for 58 who had evaluable T-cell responses are reported. Their sociodemographic characteristics are similar to those previously reported for the entire cohort (Supplementary Table 2) [23]. Follow-up rates were high, with 74% attending all 4 visits, and 93% attending at least 3.
Sexual Exposure and Sexual Behavior in the Clinical Groups
STI exposure data were similar in women in the Cervix+ and Endo+ groups (Supplementary Table 3). A comparison of susceptible and follow-up–negative women revealed no difference in condom use frequency (approximately 50% of opportunities for each group). Gonococcal infection during follow-up was associated with reinfection (P = .04, by the Fisher exact test). In addition, 78% of susceptible women reported having a new sex partner during follow-up, compared with 29% of follow-up–negative women (P = .0006, by the χ2 test).
CD4+ and CD8+ T-Cell Responses Among Women in the Endo+ and Cervix+ Groups
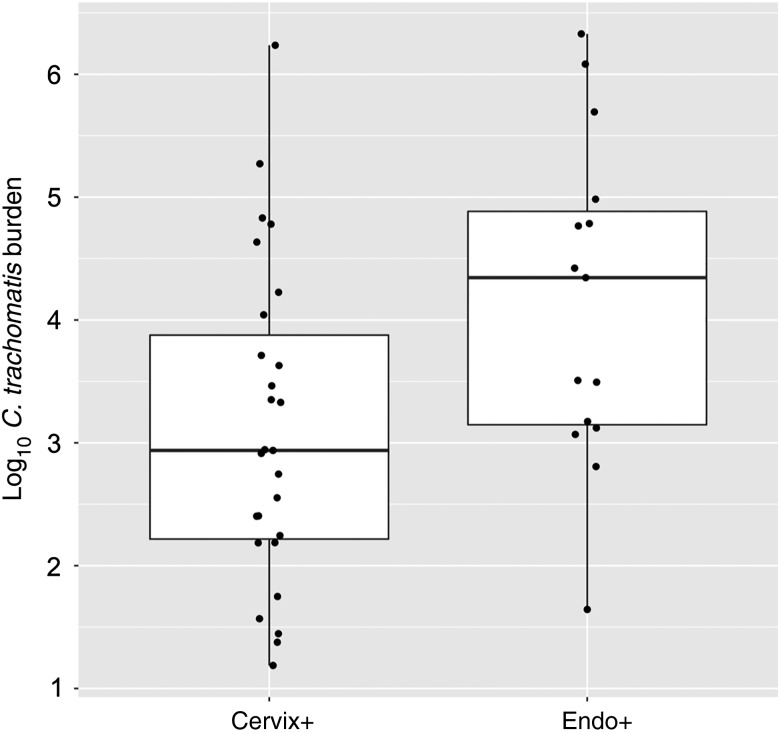
Women in the Cervix+ group also had a lower mean cervical bacterial burden when compared to women in the Endo+ group (P = .02, by the t test; Figure 1). This could indicate either a shorter duration or superior control of infection. Evaluation of questionnaires revealed that the median time between the most recent sexual encounter and the diagnosis of infection did not differ for women in the Cervix+ (9 days) and Endo+ (12 days) groups (P = .16, by the Wilcoxon test). Thus, more-recent infection was insufficient to explain the lower cervical burden in the Cervix+ group, which implied a possible role for the immune response in limiting infection.
Figure 1.
Comparison of cervical Chlamydia trachomatis burden between women with lower genital tract (the Cervix+ group) and those with upper genital tract (the Endo+ group) infection at enrollment. The lower, middle, and upper lines of the box correspond to the 25th, 50th, and 75th percentiles, respectively, for each group. The x-axis indicates the clinical groups, and the y-axis shows the log10 bacterial copies per cervical swab eluate. Each dot represents 1 subject's sample.
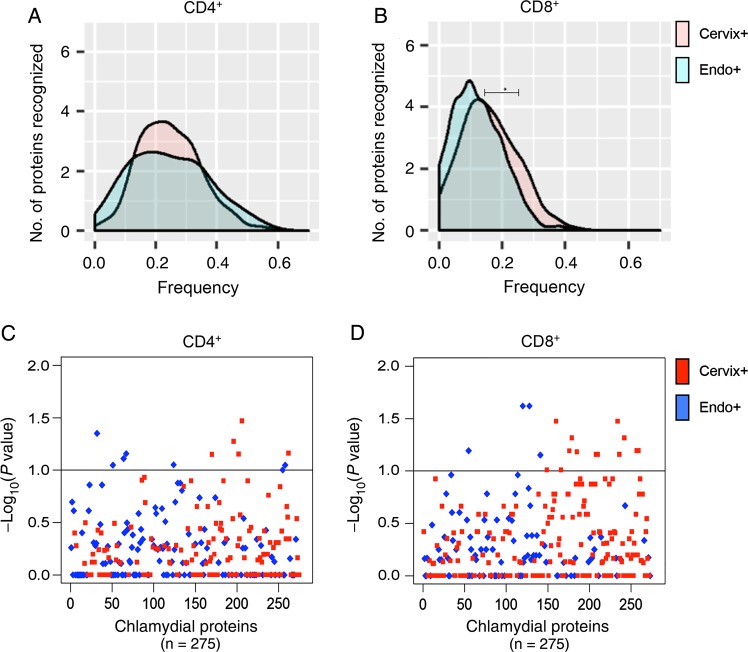
A comparison of median response frequencies for CD4+ T cells revealed no difference between the Cervix+ and Endo+ groups (25% and 23.8%, respectively; P = .53; Figure 2A). However, a higher median frequency of CD8+ T-cell responses was detected in the Cervix+ group, compared with the Endo+ group (13.8% vs 9.5%; P = .04; Figure 2B). Examination of the response frequencies to individual proteins revealed that CD4+ T cells from women in the Cervix+ group recognized 5 proteins with a significantly higher frequency (P < .10), compared with the Endo+ group (Figure 2C and Table 1), and CD8+ T cells from the Cervix+ group recognized a subset of 14 proteins with a higher frequency (P < .10) than those from the Endo+ group (Figure 2D and Table 1). Three of the antigens recognized by CD8+ T cells with increased frequency (CT461, CT511, and CT529) were among the top 20 immunodominant proteins associated with CD8+ T cells in the cohort, but none of the significant CD4+ T-cell–recognized antigens were among the immunodominant proteins associated with CD4+ T cells. These data suggest that specific antigens drive host defense mechanisms in T cells during active chlamydial infection to prevent ascension to the upper genital tract.
Figure 2.
Distribution of relative frequencies of positive CD4+ and CD8+ T-cell responses to a panel of chlamydial antigens, determined by ATLAS technology, among women with lower genital tract (Cervix+) and upper genital tract (Endo+) infection at enrollment. A and B, Per protein distributions of positive CD4+ (A) and CD8+ (B) T-cell responses to chlamydial proteins in the panel among women in the Cervix+ and Endo+ groups. The x-axis is the frequency of positive T-cell responses in the 2 groups of patients; the y-axis is the smooth fit curve of the number of proteins among the 275 proteins examined that were recognized at a particular frequency. A, The median positive response frequency of CD4+ T cells from the Cervix+ group was similar to that for the Endo+ group (25% and 23.8%, respectively; P = .53). B, CD8+ T cells from the Cervix+ group recognize a subset of chlamydial proteins more frequently than those from the Endo+ group (median response frequency, 13.8% and 9.5%, respectively; P = .04). C and D, Plots of the relative significance of the differential CD4+ (C) and CD8+ (D) T-cell responses to each protein in the panel among women in the Endo+ and Cervix+ groups. The x-axis represents chlamydial proteins in the panel, and the y-axis is the inverse log10(P value), with the dashed horizontal lines representing P values of .10. C, CD4+ T cells from women in the Cervix+ group recognized 1 chlamydial protein significantly more frequently (P < .05) and 5 proteins more frequently (P < .10) than those in the Endo+ group. D, CD8+ T cells from women in the Cervix+ group recognized 5 chlamydial proteins significantly more frequently (P < .05) and 14 proteins more frequently (P < .10) than those in the Endo+ group.
Table 1.
Antigens Associated With Control of Lower Genital Tract Infection at Enrollment
| Gene ID, by T-Cell Type | Gene | Antigen | Response Frequency | OR | P Value |
|---|---|---|---|---|---|
| CD4+ | |||||
| CT564 | yscT | Type III secretion system protein YscT | 0.132 | …a | .034 |
| CT553 | fmu | RNA methyltransferase | 0.245 | 4.84 | .053 |
| CT816 | glmS | Glucosamine-fructose-6-phosphate aminotransferase | 0.189 | 7.59 | .069 |
| CT560 | … | Hypothetical protein | 0.113 | … | .070 |
| CT490 | … | Hypothetical protein | 0.170 | 6.48 | .071 |
| CD8+ | |||||
| CT496.1 | … | Hypothetical protein | 0.191 | … | .001 |
| CT478 | oppC_2 | Oligopeptide ABC transporter permease | 0.170 | 8.28 | .034 |
| CT725 | birA | Biotin synthetase | 0.149 | 8.28 | .034 |
| CT529b | … | Hypothetical protein | 0.234 | 5.33 | .048 |
| CT739 | ftsK | DNA translocase FtsK | 0.255 | 5.33 | .048 |
| CT809 | … | Hypothetical protein | 0.170 | 7.04 | .064 |
| CT814 | … | Hypothetical protein | 0.170 | 7.04 | .064 |
| CT511Bb | rl15 | 50S ribosomal protein L15 | 0.255 | 3.90 | .064 |
| CT536 | dnaQ_2 | DNA polymerase III subunit epsilon | 0.087 | … | .066 |
| CT574 | pepP | Aminopeptidase P | 0.106 | … | .069 |
| CT582B | minD | Chromosome partitioning ATPase | 0.085 | … | .069 |
| CT729 | serS | Serine-tRNA ligase | 0.085 | … | .069 |
| CT461b | Metallophosphoesterase | 0.191 | 3.97 | .098 | |
| CT487 | yhhF | Methylase | 0.170 | 3.97 | .098 |
Abbreviations: ID, identifier; OR, odds ratio.
a The denominator is 0.
b Expresses an immunodominant protein.
CD4+ and CD8+ T-Cell Responses Among Follow-up–Negative and Susceptible Women
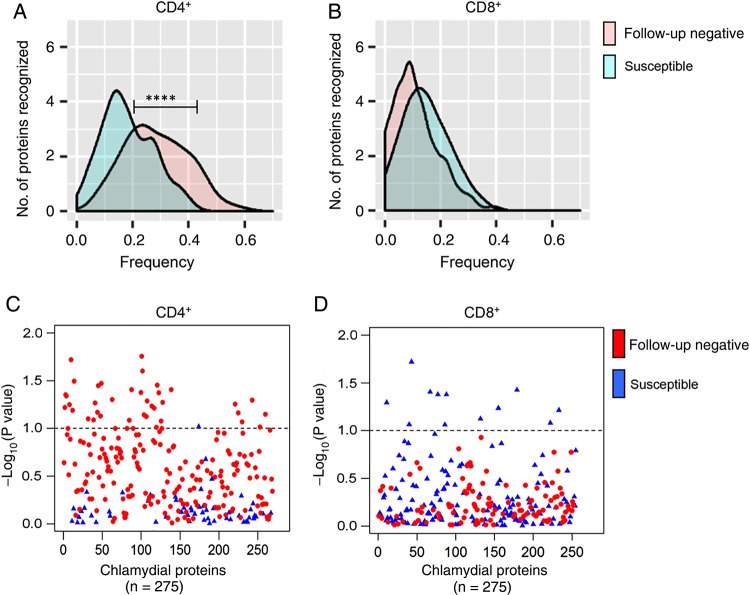
We compared the response frequencies between follow-up–negative and susceptible women and found that follow-up–negative women had a significantly greater median frequency of CD4+ T-cell responses (28.6%) as compared to susceptible women (18%; P < .0001; Figure 3A). Examination of response frequencies to individual proteins revealed that a subset of 37 proteins were recognized by CD4+ T cells of follow-up–negative women with a higher frequency (P < .10) (Figure 3C and Table 2) than observed for susceptible women. In contrast to CD4+ T-cell responses, the median positive response rate of CD8+ T cells in follow-up–negative and susceptible women did not differ (8.7% and 12.5%, respectively; P = .07; Figure 3B), and no chlamydial proteins were recognized with increased frequency by CD8+ T cells of follow-up–negative women (Figure 3D). These data suggest that a specific subset of proteins recognized by CD4+ T cells may reduce susceptibility to reinfection, and these proteins should be further explored as vaccine candidates. These proteins are summarized in Table 2. Four were among the top 20 antigens recognized by CD4+ T cells from the entire cohort: CT034, CT128, CT130, and CT368. No chlamydial proteins were recognized with increased frequency by CD8+ T cells from follow-up–negative women as compared to susceptible women.
Figure 3.
Distribution of relative frequencies of positive CD4+ and CD8+ T-cell responses to a panel of chlamydial antigens, determined by ATLAS technology, among susceptible and follow-up–negative women, as defined by infection status during 1 year of follow-up. A and B, Per protein distributions of positive CD4+ (A) and CD8+ (B) T-cell responses to chlamydial proteins in the panel among susceptible and follow-up–negative women. The x-axis is the frequency of positive T-cell responses in the 2 patient groups; the y-axis is the smooth fit curve of the number of proteins among the 275 proteins examined that were recognized at a particular frequency. A, CD4+ T cells from follow-up–negative women recognized a subset of chlamydial proteins more frequently than susceptible women (median response frequency, 28.6% and 18%, respectively; P < .0001). B, The median positive response frequencies of CD8+ T cells were similar among follow-up–negative and susceptible women (8.7% and 12.5%, respectively; P = .07). C and D, Plots of the relative significance of the differential CD4+ (C) and CD8+ (D) T-cell response to each protein in the panel among susceptible and follow-up–negative women. The x-axis represents chlamydial proteins in the panel, and the y-axis is the inverse log10(P value), with the dashed horizontal line representing P = .10. C, CD4+ T cells from follow-up–negative women recognized 14 chlamydial proteins significantly more frequently (P < .05) and 37 proteins more frequently (P < .10) than those in susceptible women. D, No proteins were recognized with increased frequency by CD8+ T cells from follow-up–negative women when compared to susceptible women.
Table 2.
Antigens Associated With Protection From Chlamydia trachomatis Infection During 1 Year of Follow-up
| Gene ID, by T Cell-Type | Gene | Antigen | Response Frequency | OR | P Value |
|---|---|---|---|---|---|
| CD4+ | |||||
| CT308 | atpA | V-type ATP synthase subunit A | 0.259 | 4.55 | .018 |
| CT020 | lepB | Signal peptidase I | 0.291 | 4.41 | .019 |
| CT313 | tal | Transaldolase | 0.327 | 3.54 | .025 |
| CT034a | ytfF | Cationic amino acid transporter | 0.370 | 2.92 | .032 |
| CT156 | … | Hypothetical protein | 0.255 | 4.08 | .034 |
| CT360 | … | Hypothetical protein | 0.182 | 4.93 | .035 |
| CT134 | … | Hypothetical protein | 0.259 | 3.31 | .036 |
| CT264 | msbA | ABC transporter ATP binding protein/permease | 0.236 | 4.77 | .039 |
| CT421 | … | Hypothetical protein | 0.283 | 2.97 | .039 |
| CT307 | atpB | V-type ATP synthase subunit B | 0.273 | 3.20 | .042 |
| CT368a | aroC | Chorismate synthase | 0.382 | 2.69 | .042 |
| CT004 | gatB | Aspartyl/glutamyl-tRNA amidotransferase subunit | 0.327 | 2.87 | .044 |
| CT005 | … | Hypothetical protein | 0.315 | 2.95 | .046 |
| CT218 | surE | 5’-nucleotidase SurE | 0.291 | 3.47 | .050 |
| CT757 | mraY | Phospho-N-acetylmuramoyl-pentapeptide transferase | 0.236 | 4.39 | .051 |
| CT319 | rI11 | 50S ribosomal protein L11 | 0.273 | 3.03 | .052 |
| CT386 | … | Metal dependent hydrolase | 0.273 | 3.42 | .054 |
| CT130a | glnQ | ABC amino acid transporter ATPase | 0.455 | 2.38 | .054 |
| CT647 | … | Hypothetical protein | 0.204 | 4.30 | .056 |
| CT017B | … | Hypothetical protein | 0.236 | 4.44 | .057 |
| CT338 | … | Hypothetical protein | 0.236 | 3.37 | .059 |
| CT002 | gatC | Glutamyl-tRNA amidotransferase subunit C | 0.236 | 3.30 | .060 |
| CT351 | … | Hypothetical protein | 0.273 | 3.32 | .061 |
| CT022 | rI31 | 50S ribosomal protein L31 | 0.291 | 2.68 | .065 |
| CT133 | … | rRNA methylase | 0.345 | 2.44 | .068 |
| CT687 | … | Cysteine desulfurase-like protein | 0.236 | 4.00 | .069 |
| CT821 | sucC | Succinyl-CoA ligase subunit beta | 0.236 | 3.18 | .071 |
| CT315 | rpoB | DNA-directed RNA polymerase subunit β | 0.309 | 2.42 | .072 |
| CT144 | … | Hypothetical protein | 0.255 | 3.12 | .074 |
| CT318 | rI1 | 50S ribosomal protein L1 | 0.309 | 3.13 | .075 |
| CT104 | Fabl | Enoyl-acyl-carrier protein reductase | 0.164 | 6.19 | .080 |
| CT128a | Adk | Adenylate kinase | 0.382 | 2.23 | .082 |
| CT252C | Lgt | Prolipoprotein diacylglycerol transferase | 0.273 | 2.68 | .082 |
| CT382 | aroG | Phospho-2-dehydro-3-deoxyheptonate aldolase | 0.259 | 3.03 | .084 |
| CT717 | … | Type III secretion system ATP synthase | 0.241 | 2.81 | .086 |
| CT808 | cafE | Rne/Rng family ribonuclease | 0.145 | 5.61 | .097 |
| CT359 | … | Hypothetical protein | 0.309 | 2.38 | .098 |
| CD8+ | … | … | … | … | … |
| None | … | … | … | … | … |
Abbreviations: ID, identifier; OR, odds ratio.
a Expresses an immunodominant protein.
DISCUSSION
In this study, we identified specific antigens recognized by CD4+ and CD8+ T cells associated with protective immunity among a high-risk cohort of women. Women in this cohort were highly sexually active, with the majority reporting irregular use of condoms (93%) and prior C. trachomatis infection (66%). The high rate of active infection (72%) and sampling of upper genital tract tissue provided an opportunity to compare responses of women in the Cervix+ and Endo+ groups.
Despite a lower mean antigen burden, T cells from women in the Cervix+ group recognized chlamydial proteins with greater (for CD8+ T cells; P = .04) or equivalent (for CD4+ T cells) frequency as compared to women in the Endo+ group. Recognition of specific proteins by CD4+ and CD8+ T cells was associated with improved control of infection. These findings indicate an important role for antigen-specific T-cell responses in preventing ascension of chlamydia to the upper genital tract. This is significant, given that chronic disease sequelae in women are primarily due to Fallopian tube scarring.
The high follow-up rate in our study provided the opportunity to compare participants who subsequently acquired C. trachomatis and those who remained uninfected over 12 months of follow-up. We found that CD4+ T cells are the key players in resistance to reinfection. Significantly greater CD4+ T-cell response frequencies were detected in women who remained uninfected, compared with susceptible women (P < .0001), and a subset of 37 proteins were recognized more frequently by CD4+ T cells from follow-up–negative women. Because having sex with a new partner is a known risk factor and was associated with acquisition (Supplementary Table 3), we adjusted for this in our statistical analysis. Nevertheless, one third of follow-up–negative women reported a new or multiple different partners during follow-up. It is possible that some of the women that we defined as follow-up negative were simply not exposed during follow-up. However, the fact that higher rates of positive CD4+ T-cell responses were coupled with enhanced responses to a specific subset of proteins in follow-up–negative women implies that these women were less susceptible because of either increased past exposure or a more robust response to a single past infection.
Our data suggest differing roles for CD8+ and CD4+ T cells in response to chlamydial genital tract infection. CD8+ T cells may contribute to limiting the bacterial burden and ascension of infection, while CD4+ T cells appear to drive protection from reinfection. In each case, limiting acute infection or preventing repeat infection was dependent on having a T-cell response to a specific subset of proteins, rather than on achieving a threshold frequency of responses to random proteins. This points toward certain proteins being better drivers of protective adaptive responses. Notably, some of these antigens are among the most frequently recognized proteins within the entire cohort.
Because the panel of proteins selected for evaluation was based on criteria derived via our prior study [21], several proteins reported in murine vaccine studies to induce protection were not assessed. Immunization with native major outer membrane protein (MOMP) protects against oviduct disease in the murine model when delivered with adjuvant [26]. This protein was not included in our current screen because in the prior screen it elicited a low frequency of T-cell responses [21], despite MOMP immunoglobulin G (IgG) seropositivity in 40% of the cohort (unpublished data). A prior study that examined T-cell IFN-γ responses to 116 recombinant chlamydial antigens in infected patients revealed responses to inner membrane and cytoplasmic proteins only; MOMP was recognized only by IgG [27]. These studies suggest that MOMP elicits poor T-cell responses in humans despite induction of antibody. Murine studies reveal T-cell responses to MOMP [28], but differences in cellular processing machinery in mice and humans at the level of the proteasome and in major histocompatibility complex lead to differences in antigen presentation [29]. Other proteins that have induced protection in mouse vaccine models that were not included in our screen included CPAF [30]; polymorphic membrane proteins E, F, and G [31, 32]; OmcB and rl16 [33]; and CT823 [34]. CrpA, IncA (an immunodominant, inclusion membrane-associated protein), and PmpD were examined in our panel, but response frequencies were not significantly different among the subgroups. A hypothetical protein, CT144, that we found to be protective when administered as a vaccine in mice [34] was recognized with a greater frequency by CD4+ T cells from follow-up–negative women (P = .07). Tarp, which has been found to induce high titers of antibodies in humans and protective immunity against upper genital tract pathologies in mice [35], was investigated and was more frequently recognized by CD8+ T cells from women in the Cervix+ group, compared with those in the Endo+ group (P = .11), although our small sample size likely limited our ability to detect statistically significant differences in these responses. Because testing was limited to one third of the proteome, we may have missed antigens important in protective immunity. However, the current panel was based on prior human screens, which allows for inclusion of proteins that might have been omitted if the literature, animal studies, and predictive algorithms were used as the determinants [21].
The proteins of recognized function associated with control or resistance to infection in our cohort are involved in protein synthesis, central metabolism, and type III secretion (Tables 1 and 2). This is consistent with our previous investigation, using ATLAS [21]. Many of the antigens identified then as inducing “effective” responses (ie, spontaneous clearance or lack of infection despite confirmed exposure) were also genes involved in central metabolism. Three proteins associated with effective immunity in that cohort (CT002, CT252, and CT757) were also associated with resistance to infection in this cohort. Additionally, CT564 and CT725, specifically associated with infection control in our TRAC cohort, were associated with effective immunity previously [21]. In the prior study, CD8+ T-cell responses were associated with effective immune responses in men and women, which is consistent with a role for these cells in prevention of ascension in this female only cohort. These proteins are essential for intracellular survival of C. trachomatis; since this pathogen lives within epithelial cells and interacts intimately with the endoplasmic reticulum, peptides of these essential proteins are likely shuttled through the vesicular system and loaded into major histocompatibility complex molecules. A limitation of the ATLAS screen is our inability to confirm that each protein was successfully expressed and presented by human APCs. Confirmation that a full-length protein is expressed in E. coli is derived from a cell-based assay in which a B3Z T-cell hybridoma is tested for recognition of a C-terminal SIINFEKL-tag inserted into each clone. The antigens and tag are processed and presented to the T cells by murine APCs [36]. However, we cannot be certain that the human APCs presented each protein in comparable fashion.
Strengths of this study included comprehensive data on demographic, clinical, and sexual characteristics, high rate of follow-up, and a stringent requirement to complete 4 follow-up visits over 1 year to be considered in the follow-up–negative subgroup. A further strength is the nonbiased evaluation of over one third of the chlamydial genome. Classifying upper and lower genital tract infection at enrollment allowed us to identify antigens important for control of infection. Additionally, following these women longitudinally over 1 year allowed us to identify antigens specifically associated with protection against subsequent infection. Several of the proteins identified were immunodominant, suggesting that this technology may be capable of identifying novel vaccine candidates with the potential for broad recognition by immunized individuals.
Supplementary Data
Supplementary materials are available at http://jid.oxfordjournals.org. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Notes
Acknowledgments. We thank the women who agreed to participate in this study; Alison Collins, Ingrid Macio, Melinda Petrina, Carol Priest, Abi Jett, and Lorna Rabe, for their efforts in the clinic and/or laboratory; and the staff at the Allegheny County Health Department STD Clinic, for their support.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases (grants U19 AI084024 and U19 AI113170).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Satterwhite CL, Torrone E, Meites E et al. Sexually transmitted infections among US women and men: prevalence and incidence estimates, 2008. Sex Transm Dis 2013; 40:187–93. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization (WHO). Global prevalence and incidence of selected curable sexually transmitted infections: overview and estimates, 2001 Geneva, Switzerland: WHO, 2001. [Google Scholar]
- 3.Miller WC, Ford CA, Morris M et al. Prevalence of chlamydial and gonococcal infections among young adults in the United States. JAMA 2004; 291:2229–36. [DOI] [PubMed] [Google Scholar]
- 4.Owusu-Edusei K Jr., Chesson HW, Gift TL et al. The estimated direct medical cost of selected sexually transmitted infections in the United States, 2008. Sex Transm Dis 2013; 40:197–201. [DOI] [PubMed] [Google Scholar]
- 5.Arno JN, Katz BP, McBride R et al. Age and clinical immunity to infections with Chlamydia trachomatis. Sex Transm Dis 1994; 21:47–52. [DOI] [PubMed] [Google Scholar]
- 6.Batteiger BE, Xu F, Johnson RE, Rekart ML. Protective immunity to Chlamydia trachomatis genital infection: evidence from human studies. J Infect Dis 2010; 201(suppl 2):S178–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geisler WM, Lensing SY, Press CG, Hook EW III. Spontaneous resolution of genital Chlamydia trachomatis infection in women and protection from reinfection. J Infect Dis 2013; 207:1850–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woolridge RL, Grayston JT, Chang IH, Cheng KH, Yang CY, Neave C. Field trial of a monovalent and of a bivalent mineral oil adjuvant trachoma vaccine in Taiwan school children. Am J Ophthalmol 1967; 63:1645–50. [DOI] [PubMed] [Google Scholar]
- 9.Woolridge RL, Grayston JT, Chang IH, Yang CY, Cheng KH. Long-term follow-up of the initial (1959–1960) trachoma vaccine field trial on Taiwan. Am J Ophthalmol 1967; 63:1650–5. [DOI] [PubMed] [Google Scholar]
- 10.Sowa S, Sowa J, Collier LH, Blyth WA. Trachoma vaccine field trials in The Gambia. J Hyg (Lond) 1969; 67:699–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rank RG, Whittum-Hudson JA. Protective immunity to chlamydial genital infection: evidence from animal studies. J Infect Dis 2010; 201(suppl 2):S168–77. [DOI] [PubMed] [Google Scholar]
- 12.O'Connell CM, Ingalls RR, Andrews CW Jr, Skurlock AM, Darville T. Plasmid-deficient Chlamydia muridarum fail to induce immune pathology and protect against oviduct disease. J Immunol 2007; 179:4027–34. [DOI] [PubMed] [Google Scholar]
- 13.Kari L, Whitmire WM, Olivares-Zavaleta N et al. A live-attenuated chlamydial vaccine protects against trachoma in nonhuman primates. J Exp Med 2011; 208:2217–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rank RG, Ramsey KH, Pack EA, Williams DM. Effect of gamma interferon on resolution of murine chlamydial genital infection. Infect Immun 1992; 60:4427–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Igietseme JU, Ramsey KH, Magee DM, Williams DM, Kincy TJ, Rank RG. Resolution of murine chlamydial genital infection by the adoptive transfer of a biovar-specific TH1 lymphocyte clone. Reg Immunology 1993; 5:317–24. [PubMed] [Google Scholar]
- 16.Gondek DC, Olive AJ, Stary G, Starnbach MN. CD4+ T cells are necessary and sufficient to confer protection against chlamydia trachomatis infection in the murine upper genital tract. J Immunol 2012; 189:2441–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morrison SG, Morrison RP. A predominant role for antibody in acquired immunity to chlamydial genital tract reinfection. J Immunol 2005; 175:7536–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morrison RP, Feilzer K, Tumas DB. Gene knockout mice establish a primary protective role for major histocompatibility complex class II-restricted responses in Chlamydia trachomatis genital tract infection. Infect Immun 1995; 63:4661–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olivares-Zavaleta N, Whitmire WM, Kari L, Sturdevant GL, Caldwell HD. CD8+ T cells define an unexpected role in live-Attenuated vaccine protective immunity against Chlamydia trachomatis infection in macaques. J Immunol 2014; 192:4648–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gervassi AL, Probst P, Stamm WE, Marrazzo J, Grabstein KH, Alderson MR. Functional characterization of class Ia- and non-class Ia-restricted Chlamydia-reactive CD8+ T cell responses in humans. J Immunol 2003; 171:4278–86. [DOI] [PubMed] [Google Scholar]
- 21.Picard MD, Bodmer JL, Gierahn TM et al. Resolution of chlamydia trachomatis infection is associated with a distinct T cell response profile. Clin Vaccine Immunol 2015; 22:1206–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grubaugh D, Flechtner JB, Higgins DE. Proteins as T cell antigens: methods for high-throughput identification. Vaccine 2013; 31:3805–10. [DOI] [PubMed] [Google Scholar]
- 23.Russell AN, Zheng X, O'Connell CM et al. Analysis of factors driving incident and ascending infection and the role of serum antibody in chlamydia trachomatis genital tract infection. J Infect Dis 2016; 213:523–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Workowski KA, Berman SM. Centers for Disease Control and Prevention sexually transmitted disease treatment guidelines. Clin Infect Dis 2011; 53(suppl 3):S59–63. [DOI] [PubMed] [Google Scholar]
- 25.Hu PQ, Tuma-Warrino RJ, Bryan MA et al. Escherichia coli expressing recombinant antigen and listeriolysin O stimulate class I-restricted CD8+ T cells following uptake by human APC. J Immunol 2004; 172:1595–601. [DOI] [PubMed] [Google Scholar]
- 26.Farris CM, Morrison SG, Morrison RP. CD4+ T cells and antibody are required for optimal major outer membrane protein vaccine-induced immunity to Chlamydia muridarum genital infection. Infect Immun 2010; 78:4374–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Follmann F, Olsen AW, Jensen KT, Hansen PR, Andersen P, Theisen M. Antigenic profiling of a Chlamydia trachomatis gene-expression library. J Infect Dis 2008; 197:897–905. [DOI] [PubMed] [Google Scholar]
- 28.Finco O, Frigimelica E, Buricchi F et al. Approach to discover T- and B-cell antigens of intracellular pathogens applied to the design of Chlamydia trachomatis vaccines. Proc Natl Acad Sci USA 2011; 108:9969–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sesma L, Alvarez I, Marcilla M, Paradela A, Lopez de Castro JA. Species-specific differences in proteasomal processing and tapasin-mediated loading influence peptide presentation by HLA-B27 in murine cells. J Biol Chem 2003; 278:46461–72. [DOI] [PubMed] [Google Scholar]
- 30.Murthy AK, Chambers JP, Meier PA, Zhong G, Arulanandam BP. Intranasal vaccination with a secreted chlamydial protein enhances resolution of genital Chlamydia muridarum infection, protects against oviduct pathology, and is highly dependent upon endogenous gamma interferon production. Infect Immun 2007; 75:666–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu H, Jiang X, Shen C et al. Chlamydia muridarum T-cell antigens formulated with the adjuvant DDA/TDB induce immunity against infection that correlates with a high frequency of gamma interferon (IFN-gamma)/tumor necrosis factor alpha and IFN-gamma/interleukin-17 double-positive CD4+ T cells. Infect Immun 2010; 78:2272–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu H, Jiang X, Shen C, Karunakaran KP, Brunham RC. Novel Chlamydia muridarum T cell antigens induce protective immunity against lung and genital tract infection in murine models. J Immunol 2009; 182:1602–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olsen AW, Theisen M, Christensen D, Follmann F, Andersen P. Protection against Chlamydia promoted by a subunit vaccine (CTH1) compared with a primary intranasal infection in a mouse genital challenge model. PloS One 2010; 5:e10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Picard MD, Cohane KP, Gierahn TM, Higgins DE, Flechtner JB. High-throughput proteomic screening identifies Chlamydia trachomatis antigens that are capable of eliciting T cell and antibody responses that provide protection against vaginal challenge. Vaccine 2012; 30:4387–93. [DOI] [PubMed] [Google Scholar]
- 35.Wang J, Chen L, Chen F et al. A chlamydial type III-secreted effector protein (Tarp) is predominantly recognized by antibodies from humans infected with Chlamydia trachomatis and induces protective immunity against upper genital tract pathologies in mice. Vaccine 2009; 27:2967–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Long D, Skoberne M, Gierahn TM et al. Identification of novel virus-specific antigens by CD4(+) and CD8(+) T cells from asymptomatic HSV-2 seropositive and seronegative donors. Virology 2014; 464–465:296–311. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.